Abstract
There is presently great interest in mechanisms of acquired resistance to EGFR inhibitors that are now being used widely in the treatment of a variety of common human cancers. To investigate these mechanisms we established EGFR inhibitor resistant clones from non-small cell lung cancer cells. A comparative analysis revealed that acquired resistance to EGFR inhibitors was associated consistently with the loss of p53 and cross-resistance to radiation. To examine the role of p53, we first knocked down p53 in sensitive parental cells and found a reduction in sensitivity to both EGFR inhibitors and radiation. Conversely, restoration of functional p53 in EGFR inhibitor resistant cells was sufficient to resensitize them to EGFR inhibitors or radiation in vitro and in vivo. Further studies indicate that p53 may enhance sensitivity to EGFR inhibitors and radiation via induction of cell cycle arrest, apoptosis and DNA damage repair. Taken together, these findings suggest a central role of p53 in the development of acquired resistance to EGFR inhibitors and prompt consideration to apply p53 restoration strategies in future clinical trials that combine EGFR inhibitors and radiation.
Keywords: EGFR, Inhibitor, Radiation, Resistance, p53
INTRODUCTION
The EGFR is a family member of the ErbB receptor tyrosine kinases and is important in oncogenesis. The activation of EGFR ultimately promotes tumor cell proliferation, survival, invasion, and angiogenesis. Hence, blockade of EGFR has emerged as a highly promising cancer therapy approach over the last decade (1, 2). To date, several EGFR targeting agents have demonstrated preclinical and clinical promise and gained FDA approval in recent years, such as cetuximab, panitumumab, gefitinib and erlotinib (3, 4). Despite a series of promising clinical trial results, increasing evidence indicates that most patients who initially respond to treatment eventually manifest tumor recurrence following EGFR therapy (5-7). These findings suggest that acquired resistance to EGFR inhibitors has developed and underscore the importance of understanding molecular mechanisms of resistance to further the advancement of EGFR inhibitor therapies in oncology.
A number of recent reports have suggested potential mechanisms for acquired resistance to EGFR inhibitors. These include mutations in EGFR, KRAS or the activation of other receptor tyrosine kinases, such as ErbB3 or c-Met to effectively bypass the effect of EGFR inhibitors (8, 9). The EGFR mutation T790M has been found in NSCLC patients with acquired resistance to gefitinib and erlotinib (10). However, these EGFR mutations appear of value to predict response to EGFR TKIs, as opposed to EGFR monocloncal antibodies, and are relatively infrequent across the spectrum of human cancer types tested to date (11). Furthermore, patients without EGFR mutations still manifest acquired resistance to EGFR inhibitors. Therefore, it remains critical to explore additional mechanisms of acquired resistance to EGFR inhibitors.
One experimental approach to explore acquired resistance involves preclinical model systems that select for EGFR inhibitor resistant clones and perform molecular screening to assess changes that occur during the evolution of resistance. In the current study, we established a series of resistant clones to two different classes of EGFR inhibitors, cetuximab and erlotinib, from sensitive cell lines without EGFR and KRAS mutations following long term EGFR inhibitor exposure. Following high throughput western-based screening, we identified a robust loss of p53 in all resistant clones. In addition, our previous studies suggested cross-resistance to radiation in those cells with acquired resistance to EGFR inhibitors (12). Since p53 has been shown to play a role in regulating response to radiation and several chemotherapeutic agents, we systematically investigated the impact of p53 in regulating acquired resistance to EGFR inhibitors and radiation in our EGFR inhibitor resistant cells.
MATERIALS AND METHODS
Reagents and antibodies are listed in the Supplementary Materials and Methods
Cell Cultures
The human lung H226 (NCI-H226), A549 and H292 tumor cell lines were obtained from ATCC. The human head and neck SCC6 (UM-SCC6) cells were kindly provided by Dr. Thomas E. Carey (University of Michigan). Both H262 and H292 were maintained in RPMI with 10% FBS. SCC6 and A549 were cultured routinely in DMEM supplemented with 10% FBS and 1 μg/ml hydrocortisone. EGFR inhibitor resistant clones of H226 and SCC6 were developed as described previously (8). All cells were authenticated by sequencing the wild type status of p53. Cell culture media and supplements were obtained from Life Technologies, Inc. (Gaithersburg, MD).
BD PowerBlot western array
Total cellular proteins from parental or resistant cells were extracted by a lysis buffer containing 10 mM Tris, pH 7.4, 0.1 mM sodium orthovanadate and 1% SDS followed by sonication. Cell extracts were then sent to BD Biosciences for analysis as described previously (13). Detailed information is provided in the Supplementary Materials and Methods.
RNA interference of p53
p53 siRNA was obtained from Dharmacon (Lafayette, CO, USA). Cells were transfected with the p53 siRNA using DharmaFECT reagent following the manufacturer's procedures.
Cell proliferation assay
Viable growing cells was determined by crystal violet staining as described previously (12).
Neutral comet assay
To detect radiation-induced DNA damage, a CometAssay kit from Trevigen (Gaithersburg, MD) was used according to the manufacturer's instructions. The comet assay performed using neutral conditions, will detect mainly double-stranded breaks (DSB) of DNA and can be useful for assessing the DNA damage following radiation. Detailed information is provided in the Supplementary Materials and Methods.
Transfection of p53
Cet-R cells were first transiently transfected with a pC53-SN plasmid (a gift from Dr. Honnavara N. Ananthaswamy, MD Anderson Cancer Center, Houston, TX) encoding human wild-type p53 using Effectene® reagent (Qiagen, Germantown, MD). To create a stable clone with inducible p53, we applied a T-REx system (Invitrogen, Carlsbad, CA) which is a Tetracyclin (Tet)-regulated mammalian expression system based on the binding of Tet to a Tet repressor and derepression of the promoter controlling the expression of p53 (14). In brief, Cet-R cells were first transfected with the pcDNA6/TR® vector expressing the Tet repressor (Invitrogen) using Effectene® reagent following manufacturer's instructions. Cells were selected with 50 μg/ml of blasticidin for 48-72 hours and then grown in 5 μg/ml of blasticidin in 96-well plates. Stable clones were selected and expanded on the basis of their expression of the Tetrepressor determined by western blotting. In parallel, the pC53-SN plasmid was digested with BamHI to release the wild-type human p53 cDNA and subcloned into the pcDNA®5/TO, (Invitrogen) tetracycline inducible vector. Cet-R cells with stable expression of the pcDNA6/TR®, were then transfected with pcDNA®5/TO-wt-hp53 using Effectene® reagent. Colonies were then selected and grown in 5 μg/ml blasticidin and 100 μg/ml hygromycin. Stable transfected clones with high levels of p53 expression upon Tet induction were then pooled and used in the experiments described below.
Immunocytochemical staining of DNA-PK
Cet-R cells were seeded on a glass chamber slide (Nalgene Nunc, Naperville, IL). Following treatment, immunofluorescent staining was performed as described in the Supplementary Materials and Methods.
Apoptosis assessment
Apoptosis was detected by flow cytometry via examination of altered plasma membrane phospholipid packing using Annexin V/PI kit from BD Biosciences Pharmingen (San Diego, CA). Detailed information is provided in the Supplementary Materials and Methods.
Determination of cetuximab and radiation response in human tumor xenografts
Athymic nude mice were obtained from Harlan Bioproducts for science (Indianapolis, IN) and maintained in a laminar air-flow cabinet under aseptic conditions. The care and treatment of experimental animals was in accordance with institutional guidelines. Cetuximab resistant cells with Tetinducible p53 (~1 × 106) were injected subcutaneously into the dorsal flank area of the mice. Tumor volume was monitored by direct measurement with calipers and calculated by the formula; π/6 × (large diameter) ×(small diameter)2. When tumor size reached ~100 mm3, mice were either fed with a regular diet or a regular diet containing doxycycline (Dox) to the end of experiment. The induction of p53 by Dox was validated by western blotting following extraction of proteins harvested from tumor specimens. Following the confirmation of p53 induction, mice were either treated with 2 mg/kg of cetuximab (i.p.) twice weekly or 2 Gy of radiation twice weekly for 3 consecutive weeks.
Radiation survival
Survival following radiation exposure was defined as the ability of the cells to maintain their clonogenic capacity and to form colonies. Briefly, after exposure to radiation, cells were trypsinized, counted, and seeded for colony formation in 35 mm dishes at 50-5000 cells/dish. Following 14-21 days, colonies were stained with crystal violet and manually counted. Colonies consisting of 50 cells or more were scored, and 4-10 replicate dishes containing 10-150 colonies/dish were counted for each treatment.
Statistical analysis
Student t-test was used to evaluate the significance of change in cetuximab response, radiation-induced DNA damage and apoptosis. Differences between clones were considered statistically significant when p < 0.05.
RESULTS
Loss of p53 in EGFR inhibitor resistant clones
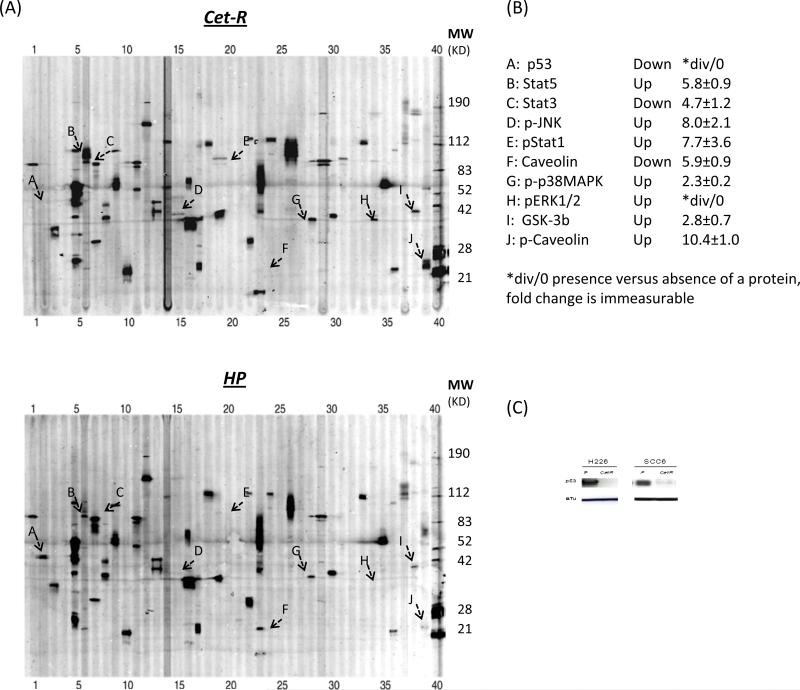
We previously established acquired resistant clones to two distinct classes of EGFR inhibitor as a model system to examine potential mechanisms of acquired resistance to EGFR inhibitors (8). Following the development of cetuximab-resistant (Cet-R) and erlotinib-resistant (Erl-R) clones of H226, we performed a western-based high-throughput screening to compare the levels of cellular signaling proteins between resistant and corresponding sensitive parental H226 cells (HP). This customized BD PowerBlot western array applied antibodies to detect phosphorylated and total levels of 42 key cellular signaling proteins which included EGFR related ERK, AKT, JNK and STAT signaling molecules. As highlighted in the images of the two PowerBlots in Fig 1A, we found that several proteins levels and activity showed significant up- or down-regulation in the Cet-R cells when compared to HP cells. Fig 1B lists the proteins with a greater than 2-fold change (up or down) in Cet-R with level 10 confidence following quantification analysis as described in the “Materials and Methods”. We found that p53 was the most profoundly down-regulated and p-ERK1/2 the most up-regulated cellular signaling protein in Cet-R cells when compared to parental HP cells. Similar results were found in the erlotinib resistant clones (Supplementary Fig. S1) derived from H226. As p53 is a key molecule in regulating cellular radiation response and has recently been shown to influence cetuximab response, we validated the down-regulation of p53 in another established Cet-R line using conventional western blotting. Consistent with the PowerBlot results, we found that p53 was significantly reduced in Cet-R clones of H226 and SCC6 (Fig. 1C). This robust loss of p53 in not only all cetuximab resistant clones, but also in our erlotinib resistant clones, led us to hypothesize that p53 may play an important role in regulating acquired resistance to EGFR inhibitors and radiation.
Fig. 1. Changes of cellular signaling proteins in cetuximab-resistant cells by western array.
(A) shows representative images of the western-based PowerBlot array from HP and Cet-R cells. The numbers above and below the array indicate the lane number of 41 channels. Several proteins showing significant expression differences between HP and Cet-R are highlighted by characters A~J. (B) lists those signaling proteins in Cet-R demonstrating up- or down-expression greater than 2 fold when compared to HP. (C) conventional western blotting validates the reduced level of p53 in Cet-R clones from H226 and SCC6 compared with the corresponding parental cells (P). The α-Tubulin (α-Tu) serves as a loading control.
Silencing p53 in parental H226 cells reduces sensitivity to cetuximab and radiation
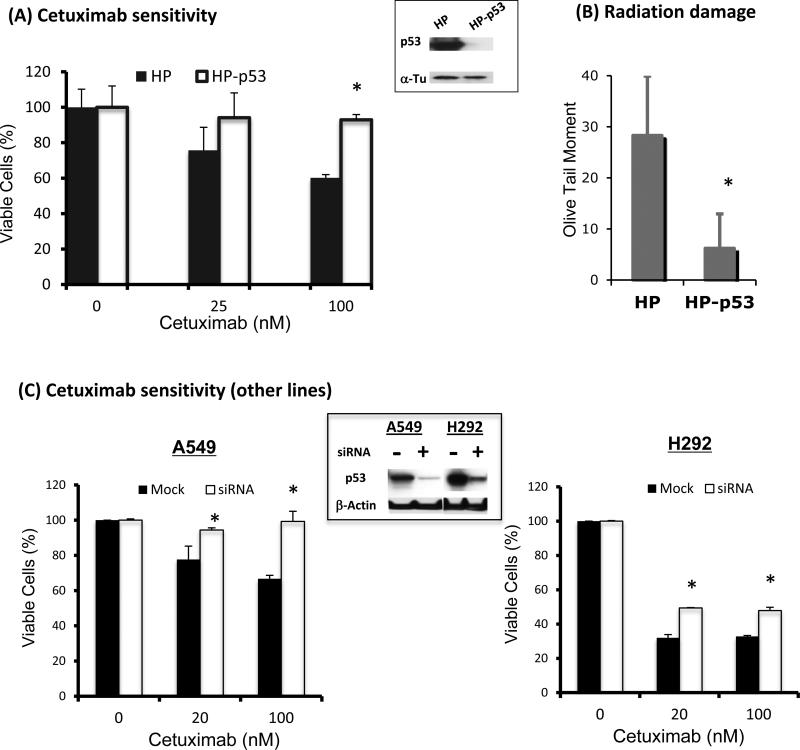
To investigate the role of p53 in regulating acquired resistance to EGFR inhibitors and radiation, we first knocked down p53 using siRNA in the H226 parental cells to examine the resultant cetuximab and/or radiation response profile. As shown in Fig. 2A, p53 levels were significantly reduced 48 hrs following knock down of p53. Thereafter, treatment with cetuximab did not significantly inhibit the growth of cells without p53. In contrast, treatment with cetuximab in HP cells with p53 resulted in significant growth inhibition in a dose dependent manner. Similarly, we observed a significant decrease in the comet tail moment by neutral comet assay in HP cells without p53 following radiation treatment (Fig 2B). Since neutral comet assay detects primarily double-stranded DNA breaks, these results suggest that knock down of p53 in the sensitive parental HP cells induces resistance to not only the EGFR inhibitor, but also to radiation. To further explore this finding, we knocked down p53 in two cetuximab-sensitive lung cancer cell lines, A549 and H292. Consistent with our finding with H226, the sensitivity to cetuximab was significantly reduced in these two cell lines following p53 knock down, as shown in Fig. 2C. These results suggest a role of p53 in regulating tumor response to cetuximab.
Fig. 2. Effects of p53 knock down on cetuximab and radiation response cells.
p53 was knocked down by siRNA in the sensitive parental tumor cells as described in ‘Material and Methods”. Following siRNA, p53 level was examined by western blotting as shown in the middle panel. Thereafter, tumor cells with or without p53 siRNA were exposed to cetuximab for 72 hrs or 6 Gy radiation. The cetuximab response was determined by cell proliferation assay and the radiation response was determined by comet assay 1 hr following radiation. (A) & (B) depict the cetuximab and radiation response following p53 knock down in H226 cells. (C) confirms the reduced cetuximab sensitivity following p53 knock down in additional cetuximab sensitive lung cancer cell lines. Data points are represented as mean ± SEM. *p<0.05
Transfection of p53 enhances sensitivity to cetuximab and radiation in cetuximab resistant cells
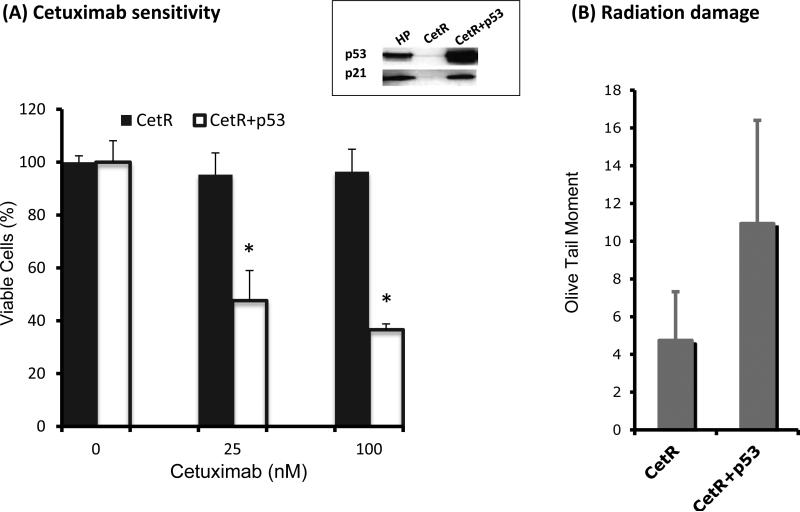
Since p53 was lost when tumor cells acquired resistance to EGFR inhibitors and radiation, we reconstituted a functional p53 in these resistant cells to examine whether the sensitivity to cetuximab and/or radiation could be restored. Although a single 1992 report noted a point mutation of p53 in codon 158 of exon 5 in H226 cells (15), we and several other investigators have confirmed the wt status of p53 without a point mutation in codon 158 in the H226 line (Supplementary Fig. S2) (16-18). We therefore transiently transfected a full length wt p53 into the Cet-R cells using a pC53-SN plasmid. As shown in Fig. 3, a functional p53 exogenous gene was expressed 48 hrs following transfection as shown by the consistent up-regulation of p21. Using a cell proliferation assay, we observed an increase of cetuximab sensitivity in the Cet-R clones following p53 transfection as shown in Fig 3A. The increased growth inhibition of cetuximab in p53-transfected Cet-R may result from induction of p21 which triggers cell cycle arrest and apoptosis as described previously (19). Similarly, using the comet analysis, we found enhancement of the radiation-induced comet tail moments in Cet-R cells following restoration of p53 (Fig. 3B). These results suggest that Cet-R cells regain sensitivity to cetuximab and radiation following reconstitution of a functional p53.
Fig. 3. Effects of p53 restoration on cetuximab and radiation response in cetuximab resistant cells in vitro.
p53 was transfected into the Cet-R cells using a pC53-SN plasmid as described in “Material and Methods”. Following transfection, restoration of functional p53 was determined by examining the expression of p53 and downstream effector p21 (shown in upper box). Thereafter, Cet-R cells with or without p53 transfection were either exposed to serial concentrations of cetuximab for 72 hrs or 6 Gy radiation. The cetuximab response (A) was determined by cell proliferation assay and the radiation response (B) was determined by comet assay 2 hours following radiation. Data points are represented as mean ± SEM. *p<0.05
Radiation response of cetuximab resistant clones with Tetracycline-inducible p53
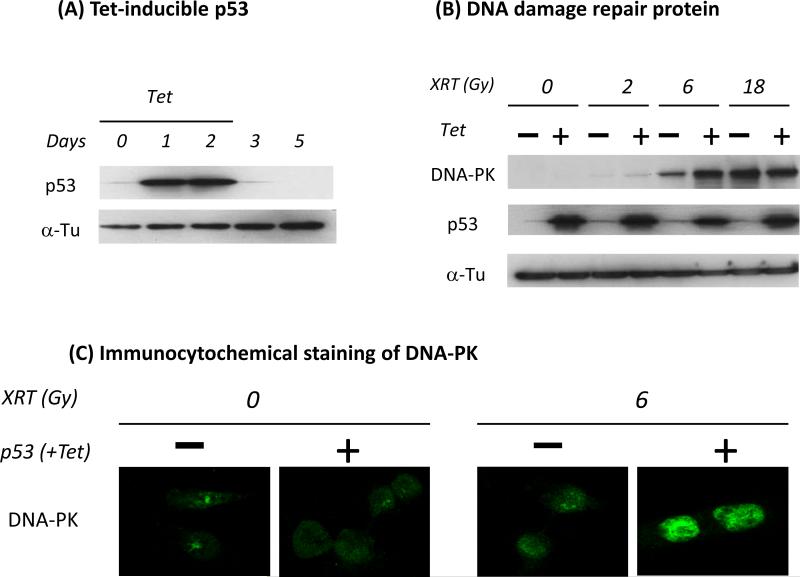
Although there was a trend towards increased radiation damage in Cet-R cells following transient transfection with p53, the difference as manifest by the comet assay was not statistically significant (Fig 3B). We generated a stable Cet-R clone with a Tet-inducible p53 system (TREx) that allowed for examination of cetuximab and radiation response in both culture and animal model systems. As shown in Fig 4A, p53 in the Cet-R cells with the T-REx system could be induced 1 day after Tet treatment. Thereafter, p53 expression was maintained once Tet was present in the medium up to day 2. Further, p53 was lost once Tet was removed from day 2 to day 5. This inducible and reversible p53 expression system may help reduce potential cytotoxic effects induced by overexpression of p53 during culture maintenance. Following generation of the Cet-R with inducible p53, we first examined the expression of DNA-PK which is a critical molecule involved in EGFR-regulated DNA damage repair and radiation response. As expected (Fig. 4B), DNA-PK was induced by radiation in a dose dependent manner in the original Cet-R cells. However, DNA-PK expression was further enhanced when p53 was induced by Tet in these Cet-R cells. A significant difference in DNA-PK expression was observed between the original and p53-induced Cet-R cells when they were exposed to 6 Gy radiation. To confirm this observation, we performed immunocytochemical staining to detect DNA-PK in individual cells. We found a consistent enhancement of DNA-PK expression in the p53-induced Cet-R cells following 6 Gy radiation exposure as shown in Fig 4C. These results indicate that radiation-induced DNA damage is increased in the Cet-R cells when p53 is re-introduced.
Fig. 4. Increased DNA damage following radiation in cetuximab resistant cells with induced p53.
Cet-R cells were stably transfected with a tetracycline (Tet)-inducible p53 as described in “Materials and Methods”. (A) shows the reversible induction of p53 by 2 μg/ml of Tet. (B) Following Tet treatment, cells were then treated with serial doses of radiation. The expression of DNA-PK was then determined by western blotting 24 hrs following radiation. (C) Transfected Cet-R cells were treated with or without 2 μg/ml of Tet followed by 6 Gy radiation. Twenty-four hrs later, the expression of DNA-PK was detected by immunocytochemical staining as described in “Materials and Methods”. Similar results were obtained in replicate experiments.
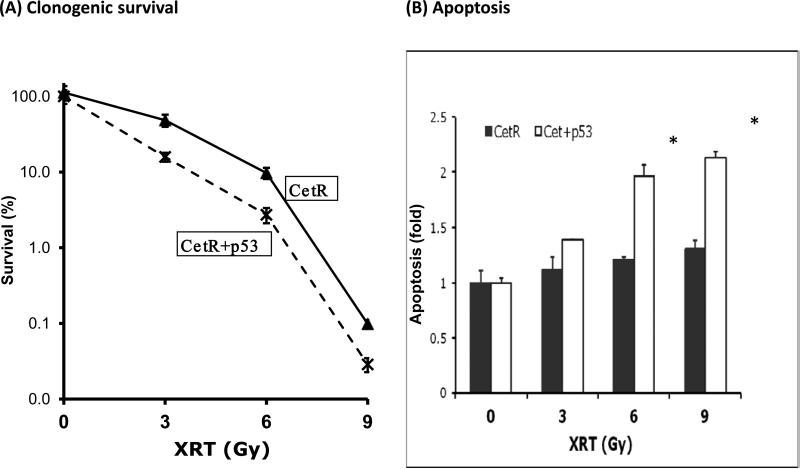
To further examine the role of p53 in regulating radiation response in Cet-R cells, we evaluated the radiosensitivity of Cet-R with or without p53 using clonogenic survival analysis. As shown in Fig 5A, Cet-R cells with p53 showed a lower survival rate than cells without p53 following exposure to 3, 6, or 9 Gy radiation. The enhanced cell death in these Cet-R cells with p53 was further confirmed by evaluating the apoptosis profile of tumor cells following radiation treatment. As shown in Fig. 5B, treatment with radiation resulted in a significant induction of apoptosis in a dose dependent manner in the Cet-R cells with p53, but not in cells without p53. Taken together, these results indicate that Cet-R cells with restoration of p53 manifest enhanced radiosensitivity, possibly by facilitating the induction of apoptosis.
Fig. 5. Increased radiosensitivity and radiation-induced apoptosis in cetuximab resistant cells with induced p53.
(A) Radiosensitivity of Cet-R with or without induced p53 was examined by clonogenic survival analysis following 3, 6 or 9 Gy radiation exposure as described in “Materials and Methods”. A stable expression of p53 was achieved by continuous exposure to 2 μg/ml of Tet during the 14-day period for colony formation. Results were expressed as the percentage of colony formation relative to controls without radiation treatment. Data points are represented as mean ± SD (B) Apoptosis of Cet-R cells with or without induced p53 was examined 72 hours following radiation by flow cytometry using Annexin and PI staining as described in “Materials and Methods”. The fold increase of cells in the early apoptotic population was determined by Cell Quest software (Becton Dickinson, San Diego, CA). Data points are represented as mean ± SEM. *p<0.05. Similar results were obtained in replicate experiments.
Restoration of p53 restores cetuximab and radiation sensitivitv in cetuximab resistant tumor xenografts
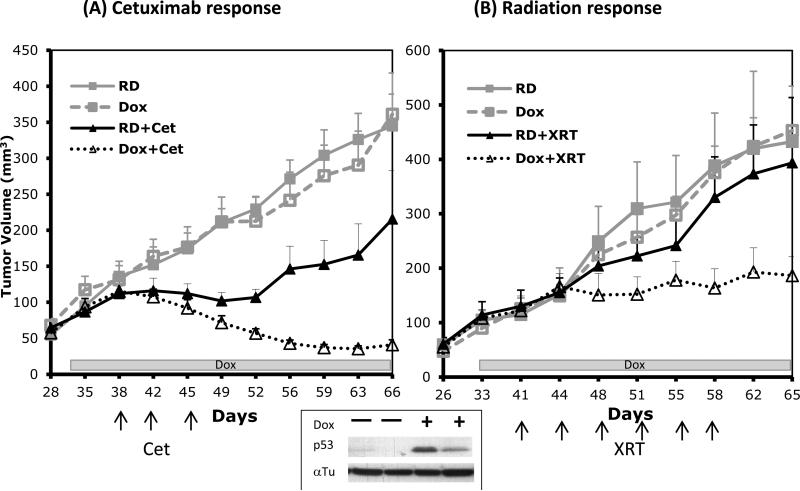
Following the establishment of a stable Cet-R clone with Tet-inducible p53, we then inoculated these cells into athymic mice to investigate the role of p53 in regulating resistance to cetuximab and radiation in vivo. When tumors reached 100 mm3 (30~33 days), mice were then fed either a regular diet (RD) or a regular diet with doxycycline supplementation (Dox) to the end of the experiment. As shown at the bottom of Fig 6A, a significant induction of p53 expression was observed in the tumor extracts from mice that received Dox for 8 days. In contrast, there was no p53 tumor expression in the mice receiving a regular diet. Following the confirmation of p53 in Dox-treated tumors, mice were then treated with 2 mg/kg of cetuximab twice weekly. As expected, tumors responded modestly to cetuximab in mice receiving a regular diet as shown by the solid lines in Fig 6A. However, tumors in the Dox-treated mice showed profound tumor regression following the same treatment with cetuximab. Indeed, tumors in the Dox-treated mice showed 5-fold volume reduction compared with Cet-treated tumors without p53 induction. A similar response pattern was observed when mice were treated with fractionated radiation as shown in Fig. 6B. These results confirm our in vitro data which indicate that restoration of p53 can restore sensitivity to cetuximab and radiation in Cet-R cells. In conjunction with the previous study that used transient tranfection of p53 (Fig 3), this animal xenograft model, with a stable p53-inducible tumor, provides additional evidence supporting a key role for p53 in regulating cetuximab and radiation response.
Fig. 6. Change in cetuximab and radiation response in cetuximab resistant tumors following induction of p53 in vivo.
Cet-R cells with Tet-inducible p53 were inoculated into athymic mice. Following the establishment of tumors, mice were fed with either a regular diet (RD) or a regular diet containing doxycycline (Dox) to induce p53 followed by cetuximab or radiation treatment as described in “Materials and Methods”. The specific days and treatment intervals for Dox, cetuximab and radiation are indicated by the box and arrow in each figure. The lower box shows the validation of p53 induction by western blotting in 2 representative mice receiving Dox for 8 days. Values represent mean tumor size (mm3) ± SEM (n=10 per group).
DISCUSSION
The development of acquired resistance to EGFR inhibitors is emerging as a potential treatment barrier for the optimization of EGFR targeted therapy. Similar to the development of acquired resistance to other molecular targeting agents, such as imatinib (Gleevec®), acquired resistance to EGFR inhibitors also exhibits cross-resistance to other therapeutic cancer drugs and radiation (20). To explore underlying mechanisms for acquired resistance to EGFR inhibitors and radiation, we screened for differences in the expression and activity of 42 key cellular signaling proteins between EGFR inhibitor resistant cells and their corresponding parental cells. Surprisingly, we found a robust loss of p53 in all resistant clones. This consistent loss of p53 in the resistant clones to two distinct classes of EGFR targeting agents drew our attention to investigate the role of p53 in regulating acquired resistance to EGFR inhibitors and radiation. Using two different approaches to either knock down p53 in the parental cells or restore functional p53 in the resistant cells, we found that the response to cetuximab and radiation can be regulated following manipulation of p53 expression. In addition, we found that p53 may affect response to EGFR inhibitors and radiation via regulation of cell cycle arrest, apoptosis and DNA damage repair. These results suggest that p53 plays a central role in regulating acquired resistance to EGFR inhibitors and radiation.
p53 is a tumor suppressor known to suppress cancer progression through the induction of cell cycle arrest, apoptosis or senescence in response to a variety of cellular stimuli. Hence, loss of p53 function in cells, either through mutation or post-translational modification might therefore be expected to lead to unchecked proliferation, tumor growth and therapeutic resistance (21, 22). A substantial number of clinical and preclinical studies identify an association of p53 mutation with poor prognosis and drug resistance (17, 23) for a variety of malignancies. In breast and colorectal tumors, p53 mutations are reported to predict resistance to a host of chemotherapeutic drugs including doxorubicin, cisplatin and 5-FU (24, 25). In contrast, reports have indicated that the expression of wild-type p53 is required for the efficacy of radiation and chemotherapy.
In addition to DNA damaging drugs and radiation, increasing evidence indicates the importance of p53 in regulating the response to several molecular targeted agents, including EGFR inhibitors. Previous studies demonstrating that cetuximab inhibits the growth of wild-type p53, but not mutated p53, cancer cells fostered the hypothesis that resistance to cetuximab may relate to p53 mutation (26). In addition, studies reported that gefitinib induced apoptosis through a p53-dependent signaling pathway and p53 mutation in combination with p21 expression in colorectal cancer was a predictor of resistance to gefitinib (27, 28). By comparing cell lines with different levels of p53, Rho et al. reported that the NSCLC H1299 cell line with a p53-null genotype was more resistant to gefitinib than cells with wild type p53 expression. Interestingly, following Tet-inducible transfection of p53, no significant change in cell growth doubling time and morphology was observed in H1299 cells. However restoration of p53 enhanced the sensitivity to gefitinib via induction of Fas-regulated apoptosis (29). Consistent with these findings, we found that restoration of functional p53 in our cetuximab-resistant cells did not induce cell cycle arrest, but did induce apoptosis (Supplementary Fig. S3). In addition, we found that apoptosis plays an important role in regulating the response to EGFR inhibitor and radiation in p53-transfected cetuximab-resistant cells (Fig. 5B). However, our model cannot exclude the possible role of p53-induced cell cycle arrest in regulating acquired resistance to EGFR inhibitors. Indeed, several different mechanisms have been suggested using distinct tumor types/models following restoration of p53 in recent studies. Ventura et al. showed that restoration of p53 induced apoptotic cell death in lymphoma, but cell cycle arrest and senescence in sarcoma (30). Xue et al. found that p53 induction in their hepatocarcinoma model led to growth arrest with senescence rather than apoptotic cell death (31). Hence, further studies with additional cell lines and model systems will be required to clarify the underlying mechanism of p53 in regulating acquired resistance to EGFR inhibitors and radiation.
Although cell cycle arrest and apoptosis are traditionally thought of as the major functions of p53 for tumor suppression, increasing evidence indicates that p53 may also impact signaling pathways that are involved in cell growth and transformation, including the PI3K/AKT and ERK pathways. It has been shown that the ERK pathway regulates the transcriptional function and subcellular localization of p53 (32). A recent report showed that mutant p53 initiated a feedback loop that involves ERK-mediated transcription of Egr-1 (Early growth response-1), which in turn increased the secretion of EGFR ligands with stimulation of EGFR signaling (33). Additional data showed that doxorubicin resistance was associated with decreased p53 and activated ERK signaling in leukemias (34). Beyond ERK, studies also indicate that p53 was involved in the regulation of survival PI3K/AKT pathway since induction of p53 resulted in decreased AKT activity in epithelial tumors (35, 36). Using non-viral gene transfer of p53 in PC3 prostate tumor cells, Bouali et al. also found a significant inhibition of AKT and ERK pathway following p53 transfection (37). In addition, exposure of p53-transfected cells to cetuximab further inhibited these two pathways. Consistent with these observations, we found a significant up-regulation of p-ERK along with p53 loss in our cetuximab-resistant cells (Fig 1). Following p53 transfection, a significant decrease of p-ERK was observed (Supplementary Fig. S3). These results suggest that p53 may regulate sensitivity to EGFR inhibitor by modulating EGFR downstream signaling functionality and apoptosis induction. These findings also support our observation that loss of p53 in EGFR inhibitor resistant cells associates with resistance to EGFR inhibitor and restoration of functional p53 can re-establish sensitivity to EGFR inhibitor.
Interestingly, a recent correlative clinical study suggested that p53 mutations predict response, not resistance to cetuximab in metastatic colorectal cancer. Oden-Gangloff and colleagues assessed tumor specimens from 64 patients with chemotherapy-refractory metastatic colorectal cancer who were treated with cetuximab-based chemotherapy (38). They unexpectedly found an association between p53 mutations and improved clinical outcome, particularly in patients without KRAS mutations. Variable data regarding the association of p53 mutations with EGFR inhibitor response highlights the complexity of p53 mutations. Unlike conventional tumor suppressors that are typically affected by nonsense frameshift mutations, some 80% of p53 mutations found in tumors are missense mutations (39). These mutations may encode oncogenic activities that are distinct from wild type and simple dominant-negative variants. The association between specific p53 mutations and drug resistance has been explored (40, 41). However, it remains unclear which property of mutant p53, such as loss of wild type p53 function, the acquisition of dominant-negative properties, the gain of new oncogenic function or perhaps a combination of the above is primarily involved in drug response and resistance. Therefore it remains important to examine the impact of different p53 mutations among a wide spectrum of tumor samples to sharpen our understanding of p53 mutation in relation to treatment response. Nonetheless, our study suggests an important role of p53 in acquired resistance to EGFR inhibitors in tumor cells with wild type p53. By examining changes in p53 from cells with the same p53 genotype, our resistant clones provide a simple platform to further investigate the role of p53 in regulating acquired resistance to EGFR inhibitors. To further extrapolate the applicability of these results to human tumors, it would be valuable to identify a cohort of patients that manifest acquired resistance to EGFR inhibitor therapy, and evaluate the p53 status of their tumors before the commencement of EGFR inhibitor therapy, and at the time of disease progression during therapy.
Acquired resistance to EGFR inhibitors presents a clinical problem not only due to the development of resistance to EGFR molecular targeting agents, but also due to potential manifestation of resistance to other drug or treatment modalities. Data from the current study suggests that the loss of p53 contributes to acquired resistance to both EGFR inhibitors and to radiation. Hence, restoring p53 may provide a valuable treatment strategy for tumors that manifest EGFR inhibitor resistance. Several approaches to restore functional p53 are in various stages of development (22). Efforts to restore p53 function include viral delivery of the wild type p53 gene to cancer cells, restoration of wild-type function to mutant p53, inhibition of p53 degradation, and activation of other p53 family members to substitute for mutant p53 and the resulting loss of wild-type p53 (22, 42). Although there remain many challenges, our current study may stimulate interest to apply p53 restoration approaches to enhance the efficacy of EGFR therapy and radiation.
In conclusion, we provide experimental evidence demonstrating a profound loss of p53 during the development of acquired resistance to EGFR inhibitors. Furthermore, we show that knock down of wild type p53 can increase resistance to EGFR inhibitors and radiation, while reconstitution of functional p53 sensitizes tumor cells to both treatments. Our acquired EGFR inhibitor resistant clones provide a valuable resource to further examine the role of p53 in EGFR targeting therapy. Results from current study also suggest opportunities for future clinical treatment strategies that incorporate EGFR-radiation combinations.
Supplementary Material
Acknowledgments
Grant support: This work was supported in part by NIH R01 CA 113448-01 (PMH)
REFERENCES
- 1.Mendelsohn J. The epidermal growth factor receptor as a target for cancer therapy. Endocr Relat Cancer. 2001;8:3–9. doi: 10.1677/erc.0.0080003. [DOI] [PubMed] [Google Scholar]
- 2.Harari PM, Allen GW, Bonner JA. Biology of Interactions: Antiepidermal Growth Factor Receptor Agents. J Clin Oncol. 2007;25:4057–65. doi: 10.1200/JCO.2007.11.8984. [DOI] [PubMed] [Google Scholar]
- 3.Ciardiello F, Tortora G. EGFR Antagonists in Cancer Treatment. N Engl J Med. 2008;358:1160–74. doi: 10.1056/NEJMra0707704. [DOI] [PubMed] [Google Scholar]
- 4.Bonner JA, Harari PM, Giralt J, et al. Radiotherapy plus cetuximab for locoregionally advanced head and neck cancer: 5-year survival data from a phase 3 randomised trial, and relation between cetuximab-induced rash and survival. The Lancet Oncol. 2010;11:21–8. doi: 10.1016/S1470-2045(09)70311-0. [DOI] [PubMed] [Google Scholar]
- 5.Engelman JA, Settleman J. Acquired resistance to tyrosine kinase inhibitors during cancer therapy. Curr Opin Genet Dev. 2008;18:73–9. doi: 10.1016/j.gde.2008.01.004. [DOI] [PubMed] [Google Scholar]
- 6.Jackman D, Pao W, Riely GJ, et al. Clinical Definition of Acquired Resistance to Epidermal Growth Factor Receptor Tyrosine Kinase Inhibitors in Non-Small-Cell Lung Cancer. J Clin Oncol. 2010;28:357–60. doi: 10.1200/JCO.2009.24.7049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wheeler DL, Dunn EF, Harari PM. Understanding resistance to EGFR inhibitors[mdash]impact on future treatment strategies. Nat Rev Clin Oncol. 2010;7:493–507. doi: 10.1038/nrclinonc.2010.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wheeler DL, Huang S, Kruser TJ, et al. Mechanisms of acquired resistance to cetuximab: role of HER (ErbB) family members. Oncogene. 2008;27:3944–56. doi: 10.1038/onc.2008.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Engelman JA, Janne PA. Mechanisms of Acquired Resistance to Epidermal Growth Factor Receptor Tyrosine Kinase Inhibitors in Non-Small Cell Lung Cancer. Clin Cancer Res. 2008;14:2895–9. doi: 10.1158/1078-0432.CCR-07-2248. [DOI] [PubMed] [Google Scholar]
- 10.Pao W, Miller VA, Politi KA, et al. Acquired Resistance of Lung Adenocarcinomas to Gefitinib or Erlotinib Is Associated with a Second Mutation in the EGFR Kinase Domain. PLoS Med. 2005;2:1–11. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pines G, K^stler WJ, Yarden Y. Oncogenic mutant forms of EGFR: Lessons in signal transduction and targets for cancer therapy. FEBS Letters. 2010;584:2699–706. doi: 10.1016/j.febslet.2010.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Benavente S, Huang S, Armstrong EA, et al. Establishment and Characterization of a Model of Acquired Resistance to Epidermal Growth Factor Receptor Targeting Agents in Human Cancer Cells. Clin Cancer Res. 2009;15:1585–92. doi: 10.1158/1078-0432.CCR-08-2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shen J, Behrens C, Wistuba II, et al. Identification and Validation of Differences in Protein Levels in Normal, Premalignant, and Malignant Lung Cells and Tissues Using High-Throughput Western Array and Immunohistochemistry. Cancer Res. 2006;66:11194–206. doi: 10.1158/0008-5472.CAN-04-1444. [DOI] [PubMed] [Google Scholar]
- 14.Yao F, Svensjo T, Winkler T, et al. Tetracycline repressor, tetR, rather than the tetR-mammalian cell transcription factor fusion derivatives, regulates inducible gene expression in mammalian cells. Hum Gene Ther. 1998;9:1939–50. doi: 10.1089/hum.1998.9.13-1939. [DOI] [PubMed] [Google Scholar]
- 15.Mitsudomi T, Steinberg SM, Nau MM, et al. p53 gene mutations in non-small-cell lung cancer cell lines and their correlation with the presence of ras mutations and clinical features. Oncogene. 1992;7:171–80. [PubMed] [Google Scholar]
- 16.Ikediobi ON, Davies H, Bignell G, et al. Mutation analysis of 24 known cancer genes in the NCI-60 cell line set. Mol Cancer Ther. 2006;5:2606–12. doi: 10.1158/1535-7163.MCT-06-0433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Petitjean A, Mathe E, Kato S, et al. Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: lessons from recent developments in the IARC TP53 database. Hum Mutat. 2007;28:622–9. doi: 10.1002/humu.20495. [DOI] [PubMed] [Google Scholar]
- 18.Faur N, Araud L, Laroche-Clary A, et al. The association between the T309G polymorphism of the MDM2 gene and sensitivity to anticancer drug is dependent on the p53 mutational status in cellular models. Br J Cancer. 2009;101:350–6. doi: 10.1038/sj.bjc.6605096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abdulkarim B, Sabri S, Deutsch E, et al. Antiviral agent Cidofovir restores p53 function and enhances the radiosensitivity in HPV-associated cancers. Oncogene. 2002;21:2334–46. doi: 10.1038/sj.onc.1205006. [DOI] [PubMed] [Google Scholar]
- 20.Ellis LM, Hicklin DJ. Resistance to Targeted Therapies: Refining Anticancer Therapy in the Era of Molecular Oncology. Clin Cancer Res. 2009;15:7471–8. doi: 10.1158/1078-0432.CCR-09-1070. [DOI] [PubMed] [Google Scholar]
- 21.Levine AJ, Oren M. The first 30 years of p53: growing ever more complex. Nat Rev Cancer. 2009;9:749–58. doi: 10.1038/nrc2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brown CJ, Lain S, Verma CS, et al. Awakening guardian angels: drugging the p53 pathway. Nat Rev Cancer. 2009;9:862–73. doi: 10.1038/nrc2763. [DOI] [PubMed] [Google Scholar]
- 23.Brosh R, Rotter V. When mutants gain new powers: news from the mutant p53 field. Nat Rev Cancer. 2009;9:701–13. doi: 10.1038/nrc2693. [DOI] [PubMed] [Google Scholar]
- 24.Royds JA, Iacopetta B. p53 and disease: when the guardian angel fails. Cell Death Differ. 2006;13:1017–26. doi: 10.1038/sj.cdd.4401913. [DOI] [PubMed] [Google Scholar]
- 25.Lin X, Howell SB. DNA mismatch repair and p53 function are major determinants of the rate of development of cisplatin resistance. Mol Cancer Ther. 2006;5:1239–47. doi: 10.1158/1535-7163.MCT-05-0491. [DOI] [PubMed] [Google Scholar]
- 26.Huether A, H^pfner M, Baradari V, et al. EGFR blockade by cetuximab alone or as combination therapy for growth control of hepatocellular cancer. Biochem Pharmacol. 2005;70:1568–78. doi: 10.1016/j.bcp.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 27.Chang G-C, Yu C-TR, Tsai C-H, et al. An epidermal growth factor inhibitor, Gefitinib, induces apoptosis through a p53-dependent upregulation of pro-apoptotic molecules and downregulation of anti-apoptotic molecules in human lung adenocarcinoma A549 cells. Eur J Pharmacol. 2008;600:37–44. doi: 10.1016/j.ejphar.2008.10.024. [DOI] [PubMed] [Google Scholar]
- 28.Ogino S, Meyerhardt JA, Cantor M, et al. Molecular Alterations in Tumors and Response to Combination Chemotherapy with Gefitinib for Advanced Colorectal Cancer. Clin Cancer Res. 2005;11:6650–6. doi: 10.1158/1078-0432.CCR-05-0738. [DOI] [PubMed] [Google Scholar]
- 29.Rho JK, Choi YJ, Ryoo B-Y, et al. p53 Enhances Gefitinib-Induced Growth Inhibition and Apoptosis by Regulation of Fas in Non-Small Cell Lung Cancer. Cancer Res. 2007;67:1163–9. doi: 10.1158/0008-5472.CAN-06-2037. [DOI] [PubMed] [Google Scholar]
- 30.Ventura A, Kirsch DG, McLaughlin ME, et al. Restoration of p53 function leads to tumour regression in vivo. Nature. 2007;445:661–5. doi: 10.1038/nature05541. [DOI] [PubMed] [Google Scholar]
- 31.Xue W, Zender L, Miething C, et al. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature. 2007;445:656–60. doi: 10.1038/nature05529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kojima K, Konopleva M, Samudio IJ, et al. Mitogen-Activated Protein Kinase Kinase Inhibition Enhances Nuclear Proapoptotic Function of p53 in Acute Myelogenous Leukemia Cells. Cancer Res. 2007;67:3210–9. doi: 10.1158/0008-5472.CAN-06-2712. [DOI] [PubMed] [Google Scholar]
- 33.Sauer L, Gitenay D, Vo C, et al. Mutant p53 initiates a feedback loop that involves Egr-1/EGF receptor/ERK in prostate cancer cells. Oncogene. 2010;29:2628–37. doi: 10.1038/onc.2010.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McCubrey JA, Abrams SL, Ligresti G, et al. Involvement of p53 and Raf//MEK//ERK pathways in hematopoietic drug resistance. Leukemia. 2008;22:2080–90. doi: 10.1038/leu.2008.207. [DOI] [PubMed] [Google Scholar]
- 35.Singh B, Reddy PG, Goberdhan A, et al. p53 regulates cell survival by inhibiting PIK3CA in squamous cell carcinomas. Genes Dev. 2002;16:984–93. doi: 10.1101/gad.973602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zwang Y, Sas-Chen A, Drier Y, et al. Two Phases of Mitogenic Signaling Unveil Roles for p53 and EGR1 in Elimination of Inconsistent Growth Signals. Molecular Cell. 2011;42:524–35. doi: 10.1016/j.molcel.2011.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bouali S, Chretien AS, Ramacci C, et al. P53 and PTEN expression contribute to the inhibition of EGFR downstream signaling pathway by cetuximab. Cancer Gene Ther. 2009;16:498–507. doi: 10.1038/cgt.2008.100. [DOI] [PubMed] [Google Scholar]
- 38.Oden-Gangloff A, Di Fiore F, Bibeau F, et al. TP53 mutations predict disease control in metastatic colorectal cancer treated with cetuximab-based chemotherapy. Br J Cancer. 2009;100:1330–5. doi: 10.1038/sj.bjc.6605008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Olivier M, Hollstein M, Hainaut P. TP53 mutations in human cancers: origins, consequences, and clinical use. Cold Spring Harb Perspect Biol. 2010;2:a001008. doi: 10.1101/cshperspect.a001008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bergamaschi D, Gasco M, Hiller L, et al. p53 polymorphism influences response in cancer chemotherapy via modulation of p73-dependent apoptosis. Cancer Cell. 2003;3:387–402. doi: 10.1016/s1535-6108(03)00079-5. [DOI] [PubMed] [Google Scholar]
- 41.Olivier M, Petitjean A, Marcel V, et al. Recent advances in p53 research: an interdisciplinary perspective. Cancer Gene Ther. 2009;16:1–12. doi: 10.1038/cgt.2008.69. [DOI] [PubMed] [Google Scholar]
- 42.Lu C, El-Deiry WS. Targeting p53 for enhanced radio- and chemo-sensitivity. Apoptosis. 2009;14:597–606. doi: 10.1007/s10495-009-0330-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.