Abstract
The molecular mechanisms that modulate the activity of the signal transducers and activators of transcription 5 (Stat5) during the progression of breast cancer remain elusive. Here, we present evidence that the calcineurin/nuclear factor of activated T cells (NFAT) pathway negatively regulates the activation of Stat5, and vice versa in breast cancer. NFAT1 interacts with Stat5 in breast cancer cells, and their physical association is mediated by the DNA binding and transactivation domains of Stat5. Ectopically expressed NFAT1 is capable of inhibiting Stat5-dependent functions, including Stat5 transactivation, Stat5-mediated transcription of the downstream target gene expression, and binding of Stat5a to the Stat5 target promoter. By contrast, overexpression of a selective NFAT inhibitor VIVIT reversed NFAT1-mediated suppression of Stat5-dependent gene expression, whereas silencing of NFAT1 through RNA interference enhanced prolactin-induced, Stat5-mediated gene transcription, and breast cancer cell proliferation. A reciprocal inhibitory effect of Stat5 activity on NFAT1 signaling was also observed, implying these two signaling cascades antagonize each other in breast cancer. Importantly, analysis of a matched breast cancer progression tissue microarray revealed a negative correlation between levels of NFAT1 and Stat5 (pY694) during the progression of breast cancer. Taken together, these studies highlight a novel negative cross talk between the NFAT1- and Stat5-signaling cascades that may affect breast tumor formation, growth, and metastasis.
The signal transducers and activators of transcription (Stat) proteins play a central role in the regulation of a variety of fundamental biological processes (1, 2). Stat5, one member of the Stat family proteins, consists of two highly related homologs, STAT5a and b, and is crucial for lymphocyte and mammary gland development (3, 4). Stat5 also participates in oncogenesis by promoting cell proliferation and/or preventing apoptosis (5). In breast cancer, data from a large set of primary malignancies and lymph node metastasis revealed that Stat5 phosphorylation and nuclear localization was associated with favorable-prognosis breast cancers, whereas a loss of Stat5 activation was observed during tumor progression (6). Thus, Stat5 may play a dual role in breast cancer functioning as a growth promoter in the early-stage breast lesions, while suppressing metastasis in its later stages (7). A tumor-promoting role for Stat5 has been supported by in vitro studies that Stat5 induced the expression of cyclin D1, a critical regulator of cell cycle progression (8). Additional evidence in support of this are in vivo mouse models of breast cancer in which hemizygous deletion of the Stat5a demonstrated a significant reduction of tumor incidence, delayed tumor formation, and reduced tumor size in SV40-T antigen transgenic mice (9). With regard to its metastasis-suppressive role in breast cancer, Stat5 has been demonstrated to inhibit breast cancer cell migration, invasion, and matrix metalloproteinase secretion and to up-regulate cell surface E-cadherin and β-catenin (10). However, the molecular mechanisms responsible for this dual nature of Stat5 activation in breast cancer have not been fully elucidated.
With the use of a novel screening technology, namely transcription factor (TF)-TF array, we have identified several potential transcription factors that interact with Stat5a in the T47D human breast cancer cells. One of them, the pro-oncogene c-myb, has been characterized as a positive regulator for Stat5 activity (11). Another novel Stat5 interactor that was identified from this screen was the nuclear factor of activated T cells (NFAT) 1, a member of the family of NFAT transcription factors. In contrast to Stat5, which functions as a promoter of tumor growth and a metastasis suppressor in breast cancer, NFAT1 has opposite effects, inhibiting tumor growth and promoting tumor metastasis (12, 13). The opposing actions of these transcription factors prompted us to investigate whether NFAT1 could modulate Stat5 activity, or vice versa in breast cancer.
In this study, we demonstrated that NFAT1 negatively regulates Stat5 activity, as evidenced by decreased tyrosine phosphorylation of Stat5, reduced expression from Stat5-dependent synthetic and natural gene-based luciferase reporters, and attenuated binding of Stat5 to the promoter of target gene. We also observed a reciprocal inhibitory effect of Stat5 activity on NFAT1 signaling in breast cancer cells. Furthermore, analysis of a matched breast cancer progression tissue microarray (TMA) by anti-NFAT1 or tyrosine-phosphorylated Stat5 immunohistochemistry demonstrated a negative correlation between levels of NFAT1 and Stat5 (pY694) during the progression of breast cancer. Thus, these data reveal that NFAT1-Stat5 pathways antagonize each other in breast cancer, and their negative cross talk may contribute to the pathogenesis of breast cancer.
Results
Stat5 interacts with NFAT1
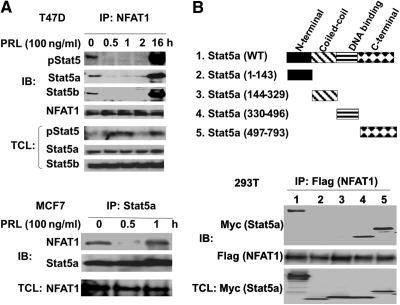
To screen for transcription factors that could be involved in the regulation of Stat5 function, we performed a TF-TF screen as previously reported (11). This led to the identification of NFAT1 as a novel Stat5a-interacting protein. To further analyze and confirm this Stat5a-NFAT1 interaction, coimmunoprecipitation analysis was performed. Whole-cell lysates derived from resting and prolactin (PRL)-stimulated T47D cells were subjected to anti-NFAT1 immunoprecipitation followed by immunoblotting with anti-pStat5, Stat5a, and Stat5b antibodies. As shown in Fig. 1A (upper panel), a complex of NFAT1 and Stat5 was observed in the resting cells. This complex dissociated when cells were stimulated with PRL and then markedly reassociated after prolonged PRL stimulation. These findings were repeated in a second breast cancer cell line, MCF7 (Fig. 1A, lower panel), which also demonstrated the reversible disassociation between NFAT1 and Stat5, but with reassociation kinetics faster than T47D. Surprisingly, as shown in Fig. 1A, the corresponding NFAT1 interaction with pStat5 was observed in the resting cells [possibly due to PRL secreted in an auto/paracrine manner from breast cancer cells (14)] and in prolonged PRL-stimulated cells.
Fig. 1.
NFAT1 interacts with Stat5. A, NFAT1 is associated with Stat5a and 5b. Overnight serum-starved T47D cells (upper panel) or MCF7 cells overexpressing NFAT1 (lower panel) were stimulated with PRL as indicated. Cell lysates were immunoprecipitated with anti-NFAT1 or anti-pStat5/Stat5a/Stat5b antibody, and analyzed by immunoblot analyses with the indicated antibodies. B, NFAT1 is associated with Stat5a through its DNA-binding and C-terminal domains. HEK293T cells were cotransfected with Stat5a-myc deletion mutants (upper panel) and NFAT1 overexpression construct. Transfectant lysates were immunoprecipitated with anti-NFAT1 antibody and sequentially immunoblotted with anti-myc (Stat5a mutants) and anti-Flag (NFAT1) antibodies (lower panel). IP, Immunoprecipitation; IB, immunoblotting; TCL, total cell lysates.
Stat5 consists of N-terminal, coiled-coil, central DNA-binding, and C-terminal (including SH2 and transactivation) domains (1). To delineate the domains within Stat5a that were responsible for the interaction with NFAT1, a series of Stat5a mutants carrying a C-terminal myc tag were transfected into human embryonic kidney (HEK)293T cells and tested for their interactions with NFAT1 using coimmunoprecipitation assay. As shown in Fig. 1B, the Stat5a mutants with the DNA-binding or C-terminal domains were associated with NFAT1, whereas the N-terminal and coiled-coil domains were incapable of interacting with NFAT1. These data indicate that the DNA-binding and C-terminal domains of Stat5a contribute to its interaction with NFAT1.
NFAT1 inhibits PRL-induced tyrosine phosphorylation of nuclear Stat5
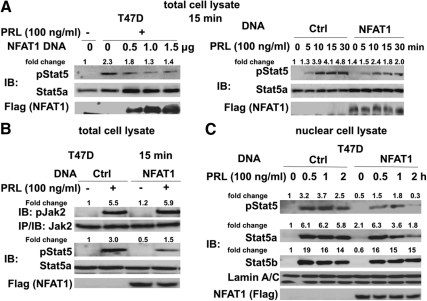
The identification of the NFAT1 and Stat5a association prompted us to investigate the role of NFAT1 in regulating Stat5 functions. First, we tested the effect of NFAT1 overexpression on the tyrosine phosphorylation of Stat5. To this end, cells were transfected with flag-tagged NFAT1, and transfectants were left unstimulated or stimulated with PRL. Whole-cell lysates were subjected to immunoblotting with antiphosphotyrosine Stat5 and anti-Stat5a antibodies. As shown in Fig. 2A, tyrosine phosphorylation of Stat5 was reduced in a dose (left panel)- and time (right panel)-dependent pattern in T47D cells transiently transfected with a NFAT1 expression construct.
Fig. 2.
Ectopically expressed NFAT1 inhibits PRL-induced tyrosine phosphorylation of Stat5. A, dose- and time-dependent inhibition of PRL-induced Stat5 phosphorylation by NFAT1. T47D cells were transfected with control (Ctrl) vector or NFAT1 overexpression construct as indicated. After 24-h starvation, transfectants were treated with PRL as indicated time. Transfectant lysates were subjected to immunoblotting with antibodies as indicated. B, Jak2 tyrosine phosphorylation is not affected by NFAT1. T47D cells were treated as described above. Transfectant lysates and immunoprecipitates were probed in immunoblots with the indicated antibodies. C, NFAT1 blocks PRL-induced nuclear accumulation of Stat5a but not Stat5b. T47D cells were transfected with Ctrl vector or NFAT1 overexpression construct, starved overnight, and stimulated with PRL at indicated time. Nuclear fraction of transfectant lysates was purified and analyzed by Western blot analysis with antibodies as indicated. Expression levels of transfected NFAT1 were assessed by reprobing the membrane with anti-Flag antibody, and nuclear fractions were verified by probing fractions with antilamin A/C antibody. IB, Immunoblotting.
To further rule out the possibility that NFAT1 inhibited Stat5 activity by blocking upstream signals from the Jak2 tyrosine kinase, the effects of NFAT1 overexpression on PRL-induced Jak2 tyrosine phosphorylation were examined. As shown in Fig. 2B, PRL-induced Jak2 phosphorylation was not reduced by NFAT1 overexpression.
To determine whether NFAT1 overexpression could affect Stat5 nuclear translocation, T47D cells were stimulated with PRL for 0–2 h, and purified nuclear fractions were analyzed by Western blot analysis. As shown in Fig. 2C, ectopic expression of NFAT1 protein resulted in decreased nuclear Stat5 phosphorylation at all time points and a subsequent reduction in Stat5a nuclear accumulation. Interestingly, PRL-stimulated Stat5b intranuclear accumulation was not affected by NFAT1 overexpression, although the association kinetics of Stat5a and Stat5b with NFAT1 upon PRL stimulation was nearly identical.
NFAT1 blocks Stat5-dependent functions
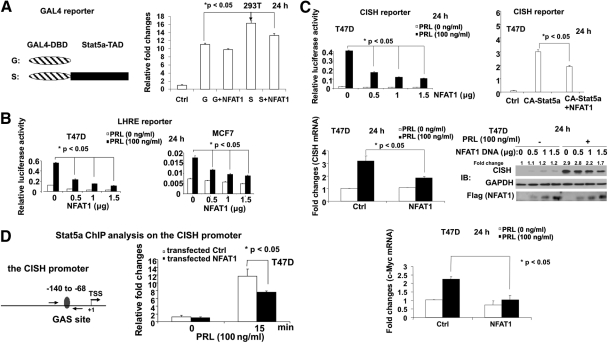
The activation of Stat5 can be measured by its tyrosine phosphorylation; thus we reasoned that NFAT1 could negatively regulate Stat5 activity. To test this, several luciferase-based reporter assays, as shown in Fig. 3, were performed. To test the direct effects of NFAT1 overexpression on Stat5 transactivation potential, a GAL4-based luciferase reporter assay was conducted in which the transactivation domain of Stat5a was fused to the GAL4 DNA-binding domain (termed “GAL4-Stat5TAD”; left panel in Fig. 3A). Transfection of the GAL4-Stat5TAD construct in HEK293T cells resulted in a significant increase of gene expression from the GAL-luciferase reporter, which contained four copies of the GAL4-binding site linked to the luciferase gene. In contrast, cotransfection of NFAT1 with the GAL4-Stat5TAD construct significantly repressed this activity (right panel in Fig. 3A).
Fig. 3.
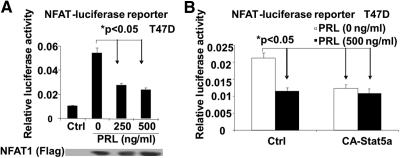
NFAT1 overexpression inhibits Stat5-mediated functions. A, NFAT1 represses Stat5 transactivation by targeting its transactivation domain. HEK293T cells were cotransfected with GAL4-Stat5TAD (transactivation domain of Stat5a fused to the GAL4 DNA-binding domain), or GAL-DBD control construct, GAL4-luc reporter, and NFAT1 overexpression construct [or control (Ctrl) vectors]. After 2 d, transfectants were lysed for luciferase assay. B, NFAT1 represses Stat5-mediated transcriptional activation. T47D (left panel) and MCF7 (right panel) cells were cotransfected with LHRE luciferase reporter with the increasing amount of NFAT1 overexpression construct as indicated. After 24 h stimulation with PRL, transfectants were lysed and assayed for luciferase activity. When normalized to luciferase activity noted in the nonstimulated MCF7 cells, PRL-induced reporter activity remained significant (P < 0.05). C, NFAT1 blocks the Stat5 target gene expression-CISH. Upper panel illustrates the CISH luciferase reporter assay: T47D cells were cotransfected with CISH reporter with the increasing amount of NFAT1 (upper left panel) or CA-Stat5a (upper right panel) overexpression constructs as indicated. Transfectants were lysed for luciferase assay. Middle-lower panel shows the CISH/c-Myc mRNA and protein analysis: T47D cells were transfected with Ctrl vector or NFAT1 overexpression construct as indicated. After 24-h treatment with PRL, changes in CISH and c-Myc mRNA levels were analyzed by real-time quantitative RT-PCR (middle left and lower panels), and altered CISH protein expression was determined by Western blot analysis with the indicated antibodies (middle right panel). When normalized to luciferase activity noted in the nonstimulated cells, PRL-induced reporter and gene expression remained significant (P < 0.05; upper and middle left and lower panels). D, NFAT1 inhibits Stat5a DNA binding to the CISH gene promoter. T47D cells were transfected with Ctrl vector or NFAT1 overexpression construct as indicated. Overnight serum-starved transfectants were stimulated with PRL for 15 min, and then subjected to ChIP analysis using Stat5a-specific antibody. Amplicon (left panel) designated for the CISH gene promoter was analyzed by real-time quantitative PCR. The values of the ChIP data were normalized to that of the input and shown as the fold increase compared with untreated control (right panel).
As expected, a similar but more pronounced repressive effect of NFAT1 on transcription was observed when using a synthetic lactogenic hormone response element (LHRE)-based reporter, which contains five tandem Stat5-binding elements (Fig. 3B), or a natural Stat5 target (CISH, cytokine-inducible SH2-containing protein) promoter-based reporter when cells were either stimulated with PRL (Fig. 3C, upper left panel) or transfected with CA-Stat5a (a constitutively active form of Stat5a, not dependent on Jak2 phosphorylation) (Fig. 3C, upper right panel). Thus, the inhibition of Stat5 activity in this setting may result from actions of NFAT that include and do not include the repression of Stat5 phosphorylation.
To further explore the regulatory role of NFAT1 on Stat5-mediated gene expression in vitro, we examined the effects of NFAT1 on the expression of the Stat5 target genes such as CISH and c-Myc. Given the above data, we hypothesized that NFAT1 overexpression should result in decreased gene expression of CISH and c-Myc through inhibiting Stat5 activation. Indeed, as shown in Fig. 3C (middle and lower panels), NFAT1 repressed endogenous CISH and c-Myc gene expression as measured by mRNA and/or protein in T47D cells. It is important to note, that although NFAT overexpression did modestly reduce basal levels from Stat5-responsive reporters and endogenous genes, a significantly greater fold reduction in PRL-induced expression was observed (i.e. even after normalization; P < 0.05).
To demonstrate that the above NFAT1 action on CISH gene expression may be the result of attenuation of the Stat5 binding to its cognate elements on the CISH promoter, binding of Stat5a to the CISH promoter in vivo was analyzed by chromatin immunoprecipitation (ChIP) analysis with a specific antibody against Stat5a. Real-time PCR analysis of these ChIP revealed that the recruitment of Stat5 to the CISH promoter was significantly blocked by transiently expressed NFAT1 (Fig. 3D). A similar inhibition of Stat5 binding to the γ-interferon activated sequence 2 DNA element from the CISH promoter by NFAT1 was also demonstrated by EMSA (data not shown). Taken together, these results suggest that NFAT1 antagonizes Stat5-mediated functions, including Stat5 transactivation, Stat5-mediated transcription of the downstream target gene expression, and Stat5 binding to the Stat5 target promoter.
VIVIT treatment reverses NFAT1-mediated suppression of Stat5 activity and NFAT1 knockdown enhances PRL-induced, Stat5-mediated gene expression and cell proliferation
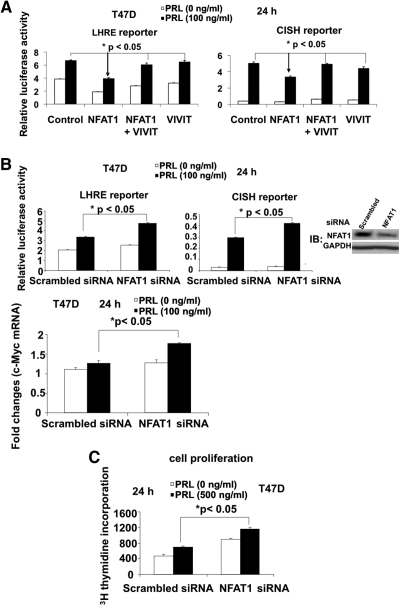
To determine whether the activation of NFAT1 is needed for its negative effects on Stat5 activation, T47D cells were cotransfected with LHRE reporter (or CISH reporter) and NFAT1 overexpression construct, together with or without a construct expressing VIVIT, a peptide aptamer derived from NFAT, which specifically inhibits the activation of NFAT by disrupting the NFAT-docking site of calcineurin (15). This aptamer has been shown to inhibit NFAT1 dephosphorylation by calcineurin, thereby blocking its activation. As shown in Fig. 4A, NFAT1-mediated repression of Stat5-dependent transcription was significantly reversed by VIVIT overexpression. These data indicated that the activation of NFAT1 is required to negatively regulate Stat5 function.
Fig. 4.
Ablation of NFAT1 activity by VIVIT reverses NFAT1-mediated suppression of Stat5 transcriptional activation, and siRNA-mediated depletion of NFAT1 protein enhances PRL-induced, Stat5-mediated transcription and breast cancer cell proliferation. A, Overexpression of VIVIT completely reverses NFAT1-mediated inhibition of Stat5a transcriptional activation. T47D cells were cotransfected with LHRE reporter (left panel) or CISH reporter (right panel) with Ctrl vector or NFAT1 overexpression construct (± VIVIT overexpression construct) as indicated. After stimulation with PRL for 24 h, luciferase assays were performed. B, siRNA-mediated NFAT1 knockdown enhances PRL-induced, Stat5-mediated transcription. T47D cells were transfected with NFAT1 siRNA or scrambled control siRNA. Transfectants were transfected 24 h after transfection with LHRE reporter (upper leftpanel) or CISH reporter (upper middle panel) as indicated. After 24 h stimulation with PRL, transfectants were assayed for luciferase activity. The efficiency of NFAT1 knockdown was evaluated by Western blot analysis with anti-NFAT1 antibody (upper right panel). Changes in c-Myc mRNA level were determined by real-time quantitative RT-PCR (lower panel). C, NFAT1 knockdown enhances PRL-induced proliferation of T47D cells. T47D cells were transfected with NFAT1 siRNA or scrambled control siRNA. Transfectants were stimulated with PRL (500 ng/ml) for 3 d and [3H] thymidine incorporation was measured by radiometry. When the data from Fig. 4, A–C, were normalized (induced/uninduced), comparable fold increases were noted in the VIVIT or siRNA populations. IB, Immunoblotting.
To further examine the effects of reducing the levels of NFAT1 protein on Stat5 functions, T47D cells were transfected with NFAT1 small interfering RNA (siRNA), and the reduced NFAT1 protein was confirmed by immunoblotting with anti-NFAT1 antibody (upper right panel in Fig. 4B). As shown in Fig. 4B (upper and lower panels), the suppression of endogenous NFAT1 protein by siRNA led to a significant increase of Stat5-mediated gene expression as measured by LHRE, CISH reporter, and c-Myc mRNA expression analysis. These findings further underline a negative regulatory role for NFAT1 in regulating Stat5-mediated transcription.
To evaluate the biological consequence of NFAT1-mediated suppression of Stat5 signaling, analysis of effects of NFAT1 knockdown on breast cancer cell proliferation was performed. As shown in Fig. 4C, siRNA-mediated reduction of NFAT1 protein resulted in a significant enhancement of both basal and PRL-induced cell proliferation in T47D cells.
When the data from Fig. 4 are analyzed as a whole, it is interesting to observe that inhibition of NFAT activity/expression altered both basal and PRL-induced expression and proliferation to a comparable degree. There are possible explanations for this that include: 1) the previously demonstrated autocrine/paracrine elaboration of PRL by both T47D and MCF7 make them susceptible to comparable reductions in both basal and PRL-augmented states, and/or 2) because NFAT is associated with Stat5 in the uninduced state, its loss could alter basal promoter function from Stat5-driven genes (i.e. NFAT may act in a context-dependent manner as a coactivator vs. corepressor; NFAT may function differentially on monomeric vs. dimeric Stat5, etc.).
Stat5 inhibits NFAT1 signaling
Given that NFAT1 suppresses Stat5-dependent functions, we next investigated whether Stat5 reciprocally inhibited NFAT1 signaling. For this, T47D cells were transiently transfected with NFAT1 overexpression construct and NFAT-responsive luciferase reporter, which contains tandem repeats of the NFAT-binding elements, and overnight-starved transfectants were stimulated with PRL. As shown in Fig. 5A, PRL treatment led to the inhibition of NFAT-responsive luciferase reporter activity in a dose-dependent manner. To further confirm that the repression of NFAT1 signaling depends on PRL-induced Stat5 activation, T47D cells were transfected with CA-Stat5a (a constitutively active form of Stat5a) in addition to NFAT-responsive luciferase reporter and NFAT1 overexpression construct. As demonstrated in Fig. 5B, transiently expressed CA-Stat5a protein alone is sufficient to block gene expression from NFAT-responsive luciferase reporter. These data suggested that Stat5 activation reciprocally suppresses NFAT1 signaling.
Fig. 5.
Stat5 inhibits NFAT1 signaling. A, PRL inhibits NFAT1-mediated transcriptional activation in a dose-dependent manner. T47D cells were cotransfected with NFAT luciferase reporter and control (Ctrl) vector or NFAT1 overexpression construct as indicated. Transfectants were treated for 24 h with PRL as indicated. Luciferase activity was measured. B, The inhibition of NFAT1-dependent transcription depends on Stat5 activity. T47D cells were cotransfected with NFAT luciferase reporter, and Ctrl, NFAT1, or CA-Stat5a overexpression construct. After 24 h stimulation with PRL, transfectants were lysed for luciferase assay.
Negative correlation between levels of NFAT1 and Stat5 (pY694) in a matched breast cancer progression TMA
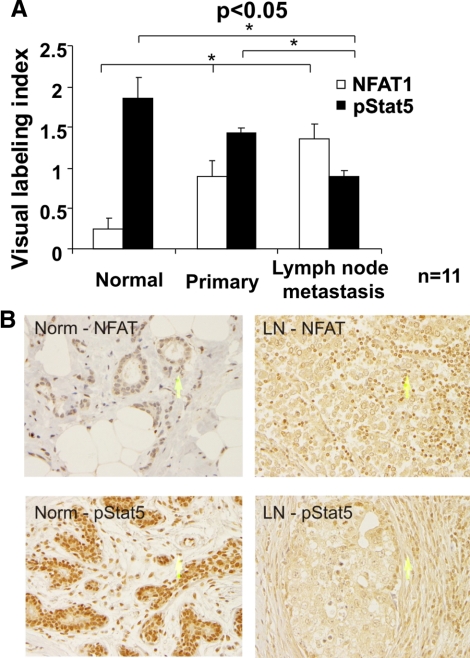
The study presented above outlines a novel negative regulatory cross talk between the NFAT1- and Stat5-signaling pathways in human breast cancer cells. To examine the relevance of this NFAT-Stat5 transcriptional complex in breast cancer patients, expression levels of NFAT1- and tyrosine-phosphorylated form of Stat5 were evaluated by immunohistochemical staining on tissue sections from a breast cancer progression TMA with patient-matched normal adjacent tissue, primary tumor, and metastatic lymph nodes. As shown in Fig. 6, A and B, expression of pStat5 in lymph node metastasis was significantly lower than that in normal adjacent tissue and primary tumor, whereas a significant increase of NFAT1 expression was observed in lymph node metastasis vs. normal adjacent tissue and primary tumor. Thus, coupled with the above in vitro studies, these in vivo association data provide further evidence for a negative regulatory cross talk between the NFAT1- and Stat5-signaling cascades in breast cancer, as seen in the inverse expression of these transcription factors during the pathogenesis of this disease.
Fig. 6.
Stat5 activation is negatively correlated with NFAT1 protein in a cohort of primary breast carcinoma specimens with adjacent normal tissues and matched metastasis in lymph node. A, The expression of pStat5 and NFAT1 in primary breast tumor, adjacent normal tissue, and matched lymph node metastasis was evaluated by immunohistochemical analysis. The immunostaining intensity was scored as 0 (absent), 1 (dim), 2 (bright), and 3 (very bright). Visual labeling index is expressed as the average immunostaining scores ± se for each group. B, Photomicrographs of antiphospho-Stat5 and anti-NFAT1 immunohistochemistry of normal (Norm) and metastatic breast cancer to lymph node (LN). Magnification, ×400.
Discussion
The contributions of Stat transcriptional factors to cytokine and growth factor signaling in normal biological processes are well documented (1, 2, 16). Beyond that, recent studies have demonstrated that these proteins also play an important role in many pathological processes including oncogenesis (5). In breast cancer, it seems that Stat5 exhibits a dual role, promoting breast tumor growth and inhibiting metastasis (6–10). To understand molecular mechanisms at the transcriptional level that modulate this dual nature of Stat5 function during the progression of breast cancer, four central questions should be addressed: 1) what are the novel transcription factors that potentially interact with Stat5 in breast cancer? 2) how do these transcription factors regulate Stat5 activity, and do they function as corepressors or coactivators, or recruit other chromatin-remodeling factors? 3) what are the dynamic associations of these transcription factors with Stat5-responsive promoters during gene activation and repression, and how does this affect tumor growth or metastasis suppression? 4) what are the correlations between the levels of active Stat5 and these transcription factors in normal breast tissues, primary breast carcinomas, and metastasis in lymph nodes or other organs?
Transcription factors can control gene expression by remodeling chromatin structure through recruiting coactivators and/or corepressors. Stat5 has been shown to interact with other transcription factors to synergistically enhance Stat5 target gene expression, such as c-Myb (11), Oct-1 (17), centrosomal P4. 1-associated protein (18), and Nmi (19). On the other hand, Runx and silencing mediator of retinoid and thyroid hormone receptor have been shown to suppress Stat5 target gene expression by binding to Stat5 as corepressors (20, 21). The role of many of these coactivators and corepressors in cancer, however, remains to be established. To analyze the full spectrum of protein interactions that may regulate Stat5 activity, we performed a TF-TF array analysis in the T47D human breast cancer cells that identified NFAT1 as a potential interactor. To confirm this, combined cellular, biochemical, and immunohistochemical studies validated this interaction and revealed a novel negative cross talk between the NFAT1- and Stat5-signaling pathways. First, the association kinetics data implied that NFAT1 must be dissociated from Stat5 for the full induction of Stat5 signaling, and that the reassembly of the NFAT1/Stat5 complex may play a role in controlling the duration and the magnitude of Stat5 activation. Mapping of the NFAT1-binding domain on the Stat5a protein revealed that their physical association was mediated by the DNA-binding and transactivation domains of Stat5a. The presence of two interacting domains may allow NFAT1 to effectively regulate Stat5 signaling by the formation of a stable and functional NFAT1-Stat5 transcription complex. A similar phenomenon has been observed in other Stat protein interactions such as Stat1/cAMP response element binding protein-binding protein p300 and Stat5a/Runx (21, 22). Second, NFAT1 overexpression was able to block PRL-induced tyrosine phosphorylation of nuclear Stat5, inhibit the transcriptional activity of both wild-type and constitutively active forms of Stat5 with a subsequent suppression of Stat5 target gene expression, and impede Stat5a binding to its target gene promoter in breast cancer cells. This negative regulatory role for NFAT1 in modulating Stat5 signaling was further corroborated by the studies demonstrating that overexpression of NFAT inhibitor was capable to reverse these negative effects of NFAT1, and siRNA-mediated NFAT1 knockdown significantly enhanced PRL-induced, Stat5-mediated gene expression and cell proliferation. Third, a reciprocal inhibitory effect of Stat5 on NFAT1 signaling was also observed, suggesting that these two signaling cascades may antagonize each other in breast cancer. Indeed, such mutual suppressive cross talk has been reported among many other transcriptional networks such as Smad/Stat5, Runx/Stat5, and the p53/the glucocorticoid receptor complexes (21, 23, 24). Further studies are needed to determine whether physical interaction between NFAT1 and Stat5 is required for their reciprocal inhibition. Finally, the clinical significance of antagonistic cross talk between the NFAT1- and Stat5-signaling pathways was emphasized by our immunohistochemical observations that reveal a negative correlation between levels of NFAT1 and active Stat5 proteins in a matched breast cancer progression TMA, although the direct biological readout of this cross talk may require further investigation using appropriate xenograft breast cancer murine models that represent various disease progression stages (25). Therefore, these data collectively provide the first in vitro and in vivo evidence for a novel negative regulatory cross talk between the NFAT1- and Stat5-signaling pathways in breast cancer.
The NFAT family of transcription factors comprises five members, NFAT1–5, and plays an important role for many physiological and pathological processes including tumorigenesis (26). In contrast to Stat5, which functions as a promoter of tumor growth and a metastasis suppressor in breast cancer, NFAT1 has opposite effects, inhibiting tumor growth and promoting metastasis. With respect to its tumor-suppressive role, it has been demonstrated that NFAT1-deficient mice display a high propensity to develop chemical carcinogen-induced tumors (12), whereas isolated NFAT1−/− cartilage cells from knockout mice have the ability to form tumors in syngeneic mice (27). When overexpressed in T47D cells, we found that NFAT1 was able to suppress breast cancer cell proliferation in vitro (data not shown). In contrast, NFAT1 is believed to contribute to metastatic progression, which is evidenced by the studies demonstrating that NFAT1 induces expression of the prometastatic autotoxin/ENPP2 and cyclooxygenase-2 genes, and promotes cell migration and invasion of MDA231 (13, 28) and T47D (Zheng, J. and C. V. Clevenger, unpublished data). Our identification of mutual repression between the NFAT1 and Stat5 signaling cascades may well explain their opposing effects on the progression of breast cancer, as evidenced by the inverse expression of phospho-Stat5 and NFAT1 in a matched breast cancer progression TMA.
The mechanisms of how NFAT1 and Stat5 proteins mutually down-regulate the signaling require additional investigation. Three potential mechanisms have been considered to explain the suppressive effects of NFAT1 on Stat5 function. First, NFAT1 might interfere with the upstream mediators of Stat5 activation such as Jak2. Our results did not support this, however, because NFAT1 overexpression did not reduce Jak2 tyrosine phosphorylation.
A second potential mechanism of NFAT1 inhibition of Stat5 activity was based on our observation that NFAT1 overexpression can inhibit the binding of Stat5 to the CISH promoter as demonstrated by EMSA and ChIP analysis. The molecular mechanism by which NFAT1 inhibits DNA binding remains unclear. Classically, phosphorylated Stat5 can form homo- or heterodimers via its SH2 domain, and then translocate to nucleus, where these dimers bind the γ-interferon activated sequence element of the target gene and regulate its transcription (1, 16). In this context, we reasoned that the decreased DNA binding of Stat5 by NFAT1 is at least partly due to decreased tyrosine phosphorylation, because nuclear Stat5 can be dephosphorylated and deactivated by the nuclear tyrosine phosphatase TC-PTP (29). Indeed, NFAT1 overexpression results in nuclear Stat5 dephosphorylation and subsequent reduction of Stat5a intranuclear accumulation. Thus, it seems reasonable to envision that reduced intranuclear accumulation of Stat5a in NFAT1 transfectants may be the result of the actions of intranuclear phosphatases, which are enhanced by NFAT1 overexpression. Whether this occurs by direct association (i.e. a Stat5-NFAT1-phosphatase complex) or indirect action (i.e. NFAT up-regulation of phosphatase expression) will require additional investigation. However, the kinetics of NFAT1-Stat5 association (disassociation after PRL stimulation, followed by reassociation) would argue that some of the actions of NFAT are indirect. Given that NFAT1 associates with Stat5 through its DNA-binding domain, we also speculate that NFAT1 might inhibit DNA binding by masking a region of DNA-binding surface of Stat5, at those times at which these two molecules are complexed. Further investigations are required to examine whether NFAT1 can directly block DNA binding of Stat5 in the nucleus.
A third potential mechanism for NFAT1 regulation of Stat5 may involve the competition by NFAT for coregulators of Stat5, given that NFAT1 interacts with the C terminus of Stat5a, a site for association of other Stat5 coregulators, such as Oct-1 and centrosomal P4.1-associated protein (17, 18). Such a mechanism is possible, given our findings that coexpression of NFAT1 with GAL4-Stat5TAD construct significantly represses gene expression from the GAL-luciferase reporter, and that both NFAT1 (data not shown) and Stat5 can be immunoprecipitated on the chromatin from the CISH promoter. To determine whether these factors are involved in the negative regulation of Stat5 function by NFAT1, further studies (e.g. ChIP and/or two-step ChIP) are needed to examine a dynamic association between these factors and Stat5 at the chromatin level.
Cross talk among multiple signaling pathways contributes to breast tumor progression. The data presented here have revealed a reciprocal inhibitory relationship between NFAT1 and Stat5 activity, but it is not surprising that many other transcriptional factors may also interact with them to form complex cell-signaling networks. For example, it has been suggested that activator protein (AP)-1 forms a stable complex with NFAT1 to promote breast cancer cell invasion by up-regulating COX2 expression in breast cancer cells (13). Interestingly, Stat5 activation has been reported to inhibit AP-1 activity through their physical association in breast cancer cells (30), and the levels of activated Stat5 are negatively correlated with those of AP components in PRL-induced adenocarcinomas from murine models of breast cancer (31). Thus, the dynamic association of the Stat5/NFAT1/AP-1 signaling complex may play a significant role in the pathology and progression of human breast cancer.
In summary, our data unveiled the existence of reciprocal, negative cross talk between the NFAT1- and Stat5-signaling pathways in breast cancer. The transcriptional complex NFAT1-Stat5 may therefore represent a novel mutual regulatory loop that modulates NFAT1/Stat5 functions. These observations are supported at the in vivo level by our finding that high levels of NFAT1 are associated with decreased levels of active Stat5 in breast cancer metastasis, and vice verse in primary tumor. Thus, the interplay between these two transcription factors may significantly impact the pathology and progression of human breast cancer. A further mechanistic understanding of how these two signaling cascades communicate with each other to affect breast tumor formation, growth, and metastasis could provide novel molecular insights into the regulation of this transcriptional complex that may enable the development of new targeted therapies for this disease.
Materials and Methods
Cell culture and reagents
The T47D and MCF7 human breast cancer cells and HEK293T cells were maintained in recommended conditions as described elsewhere (32). For serum starvation, cells were incubated in phenol red-free DMEM for 24 h. Recombinant human PRL was a kind gift of Dr. Anthony A. Kossiakoff (The University of Chicago). EMSA kit (Pierce Chemical Co., Rockford, IL), EZ-ChIP kit (Upstate Biotechnology, Inc., Lake Placid, NY), Trizol, Lipofectamin LTX and the dual luciferase assay kits (Invitrogen, Carlsbad, CA), and Fugene 6 (Roche, Indianapolis, IN) were used as directed. Antibodies used in these studies were obtained from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA) (CISH and lamin A/C), Invitrogen (Stat5a, catalog no. 71–2400; pStat5a; Stat5b, catalog no. 71–2500; glyceraldehyde 3-phosphate dehydrogenase, and Myc), Sigma (St. Louis, MO) (Flag), and BD Biosciences (Palo Alto, CA) (NFAT1). Monoclonal antiphosphoTyr-Stat5 antibody AX1 (Advantex BioReagents, El Paso, TX) and NFAT1 antibody (Sigma) were used in the immunohistochemical analysis of TMA.
Immunoprecipitation and Western blot analysis
Cell lysates were resolved by SDS-PAGE and immunoblotted with the indicated antibodies. For immunoprecipitation, cell lysates were incubated for 24 h with the appropriate antibody (2 μg) followed by incubation for 2 h with protein A/G agarose beads (Invitrogen) at 4 C. The immune complexes were resolved by SDS-PAGE and immunoblotted with the appropriate antibodies as indicated. Target proteins were visualized using enhanced chemiluminescence (ECL) or ECL-plus (GE Healthcare), and images were captured using Fujifilm LAS-3000 system. The band intensities were quantified by densitometry using ImageQuant and normalized to those of their respective control bands. Data were expressed as fold change compared with an appropriate control.
RNA expression and real-time quantitative RT-PCR analysis
Total RNA was extracted from cells using Trizol. Total RNA was reversely transcribed using ThermoScript Reverse Transcriptase (Invitrogen). Real-time quantitative RT-PCR was performed in a 25-μl reaction volume containing 1 μl cDNA template, 23 μl 2× Power SYBR mastermix and 1 μl forward/reverse primers (20 nm) using ABI 7900HT Thermocycler (Applied Biosystems, Foster City, CA) as described previously (11). The relative amount of target mRNA (CISH and c-Myc) was calculated by the 2-ΔΔCt method, relatively to control mRNA (glyceraldehyde 3-phosphate dehyrogenase).
Chromatin immunoprecipitation (ChIP) analysis
ChIP analysis was performed using EZ-ChIP as described previously (11). In brief, starved cells were stimulated with PRL for 15 min. Rabbit anti-Stat5 was used to immunoprecipitate DNA from cell lysate. The pull-down DNA and input DNA was analyzed by real-time PCR analysis as previously described (11). The fold change was normalized to non-PRL treatment control using the ratio of the pull-down DNA to the input DNA.
Plasmids, siRNA, transient transfection, and luciferase reporter assay
The LHRE and CISH luciferase reporters were previously described (32, 33). The NFAT1 overexpression construct and the NFAT-responsive luciferase reporter were obtained from Dr. L. Glimcher (Harvard School of Public Health, Boston, MA). Stat5 deletion mutants were kindly provided by Dr. K. Ikuta (21); the CA-Stat5 mutant, which is constitutively active when expressed in cells due to two-point mutations (H299R and S711F), was kindly provided by Dr. T. Kitamura (34). The VIVIT expression vector was previously described (15). The transactivation domain of Stat5 was assembled into the GAL4-DNA-binding domain vector using standard PCR techniques, and termed as GAL4-Stat5 TAD. All DNA constructs were verified by DNA sequencing. A NFAT1-specific siRNA (35) and a scrambled control siRNA were purchased from Integrated DNA Technologies (Coralville, IA).
For luciferase assay, cells were transiently cotransfected with LHRE or CISH reporters and its internal control (Renilla) in addition to the empty vector, the NFAT1 over-expression construct, the scrambled control siRNA, or the NFAT1-siRNA using Lipofectamin LTX and Lipofectamin RNAiMAX (Invitrogen) or FuGENE 6 (Roche) transfection reagents as suggested. Overnight-starved cells were stimulated with PRL as indicated. Cell lysates were assayed for luciferase activity 24 h after PRL stimulation using Victor3 1420 Multilabel Counter (PerkinElmer Corp., Wellesley, MA).
The association of Stat5a mutants with the wild type of NFAT1 was determined in HEK293T cells. In all transfection experiments, the total amount of transfected plasmid DNA was equalized with empty vector.
Cell proliferation assay
T47D cells were transiently transfected with a scrambled control siRNA or a NFAT1-specific siRNA using Lipofectamin RNAiMAX (Invitrogen) in 96-well plates. Overnight-starved transfectants were stimulated with PRL for 2 d and then pulsed with [3H]thymidine (0.5 μCi/well, Amersham Pharmacia Biotech, Piscataway, NJ) for 4 h. The incorporation of thymidine was determined by radiometry using MicroBeta TriLux scintillation counter (PerkinElmer).
Tissue microarray
A breast cancer progression TMA, as previously described (36), was used to analyze protein expression of NFAT1 and pStat5. The TMA examined included matched normal adjacent, invasive ductal carcinoma, and lymph node metastasis from 11 patients. Immunohistochemical analysis with NFAT1 and pStat5 antibodies was performed by Northwestern University's Pathology Core. In brief, after deparaffinization and hydration, antigen was retrieved by specific buffer [for NFAT1, citrate buffer (Zymed Laboratories, South San Francisco, CA), pH 6.0; for pStat5, antigen retrieval AR-10 solution (BioGenex Laboratories, Inc., San Ramon, CA)] at 95 C for 20 min. The slides were blocked with the blocking buffer (2.5% BSA and 0.1% Triton X-100) for 10 min, and then incubated overnight with the primary antibodies (NFAT1 antibody, 1:100; pStat5, 1:100) at 4 C. This was followed by a 1-h incubation at room temperature with a biotin (for pStat5) or a horseradish peroxidase (for NFAT1)-labeled secondary antibody (DAKO Corp., Carpinteria, CA). For pStat5, a DAKO EnVision- horseradish peroxidase was applied and color (for both of them) was developed using 3,3′-diaminobenzidine. Slides were counterstained with hematoxylin, coverslipped, and analyzed as previously described (37). Given that virtually 100% of the breast epithelium demonstrated some level of antiphospho-Stat5 and NFAT labeling, scoring was restricted to the assessment of label intensity on a 0–3 scale (0, absent; 1, dim; 2, bright; 3, very bright).
Statistical analysis
Data are representative of at least two independent experiments. Data were reported as the means ± se and analyzed using ANOVA or Student's t test. A P value <0.05 was considered significant.
Acknowledgments
This work was supported by National Institutes of Health grants CA69294 and CA102682, and the Lynn Sage and Avon Foundations.
Disclosure Summary: The authors have nothing to disclose.
Footnotes
- AP
- Activator protein
- CA-Stat5a
- constitutively active Stat5a
- ChIP
- chromatin immunoprecipitation
- CISH
- cytokine-inducible SH2-containing protein
- HEK
- human embryonic kidney
- LHRE
- lactogenic hormone response element
- NFAT
- nuclear factor of activated T cells
- PRL
- prolactin
- siRNA
- small interfering RNA
- Stat
- signal transducer and activator of transcription
- TMA
- tissue microarray.
References
- 1. Darnell JE., Jr 1997. STATs and gene regulation. Science 277:1630–1635 [DOI] [PubMed] [Google Scholar]
- 2. Horvath CM. 2000. STAT proteins and transcriptional responses to extracellular signals. Trends Biochem Sci 25:496–502 [DOI] [PubMed] [Google Scholar]
- 3. Liu X, Robinson GW, Wagner KU, Garrett L, Wynshaw-Boris A, Hennighausen L. 1997. Stat5a is mandatory for adult mammary gland development and lactogenesis. Genes Dev 11:179–186 [DOI] [PubMed] [Google Scholar]
- 4. Yao Z, Cui Y, Watford WT, Bream JH, Yamaoka K, Hissong BD, Li D, Durum SK, Jiang Q, Bhandoola A, Hennighausen L, O'Shea JJ. 2006. Stat5a/b are essential for normal lymphoid development and differentiation. Proc Natl Acad Sci USA 103:1000–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bromberg J. 2002. Stat proteins and oncogenesis. J Clin Invest 109:1139–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nevalainen MT, Xie J, Torhorst J, Bubendorf L, Haas P, Kononen J, Sauter G, Rui H. 2004. Signal transducer and activator of transcription-5 activation and breast cancer prognosis. J Clin Oncol 22:2053–2060 [DOI] [PubMed] [Google Scholar]
- 7. Tan SH, Nevalainen MT. 2008. Signal transducer and activator of transcription 5A/B in prostate and breast cancers. Endocr Relat Cancer 15:367–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brockman JL, Schuler LA. 2005. Prolactin signals via Stat5 and Oct-1 to the proximal cyclin D1 promoter. Mol Cell Endocrinol 239:45–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ren S, Cai HR, Li M, Furth PA. 2002. Loss of Stat5a delays mammary cancer progression in a mouse model. Oncogene 21:4335–4339 [DOI] [PubMed] [Google Scholar]
- 10. Sultan AS, Xie J, LeBaron MJ, Ealley EL, Nevalainen MT, Rui H. 2005. Stat5 promotes homotypic adhesion and inhibits invasive characteristics of human breast cancer cells. Oncogene 24:746–760 [DOI] [PubMed] [Google Scholar]
- 11. Fang F, Rycyzyn MA, Clevenger CV. 2009. Role of c-Myb during PRL-induced Stat5a signaling in breast cancer cells. Endocrinology 150:1597–1606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Robbs BK, Cruz AL, Werneck MB, Mognol GP, Viola JP. 2008. Dual roles for NFAT transcription factor genes as oncogenes and tumor suppressors. Mol Cell Biol 28:7168–7181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yiu GK, Toker A. 2006. NFAT induces breast cancer cell invasion by promoting the induction of cyclooxygenase-2. J Biol Chem 281:12210–12217 [DOI] [PubMed] [Google Scholar]
- 14. Clevenger CV, Plank TL. 1997. Prolactin as an autocrine/paracrine factor in breast tissue. J Mammary Gland Biol Neoplasia 2:59–68 [DOI] [PubMed] [Google Scholar]
- 15. Aramburu J, Yaffe MB, López-Rodriguez C, Cantley LC, Hogan PG, Rao A. 1999. Affinity-driven peptide selection of an NFAT inhibitor more selective than cyclosporin A. Science 285:2129–2133 [DOI] [PubMed] [Google Scholar]
- 16. Ihle JN. 1996. STATs: signal transducers and activators of transcription. Cell 84:331–334 [DOI] [PubMed] [Google Scholar]
- 17. Magné S, Caron S, Charon M, Rouyez MC, Dusanter-Fourt I. 2003. STAT5 and Oct-1 form a stable complex that modulates cyclin D1 expression. Mol Cell Biol 23:8934–8945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Peng B, Sutherland KD, Sum EY, Olayioye M, Wittlin S, Tang TK, Lindeman GJ, Visvader JE. 2002. CPAP is a novel stat5-interacting cofactor that augments stat5-mediated transcriptional activity. Mol Endocrinol 16:2019–2033 [DOI] [PubMed] [Google Scholar]
- 19. Zhu M, John S, Berg M, Leonard WJ. 1999. Functional association of Nmi with Stat5 and Stat1 in IL-2- and IFNγ-mediated signaling. Cell 96:121–130 [DOI] [PubMed] [Google Scholar]
- 20. Nakajima H, Brindle PK, Handa M, Ihle JN. 2001. Functional interaction of STAT5 and nuclear receptor co-repressor SMRT: implications in negative regulation of STAT5-dependent transcription. EMBO J 20:6836–6844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ogawa S, Satake M, Ikuta K. 2008. Physical and functional interactions between STAT5 and Runx transcription factors. J Biochem 143:695–709 [DOI] [PubMed] [Google Scholar]
- 22. Zhang JJ, Vinkemeier U, Gu W, Chakravarti D, Horvath CM, Darnell JE., Jr 1996. Two contact regions between Stat1 and CBP/p300 in interferon γ signaling. Proc Natl Acad Sci USA 93:15092–15096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sengupta S, Vonesch JL, Waltzinger C, Zheng H, Wasylyk B. 2000. Negative cross-talk between p53 and the glucocorticoid receptor and its role in neuroblastoma cells. EMBO J 19:6051–6064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cocolakis E, Dai M, Drevet L, Ho J, Haines E, Ali S, Lebrun JJ. 2008. Smad signaling antagonizes STAT5-mediated gene transcription and mammary epithelial cell differentiation. J Biol Chem 283:1293–1307 [DOI] [PubMed] [Google Scholar]
- 25. Tang B, Vu M, Booker T, Santner SJ, Miller FR, Anver MR, Wakefield LM. 2003. TGF-β switches from tumor suppressor to prometastatic factor in a model of breast cancer progression. J Clin Invest 112:1116–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Im SH, Rao A. 2004. Activation and deactivation of gene expression by Ca2+/calcineurin-NFAT-mediated signaling. Mol Cells 18:1–9 [PubMed] [Google Scholar]
- 27. Ranger AM, Gerstenfeld LC, Wang J, Kon T, Bae H, Gravallese EM, Glimcher MJ, Glimcher LH. 2000. The nuclear factor of activated T cells (NFAT) transcription factor NFATp (NFATc2) is a repressor of chondrogenesis. J Exp Med 191:9–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chen M, O'Connor KL. 2005. Integrin alpha6beta4 promotes expression of autotaxin/ENPP2 autocrine motility factor in breast carcinoma cells. Oncogene 24:5125–5130 [DOI] [PubMed] [Google Scholar]
- 29. Aoki N, Matsuda T. 2002. A nuclear protein tyrosine phosphatase TC-PTP is a potential negative regulator of the PRL-mediated signaling pathway: dephosphorylation and deactivation of signal transducer and activator of transcription 5a and 5b by TC-PTP in nucleus. Mol Endocrinol 16:58–69 [DOI] [PubMed] [Google Scholar]
- 30. Gutzman JH, Rugowski DE, Nikolai SE, Schuler LA. 2007. Stat5 activation inhibits prolactin-induced AP-1 activity: distinct prolactin-initiated signals in tumorigenesis dependent on cell context. Oncogene 26:6341–6348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Arendt LM, Rugowski DE, Grafwallner-Huseth TA, Garcia-Barchino MJ, Rui H, Schuler LA. 2011. Prolactin-induced mouse mammary carcinomas model estrogen resistant luminal breast cancer. Breast Cancer Res 13:R11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zheng J, Koblinski JE, Dutson LV, Feeney YB, Clevenger CV. 2008. Prolyl isomerase cyclophilin A regulation of Janus-activated kinase 2 and the progression of human breast cancer. Cancer Res 68:7769–7778 [DOI] [PubMed] [Google Scholar]
- 33. Fang F, Antico G, Zheng J, Clevenger CV. 2008. Quantification of PRL/Stat5 signaling with a novel pGL4-CISH reporter. BMC Biotechnol 8:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Onishi M, Nosaka T, Misawa K, Mui AL, Gorman D, McMahon M, Miyajima A, Kitamura T. 1998. Identification and characterization of a constitutively active STAT5 mutant that promotes cell proliferation. Mol Cell Biol 18:3871–3879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nolz JC, Fernandez-Zapico ME, Billadeau DD. 2007. TCR/CD28-stimulated actin dynamics are required for NFAT1-mediated transcription of c-rel leading to CD28 response element activation. J Immunol 179:1104–1112 [DOI] [PubMed] [Google Scholar]
- 36. Fang F, Flegler AJ, Du P, Lin S, Clevenger CV. 2009. Expression of cyclophilin B is associated with malignant progression and regulation of genes implicated in the pathogenesis of breast cancer. Am J Pathol 174:297–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. McHale K, Tomaszewski JE, Puthiyaveettil R, Livolsi VA, Clevenger CV. 2008. Altered expression of prolactin receptor-associated signaling proteins in human breast carcinoma. Mod Pathol 21:565–571 [DOI] [PubMed] [Google Scholar]