Abstract
Background
Frontotemporal lobar degeneration (FTLD) is a progressive neurodegenerative disorder that can be triggered through genetic or sporadic mechanisms. MicroRNAs (miRNAs) have become a major therapeutic focus as their pervasive expression and powerful regulatory roles in disease pathogenesis become increasingly apparent. Here we examine the role of miRNAs in FTLD patients with TAR DNA-binding protein 43 pathology (FTLD-TDP) caused by genetic mutations in the progranulin (PGRN) gene.
Results
Using miRNA array profiling, we identified the 20 miRNAs that showed greatest evidence (unadjusted P < 0.05) of dysregulation in frontal cortex of eight FTLD-TDP patients carrying PGRN mutations when compared to 32 FTLD-TDP patients with no apparent genetic abnormalities. Quantitative real-time PCR (qRT-PCR) analyses provided technical validation of the differential expression for 9 of the 20 miRNAs in frontal cortex. Additional qRT-PCR analyses showed that 5 out of 9 miRNAs (miR-922, miR-516a-3p, miR-571, miR-548b-5p, and miR-548c-5p) were also significantly dysregulated (unadjusted P < 0.05) in cerebellar tissue samples of PGRN mutation carriers, consistent with a systemic reduction in PGRN levels. We developed a list of gene targets for the 5 candidate miRNAs and found 18 genes dysregulated in a reported FTLD mRNA study to exhibit anti-correlated miRNA-mRNA patterns in affected cortex and cerebellar tissue. Among the targets is brain-specific angiogenesis inhibitor 3, which was recently identified as an important player in synapse biology.
Conclusions
Our study suggests that miRNAs may contribute to the pathogenesis of FTLD-TDP caused by PGRN mutations and provides new insight into potential future therapeutic options.
Keywords: Frontotemporal lobar degeneration, TDP-43, microRNA, progranulin
Background
Frontotemporal lobar degeneration (FTLD) is the second most common cause of early-onset dementia after Alzheimer's Disease (AD) [1]. FTLD patients are clinically characterized by personality changes and disinhibited behaviour, often combined with a gradual and progressive language dysfunction [2]. Memory impairment is typically preserved in the early phase of disease, which distinguishes them from patients with AD. Pathologically, around 40% of FTLD patients present with neuronal and/or glial tau aggregates (FTLD-tau), whereas the majority of FTLD patients show ubiquitin-immunoreactive cytoplasmic and intranuclear inclusions historically referred to as FTLD-U (FTLD with ubiquitin-positive inclusions) [3]. More recently, it was shown that hyperphosphorylated and C-terminal truncated fragments of the nuclear protein TAR DNA-binding protein 43 (TDP-43) were the main component of the pathological inclusions in FTLD-U, and the term FTLD-TDP was introduced [4,5]. Three main patterns of TDP-43 pathology are recognized in FTLD-TDP, based on the anatomical distribution, morphology, and relative proportion of distinct types of inclusions [6,7]. In this study, we will follow the nomenclature based on the Mackenzie scheme [6] where FTLD-TDP type 1 is characterized by TDP-43 positive compact neuronal cytoplasmic inclusions and short neurites, FTLD-TDP type 2 presents with long TDP-43 positive neurites and FTLD-TDP type 3 is characterized by compact and granular cytoplasmic inclusions.
In the past decade, several different genes and chromosomal loci have been associated with FTLD. Mutations in the microtubule associated protein tau (MAPT) gene were first identified as a cause of familial FTLD-tau [8-10]. More recently, our group and others discovered that heterozygous mutations in the progranulin gene (PGRN) cause FTLD-TDP through a loss of function mechanism [11,12]. Patients with PGRN mutations maintain only a single functional copy of the gene, leading to the loss of 50% of functional PGRN, causing disease through haploinsufficiency. The reduced level of PGRN, a growth factor with a key role in a variety of cellular responses, provokes neurodegeneration and associated symptomatology in FTLD patients, including deficits in behaviour, language, and movement [13-15]. Interestingly, all patients with PGRN mutations present with FTLD-TDP pathology type 1 [16,17]; however, FTLD-TDP Type 1 is also observed in a subset of FTLD-TDP patients without PGRN mutations. Although there are clear pathologic distinctions in FTLD-TDP, the molecular pathways which underlie its progression are still mostly undefined. Recent advances in our understanding of the mammalian genomes, however, have revealed novel regulatory mechanisms with critical roles in disease pathogenesis, thus offering new avenues to explore.
The recent discovery of pervasive expression for noncoding RNAs (ncRNAs) in our genomes through extensive 'transcriptomic' efforts [18-23] has significantly enhanced our fundamental knowledge of cellular signaling cascades and will likely reshape future drug discovery efforts. Indeed, PGRN mutation carriers diagnosed with FTLD exhibit a range of pathologic and phenotypic outcomes, suggesting that other contributing factors, such as ncRNAs, mediate disease progression [4,11,12,15,19,24].
The miRNA class of ncRNA, in particular, has generated a lot of interest as their widespread role in many cellular functions becomes increasingly apparent [18,25-31]. One miRNA can control the expression of hundreds of downstream gene targets, underscoring the importance of characterizing their functional roles in vitro and in vivo [25]. Over the last few years, a growing number of publications have reported dysregulation of miRNA expression in numerous diseases, including neurodegenerative disorders, such as Alzheimer's disease and Huntington's disease [28,32-34]. Recent reports have also examined miRNA regulation of PGRN, suggesting that this gene is under the control of ncRNAs, including miR-107, and miR-29b [35,36]. Furthermore, our group previously showed a functional disruption of a miR-659 binding site in FTLD patients with a common genetic variant of PGRN (rs5848) [37].
Here we profiled miRNA expression in the frontal cortex of a population of FTLD-TDP patients with PGRN mutations and compared their miRNA expression pattern with a large group of FTLD-TDP patients without PGRN mutations, with the goal to identify miRNAs responsive to PGRN haploinsufficiency. For those miRNAs showing greatest evidence of dysregulation and that were validated technically by quantitative real-time PCR in frontal cortex, we further examined their expression in the cerebellum, with the expectation that PGRN levels are globally disrupted throughout the CNS. Finally, we developed a unique list of gene targets predicted to be regulated by miRNAs dysregulated in both frontal cortex and cerebellum in our patient samples, based on previously reported microarray mRNA data as well as bioinformatic miRNA target prediction sites.
Results
miRNA array results from frontal cortex
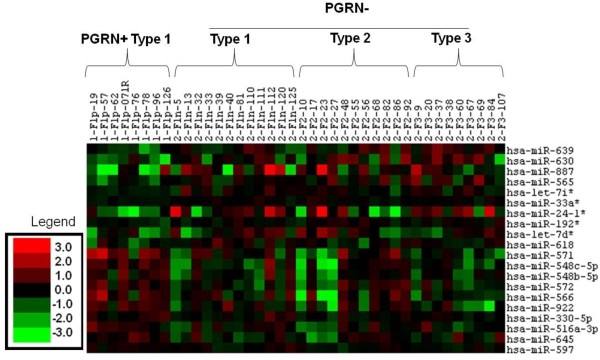
Using the ABI v2.0 arrays, we profiled the expression of 664 miRNAs from 40 FTLD-TDP patients, including 32 PGRN- FTLD-TDP and 8 PGRN+ FTLD-TDP patients (patient demographics listed in Table 1). There was detectable expression for 490 of the 664 miRNAs present on the array, and those candidate miRNAs were subjected to further analysis. We identified the 20 miRNAs which showed greatest evidence of differential expression (those with unadjusted P < 0.05) between the PGRN+ and PGRN- FTLD-TDP groups (Figure 1). None of these were statistically significant after accounting for multiple testing (overall P = 0.34) and the smallest q-value was 0.57, however we still pursued these 20 top-ranking miRNAs as the most promising candidates. Ten out of the 20 miRNAs had higher expression in the PGRN+ FTLD-TDP group (miR-571, miR548c-5p, miR-572, miR-922, miR-516a-3p, miR-566, miR-597, miR-645, miR-548b-5p, miR330-5p), while the other 10 miRNAs had lower expression in this group (miR-33a*, miR639, miR-630, miR-887, miR-565, miR-192*, miR-618, miR-24-1*, let-7d*, and let-7i*). All miRNA array results and statistical analyses are listed in Additional File 1. Graphical display of the 20 significantly changed miRNAs showed a consistent miRNA expression pattern in all three PGRN- FTLD-TDP subtypes (type 1, 2 and 3) compared to PGRN+ FTLD-TDP (Additional File 2).
Table 1.
Patient demographics for frontal cortex and cerebellar brain samples included in this study.
| Mutation status | N | FTLD-TDP Type 1 | FTLD-TDP Type 2 | FTLD-TDP Type 3 | Male/Female | Mean age at death ± SD |
|---|---|---|---|---|---|---|
| PGRN + Cortex | 8 | 8 | n/a | n/a | 3/5 | 75.5 ± 10.1 (63-87) |
|
PGRN - Cortex |
32 | 12 | 11 | 9 | 18/14 | 74.9 ± 10.5 (52-97) |
|
PGRN + Cerebellum |
10 | 10 | n/a | n/a | 7/3 | 72.3 ± 10.3 (56-87) |
|
PGRN - Cerebellum |
30 | 12 | 12 | 6 | 16/14 | 73.8 ± 10.3 (52-97) |
* There was overlap of 6 PGRN+ and 25 PGRN- samples between the cerebellum and frontal cortex analyses. n/a = not applicable
Figure 1.
Cluster diagram of Ct values for the 20 significant miRNAs from the array profiling. Each column represents a single sample, and each row represents a single miRNA. Green squares represent lower than median level of miRNA expression; black squares represent median level of miRNA expression; red squares represent higher than median level of miRNA expression. Legend units: 1.0 = differs from median probe intensity by one Ct unit. In addition to PGRN+ and PGRN- FTLD-TDP patients, 3 different FTLD-TDP pathologic subtypes are represented in this heat map: PGRN+ FTLD-TDP type 1 and PGRN- FTLD-TDP types 1, 2, and 3.
miRNA validation from frontal cortex
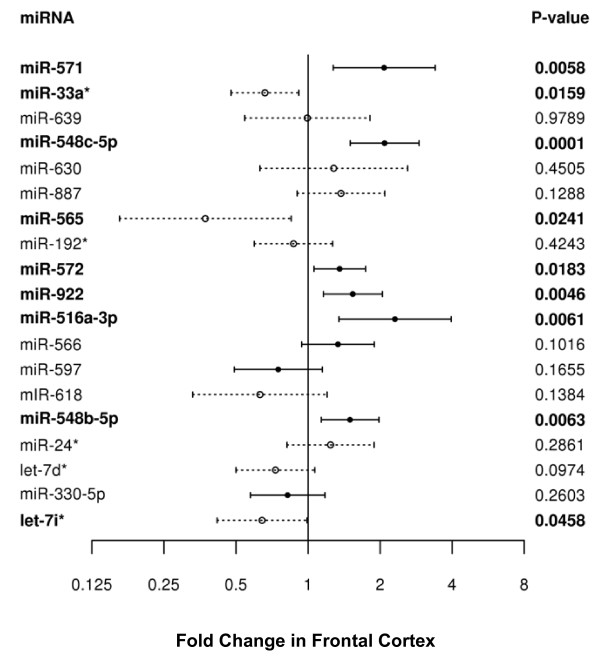
To perform technical validation of the miRNA array results, we evaluated the expression of the 20 significantly dysregulated miRNAs between the PGRN+ and PGRN- FTLD-TDP patients by qRT-PCR. One miRNA, miR-645, was undetectable using the individual miRNA assays from ABI. Of the remaining 19 detectable miRNAs, nine could be confirmed by qRT-PCR as significantly altered between the PGRN+ and PGRN- FTLD-TDP groups with P < 0.05. The validated miRNAs were miR-565, miR-33a*, let7i*, miR-922, miR-571, miR-572, miR-548b-5p, miR-548c-5p and miR-516a-3p (Figure 2).
Figure 2.
Validation of miRNA candidates in frontal cortex of FTLD patients. For each miRNA, fold change of PGRN+ FTLD-TDP patients compared to PGRN- FTLD-TDP patients with 95% confidence intervals are shown with their associated p-value. Significantly dysregulated miRNAs are shown in bold. Solid bars represent miRNAs which were upregulated in the original miRNA array, dashed bars represent miRNAs downregulated in the original miRNA array.
miRNA validation from cerebellum
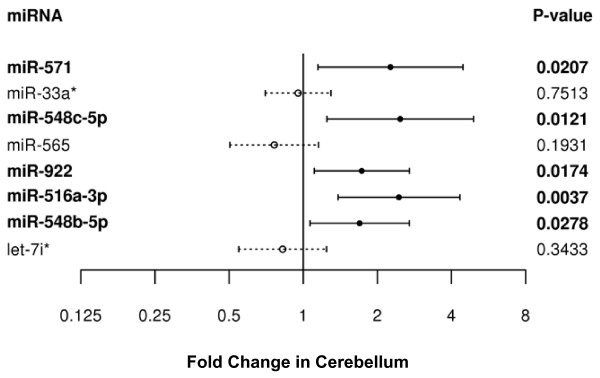
We hypothesized that the miRNAs dysregulated in the frontal cortex of the PGRN+ FTLD-TDP patients could also be differentially expressed in other brain areas as haploinsufficiency of PGRN function would not be region-specific. We therefore selected the 8 frontal cortex validated miRNAs with the largest estimated fold change and profiled their expression in the cerebellum of 10 PGRN+ FTLD-TDP and 30 PGRN- FTLD-TDP patients (patient demographics listed in Table 1, 31 patients common to frontal cortex analyses). Five of the 9 miRNAs (miR-922, miR-516a-3p, miR-571, miR-548b-5p, and miR-548c-5p), were significantly altered with P < 0.05 in the cerebellum (Figure 3).
Figure 3.
Validation of miRNA candidates in cerebellum of FTLD patients. For each miRNA, fold change of PGRN+ FTLD-TDP patients compared to PGRN- FTLD-TDP patients with 95% confidence intervals are shown with their associated p-value. Significantly dysregulated miRNAs are shown in bold. Solid bars represent miRNAs which were upregulated in the frontal cortex, dashed bars represent miRNAs downregulated in the frontal cortex.
miRNA expression in vitro
To further study the relationship between the loss of PGRN and the top five dysregulated miRNAs identified in frontal cortex and cerebellum, we performed a preliminary in vitro study in human neuroblastoma SH-SY5Y cells. In cells treated with PGRN siRNA we detected a 60% decrease in PGRN mRNA levels compared to negative control siRNA treated cells; however, no difference in miRNA expression for miR-516a-3p, miR-548b-5p and miR-548c-5p was observed (Additional File 3). Expression of miR-571 and miR-922 was too low (Ct < 35) and could not be included in the analysis.
TargetScan analysis and Ingenuity pathway analysis of miRNA targets
The overall role of miRNAs is to repress mRNA and protein expression [25,38]. To facilitate the identification of mRNA targets for the 5 PGRN+ FTLD-TDP associated miRNAs, we made use of publicly available Affymetrix mRNA arrays performed in FTLD-TDP patients with and without PGRN mutations (GEO record GDS3459) [39]. Since all 5 miRNAs were upregulated in frontal cortex and cerebellum of PGRN mutation carriers, we focused on mRNA targets which were downregulated in both frontal cortex and cerebellum of PGRN mutations carriers in the Affymetrix mRNA arrays. A total of 177 probesets showed significant downregulated expression in both the cortex and cerebellum of the PGRN+ FTLD-TDP patients (P < 0.05). When compared with the list of TargetScan predicted genes for each of the 5 PGRN+ FTLD-TDP associated miRNAs, 18 genes with anti-correlated mRNA-miRNA expression were identified (Table 2). Among the 18 genes, brain-specific angiogenesis inhibitor 3 (BAI3), glycerol kinase (GK) and solute carrier family 23, member 2 (SLC23A2) were targeted by 3 of the 5 miRNAs upregulated in the cortex and cerebellum of the PGRN+ FTLD-TDP patients. Seven genes were targeted by 2 of the 5 miRNAs, and 8 genes were targeted by 1 of the 5 miRNAs (Table 2).
Table 2.
Candidate gene targets identified in this study.
| Gene | Name | miRNA species |
|---|---|---|
| BAI3 | brain-specific angiogenesis inhibitor 3 | miR-922; miR-548b-5p; miR-548c-5p |
| GK | glycerol kinase | miR-571; miR-548b-5p; miR-548c-5p |
| SLC23A2 | solute carrier family 23 (nucleobase transporters), member 2 | miR-922; miR-548b-5p; miR-548c-5p |
| GTDC1 | glycosyltransferase-like domain containing 1 | miR-516a-3p; miR-571 |
| CNR1 | cannabinoid receptor 1 | miR-548b-5p; miR-548c-5p |
| FAM134B | family with sequence similarity 134, member B | miR-548b-5p; miR-548c-5p |
| HS2ST1 | heparan sulfate 2-O-sulfotransferase 1 | miR-548b-5p; miR-548c-5p |
| MYT1L | myelin transcription factor 1-like | miR-548b-5p; miR-548c-5p |
| NCAM1 | neural cell adhesion molecule 1 | miR-548b-5p; miR-548c-5p |
| TMEM135 | transmembrane protein 135 | miR-548b-5p; miR-548c-5p |
| ATP8A1 | ATPase, class I, type 8A, member 1 | miR-922 |
| KCNAB1 | potassium voltage-gated channel, shaker-related subfamily, beta member 1 | miR-922 |
| PTPRD | protein tyrosine phosphatase, receptor type, D | miR-922 |
| RASA1 | RAS p21 protein activator 1 | miR-922 |
| REEP1 | receptor accessory protein 1 | miR-922 |
| SNCA | synuclein, alpha | miR-922 |
| A2BP1 | ataxin-2-binding protein 1 | miR-516a-3p |
| ASTN1 | astrotactin 1 | miR-516a-3p |
List of top scoring genes predicted to be regulated by the 5 significantly upregulated miRNAs (miR-922, miR-516a-3p, miR-571, miR-548b-5p, and miR-548c-5p) from the 177 mRNAs significantly downregulated in frontal cortex and cerebellum in PGRN+ compared to PGRN- FTLD-TDP patients (GEO record GDS3459).
Next, for the 18 genes we found in common between the TargetScan and Affymetrix results, we examined their potential biological roles with Ingenuity Pathway analysis. Interestingly, neurological and cellular regulations were the most prominently represented biological roles of the significant pathways identified (P value < 0.05). In fact, 6 of the genes (astrotactin 1 (ASTN1), protein tyrosine phosphatase, receptor type, D (PTPRD), potassium voltage-gated channel, shaker-related subfamily, beta member 1 (KCNAB1), cannabinoid receptor 1 (CNR1), alpha-synuclein (SNCA), and neural cell adhesion molecule 1 (NCAM1)) were shown to have a specific role in behavioural responses, a phenotype which is consistently altered in FTLD (Additional File 4).
Discussion
Identifying the molecular events leading to pathogenic outcomes in neurodegenerative diseases, such as FTLD, may ultimately produce new avenues for prevention or treatment of these disorders. In this study, we report a novel role for ncRNAs in the molecular profile of FTLD patients with genetic mutations in the secreted growth factor PGRN. The miRNA family of ncRNAs showed distinct expression patterns in post-mortem brain tissue of FTLD-TDP patients carrying loss-of function mutations in PGRN compared to FTLD-TDP patients without known mutations, suggesting that miRNAs are potential biomarkers and therapeutic targets for genetically-linked dementia disorders.
Since the initial reports linking PGRN mutations to FTLD [11,12], the search for PGRN mediated signaling cascades has intensified, such as the recently reported associations with sortilin (SORT1) [40,41]. Through the comparison of ncRNA expression profiles from patients with genetic versus non-genetic (or sporadic) diagnosis of FTLD-TDP, we aimed to identify new pathways which are under the control of PGRN signaling in vivo. Indeed, in frontal cortex samples of FTLD-TDP patients, the expression of 3% of the detectable miRNAs from the expression arrays was significantly changed when PGRN+ and PGRN- FTLD-TDP patients were compared. Moreover, the dysregulated miRNAs showed consistent trends in all three of the PGRN- FTLD-TDP subtypes (types 1-3) compared to PGRN+ FTLD-TDP patients, suggesting the miRNA candidates we identified are unique to PGRN haploinsufficiency. In further support that the miRNAs dysregulated in our array and validation studies are under the control of systemic PGRN-mediated mechanisms, we found that 5 miRNAs (miR-548b-5p, miR-548c-5p, miR-571, miR-922, and miR-516a-3p) were also upregulated in the cerebellum of PGRN+ FTLD-TDP compared to PGRN- FTLD-TDP patients.
To further study the five candidate miRNAs, we silenced PGRN expression in SH-SY5Y cells; however, none of the 3 miRNAs detectable in SY-SY5Y cells displayed a significant difference in expression between control and PGRN-silenced cells. This finding suggests that long-term knockdown of PGRN may be necessary, consistent with the late onset of symptoms in human FTLD patients.
The mechanism by which PGRN haploinsufficiency in FTLD patients leads to altered miRNA expression is currently unclear and requires future studies. Progranulin downstream signalling involves ERK1/2 and AKT signalling and these are potential causes of altered miRNA expression. It is unlikely that the five miRNAs identified in this study are dysregulated as a result of TDP-43 aggregation since the FTLD-TDP type 1 pathology in the PGRN mutation carriers is indistinguishable from the pathology observed in sporadic FTLD-TDP patients.
It is now known that miRNAs can modulate mRNA stability and translation [25,38], therefore, we correlated publicly available mRNA expression results from sporadic (PGRN-) FTLD-TDP and PGRN+ FTLD-TDP patients with bioinformatic miRNA target predictions (TargetScan) for the 5 miRNAs upregulated in the frontal cortex and cerebellum [39]. Through this analysis, we identified 18 predicted gene targets with significantly downregulated mRNA expression profiles in PGRN+ FTLD-TDP patients. The anti-correlated expression of the upregulated miRNAs with their downregulated mRNA targets in PGRN+ patients parallels the established miRNA-mRNA regulatory relationship. Notably, Ingenuity pathway analysis of the 18 genes revealed that they have important links to biological functions involved in FTLD disease pathogenesis, including nervous system development, behavioural responses, and cell growth. Indeed, ASTN1 is known to regulate neuronal migration in cortical regions of developing brain [42], SNCA is associated with neurodegeneration and dementias, including links to FTLD-TDP in PGRN+ patients [43-46] and REEP1 has been implicated in corticospinal neurodegenerative disorders [47]. Importantly, only 3 genes are predicted to be targeted by 3 of the 5 miRNAs significantly dysregulated in both frontal cortex and cerebellum, including BAI3, a cell adhesion G protein coupled receptor. This finding is of significant interest since Bolliger et al. recently reported that C1q-like proteins can act as secreted signalling molecules that bind to BAI3 leading to the regulation of synapse formation and maintenance. In support of their finding, both C1ql and BAI3 are highly and specifically expressed in brain and are enriched in neurons [48]. Based on these findings our current data suggest that the loss of PGRN may increase the expression of miR-922, miR-548b-5p and miR-548c-5p through unknown mechanisms, leading to a decrease in the levels of BAI3, an essential protein for synapse biology.
Conclusions
Overall, our studies support a novel role for miRNAs in FTLD-TDP due to PGRN dysfunction and emphasize the value of combined miRNA and mRNA analyses. Future experiments in cell and animal models are needed to further evaluate the clinical potential of the miRNAs and gene targets identified in this study. The recent progress in human trials for miRNA-based therapeutics in non-CNS related disorders [49,50] offers hope for new alternatives for the treatment of dementias, including FTLD.
Methods
Brain samples
For the miRNA array experiment, post-mortem midfrontal cortex tissue was isolated from a collection of 40 FTLD-TDP patients selected from the Mayo Clinic Jacksonville (MCJ) brain bank. All samples were obtained with appropriate informed consent with ethical committee approval. FTLD patients included the following pathologic classifications: FTLD-TDP type 1 without PGRN mutations (PGRN- type 1, N = 12), FTLD-TDP type 2 (N = 11), FTLD-TDP type 3 (N = 9) and FTLD-TDP type 1 with PGRN mutations (PGRN+ type I, N = 8). Total RNA quantification was performed using a NanoDrop ND-1000 spectrophotometer (NanoDrop, Wilmington, DE). RNA quality was evaluated by the Agilent RNA 6000 Nano Kit (Agilent, Santa Clara, CA) and only samples with an RNA integrity value greater than 5 were included in this study. Mean RINs in frontal cortex were PGRN+ (5.6 ± 0.4), PGRN- type 1 (6.2 ± 0.6), FTLD-TDP type 2 (6.2 ± 1.0), and FTLD-TDP type 3 (6.4 ± 0.8). Mean RINs in cerebellums were PGRN+ (7.1 ± 0.6), PGRN- type 1 (7.0 ± 1.9), FTLD-TDP type 2 (7.2 ± 1.8), and FTLD-TDP type 3 (7.9 ± 0.4). Cerebellar tissue of sufficient quality for miRNA expression analyses was also available for 31 of these FTLD-TDP patients. For the miRNA expression analyses in cerebellum, 9 additional FTLD-TDP patients were obtained from the MCJ brain bank. Importantly, all PGRN mutations included in this study were clear pathogenic loss-of-function mutations, leading to haploinsufficiency. Demographic and neuropathologic information on all patients included in this study are summarized in Table 1.
miRNA array analyses
For mature miRNA expression profiling, real-time RT-PCR was performed using TaqMan Human MicroRNA Low Density Arrays Version 2.0 (Applied Biosystems) which contain 667 unique assays specific to human mature miRNAs in a two-card format. Total RNA was isolated from human frontal cortical tissue using the miRVana PARIS kit from Ambion (Applied Biosystems). Total RNA (150 ng) was reverse transcribed to cDNA for mature miRNAs using Megaplex RT Primers (Applied Biosystems) in 7.5 μls of final reaction volume. Subsequently, 2.5 μls of cDNA was pre-amplified in a 25 μl final volume with PreAmp Master Mix and Megaplex PreAmp Primers using standard conditions according to manufacturer's instructions. Preamplified cDNA was diluted in 0.1× Tris-EDTA (pH 8.0), applied to miRNA real-time array plates and mature miRNA expression was assessed using Applied Biosystems 7900HT System.
Statistical analysis of miRNA array data
There were 664 miRNAs profiled for each of the 40 samples; Ct values were obtained with the automatic baseline and manual Ct set to 0.1 threshold. Some miRNAs were only minimally expressed, and were excluded from further analyses; specifically we excluded those for which 20% or more of the samples had a missing Ct or Ct > 35. Lowess smoothing was used to normalize measures across individuals. Missing values were imputed using a K-Nearest Neighbour approach as described by Tusher et al. [51]. Any particularly extreme values for each miRNA were 'shrunk' in towards the center of the distribution so as to lessen their influence. For each comparison of two groups, two sample t-tests were used to assess nominal significance. The Westfall-Young min-P approach using 1000 permutations of group labels was used to obtain p-values adjusted for multiple testing [52]. Empirical q-values were also estimated using the permuted data.
Heat Maps and Box plots based on the miRNA array data
Normalized Ct values were adjusted by subtracting the Ct value from an arbitrary constant value of 40 so that a higher adjusted Ct value would correspond to a higher miRNA expression. The table of adjusted Ct values for the 20 significantly dysregulated miRNAs (P < 0.05) between PGRN+ and PGRN- FTLD-TDP patients was loaded in Cluster 3.0 [53]. A heat map showing the miRNA expression profiles for all the samples was generated after median centering the adjusted Ct values for each miRNA. The normalized and adjusted Ct values were summarized across groups with boxplots.
Validation of miRNA candidates in frontal cortex and cerebellum
The top 20 miRNA candidates identified in the miRNA array experiment were selected for validation by qRT-PCR in the same set of 8 PGRN+ and 32 PGRN- FTLD-TDP patient samples. In brief, 50 μls of reverse transcription primers (miRNA assay kits from ABI) for the 20 miRNAs plus RNU48 as an endogenous control were divided into 3 primer pools, lyophilized, and subsequently resuspended in water for each pool resulting in 5× multiplex RT primer pool. Total RNA (200 ng) was reverse transcribed in a 20-μl reaction volume using the miRNA Reverse Transcription Kit (Applied Biosystems) and 1 μl of cDNA was used in the Taqman miRNA assays [19]. Where duplicate Ct values differed by more than 2, the more extreme one relative to the distribution of Ct values across all samples was deleted; otherwise the mean of the duplicates was used as the final Ct for a transcript. Delta Cts were calculated by subtracting the Ct of the endogenous control RNU48. Minus delta Cts were used as the final values for analysis and assumed to represent the log base 2 of scaled expression levels. Two sample t-tests and corresponding 95% confidence intervals (CI) were used to compare groups, and the differences between means and CIs were exponentiated to provide fold change estimates under the assumption of perfect probe efficiency.
For a total of 8 miRNAs validated in frontal cortex and showing the largest fold-change, we also performed qRT-PCR using cerebellum tissue samples from 10 PGRN+ and 30 PGRN- FTLD-TDP patients, using similar techniques and analyses, as described above. Individual Ct values in the validation data sets for both the cortex and cerebellum are provided in Additional File 5.
miRNA expression in vitro
SH-SY5Y cells were cultured in a 1:1 mixture of OPTI-MEM and DMEM containing 5% heat-inactivated FBS and 1% penicillin/streptomycin. Cell cultures were kept at 37°C in a humidified atmosphere with 5% CO2. Cells were seeded at 175, 000 cells/well in 6-well plates and 24 h later transfected with siRNA against PGRN (Qiagen) or negative control siRNA at a final concentration of 25 nM using Lipofectamine2000 (Invitrogen). After 48 h of transfection, total RNA was extracted from SH-SY5Y cells and quantitative RT-PCR was performed as described above in the miRNA validation methods section.
Bioinformatics analysis
It has been extensively reported that miRNAs primarily decrease mRNA expression and repress translation [38]. For the miRNA candidates significantly dysregulated in the frontal cortex and cerebellum of PGRN+ FTLD-TDP patients, we identified their predicted mRNA targets through TargetScan. For each of those miRNAs, we compared their predicted gene targets with mRNA expression results of PGRN- and PGRN+ FTLD-TDP patients deposited in the Gene Expression [20] database. The GEO Affymetrix array dataset published by Chen-Plotkin et al. (Geo record GDS3459) profiled mRNA levels in several tissue types from controls, as well as PGRN+ and PGRN- FTLD patients [39]. The significantly dysregulated miRNAs which were anti-correlated in expression with their mRNA targets in the Affymetrix data set were further analyzed by Ingenuity software for insight into their biological roles.
Abbreviations
PGRN: progranulin; TDP-43: TAR DNA-binding protein; FTLD: frontotemporal lobar degeneration; ASTN1: astrotactin 1; PTPRD: protein tyrosine phosphatase, receptor type, D; KCNAB1: potassium voltage-gated channel, shaker-related subfamily, beta member 1; CNR1: cannabinoid receptor 1; SNCA: alpha-synuclein; NCAM1: neural cell adhesion molecule 1; BAI3: brain-specific angiogenesis inhibitor 3; GK: glycerol kinase; SLC23A2: solute carrier family 23, member 2.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
JK, JC and RR wrote the manuscript; JK, NK, MB, NF, and RR performed all experiments; JK, NK, KC, JC, and RR analyzed all results; JK, MB, NF, MDH, JG, DD, KAJ, NGR-R, BFB and RR categorized and prepared all patient samples. The final version of the manuscript is approved by all authors.
Supplementary Material
miRNA analysis from PGRN+ and PGRN- FTLD patients. A table listing all results and statistical analyses from the miRNA arrays.
Boxplot profiling of miRNAs in FTLD subtypes. This file shows comparative boxplots for all 20 significant miRNAs identified in the miRNA arrays.
Expression analysis of miRNAs in vitro with PGRN knockdown. This file shows miRNA expression in human neuroblastoma SH-SY5Y cells upon siRNA knockdown of PGRN.
Pathway analysis of mRNA targets for FTLD-associated miRNAs. This file shows Ingenuity pathway analysis of anti-correlated mRNA-miRNAs in PGRN+ FTLD-TDP patients.
qPCR data from miRNA validation studies. A table listing individual Ct values in the validation data sets for both the cortex and cerebellum.
Contributor Information
Jannet Kocerha, Email: jannet.kocerha@emory.edu.
Naomi Kouri, Email: kouri.naomi@mayo.edu.
Matt Baker, Email: baker.matt@mayo.edu.
NiCole Finch, Email: finch.nicole@mayo.edu.
Mariely DeJesus-Hernandez, Email: dejesushernandez.mariely@mayo.edu.
John Gonzalez, Email: gonzalez.john@mayo.edu.
Kumaravel Chidamparam, Email: kumarc@oceanridgebio.com.
Keith A Josephs, Email: josephs.keith@mayo.edu.
Bradley F Boeve, Email: boeve.bradley@mayo.edu.
Neill R Graff-Radford, Email: graffradford.neill@mayo.edu.
Julia Crook, Email: crook.julia@mayo.edu.
Dennis W Dickson, Email: dickson.dennis@mayo.edu.
Rosa Rademakers, Email: rademakers.rosa@mayo.edu.
Acknowledgements
This work was supported by the National Institute of Health grant numbers P50 AG16574 (Mayo Clinic Alzheimer's Disease Research Center; ADRC), R01 NS065782, R01 AG26251 and R01 AG37941, the Consortium for frontotemporal dementia (CFR), the Association for Frontotemporal Degeneration (AFTD) and the Robert and Clarice Smith Fellowship program. The authors would further like to thank Dr. David Knopman and Dr. Ronald Petersen for their continued ascertainment of FTLD patients and family members as part of the Mayo Clinic ADRC.
References
- Cardarelli R, Kertesz A, Knebl JA. Frontotemporal dementia: a review for primary care physicians. Am Fam Physician. 2010;82(11):1372–1377. [PubMed] [Google Scholar]
- McKhann GM, Albert MS, Grossman M, Miller B, Dickson D, Trojanowski JQ. Clinical and pathological diagnosis of frontotemporal dementia: report of the Work Group on Frontotemporal Dementia and Pick's Disease. Arch Neurol. 2001;58(11):1803–1809. doi: 10.1001/archneur.58.11.1803. [DOI] [PubMed] [Google Scholar]
- Morris HR, Khan MN, Janssen JC, Brown JM, Perez-Tur J, Baker M, Ozansoy M, Hardy J, Hutton M, Wood NW. et al. The genetic and pathological classification of familial frontotemporal dementia. Arch Neurol. 2001;58(11):1813–1816. doi: 10.1001/archneur.58.11.1813. [DOI] [PubMed] [Google Scholar]
- Cairns NJ, Bigio EH, Mackenzie IR, Neumann M, Lee VM, Hatanpaa KJ, White CL, Schneider JA, Grinberg LT, Halliday G. et al. Neuropathologic diagnostic and nosologic criteria for frontotemporal lobar degeneration: consensus of the Consortium for Frontotemporal Lobar Degeneration. Acta Neuropathol. 2007;114(1):5–22. doi: 10.1007/s00401-007-0237-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, Bruce J, Schuck T, Grossman M, Clark CM. et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314(5796):130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- Mackenzie IR, Baborie A, Pickering-Brown S, Du Plessis D, Jaros E, Perry RH, Neary D, Snowden JS, Mann DM. Heterogeneity of ubiquitin pathology in frontotemporal lobar degeneration: classification and relation to clinical phenotype. Acta Neuropathol. 2006;112(5):539–549. doi: 10.1007/s00401-006-0138-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampathu DM, Neumann M, Kwong LK, Chou TT, Micsenyi M, Truax A, Bruce J, Grossman M, Trojanowski JQ, Lee VM. Pathological heterogeneity of frontotemporal lobar degeneration with ubiquitin-positive inclusions delineated by ubiquitin immunohistochemistry and novel monoclonal antibodies. Am J Pathol. 2006;169(4):1343–1352. doi: 10.2353/ajpath.2006.060438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutton M, Lendon CL, Rizzu P, Baker M, Froelich S, Houlden H, Pickering-Brown S, Chakraverty S, Isaacs A, Grover A. et al. Association of missense and 5'-splice-site mutations in tau with the inherited dementia FTDP-17. Nature. 1998;393(6686):702–705. doi: 10.1038/31508. [DOI] [PubMed] [Google Scholar]
- Poorkaj P, Bird TD, Wijsman E, Nemens E, Garruto RM, Anderson L, Andreadis A, Wiederholt WC, Raskind M, Schellenberg GD. Tau is a candidate gene for chromosome 17 frontotemporal dementia. Ann Neurol. 1998;43(6):815–825. doi: 10.1002/ana.410430617. [DOI] [PubMed] [Google Scholar]
- Spillantini MG, Murrell JR, Goedert M, Farlow MR, Klug A, Ghetti B. Mutation in the tau gene in familial multiple system tauopathy with presenile dementia. Proc Natl Acad Sci USA. 1998;95(13):7737–7741. doi: 10.1073/pnas.95.13.7737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker M, Mackenzie IR, Pickering-Brown SM, Gass J, Rademakers R, Lindholm C, Snowden J, Adamson J, Sadovnick AD, Rollinson S. et al. Mutations in progranulin cause tau-negative frontotemporal dementia linked to chromosome 17. Nature. 2006;442(7105):916–919. doi: 10.1038/nature05016. [DOI] [PubMed] [Google Scholar]
- Cruts M, Gijselinck I, van der Zee J, Engelborghs S, Wils H, Pirici D, Rademakers R, Vandenberghe R, Dermaut B, Martin JJ. et al. Null mutations in progranulin cause ubiquitin-positive frontotemporal dementia linked to chromosome 17q21. Nature. 2006;442(7105):920–924. doi: 10.1038/nature05017. [DOI] [PubMed] [Google Scholar]
- Kelley BJ, Haidar W, Boeve BF, Baker M, Graff-Radford NR, Krefft T, Frank AR, Jack CR Jr, Shiung M, Knopman DS. et al. Prominent phenotypic variability associated with mutations in Progranulin. Neurobiol Aging. 2009;30(5):739–751. doi: 10.1016/j.neurobiolaging.2007.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rademakers R, Baker M, Gass J, Adamson J, Huey ED, Momeni P, Spina S, Coppola G, Karydas AM, Stewart H. et al. Phenotypic variability associated with progranulin haploinsufficiency in patients with the common 1477C-- > T (Arg493X) mutation: an international initiative. Lancet Neurol. 2007;6(10):857–868. doi: 10.1016/S1474-4422(07)70221-1. [DOI] [PubMed] [Google Scholar]
- Seelaar H, Rohrer JD, Pijnenburg YA, Fox NC, van Swieten JC. Clinical, genetic and pathological heterogeneity of frontotemporal dementia: a review. J Neurol Neurosurg Psychiatry. 2010. [DOI] [PubMed]
- Josephs KA, Ahmed Z, Katsuse O, Parisi JF, Boeve BF, Knopman DS, Petersen RC, Davies P, Duara R, Graff-Radford NR. et al. Neuropathologic features of frontotemporal lobar degeneration with ubiquitin-positive inclusions with progranulin gene (PGRN) mutations. J Neuropathol Exp Neurol. 2007;66(2):142–151. doi: 10.1097/nen.0b013e31803020cf. [DOI] [PubMed] [Google Scholar]
- Mackenzie IR. The neuropathology and clinical phenotype of FTD with progranulin mutations. Acta Neuropathol. 2007;114(1):49–54. doi: 10.1007/s00401-007-0223-8. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE. et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447(7146):799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C. et al. The transcriptional landscape of the mammalian genome. Science. 2005;309(5740):1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- Faghihi MA, Kocerha J, Modarresi F, Engstrom PG, Chalk AM, Brothers SP, Koesema E, St Laurent G, Wahlestedt C. RNAi screen indicates widespread biological function for human natural antisense transcripts. PLoS One. 2010;5(10) doi: 10.1371/journal.pone.0013177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, Nishida H, Yap CC, Suzuki M, Kawai J. et al. Antisense transcription in the mammalian transcriptome. Science. 2005;309(5740):1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E. et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129(7):1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi G, Piccoli E, Benussi L, Caso F, Redaelli V, Magnani G, Binetti G, Ghidoni R, Perani D, Giaccone G. et al. A novel progranulin mutation causing frontotemporal lobar degeneration with heterogeneous phenotypic expression. J Alzheimers Dis. 2010;23(1):7–12. doi: 10.3233/JAD-2010-101461. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollander JA, Im HI, Amelio AL, Kocerha J, Bali P, Lu Q, Willoughby D, Wahlestedt C, Conkright MD, Kenny PJ. Striatal microRNA controls cocaine intake through CREB signalling. Nature. 2010;466(7303):197–202. doi: 10.1038/nature09202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocerha J, Faghihi MA, Lopez-Toledano MA, Huang J, Ramsey AJ, Caron MG, Sales N, Willoughby D, Elmen J, Hansen HF. et al. MicroRNA-219 modulates NMDA receptor-mediated neurobehavioral dysfunction. Proc Natl Acad Sci USA. 2009;106(9):3507–3512. doi: 10.1073/pnas.0805854106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocerha J, Kauppinen S, Wahlestedt C. microRNAs in CNS disorders. Neuromolecular Med. 2009;11(3):162–172. doi: 10.1007/s12017-009-8066-1. [DOI] [PubMed] [Google Scholar]
- van Rooij E, Sutherland LB, Qi X, Richardson JA, Hill J, Olson EN. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science. 2007;316(5824):575–579. doi: 10.1126/science.1139089. [DOI] [PubMed] [Google Scholar]
- Williams AH, Valdez G, Moresi V, Qi X, McAnally J, Elliott JL, Bassel-Duby R, Sanes JR, Olson EN. MicroRNA-206 delays ALS progression and promotes regeneration of neuromuscular synapses in mice. Science. 2009;326(5959):1549–1554. doi: 10.1126/science.1181046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zernecke A, Bidzhekov K, Noels H, Shagdarsuren E, Gan L, Denecke B, Hristov M, Koppel T, Jahantigh MN, Lutgens E. et al. Delivery of microRNA-126 by apoptotic bodies induces CXCL12-dependent vascular protection. Sci Signal. 2009;2(100):ra81. doi: 10.1126/scisignal.2000610. [DOI] [PubMed] [Google Scholar]
- Hebert SS, Horre K, Nicolai L, Papadopoulou AS, Mandemakers W, Silahtaroglu AN, Kauppinen S, Delacourte A, De Strooper B. Loss of microRNA cluster miR-29a/b-1 in sporadic Alzheimer's disease correlates with increased BACE1/beta-secretase expression. Proc Natl Acad Sci USA. 2008;105(17):6415–6420. doi: 10.1073/pnas.0710263105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau P, de Strooper B. Dysregulated microRNAs in neurodegenerative disorders. Semin Cell Dev Biol. 2010;21(7):768–773. doi: 10.1016/j.semcdb.2010.01.009. [DOI] [PubMed] [Google Scholar]
- Packer AN, Xing Y, Harper SQ, Jones L, Davidson BL. The bifunctional microRNA miR-9/miR-9* regulates REST and CoREST and is downregulated in Huntington's disease. J Neurosci. 2008;28(53):14341–14346. doi: 10.1523/JNEUROSCI.2390-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao J, Herl LD, Farese RV, Gao FB. MicroRNA-29b regulates the expression level of human progranulin, a secreted glycoprotein implicated in frontotemporal dementia. PLoS One. 2010;5(5):e10551. doi: 10.1371/journal.pone.0010551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WX, Wilfred BR, Madathil SK, Tang G, Hu Y, Dimayuga J, Stromberg AJ, Huang Q, Saatman KE, Nelson PT. miR-107 regulates granulin/progranulin with implications for traumatic brain injury and neurodegenerative disease. Am J Pathol. 2010;177(1):334–345. doi: 10.2353/ajpath.2010.091202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rademakers R, Eriksen JL, Baker M, Robinson T, Ahmed Z, Lincoln SJ, Finch N, Rutherford NJ, Crook RJ, Josephs KA. et al. Common variation in the miR-659 binding-site of GRN is a major risk factor for TDP43-positive frontotemporal dementia. Hum Mol Genet. 2008;17(23):3631–3642. doi: 10.1093/hmg/ddn257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466(7308):835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen-Plotkin AS, Geser F, Plotkin JB, Clark CM, Kwong LK, Yuan W, Grossman M, Van Deerlin VM, Trojanowski JQ, Lee VM. Variations in the progranulin gene affect global gene expression in frontotemporal lobar degeneration. Hum Mol Genet. 2008;17(10):1349–1362. doi: 10.1093/hmg/ddn023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrasquillo MM, Nicholson AM, Finch N, Gibbs JR, Baker M, Rutherford NJ, Hunter TA, DeJesus-Hernandez M, Bisceglio GD, Mackenzie IR. et al. Genome-wide screen identifies rs646776 near sortilin as a regulator of progranulin levels in human plasma. Am J Hum Genet. 2010;87(6):890–897. doi: 10.1016/j.ajhg.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu F, Padukkavidana T, Vaegter CB, Brady OA, Zheng Y, Mackenzie IR, Feldman HH, Nykjaer A, Strittmatter SM. Sortilin-mediated endocytosis determines levels of the frontotemporal dementia protein, progranulin. Neuron. 2010;68(4):654–667. doi: 10.1016/j.neuron.2010.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink JM, Hirsch BA, Zheng C, Dietz G, Hatten ME, Ross ME. Astrotactin (ASTN), a gene for glial-guided neuronal migration, maps to human chromosome 1q25.2. Genomics. 1997;40(1):202–205. doi: 10.1006/geno.1996.4538. [DOI] [PubMed] [Google Scholar]
- Clinton LK, Blurton-Jones M, Myczek K, Trojanowski JQ, LaFerla FM. Synergistic Interactions between Abeta, tau, and alpha-synuclein: acceleration of neuropathology and cognitive decline. J Neurosci. 2010;30(21):7281–7289. doi: 10.1523/JNEUROSCI.0490-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs GG, Botond G, Budka H. Protein coding of neurodegenerative dementias: the neuropathological basis of biomarker diagnostics. Acta Neuropathol. 2010;119(4):389–408. doi: 10.1007/s00401-010-0658-1. [DOI] [PubMed] [Google Scholar]
- Leverenz JB, Yu CE, Montine TJ, Steinbart E, Bekris LM, Zabetian C, Kwong LK, Lee VM, Schellenberg GD, Bird TD. A novel progranulin mutation associated with variable clinical presentation and tau, TDP43 and alpha-synuclein pathology. Brain. 2007;130(Pt 5):1360–1374. doi: 10.1093/brain/awm069. [DOI] [PubMed] [Google Scholar]
- Scott DA, Tabarean I, Tang Y, Cartier A, Masliah E, Roy S. A pathologic cascade leading to synaptic dysfunction in alpha-synuclein-induced neurodegeneration. J Neurosci. 2010;30(24):8083–8095. doi: 10.1523/JNEUROSCI.1091-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCorquodale Iii D, Ozomaro U, Huang J, Montenegro G, Kushman A, Citrigno L, Price J, Speziani F, Pericak-Vance MA, Zuchner S. Mutation screening of spastin, atlastin, and REEP1 in hereditary spastic paraplegia. Clin Genet. 2010. [DOI] [PMC free article] [PubMed]
- Bolliger MF, Martinelli DC, Sudhof TC. The cell-adhesion G protein-coupled receptor BAI3 is a high-affinity receptor for C1q-like proteins. Proc Natl Acad Sci USA. 2011;108(6):2534–2539. doi: 10.1073/pnas.1019577108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery RL, van Rooij E. Therapeutic advances in MicroRNA targeting. J Cardiovasc Pharmacol. 2011;57(1):1–7. doi: 10.1097/FJC.0b013e3181f603d0. [DOI] [PubMed] [Google Scholar]
- Seto AG. The road toward microRNA therapeutics. Int J Biochem Cell Biol. 2010;42(8):1298–1305. doi: 10.1016/j.biocel.2010.03.003. [DOI] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98(9):5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westfall PH, Zaykin DV, Young SS. Multiple tests for genetic effects in association studies. Methods Mol Biol. 2002;184:143–168. doi: 10.1385/1-59259-242-2:143. [DOI] [PubMed] [Google Scholar]
- de Hoon MJ, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics. 2004;20(9):1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
miRNA analysis from PGRN+ and PGRN- FTLD patients. A table listing all results and statistical analyses from the miRNA arrays.
Boxplot profiling of miRNAs in FTLD subtypes. This file shows comparative boxplots for all 20 significant miRNAs identified in the miRNA arrays.
Expression analysis of miRNAs in vitro with PGRN knockdown. This file shows miRNA expression in human neuroblastoma SH-SY5Y cells upon siRNA knockdown of PGRN.
Pathway analysis of mRNA targets for FTLD-associated miRNAs. This file shows Ingenuity pathway analysis of anti-correlated mRNA-miRNAs in PGRN+ FTLD-TDP patients.
qPCR data from miRNA validation studies. A table listing individual Ct values in the validation data sets for both the cortex and cerebellum.