Abstract
Signal transduction via G protein-coupled receptors (GPCRs) is central for the regulation of virtually all cellular functions and has been widely implicated in human disease. Regulators of G protein Signaling (RGS proteins) belong to a diverse protein family that was originally discovered for their ability to accelerate signal termination in response to GPCR stimulation, thereby reducing the amplitude and duration of GPCR effects. All RGS proteins share a common RGS domain that interacts with G protein α subunits and mediates their biologic regulation of GPCR signaling. However, RGS proteins differ widely in size and the organization of their sequences flanking the RGS domain, which contain several additional functional domains that facilitate protein-protein (or protein-lipid) interactions. RGS proteins are subject to posttranslational modifications, and, in addition, their expression, activity, and subcellular localization can be dynamically regulated. Thus, there exist a wide array of mechanisms that facilitate their proper function as modulators and integrators of G protein signaling. Several RGS proteins have been implicated in the cardiac remodeling response and heart rate regulation, and changes in RGS protein expression and/or function are believed to participate in the pathophysiology of cardiac hypertrophy, failure and arrhythmias as well as hypertension. This review is based on recent advances in our understanding of the expression pattern, regulation and functional role of canonical RGS proteins, with a special focus on the healthy and diseased heart. In addition, we discuss their potential and promise as therapeutic targets as well as strategies to modulate their expression and function.
Keywords: RGS proteins, signal transduction, myocardium, cardiac myocytes, cardiac fibroblasts
1. Introduction
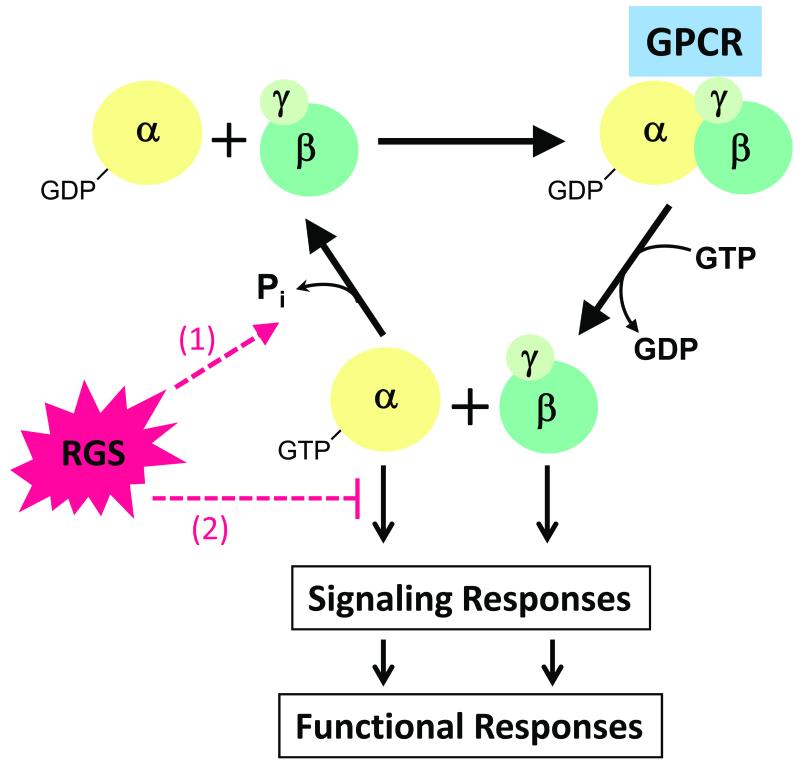
Signal transduction via G protein-coupled receptors (GPCRs) is essential for the regulation of cardiovascular function, including heart rate, growth, contraction, and vascular tone. Pertubations in GPCR signaling have pathophysiological consequences and are major contributors to cardiac disease 1. Ligand-activation of GPCRs promotes GTP-for-GDP exchange on the α subunits of heterotrimeric G proteins (Figure 1), resulting in dissociation of GTP-bound Gα from Gβγ. Both Gα and Gβγ subunits then activate (or inhibit) downstream signaling molecules (enzymes, kinases and ion channels) and thereby elicit cellular responses. Their magnitude and duration depend on how long G proteins remain activated, which is determined by a GTPase activity intrinsic to Gα. Upon GTP hydrolysis, the resulting GDP-bound inactive Gα reassociates with Gβγ and can enter a new activation cycle. Thus, the rate of GTP hydrolysis determines the duration that Gα-GTP and Gβγ are free to interact with intracellular or membrane effectors. It long remained a conundrum that the intrinsic rate of GTP hydrolysis is insufficient to account for the rate of signal termination typically observed in vivo. While some effector molecules (e.g., phospholipase C β1 2) were found to act as GTPase-activating proteins (GAPs), the discovery of RGS proteins introduced a new large and diverse protein family that leads to pronounced (up to 2000-fold) acceleration of Gα GTPase activity 3, 4, which decreases the amplitude and duration of both Gα- and Gβγ-mediated downstream signaling. Structures derived from NMR and x-ray crystallography of the RGS domain, both alone or bound to Gα subunits (in the presence of GDP and AlF4− to mimic the γ-phosphate of GTP in its transition state) provide mechanistic insight into RGS protein/Gα subunit interactions (for details see 5, 6). Binding of RGS proteins to activated Gα can also antagonize effector activation and thereby block Gα-mediated signal generation. It is generally not possible to distinguish whether RGS protein-mediated signal inhibition is due to GAP activity and/or effector antagonism, unless constitutively active GTPase-deficient Gα subunits are used for signal activation 7.
Figure 1. Regulation of G Protein-mediated Signaling by RGS proteins.
See text for detail on the G protein activation/inactivation cycle. RGS proteins regulate G protein-mediated signaling via (1) marked acceleration of Gα GTPase activity, which decreases both Gα- and Gβγ -mediated downstream effects, and (2) competition with downstream effectors for binding to activated Gα, which inhibits only Gα-mediated signal generation. Please note that this cartoon depicts the traditional view of GPCR-induced, G protein-mediated signal transduction. It does not incorporate GPCR-independent G protein activation (reviewed in 147) or G protein-independent GPCR effects (reviewed in 148). Furthermore, full dissociation of Gα and Gβγ subunits may not be required to trigger downstream effects (e.g., 149).
The RGS protein superfamily is divided into subfamilies based on sequence homology within the RGS domain and the nature and identity of non-RGS domains that facilitate protein-protein interactions, target specificity, protein stability and subcellular location (Table 1). Twenty canonical RGS proteins in 4 subfamilies share the prototypical RGS domain (app. 130 amino acids) that binds to GTP-bound Gα subunits. Nineteen other “RGS-like” proteins (i.e., GRKs, RhoGEFs, axins, D-AKAP2, nexins, RGSL) contain a RGS protein homology domain. Only some of them have been shown to interact with Gα subunits, and their GAP activity is much weaker than that of canonical RGS. Their structure and function was recently reviewed 8.
Table 1.
Structure, GAP Function, Posttranslational Modifications and Cardiac Expression of Canonical RGS Proteins
|
* Sub- family |
Gene† | Size | Non-RGS Domains | GAP for | Post-Transl. Modifications | Ventricles | Atria | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| a. acids | N-term. | C-term. | Phosphorylation | Palmitoyl. | Tissue ‡ | CM § | CF § | Tissue !! | CM !! | |||
|
R4
(B) |
RGS1 † | 209 | A. helix | None | Gi/o and Gq/11 | (+) | + | + | − | − | ||
| RGS2 | 211 | Gq/11 ≪ (Gi/o) | S46/64/73 (PKG) ? (PKC) |
C106, C116, C199 |
+ | + | + | + | + | |||
| RGS3 † | 519 | Gi/o and Gq/11 | ? (PKG) S264 (PKA) |
+ | + | + | + | + | + | |||
| RGS4 † | 205 | Gi/o and Gq/11 | S52 (PKA, PKG) | C2, C12, C95 |
(+) | (+) | − | + | + | |||
| RGS5 † | 181 | Gi/o and Gq/11 | S166 (PKC) (S84?) |
+ | + | + | + | − | ||||
| RGS8 † | 180 | Gi/o and Gq/11 | − | (+) | + | − | − | |||||
| RGS13 | 159 | Gi/o and Gq/11 | T 41 (PKA) | − | − | + | − | − | ||||
| RGS16 | 202 | Gi/o and Gq/11 | Y168 (EGFR, src, Lyn kinase) S194/S53 (? ) |
C2, C12, C98 |
+ | + | + | + | − | |||
| RGS18 | 235 | Gi/o and Gq/11 | S49 (?) | (+) | (+) | + | + | − | ||||
| RGS21 | 152 | None | None | ND | ND | ND | ND | ND | ND | |||
|
R7
(C) |
RGS6 † | 472 | DEP GGL |
None | Gi/o | + | + | + | + | + | ||
| RGS7 † | 487 | Gi/o | S434 (PKCα) | C60, C133 | − | + | − | − | − | |||
| RGS9 † | 484 | Gi/o | S427/428 (PKA) S475 (PKC) |
(+) | (+) | + | − | − | ||||
| RGS11 † | 446 | Gi/o | + | − | + | − | − | |||||
|
R12
(D) |
RGS10 | 181 | None | None | Gi/o and Gq/11 | S168 (PKA) | C60, C66 | + | + | + | + | + |
| RGS12 † | 1376 | PDZ PTB |
RBD GoLoco PM |
Gi/o | + | + | + | + | − | |||
| RGS14 † | 566 | None | RBD GoLoco |
Gi/o | S258/T494 (PKA) | −/(+) | + | + | − | − | ||
|
RZ
(A) |
RGS17 | 210 | C string (plus A. Helix RGS19) |
Gz & other Gi/o | Likely (S151) | Likely | − | + | + | + | + | |
| RGS19 † | 217 | PM | Gz & other Gi/o and Gq/11 |
S24 (CK2/PKC) S151 (ERK) |
+ | − | + | + | + | + | ||
| RGS20 † | 241 | Gz ≪ (Gi/o) | Likely (S151) | Likely | (+) | (+) | + | − | − | |||
Nomenclatures for canonical RGS protein subfamilies are based on either a prototypical subfamily member3 or arbitrary alphabetical letters 139 (in brackets)
Indicates the existence of alternatively spliced variants for the specified RGS protein genes
RGS protein expression profile obtained from studies performed in the human heart (using quantitative real-time PCR) 22 or human left ventricular myocardium (using RNase protection assays and RT-PCR) 16
RT-PCR-based RGS protein expression profile in myocytes (CM) and fibroblasts (CF) from adult rat ventricles (see Figure 2)
Single cell RT-PCR-based RGS protein expression profile established in spontaneously beating rat atrial CM 19
Domain Abbreviations: A. Helix - amphiphatic helix; CM – cardiac myocyte; CF – cardiac fibroblast; GAP – GTPase activating protein; GGL - Gγ-like domain; DEP - Disheveled-EGL10-Pleckstrin homology domain; RBD - Ras-binding domain; GoLoco - Gαi/o-Loco domain; PDZ - PSD-95 disk-large ZO-1 domain; PM - PDZ docking motif; PTB - phosphotyrosine binding domain
- For the expression profiles, + and − denote detection or lack thereof of specified isoforms and (+) indicates weak expression.
- In the phosphorylation column, the phosphorylation-specific site(s) are followed by responsible kinase(s) in brackets. Unknown sites or kinases are indicated by ?.
In the present review, we focus on the expression pattern, regulation and functional role of canonical RGS proteins in the healthy and diseased heart, as well as their potential as therapeutic drug targets. Other reviews provide further details on the structure and function of canonical RGS proteins 8, 9. Due to space constraints, only some information about the role of RGS proteins in the vasculature could be included. The reader is referred to other excellent reviews to learn more about RGS proteins in blood vessels 10, 11, the nervous system 12, 13, inflammation 14 and cancer 15 for a broader view on the importance of RGS proteins in regulating GPCR signaling and function in health and disease.
2. RGS Protein Expression in the Heart
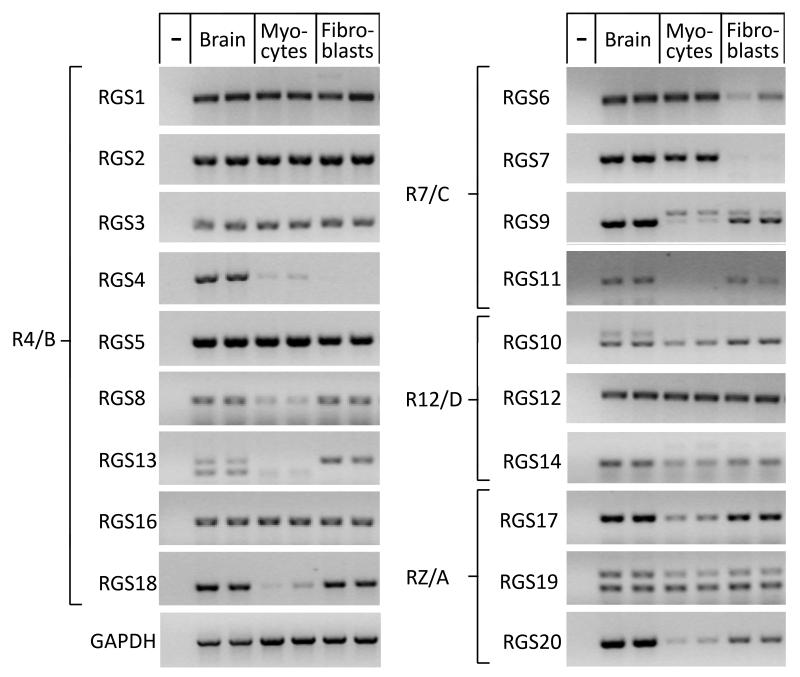
Several canonical RGS proteins are expressed in the mammalian and human myocardium 16-18 as well as in cardiac myocytes 19, 20 (see Table 1). RGS protein expression in non-myocytes has long been suggested 19, but was only recently reported for cardiac fibroblasts 21. A comprehensive, reverse transcription (RT)-PCR-based overview of canonical RGS protein expression in cardiac myocytes and fibroblasts from adult rat ventricles is shown in Figure 2 (compared to brain). Quantitative mRNA analysis revealed that RGS2, RGS3 and RGS5 are most highly expressed in the human heart 22. In contrast, RGS4, which initially garnered a lot of attention, was found only at a very low levels, consistent with Figure 2 and other studies showing lack of RGS4 in the ventricular myocardium, but enrichment in the sinoatrial node 23, 24.
Figure 2. RGS mRNA Expression Profile in Adult Rat Ventricular Myocytes and Fibroblasts.
Reverse transcription (RT) PCR analysis of freshly isolated ventricular myocytes and fibroblasts from male Sprague-Dawley rats (5 weeks old). Rat brain was used for comparison. Total RNA (1 μg) was reverse-transcribed and amplified using SuperScript One-Step RT-PCR Kit (Invitrogen, Carlsbad, CA) with RGS isoform-specific primers (primer sequences and PCR conditions available upon request). (−) denotes absence of template. GAPDH was used as internal control. RT-PCR products were visualized on ethidium bromide stained agarose gels. RGS are organized according to subfamily affiliation.
RGS protein expression profiles are most often based on Northern blots, in situ hybridizations and PCR analyses. Discrepancies between mRNA and protein levels have been reported (e.g., increased mRNA but decreased protein expression for RGS4 in breast cancer tissue 25), emphasizing the importance of protein measurements. However, protein detection has been a significant challenge in the field, because antibodies that unequivocally recognize endogenous RGS proteins are not available for many isoforms. In addition, knockout controls may be required to demonstrate specificity of protein bands of expected molecular weight 26, 27. The difficulty in detecting endogenous RGS proteins with antibodies that recognize overexpressed RGS proteins very well suggests that cellular levels of endogenous RGS proteins may be quite low. Stoichiometric information on relative G protein and RGS protein levels therefore has yet to be determined.
Several canonical RGS protein isoforms are expressed in the myocardium with regional differences between atria and ventricles (Table 1). Myocytes and fibroblasts have a unique complement of RGS proteins, so that expression studies in cardiac tissue need to be interpreted with caution. As will be described below, significant progress has been made in assigning signaling and functional roles for specific RGS protein isoforms in both major cell types in the heart, although much work remains.
3. RGS Protein Subfamilies: Structural and Functional Properties
Most R4 subfamily members (for RGS3 see below) are “small” RGS proteins with short N- and C-terminal extensions to the conservative RGS core domain. They are mostly non-discriminatory in their binding to and GAP activity for all Gi/o and Gq/11 family members. Only RGS2 generally has been considered to be selective in negatively regulating Gq/11, which has been attributed to the geometry of a Gα binding pocket that is unfavorable to Gαi/o 28. The structural determinants were recently pinpointed to three evolutionary highly conserved amino acids 29, leading the authors to speculate that RGS2 arose from the R4 subfamily to have specialized Gαq/11 GAP activity to modulate cardiovascular function. Indeed, in adult rat cardiomyocytes, RGS2 negatively regulates Gq/11 but not Gi/o-mediated signaling 30. Nevertheless, it has been reported that RGS2 interactions with Gαi/o may occur dependent on receptor-mediated Gα activation 31, 32, so that lack of interaction between recombinant RGS2 and Gi/o 33, 34 may not necessarily be indicative of a lack of regulatory interaction in cells. In fact, in cultured ventricular myocytes, a novel role of RGS2 as terminator of β2-receptor mediated Gi signaling was recently demonstrated 35. RGS2 was also shown to directly interact with and negatively regulate select adenylate cyclase (AC) isoforms (including the major cardiac isoforms ACV and ACVI) 36, 37. Gαs interaction albeit without GAP activity was reported as well 38. However, these studies were performed in HEK293 and other non-cardiac cells. In adult rat ventricular myocytes, RGS2 overexpression did not affect forskolin- or isoproterenol-induced cyclic AMP (cAMP) generation 30, suggesting that neither direct nor indirect RGS2-induced AC regulation appears to play a major role in differentiated myocytes. In neonatal rat cardiomyocytes, hypertrophy induced by β-adrenergic stimulation could be inhibited by RGS2 expression 39.
RGS3, which exists in several splice variants (reviewed in 17), is a unique R4 RGS protein in that a long N-terminus in some variants facilitates interactions with other proteins. For example, binding to Gβγ enables RGS3L (519 amino acids) to inhibit Gβγ-mediated signaling by acting as a scavenger 40 and has the ability to switch Gi/o-coupled muscarinic and adenosine receptor-induced signaling from Rac1 to RhoA activation 41. However, the switch is highly dependent on the expression level of endogenous RGS3L, which is markedly down-regulated by fibroblast growth factor 2. This mechanism could be of pathophysiological significance in the heart, but has so far only been demonstrated in H10 cells. The N-terminus of RGS3 can also interact with Smad2, Smad3 and Smad4 via their Mad homology 2 domain and inhibit Smad-mediated gene transcription by preventing Smad3/Smad4 heteromerization 42. RGS3-Smad interaction has been shown to inhibit TGFβ induced differentiation of pulmonary fibroblasts 42, and may potentially play a role in cardiac fibroblasts as well.
Other R4 RGS protein subfamily members can also regulate non-G protein signaling. For example, several isoforms can interact with the regulatory p85α subunit of phosphatidylinositol-3-OH kinase (PI3K). Subsequent inhibition of PI3K activity by inhibiting p85-Gab1/2 interactions has been shown for RGS13 in mast cells 43 and RGS16 in breast cancer cells 44. Investigations of potential RGS protein regulation of cardiac PI3K are warranted in light of its importance in modulating cell survival, growth, contractility, and metabolism 45. Furthermore, RGS13 also acts as a nuclear repressor of cAMP response element binding protein (CREB) in B lymphocytes that inhibits CREB-dependent transcription through disruption of promoter complexes 46.
R7 subfamily members are predominantly expressed in the nervous system and best known for their role in the regulation of neuronal processes, including vision, memory, motor control, reward behavior, and nociception (reviewed in 12). However, a key role of RGS6 in the heart was recently discovered (see section 5 below). Through their RGS domain, R7 RGS proteins exert GAP activity primarily on Gαi/o proteins 47. They also contain a G protein gamma-like (GGL) domain that is structurally homologous to conventional Gγ subunits but binds only with the most distant member of the Gβ family (Gβ5), an interaction that is essential for the stability and expression of all R7 RGS proteins 48. Beyond protecting R7 RGS proteins from proteolysis, the role of Gβ5 is not fully understood. It is believed to participate in determining G protein selectivity and GAP properties. The crystal structure of RGS9-Gβ5 offers some insight into potential mechanisms 49. The N-terminus of R7 RGS proteins also contains Disheveled-EGL10-Pleckstrin homology (DEP) and DEP helical extension (DHEX) domains that mediate interactions with membrane anchor proteins (i.e., RGS9 anchor protein [R9AP] and RGS7 family binding protein [R7BP]), which both play key roles in determining the catalytic activity, subcellular localization and R7 RGS protein expression levels (reviewed in 12, 50).
The R12 subfamily is comprised of members that are structurally very diverse in regions other than their RGS domain (Table 1). RGS10 lacks any additional domain, acts as GAP for Gαi/o, and Gαq/11 and is phosphorylated by protein kinase A (PKA) 51. RGS12 and RGS14 are GAPs for Gαi/o only. In addition to binding to activated Gαi/o in its activated state through their RGS domain, they can bind GDP-bound Gαi1-3 via their C-terminal GoLoco domain and act as GDP-dissociating inhibitors (GDI) 52. Inhibition of GDP-to-GTP exchange and subsequent Gα activation provide an additional GAP-independent mechanism of regulating G protein signaling through these RGS protein isoforms. Furthermore, RGS12 and RGS14 have recently emerged as integrators of G protein and Ras/Raf/ERK signaling by facilitating formation of a selective Ras-Raf-MEK-ERK multiprotein complex to promote sustained ERK activation, involving their C-terminal tandem Ras-binding domains (RBD) and for RGS12 its additional PSD-95 disk-large ZO-1 (PDZ) and phosphotyrosine binding (PTB) domains 53-55. However existence of these mechanisms in cardiac cells remains to be investigated.
Members of the RZ subfamily (reviewed in 56) are short in size and share a N-terminal cysteine string motif, which presumably provides substrate for palmitoylation for each isoform (reported so far for RGS19). Similarly, phosphorylation has been demonstrated for RGS19 at two sites, one which (S151) is conserved among subfamily members. RGS19 contains an additional C-terminal PDZ-binding motif (PM) that facilitates binding to a scaffolding protein (GIPC, GAIP-interacting protein C-terminus) that assembles receptors and signaling molecules and may promote crosstalk between G protein and non-G protein signaling pathways (reviewed 57). The RZ subfamily was originally named because RGS20 (originally known as RGSZ1) was found to selectively accelerate GTP hydrolysis of Gαz, a more distant member of the Gαi/o family that also inhibits AC and activates potassium channels. In contrast, RGS17 and RGS19 (aka GAIP) have GAP activity for all Gi/o α subunits (and Gαq/11 for RGS19). Although RGS17 is not a Gαq/11GAP in vitro, it can bind and inhibit Gq/11-mediated signaling in the cellular context through a yet undetermined mechanism 58. In the same study, despite its GAP selectivity, RGS20 blocked Gαi/o signaling. Thus, in vitro GAP activity assays are not always good predictors of function in the cellular context; effector antagonism and non-GAP mechanism are additional determinants of RGS protein function in vivo.
Taken together, canonical RGS proteins serve as GAPs for members of the Gi/o and Gq/11 families. It is generally believed that they do not serve as GAPs for Gαs; evidence to the contrary regarding RGS-PX1 59 has yet to be confirmed. GAP activity for Gα12/13 is displayed only by non-canonical “RGS-like” RhoGEFs, which are also their effectors (reviewed in 60). Although a wealth of information on the interactions between RGS proteins and Gα subunits has been collected over the past 15 years, it cannot account for the specificity with which RGS proteins regulate G protein-mediated signaling in living cells. Despite tissue- and cell-specific expression for some isoforms, most cells express several RGS proteins with diverse activities, and they are rather non-discriminatory towards G proteins. A variety of mechanisms that regulate RGS protein expression, activity, location, and interaction with other proteins are summarized below, which collectively facilitate effective and specific modulation of GPCR-induced signal transfer. Following is a brief synopsis of the current understanding of RGS protein regulation, with a special focus on mechanisms that may potentially be at play in the heart.
4. Regulation of RGS Protein Expression, Activity and Location
Expression of Different RGS Gene and Protein Products
Both alternative mRNA splicing (for specific isoforms see Table 1) and translation initiation from alternative start sites have been reported. Variations are generally not located in the core RGS domain but the additional extensions and regulatory domains, suggesting that they may play a role in fine-tune signaling responses. For example, utilization of three alternative translation start sites in human RGS2 was shown to yield proteins of different functionality in overexpression experiments, in that AC inhibition was compromised when the N-terminal AC binding site was missing, whereas GAP-mediated Gq/11 regulation was unaffected 61. However, the prevalence of these regulatory mechanisms in the cardiovascular system and their significance under physiological conditions are not known at this point.
Regulation of mRNA Expression
Numerous reports in many different cell types have shown that mRNA encoding for various RGS isoforms can be regulated by a variety of factors, including GPCR activation, second messengers and disease states. Most recently, promoter hypermethylation-dependent silencing was reported for RGS2 in human prostate cancer, suggesting epigenetic repression as a novel mechanism for regulating RGS mRNA expression 62. In the heart, many studies have been conducted in myocardial tissue and in already hypertrophied or failing hearts. In the diseased heart, a multitude of signaling changes occur, many of which are secondary to the remodeling process. Disparities regarding RGS protein expression changes between animal models of hypertrophy (e.g., 63, 64) and in humans (e.g., 16, 65) may be due to species- and model-specific differences (reviewed in 66).
Among the various RGS proteins, RGS2 has emerged as an isoform that is highly susceptible to regulation, and it also exemplifies the dynamic nature of RGS protein regulation in the heart. In response to short-term activation of the Gq/11 signaling pathway, RGS2 mRNA is transiently up-regulated in both cardiac myocytes 30, 67 and fibroblasts 21. This is generally viewed as a negative feedback mechanism in light of the role of RGS2 as a negative regulator of Gq/11 signaling 68, 69. Interestingly, acute β-adrenergic or forskolin stimulation also cause a marked increase in RGS2 mRNA 30, 39, which may point to potential cross-regulation and desensitization between Gq/11- and Gs-mediated signaling pathways. While no RGS2 regulatory effects on cAMP were detected in adult rat myocytes 30, inhibition of isoproterenol-induced hypertrophy by blunting of ERK1/2 and Akt activation was reported in neonatal myocytes 39. Importantly, in contrast to acute stimulation, marked RGS2 down-regulation has been discovered in ventricles subjected to pressure overload, myocytes from mice expressing constitutively active Gαq* 26 as well as myocytes and fibroblasts from rats subjected to prolonged angiotensin (Ang II) infusion in vivo 21 and has been implicated in exacerbating cardiac remodeling in the stressed or injured hearts 21, 26, 70. Protein kinase C (PKC)- and Ca2+-dependent changes are involved in Gq/11-mediated RGS2 mRNA regulation, but little is known so far about the precise mechanisms 68.
Regulation of Protein Stability is an alternative way to modulate RGS protein expression levels. Phosphorylation-induced slowing of RGS protein degradation has been demonstrated for some isoforms (e.g., RGS13 71, RGS16 [Y168] 72). N-end rule of degradation is another important mechanism to regulate cellular RGS protein levels (reviewed in 73). While several RGS proteins have potentially destabilizing N-terminal residues and are predicted to be degraded by this pathway, only RGS4, RGS5 and RGS16 have been confirmed so far in vitro 74 and in vivo 75. Among them, RGS4 is best characterized and can be stabilized by mutations 74 as well as palmitoylation 76 of its N-terminal C2 residue. Potential clinical relevance was suggested by detection of two potentially destabilizing mutations of RGS2 in a group of hypertensive individuals from Japan 77, one of which (Q2L) showed much reduced protein expression in HEK293 cells that was markedly enhanced by pre-treatment with a proteasome inhibitor 78. Furthermore, proteosomal degradation of RGS4 was recently linked to invasiveness of breast cancer 25.
Posttranslational Modifications
RGS isoforms from all subfamilies can be phosphorylated by a large variety of kinases (Table 1). Functional effects are diverse and include protein stabilization (see above), changes in subcellular localization (e.g., membrane translocation of RGS3, RGS4, 79; nuclear translocation of RGS10 51) and alterations in GAP activity, which can be either enhanced or reduced depending on RGS isoforms and protein kinases involved. For example, RGS2 phosphorylation by PKC leads to a reduction 80, whereas cGMP-dependent protein kinase (PKG) causes an increase 81. Several RGS proteins are also modified by palmitoylation near the N terminus and/or on conserved cysteine residue in the α4 helix of the RGS domain 82. Palmitoylation can also affect protein stabilization and membrane and lipid raft targeting (e.g., RGS7 83, RGS16 84, RGS19 85). It generally increases GAP activity, presumably as a result of increased membrane association, but this is not a requirement 86. Palmitoylation was found to be both constitutive (e.g. RGS10) and dependent on GPCR activation (e.g., RGS3) 87. The extent to which RGS protein phosphorylation and/or palmitoylation occurs in myocardial cells and its functional consequences have yet to be delineated.
Subcellular Localization
RGS protein location within the cell is diverse and depends on isoform, cell type and expression level (reviewed in 88, 89). Although most RGS proteins were predicted to be hydrophilic, many of them can be found to varying degree in the cytosol and in the nucleus. Much information on the subcellular location of RGS proteins has been derived from overexpression studies that may lead to aberrant targeting, but a few reports suggested similar localization for some endogenous RGS proteins. The location of RGS proteins in the cell is in flux and highly regulated (for specific examples, see 88, 89). Plasma membrane translocation of RGS proteins can be induced by direct recruitment by Gα-GTP or after GPCR-induced G protein activation and is facilitated by phosphorylation and palmitoylation as mentioned above. RGS proteins may not be able to freely interact with every available Gα protein, but selectively sorted by GPCRs at the plasma membrane, since GPCRs alone or in a concerted effort with their linked G proteins were shown to selectively recruit RGS proteins to the plasma membrane 90. Mechanisms proposed for nuclear targeting involve regions inside and outside the RGS domain and nuclear targeting/export signals. The function(s) of cytosolic and nuclear RGS proteins is/are not well understood. Sequestration of RGS proteins from G proteins localized at the plasma-membrane has been proposed, but additional functions are likely and appear to include regulation of transcription factors/repressors (reported for RGS13 46 and RGS6 91). RGS protein-mediated regulation of G protein signaling is also a distinct possibility in light of increasing evidence for nuclear location of functional GPCRs and G proteins (e.g., 92, 93) as well as intracrine signaling (reviewed in 94, 95). Many more studies are needed to fully validate novel interactions and putative regulatory roles and to delineate the subcellular localization of RGS proteins and its exact role in mediating canonical and emerging signaling processes in cardiac cells.
Interaction with GPCRs and Other Molecules
Although the R7 and R12 subfamilies of RGS proteins contain multiple well-established protein-protein interaction domains, the structurally simple R4 and RZ RGS proteins with short extensions to the RGS domain also display a remarkable ability to interact with many different binding partners. For example, RGS2 has been shown to interact with GPCRs, AC, PKG, TRPV channel, and tubulin via distinct regions of its N-terminus (reviewed in 96). Thus, RGS protein binding partners are diverse and range from GPCRs, effector proteins (ion channels, enzymes) and kinases to scaffold and other auxilliary proteins (reviewed in 57), so that only a few examples can be highlighted. Interaction with GPCRs (reviewed in 97) can be direct (e.g., via PDZ domains in particular RGS3 or RGS12 splice variants or the N-terminus in R4 RGS proteins) or mediated by scaffolding proteins (such as GIPC and spinophilin). Direct evidence for cellular interactions between full length GPCRs and RGS proteins in living cells has yet to be demonstrated, but many functional studies have shown selective regulation of GPCR signaling, irrespective of the particular G protein coupled (e.g., 98), demonstrating the importance of Gα- and GAP-independent mechanisms in determining selectivity of signal regulation. Interactions with several other molecules have been described, each with significant functional implications. For example, RGS3 was shown to interact with the phosphoserine-binding protein 14-3-3 via its N-terminus (S264). Since RGS3 when bound to 14-3-3 is unable to interact with G proteins, it has been proposed that 14-3-3 may act as a scavenger, regulating the amounts of RGS3 available for binding G proteins 99. Another important binding partner for several RGS isoforms (best characterized for RGS4) is the calcium sensor calmodulin (Ca2+/CaM), which binds to the well conserved α4 and α5 helices in the RGS domain without affecting GAP activity; however, Ca2+/CaM competes with phosphatidylinositol 3,4,5-trisphosphate (PIP3) binding to the same region, and PIP3 inhibits GAP function 100. Therefore, by relieving PIP3-mediated inhibition of RGS proteins, Ca2+/CaM promotes RGS-mediated inhibition of effector function. Ca2+/CaM-dependent facilitation of RGS protein action has so far been demonstrated for the modulation of intracellular Ca2+ oscillations in polarized cells 101 and voltage-dependent relaxation of IKAch (reviewed 102). Furthermore, direct binding of RGS2 to eIF2ε (eukaryotic initiation factor 2B ε subunit) via a 37 amino stretch within its RGS domain has been linked to inhibition of protein translation, implicating RGS2 as a novel regulator of protein translation 103.
5. Functional Role of RGS Proteins in the Heart
Experimental Strategies
Since the discovery of RGS proteins in the heart, overexpression strategies have been used to determine the functional capacity of cardiac RGS proteins, and, as the prototypical R4 subfamily member, RGS4 initially garnered most attention. While non-physiological interactions may occur upon overexpression, loss-of-function studies addressing the role of endogenous RGS proteins can be hampered by the presence of different RGS isoforms with potentially overlapping functions, which can result in redundancy and/or compensatory coverage. Several strategies have been utilized to reduce RGS protein expression and/or function: specific antibodies 37 or inhibitory RGS peptides 104 were successfully used to disrupt the RGS-Gα interface, while anti-sense oligonucleotides 105, ribozymes 98, or RNAi 26 were used to knock-down RGS protein expression in vitro. Conventional in vivo gene targeting strategies have been employed to generate mouse models with global deletion of select RGS isoforms (Table 2). The Neubig laboratory introduced an elegant alternative approach (Table 2), in which endogenous Gαi/o isoforms were replaced with a single amino acid point mutation in the Gα switch I region that blocks its interaction with RGS proteins and subsequent GTPase activation 106. However, it does not affect the intrinsic GTPase activity or coupling to Gβγ, receptors, and downstream effectors 107. This approach offered novel insight into the full extent of RGS protein-mediated regulation in modulating downstream effects of particular Gα subunits (unencumbered by functional redundancy among RGS) and into subtype-selective signaling by Gi/o family members. Compared to transgenic models with Gα overexpression, knock-ins of RGS-insensitive Gα mutants maintain normal Gα expression levels and reveal both Gα- and Gβγ-mediated RGS protein-sensitive responses upon GPCR-induced Gα activation. However, they cannot identify the specific RGS protein isoform(s) involved and only probes for RGS protein-mediated GAP activity regulation (and effector blockade). RGS2 effects that are mediated by their non-RGS domains will not be detected in these models.
Table 2.
Gain- and Loss-of-Function RGS Protein Models and RGS-insensitive Gα Models and their Cardiovascular Phenotypes
| Gene | Model | Cardiovascular Phenotype |
|---|---|---|
| RGS Protein Transgenic (TG) & Knockout (KO) Models: | ||
| RGS2 | KO | Hypertension and enhanced vasoconstriction due to prolonged Gq-mediated signaling and decreased cGMP- mediated relaxation 81, 110 |
| Normal basal cardiac phenotype and hypertrophic response to swimming; increased Gq signaling and hypertrophy in response to pressure overload with more rapid transition to failure and early mortality; exacerbated hypertrophy and dilation in Gαq transgenes; lack of inhibition of Gq-coupled stimuli and suppression of maladaptive hypertrophy by cGMP-selective PDE5 inhibitor sildenafil 70 | ||
| Enhanced susceptibility to atrial tachycardia/fibrillation via enhanced M3-receptor activity. 122 | ||
| RGS4 | TG | No basal phenotype; compromised adaption to pressure overload (rapid decompensation, increased mortality) 108 |
| In Gαq-expressing transgenes, delay in hypertrophy onset, but comparable end-stage hypertrophic phenotype 109 | ||
| Reduced cardiomyopathic phenotype in PPARα transgenes without change in metabolic abnormalities; resistance to streptozotocin-induced fetal gene induction 142 | ||
| No basal phenotype; reduced hypertrophic response in mice lacking GC-A receptor 143 | ||
| KO | Viable and fertile; normal neural development; subtle sensorimotor deficits. 23 | |
| Increased M2-mediated bradycardia in conscious mice and perfused hearts; lower baseline heart rate and greater increase in response to atropine in anesthetized mice; in SA nodal cells greater sensitivity to muscarinic inhibition of spontaneous action potential firing rate and decreased level of IKAch desensitization as well as slowed activation and deactivation kinetics 24 | ||
| RGS5 | TG | Attenuated hypertrophy and fibrosis response to pressure overload 112 |
| KO | Enhanced hypertrophy and fibrosis development in response to pressure overload 112 | |
| Viable and fertile; reduced blood pressure and increased heart rate; normal vasculature and remodeling response (to tumor growth and oxygen-induced retinopathy) 144 | ||
| No gross abnormalities; low blood pressure without change in heart rate; decreased body weight 145 | ||
| RGS6 | KO | Enhanced carbachol-induced bradycardia (and AV block) in conscious mice and perfused hearts; enhanced muscarinic inhibition of spontaneous action potential firing rate of SA nodal cells; reduction in time course of IKAch activation and deactivation and extent of desensitization in atrial myocytes 120, 121 |
| RGS-insensitive Gα Knock-in (KI) Model: | ||
|
Gαi2
G184S |
KI | Reduced viability, low birth weight, growth retardation, cardiac hypertrophy and increased baseline heart rate during day time, enlarged spleen, elevated neutrophil and monocyte counts, behavioral hyperactivity 146 |
| Enhanced muscarinic (but not adenosine-induced) bradycardic responses in intact mice and perfused hearts; delayed AV conduction 118, 119 | ||
| Smaller infarct size and enhanced contractile recovery after ischemia/reperfusion in perfused hearts; enhanced potency for carbachol’s negative inotropic effect after β-adrenergic stimulation in ventricular myocytes 123. | ||
RGS Proteins and Pressure Overload-induced Cardiac Remodeling
Although several RGS proteins are expressed in the heart, the R4 subfamily has so far been best characterized. The first cardiac mouse model (see Table 2) featured cardiomyocyte-specific transgenic RGS4 expression, which did not affect cardiac morphology or basal function but markedly compromised the heart’s ability to adapt to transverse aortic constriction 108 and ameliorated (although only transiently) hypertrophy and heart failure in Gαq-expressing hearts 109, suggesting that the anti-hypertrophic effect of RGS4 could be beneficial or detrimental depending on the (patho)physiological context. Mechanistic contributions of RGS4 regulation of Gi/o and/or Gq/11 pathways were not examined in this model, and the physiological significance is to be viewed in light of subsequent reports on the virtual absence of RGS4 in the working myocardium 23, 24. Investigation of RGS2 knockout mice first revealed that RGS2 plays a critical role in regulating contractile activity of vascular smooth muscle cells and blood pressure homeostasis 81, 110. PKG-mediated RGS2 phosphorylation resulting in enhanced GTPase activity was identified as a key mechanism suppressing Gq-stimulated vascular contraction 81; an increase in sympathetic tone has been proposed to potentially contribute as well 111. More recently, RGS2 was shown to be required for early myocardial compensation to pressure overload and as a mediator of anti-hypertrophic and cardioprotective cGMP-mediated effects of sildenafil, a cGMP-selective phosphodiesterase (PDE) 5 inhibitor 70. Similarly, counter-regulatory effects of ANF on Ang II-induced hypertrophic effects were shown to be dependent on guanylyl cyclase A (GC-A) receptor, PKG and RGS2 27. A role of RGS5 in protecting against cardiac hypertrophy in response to pressure overload was revealed in mice with cardiac-specific transgenic overexpression or global deletion of RGS5, presumably via regulation of MEK/ERK activation (but not JNK, p38 and Akt) 112.
While most studies to date have focused on myocyte regulation by RGS proteins, investigations into the role of RGS proteins in fibroblasts are emerging. This is particularly relevant, since cardiac fibroblasts are also important therapeutic targets 113. Exacerbation of pressure overload-induced fibrosis development has been reported for mice with global deletion of RGS5 112 or RGS2 70. Both Ang II and endothelin-1 are important profibrotic factors in human cardiac fibroblasts; and their effects are mediated via Gq/11-coupled AT1 receptors 114 and ETA receptors 115, respectively. Importantly, RGS2 was recently shown to be a functionally important and highly regulated negative regulator of Ang II-induced signaling, cell proliferation and collagen in adult ventricular fibroblasts 21. These studies suggest that RGS protein targeting could become a strategy to modulate cardiac fibroblast responses. However, in order to establish a direct (patho)physiological role of RGS2, RGS5 and potentially other RGS isoforms in regulating fibroblast behavior and fibrosis in vivo, mouse models with fibroblast-restricted deletions are required. RGS2 and RGS5 are ubiquitously expressed, and changes that occur in fibroblasts must be discerned from those in other cell types. For example, the fact that myocyte-restricted RGS5 expression markedly attenuated fibrosis in pressure overloaded hearts suggests myocyte-fibroblast crosstalk to play a major role 112. To date, gene targeting experiments have been hampered by the challenge of identifying fibroblast-specific promoter elements 116, but recent studies have shown promising results (e.g., 117).
RGS Proteins and Heart Rate Control
RGS proteins also play an essential role in regulating parasympathetic heart rate regulation, which involves M2-receptor activation of Gi/o, release of Gβγ with subsequent activation of G protein-coupled inwardly rectifying K+ (GIRK) channels, resulting in acetylcholine-activated potassium current (IKAch), and membrane hyperpolarization. In addition, vagal stimulation suppresses Gs-mediated AC activation, thereby reducing binding of cAMP to pacemaker current (If) and PKA-phosphorylation increase in L-type calcium channel current (ICa-L). The first in vivo evidence was provided in knock-in mice expressing RGS-resistant Gαi2, which displayed markedly enhanced carbachol-induced bradycardia 118 (Table 2). Direct regulation of cardiac pacemakers was subsequently suggested when isolated perfused hearts from this model showed potentiation of muscarinic inhibition of cardiac automaticity as well as atrioventricular conduction 119. Comparison of chronotropic responses of cardiomyocytes derived from embryonic stem cells with knock-in of RGS-insensitive Gαi2 or Gαo showed that endogenous RGS modulate Gi/o-coupled receptor signaling (e.g., M2, A1 and β2 receptors) in a Gα isoform-specific manner 118.
Subsequent RGS isoform-specific knockout models implicated RGS4 24 and RGS6 120, 121 as key regulators of parasympathetic heart rate control, because their loss was associated with severely exaggerated bradycardia and atrioventricular block in response to parasympathetic stimulation in vivo. The underlying mechanisms still need to be fully delineated but likely differ: while RGS4 and RGS6 can both negatively regulate Gαi/o subunits, only RGS6 has the capacity to directly interact with Gβ5 via its GGL domain and to form a complex that appears to contribute to the inactivation of IKAch 120. Both RGS4 and RGS6 were required for desensitization and rapid deactivation as well as normal activation of IKACh. Importantly, double RGS4 and RGS6 knockout mice are needed to determine whether RGS4 and RGS6 act on the same G proteins mediating GIRK regulation and whether their effects will be additive. Additional RGS isoforms may be involved in heart rate regulation. For example, enhanced susceptibility to atrial fibrillation, presumably via enhanced M3 receptor activity, was reported in RGS2 knockout mice 122.
Taken together, gain- and loss-of-function mouse models designed to interrogate RGS protein function in vivo strongly suggest that RGS proteins play important roles in the cardiovascular system in health and disease. To date, several RGS isoforms have been implicated in the regulation of blood pressure (RGS2, RGS5), cardiac automaticity and conduction (RGS4, RGS6, and potentially RGS2) and development of both hypertrophy (RGS2, RGS4, RGS5) and fibrosis (RGS2, RGS5) in response to pressure overload. Most recently, RGS proteins were also implicated to suppress Gαi2-mediated cardioprotection (Table 2) 123. Additional models targeting other RGS isoforms and in a cell-type-specific manner will be required to obtain a comprehensive picture of the functional significance of RGS proteins in regulating GPCR signaling in the heart.
6. RGS Proteins as Therapeutic Targets
GPCRs are a cell surface receptor superfamily with more than 800 genes encoding GPCRs in the human genome 124. They regulate virtually all known physiological processes in mammals and are estimated to be the target of approximately one third of approved drugs 125. In light of the vastly greater number of GPCRs (>200 the heart, 126) compared to G proteins (15 Gα, 5 Gβ and 12 Gγ subunits, 127), it has long been recognized that many different GPCRs are generally linked to the same G protein-mediated signaling pathway, but GPCRs can also functionally couple simultaneously with distinct unrelated G proteins, leading to activation of multiple intracellular effectors by a single receptor. Targeting GPCR signaling at the receptor level has yielded substantial therapeutic benefits in the cardiovascular and many other fields; yet heart failure remains a leading cause of death morbidity and mortality in the world. A long-standing alternative strategy to target GPCR signaling at the level of the G proteins has been to mitigate Gβγ signaling, initially using large peptide inhibitors and more recently small molecule inhibitors (reviewed in 128). As key regulators of G protein signaling, RGS proteins have emerged as intriguing additional therapeutic targets based on their physiological and pathophysiological importance in the heart, central nervous system, cancer biology and beyond.
Therapeutic benefits can be derived from inhibition or enhancement of RGS protein function, depending on the nature of the targeted isoform, its regulatory function and the cellular and pathophysiological context. Conceptually, RGS protein inhibitors potentiate GPCR agonist function, which would be useful for rapidly desensitizing agonists as well for minimizing GPCR agonist dosage and its side effects when given as a drug. RGS protein inhibitors could also increase the specificity of exogenous GPCR agonists, and, in addition, block effector signaling by RGS proteins. In contrast, enhancing RGS protein function could be beneficial in settings where reduction in RGS protein expression or activity is associated with pathophysiological consequences. For example, marked reduction in RGS2 in response to pressure overload and other settings with enhanced Gq signaling is known to exacerbate myocyte and cardiac hypertrophy 26, 70. Similarly, diminished RGS2 expression is associated with hypertension in mice 110 and humans 129, whereas RGS2 levels are increased in patients with Bartter’s/Gitelman’s syndrome, which is associated with reduced Ang II signaling and vasomotor tone 130. Furthermore, single nucleotide polymorphisms identified in Japanese patients with hypertension were shown to be less stable or lead to reduced plasma membrane targeting and function (reviewed in 10).
Based on current knowledge of RGS protein structure and function, strategies to target RGS protein function include altering GAP activity, steady state expression, protein or lipid interactions, posttranslational modifications and/or subcellular location. Most targeting efforts to date have focused on RGS4 as one of the best characterized isoforms. A number of peptide and small molecule inhibitors targeting RGS-Gα interaction were identified via high-throughput screening 131-133, as summarized by 134, an excellent review that also provides an overview of the strengths and limitations of the assay systems used in the quest for drugs targeting RGS proteins. As reviewed in detail elsewhere 73, the mechanism of action of one of the first RGS4 inhibitors (CCG-4986) involves covalent cysteine modifications, one of which occurs in the RGS/Gα interaction surface (aka “A site”, 135), whereas the other functionally more important one is located on the opposite face of RGS (near the “B site”) and leads to allosteric inhibition of RGS-Gα interaction 136. While CCG-4986 binds irreversibly and cannot function in cellular environment, another reversible small molecule inhibitor for RGS4 was recently introduced, which leads to similar allosteric inhibition 137. Encouraging for further drug development and a prerequisite for ultimate therapeutic utility is the fact that closely related RGS isoforms with similar sequence and structure have different responsiveness to these inhibitors. Regardless of the mechanism, disruption of RGS/Gα binding and subsequent inhibition of GAP function is expected to enhance both Gα and Gβγ-mediated effects. GAP-independent RGS protein effects that are mediated via regions outside the RGS domain could be targeted as well, particularly for isoforms with well characterized protein-protein interaction sites (e.g., RGS2 and AC 36, RGS3 and Smad 42). Stabilizing RGS protein expression is another potential strategy to enhance RGS protein function, which would affect GAP-dependent and -independent RGS protein effects. This could be achieved for R7 subfamily members by disrupting the interaction between their GGL domain and Gβ5, which is required for stable expression 48. Preventing proteosomal degradation could be another approach, particularly for isoforms that are subject to the N-end rule pathway. Intriguingly, progressive increase in invasiveness in human breast cancer was shown be tightly linked to gradual reduction in RGS4 protein (but not mRNA) due to enhanced proteosomal degradation 25.
Taken together, RGS proteins are clearly promising targets for therapeutic development. Like many GPCRs, several RGS isoforms are ubiquitously expressed. Unlike GPCR agonists/antagonists that act on the extracellular cell surface, targeting of RGS proteins requires cell-permeable compounds. Despite the significant progress already made, much work still needs to be done to develop strategies that can eventually be used successfully in vivo. At this stage, computer predictions of potential drug binding pockets indicate the back side of the RGS domain opposing Gα interaction site may be more favorable to small molecule inhibition 134. Interestingly, competitive binding of PIP3 and Ca2+/CaM 138 with implications for GAP functions (inhibited vs. no effect, respectively; see above) as well as palmitoylation leading to GAP inhibition occur in that region. In contrast to the well characterized structure of the RGS domain, little is currently known about the structure of the other domains in the N- and C-terminal extensions of RGS proteins, which could offer additional sites of intervention. Compounds that stabilize protein expression of specific RGS protein isoforms are also believed to have significant potential.
7. Conclusions and Future Perspective
The importance of GPCR signaling for determining cardiac differentiation, growth, contraction and heart rate regulation has been recognized for a very long time. After their discovery in the mid 1990s, RGS proteins were quickly appreciated as key players in the regulation of GPCR signaling. Of the 20 canonical RGS proteins, many isoforms have been detected in the heart, with a specific complement for each cell type, as shown for cardiac myocytes and fibroblasts. Many studies have been performed in various cell lines as well as primary cells from the brain and cardiovascular system, each focusing on one or a few RGS proteins. They have provided a wealth of information into the function of RGS proteins as modulators and integrators as G protein signaling. Unfortunately, it is not possible to extrapolate from one cell type to another due to the complex expression, regulatory and interaction patterns of RGS proteins with other molecules that is unique to each cell type. Studies investigating the role of RGS proteins (primarily a few isoforms from the R4 subfamily and more recently also RGS6) have demonstrated the central importance of cardiac RGS proteins in regulating myocyte function in vitro and in vivo. New evidence suggests that RGS proteins may also be important regulators of cardiac fibroblast function. Several important questions need to be addressed. For example, what is/are the functional role(s) of each RGS isoform expressed in the two major cell types in the myocardium? Investigating RGS proteins individually (and in each cell type) is obviously a daunting task. The RGS-insensitive Gα mutants will continue to be an essential tool to investigate global RGS protein-mediated inhibition of Gα-mediated signaling in cells and animal models, and expansion to Gα subunits beyond Gαi2 and Gαo is eagerly anticipated. Nevertheless, identifying (the) particular RGS isoform(s) that regulate(s) specific cell signaling and functional responses will require targeted deletion of individual RGS proteins, ideally in a cell type-specific manner. The roles of RGS proteins in the other myocardial cell types that participate in maintaining normal cardiac function and determine the response to stress (e.g., endothelial cells and inflammatory cells) also need to be addressed. Collectively, future investigations in these areas will advance our understanding of the physiological role of RGS proteins in regulating signal transduction and cell functions in the heart as well as their contributions to the development of cardiovascular disease. Studies in larger animal models and healthy and diseased human hearts will be essential for clinical translation.
A variety of mechanisms (such as GTPase acceleration, posttranslational modifications, protein-protein/lipid interactions and spatiotemporal-specific expression) are believed to enable RGS proteins to serve effectively as multifunctional signal regulators. This is evident by the fact that despite functional redundancy in vitro, specificity in RGS protein-mediated regulation of signal transduction and cellular function exists in cellular context and in vivo. Since many of the regulatory mechanisms were discovered in biochemical or overexpression studies, it must be determine which of them are of functional relevance under physiological conditions and what the mechanisms controlling them are. Furthermore, it is not clear at this point what regulatory mechanisms play a role in human disease and if and how they can be targeted therapeutically. In order to obtain insights into the regulation of endogenous RGS proteins at the protein level in primary cardiac cells and tissue, the sensitivity for RGS protein detection must be increased. Additional very useful reagents will be RGS isoform-specific inhibitors/enhancers, because they will open avenues for mechanistic studies akin to the way GPCR agonists/antagonists facilitated research into GPCR function. Although developing these reagents is a challenging task, substantial progress has already been made. With regard to the potential therapeutic use of RGS protein inhibitors or enhancers, it is hoped that further development of reversible small molecules or other compounds and validation of their properties in cells and animal models will eventually allow investigators to test the potential of targeting RGS protein expression and/or activity in vivo for the treatment of cardiac hypertrophy, failure and/or heart rate irregularities.
ACKNOWLEDGEMENTS
None.
SOURCES OF FUNDING U.M. is supported by grants from the National Heart Lung and Blood Institute HL-80127 and Established Investigator Award 0740098N from the American Heart Association. P.Z. is supported by a grant from the Rhode Island Foundation.
Non-standard Abbreviations and Acronyms
- AC
Adenylate cyclase
- Ang II
Angiotensin II
- Ca2+/CaM
Calcium Calmodulin
- CREB
cAMP response element binding protein
- DEP
Disheveled-EGL10-Pleckstrin homology domain
- EGFR
Epidermal growth factor receptor
- ERK
Extracellular signal-regulated kinase
- GAIP
Gα interacting protein
- GAP
GTPase-activating protein
- GEF
Guanine nucleotide exchange factor
- GIPC
GAIP-interacting protein C-terminus
- GIRK
G protein-coupled inwardly rectifying K+
- GGL
Gγ-like domain
- GoLoco
Gαi/o-Loco
- GPCR
G protein-coupled receptor
- MEK
MAPK/ERK kinase
- PDE
Phosphodiesterase
- PDZ
PSD-95 disk-large ZO-1 domain
- PI3K
Phosphatidylinositol-3-OH kinase
- PIP3
Phosphatidylinositol 3,4,5-trisphosphate
- PKA
Protein kinase A
- PKC
Protein kinase C
- PKG
Protein kinase G
- PM
PDZ docking motif
- PTB
Phosphotyrosine binding domain
- RGS
Regulators of G protein signaling
Footnotes
DISCLOSURES None.
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Salazar NC, Chen J, Rockman HA. Cardiac GPCRs: GPCR signaling in healthy and failing hearts. Biochim Biophys Acta. 2007;1768:1006–1018. doi: 10.1016/j.bbamem.2007.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chidiac P, Ross EM. Phospholipase C-β1 directly accelerates GTP hydrolysis by Gαq and acceleration is inhibited by Gβγ subunits. J Biol Chem. 1999;274:19639–19643. doi: 10.1074/jbc.274.28.19639. [DOI] [PubMed] [Google Scholar]
- 3.Ross EM, Wilkie TM. GTPase-activating proteins for heterotrimeric G proteins: regulators of G protein signaling (RGS) and RGS-like proteins. Annu Rev Biochem. 2000;69:795–827. doi: 10.1146/annurev.biochem.69.1.795. [DOI] [PubMed] [Google Scholar]
- 4.Berman DM, Wilkie TM, Gilman AG. GAIP and RGS4 are GTPase-activating proteins for the Gi subfamily of G protein α subunits. Cell. 1996;86:445–452. doi: 10.1016/s0092-8674(00)80117-8. [DOI] [PubMed] [Google Scholar]
- 5.Tesmer JJ, Berman DM, Gilman AG, Sprang SR. Structure of RGS4 bound to AlF4−-activated Giα1: stabilization of the transition state for GTP hydrolysis. Cell. 1997;89:251–261. doi: 10.1016/s0092-8674(00)80204-4. [DOI] [PubMed] [Google Scholar]
- 6.Soundararajan M, Willard FS, Kimple AJ, Turnbull AP, Ball LJ, Schoch GA, Gileadi C, Fedorov OY, Dowler EF, Higman VA, Hutsell SQ, Sundstrom M, Doyle DA, Siderovski DP. Structural diversity in the RGS domain and its interaction with heterotrimeric G protein α-subunits. Proc Natl Acad Sci USA. 2008;105:6457–6462. doi: 10.1073/pnas.0801508105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anger T, Zhang W, Mende U. Differential contribution of GTPase activation and effector antagonism to the inhibitory effect of RGS proteins on Gq-mediated signaling. J Biol Chem. 2004;276:3906–3915. doi: 10.1074/jbc.M309496200. [DOI] [PubMed] [Google Scholar]
- 8.Tesmer JJ. Structure and function of regulator of G protein signaling homology domains. Vol 86. Elsevier; Amsterdam, The Netherlands: 2009. [DOI] [PubMed] [Google Scholar]
- 9.Hollinger S, Hepler JR. Cellular regulation of RGS proteins: modulators and integrators of G protein signaling. Pharmacol Rev. 2002;54:527–559. doi: 10.1124/pr.54.3.527. [DOI] [PubMed] [Google Scholar]
- 10.Gu S, Cifelli C, Wang S, Heximer SP. RGS proteins: identifying new GAPs in the understanding of blood pressure regulation and cardiovascular function. Clin Sci (Lond) 2009;116:391–399. doi: 10.1042/CS20080272. [DOI] [PubMed] [Google Scholar]
- 11.Manzur M, Ganss R. Regulator of G protein signaling 5: a new player in vascular remodeling. Trends Cardiovasc Med. 2009;19:26–30. doi: 10.1016/j.tcm.2009.04.002. [DOI] [PubMed] [Google Scholar]
- 12.Anderson GR, Posokhova E, Martemyanov KA. The R7 RGS protein family: multi-subunit regulators of neuronal G protein signaling. Cell Biochem Biophys. 2009;54:33–46. doi: 10.1007/s12013-009-9052-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Traynor J. Regulator of G protein-signaling proteins and addictive drugs. Ann NY Acad Sci. 2010;1187:341–352. doi: 10.1111/j.1749-6632.2009.05150.x. [DOI] [PubMed] [Google Scholar]
- 14.Druey KM. Regulation of G-protein-coupled signaling pathways in allergic inflammation. Immunol Res. 2009;43:62–76. doi: 10.1007/s12026-008-8050-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hurst JH, Hooks SB. Regulator of G-protein signaling (RGS) proteins in cancer biology. Biochem Pharmacol. 2009;78:1289–1297. doi: 10.1016/j.bcp.2009.06.028. [DOI] [PubMed] [Google Scholar]
- 16.Mittmann C, Chung CH, Hoppner G, Michalek C, Nose M, Schuler C, Schuh A, Eschenhagen T, Weil J, Pieske B, Hirt S, Wieland T. Expression of ten RGS proteins in human myocardium: functional characterization of an upregulation of RGS4 in heart failure. Cardiovasc Res. 2002;55:778–786. doi: 10.1016/s0008-6363(02)00459-5. [DOI] [PubMed] [Google Scholar]
- 17.Wieland T, Mittmann C. Regulators of G-protein signalling: multifunctional proteins with impact on signalling in the cardiovascular system. Pharmacol Ther. 2003;97:95–115. doi: 10.1016/s0163-7258(02)00326-1. [DOI] [PubMed] [Google Scholar]
- 18.Riddle EL, Schwartzman RA, Bond M, Insel PA. Multi-tasking RGS proteins in the heart: the next therapeutic target? Circ Res. 2005;96:401–411. doi: 10.1161/01.RES.0000158287.49872.4e. [DOI] [PubMed] [Google Scholar]
- 19.Doupnik CA, Xu T, Shinaman JM. Profile of RGS expression in single rat atrial myocytes. Biochim Biophys Acta. 2001;1522:97–107. doi: 10.1016/s0167-4781(01)00342-6. [DOI] [PubMed] [Google Scholar]
- 20.Kardestuncer T, Wu H, Lim AL, Neer EJ. Cardiac myocytes express mRNA for ten RGS proteins: changes in RGS mRNA expression in ventricular myocytes and cultured atria. FEBS Lett. 1998;438:285–288. doi: 10.1016/s0014-5793(98)01319-2. [DOI] [PubMed] [Google Scholar]
- 21.Zhang P, Su J, King ME, Maldonado AE, Park C, Mende U. Regulator of G protein signaling 2 is a functionally important negative regulator of Angiotensin II-induced cardiac fibroblast responses. Am J Physiol Heart Circ Physiol. 2011 doi: 10.1152/ajpheart.00026.2011. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Larminie C, Murdock P, Walhin JP, Duckworth M, Blumer KJ, Scheideler MA, Garnier M. Selective expression of regulators of G-protein signaling (RGS) in the human central nervous system. Brain Res Mol Brain Res. 2004;122:24–34. doi: 10.1016/j.molbrainres.2003.11.014. [DOI] [PubMed] [Google Scholar]
- 23.Grillet N, Pattyn A, Contet C, Kieffer BL, Goridis C, Brunet JF. Generation and characterization of Rgs4 mutant mice. Mol Cell Biol. 2005;25:4221–4228. doi: 10.1128/MCB.25.10.4221-4228.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cifelli C, Rose RA, Zhang H, Voigtlaender-Bolz J, Bolz SS, Backx PH, Heximer SP. RGS4 regulates parasympathetic signaling and heart rate control in the sinoatrial node. Circ Res. 2008;103:527–535. doi: 10.1161/CIRCRESAHA.108.180984. [DOI] [PubMed] [Google Scholar]
- 25.Xie Y, Wolff DW, Wei T, Wang B, Deng C, Kirui JK, Jiang H, Qin J, Abel PW, Tu Y. Breast cancer migration and invasion depend on proteasome degradation of regulator of G-protein signaling 4. Cancer Res. 2009;69:5743–5751. doi: 10.1158/0008-5472.CAN-08-3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang W, Anger T, Su J, Hao J, Xu X, Zhu M, Gach A, Cui L, Liao R, Mende U. Selective loss of fine tuning of Gq/11 signaling by RGS2 protein exacerbates cardiomyocyte hypertrophy. J Biol Chem. 2006;281:5811–5820. doi: 10.1074/jbc.M507871200. [DOI] [PubMed] [Google Scholar]
- 27.Klaiber M, Kruse M, Volker K, Schroter J, Feil R, Freichel M, Gerling A, Feil S, Dietrich A, Londono JE, Baba HA, Abramowitz J, Birnbaumer L, Penninger JM, Pongs O, Kuhn M. Novel insights into the mechanisms mediating the local antihypertrophic effects of cardiac atrial natriuretic peptide: role of cGMP-dependent protein kinase and RGS2. Basic Res Cardiol. 2010;105:583–595. doi: 10.1007/s00395-010-0098-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heximer SP, Srinivasa SP, Bernstein LS, Bernard JL, Linder ME, Hepler JR, Blumer KJ. G protein selectivity is a determinant of RGS2 function. J Biol Chem. 1999;274:34253–34259. doi: 10.1074/jbc.274.48.34253. [DOI] [PubMed] [Google Scholar]
- 29.Kimple AJ, Soundararajan M, Hutsell SQ, Roos AK, Urban DJ, Setola V, Temple BR, Roth BL, Knapp S, Willard FS, Siderovski DP. Structural determinants of G-protein α subunit selectivity by regulator of G-protein signaling 2 (RGS2) J Biol Chem. 2009;284:19402–19411. doi: 10.1074/jbc.M109.024711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hao J, Michalek C, Zhang W, Zhu M, Xu X, Mende U. Regulation of cardiomyocyte signaling by RGS proteins: Differential selectivity towards G proteins and susceptibility to regulation. J Mol Cell Cardiol. 2006;41:51–61. doi: 10.1016/j.yjmcc.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 31.Ingi T, Krumins AM, Chidiac P, Brothers GM, Chung S, Snow BE, Barnes CA, Lanahan AA, Siderovski DP, Ross EM, Gilman AG, Worley PF. Dynamic regulation of RGS2 suggests a novel mechanism in G-protein signaling and neuronal plasticity. J Neurosci. 1998;18:7178–7188. doi: 10.1523/JNEUROSCI.18-18-07178.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cladman W, Chidiac P. Characterization and comparison of RGS2 and RGS4 as GTPase-activating proteins for m2 muscarinic receptor-stimulated Gi. Mol Pharmacol. 2002;62:654–659. doi: 10.1124/mol.62.3.654. [DOI] [PubMed] [Google Scholar]
- 33.Chen C, Zheng B, Han J, Lin SC. Characterization of a novel mammalian RGS protein that binds to Gα proteins and inhibits pheromone signaling in yeast. J Biol Chem. 1997;272:8679–8685. doi: 10.1074/jbc.272.13.8679. [DOI] [PubMed] [Google Scholar]
- 34.Heximer SP, Watson N, Linder ME, Blumer KJ, Hepler JR. RGS2/G0S8 is a selective inhibitor of Gqα function. Proc Natl Acad Sci USA. 1997;94:14389–14393. doi: 10.1073/pnas.94.26.14389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chakir K, Zhu W, Tsang S, Woo AY, Yang D, Wang X, Zeng X, Rhee MH, Mende U, Koitabashi N, Takimoto E, Blumer KJ, Lakatta EG, Kass DA, Xiao RP. RGS2 is a primary terminator of β2-adrenergic receptor-mediated Gi signaling. J Mol Cell Cardiol. 2011 doi: 10.1016/j.yjmcc.2011.01.015. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Salim S, Sinnarajah S, Kehrl JH, Dessauer CW. Identification of RGS2 and type V adenylyl cyclase interaction sites. J Biol Chem. 2003;278(18):15842–15849. doi: 10.1074/jbc.M210663200. [DOI] [PubMed] [Google Scholar]
- 37.Sinnarajah S, Dessauer CW, Srikumar D, Chen J, Yuen J, Yilma S, Dennis JC, Morrison EE, Vodyanoy V, Kehrl JH. RGS2 regulates signal transduction in olfactory neurons by attenuating activation of adenylyl cyclase III. Nature. 2001;409:1051–1055. doi: 10.1038/35059104. [DOI] [PubMed] [Google Scholar]
- 38.Roy AA, Baragli A, Bernstein LS, Hepler JR, Hebert TE, Chidiac P. RGS2 interacts with Gs and adenylyl cyclase in living cells. Cell Signal. 2006;18:336–348. doi: 10.1016/j.cellsig.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 39.Nunn C, Zou MX, Sobiesiak AJ, Roy AA, Kirshenbaum LA, Chidiac P. RGS2 inhibits β-adrenergic receptor-induced cardiomyocyte hypertrophy. Cell Signal. 2010;22:1231–1239. doi: 10.1016/j.cellsig.2010.03.015. [DOI] [PubMed] [Google Scholar]
- 40.Shi CS, Lee SB, Sinnarajah S, Dessauer CW, Rhee SG, Kehrl JH. Regulator of G-protein signaling 3 (RGS3) inhibits Gβ1γ2-induced inositol phosphate production, mitogen-activated protein kinase activation, and Akt activation. J Biol Chem. 2001;276:24293–24300. doi: 10.1074/jbc.M100089200. [DOI] [PubMed] [Google Scholar]
- 41.Vogt A, Lutz S, Rumenapp U, Han L, Jakobs KH, Schmidt M, Wieland T. Regulator of G-protein signalling 3 redirects prototypical Gi-coupled receptors from Rac1 to RhoA activation. Cell Signal. 2007;19:1229–1237. doi: 10.1016/j.cellsig.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 42.Yau DM, Sethakorn N, Taurin S, Kregel S, Sandbo N, Camoretti-Mercado B, Sperling AI, Dulin NO. Regulation of Smad-mediated gene transcription by RGS3. Mol Pharmacol. 2008;73:1356–1361. doi: 10.1124/mol.108.044990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bansal G, Xie Z, Rao S, Nocka KH, Druey KM. Suppression of immunoglobulin E-mediated allergic responses by regulator of G protein signaling 13. Nat Immunol. 2008;9:73–80. doi: 10.1038/ni1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liang G, Bansal G, Xie Z, Druey KM. RGS16 inhibits breast cancer cell growth by mitigating phosphatidylinositol 3-kinase signaling. J Biol Chem. 2009;284:21719–21727. doi: 10.1074/jbc.M109.028407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Oudit GY, Penninger JM. Cardiac regulation by phosphoinositide 3-kinases and PTEN. Cardiovasc Res. 2009;82:250–260. doi: 10.1093/cvr/cvp014. [DOI] [PubMed] [Google Scholar]
- 46.Xie Z, Geiger TR, Johnson EN, Nyborg JK, Druey KM. RGS13 acts as a nuclear repressor of CREB. Mol Cell. 2008;31:660–670. doi: 10.1016/j.molcel.2008.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hooks SB, Waldo GL, Corbitt J, Bodor ET, Krumins AM, Harden TK. RGS6, RGS7, RGS9, and RGS11 stimulate GTPase activity of Gi family G-proteins with differential selectivity and maximal activity. J Biol Chem. 2003;278:10087–10093. doi: 10.1074/jbc.M211382200. [DOI] [PubMed] [Google Scholar]
- 48.Chen CK, Eversole-Cire P, Zhang H, Mancino V, Chen YJ, He W, Wensel TG, Simon MI. Instability of GGL domain-containing RGS proteins in mice lacking the G protein β-subunit Gβ5. Proc Natl Acad Sci USA. 2003;100:6604–6609. doi: 10.1073/pnas.0631825100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cheever ML, Snyder JT, Gershburg S, Siderovski DP, Harden TK, Sondek J. Crystal structure of the multifunctional Gβ5-RGS9 complex. Nat Struct Mol Biol. 2008;15:155–162. doi: 10.1038/nsmb.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jayaraman M, Zhou H, Jia L, Cain MD, Blumer KJ. R9AP and R7BP: traffic cops for the RGS7 family in phototransduction and neuronal GPCR signaling. Trends Pharmacol Sci. 2009;30:17–24. doi: 10.1016/j.tips.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Burgon PG, Lee WL, Nixon AB, Peralta EG, Casey PJ. Phosphorylation and nuclear translocation of a regulator of G protein signaling (RGS10) J Biol Chem. 2001;276:32828–32834. doi: 10.1074/jbc.M100960200. [DOI] [PubMed] [Google Scholar]
- 52.Kimple RJ, De Vries L, Tronchere H, Behe CI, Morris RA, Farquhar MG, Siderovski DP. RGS12 and RGS14 GoLoco motifs are Gαi interaction sites with guanine nucleotide dissociation inhibitor activity. J Biol Chem. 2001;276:29275–29281. doi: 10.1074/jbc.M103208200. [DOI] [PubMed] [Google Scholar]
- 53.Willard MD, Willard FS, Li X, Cappell SD, Snider WD, Siderovski DP. Selective role for RGS12 as a Ras/Raf/MEK scaffold in nerve growth factor-mediated differentiation. Embo J. 2007;26:2029–2040. doi: 10.1038/sj.emboj.7601659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Willard FS, Willard MD, Kimple AJ, Soundararajan M, Oestreich EA, Li X, Sowa NA, Kimple RJ, Doyle DA, Der CJ, Zylka MJ, Snider WD, Siderovski DP. Regulator of G-protein signaling 14 (RGS14) is a selective H-Ras effector. PLoS One. 2009;4:e4884. doi: 10.1371/journal.pone.0004884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shu FJ, Ramineni S, Hepler JR. RGS14 is a multifunctional scaffold that integrates G protein and Ras/Raf MAPkinase signalling pathways. Cell Signal. 2010;22:366–376. doi: 10.1016/j.cellsig.2009.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nunn C, Mao H, Chidiac P, Albert PR. RGS17/RGSZ2 and the RZ/A family of regulators of G-protein signaling. Semin Cell Dev Biol. 2006;17:390–399. doi: 10.1016/j.semcdb.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 57.Abramow-Newerly M, Roy AA, Nunn C, Chidiac P. RGS proteins have a signalling complex: interactions between RGS proteins and GPCRs, effectors, and auxiliary proteins. Cell Signal. 2006;18:579–591. doi: 10.1016/j.cellsig.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 58.Mao H, Zhao Q, Daigle M, Ghahremani MH, Chidiac P, Albert PR. RGS17/RGSZ2, a novel regulator of Gi/o, Gz and Gq signaling. J Biol Chem. 2004;279:26314–26322. doi: 10.1074/jbc.M401800200. [DOI] [PubMed] [Google Scholar]
- 59.Zheng B, Ma YC, Ostrom RS, Lavoie C, Gill GN, Insel PA, Huang XY, Farquhar MG. RGS-PX1, a GAP for Gαs and sorting nexin in vesicular trafficking. Science. 2001;294:1939–1942. doi: 10.1126/science.1064757. [DOI] [PubMed] [Google Scholar]
- 60.Suzuki N, Hajicek N, Kozasa T. Regulation and physiological functions of G12/13-mediated signaling pathways. Neurosignals. 2009;17:55–70. doi: 10.1159/000186690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gu S, Anton A, Salim S, Blumer KJ, Dessauer CW, Heximer SP. Alternative translation initiation of human regulators of G-protein signaling-2 yields a set of functionally distinct proteins. Mol Pharmacol. 2008;73:1–11. doi: 10.1124/mol.107.036285. [DOI] [PubMed] [Google Scholar]
- 62.Wolff DW, Xie Y, Deng C, Gatalica Z, Yang M, Wang B, Wang J, Lin MF, Abel PW, Tu Y. Epigenetic repression of regulator of G-protein signaling 2 promotes androgen-independent prostate cancer cell growth. Int J Cancer. 2011 doi: 10.1002/ijc.26138. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jalili T, Takeishi Y, Song G, Ball NA, Howles G, Walsh RA. PKC translocation without changes in Gαq and PLC-β protein abundance in cardiac hypertrophy and failure. Am J Physiol. 1999;277:H2298–2304. doi: 10.1152/ajpheart.1999.277.6.H2298. [DOI] [PubMed] [Google Scholar]
- 64.Zhang S, Watson N, Zahner J, Rottman JN, Blumer KJ, Muslin AJ. RGS3 and RGS4 are GTPase activating proteins in the heart. J Mol Cell Cardiol. 1998;30:269–276. doi: 10.1006/jmcc.1997.0591. [DOI] [PubMed] [Google Scholar]
- 65.Owen VJ, Burton PB, Mullen AJ, Birks EJ, Barton P, Yacoub MH. Expression of RGS3, RGS4 and Gαi2 in acutely failing donor hearts and end-stage heart failure. Eur Heart J. 2001;22:1015–1020. doi: 10.1053/euhj.2000.2578. [DOI] [PubMed] [Google Scholar]
- 66.Hendriks-Balk MC, Peters SL, Michel MC, Alewijnse AE. Regulation of G protein-coupled receptor signalling: focus on the cardiovascular system and regulator of G protein signalling proteins. Eur J Pharmacol. 2008;585:278–291. doi: 10.1016/j.ejphar.2008.02.088. [DOI] [PubMed] [Google Scholar]
- 67.Zou MX, Roy AA, Zhao Q, Kirshenbaum LA, Karmazyn M, Chidiac P. RGS2 is upregulated by and attenuates the hypertrophic effect of α1-adrenergic activation in cultured ventricular myocytes. Cell Signal. 2006;18:1655–1663. doi: 10.1016/j.cellsig.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 68.Kehrl JH, Sinnarajah S. RGS2: a multifunctional regulator of G-protein signaling. Int J Biochem Cell Biol. 2002;34:432–438. doi: 10.1016/s1357-2725(01)00141-8. [DOI] [PubMed] [Google Scholar]
- 69.Tsang S, Woo AY, Zhu W, Xiao RP. Deregulation of RGS2 in cardiovascular diseases. Front Biosci (Schol Ed) 2010;2:547–557. doi: 10.2741/s84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Takimoto E, Koitabashi N, Hsu S, Ketner EA, Zhang M, Nagayama T, Bedja D, Gabrielson KL, Blanton R, Siderovski DP, Mendelsohn ME, Kass DA. Regulator of G protein signaling 2 mediates cardiac compensation to pressure overload and antihypertrophic effects of PDE5 inhibition in mice. J Clin Invest. 2009;119:408–420. doi: 10.1172/JCI35620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xie Z, Yang Z, Druey KM. Phosphorylation of RGS13 by the cyclic AMP-dependent protein kinase inhibits RGS13 degradation. J Mol Cell Biol. 2010;2:357–365. doi: 10.1093/jmcb/mjq031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Derrien A, Zheng B, Osterhout JL, Ma YC, Milligan G, Farquhar MG, Druey KM. Src-mediated RGS16 tyrosine phosphorylation promotes RGS16 stability. J Biol Chem. 2003;278:16107–16116. doi: 10.1074/jbc.M210371200. [DOI] [PubMed] [Google Scholar]
- 73.Sjoegren B, Neubig RR. Thinking outside of the “RGS box”: new approaches to therapeutic targeting of regulators of G protein signaling. Mol Pharmacol. 2010;78:550–557. doi: 10.1124/mol.110.065219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Davydov IV, Varshavsky A. RGS4 is arginylated and degraded by the N-end rule pathway in vitro. J Biol Chem. 2000;275:22931–22941. doi: 10.1074/jbc.M001605200. [DOI] [PubMed] [Google Scholar]
- 75.Lee MJ, Tasaki T, Moroi K, An JY, Kimura S, Davydov IV, Kwon YT. RGS4 and RGS5 are in vivo substrates of the N-end rule pathway. Proc Natl Acad Sci USA. 2005;102:15030–15035. doi: 10.1073/pnas.0507533102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang J, Xie Y, Wolff DW, Abel PW, Tu Y. DHHC protein-dependent palmitoylation protects regulator of G-protein signaling 4 from proteasome degradation. FEBS Lett. 2010;584:4570–4574. doi: 10.1016/j.febslet.2010.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yang J, Kamide K, Kokubo Y, Takiuchi S, Tanaka C, Banno M, Miwa Y, Yoshii M, Horio T, Okayama A, Tomoike H, Kawano Y, Miyata T. Genetic variations of regulator of G-protein signaling 2 in hypertensive patients and in the general population. J Hypertens. 2005;23:1497–1505. doi: 10.1097/01.hjh.0000174606.41651.ae. [DOI] [PubMed] [Google Scholar]
- 78.Bodenstein J, Sunahara RK, Neubig RR. N-terminal residues control proteasomal degradation of RGS2, RGS4, and RGS5 in human embryonic kidney 293 cells. Mol Pharmacol. 2007;71:1040–1050. doi: 10.1124/mol.106.029397. [DOI] [PubMed] [Google Scholar]
- 79.Pedram A, Razandi M, Kehrl J, Levin ER. Natriuretic peptides inhibit G protein activation. Mediation through cross-talk between cyclic GMP-dependent protein kinase and regulator of G protein signaling proteins. J Biol Chem. 2000;275:7365–7372. doi: 10.1074/jbc.275.10.7365. [DOI] [PubMed] [Google Scholar]
- 80.Cunningham ML, Waldo GL, Hollinger S, Hepler JR, Harden TK. Protein kinase C phosphorylates RGS2 and modulates its capacity for negative regulation of Gα11 signaling. J Biol Chem. 2001;276:5438–5444. doi: 10.1074/jbc.M007699200. [DOI] [PubMed] [Google Scholar]
- 81.Tang M, Wang G, Lu P, Karas RH, Aronovitz M, Heximer SP, Kaltenbronn KM, Blumer KJ, Siderovski DP, Zhu Y, Mendelsohn ME. Regulator of G-protein signaling-2 mediates vascular smooth muscle relaxation and blood pressure. Nat Med. 2003;9:1506–1512. doi: 10.1038/nm958. [DOI] [PubMed] [Google Scholar]
- 82.Jones TL. Role of palmitoylation in RGS protein function. Methods Enzymol. 2004;389:33–55. doi: 10.1016/S0076-6879(04)89003-7. [DOI] [PubMed] [Google Scholar]
- 83.Takida S, Fischer CC, Wedegaertner PB. Palmitoylation and plasma membrane targeting of RGS7 are promoted by αo. Mol Pharmacol. 2005;67:132–139. doi: 10.1124/mol.104.003418. [DOI] [PubMed] [Google Scholar]
- 84.Hiol A, Davey PC, Osterhout JL, Waheed AA, Fischer ER, Chen CK, Milligan G, Druey KM, Jones TL. Palmitoylation regulates regulators of G-protein signaling (RGS) 16 function. I. Mutation of amino-terminal cysteine residues on RGS16 prevents its targeting to lipid rafts and palmitoylation of an internal cysteine residue. J Biol Chem. 2003;278:19301–19308. doi: 10.1074/jbc.M210123200. [DOI] [PubMed] [Google Scholar]
- 85.De Vries L, Elenko E, Hubler L, Jones TL, Farquhar MG. GAIP is membrane-anchored by palmitoylation and interacts with the activated (GTP-bound) form of Gαi subunits. Proc Natl Acad Sci USA. 1996;93:15203–15208. doi: 10.1073/pnas.93.26.15203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Druey KM, Ugur O, Caron JM, Chen CK, Backlund PS, Jones TL. Amino-terminal cysteine residues of RGS16 are required for palmitoylation and modulation of Gi- and Gq-mediated signaling. J Biol Chem. 1999;274:18836–18842. doi: 10.1074/jbc.274.26.18836. [DOI] [PubMed] [Google Scholar]
- 87.Castro-Fernandez C, Janovick JA, Brothers SP, Fisher RA, Ji TH, Conn PM. Regulation of RGS3 and RGS10 palmitoylation by GnRH. Endocrinology. 2002;143:1310–1317. doi: 10.1210/endo.143.4.8713. [DOI] [PubMed] [Google Scholar]
- 88.Chidiac P, Roy AA. Activity, regulation, and intracellular localization of RGS proteins. Receptors Channels. 2003;9:135–147. [PubMed] [Google Scholar]
- 89.Sethakorn N, Yau DM, Dulin NO. Non-canonical functions of RGS proteins. Cell Signal. 2010;22:1274–1281. doi: 10.1016/j.cellsig.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Roy AA, Lemberg KE, Chidiac P. Recruitment of RGS2 and RGS4 to the plasma membrane by G proteins and receptors reflects functional interactions. Mol Pharmacol. 2003;64:587–593. doi: 10.1124/mol.64.3.587. [DOI] [PubMed] [Google Scholar]
- 91.Liu Z, Fisher RA. RGS6 interacts with DMAP1 and DNMT1 and inhibits DMAP1 transcriptional repressor activity. J Biol Chem. 2004;279:14120–14128. doi: 10.1074/jbc.M309547200. [DOI] [PubMed] [Google Scholar]
- 92.Boivin B, Lavoie C, Vaniotis G, Baragli A, Villeneuve LR, Ethier N, Trieu P, Allen BG, Hebert TE. Functional β-adrenergic receptor signalling on nuclear membranes in adult rat and mouse ventricular cardiomyocytes. Cardiovasc Res. 2006;71:69–78. doi: 10.1016/j.cardiores.2006.03.015. [DOI] [PubMed] [Google Scholar]
- 93.Wright CD, Chen Q, Baye NL, Huang Y, Healy CL, Kasinathan S, O’Connell TD. Nuclear α1-adrenergic receptors signal activated ERK localization to caveolae in adult cardiac myocytes. Circ Res. 2008;103:992–1000. doi: 10.1161/CIRCRESAHA.108.176024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gobeil F, Fortier A, Zhu T, Bossolasco M, Leduc M, Grandbois M, Heveker N, Bkaily G, Chemtob S, Barbaz D. G-protein-coupled receptors signalling at the cell nucleus: an emerging paradigm. Can J Physiol Pharmacol. 2006;84:287–297. doi: 10.1139/y05-127. [DOI] [PubMed] [Google Scholar]
- 95.Kumar R, Singh VP, Baker KM. The intracellular renin-angiotensin system: implications in cardiovascular remodeling. Curr Opin Nephrol Hypertens. 2008;17:168–173. doi: 10.1097/MNH.0b013e3282f521a8. [DOI] [PubMed] [Google Scholar]
- 96.Heximer SP, Blumer KJ. RGS proteins: Swiss army knives in seven-transmembrane domain receptor signaling networks. Sci STKE. 2007;2007:pe2. doi: 10.1126/stke.3702007pe2. [DOI] [PubMed] [Google Scholar]
- 97.Neitzel KL, Hepler JR. Cellular mechanisms that determine selective RGS protein regulation of G protein-coupled receptor signaling. Semin Cell Dev Biol. 2006;17:383–389. doi: 10.1016/j.semcdb.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 98.Wang Q, Liu M, Mullah B, Siderovski DP, Neubig RR. Receptor-selective effects of endogenous RGS3 and RGS5 to regulate mitogen-activated protein kinase activation in rat vascular smooth muscle cells. J Biol Chem. 2002;277:24949–24958. doi: 10.1074/jbc.M203802200. [DOI] [PubMed] [Google Scholar]
- 99.Niu J, Scheschonka A, Druey KM, Davis A, Reed E, Kolenko V, Bodnar R, Voyno-Yasenetskaya T, Du X, Kehrl J, Dulin NO. RGS3 interacts with 14-3-3 via the N-terminal region distinct from the RGS (regulator of G-protein signalling) domain. Biochem J. 2002;365:677–684. doi: 10.1042/BJ20020390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Popov SG, Krishna UM, Falck JR, Wilkie TM. Ca2+/Calmodulin reverses phosphatidylinositol 3,4,5-trisphosphate-dependent inhibition of regulators of G protein-signaling GTPase-activating protein activity. J Biol Chem. 2000;275:18962–18968. doi: 10.1074/jbc.M001128200. [DOI] [PubMed] [Google Scholar]
- 101.Luo X, Popov S, Bera AK, Wilkie TM, Muallem S. RGS proteins provide biochemical control of agonist-evoked [Ca2+]i oscillations. Mol Cell. 2001;7:651–660. doi: 10.1016/s1097-2765(01)00211-8. [DOI] [PubMed] [Google Scholar]
- 102.Hibino H, Inanobe A, Furutani K, Murakami S, Findlay I, Kurachi Y. Inwardly rectifying potassium channels: their structure, function, and physiological roles. Physiol Rev. 2010;90:291–366. doi: 10.1152/physrev.00021.2009. [DOI] [PubMed] [Google Scholar]
- 103.Nguyen CH, Ming H, Zhao P, Hugendubler L, Gros R, Kimball SR, Chidiac P. Translational control by RGS2. J. Cell Biol. 2009;186:755–765. doi: 10.1083/jcb.200811058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Jin Y, Zhong H, Omnaas JR, Neubig RR, Mosberg HI. Structure-based design, synthesis, and pharmacologic evaluation of peptide RGS4 inhibitors. J Pept Res. 2004;63:141–146. doi: 10.1111/j.1399-3011.2003.00114.x. [DOI] [PubMed] [Google Scholar]
- 105.Dulin NO, Sorokin A, Reed E, Elliott S, Kehrl JH, Dunn MJ. RGS3 inhibits G protein-mediated signaling via translocation to the membrane and binding to Gα11. Mol Cell Biol. 1999;19:714–723. doi: 10.1128/mcb.19.1.714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lan KL, Sarvazyan NA, Taussig R, Mackenzie RG, DiBello PR, Dohlman HG, Neubig RR. A point mutation in Gαo and Gα1 blocks interaction with regulator of G protein signaling proteins. J Biol Chem. 1998;273:12794–12797. doi: 10.1074/jbc.273.21.12794. [DOI] [PubMed] [Google Scholar]
- 107.Fu Y, Zhong H, Nanamori M, Mortensen RM, Huang X, Lan K, Neubig RR. RGS-insensitive G-protein mutations to study the role of endogenous RGS proteins. Methods Enzymol. 2004;389:229–243. doi: 10.1016/S0076-6879(04)89014-1. [DOI] [PubMed] [Google Scholar]
- 108.Rogers JH, Tamirisa P, Kovacs A, Weinheimer C, Courtois M, Blumer KJ, Kelly DP, Muslin AJ. RGS4 causes increased mortality and reduced cardiac hypertrophy in response to pressure overload. J Clin Invest. 1999;104:567–576. doi: 10.1172/JCI6713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Rogers JH, Tsirka A, Kovacs A, Blumer KJ, Dorn GW, 2nd, Muslin AJ. RGS4 reduces contractile dysfunction and hypertrophic gene induction in Gαq overexpressing mice. J Mol Cell Cardiol. 2001;33:209–218. doi: 10.1006/jmcc.2000.1307. [DOI] [PubMed] [Google Scholar]
- 110.Heximer SP, Knutsen RH, Sun X, Kaltenbronn KM, Rhee MH, Peng N, Oliveira-dos-Santos A, Penninger JM, Muslin AJ, Steinberg TH, Wyss JM, Mecham RP, Blumer KJ. Hypertension and prolonged vasoconstrictor signaling in RGS2-deficient mice. J Clin Invest. 2003;111:445–452. doi: 10.1172/JCI15598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Gross V, Tank J, Obst M, Plehm R, Blumer KJ, Diedrich A, Jordan J, Luft FC. Autonomic nervous system and blood pressure regulation in RGS2-deficient mice. Am J Physiol Regul Integr Comp Physiol. 2005;288:R1134–1142. doi: 10.1152/ajpregu.00246.2004. [DOI] [PubMed] [Google Scholar]
- 112.Li H, He C, Feng J, Zhang Y, Tang Q, Bian Z, Bai X, Zhou H, Jiang H, Heximer SP, Qin M, Huang H, Liu PP, Huang C. Regulator of G protein signaling 5 protects against cardiac hypertrophy and fibrosis during biomechanical stress of pressure overload. Proc Natl Acad Sci USA. 2010;107:13818–13823. doi: 10.1073/pnas.1008397107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Brown RD, Ambler SK, Mitchell MD, Long CS. The cardiac fibroblast: therapeutic target in myocardial remodeling and failure. Annu Rev Pharmacol Toxicol. 2005;45:657–687. doi: 10.1146/annurev.pharmtox.45.120403.095802. [DOI] [PubMed] [Google Scholar]
- 114.Kawano H, Do YS, Kawano Y, Starnes V, Barr M, Law RE, Hsueh WA. Angiotensin II has multiple profibrotic effects in human cardiac fibroblasts. Circulation. 2000;101:1130–1137. doi: 10.1161/01.cir.101.10.1130. [DOI] [PubMed] [Google Scholar]
- 115.Hafizi S, Wharton J, Chester AH, Yacoub MH. Profibrotic effects of endothelin-1 via the ETA receptor in cultured human cardiac fibroblasts. Cell Physiol Biochem. 2004;14:285–292. doi: 10.1159/000080338. [DOI] [PubMed] [Google Scholar]
- 116.Porter KE, Turner NA. Cardiac fibroblasts: at the heart of myocardial remodeling. Pharmacol Ther. 2009;123:255–278. doi: 10.1016/j.pharmthera.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 117.Takeda N, Manabe I, Uchino Y, Eguchi K, Matsumoto S, Nishimura S, Shindo T, Sano M, Otsu K, Snider P, Conway SJ, Nagai R. Cardiac fibroblasts are essential for the adaptive response of the murine heart to pressure overload. J Clin Invest. 2010;120:254–265. doi: 10.1172/JCI40295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Fu Y, Huang X, Zhong H, Mortensen RM, D’Alecy LG, Neubig RR. Endogenous RGS proteins and Gα subtypes differentially control muscarinic and adenosine-mediated chronotropic effects. Circ Res. 2006;98:659–666. doi: 10.1161/01.RES.0000207497.50477.60. [DOI] [PubMed] [Google Scholar]
- 119.Fu Y, Huang X, Piao L, Lopatin AN, Neubig RR. Endogenous RGS proteins modulate SA and AV nodal functions in isolated heart: implications for sick sinus syndrome and AV block. Am J Physiol Heart Circ Physiol. 2007;292:H2532–2539. doi: 10.1152/ajpheart.01391.2006. [DOI] [PubMed] [Google Scholar]
- 120.Posokhova E, Wydeven N, Allen KL, Wickman K, Martemyanov KA. RGS6/Gβ5 complex accelerates IKACh gating kinetics in atrial myocytes and modulates parasympathetic regulation of heart rate. Circ Res. 2010;107:1350–1354. doi: 10.1161/CIRCRESAHA.110.224212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Yang J, Huang J, Maity B, Gao Z, Lorca RA, Gudmundsson H, Li J, Stewart A, Swaminathan PD, Ibeawuchi SR, Shepherd A, Chen CK, Kutschke W, Mohler PJ, Mohapatra DP, Anderson ME, Fisher RA. RGS6, a modulator of parasympathetic activation in heart. Circ Res. 2010;107:1345–1349. doi: 10.1161/CIRCRESAHA.110.224220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Tuomi JM, Chidiac P, Jones DL. Evidence for enhanced M3 muscarinic receptor function and sensitivity to atrial arrhythmia in the RGS2-deficient mouse. Am J Physiol Heart Circ Physiol. 2009;298:H554–H561. doi: 10.1152/ajpheart.00779.2009. [DOI] [PubMed] [Google Scholar]
- 123.Waterson RE, Thompson C, Mabe N, Kaur K, Talbot JN, Neubig RR, Rorabaugh BR. Gαi2-mediated protection from ischemic injury is modulated by endogenous RGS proteins in the mouse heart. Cardiovasc Res. 2011 doi: 10.1093/cvr/cvr054. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Fredriksson R, Lagerstrom MC, Lundin LG, Schioth HB. The G-protein-coupled receptors in the human genome form five main families. Phylogenetic analysis, paralogon groups, and fingerprints. Mol Pharmacol. 2003;63:1256–1272. doi: 10.1124/mol.63.6.1256. [DOI] [PubMed] [Google Scholar]
- 125.Hopkins AL, Groom CR. The druggable genome. Nat Rev Drug Discov. 2002;1:727–730. doi: 10.1038/nrd892. [DOI] [PubMed] [Google Scholar]
- 126.Tang CM, Insel PA. GPCR expression in the heart; “new” receptors in myocytes and fibroblasts. Trends Cardiovasc Med. 2004;14:94–99. doi: 10.1016/j.tcm.2003.12.007. [DOI] [PubMed] [Google Scholar]
- 127.Wettschureck N, Offermanns S. Mammalian G proteins and their cell type specific functions. Physiol. Rev. 2005;85:1159–1204. doi: 10.1152/physrev.00003.2005. [DOI] [PubMed] [Google Scholar]
- 128.Kamal FA, Smrcka AV, Blaxall BC. Taking the heart failure battle inside the cell: Small molecule targeting of Gβγ subunits. J Mol Cell Cardiol. 2011 doi: 10.1016/j.yjmcc.2011.01.006. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Semplicini A, Lenzini L, Sartori M, Papparella I, Calo LA, Pagnin E, Strapazzon G, Benna C, Costa R, Avogaro A, Ceolotto G, Pessina AC. Reduced expression of regulator of G-protein signaling 2 (RGS2) in hypertensive patients increases calcium mobilization and ERK1/2 phosphorylation induced by angiotensin II. J Hypertens. 2006;24:1115–1124. doi: 10.1097/01.hjh.0000226202.80689.8f. [DOI] [PubMed] [Google Scholar]
- 130.Calo LA, Pagnin E, Davis PA, Sartori M, Ceolotto G, Pessina AC, Semplicini A. Increased expression of regulator of G protein signaling-2 (RGS-2) in Bartter’s/Gitelman’s syndrome. A role in the control of vascular tone and implication for hypertension. J Clin Endocrinol Metab. 2004;89:4153–4157. doi: 10.1210/jc.2004-0498. [DOI] [PubMed] [Google Scholar]
- 131.Young KH, Wang Y, Bender C, Ajit S, Ramirez F, Gilbert A, Nieuwenhuijsen BW. Yeast-based screening for inhibitors of RGS proteins. Methods Enzymol. 2004;389:277–301. doi: 10.1016/S0076-6879(04)89017-7. [DOI] [PubMed] [Google Scholar]
- 132.Roman DL, Talbot JN, Roof RA, Sunahara RK, Traynor JR, Neubig RR. Identification of small-molecule inhibitors of RGS4 using a high-throughput flow cytometry protein interaction assay. Mol Pharmacol. 2007;71:169–175. doi: 10.1124/mol.106.028670. [DOI] [PubMed] [Google Scholar]
- 133.Wang Y, Lee Y, Zhang J, Young KH. Identification of peptides that inhibit regulator of G protein signaling 4 function. Pharmacology. 2008;82:97–104. doi: 10.1159/000138387. [DOI] [PubMed] [Google Scholar]
- 134.Sjoegren B, Blazer LL, Neubig RR. Regulators of G protein signaling proteins as targets for drug discovery. Prog Mol Biol Transl Sci. 2010;91:81–119. doi: 10.1016/S1877-1173(10)91004-1. [DOI] [PubMed] [Google Scholar]
- 135.Zhong H, Neubig RR. Regulator of G protein signaling proteins: novel multifunctional drug targets. J Pharmacol Exp Ther. 2001;297:837–845. [PubMed] [Google Scholar]
- 136.Roman DL, Blazer LL, Monroy CA, Neubig RR. Allosteric inhibition of the regulator of G protein signaling-Gα protein-protein interaction by CCG-4986. Mol Pharmacol. 2010;78:360–365. doi: 10.1124/mol.109.063388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Blazer LL, Roman DL, Chung A, Larsen MJ, Greedy BM, Husbands SM, Neubig RR. Reversible, allosteric small-molecule inhibitors of regulator of G protein signaling proteins. Mol Pharmacol. 2011;78:524–533. doi: 10.1124/mol.110.065128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ishii M, Fujita S, Yamada M, Hosaka Y, Kurachi Y. Phosphatidylinositol 3,4,5-trisphosphate and Ca2+/calmodulin competitively bind to the regulators of G-protein-signalling (RGS) domain of RGS4 and reciprocally regulate its action. Biochem J. 2005;385:65–73. doi: 10.1042/BJ20040404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Zheng B, De Vries L, Farquhar M Gist. Divergence of RGS proteins: evidence for the existence of six mammalian RGS subfamilies. Trends Biochem Sci. 1999;24:411–414. doi: 10.1016/s0968-0004(99)01474-7. [DOI] [PubMed] [Google Scholar]
- 140.von Buchholtz L, Elischer A, Tareilus E, Gouka R, Kaiser C, Breer H, Conzelmann S. RGS21 is a novel regulator of G protein signalling selectively expressed in subpopulations of taste bud cells. Eur J Neurosci. 2004;19:1535–1544. doi: 10.1111/j.1460-9568.2004.03257.x. [DOI] [PubMed] [Google Scholar]
- 141.Li X, Chen L, Ji C, Liu B, Gu J, Xu J, Zou X, Gu S, Mao Y. Isolation and expression pattern of RGS21 gene, a novel RGS member. Acta Biochim Pol. 2005;52:943–946. [PubMed] [Google Scholar]
- 142.Harris IS, Treskov I, Rowley MW, Heximer S, Kaltenbronn K, Finck BN, Gross RW, Kelly DP, Blumer KJ, Muslin AJ. G-protein signaling participates in the development of diabetic cardiomyopathy. Diabetes. 2004;53:3082–3090. doi: 10.2337/diabetes.53.12.3082. 2004. [DOI] [PubMed] [Google Scholar]
- 143.Tokudome T, Kishimoto I, Horio T, Arai Y, Schwenke DO, Hino J, Okano I, Kawano Y, Kohno M, Miyazato M, Nakao K, Kangawa K. Regulator of G-protein signaling subtype 4 mediates antihypertrophic effect of locally secreted natriuretic peptides in the heart. Circulation. 2008;117:2329–2339. doi: 10.1161/CIRCULATIONAHA.107.732990. [DOI] [PubMed] [Google Scholar]
- 144.Nisancioglu MH, Mahoney WM, Jr., Kimmel DD, Schwartz SM, Betsholtz C, Genove G. Generation and characterization of rgs5 mutant mice. Mol Cell Biol. 2008;28:2324–2331. doi: 10.1128/MCB.01252-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Cho H, Park C, Hwang IY, Han SB, Schimel D, Despres D, Kehrl JH. Rgs5 targeting leads to chronic low blood pressure and a lean body habitus. Mol Cell Biol. 2008;28:2590–2597. doi: 10.1128/MCB.01889-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Huang X, Fu Y, Charbeneau RA, Saunders TL, Taylor DK, Hankenson KD, Russell MW, D’Alecy LG, Neubig RR. Pleiotropic phenotype of a genomic knock-in of an RGS-insensitive G184S Gnai2 allele. Mol. Cell. Biol. 2006;26:6870–6879. doi: 10.1128/MCB.00314-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Blumer JB, Smrcka AV, Lanier SM. Mechanistic pathways and biological roles for receptor-independent activators of G-protein signaling. Pharmacology & Therapeutics. 2007;113:488–506. doi: 10.1016/j.pharmthera.2006.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Noor N, Patel CB, Rockman HA. β-arrestin: a signaling molecule and potential therapeutic target for heart failure. J Mol Cell Cardiol. 2011 doi: 10.1016/j.yjmcc.2010.11.005. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Buenemann M, Frank M, Lohse MJ. Gi protein activation in intact cells involves subunit rearrangement rather than dissociation. Proc Natl Acad Sci USA. 2003;100:16077–16082. doi: 10.1073/pnas.2536719100. [DOI] [PMC free article] [PubMed] [Google Scholar]