Abstract
Imidazolonepropionase (HutI) (imidazolone-5-propanote hydrolase; EC 3.5.2.7) is a member of amidohydrolase superfamily and catalyzes the conversion of imidazolone-5-propanoate to N-formimino -L-glutamate in the histidine degradation pathway. We have determined the three dimensional crystal structures of HutI from A. tumefaciens (At-HutI) and an environmental sample from the Sargasso Sea Ocean Going Survey (Es-HutI) bound to the product [N-formimino-L-glutamate (NIG)] and an inhibitor [3-(2,5-dioxoimidazolidin-4yl)-propionic acid (DIP), respectively. In both structures the active site is contained within each monomer and its organization displays the landmark feature of amidohydrolase superfamily showing a metal ligand (iron), four histidines and one aspartic acid. A catalytic mechanism involving His265 is proposed based on the inhibitor bound structure. This mechanism is applicable to all HutI.
Keywords: AHS, amidohydrolases, NIG, DIP, At-HutI, Es-HutI
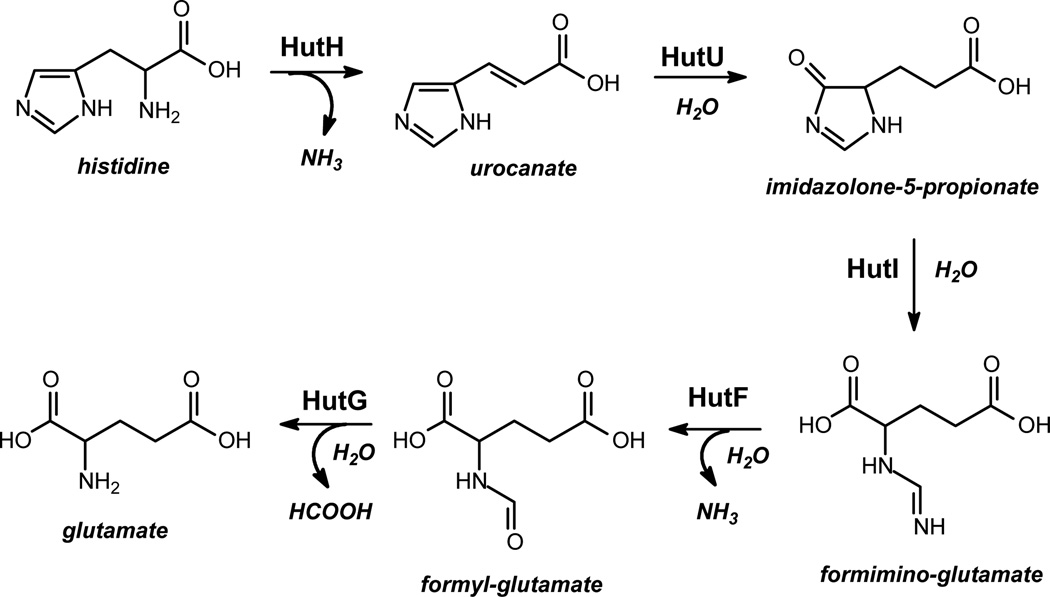
In certain eubacteria, such as Agrobacterium tumefaciens and Pseudomonas aeruginosa, L-histidine can be metabolized to L-glutamate via catalytic transformation by five enzymes (1). In the first step, L-histidine is deaminated by histidine ammonia lyase (HutH)1 to form urocanate, which is subsequently hydrated in a mechanistically complex reaction by urocanase (HutU) to produce imidazolone-5-propionate. This product is in turn hydrolyzed by imidazolonepropionase (HutI) to generate N-formimino-L-glutamate. The N-formimino substituent is then hydrolyzed by N-formimino-L-glutamate deiminase (HutF) to form N-formyl-L-glutamate and ammonia. In the final step, N-formyl-L-glutamate amidohydrolase (HutG) catalyzes hydrolysis of N-formyl-L-glutamate to L-glutamate and formate. In some organisms, the N-formimino-L-glutamate is hydrolyzed directly to formamide and L-glutamate and in others the formimino group is transferred to tetrahydrofolate (2). The pathway for the degradation of histidine is summarized in Figure1.
Figure 1.
Histidine degradation pathway (scheme 1).
To date, four of seven enzymes comprising the histidine utilization pathway have been structurally characterized. HutH from Pseudomonas putida (PDB code: 1XFL) contains an unusual 4-methylideneimidazol-5-one cofactor formed by post-translational rearrangement of the amino acid segment –Ala-Ser-Gly– within this enzyme (3). In the active site of HutU from P. putida, there is a tightly bound NAD+ that is utilized as an unusual nonredox electrophilic cofactor for hydration of urocanate (PDB code: 1UWK) (4). Its mechanism of action has been elucidated earlier by biochemical methods (5, 6). One of three enzyme variants that can act upon the product of the HutI reaction, N-formimino-L-glutamate hydrolase, has been crystallized and its structure determined to high resolution (PDB code: 1XFK). More recently, the structure of HutI has been determined from Bacillus subtilis (PBD code: 2G3F), A. tumefaciens (PDB code: 2GOK), and an environmental sample from the Sargasso Sea (PDB code: 2OOF) (7, 8). These structures of HutI have demonstrated that it is a member of the amidohydrolase superfamily (AHS).
The AHS superfamily was first recognized by Sander and Holm based upon the structural and mechanistic similarities among adenosine deaminase, phosphotriesterase, and urease (9). All enzymes comprising this superfamily adopt a (β/α)8-barrel structural fold with an active site located at the C-terminus of the β-barrel, and all structurally characterized superfamily members possess mono- or bi-nuclear metal centers within their active sites (10). Nearly all functionally characterized AHS catalyze hydrolysis of C-O, C-N, or P-O bonds within esters, amides, or aromatic amines via activation of water or hydroxide by the metal center. However, members of this superfamily also catalyze decarboxylation reactions, addition of water to activated double bonds, and isomerization of aldose and ketose sugars (10). Well characterized members of this superfamily include cytosine deaminase (11), dihydroorotase(12), isoaspartyl dipeptidase (13), N-formimino-L-glutamate deiminase (14), N-acetyl-D-glucosamine-6-phosphate deacetylase (15), and uronate isomerise (16).
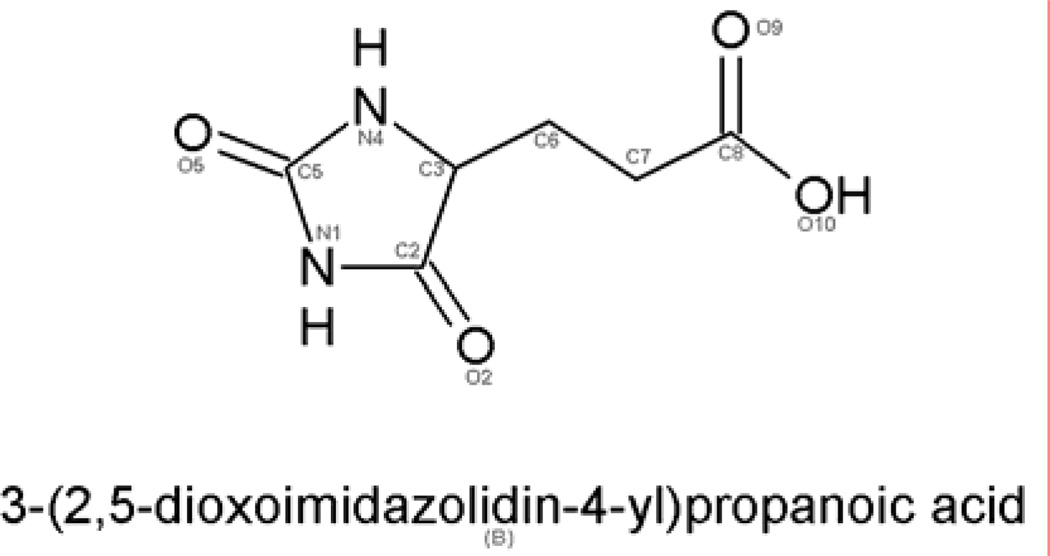
The three-dimensional X-ray structures of HutI from Bacillus subtilis, A. tumefaciens, and environmental sample demonstrate that this enzyme has a single metal ion bound to the α-site. Most of the enzymes in the AHS with an α-mononuclear metal center catalyze the deamination of an aromatic amine. Examples include adenosine deaminase (17), guanine deaminase (18), cytosine deaminase (11), and SAH deaminase(19). The two notable exceptions are HutI and HutF. The structure of HutI from B. subtilis (Bs-HutI) has been determined in the presence of imidazole-4-acetic acid, an inhibitor that remotely resembles the substrate (8). From this HutI-ligand complex a mechanism has been proposed that utilizes the zinc in the active site to activate the hydrolytic water/hydroxide and a semi-conserved glutamate at the end of β-strand 5 to facilitate proton transfer reactions. Herein, we report three dimensional x-ray structures of HutI from A. tumefaciens (At-HutI) and an environmental sample from the Sargasso Sea Ocean Going Survey (Es-HutI) bound to the product [N-formimino-L-glutamate (NIG)] and an inhibitor [3-(2,5-dioxoimidazolidin-4yl)-propionic acid (DIP) (also known as 3-ureidopropionate)], respectively (Fig. 2). From these protein-ligand structures an enzyme mechanism has been proposed for conversion of imidazolone-5-propionate to N-formimino-L-glutamate by HutI.
Figure 2.
3-(2,5-dioxoimidazolidin-4-yl)-propionic acid (DIP) inhibitor of HutI.
Experimental Procedures
Protein Production - At-HutI and Es-HutI
The At-HutI gene was amplified using polymerase chain reaction (PCR) from Agrobacterium tumefaciens genomic DNA using a Forward (CCAGGGAACAATTCTGCGAAGG) and a reverse (CTGGAGAAACCTTCTGTCCCTTG) primer. For Es-HutI, PCR from a Sargasso Sea environmental sample codon optimized synthetic gene using a Forward (AATTGCGAACGTGTGTGGCTGAAC) and a reverse (CCCCATGCAGTGTTTCTTCACCG) primer resulted the final target gene. The amplified genes were gel purified and cloned into pSGX3 (BC) vector designed to express the protein of interest with a C-terminal hexa-histidine affinity tag. Protein expression/purification utilized previously published protocols (20).
Preparation of protein – ligand complexes
The apo-enzyme crystals for Es-HutI and At-HutI were obtained in the P3221 and and C2 space group via sitting drop vapor diffusion at 20°C against a reservoir solution containing 1.8 M Tri-ammonium citrate, pH 7.0 and 25% (w/v) PEG 3350, 0.1 M Bis-Tris pH 5.5, and 0.2M MgCl2 respectively. The Es-HutI apo-crystals were used for the inhibitor 3-(2,5-dioxoimidazolidin-4yl)-propionic acid (DIP) soaking experiments. The crystals were soaked in various concentrations of DIP (2mM, 3mM, 5mM, 8mM and 10 mM) in reservoir solution (1.8 M Tri-ammonium citrate, pH 7.0) and incubated for various time intervals ranging from 10 min to 12 hours. The product, N-formimino-L-glutamate (NIG) was co-crystallized with At-HutI via hanging drop vapor diffusion at 20°C against a reservoir solution containing 25% (w/v) PEG 3350, 0.1 M Bis-Tris pH 5.5, and 0.2M MgCl2 (2µL of protein at 30 mg/mL plus 2µL of reservoir solution). The protein solution contained six different molar ratios ranging from 1:1 to 1:6 of protein:product.
Data collection, structure determination, and refinement
Crystals soaked in 10 mM inhibitor concentration for 12 hours or produced via co-crystallization with a 1:3 protein:product molar ratio were flash frozen by direct immersion in liquid nitrogen following cryoprotection with addition of 20% glycerol to the crystallization reservoir solution. High resolution diffraction data (enzyme-inhibitor complex: 1.97Å; enzyme-product complex: 1.83 Å), were recorded under standard cryogenic conditions using NSLS Beamline X12C (National Synchrotron Light Source, Brookhaven National Laboratory) and diffraction data were processed using HKL2000 (21). Molecular replacement using native model (PDB code: 2OOF) yielded the starting model for the inhibitor bound structure and was refined using CNS (22). The product bound structure was phased and refined using the native structure (PDB code: 2GOK) in CNS (22). Difference Fourier syntheses revealed interpretable electron density features in the vicinity of the metal centers for 3-(2,5-dioxoimidazolidin-4yl)-propionic acid (DIP) and N-formimino-L-glutamate (NIG), respectively, in each co-crystal structure. During refinement stereochemical quality of both the models was monitored in PROCHECK (23). The final atomic coordinates and structure factors of both have been deposited with the Protein Data Bank (www.rcsb.org; PDB codes: 2Q09 and 2PUZ).
X-ray Fluorescence Scan and Inductively Coupled Plasma Mass Spectrometry Analyses
To identify the nature of bound metal ion, fluorescence scanning for Zn, Fe, and Mg were performed with the protein solutions used for the crystallization before the data collection at beamline X12C (data not shown). The nature of bound metal ion was also confirmed with Inductively Coupled Plasma Mass Spectrometry (ICP-MS) analysis. Metal determination and quantification were performed with an Elan DRC II ICP-MS from Perkin Elmer. An analog detection mode was used with three averaged replicates per reading. External calibration standards were prepared through the serial dilution of a single 10 ppm stock mixture of Zn, Cd, Co, Cu, Mn, Ni and Fe in 2% nitric acid. Freshly prepared standards generally contained 2, 20 and 200 ppb of the metal ions in 1% Trace Select nitric acid from Fluka, diluted in MilliQ deionized water. The masses of the isotopes detected were 55Mn, 57Fe, 59Co, 60Ni, 66Zn and 111Cd. 115In was used as an internal standard for 111Cd whereas 69Ga was used as an internal standard for all other isotopes. The results showed the metal content as Fe (77%), Zn (10%) and Ni (13%).
Results and Discussion
Structure determination of Inhibitor bound complex
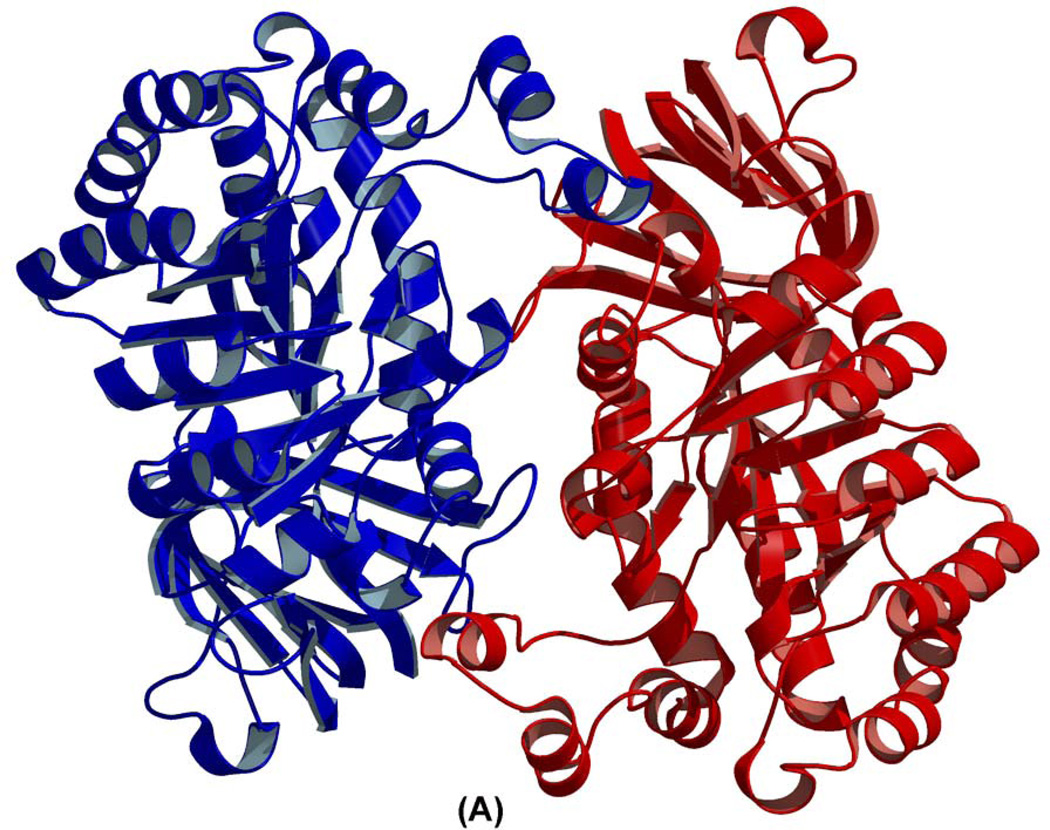
The enzyme-inhibitor crystals belong to the trigonal space group P3221 (Unit Cell dimensions: a=95.9Å, b=95.9Å, c=115.5Å) with one complex/asymmetric unit, which is consistent with the protein monomer observed in solution via analytical gel-filtration (data not shown). The atomic model was refined to a crystallographic R-value of 19.4%, with Rfree=21.4%. The stereochemistry of the model was of high quality as documented by PROCHECK (23), with ~90% residues found in the most favorable region of the Ramachandran plot. His265 and Asp23, both with well defined electron density, adopt disallowed (φ,ψ) values. His265 is one of the active site residues. The final refined atomic model contains a single chain (residues 4–407) (Fig. 3(A)), an iron molecule and one DIP molecule, plus 275 water molecules (Table 1).
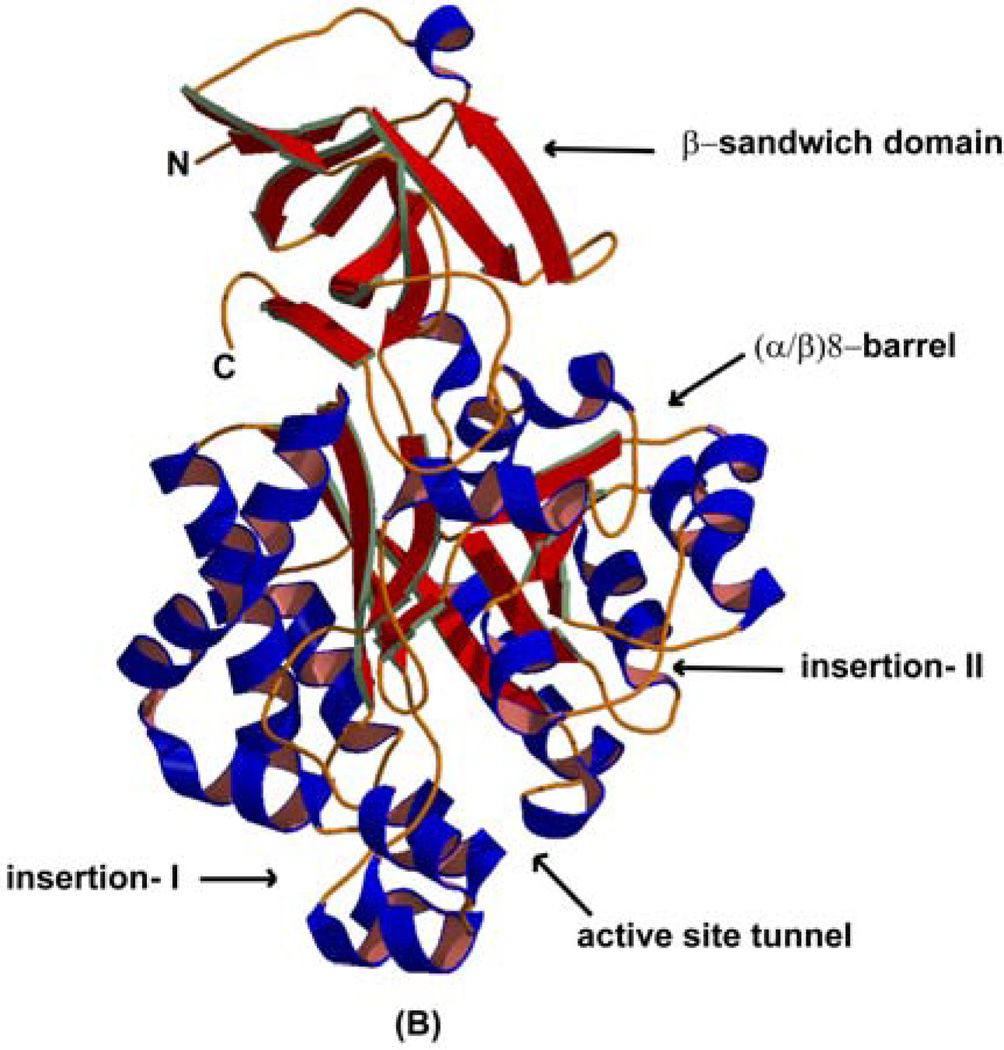
Figure 3.
(A) Structure of the 2PUZ dimer. (B) overall monomer structures of 2Q09 and 2 PUZ, showing secondary structure elements.
Table 1.
Data and refinement statistics
| Data set | Product bound (2PUZ) | Inhibitor bound (2Q09) |
|---|---|---|
| Cell dimensions |
a=140.9, b=64.4, c=103.9; |
a=95.9, b=95.9, c=115.5; |
| β = 112.3° | β = 120.0° | |
| Space group | C2 | P3221 |
| Data Statistics | Collection | |
| Wavelength (Å) | 1.1 | 1.1 |
| Temperature (K) | 100 | 100 |
| Resolution range | 50.0-1.83 | 50-1.97 |
| Unique reflections | 75466(7337) | 42964(3562) |
| Completeness (%) | 99.5(97.4) | 97.2(81.6) |
| Mean I/σ(I) | 27.9(2.1) | 19.7(1.9) |
| Redundancy | 6.9(5.7) | 17.6(5.8) |
| Rmerge1 | 0.041(0.169) | 0.063(0.391) |
| Refinement Statistics | ||
| No. of reflections (work) | 74509 | 41444 |
| No. of reflections (test) | 3784 | 1251 |
| 2Rfactor/3Rfree | 0.198 /0.225 | 0.194/0.214 |
| Resolution range (Å) | 30.0-1.83 | 31.0-1.97 |
| RMSD for bond length (Å) | 0.005 | 0.006 |
| RMSD bond angles (°) | 1.30 | 1.4 |
| <B-values> | ||
| Main-chain (Å2) | 29.8 | 30.8 |
| Side-chain (Å2) | 31.2 | 32.4 |
| Number of non-H atoms | ||
| No. of heteroatoms | 16 | 13 |
| No of water molecules | 587 | 275 |
Values for the highest resolution shell are given within parentheses.
Rmerge= Σ|Ii-〈I〉| /Σ|Ii| where Ii is the intensity of the ith measurement, and 〈I〉 is the mean intensity for that reflection.
Rfactor=Σ||Fobs|−|Fcalc||/Σ|Fobs|where|Fcalc| and |Fobs| are the calculated and observed structure factor amplitudes, respectively.
Rfree= as for Rfactor, but for 5% of the total reflections chosen at random and omitted from refinement.
Structure determination of the product bound complex
The enzyme-product co-crystals belong to the monoclinic space group C2 (Unit Cell dimensions: a=140.9Å, b=64.4Å, c=103.9Å) with two complexes/asymmetric unit. The atomic model was refined to a crystallographic R-value of 19.8% with Rfree=22.5%. The stereochemistry of the model was of high quality as documented by PROCHECK(23), with >90% residues found in the most favorable region of the Ramachandran plot. His279, with unambiguous electron density in both protomers, located in the active site at the C-terminus of a β-strand before a β-turn adopts disallowed (φ,ψ) values. The final refined atomic model contains two structurally similar polypeptide chains (residues 17–420) plus two iron molecule, one magnesium (Mg2+) cation, one chloride anion, and one NIG molecule, plus 587 water molecules (Fig. 3(A); Table 1). The electron density for NIG molecule was observed only in one monomer. The partially filled active site with the ligand is not uncommon. A root-mean-square-deviation (R.M.S.D.) of 0.33Å was calculated for 404 common α carbon atomic pairs comprising the protein dimer. These two protomers are related by a non-crystallographic 2-fold axis, with each monomer burying ~1,770Å2 of solvent accessible area upon dimerization, which exceeds values typically found in interacting surfaces for a protein of this size (24). However, this dimerization is an artifact of crystal packing as shown earlier (7). At least 33 residues from each monomer are involved in dimer formation, with 12 direct hydrogen bonding interactions stabilizing the dimer interface.
Overall structure of the enzyme-ligand complexes
The overall monomer structures of both enzyme-ligand complexes are very similar with secondary structural compositions of ~20% β-strand, ~35% α-helix, and ~45% random coil. Each monomer is composed of a small β-sandwich N/C-terminal domain (At-HutI: residues 17–80 and 364–420; Es-HutI: residues 4–66 and 351–407) and a large middle domain, which adopts the (α/β)8 barrel characteristic of AHS (At-HutI: residues 81–379; Es-HutI: 67–365) (Fig. 3(B)). The (α/β)8 barrel is interrupted twice by short segments of random coil and α-helix. The insert-I is between residues 85–128 in At-HutI or 71–114 in Es-HutI, which are inserted between the β1 strand and the α1 helix, whereas insert-II is between residues 301–316 in At-HutI or 287–302 in Es-HutI between β7 and α7 (Fig. 3(B)). These two insertions form the peptide flaps that border the entrance to the active site tunnel (barrel pore) at the C-terminal ends of its β-strands. A HINGEprot (25) analysis showed the role of two hinge residues (85 and 124; At-HutI or 71 and 110; Es-HutI), in opening and closing of this peptide flaps (7). The active site with its bound metal ion and ligands, is located immediately behind these helical flaps. The (α/β)8 barrel fold is conserved among amidohydrolase superfamily and overall structure of HutI has the highest degree of structural homology to cytosine deaminase (CDA) (11) with an overall root-mean-square difference (rmsd) between 353 α-carbon positions of 2.8 Å as reported by the DALI server (26). This structural similarity between enzymes of similar function is undetectable by BLAST searches due to low sequence identity between CDA and HutI (18%) or through available fold recognition servers, however, Holm and Sander had previously predicted on the basis of functional similarities and evolutionary relationship in the aminohydrolase superfamily (9). It has been shown earlier that the β-strands of the (α/β)8 barrel and the metal-binding residues of the active site correspond closely (R.M.S.D.=0.3Å for 5 residues metal coordinating residues), whereas the external helices of the barrel and surface-exposed loops diverge significantly (7). The β-sandwich domain lies at the end of the barrel away from the active site, and is formed by two separate peptide segments (residues 17–80 and 364–420 in At-HutI; residues 4–66 and 351–407 in Es-HutI), derived from opposite ends of the HutI primary sequence flanking the (α/β)8 barrel forming sequence. This small β-sandwich domain is connected to the barrel by two short peptide linkers: the first is a random coil (residues 80–82; At-HutI; 66–88; Es-HutI), while the second is a helix (residues 357–372, At-HutI; 344–359, Es-HutI).
Architecture of product- and inhibitor-bound active site
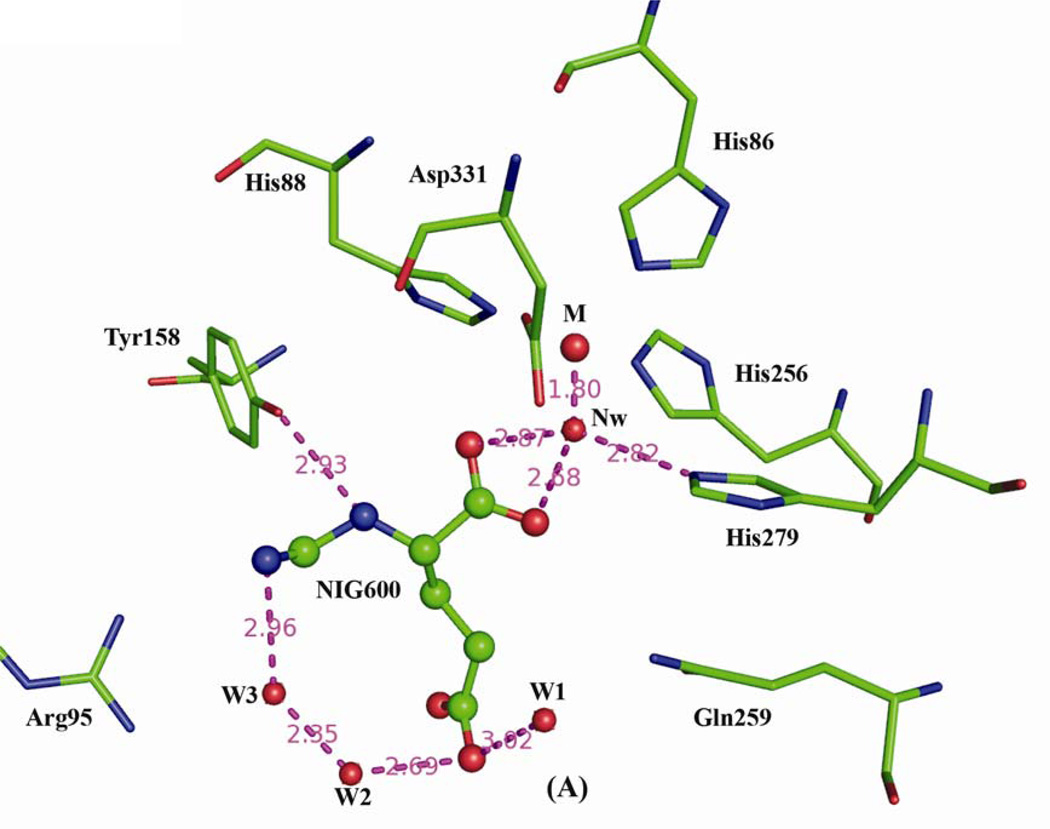
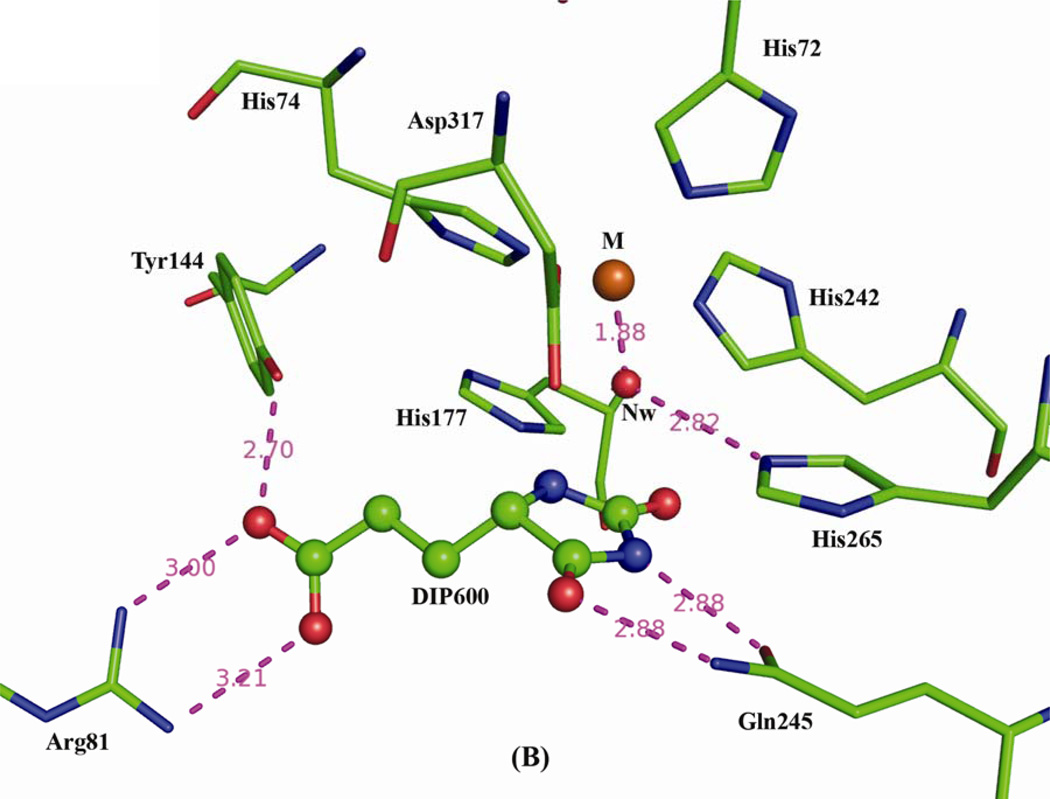
The HutI active site is located within the negatively charged cavity of the (α/β)8 barrel domain and contains a single bound iron in both the structures. The presence of iron was initially identified with x-ray fluorescence analysis performed at the NSLS X12C beamline and confirmed via Inductively Coupled Plasma Mass Spectrometry (ICP-MS) analysis. It is remarkable that the enzyme from Bacillus subtilis (Bs-HutI) contains a zinc metal ion instead of iron (8). In our product bound structure, the iron is directly coordinated by His86, Nε2--2.12Å; His88, Nε2--2.3Å; His256, Nε2--2.4Å, and Asp331, Oδ1--2.3Å) and a water nucleophile (1183--1.8Å) (Fig. (4)A) and similar coordination was observed in inhibitor bound complex with slight variations in metal ion coordination distances; His72, Nε2--2.1Å; His74, Nε2--2.1Å; His242, Nε2--2.3Å, and Asp317, Oδ1--2.3Å) and a water nucleophile (260--1.8Å) (Fig. 4(B)). His279 in the product complex and His265 in the inhibitor complex both interact with the nucleophilic water (ion-sidechain separation~2.8Å). The catalytic metal ion and all surrounding sidechains are well ordered in the electron density maps
Figure 4.
Active sites of the (A) Enzyme-Product and (B) Enzyme-Inhibitor complexes, shown in similar orientations. Nearby protein residues are shown as color-coded stick figures and ligand atoms as color-coded ball-and-stick figures (C: green, N: blue; O: red) with iron (M: orange) and water molecules (red) as spheres. Nw represents nucleophilic water, while W1-W3 represent structural waters stabilizing product binding.
Unbiased Fourier difference syntheses calculated with diffraction data from product co-crystallized and inhibitor soaked crystals display clear electron density for the bound product/inhibitor in close proximity to (α/β)8 barrel (Fig. 5(A) and 5(B)). Both electron density maps were calculated prior to the inclusion of ligands in the crystallographic model. The O1 and O2 atoms of the product NIG600 are 3.3Å and 3.9Å from the iron. The product also interacts with His88, Tyr158, nucleophilic water 1183, and waters W3, and W2 (Fig. 4(A)). The N1 atom of the inhibitor DIP600, is 4.9Å from the iron, and is little farther than the product (Fig. 4(B)). Inhibitor DIP600 makes salt bridges with Arg81. In addition, DIP600 interacts with Gln245, and Tyr144 (Fig. 4(B)). His72/86, His74/88 and Arg81/95 are from the Insertion-I region; His242/256, and His-265/279 are located at the C terminus of β-strands 5 and 6, respectively; and Tyr144/158, Gln245/259, and Asp317/331 are located in the β-α-loops 2, 5, and 8, respectively. Insertion-I acts as a flap on the entrance of central cavity (Fig. 3(B)), making the metal ion and substrate binding site deeply buried in the molecule. The two co-crystal structures superimpose well in most of the regions, except between flap region from Insertion-I (Ala96 to Lys114; At-HutI numbering).
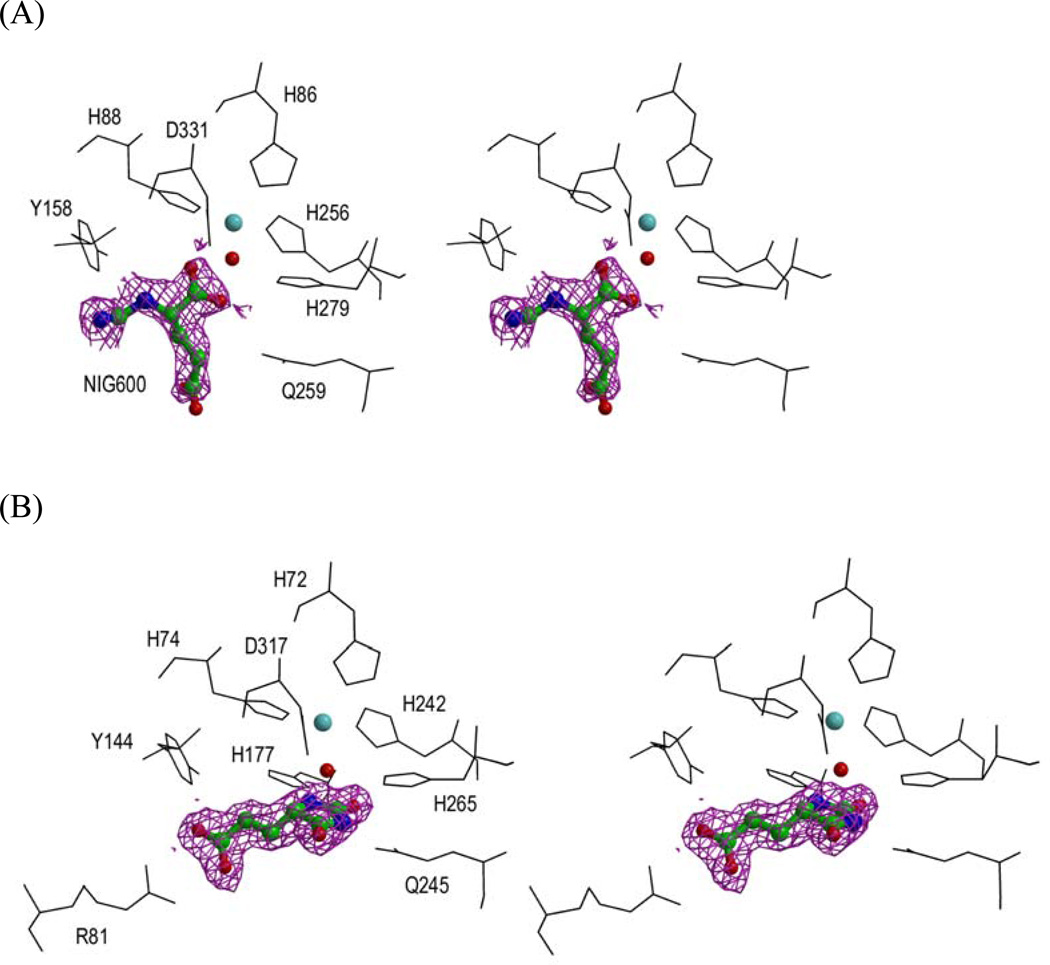
Figure 5.
(A) Stereoview of the active site along with metal ion (cyan), nucleophilic water (red), bound product NIG and (B) inhibitor DIP. Both NIG and DIP are shown with their 2Fo-Fc electron density map contoured to 1σ level.
Mechanism of Action
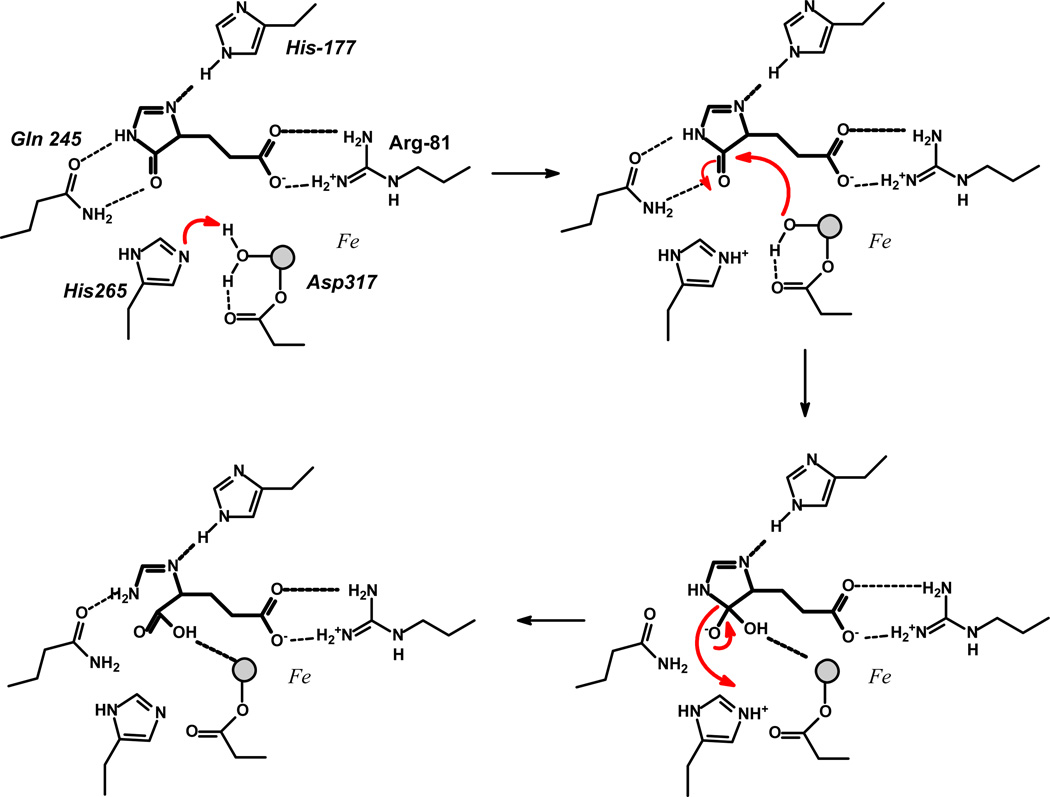
The structures of HutI bound to product and an inhibitor in the active site enable a chemical reaction mechanism for this enzyme to be proposed. In this regard the structure of HutI from the Sargasso Sea with bound inhibitor is more useful than is the complex of HutI from A. tumefaciens with product. It appears that the complex of the product shows the product as it begins to depart from the active site. Thus, a water has now entered the coordination environment of the metal ion and that the carboxylate group of the original substrate is no longer ion-paired with Arg95/Arg81 and that the N-formimino group has rotated away from the initial cleavage site and appears H-bonded with Tyr158/Tyr144. The proposed reaction mechanism presented in Figure 6 is based on the inhibitor bound structure. In the apo-protein active site the lone metal ion is ligated to the protein via electrostatic interactions involving His-72, His-74, His-242, and Asp-317. The fifth ligand to the metal is a water/hydroxide molecule that is in turn hydrogen bonded to the sidechain carboxylate group of Asp-317 and the sidechain imidazole of His-265, which is conserved in all HutI (Fig. 7). An identical set of ligands with the same geometry is observed in the active sites of adenosine deaminase (16), guanine deaminase (18), cytosine deaminase (9, 11, 26), and SIH deaminase (19).
Figure 6.
Reaction mechanism for HutI.
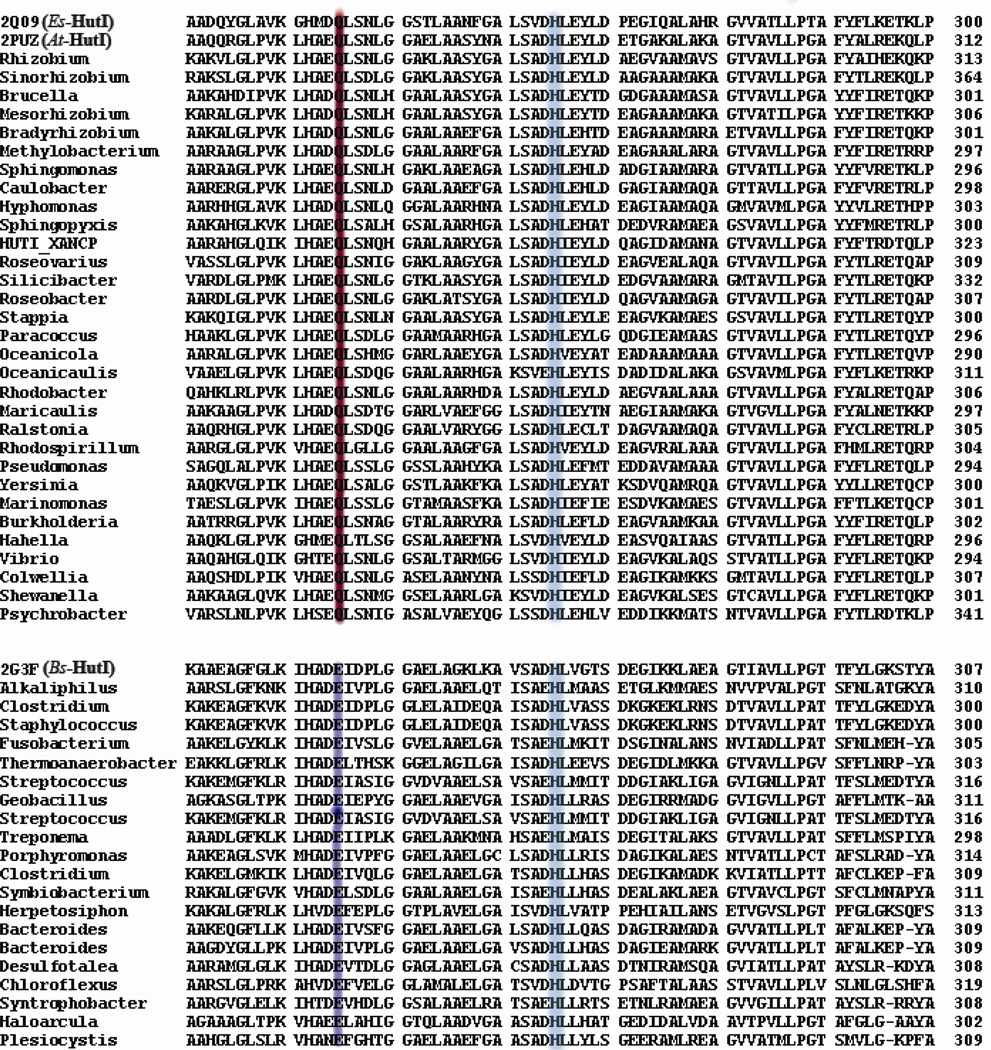
Figure 7.
Multiple sequence alignment of imidazolonepropionase sequences from various organisms showing two different classes of catalytic residues (Gln: red; Glu: blue) at positions 245 in Es-HutI (2Q09) or 259 in At-HutI (2PUZ) and 252 in Bs-HutI (2G3F). The conserved histdine [His265 (Es-HutI) and His279 (At-HutI)] is shaded in sky-blue color.
When substrate binds to the active site, the carboxylate functional group forms an ion pair with the sidechain of invariant Arg-81. The nitrogen attached to the α-carbon of the substrate forms a hydrogen bond to His-177 and the carbonyl oxygen of the bond to be hydrolyzed forms a hydrogen bond to the carboxamide group of Gln-245 and possibly with Thr-322. These hydrogen bonding interactions serve to activate the carbonyl group for nucleophilic attack. There is no evidence in HutI, or in any of the other α-mononuclear metal activated enzymes from the amidohydrolase superfamily, that the metal functions to polarize the carbonyl group (10). Catalysis is initiated by a proton transfer from the metal bound water molecule to His-265. The carbonyl carbon of the substrate is attacked by the metal bound hydroxide to form a tetrahedral intermediate that is stabilized by the hydrogen bonding interactions with Gln-245. This intermediate then collapses and the product forms via a proton transfer from the charged His-265 to the terminal nitrogen of the product.
Alternative mechanisms to the one presented in Figure 6 can also be drawn. For example, a second proton transfer could occur from the metal bound water/hydroxide to the side chain carboxylate of Asp-317. This acid could then transfer this proton to the leaving group nitrogen upon collapse of the tetrahedral intermediate. In the reaction catalyzed by dihydroorotase, a member of the amidohydrolase superfamily with a binuclear metal center, there is excellent structural evidence for participation of the equivalent aspartate carboxylate group in proton transfer from the bridging hydroxide to the leaving group nitrogen (12). In addition, a catalytic mechanism can be devised wherein the final proton transfer is initiated by His-177 to the nitrogen attached to the α-carbon. In this alternative mechanism, a different tautomer of the product would be formed and His-177 would have to be initially protonated rather than neutral.
The mechanism proposed here for the hydrolysis of imidazole-5-propionate by HutI differs from that proposed earlier for HutI from B. subtilis (8). In the B. subtilis enzyme the residue equivalent to Gln-245 is a glutamate (Fig. 7). Yu et al. have postulated that the leaving group nitrogen is protonated from this glutamate residue (Glu-252) and that the other nitrogen is also protonated by a group that is ambiguously suggested as being the equivalent to His-177 (His-185). The mechanism by Yu et al. suggests no role for the fully conserved residue equivalent to His-265 (His-272) (Fig. 7). We believe that the mechanism proposed by Yu et al. is not applicable to any of the variants of HutI bearing glutamine residue at the position equivalent to Gln-245 from Es-HutI, since a glutamine is quite unlikely to support proton transfer as depicted for Bs-HutI (Fig. 7). However, all of the variants mentioned in Figure 1 that have a glutamate at this residue position could operate by the mechanism proposed here for At-HutI and Es-HutI. A protonated glutamic acid could polarize the carbonyl group of the substrate and stabilize the tetrahedral adduct upon nucleophilic attack by the metal borne hydroxide.
Acknowledgment
We gratefully acknowledge data collection support from beamline X12C (NSLS)‥ We thank Robert Hall for the metal analysis.
Footnotes
Research was supported by a National Institutes of Health Co-operative Agreement award to SGX Pharmaceuticals, Inc. (GM074945; Principal Investigator: Stephen K. Burley) under DOE Prime Contract No. DEAC02-98CH10886 with Brookhaven National Laboratory. F.R. was supported by GM071790. Financial support for X12C beamline (NSLS) comes principally from the Offices of Biological and Environmental Research and of Basic Energy Sciences of the US Department of Energy, and from the National Center for Research Resources of the National Institutes of Health.
Abbreviations used: HutI, imidazolonepropionase; HutH, histidine ammonia lyase; HutU, urocanase; NIG, the N-formimino-L-glutamate; DIP, [3-(2,5-dioxoimidazolidin-4yl)-propionic acid; AT, Agrobacterium tumefaciens; AHS, amidohydrolase superfamily; ES, Sargasso Sea Ocean Going Survey.
References
- 1.Borek BA, Waelsch H. The enzymatic degradation of histidine. J Biol Chem. 1953;205:459–474. [PubMed] [Google Scholar]
- 2.Magasanik B, Bowser HR. The degradation of histidine by Aerobacter aerogenes. J. Biol. Chem. 1955;213:571–580. [PubMed] [Google Scholar]
- 3.Schwede TF, Retey J, Shultz GE. Crystal Structure of Histidine Ammonia-Lyase Revealing a Novel Polypeptide Modification as the Catalytic Electrophile. Biochemistry. 1999;38:5355–5361. doi: 10.1021/bi982929q. [DOI] [PubMed] [Google Scholar]
- 4.Kessler D, Rétey J, Schulz GE. Structure and Action of Urocanase. J. Mol. Biol. 2004;342:183–194. doi: 10.1016/j.jmb.2004.07.028. [DOI] [PubMed] [Google Scholar]
- 5.Schubert C, Rottele H, Spraul M, Retey J. On the mechanism of action of urocanase: Observation of the enzyme-bound NAD+-inhibitor adduct by 13C NMR spectroscopy. Angew. Chem. Int. Ed. Engl. 1995;34:652–654. [Google Scholar]
- 6.Schubert C, Zhao Y, Shin H-H, Retey J. On the mechanism of the urocanase reaction: Cofirmation of the structure of the NAD+-inhibitor adduct by direct 13C-13C coupling. Angew. Chem. Int. Ed. Engl. 1994;33:1279–1280. [Google Scholar]
- 7.Tyagi R, Kumaran D, Burley SK, Swaminathan S. X-ray structure of imidazolonepropionase from Agrobacterium tumefaciens at 1.87 Å resolution. Proteins. 2007;69:652–658. doi: 10.1002/prot.21559. [DOI] [PubMed] [Google Scholar]
- 8.Yu Y, Liang Y-H, Brostromer E, Quan J-M, Panjikar S, Dong Y-H, Su X-D. A Catalytic Mechanism Revealed by the Crystal Structures of the Imidazolonepropionase from Bacillus subtilis. J Biol Chem. 2006;281:36929–36936. doi: 10.1074/jbc.M607703200. [DOI] [PubMed] [Google Scholar]
- 9.Holm L, Sander C. An evolutionary treasure: unification of a broad set of amidohydrolases related to urease. Proteins. 1997;281:72–82. [PubMed] [Google Scholar]
- 10.Seibert CM, Raushel FM. Structural and catalytic diversity within the amidohydrolase superfamily. Biochemistry. 2005;44:6383–6391. doi: 10.1021/bi047326v. [DOI] [PubMed] [Google Scholar]
- 11.Ireton GC, McDermott G, Black ME, Stoddard BL. The structure of Escherichia coli Cytosine Deaminase. J Mol Biol. 2002;315:687–697. doi: 10.1006/jmbi.2001.5277. [DOI] [PubMed] [Google Scholar]
- 12.Porter TN, Li Y, Raushel FM. Mechanism for the Dihydroorotase Reaction. Biochemistry. 2004;43:16285–16292. doi: 10.1021/bi048308g. [DOI] [PubMed] [Google Scholar]
- 13.Marti-Arbona R, Fresquet V, Thoden JB, Davis ML, Holden HM, Raushel FM. Mechanism of the Reaction Catalyzed by Isoaspartyl Dipeptidase from Escherichia coli. Biochemistry. 2005;44:7115–7124. doi: 10.1021/bi050008r. [DOI] [PubMed] [Google Scholar]
- 14.Marti-Arbona R, Raushel FM. Mechanistic Characterization of N-Formimino-L-Glutamate Iminohydrolase from Pseudomonas aeruginosa. Biochemistry. 2006;45:14256–14262. doi: 10.1021/bi061673i. [DOI] [PubMed] [Google Scholar]
- 15.Hall RS, Xiang DF, Xu C, Raushel FM. N-Acetyl-d-Glucosamine-6-Phosphate: Substrate Activation via a Single Divalent Metal Ion. Biochemistry. 2007;46:7942–7952. doi: 10.1021/bi700543x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Williams L, Nguyen T, Li Y, Porter TN, Raushel FM. Uronate Isomerase: A Nonhydrolytic Member of the Amidohydrolase Superfamily with an Ambivalent Requirement for a Divalent Metal Ion. Biochemistry. 2006;45:7453–7462. doi: 10.1021/bi060531l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilson DK, Rudolph FB, Quiocho FA. Atomic Structure of Adenosine Deaminase Complexed with a Transition State Analog: Understanding Catalysis and Immunodeficiency Mutations. Science. 1991;252:1278–1284. doi: 10.1126/science.1925539. [DOI] [PubMed] [Google Scholar]
- 18.Liaw SH, Chang YJ, Lai CT, Chang HC, Chang GG. Crystal Structure of Bacillus subtilis Guanine Deaminase. The first domain-swapped structure in the cytidine deaminase superfamil. J. Biol. Chem. 2004;279:35479–35485. doi: 10.1074/jbc.M405304200. [DOI] [PubMed] [Google Scholar]
- 19.Hermann J, Marti-Arbona R, Fedorov E, Fedorov A, Almo S, Shoichet BK, Raushel FM. Structure-Based Activity Prediction for an Enzyme of Unknown Function. Nature. 2007;448:775–779. doi: 10.1038/nature05981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rao KN, Bonanno JB, Burley SK, Swaminathan S. Crystal Structure of Glycerophosphodiester Phosphodiesterase From Agrobacterium tumefaciens by SAD With a Large Asymmetric Unit. Proteins. 2006;65:514–518. doi: 10.1002/prot.21079. [DOI] [PubMed] [Google Scholar]
- 21.Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods Enzymol. 1997;276:307–648. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 22.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 23.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PRO-CHECK: a program to check the stereochemical quality of protein structures. J Appl Cryst. 1993;26:283–291. [Google Scholar]
- 24.Jones S, Thornton JM. Principles of protein-protein interactions. 1996;93:13–20. doi: 10.1073/pnas.93.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.HingeProt [Google Scholar]
- 26.Holm L, Sander C. Protein structure comparison by alignment of distance matrices. J Mol Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]