Abstract
Coronavirus (CoV) RNA synthesis includes the replication of the viral genome, and the transcription of sgRNAs by a discontinuous mechanism. Both processes are regulated by RNA sequences such as the 5′ and 3′ untranslated regions (UTRs), and the transcription regulating sequences (TRSs) of the leader (TRS-L) and those preceding each gene (TRS-Bs). These distant RNA regulatory sequences interact with each other directly and probably through protein-RNA and protein-protein interactions involving viral and cellular proteins.1 By analogy to other plus-stranded RNA viruses, such as polioviruses, in which translation and replication switch involves a cellular factor (PCBP) and a viral protein (3CD),2 it is conceivable that in CoVs the switch between replication and transcription is also associated with the binding of proteins that are specifically recruited by the replication or transcription complexes. Complexes between RNA motifs such as TRS-L and the TRS-Bs located along the CoV genome are probably formed previously to the transcription start, and most likely promote template-switch of the nascent minus RNA to the TRS-L region.3 Many cellular proteins interacting with regulatory CoV RNA sequences4 are members of the heterogeneous nuclear ribonucleoprotein (hnRNP) family of RNA-binding proteins, involved in mRNA processing and transport, which shuttle between the nucleus and the cytoplasm. In the context of CoV RNA synthesis, these cellular ribonucleoproteins might also participate in RNA-protein complexes to bring into physical proximity TRS-L and distant TRS-B, as proposed for CoV discontinuous transcription.5–7 In this review, we summarize RNA-RNA and RNA-protein interactions that represent modest examples of complex quaternary RNA-protein structures required for the fine-tuning of virus replication. Design of chemically defined replication and transcription systems will help to clarify the nature and activity of these structures.
Key words: coronavirus, plus-strand RNA virus, replication, transcription, RNA-RNA interactions, RNA-protein interactions
Introduction
The replication and transcription strategies of RNA viral genomes involve intramolecular interactions between physically close and distal domains, or even intermolecular regulatory contacts. The long-range and local RNA-RNA contacts result in tertiary structures that modulate the function of enhancers, promoters and silencers. In addition to the direct interactions within the viral RNA, a wide spectrum of contacts between viral and cellular proteins has been identified that leads to a complex interaction network required for the fine-tuning of virus replication. This network conforms a higher order RNA-protein structure. Nowadays, limited technological procedures have studied compartmentalized aspects of these structures that represent frozen states in a dynamic equilibrium. A widely used strategy to approach RNA structure is the application of predictive algorithms that consider the viral RNA genomes free in solution.8–10 Although these theoretical predictions are useful to approach scientific questions, the results withdrawn are necessarily limited to small RNA domains. More refined studies predict RNA structure as it is being synthesized,11 but the real situation, the RNA in contact with viral and cellular proteins, is only addressed using reduced RNA motifs.
RNA viruses are classified according to the nature of their RNA genomes into plus-strand, minus-strand and double-stranded RNA viruses. Of these three classes, the most abundant are the plus-stranded RNA viruses. The first replication step in these viruses is the translation of their genome that acts as a viral mRNA. Viral RNA replication takes place in replication complexes generally associated to intracellular membranes. Replication and transcription of several plus-strand RNA viruses require RNA motifs frequently located within the non-coding domains of the 5′ and 3′ ends of the genome. Plus-stranded RNA viruses replicate their genome through a continuous process that starts with the synthesis of an intermediate full-length minus-strand RNA, which then serves as the template for the asymmetric synthesis of progeny plus-strand genomes. During replication, cis acting replication elements (CREs) residing within the coding sequence,12–16 or genome cyclization may be needed.17,18 The cyclization of RNA viral genomes may be mediated by RNA interactions and also by RNA-protein and protein-protein interactions. Interestingly, long-range and local RNA-RNA interactions may be mutually exclusive, and may function as molecular switches controlling critical aspects, such as changes between replication, transcription and translation.19–22
Nidoviruses include the Coronaviridae family with two subfamilies, Coronavirinae and Torovirinae. The Coronavirinae incorporates three genera: α, β and γ; see talk.ictvonline.org/media/g/vertebrate-2008/default.aspx for official coronavirus taxonomy. The CoVs have the largest known viral RNA genome, a plus-strand RNA of 27 to 31 kb. Both viral genome and sgRNAs share common 5′ and 3′ termini that could enable these molecules to amplify by a partially characterized mechanism (Fig. 1). CoV genome includes a 5′ end CAP and a 3′ end poly(A) that may also promote 5′ to 3′ interactions, mediated by viral and cellular proteins. These interactions may be involved in the timely switches controlling CoV replication and transcription. These activities include the synthesis of minus-strand RNAs associated to double-layered membranes (DMVs).23–25
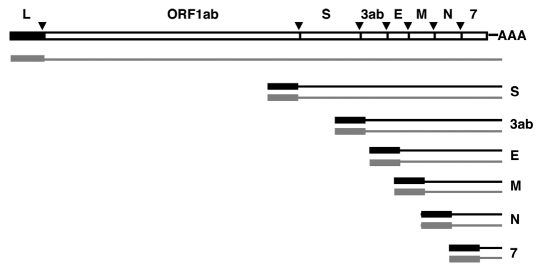
Figure 1.
CoV genome structure and strategy of sgRNA expression. The upper line represents the TGEV genome structure. Letters above the bar indicate the gene names. L, leader sequence. AAA, poly(A). The shorter lines represent the viral mRNAs. Letters to the right of these lines indicate the gene encoded by each mRNA. Black lines represent the positive stranded RNAs, whereas gray lines represent the negative stranded RNAs. The triangles indicate the positions of the conserved core sequences (CSs) located upstream of each gene.
Replication and transcription processes require the translation and processing of a large protein complex, encoded by the 20 kb replicase gene, into up to 16 non-structural proteins (nsps).7 During nidovirus transcription, a 3′ nested set of sgRNAs of increasing size is synthesized (Fig. 1). Each sgRNA contains a 5′ leader segment connected to a body sequence that is identical to the 3′ end of the genome. CoV transcription requires discontinuous RNA synthesis that involves highly specific long distance interactions promoting a template switch leading to a high frequency recombination.26,27 This process is guided by transcription regulating sequences located at the 3′ end of the leader (TRS-L) and preceding the body (TRS-B) of each gene. Accumulating evidence supports a model in which the discontinuous step in sgRNA synthesis occurs during minus-strand RNA synthesis.3,27–31
CoV genome 5′- and 3′ ends and internal structural motifs involved in virus genome replication, and their interaction with viral and cell proteins are being summarized in this review, in comparison with similar long-distance and local RNA-RNA and RNA-protein interactions necessary for the replication of other RNA virus genomes. This review can be complemented with previously published ones.4,6,32–34 Due to the limited space, we have only reported selected representative examples of the different types of interactions described in plus-strand RNA viruses.
Cis-acting RNA Motifs Involved in CoV RNA Synthesis
Many features used by CoVs for genome replication and transcription have been established using natural and artificially constructed defective interfering (DI) RNAs that conserve the cis-acting signals required for their replication in trans by a helper virus.35–41 Recently, reverse genetic systems have been developed for several CoVs, allowing the study of CoV replication and transcription mechanisms in the context of the viral genome.42–47
The minimal sequences required for genome replication have been defined in the three CoV genera by DI RNA deletion mapping analysis and, in all cases these sequences were located at both ends of the genome. For transmissible gastroenterities virus (TGEV), 35,48 from genus α, mouse hepetitis (MHV) from genus β, and infectious bronchitis virus (IBV) from genus γ, these sequences expanded the first 466 to 649 nt at the 5′ end of the genome, and the 388 to 493 nt plus the poly(A) tail at the 3′ end. These regions of the genome contain secondary and higher-order RNA structures, named cis-acting RNA elements that interact with RNA motifs or proteins, and are located within the highly structured 5′ and 3′ UTRs of the CoV genome although, sometimes, they are also embedded into the adjacent coding sequences.
5′ cis-acting RNA elements.
Higher-order RNA structures in the 5′ end of the CoV genome were first studied in the bovine coronavirus (BCoV), where six stem-loop structures, denoted SL-I to SL-VI (Fig. 2), were identified and confirmed by enzymatic probing and functional mutational analysis using DI RNAs.38,53–57 The first four stem-loops mapped within the 5′ UTR, while the SL-V and -VI mapped into the adjacent ORF 1a coding sequence. A consensus secondary structural model for the first 5′ UTR 140 nt, based on the sequence of nine CoVs representing the three genera, has been proposed.58 In this model, the region comprising the original SL-I contains two stem-loops that within this review have been named SL-Ia and SL-Ib, in order to maintain the original nomenclature proposed in previous reports for the next stem-loops (Fig. 2). Recently, it has been predicted that stem-loops SL-Ia and SL-Ib are structurally conserved in all known CoVs,59 and their structures have been confirmed by nuclear magnetic resonance (NMR) spectroscopy in MHV and human CoV (HCoV)-OC43.60–62 A general feature of SL-Ia is that it adopts a bipartite structure and, at least in MHV, the upper region of the stem-loop needs to form a double stranded stem to support viral replication. The lower portion of this stem loop has an optimized lability that probably mediates a long-range interaction between the 5′ and 3′ ends of the genome critical for sgRNA synthesis, but not required for genomic minus-strand synthesis.60 SL-Ib is the most conserved structure in the CoV 5′ UTR. It is composed of a 5 bp stem and a highly conserved loop sequence that adopts a YNMG-type tetra-loop conformation, which has an important role in MHV sgRNA synthesis.61,62
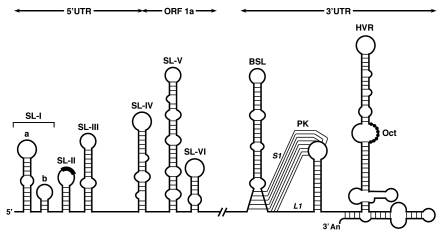
Figure 2.
Cis-acting RNA elements at the 5′- and 3′- genome ends of genus β CoVs. The higher-order RNA structures indicated in the diagram are mainly based on studies done with BCoV and MHV. SL, stem-loop. The core sequence within the leader TRS is shown as a black box on the top of SL-II. SL-1a and SL-1b correspond to SL-I and SL-II described in MHV.60–62 BSL, bulged stem-loop; PK, pseudoknot; S1 and L1, stem 1 and loop 1 of the pseudoknot; HVR, hypervariable region; Oct, 5′-GGA AGA GG-3′ conserved octanucleotide.
The predicted SL-II in the BCoV (named SL-III in other studies),58,59,62 which includes the leader core sequence (CS) into the terminal loop, has a low free energy and is poorly conserved among CoVs.54,62 In fact, SL-II seems non-structured in other CoVs.59,62 Downstream to the leader CS is located SL-III, an RNA motif structurally conserved in all CoVs.59 The presence of a large number of co-variations supports the existence of this stem-loop and particularly the upper half (designated SL-III in the BCoV model),55 which has been confirmed by enzyme structure-probing and is essential for replication of BCoV DI RNA.55 A bulged stem-loop (BSL) SL-IV has been predicted in all CoVs.59 This BSL contains the start codon of ORF 1a preceding the downstream arm of this stem. The existence of this higher-order structure was demonstrated in BCoV by RNase mapping and, similarly to SL-III, the structure rather than the primary sequence was important for DI RNA replication.56 In addition, both SL-III and SL-IV are targets for the binding of viral and cellular proteins, which may play a role in the function of both stem-loops.55,56
Recent studies with BCoV DI RNAs have shown that the 5′-terminal 186 nt of ORF 1a coding region were required for DI RNA replication.53 This region contained two predicted stem-loops, named SL-V and -VI (Fig. 2), with structures confirmed by RNase probing and supported by nucleotide co-variation among closely related genus β CoVs.53,57 In the same study, several cellular proteins binding both stem-loops in vitro were identified, although the functional role of these interactions has not been proven.
3′ cis-acting RNA elements.
The cis-acting RNA elements of the CoV 3′ UTR have been extensively studied in the genus β CoVs MHV and BCoV, and three higher-order structures have been demonstrated by chemical and enzymatic probing and by genetic studies with DI RNAs and recombinant viruses (Fig. 2). The first structure is a BSL of around 68 nt, beginning immediately downstream of the N gene stop codon, which is essential for MHV viral RNA replication.63,64 Adjacent to the BSL there is a hairping-type pseudoknot (PK) of around 54 nt that overlaps with the BSL in 5 nt and is also required for viral RNA replication.22,65 Despite considerable primary sequence divergence, overlapping BSL and PK sequences are conserved in all genus β CoVs and are functionally equivalent.64,66,67 However, genus α CoVs contain a highly conserved PK but not a detectable BSL,65 and CoVs from genus γ have only a conserved and functionally essential BSL.39 These data indicate that CoVs from genera α and γ must have developed alternative mechanisms to those controlling genus β CoV replication. 3′-Downstream of the PK there is a hypervariable region (HVR) that is highly divergent both in primary sequence and secondary structure even among closely related CoVs like MHV and BCoV. However, the HVR contains an octanucleotide sequence, 5′-GGA AGA GG-3′, that is universally conserved among CoVs. In the case of MHV, the HVR consists in a complex multiple stem-loop structure expanding the last 160 nt of the genome (Fig. 2).68 Mutational analysis of MHV HVR indicated that this stem-loop was required for DI RNA replication.68 However, further genetic analysis in the context of the full-length genome showed that the deletion of the entire HVR, including the octanucleotide, had only a modest effect on genomic RNA replication in cell culture, although an effect in pathogenesis was observed.69 This discrepancy could be explained by the competitive nature of DI replication that could increase the effect of mutations that only have a moderate impact in the context of the intact genome. Finally, the 3′ poly(A) tail acts as a cis-replication signal for both MHV and BCoV DI RNAs.70 In this study, a correlation between DI RNA replication and the ability to bind poly(A)-binding protein (PABP) is reported.
In other plus-strand RNA viruses, similarly to CoVs, evidence for higher-order structural elements in the 5′ and 3′ UTRs, or even in coding sequences far away from genome ends have been reported. These structural elements are involved in RNA replication, transcription, translation and encapsidation and have been recently reviewed in reference 32.
A new kind of RNA secondary and higher order structures, named genome-scale ordered structures (GORS), has also been identified in the coding sequences of picornavirus, calicivirus and flavivirus genomes. These RNA structures were not associated with translation or replication, but were correlated with the ability of these viruses to persist in their natural host.71,72
Short-Distance RNA-RNA Interactions in CoV RNA Synthesis
The cis-acting elements in viral RNA genomes have been conventionally viewed as static structures. However, recent research has changed this point of view providing evidence for cooperative activity involving short and long-distance RNA-RNA interactions, as well as conformational changes of these structures to control viral replication and transcription.73,74
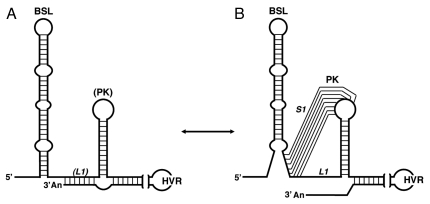
Short-distance RNA-RNA interactions and RNA conformational changes have been reported at the 3′ UTR of MHV. In addition to the PK described above, a predicted interaction between PK loop 1 (L1) and the 3′ end of the genome has been reported (Fig. 3).75 In the same study, evidence of an interaction of the replicase products nsp8 and nsp9 with the PK is presented. With this information, a model for the initiation of CoV minus-strand RNA synthesis has been proposed. According to this model, in the initial structure of the 3′ UTR, the BSL is folded and the 3′ end of the genome is annealed to the base of PK loop (L1) (Fig. 3A). The formed stem-loop binds to a protein complex that includes nsp8 and nsp9 causing a conformational shift that releases the 3′ end of the genome and disrupts the lower stem of the BSL, leading to the formation of the PK (Fig. 3B). The new conformation is probably recognized by the viral RdRp and associated factors, promoting minus-strand RNA synthesis. The two postulated structures cannot simultaneously fold up, leading to the proposal that the BSL and the PK are components of a molecular switch that regulates different steps of RNA synthesis.22,63
Figure 3.
Alternative RNA structures at the MHV 3′ UTR. Two RNA motifs that probably exist in a dynamic equilibrium are shown. The switch between both structures has been postulated in the initiation of CoV RNA synthesis.75 (A) 3′ UTR structure in which the BSL is folded and the 3′ end is annealed to the base of PK loop (L1). (B) 3′ UTR conformation in which the 3′ end is released and the lower stem of BSL is disrupted leading to the formation of the PK. BSL, bulged stem-loop; PK, pseudoknot; S1 and L1, stem 1 and loop 1 of the pseudoknot; HVR, hypervariable region. Acronyms as in Figure 2.
A molecular switch has also been described in the equine arteritis virus, the arterivirus prototype.76,77 A PK interaction between the arterivirus loop SL-5, located at the 3′ UTR and the hairpin of SL-4, located just upstream of SL-5, at the 3′ terminus of the N gene, are essential for viral RNA synthesis. Furthermore, it has been proposed that the formation of this PK may constitute a molecular switch that could regulate the specificity and timing of viral RNA synthesis. Similar PK interactions near the 3′ end have been predicted for all known arteriviruses.77 In the porcine reproductive and respiratory syndrome virus (PRRSV), an additional kissing loop between SL-4 and an upstream hairpin located within the N gene is required for replication.78 However, a similar kissing loop interaction has not been predicted in other arteriviruses. Similar short-distance RNA-RNA interactions and RNA conformational changes required for RNA synthesis have been observed in other RNA virus families as indicated above.
Long-Distance RNA-RNA Interactions Involved in CoV RNA Synthesis
Long-distance RNA-RNA interactions mediate CoV RNA synthesis. CoV discontinuous transcription process implies a premature termination during the synthesis of the minus stranded RNAs and a template switch of the nascent minus RNA to the leader region. This switch requires long distance RNA-RNA interactions, probably assisted by RNA-protein complexes that would bring in close proximity the 5′ end TRS-L and the TRS-B located 5′ upstream of each gene (Fig. 4). These complexes, formed previously to template switch, might contribute to the stop of the minus RNA synthesis at TRS-B sequences.3,5,31
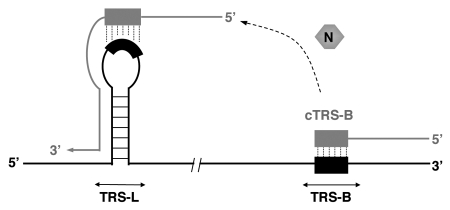
Figure 4.
Postulated template switch during CoV transcription. Nascent minus RNA strand (gray) is synthesized by the RdRp using genomic RNA (black) as a template. After template switch (indicated by the dashed arrow), nascent minus RNA strand hybridizes with the TRS-L, to resume synthesis of the subgenomic RNA. Different viral and cellular proteins may be involved in this process. N protein is required for an efficient transcription, and may help template switch by acting as an RNA chaperone.
After this template switch takes place, a copy of the leader sequence is added to the nascent RNA by a discontinuous synthesis step.29,79,80 The requirement of base pairing between the newly synthesized minus RNA and the TRS-L to facilitate transcription has been described in arteriviruses27,30 and CoVs.3,27,31,81,82 The TRSs include a conserved CS and 5′ and 3′ variable flanking sequences. The template switch takes place after copying the CS sequence. A three RNA strand intermediate could be formed before template switching takes place, including the RNA template (TRS-B), the nascent negative stranded RNA (cTRS-B) and the leader region (TRS-L). If the ΔG associated to the formation of TRS-L and cTRS-B duplex is above a minimum threshold, the template switch may take place.3,31
CoV transcription resembles a similarity assisted copy-choice RNA recombination mechanism,26,27 being the TRS-L the acceptor RNA and cTRS-B the donor RNA. This recombination mechanism requires sequence homology between the donor and acceptor RNAs and a hairpin structure present in the acceptor RNA.83–85 Recently, it has been reported that a template switch may also take place within CoV sgRNAs in the absence of the standard TRS-L structure, an innovative result that may require additional assessment.86 In torovirus, the discontinuous synthesis is also required for sgRNA2 synthesis. In this case, sequence similarity and the presence of a hairpin structure are needed.87
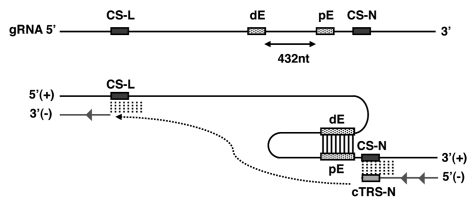
During TGEV transcription, a good correlation has been observed between the sgRNA levels and the ΔG values of the duplexes between TRS-L and each cTRS-B, with only one exception in the synthesis of N gene sgRNA. This sgRNA was the most abundant, despite its lower ΔG value.88 This exception led to the discovery in our laboratory of a cis-acting element upstream of N gene TRS-B that specifically regulated N gene transcription by a long-distance RNA-RNA interaction (Fig. 5).88 The complementarity between the proximal element (pE) located 6 nt upstream of CS-N and the distal element (dE), located 449 nt upstream of TRS-N core sequence (CS-N), was essential to increase the level of sgRNA-N. Interestingly, the ΔG value of this interaction was more relevant than the pE and dE primary sequences. This novel transcription regulatory mechanism is present in the CoV genus α, in which the N gene is not the most 3′ one at the end of viral genome. This RNA-RNA interaction most likely promotes an increase in the transcription complex stop at the TRS-B located preceding gene N, during the synthesis of the minus-strand RNA (Fig. 5). This interruption would specifically enhance the frequency of the template switch to the leader during sgRNA N synthesis. This observation is reminiscent of that previously reported by White's group for transcription in tombusviruses by a premature termination mechanism.33,74,89 A similar long-distance RNA-RNA interaction between two genomic RNAs promoting transcription was previously described in red clover necrotic mosaic virus (RCNMV).90 In flock house virus (FHV) RNA production is also regulated by an intramolecular RNA-RNA interaction.91
Figure 5.
Working model for the specific regulation of TGEV sgRNA-N transcription. The upper line represents the TGEV genome (not to scale) including the core sequence (CS-L) located at the 3′ end of the leader, the distal and proximal elements (dE and pE, respectively) and the CS of N gene (CS-N). The number below the arrow represents the distance between dE and pE elements. The lower lines represent the long distance RNA-RNA interaction within the genome that may act as a stop signal during the synthesis of the negative stranded sgRNA-N (gray line). The dashed arrow indicates the template switch step. cTRS-N, sequence complementary to the CS-N and the 3′ TRS-N flanking sequences.
Long-distance RNA-RNA interactions between RNA sequence motifs mapping at distal positions of the genome (800 nt or more) that regulate replication and transcription, have also been described in other RNA viruses. In fact, potato virus X replication requires the involvement of RNA motifs, such us a hexa-nucleotide at the 3′ end of the genome and a central conserved sequence, into different long distance interactions.92,93 In addition, plus-strand genomic and sgRNA synthesis also require interactions between an octa-nucleotide located at the genome 5′ end, and the central conserved sequences indicated above.94 In hepatitis C virus (HCV), genome replication involves RNA-RNA interactions between a cis-acting element located within the coding sequences and other structural motifs inserted up and downstream to this element.12,19
Genome cyclization is the most distal intramolecular interaction possible within two domains in a viral genome. This type of contact has been described in several RNA virus systems, including CoVs, as a prior step to genome replication and transcription. Previous studies on MHV DI RNA replication, indicate that the last 55 nt of the 3′ end plus the poly(A) tail are sufficient to promote the synthesis of the negative-strand, while both the 5′ and 3′ ends of the genome are required for plus-strand RNA synthesis.95 This observation led us to postulate an interaction between the 5′ and 3′ ends of the genome for CoV replication. A direct RNA-RNA interaction between genome ends has been predicted for MHV, TGEV and severe and acute respiratory syndrome CoV (SARS-CoV) using computer programs designed to analyze full-length genomes.96,97 Genetic evidence supporting a 5′-3′ end cross-talk that stimulates sgRNA synthesis60 came from a study on MHV, in which it was suggested that the base of stem loop SL-Ia, located at the 5′ UTR (Fig. 2), mediates a long range interaction with the 3′ end. In addition, experimental evidence reported for MHV98 and TGEV (Almazan F, Galan C and Enjuanes L, unpublished results) suggests that the interaction between 5′ and 3′ ends may also be mediated by RNA-protein interactions as described below.
Genome cyclization mediated by RNA-RNA, RNA-protein and protein-protein interactions has also been described in other positive stranded RNA viruses. In poliovirus genome circularization the PABP binds to the 3′ poly(A) tail and interacts with the protease-polymerase precursor (3CD) bound to the 5′ cloverleaf RNA.99 This circularized ribonucleoprotein is required to initiate the synthesis of the negative stranded RNA.100 In contrast, the binding of the cellular poly(rC) binding protein (PCBP) to the 5′ cloverleaf RNA promotes the translation.101 Similarly, in pestivirus such as bovine viral diarrhea virus (BVDV), genome circularization is also mediated by three proteins of the NFAR group. This circularization promotes the switch from translation to replication.102 Foot and mouth disease virus (FMDV) uses specific higher-order RNA structures located at the termini of viral genome to regulate the translation and viral RNA synthesis. The replication process requires direct 5′ to 3′ end RNA-RNA interactions mediated by the S region preceding the IRES, the IRES 5′ end and the 3′ end of the genome.103 In addition, RNA-protein interactions also play an important role connecting both ends.
In flaviviruses, genome circularization is mediated by direct long-distance RNA-RNA interactions between complementary sequence motifs present at both ends of the genome. Complementarity disruption impairs RNA synthesis, providing evidence for the requirement of circularization during the replication process.17,20,104
The long-distance RNA-RNA interactions described until now probably represent the tip of the iceberg of a complex network involving complete RNA virus genomes. These long-distance interactions probably lead to a tertiary structure of the full-length virus genome. The best paradigm of this theory is probably reflected in the pioneering studies on tomato bushy stunt virus (TBSV), for which a global RNA structure has been proposed to regulate replication, transcription and translation.33,89
RNA-Protein Interactions Involved in CoV RNA Synthesis
The identification of viral proteins participating in coronavirus RNA synthesis was traditionally based on the analysis of temperature-sensitive mutant viruses. This approach led to the identification of several replicase-encoded nsps as key factors in replication and transcription.1,105 Cell factors involved in RNA synthesis have also been identified by studying their binding to selected regions of the genome,1,106 or to replicase proteins.107,108 Recently, high-throughput assays have been used for the identification of viral and cellular factors affecting CoV replication. These technologies include genome-wide two-hybrid screenings109,110 and proteomic analyses111,112 that identify interactions between replicase components, probably relevant in CoV replication. Alternatively, strategies similar to the ones applied to other plus-stranded RNA virus genomes may also be useful to study CoV replication. This is the case of high-throughput functional assays using host cell mutants or small interfering RNAs (siRNAs) affecting viral RNA amplification and expression applied to HCV,113,114 and human immunodeficiency virus (HIV).115,116
One of the largest limitations in the identification of factors affecting replication or transcription is to distinguish between direct and indirect effects. In addition, factors affecting RNA synthesis frequently also affect translation.117 Therefore, it becomes essential to study the role of factors in defined functional assays, such as in vitro replication or transcription systems. The purification of in vitro active CoV replication-transcription complexes (RTCs) has been described in reference 118. This system could be used to analyze the role of viral and cell factors in RNA synthesis. Although both viral and cellular proteins are most likely part of the purified active complexes, the precise components that reproduce the activity still need to be defined. An alternative is to develop in vitro systems based on defined components. The key difficulty for the development of these CoV in vitro systems is the requirement of a large number of components.
Viral proteins involved in CoV RNA synthesis.
Among the viral proteins required in CoV RTC, it has been proposed that the RdRp (nsp12), the helicase (Hel, nsp13) and N protein are essential.5 In addition, other viral proteins probably contribute to the formation of this complex and the regulation of its activity. In fact, several replicase proteins such as nsp3, nsp4 and nsp6 have mainly scaffolding and membrane rearrangement functions.7,119–121 In addition, nsp3 and nsp5 act as proteases for the correct processing of replicase polyproteins.7
Arterivirus nsp1 binds RNA and has been involved in transcription,122,123 probably by modulating the relative abundance of subgenomic and genomic RNAs.124 In CoVs, the binding of nsp1 to a BCoV cis-acting replication signal has been reported in reference 57. Nevertheless, nsp1 protein has not been involved in RNA synthesis. In line with this observation, the replicase of IBV, a member of CoV genus γ, does not encode a nsp1,7,125 suggesting that CoV nsp1 is probably dispensable for CoV RNA synthesis.
Nsp2 is recruited to the RTC.126 Although nsp2 is dispensable for genus β CoV replication, a 50% reduction in viral RNA synthesis is observed when this protein is deleted.127 Therefore the role of nsp2 in CoV RNA synthesis still remains to be fully established.
Nsp3 is a multifunctional protein that binds RNA.111,128 The role of nsp3 in CoV RNA synthesis remains unclear, as most of its domains are accessory for RTC activity, although they may play a role in pathogenesis.129,130 The interaction between nsp3 and N protein has been recently described, and it has been proposed that nsp3 is involved in the location of CoV RNA genome within the RTC complex at early stages of RNA synthesis.131 SARS-CoV E protein binds the N-terminal acidic domain of nsp3, but its specific role in CoV replication remains to be stablished.132
It has been proposed that nsps 7 to 10 form a functional cassette involved in virus RNA synthesis.133 Nsp7 and nsp8 form a hexadecameric complex that has structural properties compatible with that of a processivity factor for RdRp.134 Nsp8 has RdRp activity and probably produces the primers required for nsp12 mediated RNA synthesis.135,136 Nsp9 is a single-stranded RNA binding protein that may stabilize viral RNAs during RNA synthesis and processing.137,138 Nsp9 forms a homodimer and its oligomerization is required for viral replication.139 MHV nsp8 and nsp9 interact with RdRp,140 and genetic evidence suggest that they also interact with the RNA PK at the 3′ end of the viral genome.75 Nsp10 contains two Zn-finger motifs, binds nucleic acids non-specifically,141,142 and has been implicated in minus-strand RNA synthesis.143
Nsps 14, 15 and 16 encode RNA-processing activities that are unique to the RNA virus world, as exonuclease (ExoN), endoribonuclease (EndoU) and methyltransferase (MT) activities, respectively.7 All these activities are required for efficient CoV RNA synthesis.43,144–146 It has been proposed that the presence of these activities, especially nsp14, allows the maintenance of functional CoV genomes of large size by acting as a proofreading system.125,147,148 CoV nsp14 also encodes a MT activity at its C-terminus, required for RNA synthesis.149 It is possible that MT activities of nsp14 and nsp16 act coordinately for viral mRNA capping.150
CoV N protein forms a ribonucleoprotein complex with genomic RNA. In addition to its structural role, this protein has also a prominent role in RNA synthesis. N protein colocalizes with the RTC.151,152 Initial observations suggested that N protein plays a role in RNA synthesis153–158 and additional studies confirmed that this was the case.159–161 These results are in agreement with the significant increase of infectious virus rescue from different CoV cDNA clones by providing N protein in trans.47,160,162 The N-terminal domain of CoV N protein binds specifically to TRS-L sequences and facilitates unwinding of TRS duplexes.163 CoV N protein also has RNA chaperone activity164 that enhances template switch in vitro.165 As a consequence of these activities, it has been postulated that N protein may facilitate the CoV template switch step during CoV transcription.163,165 In fact, it has been recently reported that N protein is required for an efficient transcription but is not essential for CoV RNA replication.165
Cellular proteins involved in CoV RNA synthesis.
Cellular proteins associated with CoV RNA synthesis are reviewed below according to their binding to 5′ UTR, internal TRS-Bs, 3′ UTR or to viral replicase components. Protein binding to 5′ or 3′ UTR might be involved in viral replication and also in transcription, translation and viral RNA stability. Binding to internal TRS-Bs might be related to transcription or post-transcriptional steps.
Cellular proteins binding CoV 5′ UTR. In MHV, belonging to CoV genus β, polypyrimidine-tract binding (PTB) protein, also known as hnRNP I, binds pentanucleotide repeats of leader TRS, located in the 5′ UTR (nt 56–112). Deletion of these leader repeats in DI RNAs significantly inhibited RNA transcription, suggesting that PTB might be involved in transcription regulation.166 Functional studies showed delayed virus production and reduction of viral RNA synthesis in MHV infected cells overexpressing PTB. This protein interacts with viral N protein both in vitro and in vivo, suggesting a possible contribution to the ribonucleoprotein (RNP) complex formation.167 PTB also binds to the TRS-L sequences at the 5′ end of TGEV genome106 (Sola I and Enjuanes L, unpublished results). hnRNP A1 specifically binds to MHV RNA consensus sequences present in viral genome negative RNAs cTRS-L and cTRS-B, complementary to TRS-L and TRS-B RNA motifs, respectively.168 Another member of hnRNP family, SYNCRIP or hnRNP Q, binds to MHV 5′ UTR or to its complementary sequence c5′ UTR.169 Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was also identified in association with TGEV TRS-L (Table 1). In addition, the knockdown of the GAPDH gene produced a positive effect on CoV-replication both in a TGEV replicon and full-length infectious virus systems, suggesting a potential role in the balance of cellular factors that positively influence TGEV RNA synthesis.106 This hypothesis is supported by the formation of a complex including GAPDH, the bifunctional aminoacyl-tRNA synthetase (EPRS), hnRNP Q and the ribosomal protein L13a, in an inflammatory response.170
Table 1.
Illustrative examples of cellular proteins binding CoV TRSs identified by RNA-affinity chromatography and mass spectrometry
| Biological processa | Protein name |
| mRNA splicing | hnRNP A1, hnRNP A2/B1, hnRNP A3, hnRNP I, hnRNP Q |
| Transcriptional regulation | ILF2, ILF3, SND1 |
| mRNA binding and processing | Nucleolin, DDX1, DHX15 |
| Translational regulation | eIF1α, eIF3S10 |
| Cytoskeleton-movement | Tubulin, annexin A2, moesin |
| Metabolic processes | GAPDH |
Gene ontology classification of biological processes.
Cellular proteins binding CoV TRS-B. hnRNP A1 binds to the complementary RNA strand of MHV TRS-L (cTRS-L) and of several TRS-Bs (cTRS-Bs).168,171 Mutagenesis of TRS-B sequences in a DI RNA reporter system resulted in reduced transcriptional activities that correlated with relative binding affinities of the cTRS-B sequences to hnRNP A1, suggesting that this protein may be a transcription factor in CoV RNA synthesis.168,171 Nevertheless, it cannot be ruled out that the observed effect was due to changes in the extent of the complementarity between the newly synthesized cTRS-B and TRS-L. In an in vitro reconstitution assay, hnRNP A1 mediates the formation of an RNP complex with the minus strand cTRS-L and cTRS-B.172 Furthermore, hnRNP A1 interacted both in vitro and in vivo with MHV N protein, that forms part of the RTC.173
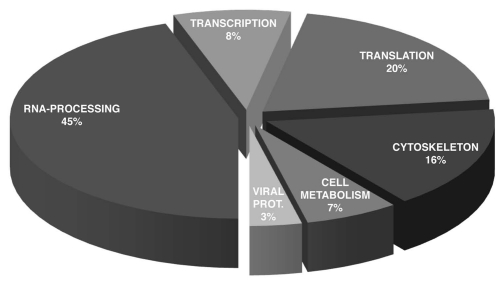
We have used TGEV CoV as a model to identify candidate cellular proteins participating in CoV transcription. Leader and body TRSs (TRS-L and TRS-B, respectively) with either positive or negative polarity, as well as RNA duplexes mimicking TRS-L/cTRS-B hybrids formed during transcription template switch, were used in RNA affinity chromatography assays. Mass spectrometry analysis led to the identification of around 40 cellular proteins interacting with TGEV TRSs. A major proportion of these proteins was associated with cellular RNA metabolism, including pre-mRNA processing (45%), transcription (8%) and translation (20%) (Fig. 6). Members of hnRNP family or the DEAD box family of cellular RNA helicases (DDX and DHX) were extensively represented. hnRNP I was found in association with the plus-stranded RNA of TRS-L and several TRS-B and, therefore, PTB may participate in the regulation of sgRNA synthesis (Sola I and Enjuanes L, unpublished results). Other identified proteins were related to cytoskeleton (16.7%) or cell metabolism processes (3.3%) (Fig. 6 and Table 1). Some of the identified proteins have already been involved in the life cycles of other plus-stranded RNA viruses and may provide novel insights into the protein machinery utilized by CoVs in RNA synthesis.174
Figure 6.
Cell proteins with affinity for TGEV RNA structural motifs. Functional classification of cellular proteins interacting with CoV TRSs. RNA-affinity chromatography assays were performed with TGEV-infected human Huh7 cell extracts and viral TRS RNAs. Forty proteins interacting with TRS RNAs were identified by mass spectrometry and categorized according to the biological processes in which they are involved.
Cellular proteins binding CoV 3′ UTR. UV-crosslinking experiments showed binding of hnRNP A1 to MHV genome 3′ UTR sites located at nt 90–170 (mapping within the HVR domain) and nt 260–350 numbered from the 3′ end of the genome (partially overlaping with the BSL).98 Overexpression of hnRNP A1 or of a dominant-negative mutant derived from this protein, accelerated or delayed, respectively, viral RNA synthesis, suggesting that hnRNP A1 participates in the formation of the RTC.175 hnRNP A1 is translocated from the nucleus to the cytoplasm of infected cells, probably as a result of specific interaction with MHV RNA.168 Other member of the hnRNP A2/B1 multigene family can substitute hnRNP A1 in MHV replication.176 hnRNP A1 binding sites are complementary to those on the negative-strand RNA that bind PTB. Partial deletion of hnRNP A1 and PTB binding sites abolished sgRNA synthesis by MHV DI RNAs.177 hnRNP A1 and PTB also bind to the complementary strands at the 5′ end of MHV RNA and mediate the in vitro formation of a RNP complex involving the 5′ and 3′ end fragments of MHV RNA.98
Other cellular proteins bind the CoV 3′ end genome and also affect virus replication. Thus, PABP binds to BCoV70 and TGEV (Marquez-Jurado S, Enjuanes L and Almazan F, unpublished results) poly(A). Furthermore, a positive effect of PABP in TGEV RNA synthesis has been observed in reference 106. Other 3′-interacting host factors such as the hnRNP Q and the EPRS also positively contribute to TGEV RNA synthesis, as shown in functional analysis with siRNAs using both a TGEV derived replicon and the infectious virus itself.106 Transcriptional coactivator p100 (SND1) specifically binds the 3′ end of the TGEV,106 and also the TRS-L and TRS-B RNA motifs (Table 1) (Sola I and Enjuanes L, unpublished results). P100 protein interacts with the nsp1 of EAV, specifically involved in mRNA synthesis.178 However, no functional studies have shown the role of p100 in arterivirus transcription. Mitochondrial proteins aconitase, heat shock protein (HSP)40, HSP60 and HSP70 bind to MHV 3′-most 42 nt of RNA genome, although their requirement for CoV replication was not directly addressed.179,180 Specific mutations in this 3′ UTR sequence led to a virus with a defect in sgRNA synthesis,181 supporting the relevance of this 3′ end sequence domain in virus transcription.
Cellular proteins binding CoV replicase subunits. An alternative approach to identify host factors that participate in CoV RNA synthesis relies on their specific binding to viral replicase subunits. Several members of the cellular DEAD box family of helicases such as DDX5 and DDX1 have been associated with CoV RNA synthesis. These helicases have been involved in several biological processes related to RNA transcriptional regulation, pre-mRNA processing, RNA degradation, RNA export, ribosome assembly and translation.182 The specific interaction of cellular DDX5 protein with the SARS-CoV helicase protein (nsp13) has been shown using yeast and mammalian two-hybrid assays and co-immunoprecipitation. Silencing of DDX5 expression by siRNAs significantly reduced viral RNA replication and virus titers.107 DDX1 interacted with IBV and SARS-CoV exonuclease (np14), as demonstrated by yeast two-hybrid and coimmunoprecipitation assays. The effect of DDX1 expression knockdown in CoV RNA replication and transcription indicated that DDX1 might be a cofactor essential for these activities.108 Interestingly, RNA helicase DDX1 has also been identified in association with TGEV TRSs (Sola I and Enjuanes L, unpublished results) and might perform common functions in CoV replication.
Perspectives
Recent advances in understanding the molecular mechanisms of replication and transcription in positive-stranded RNA viruses indicate that regulatory elements in viral genomes are dynamic complexes involving short and long-distance RNA-RNA interactions and also RNA-protein complexes including viral and cellular proteins. Transition between two structural conformations may regulate the switch between essential activities within virus cycle. Future challenges in this field rely on the development of novel methodologies to study RNA structure formation in vivo as a consequence of RNA-RNA and RNA-protein interactions in the cellular environment. In addition, in vitro reconstitution of replication-transcription complexes with defined composition will be critical to analyze the contribution of viral and cell factors to RNA synthesis.
Acknowledgements
This work was supported by grants from Ministry of Science and Innovation of Spain (BIO2007-60978 and BIO2010-16705), US National Institutes of Health (ARRA-W000151845) and the European Community Seventh Framework Programme (FP7/2007-2013) under the projects “EMPERIE” (EC Grant Agreement number 223498) and “PoRRSCon” (EC Grant Agreement number 245141). I.S. and S.Z. received contracts from the indicated EU projects, respectively. P.A.M. received a fellowship from the Ministry of Science and Innovation of Spain.
Abbreviations
- CoV
coronavirus
- sgRNA
subgenomic RNA
- UTR
untranslated region
- TRS
transcription-regulating sequences
- SL
stem loop
- DI RNA
defective interfering RNA
- ORF
open reading frame
- BSL
bulged stem-loop
- PK
pseudoknot
- CS
conserved core sequence
- ΔG
thermodynamic free energy
- nsp
non-structural protein
- RTC
replication-transcription complex
- hnRNP
heterogeneous nuclear ribonucleoprotein
- RNP complex
ribonucleoprotein complex
References
- 1.Shi ST, Lai MM. Viral and cellular proteins involved in coronavirus replication. In: Enjuanes L, editor. Curr Top Microbiol Immunol. 2005. pp. 95–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gamarnik AV, Andino R. Switch from translation to RNA replication in a positive-stranded RNA virus. Genes Develop. 1998;12:2293–2304. doi: 10.1101/gad.12.15.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sola I, Moreno JL, Zuñiga S, Alonso S, Enjuanes L. Role of nucleotides immediately flanking the transcription-regulating sequence core in coronavirus subgenomic mRNA synthesis. J Virol. 2005;79:2506–2516. doi: 10.1128/JVI.79.4.2506-2516.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu P, Leibowitz J. RNA higher-order structures within the coronavirus 5′ and 3′ untranslated regions and their roles in viral replication. In: Lal SK, editor. Molecular Biology of the SARS-Coronavirus. Berlin Heidelberg: Springer-Verlag; 2010. pp. 47–62. [Google Scholar]
- 5.Enjuanes L, Almazan F, Sola I, Zuniga S. Biochemical aspects of coronavirus replication and virus-host interaction. Annu Rev Microbiol. 2006;60:211–230. doi: 10.1146/annurev.micro.60.080805.142157. [DOI] [PubMed] [Google Scholar]
- 6.Masters PS. The molecular biology of coronaviruses. Adv Virus Res. 2006;66:193–292. doi: 10.1016/S0065-3527(06)66005-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ziebuhr J. The coronavirus replicase. In: Enjuanes L, editor. Coronavirus replication and reverse genetics. Springer; 2005. pp. 57–94. [Google Scholar]
- 8.Mathews DH, Turner DH, Zuker M. RNA secondary structure prediction. Curr Protoc Nucleic Acid Chem. 2007;11:11–12. doi: 10.1002/0471142700.nc1102s28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reuter JS, Mathews DH. RNA structure: software for RNA secondary structure prediction and analysis. BMC Bioinformatics. 2010;11:129. doi: 10.1186/1471-2105-11-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Batenburg FH, Gultyaev AP, Pleij CW. An APL-programmed genetic algorithm for the prediction of RNA secondary structure. J Theor Biol. 1995;174:269–280. doi: 10.1006/jtbi.1995.0098. [DOI] [PubMed] [Google Scholar]
- 12.Friebe P, Boudet J, Simorre JP, Bartenschlager R. Kissing-loop interaction in the 3′ end of the hepatitis C virus genome essential for RNA replication. J Virol. 2005;79:380–392. doi: 10.1128/JVI.79.1.380-392.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goodfellow I, Chaudhry Y, Richardson A, Meredith J, Almond JW, Barclay W, et al. Identification of a cis-acting replication element within the poliovirus coding region. J Virol. 2000;74:4590–4600. doi: 10.1128/jvi.74.10.4590-4600.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mason PW, Bezborodova SV, Henry TM. Identification and characterization of a cis-acting replication element (cre) adjacent to the internal ribosome entry site of foot-and-mouth disease virus. J Virol. 2002;76:9686–9694. doi: 10.1128/JVI.76.19.9686-9694.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mcknight KL, Lemon SM. The rhinovirus type 14 genome contains an internally located RNA structure that is required for viral replication. RNA. 1998;4:1569–1584. doi: 10.1017/s1355838298981006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paul AV, Rieder E, King DW, van Boom JH, Wimmer E. Identification of an RNA hairpin in poliovirus RNA that serves as the primary template in the in vitro uridylylation of VPg. J Virol. 2000;74:10359–10370. doi: 10.1128/jvi.74.22.10359-10370.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Villordo SM, Gamarnik AV. Genome cyclization as strategy for flavivirus RNA replication. Virus Res. 2009;139:230–239. doi: 10.1016/j.virusres.2008.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang B, Dong H, Stein DA, Iversen PL, Shi PY. West Nile virus genome cyclization and RNA replication require two pairs of long-distance RNA interactions. Virology. 2008;373:1–13. doi: 10.1016/j.virol.2008.01.016. [DOI] [PubMed] [Google Scholar]
- 19.Diviney S, Tuplin A, Struthers M, Armstrong V, Elliott RM, Simmonds P, et al. A hepatitis C virus cis-acting replication element forms a long-range RNA-RNA interaction with upstream RNA sequences in NS5B. J Virol. 2008;82:9008–9022. doi: 10.1128/JVI.02326-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Romero-Lopez C, Berzal-Herranz A. A long-range RNA-RNA interaction between the 5′ and 3′ ends of the HCV genome. RNA. 2009;15:1740–1752. doi: 10.1261/rna.1680809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yuan X, Shi K, Young MY, Simon AE. The terminal loop of a 3′ proximal hairpin plays a critical role in replication and the structure of the 3′ region of Turnip crinkle virus. Virology. 2010;402:271–280. doi: 10.1016/j.virol.2010.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goebel SJ, Hsue B, Dombrowski TF, Masters PS. Characterization of the RNA components of a putative molecular switch in the 3′ untranslated region of the murine coronavirus genome. J Virol. 2004;78:669–682. doi: 10.1128/JVI.78.2.669-682.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Knoops K, Kikkert M, Worm SH, Zevenhoven-Dobbe JC, van der Meer Y, Koster AJ, et al. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 2008;6:226. doi: 10.1371/journal.pbio.0060226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Snijder EJ, van der Meer Y, Zevenhoven-Dobbe J, Onderwater JJ, van der Meulen J, Koerten HK, et al. Ultrastructure and origin of membrane vesicles associated with the severe acute respiratory syndrome coronavirus replication complex. J Virol. 2006;80:5927–5940. doi: 10.1128/JVI.02501-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gosert R, Kanjanahaluethai A, Egger D, Bienz K, Baker SC. RNA replication of mouse hepatitis virus takes place at double-membrane vesicles. J Virol. 2002;76:3697–3708. doi: 10.1128/JVI.76.8.3697-3708.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brian DA, Spaan WJM. Recombination and coronavirus defective interfering RNAs. Semin Virol. 1997;8:101–111. doi: 10.1006/smvy.1997.0109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pasternak AO, van den Born E, Spaan WJ, Snijder EJ. Sequence requirements for RNA strand transfer during nidovirus discontinuous subgenomic RNA synthesis. EMBO J. 2001;20:7220–7228. doi: 10.1093/emboj/20.24.7220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baric RS, Yount B. Subgenomic negative-strand RNA function during mouse hepatitis virus infection. J Virol. 2000;74:4039–4046. doi: 10.1128/jvi.74.9.4039-4046.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sawicki SG, Sawicki DL. A new model for coronavirus transcription. Adv Exp Med Biol. 1998;440:215–219. doi: 10.1007/978-1-4615-5331-1_26. [DOI] [PubMed] [Google Scholar]
- 30.van Marle G, Dobbe JC, Gultyaev AP, Luytjes W, Spaan WJM, Snijder EJ. Arterivirus discontinuous mRNA transcription is guided by base pairing between sense and antisense transcription-regulating sequences. Proc Natl Acad Sci USA. 1999;96:12056–12061. doi: 10.1073/pnas.96.21.12056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zuñiga S, Sola I, Alonso S, Enjuanes L. Sequence motifs involved in the regulation of discontinuous coronavirus subgenomic RNA synthesis. J Virol. 2004;78:980–994. doi: 10.1128/JVI.78.2.980-994.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu Y, Wimmer E, Paul AV. Cis-acting RNA elements in human and animal plus-strand RNA viruses. Biochim Biophys Acta. 2009;1789:495–517. doi: 10.1016/j.bbagrm.2009.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu B, Pogany J, Na H, Nicholson BL, Nagy PD, White KA. A discontinuous RNA platform mediates RNA virus replication: building an integrated model for RNA-based regulation of viral processes. PLoS Pathog. 2009;5:1000323. doi: 10.1371/journal.ppat.1000323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Enjuanes L, Sola I, Zuñiga S, Moreno JL. Coronavirus RNA synthesis: Transcription. In: Thiel V, editor. Coronaviruses: Molecular and celular biology. Norfolk: Caister Academic Press; 2007. pp. 81–107. [Google Scholar]
- 35.Izeta A, Smerdou C, Alonso S, Penzes Z, Mendez A, Plana-Duran J, et al. Replication and packaging of transmissible gastroenteritis coronavirus-derived synthetic minigenomes. J Virol. 1999;73:1535–1545. doi: 10.1128/jvi.73.2.1535-1545.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Makino S, Fujioka N, Fujiwara K. Structure of the intracellular viral RNAs of defective interfering particles of mouse hepatitis virus. J Virol. 1985;54:329–336. doi: 10.1128/jvi.54.2.329-336.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Makino S, Shieh CK, Keck JG, Lai MMC. Defective-interfering particles of murine coronaviruses: mechanism of synthesis of defective viral RNAs. Virology. 1988;163:104–111. doi: 10.1016/0042-6822(88)90237-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chang RY, Hofmann MA, Sethna PB, Brian DA. A cis-acting function for the coronavirus leader in defective interfering RNA replication. J Virol. 1994;68:8223–8231. doi: 10.1128/jvi.68.12.8223-8231.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dalton K, Casais R, Shaw K, Stirrups K, Evans S, Britton P, et al. cis-acting sequences required for coronavirus infectious bronchitis virus defective-RNA replication and packaging. J Virol. 2001;75:125–133. doi: 10.1128/JVI.75.1.125-133.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Méndez A, Smerdou C, Izeta A, Gebauer F, Enjuanes L. Molecular characterization of transmissible gastroenteritis coronavirus defective interfering genomes: packaging and heterogeneity. Virology. 1996;217:495–507. doi: 10.1006/viro.1996.0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Penzes Z, Tibbles K, Shaw K, Britton P, Brown TDK, Cavanagh D. Characterization of a replicating and packaged defective RNA of avian coronavirus infectious bronchitis virus. Virology. 1994;203:286–293. doi: 10.1006/viro.1994.1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Almazan F, Gonzalez JM, Penzes Z, Izeta A, Calvo E, Plana-Duran J, et al. Engineering the largest RNA virus genome as an infectious bacterial artificial chromosome. Proc Natl Acad Sci USA. 2000;97:5516–5521. doi: 10.1073/pnas.97.10.5516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Almazan F, DeDiego ML, Galan C, Escors D, Alvarez E, Ortego J, et al. Construction of a SARS-CoV infectious cDNA clone and a replicon to study coronavirus RNA synthesis. J Virol. 2006;80:10900–10906. doi: 10.1128/JVI.00385-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.St-Jean JR, Desforges M, Almazan F, Jacomy H, Enjuanes L, Talbot PJ. Recovery of a neurovirulent human coronavirus OC43 from an infectious cDNA clone. J Virol. 2006;80:3670–3674. doi: 10.1128/JVI.80.7.3670-3674.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yount B, Denison MR, Weiss SR, Baric RS. Systematic assembly of a full-length infectious cDNA of mouse hepatitis virus strain A59. J Virol. 2002;76:11065–11078. doi: 10.1128/JVI.76.21.11065-11078.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thiel V, Herold J, Schelle B, Siddell S. Infectious RNA transcribed in vitro from a cDNA copy of the human coronavirus genome cloned in vaccinia virus. J Gen Virol. 2001;82:1273–1281. doi: 10.1099/0022-1317-82-6-1273. [DOI] [PubMed] [Google Scholar]
- 47.Casais R, Thiel V, Siddell SG, Cavanagh D, Britton P. Reverse genetics system for the avian coronavirus infectious bronchitis virus. J Virol. 2001;75:12359–12369. doi: 10.1128/JVI.75.24.12359-12369.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Escors D, Izeta A, Capiscol C, Enjuanes L. Transmissible gastroenteritis coronavirus packaging signal is located at the 5′ end of the virus genome. J Virol. 2003;77:7890–7902. doi: 10.1128/JVI.77.14.7890-7902.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim Y, Jeong Y, Makino S. Analysis of cis-acting sequences essential for coronavirus defective interfering RNA replication. Virology. 1993;197:53–63. doi: 10.1006/viro.1993.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lin YJ, Lai MMC. Deletion mapping of a mouse hepatitis virus defective interfering RNA reveals the requirement of an internal and discontinuous sequence for replication. J Virol. 1993;67:6110–6118. doi: 10.1128/jvi.67.10.6110-6118.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Luytjes W, Gerritsma H, Spaan WJM. Replication of synthetic defective interfering RNAs derived from coronavirus mouse hepatitis virus-A59. Virology. 1996;216:174–183. doi: 10.1006/viro.1996.0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.van der Most R, Luytjes W, Rutjes S, Spaan WJM. Translation but not the encoded sequence is essential for the efficient propagation of the defective interfering RNAs of the coronavirus mouse hepatitis virus. J Virol. 1995;69:3744–3751. doi: 10.1128/jvi.69.6.3744-3751.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brown CG, Nixon KS, Senanayake SD, Brian DA. An RNA stem-loop within the bovine coronavirus nsp1 coding region is a cis-acting element in defective interfering RNA replication. J Virol. 2007;81:7716–7724. doi: 10.1128/JVI.00549-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chang RY, Krishnan R, Brian DA. The UCU AAA C promoter motif is not required for high-frequency leader recombination in bovine coronavirus defective interfering RNA. J Virol. 1996;70:2720–2729. doi: 10.1128/jvi.70.5.2720-2729.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Raman S, Bouma P, Williams GD, Brian DA. Stem-loop III in the 5′ untranslated region is a cis-acting element in bovine coronavirus defective interfering RNA replication. J Virol. 2003;77:6720–6730. doi: 10.1128/JVI.77.12.6720-6730.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Raman S, Brian DA. Stem-loop IV in the 5′ untranslated region is a cis-acting element in bovine coronavirus defective interfering RNA replication. J Virol. 2005;79:12434–12446. doi: 10.1128/JVI.79.19.12434-12446.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gustin KM, Guan BJ, Dziduszko A, Brian DA. Bovine coronavirus nonstructural protein 1 (p28) is an RNA binding protein that binds terminal genomic cis-replication elements. J Virol. 2009;83:6087–6097. doi: 10.1128/JVI.00160-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kang H, Feng M, Schroeder ME, Giedroc DP, Leibowitz JL. Putative cis-acting stem-loops in the 5′ untranslated region of the severe acute respiratory syndrome coronavirus can substitute for their mouse hepatitis virus counterparts. J Virol. 2006;80:10600–10614. doi: 10.1128/JVI.00455-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen SC, Olsthoorn RC. Group-specific structural features of the 5′-proximal sequences of coronavirus genomic RNAs. Virology. 2010;401:29–41. doi: 10.1016/j.virol.2010.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li L, Kang H, Liu P, Makkinje N, Williamson ST, Leibowitz JL, et al. Structural lability in stem-loop 1 drives a 5′ UTR-3′ UTR interaction in coronavirus replication. J Mol Biol. 2008;377:790–803. doi: 10.1016/j.jmb.2008.01.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu P, Li L, Keane SC, Yang D, Leibowitz JL, Giedroc DP. Mouse hepatitis virus stem-loop 2 adopts a uYNMG(U)a-like tetraloop structure that is highly functionally tolerant of base substitutions. J Virol. 2009;83:12084–12093. doi: 10.1128/JVI.00915-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu P, Li L, Millership JJ, Kang H, Leibowitz JL, Giedroc DP. A U-turn motif-containing stem-loop in the coronavirus 5′ untranslated region plays a functional role in replication. RNA. 2007;13:763–780. doi: 10.1261/rna.261807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hsue B, Hartshorne T, Masters PS. Characterization of an essential RNA secondary structure in the 3′ untranslated region of the murine coronavirus genome. J Virol. 2000;74:6911–6921. doi: 10.1128/jvi.74.15.6911-6921.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hsue B, Masters PS. A bulged stem-loop structure in the 3′ untranslated region of the genome of the coronavirus mouse hepatitis virus is essential for replication. J Virol. 1997;71:7567–7578. doi: 10.1128/jvi.71.10.7567-7578.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Williams GD, Chang RY, Brian DA. A phylogenetically conserved hairpin-type 3′ untranslated region pseudoknot functions in coronavirus RNA replication. J Virol. 1999;73:8349–8355. doi: 10.1128/jvi.73.10.8349-8355.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wu HY, Guy JS, Yoo D, Vlasak R, Urbach E, Brian DA. Common RNA replication signals exist among group 2 coronaviruses: evidence for in vivo recombination between animal and human coronavirus molecules. Virology. 2003;315:174–183. doi: 10.1016/S0042-6822(03)00511-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Goebel SJ, Taylor J, Masters PS. The 3′ cis-acting genomic replication element of the severe acute respiratory syndrome coronavirus can function in the murine coronavirus genome. J Virol. 2004;78:7846–7851. doi: 10.1128/JVI.78.14.7846-7851.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liu Q, Johnson RF, Leibowitz JL. Secondary structural elements within the 3′ untranslated region of mouse hepatitis virus strain JHM genomic RNA. J Virol. 2001;75:12105–12113. doi: 10.1128/JVI.75.24.12105-12113.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Goebel SJ, Miller TB, Bennett CJ, Bernard KA, Masters PS. A hypervariable region within the 3′ cis-acting element of the murine coronavirus genome is nonessential for RNA synthesis but affects pathogenesis. J Virol. 2007;81:1274–1287. doi: 10.1128/JVI.00803-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Spagnolo JF, Hogue BG. Host protein interactions with the 3′ end of bovine coronavirus RNA and the requirement of the poly(A) tail for coronavirus defective genome replication. J Virol. 2000;74:5053–5065. doi: 10.1128/jvi.74.11.5053-5065.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Davis M, Sagan SM, Pezacki JP, Evans DJ, Simmonds P. Bioinformatic and physical characterizations of genome-scale ordered RNA structure in mammalian RNA viruses. J Virol. 2008;82:11824–11886. doi: 10.1128/JVI.01078-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Simmonds P, Tuplin A, Evans DJ. Detection of genome-scale ordered RNA structure (GORS) in genomes of positive-stranded RNA viruses: Implications for virus evolution and host persistence. RNA. 2004;10:1337–1351. doi: 10.1261/rna.7640104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Simon AE, Gehrke L. RNA conformational changes in the life cycles of RNA viruses, viroids and virus-associated RNAs. Biochim Biophys Acta. 2009;1789:571–583. doi: 10.1016/j.bbagrm.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Miller WA, White KA. Long-distance RNA-RNA interactions in plant virus gene expression and replication. Annu Rev Phytopathol. 2006;44:447–467. doi: 10.1146/annurev.phyto.44.070505.143353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zust R, Miller TB, Goebel SJ, Thiel V, Masters PS. Genetic interactions between an essential 3′ cis-acting RNA pseudoknot, replicase gene products and the extreme 3′ end of the mouse coronavirus genome. J Virol. 2008;82:1214–1228. doi: 10.1128/JVI.01690-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Beerens N, Snijder EJ. RNA signals in the 3′ terminus of the genome of Equine arteritis virus are required for viral RNA synthesis. J Gen Virol. 2006;87:1977–1983. doi: 10.1099/vir.0.81750-0. [DOI] [PubMed] [Google Scholar]
- 77.Beerens N, Snijder EJ. An RNA pseudoknot in the 3′ end of the arterivirus genome has a critical role in regulating viral RNA synthesis. J Virol. 2007;81:9426–9436. doi: 10.1128/JVI.00747-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Verheije MH, Olsthoorn RCL, Kroese MV, Rottier PJM, Meulenberg JJM. Kissing interaction between 3′ noncoding and coding sequences is essential for porcine arterivirus RNA replication. J Virol. 2002;76:1521–1526. doi: 10.1128/JVI.76.3.1521-1526.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lai MMC, Cavanagh D. The molecular biology of coronaviruses. Adv Virus Res. 1997;48:1–100. doi: 10.1016/S0065-3527(08)60286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sethna PB, Hung SL, Brian DA. Coronavirus subgenomic minus-strand RNAs and the potential for mRNA replicons. Proc Natl Acad Sci USA. 1989;86:5626–5630. doi: 10.1073/pnas.86.14.5626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Alonso S, Izeta A, Sola I, Enjuanes L. Transcription regulatory sequences and mRNA expression levels in the coronavirus transmissible gastroenteritis virus. J Virol. 2002;76:1293–1308. doi: 10.1128/JVI.76.3.1293-1308.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ozdarendeli A, Ku S, Rochat S, Williams GD, Senanayake SD, Brian DA. Downstream sequences influence the choice between a naturally occurring noncanonical and closely positioned upstream canonical heptameric fusion motif during bovine coronavirus subgenomic mRNA synthesis. J Virol. 2001;75:7362–7374. doi: 10.1128/JVI.75.16.7362-7374.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Draghici HK, Varrelmann M. Evidence for similarity-assisted recombination and predicted stem-loop structure determinant in potato virus X RNA recombination. J Gen Virol. 2010;91:552–562. doi: 10.1099/vir.0.014712-0. [DOI] [PubMed] [Google Scholar]
- 84.Nagy PD, Simon AE. New insights into the mechanisms of RNA recombination. Virology. 1997;235:1–9. doi: 10.1006/viro.1997.8681. [DOI] [PubMed] [Google Scholar]
- 85.Nagy PD, Zhang C, Simon AE. Dissecting RNA recombination in vitro: role of RNA sequences and the viral replicase. EMBO J. 1998;17:2392–2403. doi: 10.1093/emboj/17.8.2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wu HY, Brian DA. Subgenomic messenger RNA amplification in coronaviruses. Proc Natl Acad Sci USA. 2010;107:12257–12262. doi: 10.1073/pnas.1000378107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.van Vliet ALW, Smits SL, Rottier PJM, de Groot RJ. Discontinuous and non-discontinuous subgenomic RNA transcription in a nidovirus. EMBO J. 2002;21:6571–6580. doi: 10.1093/emboj/cdf635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Moreno JL, Zuniga S, Enjuanes L, Sola I. Identification of a coronavirus transcription enhancer. J Virol. 2008;82:3882–3893. doi: 10.1128/JVI.02622-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wang S, Mortazavi L, White KA. Higher-order RNA structural requirements and small-molecule induction of tombusvirus subgenomic mRNA transcription. J Virol. 2008;82:3864–3871. doi: 10.1128/JVI.02416-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sit TL, Vaewhongs AA, Lommel SA. RNA-mediated trans-activation of transcription from a viral RNA. Science. 1998;281:829–832. doi: 10.1126/science.281.5378.829. [DOI] [PubMed] [Google Scholar]
- 91.Lindenbach BD, Sgro JY, Ahlquist P. Long-distance base pairing in flock house virus RNA1 regulates subgenomic RNA3 synthesis and RNA2 replication. J Virol. 2002;76:3905–3919. doi: 10.1128/JVI.76.8.3905-3919.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kim KH, Hemenway CL. Long-distance RNA-RNA interactions and conserved sequence elements affect potato virus X plus-strand RNA accumulation. RNA. 1999;5:636–645. doi: 10.1017/s1355838299982006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kim KH, Hemenway C. Mutations that alter a conserved element upstream of the potato virus X triple block and coat protein genes affect subgenomic RNA accumulation. Virology. 1997;232:187–197. doi: 10.1006/viro.1997.8565. [DOI] [PubMed] [Google Scholar]
- 94.Hu B, Pillai-Nair N, Hemenway C. Long-distance RNA-RNA interactions between terminal elements and the same subset of internal elements on the potato virus X genome mediate minus- and plus-strand RNA synthesis. RNA. 2007;13:267–280. doi: 10.1261/rna.243607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lin YJ, Liao CL, Lai MMC. Identification of the cis-acting signal for minus-strand RNA synthesis of a murine coronavirus: implications for the role of minus-strand RNA in RNA replication and transcription. J Virol. 1994;68:8131–8140. doi: 10.1128/jvi.68.12.8131-8140.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sgro JY, Holt J, Zuker M, Palmenberg A. RNA folding of the complete SARS and MHV coronavirus genomes; Seventh International Symposium on Positive Strand RNA Viruses; San Francisco, California, USA. 2004. [Google Scholar]
- 97.Holt J, Sgro JY, Zuker M, Palmenberg A. Computer folding of full-length viral genomes: a new toolkit for automated analysis of RNAs longer than 10,000 bases; Seventh International Symposium on Positive Strand RNA Viruses; San Francisco, California, USA. 2004. [Google Scholar]
- 98.Huang P, Lai MMC. Heterogeneous nuclear ribonucleoprotein A1 binds to the 3′-untranslated region and mediates potential 5′-3′-end cross talks of mouse hepatitis virus RNA. J Virol. 2001;75:5009–5017. doi: 10.1128/JVI.75.11.5009-5017.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Herold J, Andino R. Poliovirus RNA replication requires genome circularization through a protein-protein bridge. Mol Cell. 2001;7:581–591. doi: 10.1016/S1097-2765(01)00205-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gamarnik AV, Andino R. Two functional complexes formed by KH domain containing proteins with the 5′ noncoding region of poliovirus RNA. RNA. 1997;3:882–892. [PMC free article] [PubMed] [Google Scholar]
- 101.Parsley TB, Towner JS, Blyn LB, Ehrenfeld E, Semler BL. Poly (rC) binding protein 2 forms a ternary complex with the 5′-terminal sequences of poliovirus RNA and the viral 3CD proteinase. RNA. 1997;3:1124–1134. [PMC free article] [PubMed] [Google Scholar]
- 102.Isken O, Grassmann CW, Sarisky RT, Kann M, Zhang S, Grosse F, et al. Members of the NF90/NFAR protein group are involved in the life cycle of a positive-strand RNA virus. EMBO J. 2003;22:5655–5665. doi: 10.1093/emboj/cdg562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Martinez-Salas E, Pacheco A, Serrano P, Fernandez N. New insights into internal ribosome entry site elements relevant for viral gene expression. J Gen Virol. 2008;89:611–626. doi: 10.1099/vir.0.83426-0. [DOI] [PubMed] [Google Scholar]
- 104.Song BH, Yun SI, Choi YJ, Kim JM, Lee CH, Lee YM. A complex RNA motif defined by three discontinuous 5-nucleotide-long strands is essential for Flavivirus RNA replication. RNA. 2008;14:1791–1813. doi: 10.1261/rna.993608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sawicki SG, Sawicki DL, Younker D, Meyer Y, Thiel V, Stokes H, et al. Functional and genetic analysis of coronavirus replicase-transcriptase proteins. PLoS Pathog. 2005;1:39. doi: 10.1371/journal.ppat.0010039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Galan C, Sola I, Nogales A, Thomas B, Akoulitchev A, Enjuanes L, et al. Host cell proteins interacting with the 3′ end of TGEV coronavirus genome influence virus replication. Virology. 2009;391:304–314. doi: 10.1016/j.virol.2009.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Chen JY, Chen WN, Poon KM, Zheng BJ, Lin X, Wang YX, et al. Interaction between SARS-CoV helicase and a multifunctional cellular protein (Ddx5) revealed by yeast and mammalian cell two-hybrid systems. Arch Virol. 2009;154:507–512. doi: 10.1007/s00705-009-0323-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Xu L, Khadijah S, Fang S, Wang L, Tay FP, Liu DX. The cellular RNA helicase DDX1 interacts with coronavirus nonstructural protein 14 and enhances viral replication. J Virol. 2010;84:8571–8583. doi: 10.1128/JVI.00392-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.von Brunn A, Teepe C, Simpson JC, Pepperkok R, Friedel CC, Zimmer R, et al. Analysis of Intraviral Protein-Protein Interactions of the SARS Coronavirus ORFeome. PLoS ONE. 2007;2:1–11. doi: 10.1371/journal.pone.0000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Pan J, Peng X, Gao Y, Li Z, Lu X, Chen Y, et al. Genome-wide analysis of protein-protein interactions and involvement of viral proteins in SARS-CoV replication. PLoS ONE. 2008;3:3299. doi: 10.1371/journal.pone.0003299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Neuman BW, Joseph JS, Saikatendu KS, Serrano P, Chatterjee A, Johnson MA, et al. Proteomics analysis unravels the functional repertoire of coronavirus non-structural protein 3. J Virol. 2008;82:5279–5294. doi: 10.1128/JVI.02631-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zhang L, Zhang ZP, Zhang XE, Lin FS, Ge F. Quantitative proteomics analysis reveals BAG3 as a potential target to suppress severe acute respiratory syndrome coronavirus replication. J Virol. 2010;84:6050–6059. doi: 10.1128/JVI.00213-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Tai AW, Benita Y, Peng LF, Kim SS, Sakamoto N, Xavier RJ, et al. A functional genomic screen identifies cellular cofactors of hepatitis C virus replication. Cell Host Microbe. 2009;5:298–307. doi: 10.1016/j.chom.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Suratanee A, Rebhan I, Matula P, Kumar A, Kaderali L, Rohr K, et al. Detecting host factors involved in virus infection by observing the clustering of infected cells in siRNA screening images. Bioinformatics. 2010;26:653–658. doi: 10.1093/bioinformatics/btq398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Brass AL, Dykxhoorn DM, Benita Y, Yan N, Engelman A, Xavier RJ, et al. Identification of host proteins required for HIV infection through a functional genomic screen. Science. 2008;319:921–926. doi: 10.1126/science.1152725. [DOI] [PubMed] [Google Scholar]
- 116.Zhou H, Xu M, Huang Q, Gates AT, Zhang XD, Castle JC, et al. Genome-scale RNAi screen for host factors required for HIV replication. Cell Host Microbe. 2008;4:495–504. doi: 10.1016/j.chom.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 117.Boguszewska-Chachulska AM, Haenni AL. RNA viruses redirect host factors to better amplify their genome. Adv Virus Res. 2005;65:29–61. doi: 10.1016/S0065-3527(05)65002-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.van Hemert MJ, van den Worm SH, Knoops K, Mommaas AM, Gorbalenya AE, Snijder EJ. SARS-coronavirus replication/transcription complexes are membrane-protected and need a host factor for activity in vitro. PLoS Pathog. 2008;4:1000054. doi: 10.1371/journal.ppat.1000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Gadlage MJ, Sparks JS, Beachboard DC, Cox RG, Doyle JD, Stobart CC, et al. Murine hepatitis virus nonstructural protein 4 regulates virus-induced membrane modifications and replication complex function. J Virol. 2010;84:280–290. doi: 10.1128/JVI.01772-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Clementz MA, Kanjanahaluethai A, O'Brien TE, Baker SC. Mutation in murine coronavirus replication protein nsp4 alters assembly of double membrane vesicles. Virology. 2008;375:118–129. doi: 10.1016/j.virol.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Oostra M, Hagemeijer MC, van Gent M, Bekker CP, te Lintelo EG, Rottier PJ, et al. Topology and membrane anchoring of the coronavirus replication complex: not all hydrophobic domains of nsp3 and nsp6 are membrane spanning. J Virol. 2008;82:12392–12405. doi: 10.1128/JVI.01219-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Tijms MA, van Dinten LC, Gorbalenya AE, Snijder EJ. A zinc finger-containing papain-like protease couples subgenomic mRNA synthesis to genome translation in a positive-stranded RNA virus. Proc Natl Acad Sci USA. 2001;98:1889–1894. doi: 10.1073/pnas.041390398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.van Dinten LC, den Boon JA, Wassenaar ALM, Spaan WJM, Snijder EJ. An infectious arterivirus cDNA clone: identification of a replicase point mutation that abolishes discontinuous mRNA transcription. Proc Natl Acad Sci USA. 1997;94:991–996. doi: 10.1073/pnas.94.3.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Nedialkova DD, Gorbalenya AE, Snijder EJ. Arterivirus Nsp1 modulates the accumulation of minus-strand templates to control the relative abundance of viral mRNAs. PLoS Pathog. 2010;6:1000772. doi: 10.1371/journal.ppat.1000772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Gorbalenya AE, Enjuanes L, Ziebuhr J, Snijder EJ. Nidovirales: evolving the largest RNA virus genome. Virus Res. 2006;117:17–37. doi: 10.1016/j.virusres.2006.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hagemeijer MC, Verheije MH, Ulasli M, Shaltiel IA, de Vries LA, Reggiori F, et al. Dynamics of coronavirus replication-transcription complexes. J Virol. 2010;84:2134–2149. doi: 10.1128/JVI.01716-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Graham RL, Sims AC, Brockway SM, Baric RS, Denison MR. The nsp2 replicase proteins of murine hepatitis virus and severe acute respiratory syndrome coronavirus are dispensable for viral replication. J Virol. 2005;79:13399–13411. doi: 10.1128/JVI.79.21.13399-13411.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Tan J, Kusov Y, Mutschall D, Tech S, Nagarajan K, Hilgenfeld R, et al. The “SARS-unique domain” (SUD) of SARS coronavirus is an oligo(G)-binding protein. Biochem Biophys Res Commun. 2007;364:877–882. doi: 10.1016/j.bbrc.2007.10.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Frieman M, Ratia K, Johnston RE, Mesecar AD, Baric RS. Severe acute respiratory syndrome coronavirus papain-like protease ubiquitin-like domain and catalytic domain regulate antagonism of IRF3 and NFkappaB signaling. J Virol. 2009;83:6689–6705. doi: 10.1128/JVI.02220-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Eriksson KK, Cervantes-Barragan L, Ludewig B, Thiel V. Mouse hepatitis virus liver pathology is dependent on ADP-ribose-1″-phosphatase, a viral function conserved in the alpha-like supergroup. J Virol. 2008;82:12325–12334. doi: 10.1128/JVI.02082-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Hurst KR, Ye R, Goebel SJ, Jayaraman P, Masters PS. An interaction between the nucleocapsid protein and a component of the replicase-transcriptase complex is crucial for the infectivity of coronavirus genomic RNA. J Virol. 2010;84:10276–10288. doi: 10.1128/JVI.01287-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Alvarez E, DeDiego ML, Nieto-Torres JL, Jimenez-Guardeno JM, Marcos-Villar L, Enjuanes L. The envelope protein of severe acute respiratory syndrome coronavirus interacts with the non-structural protein 3 and is ubiquitinated. Virology. 2010;402:281–291. doi: 10.1016/j.virol.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Deming DJ, Graham RL, Denison MR, Baric RS. Processing of open reading frame 1a replicase proteins nsp7 to nsp10 in murine hepatitis virus strain A59 replication. J Virol. 2007;81:10280–10291. doi: 10.1128/JVI.00017-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Zhai Y, Sun F, Li X, Pang H, Xu X, Bartlam M, et al. Insights into SARS-CoV transcription and replication from the structure of the nsp7-nsp8 hexadecamer. Nat Struct Mol Biol. 2005;12:980–986. doi: 10.1038/nsmb999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Imbert I, Guillemot JC, Bourhis JM, Bussetta C, Coutard B, Egloff MP, et al. A second, non-canonical RNA-dependent RNA polymerase in SARS coronavirus. EMBO J. 2006;25:4933–4942. doi: 10.1038/sj.emboj.7601368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.te Velthuis AJ, Arnold JJ, Cameron CE, van den Worm SH, Snijder EJ. The RNA polymerase activity of SARS-coronavirus nsp12 is primer dependent. Nucleic Acids Res. 2010;38:203–214. doi: 10.1093/nar/gkp904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Egloff MP, Ferron F, Campanacci V, Longhi S, Rancurel C, Dutartre H, et al. The severe acute respiratory syndrome-coronavirus replicative protein nsp9 is a single-stranded RNA-binding subunit unique in the RNA virus world. Proc Natl Acad Sci USA. 2004;101:3792–3796. doi: 10.1073/pnas.0307877101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Sutton G, Fry E, Carter L, Sainsbury S, Walter T, Nettleship J, et al. The nsp9 replicase protein of SARS-coronavirus, structure and functional insights. Structure. 2004;12:341–353. doi: 10.1016/j.str.2004.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Miknis ZJ, Donaldson EF, Umland TC, Rimmer RA, Baric RS, Schultz LW. Severe acute respiratory syndrome coronavirus nsp9 dimerization is essential for efficient viral growth. J Virol. 2009;83:3007–3018. doi: 10.1128/JVI.01505-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Brockway SM, Clay CT, Lu XT, Denison MR. Characterization of the expression, intracellular localization and replication complex association of the putative mouse hepatitis virus RNA-dependent RNA polymerase. J Virol. 2003;77:10515–10527. doi: 10.1128/JVI.77.19.10515-10527.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Joseph JS, Saikatendu KS, Subramanian V, Neuman BW, Brooun A, Griffith M, et al. Crystal structure of nonstructural protein 10 from the severe acute respiratory syndrome coronavirus reveals a novel fold with two zinc-binding motifs. J Virol. 2006;80:7894–7901. doi: 10.1128/JVI.00467-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Matthes N, Mesters JR, Coutard B, Canard B, Snijder EJ, Moll R, et al. The non-structural protein Nsp10 of mouse hepatitis virus binds zinc ions and nucleic acids. FEBS Lett. 2006;580:4143–4149. doi: 10.1016/j.febslet.2006.06.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Siddell SG, Sawicki D, Meyer Y, Thiel V, Sawicki S. Identification of the mutations responsible for the phenotype of three MHV RNA-negative ts mutants. Adv Exp Med Biol. 2001;494:453–458. doi: 10.1007/978-1-4615-1325-4_66. [DOI] [PubMed] [Google Scholar]
- 144.Ivanov KA, Hertzig T, Rozanov M, Bayer S, Thiel V, Gorbalenya AE, et al. Major genetic marker of nidoviruses encodes a replicative endoribonuclease. Proc Natl Acad Sci USA. 2004;101:12694–12699. doi: 10.1073/pnas.0403127101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Minskaia E, Hertzig T, Gorbalenya AE, Campanacci V, Cambillau C, Canard B, et al. Discovery of an RNA virus 3′ 5′ exoribonuclease that is critically involved in coronavirus RNA synthesis. Proc Natl Acad Sci USA. 2006;103:5108–5113. doi: 10.1073/pnas.0508200103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Kang H, Bhardwaj K, Li Y, Palaninathan S, Sacchettini J, Guarino L, et al. Biochemical and genetic analyses of murine hepatitis virus Nsp15 endoribonuclease. J Virol. 2007;81:13587–13597. doi: 10.1128/JVI.00547-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Eckerle LD, Becker MM, Halpin RA, Li K, Venter E, Lu X, et al. Infidelity of SARS-CoV Nsp14-exonuclease mutant virus replication is revealed by complete genome sequencing. PLoS Pathog. 2010;6:1000896. doi: 10.1371/journal.ppat.1000896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Eckerle LD, Lu X, Sperry SM, Choi L, Denison MR. High fidelity of murine hepatitis virus replication is decreased in nsp14 exoribonuclease mutants. J Virol. 2007;81:12135–12144. doi: 10.1128/JVI.01296-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Chen Y, Cai H, Pan J, Xiang N, Tien P, Ahola T, et al. Functional screen reveals SARS coronavirus nonstructural protein nsp14 as a novel cap N7 methyltransferase. Proc Natl Acad Sci USA. 2009;106:3484–3489. doi: 10.1073/pnas.0808790106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Decroly E, Imbert I, Coutard B, Bouvet M, Selisko B, Alvarez K, et al. Coronavirus nonstructural protein 16 is a cap-0 binding enzyme possessing (nucleoside-2′O)-methyltransferase activity. J Virol. 2008;82:8071–8084. doi: 10.1128/JVI.00407-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Bost AG, Prentice E, Denison MR. Mouse hepatitis virus replicase protein complexes are translocated to sites of M protein accumulation in the ERGIC at late times of infection. Virology. 2001;285:21–29. doi: 10.1006/viro.2001.0932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Verheije MH, Hagemeijer MC, Ulasli M, Reggiori F, Rottier PJ, Masters PS, et al. The coronavirus nucleocapsid protein is dynamically associated with the replication-transcription complexes. J Virol. 2010;84:11575–11579. doi: 10.1128/JVI.00569-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Baric RS, Nelson GW, Fleming JO, Deans RJ, Keck JG, Casteel N, et al. Interactions between coronavirus nucleocapsid protein and viral RNAs: implications for viral transcription. J Virol. 1988;62:4280–4287. doi: 10.1128/jvi.62.11.4280-4287.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Compton SR, Rogers DB, Holmes KV, Fertsch D, Remenick J, Mc Gowan JJ. In vitro replication of mouse hepatitis virus strain A59. J Virol. 1987;69:2313–2321. doi: 10.1128/jvi.61.6.1814-1820.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Kim Y-N, Makino S. Characterization of a murine coronavirus defective interfering RNA internal cis-acting replication signal. J Virol. 1995;69:4963–4971. doi: 10.1128/jvi.69.8.4963-4971.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Laude H, Masters PS. The coronavirus nucleocapsid protein. In: Siddell SG, editor. The coronaviridae. New York: Plenum press; 1995. pp. 141–158. [Google Scholar]
- 157.Nelson GW, Stohlman SA, Tahara SM. High affinity interaction between nucleocapsid protein and leader/intergenic sequence of mouse hepatitis virus RNA. J Gen Virol. 2000;81:181–188. doi: 10.1099/0022-1317-81-1-181. [DOI] [PubMed] [Google Scholar]
- 158.Stohlman SA, Baric RS, Nelson GN, Soe LH, Welter LM, Deans RJ. Specific interaction between coronavirus leader RNA and nucleocapsid protein. J Virol. 1988;62:4288–4295. doi: 10.1128/jvi.62.11.4288-4295.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Almazan F, Galan C, Enjuanes L. The nucleoprotein is required for efficient coronavirus genome replication. J Virol. 2004;78:12683–12688. doi: 10.1128/JVI.78.22.12683-12688.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Schelle B, Karl N, Ludewig B, Siddell SG, Thiel V. Selective replication of coronavirus genomes that express nucleocapsid protein. J Virol. 2005;79:6620–6630. doi: 10.1128/JVI.79.11.6620-6630.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Thiel V, Karl N, Schelle B, Disterer P, Klagge I, Siddell SG. Multigene RNA vector based on coronavirus transcription. J Virol. 2003;77:9790–9798. doi: 10.1128/JVI.77.18.9790-9798.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Yount B, Curtis KM, Baric RS. Strategy for systematic assembly of large RNA and DNA genomes: the transmissible gastroenteritis virus model. J Virol. 2000;74:10600–10611. doi: 10.1128/jvi.74.22.10600-10611.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Grossoehme NE, Li L, Keane SC, Liu P, Dann CE, 3rd, Leibowitz JL, et al. Coronavirus N protein N-terminal domain (NTD) specifically binds the transcriptional regulatory sequence (TRS) and melts TRS-cTRS RNA duplexes. J Mol Biol. 2009;394:544–557. doi: 10.1016/j.jmb.2009.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Zuñiga S, Sola I, Moreno JL, Sabella P, Plana-Duran J, Enjuanes L. Coronavirus nucleocapsid protein is an RNA chaperone. Virology. 2007;357:215–227. doi: 10.1016/j.virol.2006.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Zuñiga S, Cruz JL, Sola I, Mateos-Gomez PA, Palacio L, Enjuanes L. Coronavirus nucleocapsid protein facilitates template switching and is required for efficient transcription. J Virol. 2010;84:2169–2175. doi: 10.1128/JVI.02011-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Li HP, Huang P, Park S, Lai MMC. Polypyrimidine tract-binding protein binds to the leader RNA of mouse hepatitis virus and serves as a regulator of viral transcription. J Virol. 1999;73:772–777. doi: 10.1128/jvi.73.1.772-777.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Choi KS, Huang P, Lai MM. Polypyrimidine-tract-binding protein affects transcription but not translation of mouse hepatitis virus RNA. Virology. 2002;303:58–68. doi: 10.1006/viro.2002.1675. [DOI] [PubMed] [Google Scholar]
- 168.Li HP, Zhang X, Duncan R, Comai L, Lai MMC. Heterogeneous nuclear ribonucleoprotein A1 binds to the transcription-regulatory region of mouse hepatitis virus RNA. Proc Natl Acad Sci USA. 1997;94:9544–9549. doi: 10.1073/pnas.94.18.9544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Choi KS, Mizutani A, Lai MM. SYNCRIP, a member of the heterogeneous nuclear ribonucleoprotein family, is involved in mouse hepatitis virus RNA synthesis. J Virol. 2004;78:13153–13162. doi: 10.1128/JVI.78.23.13153-13162.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Jia J, Arif A, Ray PS, Fox PL. WHEP domains direct noncanonical function of glutamyl-Prolyl tRNA synthetase in translational control of gene expression. Mol Cell. 2008;29:679–690. doi: 10.1016/j.molcel.2008.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Zhang X, Lai MMC. Interactions between the cytoplasmic proteins and the intergenic (promoter) sequence of mouse hepatitis virus RNA: correlation with the amounts of subgenomic mRNA transcribed. J Virol. 1995;69:1637–1644. doi: 10.1128/jvi.69.3.1637-1644.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Zhang X, LI HP, Xue W, Lai MCC. Formation of a ribonucleoprotein complex of mouse hepatitis virus involving heterogeneous nuclear ribonucleoprotein A1 and transcription-regulatory elements of viral RNA. Virology. 1999;264:115–124. doi: 10.1006/viro.1999.9970. [DOI] [PubMed] [Google Scholar]
- 173.Wang Y, Zhang X. The nucleocapsid protein of coronavirus mouse hepatitis virus interacts with the cellular heterogeneous nuclear ribonucleoprotein A1 in vitro and in vivo. Virology. 1999;265:96–109. doi: 10.1006/viro.1999.0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Harris D, Zhang Z, Chaubey B, Pandey VN. Identification of cellular factors associated with the 3′-nontranslated region of the hepatitis C virus genome. Mol Cell Proteomics. 2006;5:1006–1018. doi: 10.1074/mcp.M500429-MCP200. [DOI] [PubMed] [Google Scholar]
- 175.Shi ST, Huang P, Li HP, Lai MMC. Heterogeneous nuclear ribonucleoprotein A1 regulates RNA synthesis of a cytoplasmic virus. EMBO J. 2000;19:4701–4711. doi: 10.1093/emboj/19.17.4701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Shi ST, Yu GY, Lai MM. Multiple type A/B heterogeneous nuclear ribonucleoproteins (hnRNPs) can replace hnRNP A1 in mouse hepatitis virus RNA synthesis. J Virol. 2003;77:10584–10593. doi: 10.1128/JVI.77.19.10584-10593.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Huang P, Lai MMC. Polypyrimidine tract-binding protein binds to the complementary strand of the mouse hepatitis virus 3′ untranslated region, thereby altering RNA conformation. J Virol. 1999;73:9110–9116. doi: 10.1128/jvi.73.11.9110-9116.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Tijms MA, Snijder EJ. Equine arteritis virus non-structural protein 1, an essential factor for viral subgenomic mRNA synthesis, interacts with the cellular transcription co-factor p100. J Gen Virol. 2003;84:2317–2322. doi: 10.1099/vir.0.19297-0. [DOI] [PubMed] [Google Scholar]
- 179.Nanda SK, Johnson RF, Liu Q, Leibowitz JL. Mitochondrial HSP70, HSP40 and HSP60 bind to the 3′ untranslated region of the Murine hepatitis virus genome. Arch Virol. 2004;149:93–111. doi: 10.1007/s00705-003-0196-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Nanda SK, Leibowitz JL. Mitochondrial aconitase binds to the 3′ untranslated region of the mouse hepatitis virus genome. J Virol. 2001;75:3352–3362. doi: 10.1128/JVI.75.7.3352-3362.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Johnson RF, Feng M, Liu P, Millership JJ, Yount B, Baric RS, et al. Effect of mutations in the mouse hepatitis virus 3′(+)42 protein binding element on RNA replication. J Virol. 2005;79:14570–14585. doi: 10.1128/JVI.79.23.14570-14585.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Tanner NK, Linder P. DExD/H box RNA helicases: from generic motors to specific dissociation functions. Mol Cell. 2001;8:251–262. doi: 10.1016/s1097-2765(01)00329-x. [DOI] [PubMed] [Google Scholar]