Abstract
Background
MicroRNAs are regulators of gene expression, which act mainly by decreasing mRNA levels of their multiple targets. Deregulated microRNA expression has been shown for acute myeloid leukemia, a disease also characterized by altered gene expression associated with distinct genomic aberrations such as nucleophosmin (NPM1) mutations. To shed further light on the role of deregulated microRNA and gene expression in cytogenetically normal acute myeloid leukemia with NPM1 mutation we performed an integrative analysis of microRNA and mRNA expression data sets.
Design and Methods
Both microRNA and gene expression profiles were investigated in samples from a cohort of adult cytogenetically normal acute myeloid leukemia patients (n=43; median age 46 years, range 23–60 years) with known NPM1 mutation status (n=23 mutated, n=20 wild-type) and the data were integratively analyzed. Putative microRNA-mRNA interactions were validated by quantitative reverse transcriptase polymerase chain reaction, western blotting and luciferase reporter assays. For selected microRNAs, sensitivity of microRNA-overexpressing cells to cytarabine treatment was tested by FACS viability and cell proliferation assays.
Results
Our integrative approach of analyzing both microRNA- and gene expression profiles in parallel resulted in a refined list of putative target genes affected by NPM1 mutation-associated microRNA deregulation. Of 177 putative microRNA – target mRNA interactions we identified and validated 77 novel candidates with known or potential involvement in leukemogenesis, such as IRF2-miR-20a, KIT-miR-20a and MN1-miR-15a. Furthermore, our data showed that deregulated expression of tumor suppressor microRNAs, such as miR-29a and miR-30c, might contribute to sensitivity to cytarabine, which is observed in NPM1 mutated acute myeloid leukemia.
Conclusions
Overall, our observations highlight that integrative data analysis approaches can improve insights into leukemia biology, and lead to the identification of novel microRNA - target gene interactions of potential relevance for acute myeloid leukemia treatment.
Keywords: microRNA, miRNA, gene expression profiling, GEP, acute myeloid leukemia, AML, NPM1 mutation
Introduction
In recent years, many chromosomal aberrations and/or gene mutations have been identified which alter normal gene function or expression, contribute to leukemic transformation and provide important diagnostic and prognostic information in acute myeloid leukemia (AML).1 About 40–50% of all cases of AML do not show chromosomal aberrations, and are therefore termed cytogenetically normal (CN-AML). Here, the identification of NPM1, FLT3 or CEBPA gene mutations allows the dissection of CN-AML into subgroups with different prognoses.2 However, the mechanisms of gene deregulation, which is commonly observed in AML, as well as the function of most of the leukemia-associated mutations, including the most common one affecting NPM1, are still not fully understood.1
NPM1 is involved in various cellular processes, and is important in the regulation of cell proliferation and apoptosis.3 Mutations in the NPM1 gene (NPM1mut) were found to be the most prevalent genetic alteration in adult AML patients. Of note, 48–64% of CN-AML patients show mutations in NPM1,2,4–5 which result in mutant proteins that are aberrantly localized to the cytoplasm of leukemic cells and thought to be critical for leukemogenesis.3 AML with NPM1mut is generally associated with a good response to induction chemotherapy.4–6 However, the mechanisms underlying this chemosensitivity are still unknown. Moreover, AML with NPM1mut shows a distinct gene expression profile (GEP),7–9 as well as a distinct microRNA signature,9–11 which both require further exploration to assess their impact on NPM1mut AML pathogenesis.
MicroRNAs (miRNAs) are non-coding RNA molecules of approximately 22 nucleotides, and function as post-transcriptional regulators of gene expression.12 There is recent evidence showing that mammalian miRNAs predominantly act by mRNA destabilization and subsequent decrease of target mRNA levels.13 Given the fact that a miRNA can target the mRNA of hundreds of genes,14–15 this might explain their widespread function in cellular processes such as differentiation, proliferation and apoptosis. Therefore, miRNAs can function both as oncogenes and tumor suppressors, and deregulated expression has been associated with various human cancers, including AML.16–17 Early studies interrelating miRNA expression and gene expression profiles already revealed interesting insights into altered gene regulation in AML, for example miR-29b mediated MCL1 regulation.18 However, large-scale integrative analyses investigating the direct impact of miRNA deregulation on gene expression have not been performed so far. In the current study, we therefore screened miRNA- and gene expression in a defined cohort of adults with CN-AML and focused our efforts on NPM1mut-associated miRNA- and gene expression changes. We then investigated the interrelation of miRNA- and gene expression signatures by an integrative analysis of the respective NPM1mut-associated profiles.
Design and Methods
The patients’ samples and cell lines used are described in the Online Supplementary Appendix, which also provides information and data concerning cell culture, miRNA expression profiling, northern blotting and quantitative reverse-transcriptase PCR (qRT-PCR) analysis for miRNA detection, gene expression profiling, data analysis of microarrays, immunoblot analysis, luciferase reporter assays and nucleofection of myeloid cells.
MicroRNA expression profiling
To screen miRNA expression in AML and leukemia cell lines, we set up a microarray platform using a commercially available oligonucleotide probe set based on version 6.0 of the Sanger miRNA database (mirVana miRNA Probe Set, Ambion). Further details on miRNA expression profiling, data analysis and validation are provided in the Online Supplementary Appendix.
Transfection with synthetic microRNAs
HeLa cells were transfected with the indicated microRNA mimics (Pre-miRs, Ambion) or a negative control RNA (Pre-miR Negative Control #1, Ambion) at a final concentration of 30 nM using the Lipofectamine 2000 transfection reagent (Invitrogen) according to the manufacturer’s protocol. Cells were harvested 24 or 48 h post-transfection. For target gene investigation and functional analyses, HEL and K-562 cells were transfected with pre-miR at a final concentration of 100 nM using the Nucleofector II Device and Cell Line Nucleofector Kit V (Lonza; see Online Supplementary Appendix for transfection efficiencies, monitored by GFP plasmid transfection).
Quantitative reverse transcriptase polymerase chain reaction for mRNA detection
Total RNA of transfected cell lines was extracted using the mirVana miRNA Isolation Kit (Ambion), and cDNA synthesis using the SuperScript III First-Strand Synthesis System (Invitrogen) was primed with random hexamer primers according to the manufacturer’s protocol. Analyses were carried out using the Fast SYBR Green Master Mix (Applied Biosystems) with a 7900HT Fast Real-Time PCR System (Applied Biosystems) in the fast mode. Primer sequences, the cycling program for the genes analyzed (n=47) and primer testing are presented in the Online Supplementary Appendix (Online Supplementary Table S3, Figures S1 and S2). Samples were assayed in duplicate and three different housekeeping genes (ACTB, LMNB1, PGK1) were analyzed for data normalization. Quantity mean values for gene expression were calculated according to standard curves, for which dilution series of cDNA of untransfected HeLa or HEL cells were used. Data were analyzed using SDS 2.3 software (Applied Biosystems) and visualized using GraphPad Prism 4 (GraphPad Software).
Cell proliferation assays and flow cytometric detection of apoptosis
Two hours post-transfection with pre-miRs, cells were either left untreated or treated with a subtoxic concentration of cytarabine [according to previously measured dose-response curves: ~80% viability in untransfected cells (measured 24 h after exposure to the drug); HEL: 0.01 μM cytarabine; K-562: 1 μM]. Cells were diluted 24 h post-transfection 1:5 in medium with or without cytarabine to prevent overgrowth of the culture. Cell proliferation was measured at 48, 72 and 96 h post-transfection using the ATP quantitating CellTiter-Glo Luminescent Cell Viability Assay (Promega). At the same time-points, apoptosis induction was measured by double-staining the cells with annexin-PE/7-AAD (BD Pharmingen) using a FACSCalibur flow cytometer (BD). FACS data were analyzed using CellQuest Pro software (BD Pharmingen).
Statistical analyses
Group-wise comparisons of the distributions of clinical variables or experimental results were performed using Fisher’s exact test, one-sample t test or unpaired t test, as appropriate. All tests were two-tailed and an effect was considered statistically significant if the P value was 0.05 or less. Data were visualized and analyzed using GraphPad Prism 4.
Results
Parallel analysis of NPM1 mutation-associated microRNA and gene expression changes in cytogenetically normal acute myeloid leukemia
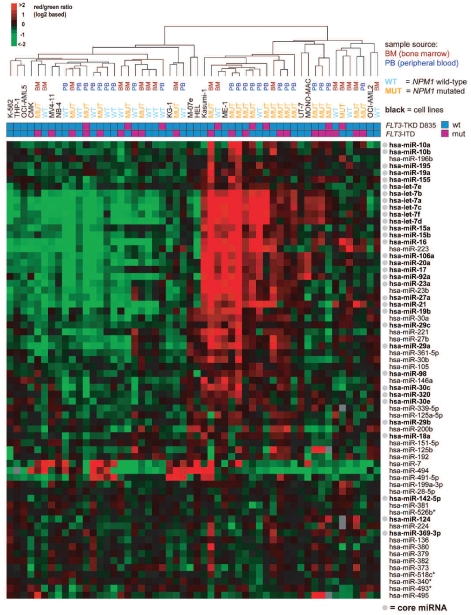
By profiling miRNA and gene expression in 43 CN-AML samples, we identified characteristic NPM1mut-associated expression patterns both at the miRNA and mRNA level. A NPM1mut-associated gene expression signature based on our data (comprising 730 genes down-regulated in NPM1mut cases and 663 up-regulated genes) was in agreement with published signatures,7–9 showing for example the characteristic up-regulation of HOX genes and an inverse correlation with MN1 and BAALC (Online Supplementary Table S4). With regard to the miRNA profiling, 66 miRNAs were differentially expressed in NPM1mut compared to NPM1 wild-type (NPM1wt) cases (Figure 1; Online Supplementary Table S5). These miRNAs comprised, among others, the oncogenic miR-17-92 cluster and miR-155, as well as miRNAs with a known tumor-suppressor role such as the members of the let-7 family, the miR-15/16 cluster, and the miR-29 family. The vast majority of the miRNAs in this signature was up-regulated in NPM1mut CN-AML and showed a broad overlap with previously reported NPM1mut miRNA signatures,9–11 based on which we defined a “core set” of 33 up-regulated miRNAs (core miRNAs defined by overlapping results of our supervised analyses and the miRNA signature published by Garzon et al.10: see Figure 1 and Online Supplementary Table S5). Consistently, hierarchical clustering based on the supervised findings was NPM1-associated (P=0.0238; Figure 1), although in line with the findings of Garzon et al., we did not observe a clear-cut separation of NPM1mut and NPM1wt cases, which might be due to additional yet unknown secondary genomic events. For exemplary miRNAs, microarray findings were validated by northern blot analysis and/or qRT-PCR (Online Supplementary Figure S3).
Figure 1.
Hierarchical clustering based on NPM1mut-associated miRNAs as determined by SAM analysis. This heatmap displays expression levels of the miRNAs associated with NPM1mut CN-AML (determined by SAM NPM1mut versus NPM1wt; false discovery rate = 0.09631). Sixty-six miRNAs (rows) and 40 AML patients with normal karyotype (23 of these with NPM1mut) and 14 NPM1wt leukemia cell lines (columns) were hierarchically clustered (arrays from 3 patients were excluded due to borderline quality). Mean-centered log2 expression ratios are depicted by the color scale, as indicated. Samples have been color coded according to the underlying molecular aberration and sample source (bone marrow or peripheral blood). MUT = NPM1 mutation; WT = NPM1 wild-type; FLT3-ITD = internal tandem duplication; TKD D835 = tyrosine kinase domain (TKD) mutation at codon D835. Cell line names are shown in black. NPM1mut and NPM1wt patients are significantly differentially distributed between the two groups (P=0.0238; Fisher’s exact test), whereas there is no significant difference for FLT3mut and FLT3wt samples (P=0.2756). Sample source (bone marrow versus peripheral blood) did not result in significant miRNA expression pattern differences (P=0.7496). The core 33 upregulated miRNAs used for the subsequent integrated analysis are printed in bold and marked with a gray dot.
In order to identify suitable cell lines for future functional analyses with the aim of over-expressing the miRNAs highly expressed in NPM1mut AML, we profiled miRNA expression of 14 NPM1wt leukemia cell lines. We identified nine cell lines that co-clustered with the NPM1wt-predominant subgroup (Figure 1), which could serve as potential models for functional analyses.
Integrative analysis of NPM1 mutation-associated microRNA and gene expression signatures
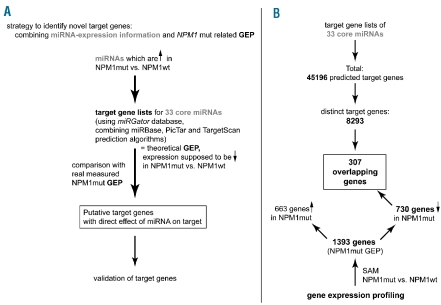
To identify putative target genes of NPM1mut-associated miRNAs, an integrative analysis of miRNA expression and gene expression data was performed (Figure 2). First, we generated target gene lists for the core set of 33 up-regulated miRNAs by using the miRGator database,19 which can combine three different prediction tools [MicroCosm Targets (formerly miRBase Targets, M), TargetScan (T) and PicTar (P)] which are based on different algorithms and prediction stringencies.12 We included all genes that were predicted by any one of these algorithms to obtain a gene list comprising all potential targets, which resulted in 8293 unique predicted target genes (Figure 2B). The predictions by TargetScan and PicTar have a high degree of overlap because they require more stringent seed region pairing than MicroCosm Targets, and only few targets were predicted by all three algorithms (see Online Supplementary Table S6; candidates are marked M/T/P according to the respective algorithm by which they were predicted). To further refine this theoretical list (constituting a theoretical NPM1mut-associated GEP) of potential direct miRNA targets in NPM1mut AML, we filtered for genes down-regulated in NPM1mut versus NPM1wt cases, as a recent study showed that mammalian miRNAs predominantly act to decrease target mRNA levels.13 Thus, we correlated NPM1mut-associated miRNA and mRNA expression, both obtained from the same patients to narrow down putative target genes of which mRNA levels might have been directly affected by the respective miRNAs. As many factors can have an impact on gene expression levels, such an approach can only reveal direct targets in which no influencing factors obscure the direct miRNA-mRNA interaction, and in addition will likely reveal many false positive interactions. Nevertheless, we hypothesized that such an approach might allow us to identify and validate novel miRNA-mRNA interaction partners, although the potential direct miRNA targets were not statistically significantly enriched in the NPM1mut-associated mRNA signature, most likely due to fact that gene expression is influenced by many factors. For 307 NPM1mut-associated down-regulated genes the putative targeting miRNA was up-regulated (genes highlighted in Online Supplementary Table S4; core miRNAs predicted to target the respective genes are listed in Online Supplementary Table S7). These included candidate genes implicated in tumorigenesis and/or leukemogenesis, such as APP, CCND1, IRF2, SPARC, MN1, SERPINB9 and KIT. Interestingly, many of these genes are also putative targets of not only one, but several miRNAs of the NPM1mut-associated miRNA signature, and/or have several binding sites for the respective individual miRNAs.
Figure 2.
Integrative data analysis strategy and results. (A) Strategy description of integrative analysis of miRNA- and gene-expression data. (B) Results of integrative analysis. Target gene lists of 33 core miRNAs resulted in a total of 45196 target genes (multiple entries per gene, because of being predicted targets by several miRNAs with several target sites), which represent 8293 distinct gene names).
Validation of target genes by quantitative reverse transcriptase polymerase chain reaction in cells transfected with synthetic microRNAs
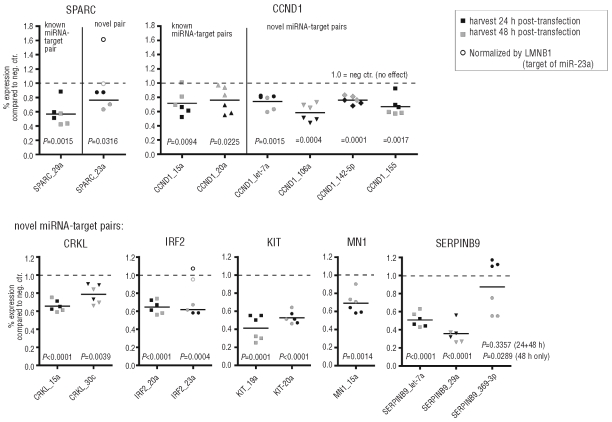
To further investigate individual miRNA-target gene relations, we first studied the impact of transfection of synthetic miRNA mimics on target gene mRNA levels by qRT-PCR. As testing all 33 miRNAs and 307 putative direct target genes was not feasible, we focused on 11 miRNAs representative of most miRNA families present in the NPM1mut signature and selected 42 genes of potential relevance for leukemogenesis and/or tumorigenesis (n=177 different miRNA-target gene pairs predicted by at least one of the three prediction algorithms M/T/P, see Online Supplementary Table S6). Expression of the putative target genes was analyzed 24 and 48 h post-transfection (exemplary genes are depicted in Figure 3; detailed overview of results in Online Supplementary Table S6). For data normalization three different housekeeping genes (ACTB, LMNB1, PGK1) were utilized as endogenous controls (Figure 3), because housekeeping genes themselves can be targeted by miRNAs (Online Supplementary Table S6). For example, normalization of the measurements for SPARC and IRF2 using LMNB1, known to be targeted by miR-23a,20 might lead to misinterpretation of results if compared to normalization with the other housekeeping genes (Figure 3, open symbols; Online Supplementary Table S8).
Figure 3.
Validation of predicted miRNA targets by qRT-PCR. Normalized qRT-PCR analyses of selected miRNA target genes in HeLa cells which were transfected with the indicated miRNA mimic or negative control and harvested 24 or 48 h post-transfection. Each data point represents the normalization with one of three housekeeping genes (ACTB, LMNB1, PGK1) and is the mean of two independent transfection experiments (with 2 technical replicates each). Each value is calculated as percentage expression of target gene mRNA of miRNA-of-interest transfected cells compared to negative control (1.0 = 100%) transfected cells. The horizontal lines show mean reduction. All effects were tested for significance (mean different from 1; one-sample t test). For SPARC and IRF2, outliers (open symbols) are the values normalized by LMNB1 (a known target of miR-23a) and were therefore excluded for calculation of the mean and for statistical testing.
As expected, target gene validation was successful for known miRNA-target gene pairs (Figure 3; Online Supplementary Table S6, blue shaded boxes), which were almost all detected by our qRT-PCR screen (with two exceptions out of 11), thereby validating our experimental approach of monitoring mRNA changes to identify novel miRNA-target gene interactions. Examples include E2F1 regulated by miR-20a,21 SPARC targeted by miR-29a,22 and CCND1 regulated by miR-15a and miR-20a.23–24 In addition, many novel pairs were identified, including SPARC, which was also significantly affected by miR-23a, and the cell cycle regulator CCND1, which was influenced by several miRNAs of the signature. Further novel target genes included interferon regulatory factor-2 (IRF2), regulated by miR-20a and miR-23a, the receptor tyrosine kinase KIT (regulated by miR-19a and miR-20a) and SERPINB9 (regulated by let-7a, miR-29a and miR-369-3p). In total, 35 of the 177 miRNA-target gene relations (19.8%) showed a mRNA level reduction of more than 30% after miRNA transfection, a cut-off defined by effects that in general were highly significant (P<0.001; Online Supplementary Table S6, green shading, and Online Supplementary Figure S4), and 42 of 177 relations (23.7%) showed a weaker but consistent and significant mRNA reduction in the range of 10–30% (P<0.05; Online Supplementary Table S6, yellow shading). Thus, in total we observed for 43.5% of miRNA-target gene pairs (77 of 177) an effect of miRNA transfection on mRNA levels.
Furthermore, selected miRNA-target gene interactions (n=13) were validated in two different myeloid cell lines (HEL and K-562; Online Supplementary Figure S5).
Validation of targets on protein level and by luciferase reporter experiments
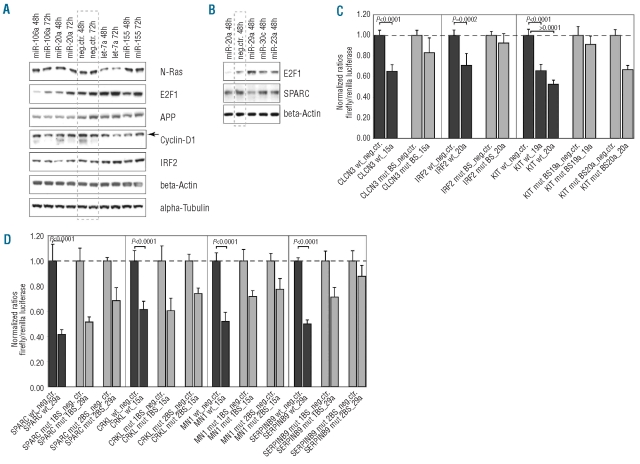
Following qRT-PCR validation of miRNA-target gene pairs, we looked at protein expression of selected candidates in HeLa cells transfected with miRNA mimics. Western blot analysis of targets showed that the transfected miRNAs also led to a down-regulation of the respective target protein levels (compared to negative control RNA transfected cells), such as the positive controls E2F1, targeted by miR-20a and miR-106a,21 and NRAS, targeted by let-7a (Figures 4A-B).25 APP protein levels were reduced by miR-20a and miR-106a transfection, as previously reported.26–27 As seen in the qRT-PCR analyses, cyclin-D1 levels were reduced by several miRNAs of the signature, here shown for miR-20a (48 h post-transfection, 22% reduction compared to negative control),24 and the novel regulators miR-106a, let-7a (27% and 34% reduction at 48 h, and 35% and 54% reduction at 72 h, respectively) and miR-155 (44% reduction at 48 h). Moreover, the newly identified miR-20a target IRF2 was reduced on protein level, supporting the qRT-PCR results. As previously described, SPARC levels were strongly down-regulated by miR-29a (46% reduction),22 and also, to a lesser extent (9%), by miR-23a, again as seen in the qRT-PCR analysis.
Figure 4.
Validation of miRNA targets on protein level and by luciferase reporter experiments. (A) and (B) Protein expression analysis of selected known and novel miRNA targets by western blots in HeLa cells, which were transiently transfected with the indicated miRNA mimics or a negative control RNA (lanes marked by dashed gray line) and harvested at the indicated time points (48 or 72 h post-transfection). β-Actin and α-tubulin served as loading controls. (C) and (D) Normalized ratios firefly/renilla luciferase as determined by reporter assays in HeLa cells which were transfected with the indicated miRNA mimics or a negative control (neg.ctr.; ratio set to 1) and co-transfected with the pMIR-REPORT vector containing the 3‘UTR of candidate target genes downstream of the firefly luciferase reporter gene. (C) 3’UTRs containing one putative miRNA binding site, (D) 3’UTRs containing two putative miRNA binding sites. MiRNA binding sites were either wild-type (wt; dark gray bars) or mutated (mut 1BS = 1 binding site mutated, mut 2BS = both binding sites mutated; light gray bars). Bars represent the mean of two independent experiments with transfections performed in triplicate, error bars depict the standard deviation (SD). All effects observed were highly significant (P<0.001; unpaired t test).
To further validate the direct regulatory effect of miRNAs of the signature on their target genes, we performed luciferase reporter assays. Compared to negative control-transfected cells, the firefly/renilla luciferase ratio was strongly reduced in the miRNA-transfected cells for the positive control SPARC plus miR-29a (58% reduction). A significant reduction was also apparent for the newly identified miRNA-target gene pairs CLCN3–miR-15a, CRKL–miR-15a, IRF2-miR-20a, KIT–miR-19a and -miR-20a, MN1–miR-15a as well as SERPINB9-miR-29a (29–50% reduction, P<0.001; dark gray bars in Figures 4 C-D). Here, the mutant binding sites were able to attenuate repression (Figures 4 C-D, light gray bars and Online Supplementary Figure S6), suggesting a direct regulation of the target by the respective miRNA. For 3’UTR with two predicted miRNA binding sites, mutation of both sites was necessary to diminish repression of luciferase effectively (Figure 4D).
NPM1 mutation-associated microRNAs might contribute to chemosensitivity
With our integrative approach we identified a variety of novel miRNA- target gene pairs of our NPM1mut-associated miRNA signature, consisting of various miRNAs whose expression was significantly up-regulated in NPM1mut compared to NPM1wt cases. To analyze the biological role of these miRNAs, we looked at the effects of ectopic miRNA expression in two NPM1wt leukemic cell lines (HEL and K-562). As expected, both cell lines expressed the miRNAs of the signature at a low level (Figure 1), and were therefore suitable for studying the effects of miRNA over-expression.
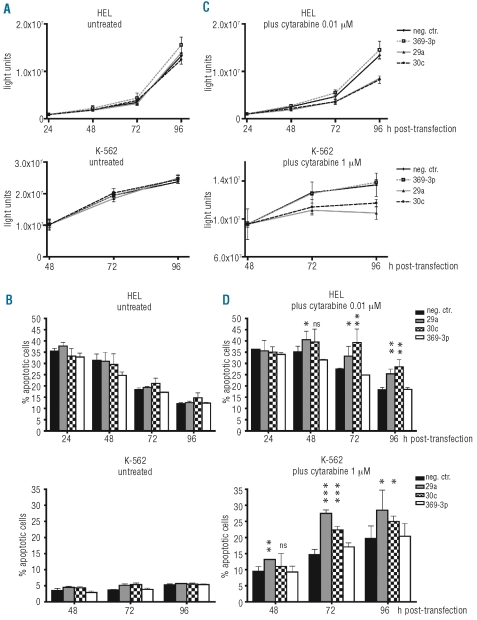
In our experimental set-up, we did not observe a significant growth inhibitory effect for miR-29a, miR-30c or miR-369-3p transfection in HEL and K-562 cells, if the cells were left untreated after transfection and were not exposed to additional stress (Figure 5A). In agreement, there was also no apoptosis induction effect following miRNA over-expression in either cell line (Figure 5B).
Figure 5.
Functional effect of over-expression of miRNAs of the NPM1mut signature. (A) and (C) Growth curves of HEL cells (upper panels) or K-562 cells (lower panels) transfected with miRNA mimics of miR-29a, -30c, -369-3p or a negative control. Cells were either left untreated (A) or were exposed to subtoxic concentrations of cytarabine (C) after transfection (HEL: 0.01 μM cytarabine; K-562: 1 μM cytarabine). Cell growth was monitored by CellTiter-Glo assay at the indicated time-points post-transfection. (B) and (D) Monitoring of apoptosis induction by double-staining of cells with annexin-PE/7-AAD at the indicated time-points post-transfection. Cells were either left untreated (B) or were exposed to subtoxic concentrations of cytarabine (D), as mentioned above. The results (mean of 3 independent experiments; error bars show standard deviation) are shown as percentage of apoptotic cells (annexin-PE positive plus annexin-PE/7-AAD double-positive cells) and were tested for significance (compared to neg.ctr.).
Recently, it was shown for breast cancer cells that miRNA expression can increase sensitivity of cells for example to doxorubicin- or VP-16-induced loss of viability.28–29 Therefore, we next tested for a possible sensitization effect by these miRNAs in our leukemic cell line models. Transfected cells were exposed to subtoxic concentrations of cytarabine, which constitutes the backbone of chemotherapy in AML treatment. Interestingly, after cytarabine treatment a growth inhibitory effect was observed in miR-29a as well as in miR-30c over-expressing cells, but not in miR-369-3p over-expressing cells (Figure 5C). Moreover, in cytarabine-treated cells, a remarkable induction of apoptosis could be observed for miR-29a and miR-30c beginning 48 h post-transfection and treatment start and peaking at 72-96 h post-transfection (87% increase in apoptotic cells by miR-29a and 52% by miR-30c for K-562 cells at 72 h; Figure 5D). For HEL cells, the induction of apoptosis by miR-30c was even more pronounced than that by miR-29a, peaking at 96 h after transfection (56% increase in apoptotic cells by miR-30c and 39% by miR-29a at 96 h). Thus, the over-expression of distinct miRNAs of the NPM1mut signature can lead to sensitization to cytarabine.
Discussion
Recently, several miRNA profiling studies in AML revealed an up-regulation of distinct miRNAs in NPM1mut versus NPM1wt CN-AML cases as one of the most consistent signatures in AML.9–10 In accordance, our profiling approach also revealed a strong NPM1mut-associated signature of up-regulated miRNAs, which substantially overlapped with published data and comprised many miRNAs known to be implicated in tumorigenesis and/or leukemogenesis. Exemplary miRNAs with an oncogenic potential included miR-155,30 as well as miR-17-92 cluster members.31 Furthermore, our core miRNA signature also comprised tumor-suppressor miRNAs including the miR-29 and miR-30 family. Here, one might speculate that this could in part account for the favorable response to therapy in NPM1mut cases, or vice versa, low expression of the respective tumor-suppressor miRNAs in NPM1wt cases might contribute to an inferior outcome. Therefore, we hypothesize that over-expression of a distinct set of miRNAs might play an important role in NPM1mut CN-AML, resulting in deregulated expression of target genes possibly contributing to leuke-mogenesis. So far, this issue has not been addressed systematically for NPM1mut CN-AML.
In order to identify putative target genes of the NPM1mut-associated miRNAs, miRNA target gene lists can be generated using prediction algorithms, such as the miRGator database, which usually reveal a large number of candidates. This list could be effectively narrowed down to a set of promising candidate genes potentially implicated in NPM1mut-associated leukemogenesis by integrating this theoretical NPM1mut-associated target gene list with GEP data. Similar to our approach, other studies in AML have combined miRNA and gene expression information; however, many analyzed only a limited number of miRNAs and the expression of their putative target genes at a time (such as miR-181a and the HOX-associated miRNA miR-196a, -10a and -10b).32–33 Thus, in addition to a recent study by Havelange et al., who integrated miRNA and mRNA expression from 48 AML cases,34 our study represents an additional large integrative approach that combines miRNA and gene expression data from the same patient cohort, with the purpose to understand the complex regulatory network of miRNA and associated target genes involved in the pathogenesis and clinical characteristics of NPM1mut AML.
Recently, it has been shown that mammalian miRNAs act mainly via mRNA-destabilization and subsequent decrease of target mRNA levels and not only, as previously thought, via translational repression.13,15 We, therefore, evaluated putative miRNA-target gene interactions following over-expression of the respective miRNAs in cell line models by mRNA expression analysis using qRT-PCR, and in contrast to Havelange et al. focused less on gene ontology and network analysis.34 This approach allowed us to screen a comprehensive list of target genes of representatives of the miRNA families deregulated in NPM1mut AML. Members of these miRNA families mostly have the same predicted targets due to identical seed region sequences.12 We were not only able to validate known miRNA-target gene interactions such as miR-20a-E2F1 and miR-15a-CCND1, but also to identify a variety of novel miRNA targets. Thus, while our mRNA measurement-based approach might miss single target genes, monitoring mRNA levels nevertheless constitutes a feasible and fast method to identify novel miRNA targets, especially as candidates could also be validated by western blotting and luciferase reporter assays.
One example of a newly validated miRNA target is IRF2, coding for a transcriptional suppressor of type I interferon (IFN) signaling. Mice deficient for IRF2 show a substantial decrease of hematopoietic stem cells (HSCs) and a significant increase of immature progenitor cells.35 Being essential for the preservation of the HSC pool, IRF2 normally suppresses IFN signaling in wild-type HSCs, thereby maintaining HSCs mostly in a quiescent state. In accordance, IFN-α stimulates the proliferation and turnover of dormant HSCs in vivo, and the cycling HSCs then become sensitive to antiproliferative chemotherapeutic agents, which are not effective for dormant HSCs.35–36 This mechanism might in part contribute to the chemosensitivity of NPM1mut AML cases. Also, IRF2 knock-down associated growth inhibition and induction of differentiation in a leukemia cell line further emphasizes the role of IRF2 deregulation in leukemogenesis.37 Moreover, the receptor tyrosine kinase KIT, important for normal hematopoiesis,38 was validated as a target of several NPM1mut-associated miRNAs. KIT is crucial for maintaining HSC self-renewal and quiescence and was shown to be down-regulated in IRF2 deficient cells.35 Similarly, the correlation of low MN1 expression and NPM1mut might be explained by miRNA deregulation, and in line with our findings a recent MN1-associated miRNA signature showed a negative correlation of MN1 expression and the miR-15/16 cluster.39 Functioning as an oncogenic transcriptional co-activator, high expression of MN1 is associated with poor outcome in CN-AML, and especially with a poor response to induction chemotherapy.40 In accordance with this, there is recent evidence from a murine AML model showing that MN1 over-expression leads to resistance to cytarabine and doxorubicin.41 Thus, the down-regulation of genes involved in chemosensitivity such as MN1, KIT and IRF2 by NPM1mut-associated miRNAs might be linked to the favorable response to chemotherapy observed for NPM1mut AML.
In addition, several studies have shown that miRNAs can predict sensitivity or resistance to anticancer-treatment: for example, miR-30c down-regulation has been associated with drug-resistant ovarian cancer cells, and miRNAs were also reported to influence sensitivity to chemotherapy.29 In accordance with these findings, our analysis of the biological role of selected up-regulated NPM1mut-associated miRNAs showed that over-expression of miR-29a and miR-30c can sensitize cells to cytarabine, leading to growth inhibition and apoptosis induction following low-dose cytara-bine treatment. This tumor-suppressive role for the miR-29 family has also been shown in a xenograft leukemia model,18 and our results in cell lines indicate that over-expression of miR-30c has a similarly pronounced growth inhibitory and apoptosis-inducing effect as miR-29a. Thus, the over-expression of tumor-suppressive miRNAs in NPM1mut AML, in addition to the influence of miRNAs on single genes as mentioned above, might contribute to the favorable response to chemotherapy. On the other hand, down-regulation of the miR-29 and miR-30 family members in NPM1wt cases might play an important role in leuke-mogenesis. Furthermore, a recent study inferring miRNA networks from miRNA expression data in normal tissue or cancer also suggested a link between the miR-29 and miR-30 family.42 In normal tissue as well as in AML, the expression patterns of miR-29 and miR-30 family members are closely related, thereby implying co-regulation and a potential collaborative tumor-suppressor mechanism. Therefore, a miRNA-based therapy using miR-30c, as already suggested for the miR-29 family,18 and its ability to induce chemosensitivity, might be an interesting approach for the treatment of AML.
In conclusion, our integrative data analysis approach combining miRNA and gene expression information led to the identification of a valuable panel of novel miRNA-target gene interactions that will benefit future investigations of the biology of NPM1mut AML. Further studies will be necessary to explore the functional impact of the respective interconnections in more depth and should also focus on the mechanisms underlying the miRNA deregulation. Nevertheless, the chemosensitivity-associated candidates identified in our study may contribute to an improved understanding of chemoresistance in NPM1wt cases and AML in general and ultimately result in novel therapeutic strategies for adverse-risk AML.
Acknowledgments
The authors would like to thank the staff of the Microarray Facility of the University of Ulm for providing high-quality microarrays. Furthermore, we thank Sabrina Heinrich for excellent technical assistance, and Stefan Fröhling for his critical review of the manuscript. We thank all the members of the German-Austrian AMLSG for their continuous support of the study group and its treatment protocols.
Footnotes
Funding: this work was supported in part by the Deutsche José Carreras Stiftung e.V. (DJCLS R 06/41v). LB was supported in part by the Deutsche Forschungsgemeinschaft (Heisenberg-Stipendium BU 1339/3-1). SS was supported by the International Graduate School in Molecular Medicine Ulm.
The online version of this article has a Supplementary Appendix.
Authorship and Disclosures
The information provided by the authors about contributions from persons listed as authors and in acknowledgments is available with the full text of this paper at www.haematologica.org.
Financial and other disclosures provided by the authors using the ICMJE (www.icmje.org) Uniform Format for Disclosure of Competing Interests are also available at www.haematologica.org.
References
- 1.Dohner H, Estey EH, Amadori S, Appelbaum FR, Buchner T, Burnett AK, et al. Diagnosis and management of acute myeloid leukemia in adults: recommendations from an interna-tional expert panel, on behalf of the European LeukemiaNet. Blood. 2010;115(3):453–74. doi: 10.1182/blood-2009-07-235358. [DOI] [PubMed] [Google Scholar]
- 2.Schlenk RF, Dohner K, Krauter J, Frohling S, Corbacioglu A, Bullinger L, et al. Mutations and treatment outcome in cytogenetically normal acute myeloid leukemia. N Engl J Med. 2008;358(18):1909–18. doi: 10.1056/NEJMoa074306. [DOI] [PubMed] [Google Scholar]
- 3.Falini B, Bolli N, Liso A, Martelli MP, Mannucci R, Pileri S, et al. Altered nucle-ophosmin transport in acute myeloid leukaemia with mutated NPM1: molecular basis and clinical implications. Leukemia. 2009;23(10):1731–43. doi: 10.1038/leu.2009.124. [DOI] [PubMed] [Google Scholar]
- 4.Falini B, Mecucci C, Tiacci E, Alcalay M, Rosati R, Pasqualucci L, et al. Cytoplasmic nucleophosmin in acute myelogenous leukemia with a normal karyotype. N Engl J Med. 2005;352(3):254–66. doi: 10.1056/NEJMoa041974. [DOI] [PubMed] [Google Scholar]
- 5.Dohner K, Schlenk RF, Habdank M, Scholl C, Rucker FG, Corbacioglu A, et al. Mutant nucleophosmin (NPM1) predicts favorable prognosis in younger adults with acute myeloid leukemia and normal cytogenetics: interaction with other gene mutations. Blood. 2005;106(12):3740–6. doi: 10.1182/blood-2005-05-2164. [DOI] [PubMed] [Google Scholar]
- 6.Falini B, Martelli MP, Bolli N, Sportoletti P, Liso A, Tiacci E, et al. Acute myeloid leukemia with mutated nucleophosmin (NPM1): is it a distinct entity? Blood. 2011;117(4):1109–20. doi: 10.1182/blood-2010-08-299990. [DOI] [PubMed] [Google Scholar]
- 7.Alcalay M, Tiacci E, Bergomas R, Bigerna B, Venturini E, Minardi SP, et al. Acute myeloid leukemia bearing cytoplasmic nucleophosmin (NPMc+ AML) shows a distinct gene expression profile characterized by up-regulation of genes involved in stem-cell maintenance. Blood. 2005;106(3):899–902. doi: 10.1182/blood-2005-02-0560. [DOI] [PubMed] [Google Scholar]
- 8.Verhaak RG, Goudswaard CS, van Putten W, Bijl MA, Sanders MA, Hugens W, et al. Mutations in nucleophosmin (NPM1) in acute myeloid leukemia (AML): association with other gene abnormalities and previously established gene expression signatures and their favorable prognostic significance. Blood. 2005;106(12):3747–54. doi: 10.1182/blood-2005-05-2168. [DOI] [PubMed] [Google Scholar]
- 9.Becker H, Marcucci G, Maharry K, Radmacher MD, Mrozek K, Margeson D, et al. Favorable prognostic impact of NPM1 mutations in older patients with cytogeneti-cally normal de novo acute myeloid leukemia and associated gene- and microRNA-expression signatures: a Cancer and Leukemia Group B study. J Clin Oncol. 2010;28(4):596–604. doi: 10.1200/JCO.2009.25.1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garzon R, Garofalo M, Martelli MP, Briesewitz R, Wang L, Fernandez-Cymering C, et al. Distinctive microRNA signature of acute myeloid leukemia bearing cytoplasmic mutated nucleophosmin. Proc Natl Acad Sci USA. 2008;105(10):3945–50. doi: 10.1073/pnas.0800135105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jongen-Lavrencic M, Sun SM, Dijkstra MK, Valk PJ, Lowenberg B. MicroRNA expression profiling in relation to the genetic heterogeneity of acute myeloid leukemia. Blood. 2008;111(10):5078–85. doi: 10.1182/blood-2008-01-133355. [DOI] [PubMed] [Google Scholar]
- 12.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466(7308):835–40. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455(7209):58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 15.Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455(7209):64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10(10):704–14. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marcucci G, Mrozek K, Radmacher MD, Garzon R, Bloomfield CD. The prognostic and functional role of microRNAs in acute myeloid leukemia. Blood. 2011;117(4):1121–9. doi: 10.1182/blood-2010-09-191312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garzon R, Heaphy CE, Havelange V, Fabbri M, Volinia S, Tsao T, et al. MicroRNA 29b functions in acute myeloid leukemia. Blood. 2009;114(26):5331–41. doi: 10.1182/blood-2009-03-211938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nam S, Kim B, Shin S, Lee S. miRGator: an integrated system for functional annotation of microRNAs. Nucleic Acids Res. 2008;36(Database issue):D159–64. doi: 10.1093/nar/gkm829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin ST, Fu YH. miR-23 regulation of lamin B1 is crucial for oligodendrocyte development and myelination. Dis Model Mech. 2009;2(3–4):178–88. doi: 10.1242/dmm.001065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435(7043):839–43. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 22.Kapinas K, Kessler CB, Delany AM. miR-29 suppression of osteonectin in osteoblasts: regulation during differentiation and by canonical Wnt signaling. J Cell Biochem. 2009;108(1):216–24. doi: 10.1002/jcb.22243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bonci D, Coppola V, Musumeci M, Addario A, Giuffrida R, Memeo L, et al. The miR-15a-miR-16-1 cluster controls prostate cancer by targeting multiple oncogenic activities. Nat Med. 2008;14(11):1271–7. doi: 10.1038/nm.1880. [DOI] [PubMed] [Google Scholar]
- 24.Yu Z, Wang C, Wang M, Li Z, Casimiro MC, Liu M, et al. A cyclin D1/microRNA 17/20 regulatory feedback loop in control of breast cancer cell proliferation. J Cell Biol. 2008;182(3):509–17. doi: 10.1083/jcb.200801079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120(5):635–47. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 26.Hebert SS, Horre K, Nicolai L, Bergmans B, Papadopoulou AS, Delacourte A, et al. MicroRNA regulation of Alzheimer's Amyloid precursor protein expression. Neurobiol Dis. 2009;33(3):422–8. doi: 10.1016/j.nbd.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 27.Patel N, Hoang D, Miller N, Ansaloni S, Huang Q, Rogers JT, et al. MicroRNAs can regulate human APP levels. Mol Neurodegener. 2008;3:10. doi: 10.1186/1750-1326-3-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liang Z, Wu H, Xia J, Li Y, Zhang Y, Huang K, et al. Involvement of miR-326 in chemotherapy resistance of breast cancer through modulating expression of multidrug resistance-associated protein 1. Biochem Pharmacol. 2010;79(6):817–24. doi: 10.1016/j.bcp.2009.10.017. [DOI] [PubMed] [Google Scholar]
- 29.Hummel R, Hussey DJ, Haier J. MicroRNAs: predictors and modifiers of chemo- and radiotherapy in different tumour types. Eur J Cancer. 2010;46(2):298–311. doi: 10.1016/j.ejca.2009.10.027. [DOI] [PubMed] [Google Scholar]
- 30.O'Connell RM, Rao DS, Chaudhuri AA, Boldin MP, Taganov KD, Nicoll J, et al. Sustained expression of microRNA-155 in hematopoietic stem cells causes a myelopro-liferative disorder. J Exp Med. 2008;205(3):585–94. doi: 10.1084/jem.20072108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Olive V, Jiang I, He L. mir-17-92, a cluster of miRNAs in the midst of the cancer network. Int J Biochem Cell Biol. 2010;42(8):1348–54. doi: 10.1016/j.biocel.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Debernardi S, Skoulakis S, Molloy G, Chaplin T, Dixon-McIver A, Young BD. MicroRNA miR-181a correlates with morphological subclass of acute myeloid leukaemia and the expression of its target genes in global genome-wide analysis. Leukemia. 2007;21(5):912–6. doi: 10.1038/sj.leu.2404605. [DOI] [PubMed] [Google Scholar]
- 33.Lutherborrow M, Bryant A, Jayaswal V, Agapiou D, Palma C, Yang YH, et al. Expression profiling of cytogenetically normal acute myeloid leukemia identifies microRNAs that target genes involved in monocytic differentiation. Am J Hematol. 2011;86(1):2–11. doi: 10.1002/ajh.21864. [DOI] [PubMed] [Google Scholar]
- 34.Havelange V, Stauffer N, Heaphy CC, Volinia S, Andreeff M, Marcucci G, et al. Functional implications of microRNAs in acute myeloid leukemia by integrating microRNA and messenger RNA expression profiling. Cancer. 2011 Mar 31; doi: 10.1002/cncr.26096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sato T, Onai N, Yoshihara H, Arai F, Suda T, Ohteki T. Interferon regulatory factor-2 protects quiescent hematopoietic stem cells from type I interferon-dependent exhaustion. Nat Med. 2009;15(6):696–700. doi: 10.1038/nm.1973. [DOI] [PubMed] [Google Scholar]
- 36.Essers MA, Offner S, Blanco-Bose WE, Waibler Z, Kalinke U, Duchosal MA, et al. IFNalpha activates dormant haematopoietic stem cells in vivo. Nature. 2009;458(7240):904–8. doi: 10.1038/nature07815. [DOI] [PubMed] [Google Scholar]
- 37.Choo A, Palladinetti P, Holmes T, Basu S, Shen S, Lock RB, et al. siRNA targeting the IRF2 transcription factor inhibits leukaemic cell growth. Int J Oncol. 2008;33(1):175–83. [PubMed] [Google Scholar]
- 38.Lennartsson J, Jelacic T, Linnekin D, Shivakrupa R. Normal and oncogenic forms of the receptor tyrosine kinase kit. Stem Cells. 2005;23(1):16–43. doi: 10.1634/stemcells.2004-0117. [DOI] [PubMed] [Google Scholar]
- 39.Langer C, Marcucci G, Holland KB, Radmacher MD, Maharry K, Paschka P, et al. Prognostic importance of MN1 transcript levels, and biologic insights from MN1-associated gene and microRNA expression signatures in cytogenetically normal acute myeloid leukemia: a cancer and leukemia group B study. J Clin Oncol. 2009;27(19):3198–204. doi: 10.1200/JCO.2008.20.6110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Heuser M, Beutel G, Krauter J, Dohner K, von Neuhoff N, Schlegelberger B, et al. High meningioma 1 (MN1) expression as a predictor for poor outcome in acute myeloid leukemia with normal cytogenetics. Blood. 2006;108(12):3898–905. doi: 10.1182/blood-2006-04-014845. [DOI] [PubMed] [Google Scholar]
- 41.Pardee T, Mascenik T, Bolemon BH, Cook GJ. Over expression of MN1 confers resistance to chemotherapy in vitro and in vivo. Blood (ASH Annual Meeting Abstracts) 2010;116:969. [Google Scholar]
- 42.Volinia S, Galasso M, Costinean S, Tagliavini L, Gamberoni G, Drusco A, et al. Reprogramming of miRNA networks in cancer and leukemia. Genome Res. 2010;20(5):589–99. doi: 10.1101/gr.098046.109. [DOI] [PMC free article] [PubMed] [Google Scholar]