Abstract
Glycan-binding proteins are commonly used as analytical reagents to detect the levels of specific glycan structures in biological samples. A detailed knowledge of the specificities of glycan-binding proteins is required for properly interpreting their binding data. A powerful technology for characterizing glycan-binding specificity is the glycan array. However, the interpretation of glycan-array data can be difficult due to the complex fine specificities of certain glycan-binding proteins. We developed a systematic approach, called outlier-motif analysis, for extracting fine-specificity information from glycan-array data, and we applied the method to the study of four commonly used lectins: two mannose binders (concanavalin A and Lens culinaris) and two galactose binders (Bauhinia purpurea and peanut agglutinin). The study confirmed the known, primary specificity of each lectin and also revealed new insights into their binding preferences. Lens culinaris's main specificity may be non-terminal, α-linked mannose with a single linkage at its 2′ carbon, which is more restricted than previous definitions. We found broader specificity for bauhinea purpurea (BPL) than previously reported, showing that BPL can bind terminal N-acetylgalactosamine (GalNAc) and penultimate β-linked galactose under certain limitations. Peanut agglutinin may bind terminal Galβ1,3Gal, a glycolipid motif, in addition to terminal Galβ1,3GalNAc, a common O-linked glycoprotein motif. These results could be used to more accurately interpret data obtained using these well-studied lectins. Furthermore, this study demonstrates a systematic and general approach for extracting fine-specificity information from glycan-array data.
Keywords: glycan arrays, lectin, motif segregation, outlier analysis
Introduction
Glycan detection using lectins and glycan-binding antibodies is an important complement to other glycobiology analytical methods involving mass spectrometry, glycosidase digestion and chromatography. In order to most effectively use glycan-binding proteins as analytical reagents, it is important to fully understand their specificities. Knowledge of the specificity of analytical reagents could help in the selection of reagents to detect particular glycan targets and in the interpretation of measurements acquired using those reagents. A valuable tool for characterizing the nature of interactions between glycans and proteins is the glycan microarray (Drickamer and Taylor 2002; Blixt et al. 2004; Manimala et al. 2006; Yue and Haab 2009; Bathe et al. 2010; Lee et al. 2010; Lo et al. 2010). Various glycan-array platforms have been established, which use either chemically synthesized glycans or glycans purified from biological material. These data could provide rich insights into biological processes involving protein–glycan interactions and also could provide better understanding of the specificities of glycan-binding reagents. For example, a glycan-array study revealed significant diversity in the off-target binding activities of various glycan-binding antibodies (Manimala et al. 2007).
While glycan-array data provide a valuable resource for understanding glycan-binding specificity, its interpretation remains challenging in certain cases. The difficulty is particularly great for proteins that bind multiple structures or that have varying affinity depending on the presentation or overall context of a particular structure. For many proteins, the primary glycan-binding specificity is known, but details about the fine specificity, such as preferred presentations of binding determinants or potentially blocking side chains, are not clear. Previously we introduced motif segregation analysis as an approach to systematizing and automating the analysis of glycan-array data (Porter et al. 2010). Motif segregation is accurate for extracting the primary binding specificities of a wide variety of glycan-binding proteins. However, in the case of glycan-binding proteins with complex behaviors, the results from motif segregation may not capture the details of the fine specificity of binding. Motif segregation functions by identifying the motifs (component parts of oligosaccharides) that are selectively present in the glycans on a glycan array that are strongly bound by a particular lectin (Figure 1A). In the case when the stock set of pre-defined motifs does not include the precise determinant of a particular lectin, highly accurate information will not be extracted from the glycan-array data. The goal of this work was to enable a systematic extraction of fine-specificity information from glycan-array data and to use that approach to describe the binding of several commonly used lectins.
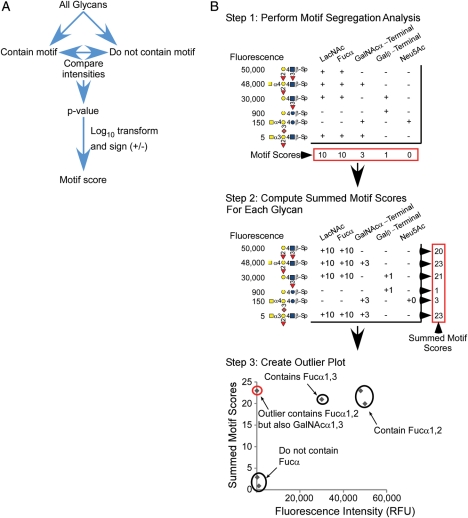
Fig. 1.
OMA. (A) Flow chart depicting the motif segregation method used to derive the motif scores. (B) OMA using demonstration data. The process begins with motif segregation analysis to assign a score to each motif (top). The next step is to sum the significant motif scores (>3.0) in each glycan to give a summed motif score for each glycan (middle). Next, the summed motif scores are plotted with respect to fluorescence intensity for each glycan. The graphical glycan notation conforms to the convention used by the CFG.
Several approaches could be pursued to address the goal of extracting detailed binding information from glycan-array data. One approach would be to greatly increase the number of pre-defined motifs in motif segregation analysis. However, given the numerous ways in which binding determinants could possibly arise, some of which may be unforeseen, the pre-definition of motifs likely always will be subject to human bias. Another approach may be to divide the glycans according to whether they are bound or not bound by a given lectin [referred to as “Intensity Segregation” in our previous work (Porter et al. 2010)] and then seek to identify component glycans (or motifs) that are present in one group but not the other. Several glycan-pattern recognition algorithms are in development (Aoki-Kinoshita and Kanehisa 2006). This approach has promise and is worthy of pursuit but still is unproven for the identification of fine specificities.
In this work, we developed an alternate approach to this problem. Our approach is to use the pre-defined motifs as “seeds” in order to identify primary specificities using our previous motif segregation analysis and then to build on that information to further define the binding determinants. This process is enabled by the identification of “outlier” glycans for which the binding of a lectin is not accurately described by the stock set of motifs. The outlier glycans are examined to identify new motifs that potentially define the true binding determinant, and the process is repeated using the new motifs. The entire process is repeated until motifs are defined that accurately describe the binding patterns of a given lectin.
We used this method to describe the fine specificity of four different lectins that are commonly used as analytical reagents. It is important to have an accurate understanding of such reagents, since they are used to measure glycan levels in a wide range of applications (Hirabayashi 2004; Sharon 2007), including various microarray platforms (Angeloni et al. 2005; Kuno et al. 2005; Pilobello et al. 2005; Patwa et al. 2006; Chen et al. 2007; Li et al. 2009; Yue et al. 2009; Yue and Haab 2009). We analyzed two lectins with primary specificity to mannose (concanavalin A and Lens culinaris) and two lectins with primary specificity to galactose (Bauhinia purpurea and peanut agglutinin). The use of two different lectins for each primary specificity allowed us to compare within a category, and the use of two different primary specificities allowed us to test the method over a variety of lectin types. Since these lectins are widely used to detect the presence of their target glycans in biological samples, a more detailed understanding of their specificities would be valuable. Here, we present insights into the fine specificities of these lectins and also demonstrate that outlier-motif analysis (OMA) is effective for refining and precisely defining the binding specificities of glycan-binding proteins.
Results
Outlier-motif analysis
To enable the systematic extraction of detailed binding specificities from glycan-array data, we extended motif segregation analysis (Porter et al. 2010; Figure 1A) with a method called OMA (Figure 1B). We begin with the pre-defined motifs as a starting point to get an idea of the primary binding specificity and then work from those structures to refine the definition of the binding determinant based on the data. The first step is to characterize how well the pre-defined motifs describe the binding intensities in the glycan-array data. If a particular glycan contains the motifs that represent the binding determinant, then the fluorescence signal at that glycan should be high. Likewise, if the motifs representing the binding determinant are not present in a particular glycan, the signal should be low. To look at this relationship, the following steps are taken (Figure 1B). We first compute the motif segregation scores for each pre-defined motif, according to our previously described method. Next, we sum the significant motif scores for each glycan. (Each individual motif score was thresholded to convert values <3.0 to zero, which removed contributions from insignificant scores and improved the interpretation of the data.) The summed motif score provides a summary of the number and strength of the motifs present in each glycan. Glycans containing several motifs with good scores will have high summed scores, and those with few or no high-scoring motifs will have low summed scores. The collection of individual motif scores and summed motif scores for each glycan are available in the Supplementary data. We then look at the correlation between the signal intensities and the summed motif scores for all glycans on the array.
Ideally, the summed motif scores and signal intensities correlate over all the glycans. Perfect correlation may not occur in some cases; for example, if the summed score includes contributions from motifs that are subsets of a broader motif. The correlation plot allows the identification of “outlier” glycans for which the summed motif scores have a significant deviation from correlating with binding intensity. The two types of outliers are: (i) glycans that have low motif scores (they do not contain the motifs predicted to represent the binding determinant) yet have high binding; and (ii) glycans that have high motif scores (they contain the motifs predicted to be the binding determinant) yet have low binding. The first type of outlier indicates that for certain glycans, the binding determinant is not represented in the pre-defined motifs, and the second type of outlier indicates that for other glycans, binding does not occur even though a high-scoring motif is present. In order to determine which glycans are considered outliers, we set thresholds along the x- and the y-axes (Supplementary data, Figures S1–S4). Thresholds were set by examining the distributions of the fluorescence values and summed motif scores. Bimodal distributions containing distinct high and low groups were apparent for each lectin, which allowed us to set a lower threshold that captured all slightly elevated glycans and an upper threshold that captured highly elevated glycans. Glycans above the upper threshold in one axis but below the lower threshold in the other axis were considered clear outliers, and glycans above the upper threshold in one axis but between the upper and lower threshold in the other axis were potential outliers. This latter group of outliers was bound by the lectin, as indicated by their moderate to high levels of fluorescence [∼1000 to ∼30,000 relative fluorescence units (RFUs)]. However, due to potential non-specific interactions related to the spacers, and in order to maintain a clear interpretation of specific interactions, these glycans were not included in the determination of the lectins' primary binding specificity.
After the identification of outlier glycans based on these thresholds, the next step is to compare the outlier glycans to the non-outlier glycans to identify potential new motifs or modifications to motifs that could more accurately describe the binding determinant. The process is then repeated with the inclusion of the new motifs. The reduction in outlier glycans in the repeated analysis is an indication that the new motifs more accurately describe the binding determinant. We applied this process to the study of two lectins each from two classes of binding specificities: mannose binders (concanavalin A and L. culinaris) and galactose binders (B. purpurea and peanut agglutinin).
Binding specificities of the mannose binders ConA and LCA
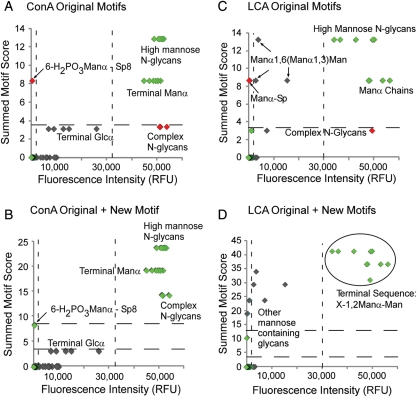
OMA was performed starting with the list of 63 motifs that were defined in our previous development of motif segregation (Porter et al. 2010). The analysis was applied to glycan-array data for concanavalin A (ConA) and lens culinaris (LCA) from the Consortium for Functional Glycomics (CFG) array version 2.0, which contained 264 uniquely printed targets (258 glycans and 6 glycoproteins). For ConA, the top significant motifs generated by motif segregation were “terminal Manα” (score = 8.5), “N-glycan, high mannose” (score = 4.6), “N-glycan, complex” (score = 3.4) and “terminal Glcα” (score = 3.2). “N-glycan, hybrid” is not represented on the version 2.0 array so could not be scored. The glycans containing these motifs clearly had higher binding intensities than the other glycans (Figure 2A), indicating that these motifs accurately describe the binding determinant of ConA. However, one outlier was identified with a high summed motif score but low fluorescence signal, 6-H2PO3Manα-Sp8. This glycan had been classified as containing the “terminal Manα” motif and for that reason had a high summed motif score. Although firm conclusions cannot be made from one glycan (no other phosphorylated mannose structures were on the array), the phosphate group likely blocks ConA binding.
Fig. 2.
OMA of the mannose binders ConA and LCA. The summed motif scores for each glycan, after analysis with either the original or the updated motifs, are plotted with respect to fluorescence intensity after detection with ConA or LCA. (A) ConA using the original stock set of 63 motifs. (B) ConA after the inclusion of the “unphosphorylated Manα” motif. (C) LCA using the original motifs. (D) LCA after the inclusion of the new motifs. The dashed lines represent thresholds for defining outliers, based on the distributions from all the glycans (Supplementary data, Figures S1–S4). Symbols in the upper right sectors are the glycans in expected regions, symbols in the upper left and lower right indicate the outliers, and intermediate symbols indicate the bound glycans that may not represent the primary specificity of the lectin. Sp, spacer.
We accounted for this outlier by defining a new motif, “unphosphorylated Manα—terminal or sub-terminal”. This new motif gave the strongest motif score of 11, and adding this motif into the outlier analysis enhanced the division between strong ConA binders (glycans containing Manα) and weaker ones (glycans containing terminal Glcα) (Figure 2B). The single glycan containing phosphorylated mannose is no longer an outlier based on the upper threshold. Therefore, this analysis confirmed the known specificity of ConA for both sub-terminal and terminal mannose, a weaker affinity for terminal glucose, and also suggested a requirement for a lack of phosphate on the mannose.
Several glycans had weak but measurable binding above the lower threshold, yet a zero summed motif score. We did not further characterize motifs for this latter group of structures due to evidence of non-glycan mediated binding. For example, the fluorescence of Galβ1,4GlcNAcβ-Sp8 was much greater than the same disaccharide attached to “-Sp0” (6219 vs 323 RFUs, respectively; Supplementary data, Table SI), suggesting the longer spacer induced ConA binding.
For LCA, the top significant motifs contained mannose, but the OMA graph showed many outlier glycans that had high summed motif scores but low fluorescence (Figure 2C). Comparing these low-binding outlier glycans to the high-binding glycans, we observed an enrichment in X-1,2-Manα-Man sequences among the high-binding group, where X could be the single addition of a Manα or GlcNAcβ, and the 2′ substituted Manα could be in any linkage with either a Manα or Manβ. Further chain extension was associated with low LCA binding. This motif represents a single-saccharide extension off the core pentasaccharide of N-linked glycans.
Previous studies using frontal affinity chromatography have found core fucosylation (fucose in α1,6 linkage to the reducing GlcNAc of the N-glycan chitobiose core) to be the main binding determinant of LCA (Kinoshita et al. 1991; Matsumura et al. 2007; Tateno et al. 2009). The glycan array used in this analysis lacked glycans containing core fucose, yet several glycans displayed strong-binding signals from LCA interacting with both N-glycans and short mannose oligosaccharides. This finding suggests that the main binding determinant of LCA is in the mannose branches of N-glycans and that core fucosylation may serve to enhance this interaction. This secondary affinity for core fucosylation is supported by the original analysis of LCA specificity, which found that while core-fucosylated, N-glycan containing peptides were more strongly retained on an LCA-Sepharose column, these peptides could not be eluted by free fucose (Kornfeld et al. 1981).
Another low-binding outlier was Manα-Sp, indicating that, unlike ConA, LCA's structural preference may be sub-terminal and not terminal mannose. Based on these observations, we defined the new motifs “sub-terminal Manα” (score = 10), “mannose” (score = 10) and “X-1,2-Manα-Man” (defined above, score = 7.2). The third motif seems to accurately describe the primary binding determinant of LCA, since an outlier plot using the new motifs clearly segregates binding glycans from non-binding and low-binding glycans based on the presence or the absence of that motif (Figure 2D). A few mannose-containing glycans show lower scores and fluorescence, indicating that weak binding to mannose may be possible in other configurations. Four glycans were outliers with very low fluorescence but elevated summed motif scores. These glycans contained mannose in a configuration that did not allow LCA binding. Because the broad motif of “mannose” had a good score for LCA, glycans containing that motif also had a somewhat high summed motif score. However, the scores are not as high as the glycans containing the primary determinant, which shows the good separation achieved through the summed motif score. Additional experimental work will be required to validate this information about the specificity of LCA, but this analysis shows the value of OMA for describing glycan-array data to provide insights into fine specificities of protein–glycan interactions.
Binding specificities of the galactose binders BPL and PNA
Using data from CFG glycan array version 2.0, the most significant motifs in the initial analysis of bauhinea purpurea (BPL) were “terminal Galβ1,3” (score = 11), “terminal Galβ” (score = 11) and “terminal N-acetyllactosamine” (score = 3.0), which is consistent with the known primary specificity. The outlier plot revealed several outliers in both the high-sum/low-fluorescence and low-sum/high-fluorescence categories (Figure 3A). The high-sum/low-fluorescence outliers all were terminal Galβ glycans that contained various sites of sulfation. Our definition of “terminal Galβ” encompassed sulfated Galβ, so sulfated glycans had a high summed motif score even though they did not bind BPL. Therefore, we accounted for this type of outlier by defining a new motif, “terminal Galβ, unsulfated.”
Fig. 3.
OMA of the galactose binders BPL and PNA. The summed motif scores for each glycan, after analysis with either the original or the updated motifs, are plotted with respect to fluorescence intensity after detection with BPL or PNA. (A) BPL using the original motifs. (B) BPL using the original plus new motifs. (C) PNA using the original motifs. (D) PNA using the original plus new motifs. The dashed lines represent the thresholds for defining outliers, based on the distributions from all the glycans (Supplementary data, Figures S1–S4). Symbols in the upper right sectors are the glycans in expected regions, symbols in the upper left and lower right indicate the outliers, and intermediate symbols indicate the bound glycans that may not represent the primary specificity of the lectin. Sp, spacer.
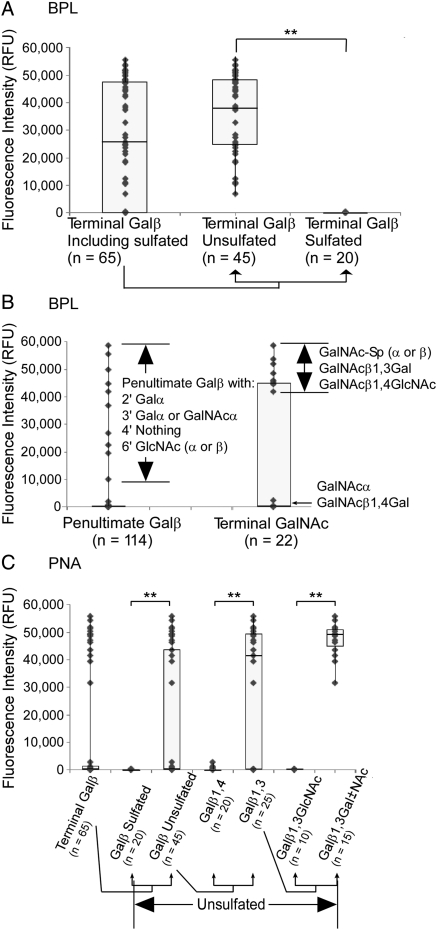
Accounting for the outliers in the low-sum/high-fluorescence group was less straightforward. These glycans contained terminal Galα, GalNAcα/β or GlcNAcα/β. In most of these glycans, the penultimate sugar was a Galβ. In the glycans with no sub-terminal Galβ, the terminal sugar was exclusively GalNAc. The binding to these features could be examined by directly comparing the signal levels of glycans containing or not containing particular motifs (Figure 4). (This analysis is in contrast to our standard motif segregation analysis, which compares glycans containing a motif to all other glycans on the array.) Directly comparing sulfated to non-sulfated terminal Galβ clearly shows the blocking effect of sulfation on BPL binding (Figure 4A). An examination of the glycans containing penultimate Galβ suggested that binding by BPL was allowed only when the penultimate Galβ had the following substituents: 2′ Galα, 3′ Galα or GalNAcα, 4′ unsubstituted and/or 6′ GlcNAcα/β (Figure 4B). The glycans containing penultimate Galβ but not bound by BPL had the following substituents: 2′ Fucα, 3′ GlcNAcα/β, a charged substituent (GlcA, sialic acid or sulfate) at any location or 4′ anything. These observations suggest that BPL can bind penultimate Galβ only with certain α-linked groups at 2′ or 3′, nothing at 4′ and nothing charged. These rules may be further generalized or simplified upon experimentation with a greater diversity of glycans.
Fig. 4.
Motif comparisons of BPL and PNA. Each point on the scatter plots represents a glycan on the glycan array. The fluorescence after detection with BPL or PNA is plotted for glycans containing particular motifs. (A) The effect of sulfation on BPL binding. The fluorescence is plotted for glycans that contain terminal Galβ and separately for those that are sulfated or unsulfated. The difference between BPL binding to unsulfated and sulfated glycans (indicated by double asterisks) was highly significant (P << 0.001). (B) BPL binding to penultimate Galβ and terminal GalNAc. The scatter plots show the fluorescence signals of all glycans containing either of these two motifs. Additional features of the glycans in each group are indicated. (C) Iterative comparison of motifs bound by PNA. Each group of glycans is a subset of that to the left, beginning with all glycans containing terminal Galβ. PNA showed significantly higher binding to terminal Galβ when the glycan was not sulfated (P = 0.003), followed by significantly higher binding when the glycan was unsulfated and the Galβ was in 1,4 linkage to the penultimate sugar (P < 0.001), followed by significantly higher binding still when the glycan was unsulfated and the Galβ was in 1,4 linkage to a Gal or GalNAc and not a GlcNAc (P << 0.001). All P-values were calculated using the student's t-test.
In a similar way, a clear division existed among glycans containing terminal GalNAc (Figure 4B). BPL binding of terminal GalNAc was detectable only if the glycans contained the following motifs: GalNAcβ1,3Gal, GalNAcβ1,4GlcNAc or GalNAcα/β-spacer. Terminal αGalNAc linked to any other glycan was not bound by BPL. Thus, it appears that BPL can bind terminal, β-linked GalNAc that is bound to certain penultimate glycans.
To account for all of these observations for BPL, two new motifs were created, “terminal Galβ, unsulfated” (score = 19), and another that encompassed the aforementioned rules for terminal/sub-terminal Galβ and terminal GalNAc binding, which produced the highest motif score of 24. Adding these new motifs to the OMA raised the summed motif scores of all of the strong-binding glycans while leaving the scores of the low-binding glycans unaffected (Figure 3B). The improved correlation between fluorescence signal and summed motif score indicates that the new motifs more accurately describe the binding determinants of BPL. While the upper threshold was not reached for sub-terminal Galβ and terminal GalNAc containing glycans, the new motifs elevated the summed motif scores for these glycans above the sulfated non-binding outliers along the y-axis. As with ConA, the remaining weak binding glycans, just above the lower fluorescence threshold, did not show consistent features and were possibly due to non-specific binding.
OMA of peanut agglutinin (PNA) revealed similarities and differences with BPL. The strongest individual motif scores were “terminal Galβ1,3” (score = 6.6), “terminal Galβ” (score = 4.0) and “O-glycan core 2” (score = 3.1), consistent with the previously characterized specificity. The outlier plot showed outliers only in the high-sum/low-fluorescence category, with a broad division between outliers and binders (Figure 3C). As with BPL, sulfated Galβ blocked binding (Figure 4C). However, some glycans containing unsulfated terminal Galβ were not bound by PNA. Subdividing these glycans into terminal Galβ1,3 and terminal Galβ1,4 revealed the high preference of PNA for terminal Galβ1,3 (Figure 4C). Again, not all terminal Galβ1,3 was bound by PNA, so we further subdivided these glycans into Galβ1,3Gal/GalNAc and Galβ1,3GlcNAc, which showed the binding specificity of PNA to be “terminal unsulfated Galβ1,3Gal/GalNAc” (Figure 4C). This new motif gave the strongest motif score (score = 9.7) and after addition to the original summed motif scores cleanly separated the outliers from the true binders (Figure 3D).
Summary of findings
Taken together, these analyses confirm the primary specificities of the lectins but also uncover some unexpected binding characteristics. Table I provides a summary of our findings when compared with the specificities that are commonly reported as the primary binding determinants of the lectins (e.g. in vendor catalogs). In the cases of ConA and PNA, the primary specificities are highly accurate. However, our findings indicate that BPL and LCA have more complex specificities than commonly cited. BPL binds the primary determinant of terminal, β-linked galactose, but it also appears to bind sub-terminal galactose and terminal GalNAc in particular situations. The exact rules determining the additional binding capability of BPL are not identifiable from the available glycan-array data. In the case of LCA, our analyses indicate a lack of binding to terminal mannose, in contrast to its reported specificity. LCA apparently mainly binds the “X-1,2-Manα-Man” motif. The role of core fucose in LCA binding was not determinable from these data, but our analyses show that core fucose is not required for LCA binding.
Table I.
Summary of refined lectin specificities
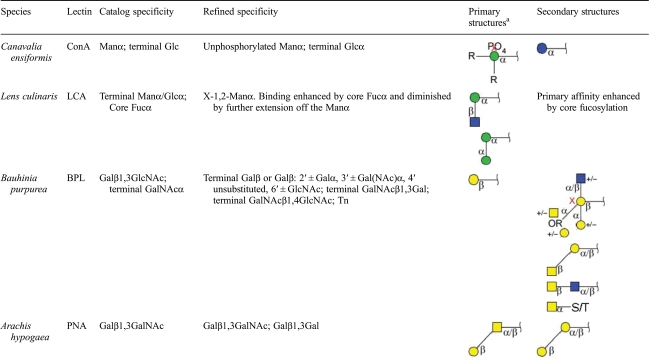 |
The graphical glycan notation is the system used by the CFG.
aR indicates any glycan, and X indicates no attachment allowed.
Discussion
This work addresses the need for more-detailed characterizations of the specificities of glycan-binding proteins and the need for a systematic approach to extract that information from glycan-array data. Our analytical approach is built on the iterative process of scoring motifs, identifying outlier glycans and redefining motifs. This method should be generally useful for any type of glycan-array data, both for assessing how well certain motifs represent the true binding determinant and for uncovering complex and fine specificities. We applied the method to the study of four commonly used lectins and uncovered some unexpected features of glycan-binding specificity.
ConA
ConA's binding structure and specificity have been well determined for mannose-containing structures (Hardman and Ainsworth 1972; Naismith and Field 1996; Gupta et al. 1997), including recognition of biantennary, complex N-glycans (Moothoo and Naismith 1998), and for terminal glucose (Gupta et al. 1997). OMA confirmed the main specificity for mannose and the secondary specificity for terminal glucose. The value of the approach for finding exceptions was shown in identifying a single high sum, low-binding outlier glycan, which contained phosphorylated mannose. A general rule cannot be derived from one glycan, but the identification of such exceptions provides useful insights and suggests that phosphate inhibits ConA binding.
LCA
OMA uncovered some new insights into LCA specificity. Our analysis showed that strong binding is possible in the absence of core fucosylation and that the main determinant may be the X-1,2-Manα-Man motif. This X-1,2-Manα-Man structure involves substitution of the Manα at two axial positions. The position of two large axial groups is likely to cause a high amount of strain forcing the ring into an altered conformation in order to reach a lower energy state. This conformation may be recognized by LCA. The fact that LCA does not bind terminal mannose supports this interpretation. Experimental support for our defined motif comes from the finding that glycopeptides with GlcNAc substitution of the tri-mannose, N-glycan core (Man3GlcNAc2), create the strongest inhibitors of LCA red blood cell agglutination (Kornfeld et al. 1971). We also found that further extensions off the X-1,2-Manα-Man motif seem to be inhibitory. These findings reveal a binding specificity for LCA that is more restricted than previous definitions, since it precludes terminal mannose and extended structures.
Core fucosylation may strengthen the LCA binding to N-glycans, as suggested from studies using frontal-affinity chromatography (Kinoshita et al. 1991; Matsumura et al. 2007; Tateno et al. 2009). We were not able to study the contribution of core fucose because glycans with core fucosylation were not present on glycan array version 2.0, and no higher array versions with sufficient signal quality for a reliable analysis were available. Structural principles support an enhancement of LCA binding by core fucose, since core fucose may interact favorably with a hydrophobic region outside of the mannose-binding pocket and orient the lectin for proper binding to the X-1,2-Manα-Man motif. A molecular modeling study provided some insight an enhanced interaction with core-fucosylated N-glycans (Sokolowski et al. 1997). Core-fucosylated glycans showed three extra potential hydrogen bond interactions with LCA and the in a secondary binding site, whereas non-fucosylated glycans showed an interaction between the opposing Manα arm and the secondary binding site. However, experimental evidence to support the mannose motif as the primary binder was provided by early experiments showing that fucose is unable to elute core-fucosylated glycans from an LCA-Sepharose column (Kornfeld et al. 1981). Therefore, the main determinant for LCA may be X-1,2-Manα-Man, with an enhancement by core fucose. Glycan microarrays containing a variety of core-fucosylated glycans and addition binding experiments could give more information on the binding preferences of LCA.
BPL
New insights also were obtained for BPL. Our analysis showed BPL to be a fairly flexible galactose binder. Unless the glycan was sulfated, BPL was able to bind all terminal Galβ linkages, terminal GalNAcβ1,4GlcNAc, terminal GalNAcβ1,3Gal and GalNAcα when linked to a spacer. These findings are consistent with the literature (Wu et al. 2004). The novel finding for BPL binding is the recognition of penultimate Galβ. The glycans containing penultimate Galβ but not bound by BPL had the following substituents: 2′ Fucα, 3′ GlcNAcα/β, a charged substituent (GlcA, sialic acid or sulfate) at any location or 4′ anything. These observations suggest that BPL can bind penultimate Galβ only with certain α-linked groups at 2′ or 3′, nothing at 4′ and nothing charged. These rules may be further generalized or simplified upon experimentation with a greater diversity of glycans. These additional binding rules should be taken into account when interpreting results obtained using BPL.
PNA
Our results for PNA were consistent with the literature in that the binding specificity was found to be terminal Galβ1,3GalNAc (Lotan et al. 1975; Iskratsch et al. 2009). We did find that an additional, terminal glycan structure, Galβ1,3Gal was also capable of being bound by PNA. This additional recognition motif may need to be taken into consideration if PNA is used in glycolipid identification by the confounding presence of muco- (Galβ1,3Gal) and ganglio- (Galβ1,3GalNAc) terminating glycolipids.
Previous research using co-crystalization (Kundhavai Natchiar et al. 2004) and agglutination assays (Lotan et al. 1975) suggested that lactose (Galβ1,4Glcβ) may be a weak PNA ligand in addition to Galβ1,3-containing glycans. In our glycan-array analysis, the Galβ1,4Glcβ glycans showed little to no fluorescence with PNA detection and were just barely above our lower threshold. The lack of binding to lactose could be attributed to lactose not representing a primary specificity of PNA. Alternatively, PNA may bind free lactose better than covalently attached, constrained lactose.
The accuracy of the results achieved here inherently depends on how many and what types of glycans are present on the array. If only a few glycans contain a given motif, the ability to test the contribution of that motif to binding is correspondingly low. The validation of the accuracy of these results could be achieved with additional glycans not represented on the arrays used here, or through additional experiments in other formats. Additional experiments could test new motifs by measuring the binding of lectins to glycans designed around the presence or the absence of the new motifs. This strategy would account for the possibility of “overfitting” the data, or developing rules that describe the existing data but are not generally true. Another approach to further validating glycan-array findings is to compare with structural information, as suggested earlier (Taylor and Drickamer 2009). The proposed motifs from these analyses could be docked to the lectin structures to see if the structural fit correlates with the predicted binding strength of the motif. Such structural analyses should provide very useful and complementary insights into the motif information derived from glycan-array data (Taylor and Drickamer 2007; Chandrasekaran et al. 2008; DeMarco and Woods 2008; Taylor and Drickamer 2009).
A future goal will be to automate the outlier analysis, as opposed to the manual inspection of outlier glycans that was used here. Manual inspection was necessary for the initial study to test the method, but automated generation of refined motifs will speed up the analysis and potentially remove bias from user intervention. The first step in automation of OMA would be to establish set rules for setting the thresholds, which could be done by determining the quantitative relationships between the distributions of low and high glycans (Supplementary data, Figures S1–S4), given enough data sets. Once outliers are identified, it would be possible to scan through potential improved motifs to search for those that account for the outliers. An approach to generating new motifs could be built on the use of using logical operators to combine motifs. AND, OR and NOT operators could be used to combine motifs to define the more complex motifs that were necessary to accurately describe binding determinants. Such an approach also could be combined with pattern recognition methods to identify features that are unique to the outlier glycans.
Another improvement to this approach will be to develop additional statistics for the motif comparisons. The Mann–Whitney test used here may lead to misleading results under certain circumstances, such as in the case of very moderate changes in fluorescence or in comparisons of motifs with greatly divergent levels of representation. Another good option for statistically comparing the binding between groups of glycans is the area under the curve in receiver-operator-characteristic analysis, which examines the amount of separation at various thresholds between two groups of measurements. This test would be valuable to examine the degree to which two groups of glycans are separated without regard to normality or rank within each group. Furthermore, the test would remove potential bias introduced from greatly different numbers of glycans in the groups that contain or do not contain a motif. Future implementations of motif segregation will explore this and other alternate methods.
This approach also will be useful for supplementing biological studies of processes involving protein–glycan interactions. Interactions between proteins and glycans are an important component of multiple biological functions, including cell signaling, cell migration, immune recognition, protein processing and quality control and the regulation of extracellular spaces (Dube and Bertozzi 2005; Varki et al. 1999). Characterizing the nature of these interactions is important for understanding the biology of these systems and for developing approaches for treating diseases that are affected by protein–glycan intercommunication (Fuster and Esko 2005). Detailed analyses of glycan-array data could assist those efforts.
Materials and methods
Data source
The glycan-array data were obtained from the CFG (www.functionalglycomics.org). All experiments were run on the printed array version 2.0. The data sets used were as follows: ConA, primscreen_PA_v2_700_09262005; LCA, primscreen_PA_v2_710_09262005; BPL, primscreen_PA_v2_699_09262005; and PNA, primscreen_PA_v2_719_09262005.
Generation of glycan-array data
The glycan-array experiments were performed by Core D of the CFG, as described previously (Blixt et al. 2004). A brief summary is given here. Synthetic glycans were functionalized with a spacer containing a terminal NH2 group and spotted onto N-hydroxysuccinimide-activated microscope slides (Slide H, Schott Nexterion, Elmsford, NY) using a robotic microarrayer. Lectins at a concentration of 0.1–200 µg/mL in a buffer of PBS containing 0.005–0.5% Tween-20 were incubated on the arrays for 30–60 min. The lectins were tagged with either a fluorophore (Alexa Fluor 488, Life Technologies Corporation, Carlsbad, CA) or biotin. If fluorophore-labeled analytes were used, the arrays were washed and immediately scanned for fluorescence using a microarray scanner. Biotinylated analytes were detected with an incubation of streptavidin–fluorescein isothiocyanate (FITC) followed by washing and scanning. Image analysis software was used to quantify the fluorescence intensities at each glycan spot. The data from six replicate spots were averaged to achieve a final value.
Data analysis
The primary data analysis and calculations were performed with Microsoft Office Excel 2007, and the figures were created in Deneba Canvas X. The motif segregation method used a calculation of the statistical difference between the intensities of the glycans containing a particular motif and intensities of the glycans not containing the motif (Figure 1A). We used the P-value from the Mann–Whitney non-parametric test for that purpose. For the purpose of graphically representing the scores, we log-transformed (base 10) the P-values. To indicate whether the motif-containing or the non-motif-containing glycans had the higher values, we multiplied the log-transformed P-values by the sign of the z-score. The logged and signed P-values represented the “motif score” for that particular motif, and values >3.0 (P < 0.001) were considered significant. The “summed motif score” was calculated for each glycan structure by summing each of the significant motif scores contained in each glycan (Figure 1B). Table I was also generated in Microsoft Office Excel 2007. The catalog specificities were obtained from Vector Labs and Sigma catalogs provided for each given lectin. The glycan structures were created using Glycanbuilder (Ceroni et al. 2007).
Supplementary data
Supplementary data for this article is available online at http://glycob.oxfordjournals.org/.
Funding
We gratefully acknowledge support of this work from the Frederik and Lena Meijer Student Internship Program (to D.L.), the NCI (R33 CA122890 to B.B.H.), the NIGMS (The Consortium for Functional Glycomics, GM62116, Bridging grant to B.B.H.) and the Van Andel Institute.
Conflict of interest
None declared.
Abbreviations
BPL, bauhinea purpurea; CFG, Consortium for Functional Glycomics; ConA, concanavalin A; FITC, fluorescein isothiocyanate; GalNAc, N-acetylgalactosamine; LCA, lens culinaris; OMA, outlier-motif analysis; PNA, peanut agglutinin; RFU, relative fluorescence units.
Supplementary Material
References
- Angeloni S, Ridet JL, Kusy N, Gao H, Crevoisier F, Guinchard S, Kochhar S, Sigrist H, Sprenger N. Glycoprofiling with micro-arrays of glycoconjugates and lectins. Glycobiology. 2005;15:31–41. doi: 10.1093/glycob/cwh143. doi:10.1093/glycob/cwh143. [DOI] [PubMed] [Google Scholar]
- Aoki-Kinoshita KF, Kanehisa M. Bioinformatics approaches in glycomics and drug discovery. Curr Opin Mol Ther. 2006;8:514–520. [PubMed] [Google Scholar]
- Bathe OF, Shaykhutdinov R, Kopciuk K, Weljie AM, McKay A, Sutherland FR, Dixon E, Dunse N, Sotiropoulos D, Vogel HJ. Feasibility of identifying pancreatic cancer based on serum metabolomics. Cancer Epidemiol Biomarkers Prev. 2010;20:140–147. doi: 10.1158/1055-9965.EPI-10-0712. [DOI] [PubMed] [Google Scholar]
- Blixt O, Head S, Mondala T, Scanlan C, Huflejt ME, Alvarez R, Bryan MC, Fazio F, Calarese D, Stevens J, et al. Printed covalent glycan array for ligand profiling of diverse glycan binding proteins. Proc Natl Acad Sci USA. 2004;101:17033–17038. doi: 10.1073/pnas.0407902101. doi:10.1073/pnas.0407902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceroni A, Dell A, Haslam SM. The GlycanBuilder: A fast, intuitive and flexible software tool for building and displaying glycan structures. Source Code Biol Med. 2007;2:3. doi: 10.1186/1751-0473-2-3. doi:10.1186/1751-0473-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasekaran A, Srinivasan A, Raman R, Viswanathan K, Raguram S, Tumpey TM, Sasisekharan V, Sasisekharan R. Glycan topology determines human adaptation of avian H5N1 virus hemagglutinin. Nat Biotechnol. 2008;26:107–113. doi: 10.1038/nbt1375. doi:10.1038/nbt1375. [DOI] [PubMed] [Google Scholar]
- Chen S, LaRoche T, Hamelinck D, Bergsma D, Brenner D, Simeone D, Brand RE, Haab BB. Multiplexed analysis of glycan variation on native proteins captured by antibody microarrays. Nat Methods. 2007;4:437–444. doi: 10.1038/nmeth1035. [DOI] [PubMed] [Google Scholar]
- DeMarco ML, Woods RJ. Structural glycobiology: A game of snakes and ladders. Glycobiology. 2008;18:426–440. doi: 10.1093/glycob/cwn026. doi:10.1093/glycob/cwn026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drickamer K, Taylor ME. Glycan arrays for functional glycomics. Genome Biol. 2002;3:REVIEWS1034. doi: 10.1186/gb-2002-3-12-reviews1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dube DH, Bertozzi CR. Glycans in cancer and inflammation–potential for therapeutics and diagnostics. Nat Rev Drug Discov. 2005;4:477–488. doi: 10.1038/nrd1751. doi:10.1038/nrd1751. [DOI] [PubMed] [Google Scholar]
- Fuster MM, Esko JD. The sweet and sour of cancer: Glycans as novel therapeutic targets. Nat Rev Cancer. 2005;5:526–542. doi: 10.1038/nrc1649. doi:10.1038/nrc1649. [DOI] [PubMed] [Google Scholar]
- Gupta D, Dam TK, Oscarson S, Brewer CF. Thermodynamics of lectin-carbohydrate interactions. Binding of the core trimannoside of asparagine-linked carbohydrates and deoxy analogs to concanavalin A. J Biol Chem. 1997;272:6388–6392. doi:10.1074/jbc.272.10.6388. [PubMed] [Google Scholar]
- Hardman KD, Ainsworth CF. Structure of concanavalin A at 2.4-A resolution. Biochemistry. 1972;11:4910–4919. doi: 10.1021/bi00776a006. doi:10.1021/bi00776a006. [DOI] [PubMed] [Google Scholar]
- Hirabayashi J. Lectin-based structural glycomics: Glycoproteomics and glycan profiling. Glycoconj J. 2004;21:35–40. doi: 10.1023/B:GLYC.0000043745.18988.a1. doi:10.1023/B:GLYC.0000043745.18988.a1. [DOI] [PubMed] [Google Scholar]
- Iskratsch T, Braun A, Paschinger K, Wilson IB. Specificity analysis of lectins and antibodies using remodeled glycoproteins. Anal Biochem. 2009;386:133–146. doi: 10.1016/j.ab.2008.12.005. doi:10.1016/j.ab.2008.12.005. [DOI] [PubMed] [Google Scholar]
- Kinoshita N, Ohno M, Nishiura T, Fujii S, Nishikawa A, Kawakami Y, Uozumi N, Taniguchi N. Glycosylation at the Fab portion of myeloma immunoglobulin G and increased fucosylated biantennary sugar chains: Structural analysis by high-performance liquid chromatography and antibody-lectin enzyme immunoassay using Lens culinaris agglutinin. Cancer Res. 1991;51:5888–5892. [PubMed] [Google Scholar]
- Kornfeld K, Reitman ML, Kornfeld R. The carbohydrate-binding specificity of pea and lentil lectins. Fucose is an important determinant. J Biol Chem. 1981;256:6633–6640. [PubMed] [Google Scholar]
- Kornfeld S, Rogers J, Gregory W. The nature of the cell surface receptor site for Lens culinaris phytohemagglutinin. J Biol Chem. 1971;246:6581–6586. [PubMed] [Google Scholar]
- Kundhavai Natchiar S, Arockia Jeyaprakash A, Ramya TN, Thomas CJ, Suguna K, Surolia A, Vijayan M. Structural plasticity of peanut lectin: An X-ray analysis involving variation in pH, ligand binding and crystal structure. Acta Crystallogr D Biol Crystallogr. 2004;60:211–219. doi: 10.1107/S090744490302849X. doi:10.1107/S090744490302849X. [DOI] [PubMed] [Google Scholar]
- Kuno A, Uchiyama N, Koseki-Kuno S, Ebe Y, Takashima S, Yamada M, Hirabayashi J. Evanescent-field fluorescence-assisted lectin microarray: A new strategy for glycan profiling. Nat Methods. 2005;2:851–856. doi: 10.1038/nmeth803. doi:10.1038/nmeth803. [DOI] [PubMed] [Google Scholar]
- Lee SM, Chan RW, Gardy JL, Lo CK, Sihoe AD, Kang SS, Cheung TK, Guan YI, Chan MC, Hancock RE, et al. Systems-level comparison of host responses induced by pandemic and seasonal influenza A H1N1 viruses in primary human type I-like alveolar epithelial cells in vitro. Respir Res. 2010;11:147. doi: 10.1186/1465-9921-11-147. doi:10.1186/1465-9921-11-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Simeone D, Brenner D, Anderson MA, Shedden K, Ruffin MT, Lubman DM. Pancreatic cancer serum detection using a lectin/glyco-antibody array method. J Proteome Res. 2009;8:483–492. doi: 10.1021/pr8007013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo JF, Yu CC, Chiou SH, Huang CY, Jan CI, Lin SC, Liu CJ, Hu WY, Yu YH. The epithelial-mesenchymal transition mediator S100A4 maintains cancer initiating cells in head and neck cancers. Cancer Res. 2010;71:1912–1923. doi: 10.1158/0008-5472.CAN-10-2350. [DOI] [PubMed] [Google Scholar]
- Lotan R, Skutelsky E, Danon D, Sharon N. The purification, composition, and specificity of the anti-T lectin from peanut (Arachis hypogaea) J Biol Chem. 1975;250:8518–8523. [PubMed] [Google Scholar]
- Manimala JC, Roach TA, Li Z, Gildersleeve JC. High-throughput carbohydrate microarray analysis of 24 lectins. Angew Chem Int Ed Engl. 2006;45:3607–3610. doi: 10.1002/anie.200600591. doi:10.1002/anie.200600591. [DOI] [PubMed] [Google Scholar]
- Manimala JC, Roach TA, Li Z, Gildersleeve JC. High-throughput carbohydrate microarray profiling of 27 antibodies demonstrates widespread specificity problems. Glycobiology. 2007;17:17C–23C. doi: 10.1093/glycob/cwm047. doi:10.1093/glycob/cwm047. [DOI] [PubMed] [Google Scholar]
- Matsumura K, Higashida K, Ishida H, Hata Y, Yamamoto K, Shigeta M, Mizuno-Horikawa Y, Wang X, Miyoshi E, Gu J, et al. Carbohydrate binding specificity of a fucose-specific lectin from Aspergillus oryzae: A novel probe for core fucose. J Biol Chem. 2007;282:15700–15708. doi: 10.1074/jbc.M701195200. doi:10.1074/jbc.M701195200. [DOI] [PubMed] [Google Scholar]
- Moothoo DN, Naismith JH. Concanavalin A distorts the β-GlcNAc-(1–>2)-Man linkage of β-GlcNAc-(1–>2)-α-Man-(1–>3)-[β-GlcNAc-(1–>2)-α-Man- (1–>6)]-Man upon binding. Glycobiology. 1998;8:173–181. doi: 10.1093/glycob/8.2.173. doi:10.1093/glycob/8.2.173. [DOI] [PubMed] [Google Scholar]
- Naismith JH, Field RA. Structural basis of trimannoside recognition by concanavalin A. J Biol Chem. 1996;271:972–976. doi: 10.1074/jbc.271.2.972. doi:10.1074/jbc.271.2.972. [DOI] [PubMed] [Google Scholar]
- Patwa TH, Zhao J, Anderson MA, Simeone DM, Lubman DM. Screening of glycosylation patterns in serum using natural glycoprotein microarrays and multi-lectin fluorescence detection. Anal Chem. 2006;78:6411–6421. doi: 10.1021/ac060726z. doi:10.1021/ac060726z. [DOI] [PubMed] [Google Scholar]
- Pilobello KT, Krishnamoorthy L, Slawek D, Mahal LK. Development of a lectin microarray for the rapid analysis of protein glycopatterns. Chembiochem. 2005;6:985–989. doi: 10.1002/cbic.200400403. doi:10.1002/cbic.200400403. [DOI] [PubMed] [Google Scholar]
- Porter A, Yue T, Heeringa L, Day S, Suh E, Haab BB. A motif-based analysis of glycan array data to determine the specificities of glycan-binding proteins. Glycobiology. 2010;20:369–380. doi: 10.1093/glycob/cwp187. doi:10.1093/glycob/cwp187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon N. Lectins: Carbohydrate-specific reagents and biological recognition molecules. J Biol Chem. 2007;282:2753–2764. doi: 10.1074/jbc.X600004200. doi:10.1074/JBC.X600004200. [DOI] [PubMed] [Google Scholar]
- Sokolowski T, Peters T, Perez S, Imberty A. Conformational analysis of biantennary glycans and molecular modeling of their complexes with lentil lectin. J Mol Graph Model. 1997;15:37–42. doi: 10.1016/s1093-3263(97)00011-9. , 54. [DOI] [PubMed] [Google Scholar]
- Tateno H, Nakamura-Tsuruta S, Hirabayashi J. Comparative analysis of core-fucose-binding lectins from Lens culinaris and Pisum sativum using frontal affinity chromatography. Glycobiology. 2009;19:527–536. doi: 10.1093/glycob/cwp016. doi:10.1093/glycob/cwp016. [DOI] [PubMed] [Google Scholar]
- Taylor ME, Drickamer K. Paradigms for glycan-binding receptors in cell adhesion. Curr Opin Cell Biol. 2007;19:572–577. doi: 10.1016/j.ceb.2007.09.004. doi:10.1016/j.ceb.2007.09.004. [DOI] [PubMed] [Google Scholar]
- Taylor ME, Drickamer K. Structural insights into what glycan arrays tell us about how glycan-binding proteins interact with their ligands. Glycobiology. 2009;19:1155–1162. doi: 10.1093/glycob/cwp076. doi:10.1093/glycob/cwp076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varki A, Cummings R, Esko J, Freeze H, Hart G, Marth J. Essentials of Glycobiology. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1999. [PubMed] [Google Scholar]
- Wu AM, Wu JH, Liu JH, Singh T. Recognition profile of Bauhinia purpurea agglutinin (BPA) Life Sci. 2004;74:1763–1779. doi: 10.1016/j.lfs.2003.08.031. doi:10.1016/j.lfs.2003.08.031. [DOI] [PubMed] [Google Scholar]
- Yue T, Goldstein IJ, Hollingsworth MA, Kaul K, Brand RE, Haab BB. The prevalence and nature of glycan alterations on specific proteins in pancreatic cancer patients revealed using antibody-lectin sandwich arrays. Mol Cell Proteomics. 2009;8:1697–1707. doi: 10.1074/mcp.M900135-MCP200. doi:10.1074/mcp.M900135-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue T, Haab BB. Microarrays in glycoproteomics research. Clin Lab Med. 2009;29:15–29. doi: 10.1016/j.cll.2009.01.001. doi:10.1016/j.cll.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.