Abstract
Heparan sulfate (HS) and heparin are highly sulfated polysaccharides exhibiting essential physiological functions. The sulfation patterns determine the functional selectivity for HS and heparin. Chemical synthesis of HS, especially those larger than a hexasaccharide, remains challenging. Enzymatic synthesis of HS has recently gained momentum. Here we describe the divergent assembly of HS heptasaccharides and nonasaccharides from a common hexasaccharide precursor. The hexasaccharide precursor was synthesized via a chemical method. The subsequent elongation, sulfation and epimerization were completed by glycosyltransferases, HS sulfotransferases and epimerase. Using the synthesized heptasaccharides, we discovered that the iduronic acid is critical for binding to fibroblast growth factor-2. We also designed a synthetic path to prepare a nonasaccharide with an antithrombin-binding affinity of 3 nM. Our method demonstrated the feasibility of combining chemical and enzymatic synthesis to prepare structurally defined HS oligosaccharides with desired biological activities.
Keywords: antithrombin, FGF, heparan sulfate, oligosaccharides, sulfotransferases
Introduction
Heparan sulfate (HS) is a unique class of macromolecular natural product that is present in large quantities on the mammalian cell surface and in the extracellular matrix. The roles of HS in regulating blood coagulation, embryonic development and the inflammatory responses and assisting viral/bacterial infections have been documented extensively. The structural unit of HS consists of a disaccharide repeat of glucuronic acid (GlcUA) or iduronic acid (IdoUA) and glucosamine (GlcN) carrying sulfo groups (Peterson et al. 2009). Sulfation types and the distribution of IdoUA dictate the biological activity of HS (Gama et al. 2006; de Paz and Seeberger 2008). Heparin, a widely used anticoagulant drug, is a specialized highly sulfated form of HS. The diverse biological functions present considerable opportunities for exploiting HS or HS-protein conjugates for developing new classes of anticancer (Shriver et al. 2004), antiviral (Baleux et al. 2009) and improved anticoagulant drugs (Chen et al. 2007). A method to construct HS is, therefore, critical for developing HS-based therapeutics. Total chemical synthesis is fully capable of preparing short fragments of HS. The most successful example is the synthesis of an antithrombin (AT)-binding heparin pentasaccharide analog known as fondaparinux (Petitou and van Boeckel 2004). However, the chemical synthesis of oligosaccharides larger than a hexasaccharide is extremely challenging (Petitou and van Boeckel 2004). In addition to the size, the number of oligosaccharides with diverse sulfation patterns required for structure–activity relationship studies presents another obstacle because the syntheses do not usually follow a single route. Typically, different sets of precursor structures are needed depending on the sulfation patterns of the target compounds. The preparations of an array of precursors are tedious and time-consuming. It is highly desirable for a divergent approach, where a single precursor can lead to various sulfated oligosaccharides. Chemo-enzymatic methods offer a promising solution.
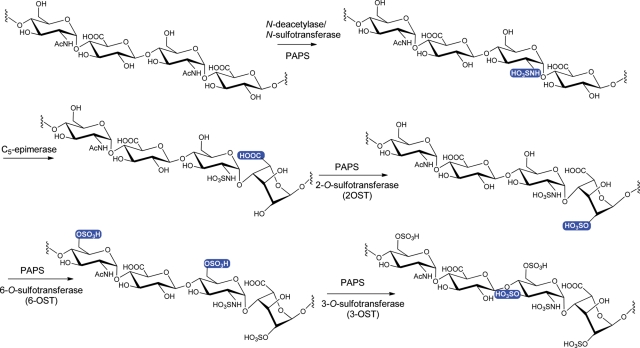
The biosynthesis of HS involves numerous specialized enzymes, including HS polymerase, epimerase and sulfotransferases (Figure 1). HS polymerase is responsible for building the polysaccharide backbone, containing the repeating unit of -GlcUA-GlcNAc-. The backbone is then modified by N-deacetylase/N-sulfotransferase (NST) (having two separate domains exhibiting the activity of N-deacetylase and NST, respectively), C5-epimerase (C5-epi, converting GlcUA to IdoUA), 2-O-sulfotransferase (2-OST), 6-OST and 3-OST to produce the fully elaborated HS. Using HS sulfotransferases and C5-epi, we previously developed a method to synthesize HS polysaccharides (Chen et al. 2005, 2007; Zhang et al. 2008) as well as oligosaccharides (Liu et al. 2010).
Fig. 1.
Biosynthetic pathway of HS. The HS biosynthetic pathway includes the biosynthesis of backbone and modifications. The synthesis of the backbone, a repeating unit of -GlcUA-GlcNAc without sulfo groups nor IdoUA units, is not shown. The modification site at each step is highlighted in a blue box.
In this article, we exploit to interface chemical and enzymatic synthesis to prepare different biologically active HS oligosaccharides from a single hexasaccharide precursor. The synthesis of hexasaccharide precursor (GlcNH2-GlcUA-Glc NH2-GlcUA-GlcNH2-GlcUA-OEt, hexasaccharide 1) was achieved by chemical synthesis in high yields. Sulfation and epimerization of GlcUA to IdoUA were achieved by enzymes. We demonstrated the synthesis of heptasaccharides carrying different sulfation patterns in less than three or four steps. The heptasaccharides were used to determine the critical structural elements for binding to fibroblast growth factor-2 (FGF-2) having no affinity to AT. Enzymatically extending the hexasaccharide precursor to a nonasaccharide, we synthesized a nonasaccharide that binds to AT with high affinity. Our results demonstrate that the combination of chemical and enzymatic synthesis provides the flexibility for the synthesis of a wide range of HS oligosaccharides. This technique could become a powerful tool for the on-demand synthesis of HS oligosaccharides to elucidate the structure and activity relationship of HS.
Results
Chemical synthesis of hexasaccharide 1
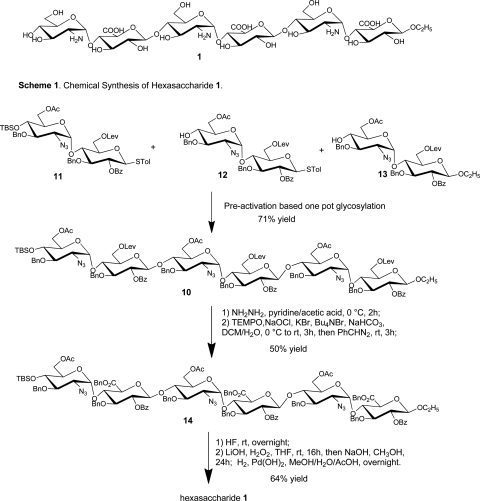
Chemical synthesis of the fully protected hexasaccharide 10 was performed using the pre-activation-based one-pot oligosaccharide synthesis method (Figure 2; Huang et al. 2004; Wang et al. 2010). Starting from three disaccharide building blocks 11–13 (Wang et al. 2010), hexasaccharide 10 was obtained in 71% yield in one pot. Deprotection of the hexasaccharide 10 was performed by first removing the three levulinoyl protecting groups with hydrazine in pyridine and acetic acid, followed by 2,2,6,6-tetramethyl-1-piperidinyloxy and sodium hypochloride oxidization (Lee et al. 2004; Huang and Huang 2007) and benzyl ester formation producing compound 14 in 50% yield. Subsequent silyl ether protective group removal, saponification and hydrogenation provided the fully deprotected hexasaccharide 1 in 64% overall yield. All the synthetic hexasaccharides were fully characterized by NMR and mass spectrometry (MS) (Supplementary data).
Fig. 2.
Synthetic scheme of hexasaccharide 1.
Enzymatic synthesis of heptasaccharides with different sulfation patterns
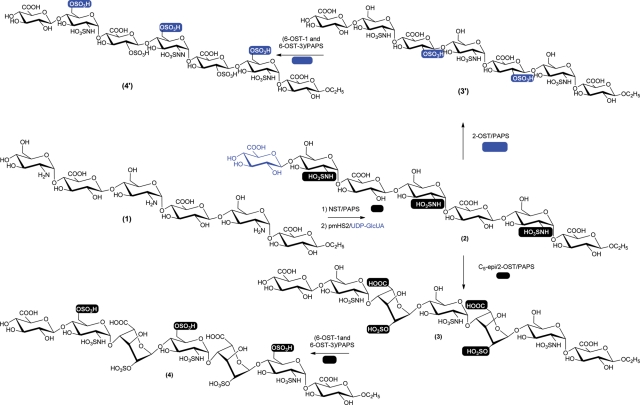
Using hexasaccharide 1 as a starting material, we synthesized five heptasaccharide derivatives differing in sulfations and the presence of IdoUA residues (Figure 3). Hexasaccharide 1 was first N-sulfated using NST to form N-sulfated hexasaccharide. The resultant hexasaccharide was then elongated to heptasaccharide 2 by Pasteurella multocida heparosan synthase-2 (pmHS2) using a UDP-GlcUA as a sugar donor. The N-sulfation at the nonreducing end is necessary for extension to a heptasaccharide as hexasaccharide 1 was unreactive to pmHS2.
Fig. 3.
Synthetic scheme of heptasaccharides. Hexasaccharide 1 (hexasaccharide precursor) was first N-sulfated using NST followed by the extension using pmHS2 and UDP-GlcUA to yield heptasaccharide 2. Heptasaccharide 2 was converted to heptasaccharide 3 by C5-epimerase and 2-OST and subsequently converted to heptasaccharide 4 by 6-OST-1 and 6-OST-3. Heptasaccharide 2 was also converted to heptasaccharide 3′ by 2-OST (without C5-epimerase) and subsequently converted to heptasaccharide 4′ by 6-OST-1 and 6-OST-3. The sites of modification at each step are highlighted in black (for the synthesis of heptasaccharides 2–4) or blue (for the synthesis of heptasaccharides 3′ and 4′) background.
Heptasaccharide 2 was then subjected to additional modifications to yield different products. In one modification, C5-epi was included at the 2-O-sulfation step to synthesize heptasaccharide 3, containing two IdoUA2S residues. Heptasaccharide 3 was subsequently modified by 6-OST-1 and 6-OST-3 to form heptasaccharide 4. In another modification, C5-epi was excluded in order to prepare heptasaccharide 3′, containing two GlcUA2S residues. Heptasaccharide 3′ was then modified by 6-OST-1 and 6-OST-3 to lead to heptasaccharide 4′.
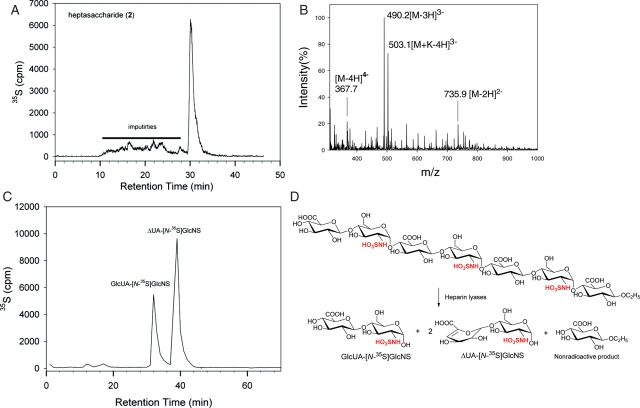
Structural analyses of heptasaccharides 2, 3 and 4 were completed using electrospray ionization (ESI) MS and disaccharide analysis. After N-[35S]sulfation and extension by pmHS2, heptasaccharide 2 displayed a major symmetric peak (30.5 min) by polyamine-based anion exchange (PAMN)-high-performance liquid chromatography (HPLC) with minor impurities (Figure 4A). We then resynthesized nonradioactively labeled heptasaccharide 2 under the identical condition. The product was purified by HPLC for MS analysis. ESI-MS analysis revealed the molecular weight of the product to be 1474.6 ± 0.2 Da, very close to the calculated molecular (1474.2 Da; Figure 4B). The structure of heptasaccharide 2 was further proved by the disaccharide analysis of heparin lyase digestion, a common approach used in analyzing the structure of HS oligosaccharides (Venkataraman et al. 1999; Zaia and Costello 2001; Lawrence et al. 2008; McCullough et al. 2010; Wei et al. 2011). To this end, 35S-labeled heptasaccharide 2 was digested to disaccharides as illustrated in Figure 4D. The resultant disaccharides were resolved by reverse-phase ion pairing (RPIP)-HPLC, showing two 35S-labeled disaccharides (Figure 4C). Coeluting with the disaccharide standards, we confirmed the 35S-peak eluted at 35 min is GlcUA-GlcNS and the 35S-peak eluted at 40 min is ΔUA-GlcNS. The measured ratio of GlcUA-GlcNS and ΔUA-GlcNS was 1:1.9, which is close to the theoretical value (1:2) of heptasaccharide 2 digested by heparin lyases. Taken together, our results proved the structure of heptasaccharide 2 as shown in Figure 3. Using similar techniques, we proved the successful synthesis of heptasaccharides 3 and 4. The data for the structural characterization of heptasaccharides 3 and 4 are shown in Supplementary data, Figures 1S and 2S. A 35S-labeled disaccharide from nitrous acid-degraded 2-O-[35S]sulfo heptasaccharide 3 coeluted with the disaccharide standard, IdoUA2S-AnMan (but not GlcUA2S-AnMan), confirming that heptasaccharide 3 contains IdoUA2S residues (Supplementary data, Figure 1SE). Furthermore, we confirmed the nonreducing terminus of heptasaccharide 4 to be a GlcUA because the heptasaccharide was susceptible to the digestion of β-glucuronidase (Supplementary data, Figure 3S).
Fig. 4.
Structural characterization of heptasaccharide 2. (A) The HPLC chromatogram of 35S-labeled heptasaccharide 2 using a PAMN column. Solid bar indicates the impurities. (B) The MS spectrum of HPLC-purified heptasaccharide 2. (C) The HPLC of the disaccharide analysis of heparin lyase-digested 35S-labeled heptasaccharide 2. The disaccharide analysis was carried out on a C18-column eluted under RPIP-HPLC conditions. The elution positions of the disaccharides were confirmed with appropriate disaccharide standards. (D) The reactions involved in the digestions of 35S-labeled heptasaccharide 2 using heparin lyases. The 35S-labeling sites are colored in red. A nonradioactive monosaccharide was also formed during the digestion; however, it was not detectable.
The structures of heptasaccharides 3′ and 4′ were proved by the disaccharide analysis. Here, we did not prepare sufficient amount of samples for MS analysis. For heptasaccharide 3′, we demonstrated the presence of GlcUA2S (Supplementary data, Figure 4S). As expected, the disaccharide analysis of heptasaccharide 4′ revealed the disaccharides GlcUA-GlcNS 6S and ΔUA2S-GlcNS6S (Supplementary data, Figure 5S).
Determination of the contribution of sulfation types and IdoUA residue to the binding of FGF-2
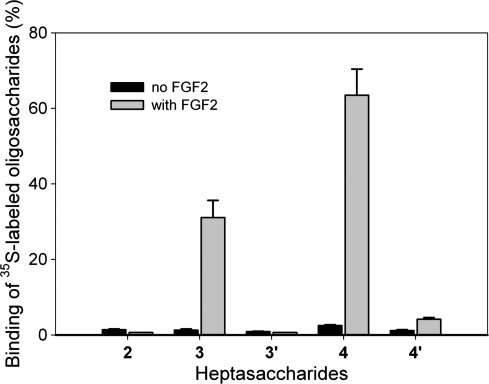
HS is known to play an important role in binding to growth factors and growth factor receptors to regulate the cell growth process (Bai and Esko 1996). It has been widely accepted that sulfation patterns dominate the binding affinity between HS and growth factors (Kreuger et al. 2006). Here, we examined the binding of FGF-2 to the heptasaccharides with different sulfation patterns using a filter-trapping method described by Maccarana et al. (1993). In this experiment, 35S-labeled heptasaccharides were incubated with purified FGF-2 (Figure 5). The mixture was then spotted onto a nitrocellulose membrane, which binds to protein nonspecifically, permitting to capture the complex of FGF-2 and heptasaccharides. The binding was quantified via scintillation counting. As expected, both heptasaccharides 3 and 4 bound to FGF-2 very well, while neither heptasaccharide 3′ nor heptasaccharide 4′ displayed significant binding to FGF-2. It is important to note that heptasaccharides 3 and 3′, as well as heptasaccharides 4 and 4′, carry identical numbers of sulfo groups. The structural difference is whether it contains the IdoUA residue. Thus, our data demonstrate that the IdoUA residue is essential for binding to FGF-2. We also observed ∼2-fold difference between heptasaccharides 3 and 4 in binding to FGF-2, suggesting that additional 6-O-sulfo groups enhance the binding to FGF-2.
Fig. 5.
The binding of 35S-labeled heptasaccharides to FGF-2. FGF-2 (1 μg) was incubated with 5000 cpm of 35S-labeled oligosaccharides in the PBS buffer. The mixture was spotted onto a nitrocellulose membrane. The membrane was then washed with a PBS buffer containing 300 mM NaCl. The spotted membrane was then mixed with scintillation fluid to measure the amount 35S-counts in order to determine the percentage of 35S-labeled oligosaccharides bound to FGF-2. The data presented represent the average of two experiments. Error bars indicate the range.
To compare the FGF-2 binding affinity of the synthetic heptasaccharide to the full-length HS, we determined the binding affinity (Kd) of heptasaccharide 4 that carries the IdoUA residue as well as 6-O- and 2-O-sulfo groups (Supplementary data, Figure 6S). The binding affinity was determined to be 202 ± 14 nM (average of two determinations ± range) using affinity coelectrophoresis. The Kd of FGF-2 for a full-length HS was determined to be 35 nM using a surface plasmon resonance method (Chen et al. 2005). The data suggest that heptasaccharide has somewhat lower affinity than a full-length polysaccharide.
Synthesis of AT-binding nonasaccharide
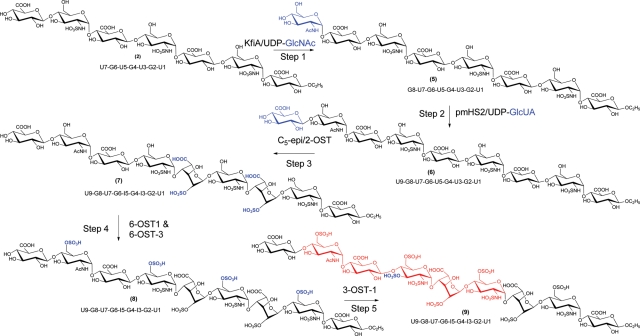
Our next goal was to prepare an oligosaccharide that binds to AT from heptasaccharide 2. The synthesis involved in extension to a nonasaccharide followed by the modifications by C5-epimerase and sulfotransferases as illustrated in Figure 6. It is worthwhile to note that the extension to nonasaccharide is necessary to confer the AT-binding affinity as the modification of heptasaccharide 4 with 3-OST-1 did not render AT-binding affinity (Table I). This experiment exemplified the use of enzymes to design the oligosaccharide structures with a desired biological activity. The target oligosaccharide, nonasaccharide 9, should consist of a GlcUA residue at the U7 position and a 3-O-sulfo group at the G6 GlcN because both GlcUA and 3-O-sulfated GlcN residues are part of the AT-binding site (Zhang et al. 1999). To avoid the conversion of the GlcUA residue at the U7 position by C5-epi, a GlcNAc residue was introduced at the G8 position (Liu et al. 2010). The GlcNAc residue of G8 should also be 6-O-sulfated as it is known that the 6-O-sulfation is required to display high affinity to AT-binding in the pentasaccharide context (Atha et al. 1985). We then introduced another GlcUA residue at the U9 position to ensure the 6-O-sulfation at the G8 position by 6-OST. The 3-O-sulfation at the G6 position was carried out by 3-OST-1 as it sulfates the GlcNS residue with the disaccharide sequence of -GlcUA-GlcNS6S- (U7-G6), but not in the sequences of -IdoUA2S-GlcNS6S- (I5-G4-I3-G2) or -GlcUA-GlcNAc6S- (U9-G8; Xu et al. 2008).
Fig. 6.
Scheme for the synthesis of AT-binding nonasaccharide 9. The reaction sites are colored in blue. The potential AT-binding domain in nonasaccharide 9 is colored in red.
Table I.
The binding of HS polysaccharide and oligosaccharides to AT
| Samples | AT binding (%)a |
|---|---|
| 3-O-[35S]-sulfated HSb | 37 ± 13 |
| N-[35S]-sulfated HSc | 1.5 ± 1.5 |
| Nonasaccharide 9 | 70 ± 5 |
| 3-O-[35S]-sulfated heptasaccharide 4d | 2.8 ± 2.5 |
aThe binding percentage of the samples to AT was determined using an affinity column as described in Experimental procedures.
bThe 3-O-[35S]-sulfated HS was prepared by incubating HS from bovine kidney with purified 3-OST-1 and [35S]PAPS.
cThe N-[35S]-sulfated HS was prepared by incubating HS from bovine with purified NST and [35S]PAPS.
dThe 3-O-[35S]-sulfated heptasaccharide 4 was prepared by incubating heptasaccharide 4 with purified 3-OST-1 and [35S]PAPS.
A series of HPLC analysis and the disaccharide analysis were conducted to prove the structures of the final product and the intermediate products. The successful extension of heptasaccharide 2 to octasaccharide 5 and nonasaccharide 6 was detected by observing the shift in retention time on diethylaminoethyl (DEAE)-HPLC analysis (Supplementary data, Figure 7S). The desired 2-O-sulfation/epimerization of nonasaccharide 7 was confirmed by the site-specific 35S-labeling technique followed by a disaccharide analysis of heparin lyase digested nonasaccharide 7 as shown in Supplementary data, Figure 8S. Furthermore, for the 2-O-[35S]-labeled nonasaccharide 7, we proved that the IdoUA residues carry 2-O-sulfo groups (Supplementary data, Figure 9S). Taken together, our data confirmed the structure of nonasaccharide 7.
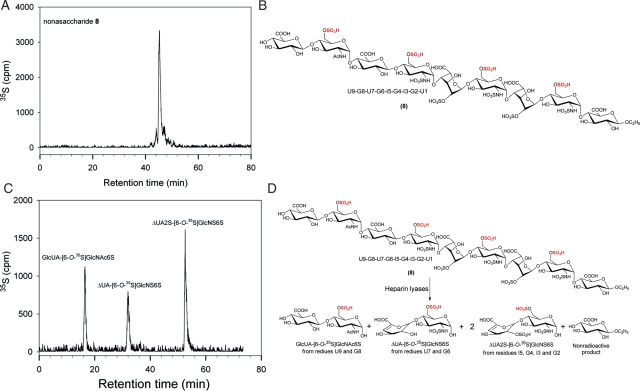
The structure of nonasaccharide 8 was also proved by the disaccharide analysis of the compound after the digestion with heparin lyases. The 6-O-[35S]-labeled nonasaccharide displayed a predominantly single 35S-labeled peak, suggesting that the preparation was pure (Figure 7A). Digestion of the nonasaccharide should yield three 35S-labeled disaccharides, GlcUA-GlcNAc6S, ΔUA-GlcNS6S and ΔUA2S-GlcNS6S, as illustrated in Figure 7D. As expected, the disaccharide analysis of heparin lyases-digested nonasaccharide showed three anticipated disaccharides with the ratio of 1.1:1:1.7, very close to the theoretically calculated value of 1:1:2. Therefore, the data confirmed the structure of nonasaccharide 8.
Fig. 7.
Structural characterization of nonasaccharide 8. (A) DEAE-HPLC chromatogram of 35S-labeled nonasaccharide. (B) The structure of nonasaccharide 8. (C) The RPIP-HPLC chromatogram of the disaccharides of heparin lyase-digested N-[35S]sulfo nonasaccharide 8. (D) The reaction involved in the digestion of nonasaccharide 8 by heparin lyases. A nonradioactive monosaccharide was formed during the digestion; however, it was not detectable.
The synthesis of nonasaccharide 9 was accomplished using 3-OST-1 enzyme, which sulfates the saccharide residue G6. DEAE-HPLC analysis of nonasaccharide 9 suggested that the product was pure (Supplementary data, Figure 10SA). Because 3-O-sulfation is known to prevent the digestion of oligosaccharides to disaccharides by heparin lyases (Yamada et al. 1993; Sundaram et al. 2003), we chose to use two-step nitrous acid degradation to convert nonasaccharide 9 to the disaccharide as illustrated in Supplementary data, Figure 10SD. As expected, we observed the presence of disaccharide of GlcUA-AnMan3S6S.
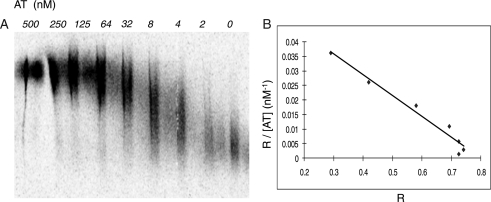
The binding of nonasaccharide to AT was carried out using an AT-affinity column (Table I). Here, two polysaccharide controls were included, 3-O-[35S]-sulfated HS and N-[35S]-sulf ated HS. As expected, ∼37% of the 3-O-[35S]-sulfated HS binds to AT, while the N-[35S]-sulfated HS did not display significant binding to AT due to the lack of the critical 3-O-sulfo group as demonstrated in our previous publication (Xu et al. 2008). Comparing with the polysaccharide controls, nonasaccharide 9 shows excellent binding (70%) to AT. In theory, 100% of nonasaccharide 9 should bind to AT. Lower than 100% binding is due to the fact the product recovery from an AT-affinity column was not complete. A similar case has been reported in our previous publication (Xia et al. 2002). In contrast, the 3-O-[35S]-sulfated heptasaccharide 4 does not bind to AT (Table I). 3-O-sulfated heptasaccharide 4 is expected to have the following structure: GlcA-GlcNS6S3S-Ido A2S-GlcNS6S-IdoA2S-GlcNS6S-GlcA-O-Et. It is known that the actual AT-binding site should include the following structure: -GlcNS(or NAc)6S-GlcA-GlcNS6S3S-IdoA2S-GlcNS6S-. Comparing these two structures, one can find that a GlcNAc6S (or GlcNS6S) residue is missing from the nonreducing end in 3-O-sulfated 4, which is known to be essential for binding to AT (Atha et al. 1985). Furthermore, the binding affinity (Kd) of nonasaccharide 9 to AT was determined to be 3.0 ± 1.1 nM (average of three determination ± SD). The representative gel picture and data analysis are shown in Figure 8. A Kd value of fondaparinux, an approved pentasaccharide anticoagulant drug, was determined to be 5.9 ± 1.5 nM using this method, suggesting that nonasaccharide 9 has slightly higher affinity than fondaparinux (10). The biological significance for nonasaccharide 9, such as its in vitro and in vivo anticoagulant activities, remains to be investigated largely because the study requires substantial amount of sample.
Fig. 8.
Determination of the binding affinity of nonasaccharide 9 to AT. (A) The representative autoradiography of the agarose gel in which purified 3-O-[35S]-sulfated nonasaccharide 9 was subjected to electrophoresis through separation zones containing AT at concentrations indicated. Approximately 8000 cpm/lane of nonasaccharide was loaded into each zone. (B) The plot of R/[AT] vs R, where R = (M0 − M)/M0. M0 is the migration of free 35S-labeled nonasaccharide and M is the observed migration of the nonasaccharide in the presence of AT at various concentrations. The plot yields a straight line with a slope of −1/Kd. The linear coefficient (r2) was determined to be 0.97.
Discussion
In this manuscript, we describe a unique approach to prepare HS oligosaccharides by combining chemical and enzymatic synthesis. A hexasaccharide precursor was chemically synthesized which was used as a key intermediate for divergent synthesis. The hexasaccharide was further modified with enzymes, including glycosyltransferases, HS C5-epimerase and sulfotransferases, to produce the oligosaccharides with different sulfation patterns and sizes. Our method can elongate an oligosaccharide to the desirable size via the action of glycosyltransferase. The types of sulfation can be controlled by selecting the number of modification steps. The position of N-sulfo GlcN controls the position and number of the IdoUA2S residues as demonstrated in the synthesis of heptasaccharide 4 and nonasaccharide 9. Consequently, the modification directed the biological activities of the oligosaccharide products. Clearly, the chemoenzymatic method offers the flexibility and throughput to investigate the structure–function relationship of HS in a given biological process. It should be noted that the project was aimed to demonstrate the feasibility of using HS biosynthetic enzymes to generate the oligosaccharides with distinct biological functions from a single starting hexasaccharide (1). Using this hexasaccharide (6 mg), we synthesized up to 11 oligosaccharides, including the final products and intermediates. It is not completely surprising that the scale of the synthesis demonstrated in this study was small. In this study, we did not measure the yields at each reaction step. As described in our previous publication (Liu et al. 2010), the conversion to the desired product at each step is nearly quantitative, and the product loss predominantly occurs at the purification step with a yield of 30–50%. In a more targeted synthesis, the scale of the synthesis can be readily enlarged to milligrams or even higher as demonstrated in our recent publications (Sheng et al. 2011; Xu et al. 2011).
Using enzymatically synthesized oligosaccharides, we revealed the structural selectivity of HS oligosaccharides for binding to FGF-2. The interaction of HS and FGF-2 has been implicated in regulating cell growth and angiogenesis processes (Kreuger et al. 2006). Although the contribution of sulfo groups to the binding affinity to FGF-2 has been reported (Maccarana et al. 1993; Ashikari-Hada et al. 2004, 2009; Wang et al. 2010), the binding affinity between HS and FGF-2 is perceived to be mainly governed by the density of negative charges, rather by specific monosaccharide sequences (Kreuger et al. 2006). In this study, five heptasaccharides were successfully prepared. These oligosaccharides permitted us to dissect the contribution of 2-O-sulfation, 2,6-O-sulfation as well as the presence of an IdoUA residue to the binding to FGF-2. We observed that the presence of IdoUA significantly enhances the percentage of the binding to FGF-2, despite the fact that the oligosaccharides carry identical number of sulfo groups. Indeed, structural analysis of the binding of FGF-2 and oligosaccharides using NMR revealed that an IdoUA residue provides the structural flexibility to an oligosaccharide (Guglieri et al. 2008). Such structural flexibility allows the oligosaccharides to position the sulfo groups toward FGF-2 to enhance the binding affinity (Raman et al. 2003). Using chemically synthesized HS hexasaccharides, we previously demonstrated that hexasaccharides containing a trisaccharide domain containing -GlcNS-IdoUA2S-GlcNS- inhibited the binding of FGF-2 to HS. Replacing the IdoUA2S residue with GlcUA2S abolished the inhibition effect on the binding of HS to FGF-2 (Wang et al. 2010). Therefore, our data are consistent with the previous findings, namely, an IdoUA residue plays a critical role for binding to FGF-2.
The role of the IdoUA residue in polysaccharides in stimulating FGF-2/fibroblast growth factor receptors (FGFR)-mediated cell proliferation was reported. It was shown that the enzymatically synthesized polysaccharide lacking the IdoUA has significantly reduced activity in promoting cell proliferation, consistent with its contribution to the FGF-binding (Chen et al. 2007). However, a reported published by Borgenstrom et al. (2003) concluded that heavily sulfated heparosan binds to FGF-1 and FGF-2, despite the fact that this polysaccharide contains no IdoUA residues. It should be noted that a significant level of the 3-O-sulfated GlcUA residue, an unnatural structural unit, has been found in chemically sulfated heparosan (Lindahl et al. 2005). Therefore, whether the presence of GlcUA3S in those chemically sulfated heparosan could contribute to the binding to FGF-1 or FGF-2 remains to be answered. We also noted that chemically sulfated heparosan is a polysaccharide (>50 saccharide residues), whereas the effect of IdoUA on FGF-2 binding was observed on a heptasaccharide. The size of the polysaccharide may contribute to the binding affinity to FGF-2.
Our method also demonstrated the synthesis of an AT-binding oligosaccharide from the common intermediate hexasaccharide 1. The synthesis was achieved by introducing a GlcNAc residue, which is used to control the extent of epimerization and serve as an acceptor site for 6-O-sulfation. This effort is an excellent example of designing oligosaccharide structures for specific biological activities. Because the AT-binding site consists of a pentasaccharide sequence of -GlcNAc (or NS)6S-GlcUA-GlcNS3S6S-IdoUA2S-GlcNS6 S-, both GlcNAc(or NS)6S and GlcUA are essential for the affinity to AT (Figure 6). The GlcNAc residue (of G8 in nonasaccharides) preserves the GlcUA at the U7 position in nonasaccharide 8 after the modification by C5-epi and 2-OST. The GlcNAc residue of G8 is also important to serve as the acceptor for the 6-O-sulfation because the GlcNAc6S is part of the AT-binding site (Atha et al. 1985). Without the GlcNAc residue of G8, the resultant oligosaccharide does not bind to AT even it carries a 3-O-sulfo GlcN unit.
In summary, we demonstrate the feasibility of chemoenzymatic synthesis of multiple oligosaccharides from a single hexasaccharide precursor. The divergent design of our approach allowed us to access diverse HS oligosaccharide structures with the desired binding affinities to the target proteins. Although higher binding affinities are not always equal to stronger biological activities, our results clearly demonstrate the structural selectivity of HS toward the interactions with AT and FGF-2. Our next goal is to prepare a large number of oligosaccharides with different sulfation patterns and to synthesize targeted oligosaccharides in large quantity to construct HS-based microarray and conduct further biological evaluation.
Experimental procedures
Chemical synthesis of hexasaccharide 1
The synthetic scheme for hexasaccharide 1 is shown in Figure 1. Briefly, a one-pot glycosylation reaction involved three disaccharide precursors, 11, 12 and 13, was performed to yield the fully protected hexasaccharide 10. The hexasaccharide 10 was then deprotected to produce hexasaccharide 1. The detailed procedures for the synthesis of hexasaccharide 1 and NMR and MS analysis data are described in Supplementary data.
Expression and purification of HS biosynthetic enzymes
In total, eight enzymes were employed for the synthesis of sulfated oligosaccharides, including NST, C5-epi, 2-OST, 6-OST-1, 6-OST-3, 3-OST-1, N-acetylglucosaminyltransferase (KfiA) from Escherichia coli K5 and pmHS2. All of these enzymes were expressed in E. coli using different fusion proteins as described previously (Chen et al. 2005, 2007). For C5-epi, 2-OST, 6-OST-1 and 6-OST-3, maltose-binding fusion proteins were used, and the fusion proteins were purified by an amylose column (New England Biolab, MA, USA). For NST, a glutathione S-transferase fusion was prepared and purified by a glutathione Sepharose column (GE Healthcare, Uppsala, Sweden; Kakuta et al. 1997). For 3-OST-1, KfiA and pmHS2, an N-terminal 6× His-tagged protein was prepared and purified by a Ni-agarose column (GE Healthcare; Edavettal et al. 2004; Liu et al. 2010).
Enzymatic preparation of sulfated oligosaccharides
The oligosaccharides were subjected to various sulfation and epimerization steps depending on the structures of the target compound. For the N-sulfation step, the oligosaccharide substrate (20–30 μg) was incubated with NST (80 μg) and 3′-phosphoadenosine 5′-phophosulfate (PAPS) (2 equiv.) in 300 μL buffer containing 50 mM 2-(N-morpholino)ethanesulfonic acid (MES) (pH 7.0) and 1% Triton X-100 overnight at 37°C. To prepare N-[35S]-sulfated oligosaccharides, [35S]PAPS (1–5 × 106 cpm) was included in the reaction mixture. To prepare the 2-O-, 6-O- or 3-O-sulfated oligosaccharides, nearly identical procedures were followed except for replacing NST with 2-OST, 6-OST-1 (and 6-OST-3) and 3-OST-1, respectively. The N-sulfated products were purified by a BioGel P-10 column (0.75 × 200 cm, BioRad, CA, USA) which was eluted with a buffer containing 20 mM Tris and 1 M NaCl (pH 7.5) at a flow rate of 8 mL/h. For the 2-O-, 6-O- or 3-O-sulfated oligosaccharides, the products were purified by a DEAE column after the reaction as follows. The reaction mixture (300 μL) was mixed with 1 mL of 0.01% Triton X-100 buffer at pH 5.0 containing 150 mM NaCl, 50 mM NaOAc, 3 M urea, 1 mM ethylenediaminetetraacetic acid (EDTA), then followed by four times wash with the same buffer, each time 1 mL. The column was eluted with 1 M NaCl in 0.001% Triton X-100 buffer. The purified oligosaccharides were dialyzed using molecular weight cutoff (MWCO) 3500 membrane and dried. Alternatively, the products were purified by the DEAE-HPLC method as described in HPLC analysis of oligosaccharides.
To prepare the oligosaccharides containing an IdoUA2S residue, the substrate (30 μg) was incubated with C5-epi (80 μg) in 300 μL buffer containing 50 mM MES (pH 7.0) and 1% Triton X-100, 2 mM calcium chloride. After the reaction proceeded for 30 min, 2-OST (80 μg) and PAPS (100 μM) were added and incubated overnight at 37°C. The product was recovered by a DEAE column as described in Enzymatic preparation of sulfated oligosaccharides.
HPLC analysis of oligosaccharides
Both DEAE-HPLC and PAMN-HPLC were used to purify the oligosaccharides. For DEAE-HPLC, method 1 used a DEAE-nonporous (NPR) column (Tosohaas, Japan) that was eluted with 20 mM, pH 7.0, Tris–HCl buffer for 10 min, then with a linear gradient of NaCl in 20 mM, pH 7.0, Tris–HCl from 0 to 1 M in 60 min at a flow rate of 0.4 mL/min. Method 2 used the same column that was eluted with 0.35 M NaCl and 20 mM, pH 7.0, Tris–HCl buffer for 10 min, then with a linear gradient of NaCl in 20 mM, pH 7.0, Tris–HCl buffer from 0.35 to 0.65 M in 90 min at a flow rate of 0.4 mL/min. Method 2 was used for the separation described in Supplementary data, Figures 1SA and 2SA and B, while method 1 was used for the other DEAE-HPLC. For PAMN-HPLC, a YMC-pack Polyamine II column (250 × 4.6 mm, YMC, Kyoto, Japan) was eluted with 0.4 M KH2PO4 for 10 min, then with a linear gradient of KH2PO4 from 0.4 to 1 M in 45 min at a flow rate of 0.5 mL/min.
Disaccharide analysis
The oligosaccharides were digested with a mixture of heparin lyases or by nitrous acid to yield disaccharides. For heparin lyase digestion, the oligosaccharide (100,000 cpm) was incubated with a mixture of heparin lyases I, II and III (each at ∼20 μg) in 200 μL of 50 mM Na2HPO3, pH 7.0, at 37°C overnight. The recombinant heparin lyases were expressed and purified as described previously (Duncan et al. 2006). For nitrous acid degradation, the oligosaccharide (100,000 cpm) was incubated with 20 μL of H2O and 40 μL of 1 M HNO2, which was prepared from a fresh mixture (v/v 1:1) of 0.5 M H2SO4 and 0.5 M Ba(NO2)2. The reaction was incubated on ice for 30 min with occasional vortex. The reaction was quenched by a mixture of 20 μL of 1 M Na2CO3 and 1 M NaHCO3 (v/v 1:1). Then, 20 μL of 0.5 M NaBH4 was added and incubated at 50°C for 30 min, 20 μL of 10 M acetic acid was added to stop reduction. The resultant disaccharide was mixed with 0.5 μL of 5% phenol red and purified by BioGel P-2 (BioRad) column, which was equilibrated with 0.1 M ammonium bicarbonate at a flow rate of 4 mL/h. The resultant disaccharides were analyzed using the RPIP-HPLC method as described previously (Liu et al. 1999). The identities of the disaccharides were determined by coeluting with standards.
Microdialysis of oligosaccharides
In order to improve the sensitivity of the analysis by MS, the oligosaccharides were subjected to microdialysis to remove the salt. The dialysis was carried out using hollow fiber dialysis tubing (MWCO 13,000 Da, Spectrum, CA, USA) against 20 mM ammonium acetate.
MS analysis
The analyses were performed at the ADME Mass Spectrometry Center on an Agilent 1100 MSD-trap. The oligosaccharides were dissolved in 70% acetonitrile and 10 mM imidazole. A syringe pump (Harvard Apparatus, MA, USA) was used to introduce the sample via direct infusion (10 μL/min). Experiments were carried out in negative ionization mode with the electrospray source set to 3000 V and 200°C and the compound stability set to 30%. Nitrogen was used for both nebulizer (5 L/min) and drying gas (15 psi). The MS data were acquired and processed using Bruker Trap Software 4.1.
FGF-binding assay
The binding of the oligosaccharides and FGF-2 (a gift from Dr. Linhardt, Rensselaer Polytechnic Institute) was carried out using a “filter-trapping” assay (Maccarana et al. 1993). Briefly, FGF-2 (1 μg) was incubated with 35S-labeled oligosaccharides (5000 cpm) in 200 μL of phosphate buffered saline (PBS) buffer at room temperature for 30 min. The mixture was then spotted onto nitrocellulose membrane (GE Healthcare). The spots were washed three times with 500 μL of PBS buffer containing 300 mM NaCl. The spot was cut out and mixed with scintillation fluid to measure [35S]radioactivity.
AT-binding experiment and determination of the binding affinity to AT
Approximately 1 × 105 cpm of [35S]-labeled compound was incubated with 5 μg of human AT (Cutter Biological, CA, USA) in 50 μL binding buffer containing 10 mM Tris–HCl, pH 7.5, 150 mM NaCl, 1 mM Mn2+, 1 mM Mg2+, 1 mM Ca2+, 10 μM dextran sulfate, 0.0004% Triton X-100 and 0.02% sodium azide for 30 min at room temperature. Concanavalin A-Sepharose (Sigma, 50 μL of 1:1 slurry, MO, USA) was then added and the reaction was shaken at room temperature for 1 h. The beads were then washed by 3 × 1 mL binding buffer, and the bound oligosaccharide was eluted with 1 M NaCl.
The dissociation constant (Kd) of the oligosaccharide and AT was determined using affinity co-electrophoresis (Lee and Lander 1991). Approximately 4000–5000 cpm of AT-binding 35S-labeled oligosaccharide was loaded per lane with zones of AT at concentrations 0, 8, 16, 32, 64, 128 and 256 nM. The gel was run at 400 mA for 2 h, dried and then analyzed on a PhosphoImager (Amersham Biosciences, Sweden, Storm 860). The retardation coefficient was calculated at R = (M0 − M)/M0, where M0 is the mobility of the polysaccharide through the zone without AT, and M is the mobility of the oligosaccharide through each separation zone. The retardation coefficient was then plotted against the retardation coefficient divided by its respective concentration of AT. The slope of the line represents −1/Kd.
Supplementary data
Supplementary data for this article is available online at http://glycob.oxfordjournals.org/.
Funding
This work is supported in part by National Institutes of Health (AI50050, HL094463, AI074775 and GM072667).
Conflict of interest
None declared.
Abbreviations
AT, antithrombin; DEAE, diethylaminoethyl; EDTA, ethylenediaminetetraacetic acid; ESI, electrospray ionization; FGF-2, fibroblast growth factor-2; FGFR, fibroblast growth factor receptors; GlcN, glucosamine; GlcUA, glucuronic acid; HPLC, high-performance liquid chromatography; HS, heparan sulfate; IdoUA, iduronic acid; KfiA, N-acetylglucosaminyltransferase; MES, 2-(N-morpholino)ethanesulfonic acid; MS, mass spectrometry; MWCO, molecular weight cutoff; NPR, nonporous; NST, N-sulfotransferase; 2-OST, 2-O-sulfotransferase; PAMN, polyamine-based anion exchange; PAPS, 3′-phosphoadenosine 5′-phophosulfate; PBS, phosphate buffered saline; pmHS2, Pasteurella multocida heparosan synthase-2; RPIP, reverse-phase ion pairing.
Supplementary Material
References
- Ashikari-Hada S, Habuchi H, Kariya Y, Itoh N, Reddi H, Kimata K. Characterization of the growth factor-binding structures in heparin/heparan sulfate using an octasaccharide library. J Biol Chem. 2004;279:12346–12354. doi: 10.1074/jbc.M313523200. doi:10.1074/jbc.M313523200. [DOI] [PubMed] [Google Scholar]
- Ashikari-Hada S, Habuchi H, Sugaya N, Kobayashi T, Kimata K. Specific inhibition of FGF-2 signaling with 2-O-sulfated octasaccharides of heparin sulfate. Glycobiology. 2009;19:644–654. doi: 10.1093/glycob/cwp031. doi:10.1093/glycob/cwp031. [DOI] [PubMed] [Google Scholar]
- Atha DH, Lormeau J-C, Petitou M, Rosenberg RD, Choay J. Contribution of monosaccharide residues in heparin binding to antithrombin III. Biochemistry. 1985;24:6723–6729. doi: 10.1021/bi00344a063. doi:10.1021/bi00344a063. [DOI] [PubMed] [Google Scholar]
- Bai X, Esko JD. An animal cell mutant defective in heparan sulfate hexuronic acid 2-O-sulfation. J Biol Chem. 1996;271:17711–17717. doi: 10.1074/jbc.271.30.17711. doi:10.1074/jbc.271.30.17711. [DOI] [PubMed] [Google Scholar]
- Baleux F, Loureiro-Morais L, Hersant Y, Clayette P, Arenzana-Seisdedos F, Bonnaffe D, Lortat-Jacob H. A synthetic CD4-heparan sulfate glycoconjugate inhibits CCR5 and CXCR4 HIV-1 attachment and entry. Nat Chem Biol. 2009;5:743–748. doi: 10.1038/nchembio.207. doi:10.1038/nchembio.207. [DOI] [PubMed] [Google Scholar]
- Borgenstrom M, Jalkanen M, Salmivirta M. Sulfated derivatives of Escherichia coli K5 polysaccharides as modulators of fibroblast growth factor signaling. J Biol Chem. 2003;278:49882–49889. doi: 10.1074/jbc.M304208200. doi:10.1074/jbc.M304208200. [DOI] [PubMed] [Google Scholar]
- Chen J, Avci FY, Muñoz EM, McDowell LM, Chen M, Pedersen LC, Zhang L, Linhardt RJ, Liu J. Enzymatically redesigning of biologically active heparan sulfate. J Biol Chem. 2005;280:42817–42825. doi: 10.1074/jbc.M504338200. doi:10.1074/jbc.M504338200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Jones CL, Liu J. Using an enzymatic combinatorial approach to identify anticoagulant heparan sulfate structures. Chem Biol. 2007;14:986–993. doi: 10.1016/j.chembiol.2007.07.015. doi:10.1016/j.chembiol.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Paz JL, Seeberger PH. Deciphering the glycosaminoglycan code with the help of microarrays. Mol Biosyst. 2008;4:707–711. doi: 10.1039/b802217h. doi:10.1039/b802217h. [DOI] [PubMed] [Google Scholar]
- Duncan MB, Liu M, Fox C, Liu J. Characterization of the N-deacetylase domain from the heparan sulfate N-deacetylase/N-sulfotransferase 2. Biochem Biophys Res Commun. 2006;339:1232–1237. doi: 10.1016/j.bbrc.2005.11.142. doi:10.1016/j.bbrc.2005.11.142. [DOI] [PubMed] [Google Scholar]
- Edavettal SC, Lee KA, Negishi M, Linhardt RJ, Liu J, Pedersen LC. Crystal structure and mutational analysis of heparan sulfate 3-O-sulfotransferase isoform 1. J Biol Chem. 2004;279:25789–25797. doi: 10.1074/jbc.M401089200. doi:10.1074/jbc.M401089200. [DOI] [PubMed] [Google Scholar]
- Gama C, Tully SE, Sotogaku N, Clark PM, Rawat M, Vaidehi N, Goddard WA, Nishi A, Hsieh-Wilson LC. Sulfation patterns of glycosaminoglycans encode molecular recognition and activity. Nat Chem Biol. 2006;2:467–473. doi: 10.1038/nchembio810. doi:10.1038/nchembio810. [DOI] [PubMed] [Google Scholar]
- Guglieri S, Hricovini M, Raman R, Polito L, Torri G, Casu B, Sasisekharan R, Guerrini M. Minimum FGF2 binding structural requirements of heparin and heparan sulfate oligosaccharides as determined by NMR spectroscopy. Biochemistry. 2008;47:13862–13869. doi: 10.1021/bi801007p. doi:10.1021/bi801007p. [DOI] [PubMed] [Google Scholar]
- Huang L, Huang X. Highly efficient synthesis of hyaluronic acid oligosaccharides. Chem Eur J. 2007;13:529–540. doi: 10.1002/chem.200601090. doi:10.1002/chem.200601090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Huang L, Wang H, Ye X-S. Iterative one-pot synthesis of oligosaccharides. Angew Chem Int Ed. 2004;43:5221–5224. doi: 10.1002/anie.200460176. doi:10.1002/anie.200460176. [DOI] [PubMed] [Google Scholar]
- Kakuta Y, Pedersen LG, Carter CW, Negishi M, Pedersen LC. Crystal structure of estrogen sulphotransferase. Nat Struct Biol. 1997;4:904–908. doi: 10.1038/nsb1197-904. doi:10.1038/nsb1197-904. [DOI] [PubMed] [Google Scholar]
- Kreuger J, Spillmann D, Li J-p, Lindahl U. Interactions between heparan sulfate and proteins: The concept of specificity. J Cell Biol. 2006;174:323–327. doi: 10.1083/jcb.200604035. doi:10.1083/jcb.200604035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence R, Lu H, Rosenberg R, Esko JD, Zhang L. Disaccharide structure code for the easy representation of constituent oligosaccharides from glycosaminoglycans. Nat Methods. 2008;5:291–292. doi: 10.1038/nmeth0408-291. doi:10.1038/nmeth0408-291. [DOI] [PubMed] [Google Scholar]
- Lee MK, Lander AD. Analysis affinity and structural selectivity in the binding of proteins to glycosaminoglycans: Development of a sensitive electrophoretic approach. Proc Natl Acad Sci USA. 1991;88:2768–2772. doi: 10.1073/pnas.88.7.2768. doi:10.1073/pnas.88.7.2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JC, Lu XA, Kulkarni SS, Wen YS, Hung SC. Synthesis of heparin oligosaccharides. J Am Chem Soc. 2004;126:476–477. doi: 10.1021/ja038244h. doi:10.1021/ja038244h. [DOI] [PubMed] [Google Scholar]
- Lindahl U, Li J, Kusche-Gullberg M, Salmivirta M, Alaranta S, Veromaa T, Emies J, Roberts I, Taylor C, Oreste P, et al. Generation of “neoheparin” from E. Coli K5 capsular polysaccharide. J Med Chem. 2005;48:349–352. doi: 10.1021/jm049812m. doi:10.1021/jm049812m. [DOI] [PubMed] [Google Scholar]
- Liu J, Shworak NW, Sinaÿ P, Schwartz JJ, Zhang L, Fritze LMS, Rosenberg RD. Expression of heparan sulfate d-glucosaminyl 3-O-sulfotransferase isoforms reveals novel substrate specificities. J Biol Chem. 1999;274:5185–5192. doi: 10.1074/jbc.274.8.5185. doi:10.1074/jbc.274.8.5185. [DOI] [PubMed] [Google Scholar]
- Liu R, Xu Y, Chen M, Weïwer M, Zhou X, Bridges AS, DeAngelis PL, Zhang Q, Linhardt RJ, Liu J. Chemoenzymatic design of heparan sulfate oligosaccharides. J Biol Chem. 2010;285:34240–34249. doi: 10.1074/jbc.M110.159152. doi:10.1074/jbc.M110.159152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maccarana M, Casu B, Lindahl U. Minimal sequence in heparin/heparan sulfate required for binding of basic fibroblast growth factor. J Biol Chem. 1993;268:23898–23905. [PubMed] [Google Scholar]
- McCullough BJ, Kalapthakis JM, Chin W, Taylor K, Clarke DJ, Eastwood H, Campopiano D, MacMillan D, Dorin J, Barran PE. Binding a heparin derived disaccharide to defensin inspired peptides: Insights to antimicrobial inhibition from gas-phase measurements. Phys Chem Chem Phys. 2010;12:3589–3596. doi: 10.1039/b923784d. doi:10.1039/b923784d. [DOI] [PubMed] [Google Scholar]
- Peterson SP, Frick A, Liu J. Designing of biologically active heparan sulfate and heparin using an enzyme-based approach. Nat Prod Rep. 2009;26:61–627. doi: 10.1039/b803795g. [DOI] [PubMed] [Google Scholar]
- Petitou M, van Boeckel CAA. A synthetic antithrombin III binding pentasaccharide is now a drug! What comes next? Angew Chem Int Ed. 2004;43:3118–3133. doi: 10.1002/anie.200300640. doi:10.1002/anie.200300640. [DOI] [PubMed] [Google Scholar]
- Raman R, Venkataraman G, Ernst S, Sasisekharan R. Structural specificity of heparin binding in the fibroblast growth factor family of proteins. Proc Natl Acad Sci USA. 2003;100:2357–2362. doi: 10.1073/pnas.0437842100. doi:10.1073/pnas.0437842100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng Z, Liu R, Xu Y, Liu J. The dominating role of N-deacetylase/N-sulfotransferase 1 in forming domain structures in heparan sulfate. J Biol Chem. 2011;286:19768–19776. doi: 10.1074/jbc.M111.224311. doi:10.1074/jbc.M111.224311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shriver Z, Raguram S, Sasisekharan R. Glycomics: A pathway to a class of new and improved therapeutics. Nat Rev Drug Discov. 2004;3:863–873. doi: 10.1038/nrd1521. doi:10.1038/nrd1521. [DOI] [PubMed] [Google Scholar]
- Sundaram M, Qi Y, Shriver Z, Liu D, Zhao G, Venkataraman G, Langer R, Sasisekharan R. Rational design of low-molecular weight heparins with improved in vivo activity. Proc Natl Acad Sci USA. 2003;100:651–656. doi: 10.1073/pnas.252643299. doi:10.1073/pnas.252643299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkataraman G, Shriver Z, Raman R, Sasisekharan R. Sequencing complex polysaccharides. Science. 1999;286:537–542. doi: 10.1126/science.286.5439.537. doi:10.1126/science.286.5439.537. [DOI] [PubMed] [Google Scholar]
- Wang Z, Xu Y, Yang B, Tiruchinapally G, Sun B, Liu R, Dulaney S, Liu J, Huang X. Preactivation-based one-pot combinatorial synthesis of heparin-like hexasaccharides for the analysis of heparin–protein interactions. Chem Eur J. 2010;16:8365–8375. doi: 10.1002/chem.201000987. doi:10.1002/chem.201000987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W, Ninonuevo MR, Sharma A, Dana-Leon LM, Leary JA. A comprehensive compositional analysis of heparin/heparan sulfate-derived disaccharides from human serum. Anal Chem. 2011;83:3703–3708. doi: 10.1021/ac2001077. doi:10.1021/ac2001077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia G, Chen J, Tiwari V, Ju W, Li J-P, Malmström A, Shukla D, Liu J. Heparan sulfate 3-O-sulfotransferase isoform 5 generates both an antithrombin-binding site and an entry receptor for herpes simplex virus, type 1. J Biol Chem. 2002;277:37912–37919. doi: 10.1074/jbc.M204209200. doi:10.1074/jbc.M204209200. [DOI] [PubMed] [Google Scholar]
- Xu Y, Masuko S, Takieddin M, Xu H, Liu R, Jing J, Mousa SA, Linhardt RJ, Liu J. Chemoenzymatic synthesis of homogeneous ultra-low molecular weight heparin. Science. 2011 doi: 10.1126/science.1207478. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Moon A, Song D, Pedersen LC, Liu J. Engineering sulfotransferases to modify heparan sulfate. Nat Chem Biol. 2008;4:200–202. doi: 10.1038/nchembio.66. doi:10.1038/nchembio.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada S, Yoshida K, Sugiura M, Sugahara K, Khoo KH, Morris HR, Dell A. Structural studies on the bacterial lyase-resistant tetrasaccharides derived from the antithrombin III-binding site of porcine intestinal heparin. J Biol Chem. 1993;268:4780–4787. [PubMed] [Google Scholar]
- Zaia J, Costello CE. Compositional analysis of glycosaminoglycans by electrospray mass spectrometry. Anal Chem. 2001;73:233–239. doi: 10.1021/ac000777a. doi:10.1021/ac000777a. [DOI] [PubMed] [Google Scholar]
- Zhang Z, McCallum SA, Xie J, Nieto L, Corzana F, Jiménez-Barbero J, Chen M, Liu J, Linhardt RJ. Solution structure of chemoenzymatically synthesized heparin and its precursors. J Am Chem Soc. 2008;130:12998–13007. doi: 10.1021/ja8026345. doi:10.1021/ja8026345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Yoshida K, Liu J, Rosenberg RD. Anticoagulant heparan sulfate precursor structures in F9 embryonic carcinoma cells. J Biol Chem. 1999;274:5681–5691. doi: 10.1074/jbc.274.9.5681. doi:10.1074/jbc.274.9.5681. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.