Abstract
The human DNA repair enzyme uracil DNA glycosylase (hUNG) locates and excises rare uracil bases that arise in DNA from cytosine deamination or through dUTP incorporation by DNA polymerases. Previous NMR studies of hUNG have revealed millisecond time scale dynamic transitions in the enzyme-nonspecific DNA complex, but not the free enzyme, that were ascribed to a reversible clamping motion of the enzyme as it scans along short regions of duplex DNA in its search for uracil. Here we further probe the properties of the nonspecific DNA binding surface of {2H12C}{15N}-labeled hUNG using a neutral chelate of a paramagnetic Gd3+ cosolute (Gd(HP-DO3A)). Overall, the measured paramagnetic relaxation enhancements (PREs) on R2 of the backbone amide protons for free hUNG and its DNA complex were in good agreement with those calculated based on their relative exposure observed in the crystal structures of both enzyme forms. However, the calculated PREs systematically underestimated the experimental PREs by large amounts in discrete regions implicated in DNA recognition and catalysis: active site loops involved in DNA recognition (268–274, 246–250), the uracil binding pocket (143–148, 169–170), a transient extrahelical base binding site (214–216), and a remote hinge region (129–132) implicated in dynamic clamping. These reactive hot spots were not correlated with electrostatic, structural or hydrophobic properties that might be common to these regions, leaving the possibility that the effects arise from dynamic sampling of exposed conformations that are distinct from the static structures. Consistent with this suggestion, the above regions have been previously shown to be flexible based on relaxation dispersion measurements and course-grained normal mode analysis. A model is suggested where the intrinsic dynamic properties of these regions allows sampling of transient conformations where the backbone amide groups have greater average exposure to solvent as compared to the static structures. We conclude that PREs derived from the paramagnetic cosolute reveal dynamic hot spots in hUNG and that these regions are highly correlated with substrate binding and recognition.
Keywords: Gd(HP-DO3A), gadoteridol, enzyme-DNA interaction, human uracil DNA glycosylase, NMR, paramagnetic relaxation enhancement, dynamics
Intermolecular accessibility of binding surfaces is a general requirement for the specific association of biomolecules with each other and small ligands. Although the accessibility of binding determinants may be suggested by inspection of structural models of the binding partners, such models do not reveal the pathway for forming the specific complex, nor do they address whether structural fluctuations or flexibility may be important to allow specific interactions to form. It is becoming increasingly appreciated that rapid and efficient macromolecular recognition may be viewed as a multiple step process involving collisional encounter of the binding partners, followed by intramolecular exploration of the binding surfaces that occurs within a funnel-shaped energy landscape leading to the specific complex (1, 2, 3). By necessity, such a model for macromolecular recognition requires the rapid formation and dissolution of weak intermolecular contacts that must be accessible, either statically or dynamically during the lifetime of the encounter complex, and may not necessarily be inferred from inspection of high-resolution structural models alone of the specific complexes.
The DNA repair enzyme human uracil DNA glycosylase (hUNG) is one of the most efficient enzyme catalysts, and presents an experimentally amenable system to explore how the accessibility of an enzyme’s surface relates to DNA recognition. hUNG is known to form a specific complex with uracil-containing DNA at a diffusion-controlled rate of ~ 5 × 108 M−1s−1 (4). The damage search process begins with three-dimensional diffusion to a random site in the DNA chain, followed by the formation of weak nonspecific interactions that enable the enzyme to slide and hop along the DNA backbone using nonspecific interactions until a uracil site is located, at which time substrate specific interactions can be formed (5, 6, 7, 8). Millisecond dynamic motions around the DNA binding site of hUNG have been detected in NMR relaxation dispersion measurements of the nonspecific enzyme-DNA complex, but not in the free enzyme, suggesting that upon non-specific DNA binding, the conformational energy landscape of hUNG is flattened, enabling the enzyme to populate transient structural conformers not thermally accessible to the free enzyme (8). A functional connection between the observed dynamics and DNA sliding was suggested by a course-grained normal mode analysis and by a comparison of the structures of the free enzyme with its nonspecific DNA complex. Both approaches pointed to a low energy correlated motion leading to a reversible “clamping” of hUNG around the DNA chain. We proposed that such a mode would allow rapid oscillation between a weak binding open state that would be productive for sliding, and a closed state that would allow specific interrogation of DNA base pairs for the presence of uracil (8). This interrogation event involves enzymatic capture of extrahelical uracil bases that have transiently escaped the DNA base stack by virtue of their destabilization relative to normal DNA base pairs(6, 9).
The above biophysical studies suggested that the intermolecular accessibility of some contacts essential for substrate recognition by hUNG may be modulated by conformational dynamics. Here we have exploited a freely diffusing Gd3+ paramagnet probe [Gd(HP-DO3A)] to explore the accessibility of the hUNG surface to a small molecule probe. This neutral chelate of the lanthanide metal is sufficiently small (r = 3.5 Å) to approach solvent exposed regions on the enzyme surface and smaller crevices that may be only transiently exposed by enzymatic motions. We envisioned that the strong distance modulated effect of the electron-nuclear dipole-dipole interaction on the transverse relaxation rate of nearby protons might allow the detection of subtle dynamic properties of hUNG that regulate access of DNA and other ligands to its active site. These measurements show that the active site and DNA binding regions of free and DNA-bound hUNG are hot spots for relaxation by the paramagnetic probe. This unusual sensitivity does not relate to the calculated exposure of these regions in the crystal structures of these enzymatic states, nor does it correlate with the structural, hydrophobic or electrostatic character of these sites. We conclude that the probe samples transient conformations of hUNG that have dynamic accessibility, some of which were completely undetectable by previous relaxation dispersion measurements that depend upon large differences in chemical shifts between conformational dynamic sites. The unique flexibility of these regions is likely to be important to enzyme function.
MATERIALS AND METHODS
NMR Sample Preparation
BL21 (DE3) Codon Plus cells were transformed with a PET21a plasmid containing the UNG gene under the control of an isopropyl β-D-1-thiogalactopyranoside (IPTG)-inducible T7 promoter. Isotopically enriched hUNG was produced by growing these cells in morpholine propanol sulfonic acid (MOPS) minimal media containing 15NH4Cl, perdeuterated glucose and 99% 2H2O. The over expression and purification of hUNG was described previously(10). All the isotropically enriched chemicals were purchased from Cambridge Isotope Laboratories (CIL), Inc. The palindromic DNA strand 5′-C1T2G3G4A5T6C7C8A9G10-3′ was purchased from Integrated DNA Technologies (IDT), Inc. and purified in house using a preparative Phenomenex Jupiter column. DNA concentration was measured via UV/Vis absorbance with molar extinction coefficients for duplex DNA previously derived from phosphate analysis. For all the NMR samples, a standard buffer containing 10 mM NaH2PO4 (pH 7.0), 75 mM NaCl, 1 mM EDTA and 1 mM DTT was used, as it was found to be optimal for protein solubility and stoichiometric DNA binding. The paramagnetic NMR samples (Gd3+ concentrations of 1 mM, 2.5 mM, 5 mM and 10 mM) were made by the addition of an appropriate volume of 0.5 M ProHance (Gd(HP-DO3A), Gadoteridol) stock solution obtained from Bracco Diagnostics.
PRE Measurements
All NMR experiments were performed at 20 °C on a 600 MHz Varian Inova spectrometer with a room temperature triple resonance probe. The paramagnetic relaxation enhancement (PRE) of amide protons was measured using a modified INEPT element with variable relaxation delay (11, 12) The TROSY detected spectra were acquired with 1280 points in the direct dimension (128 transients) and 120 increments in the indirect dimension (evolution time = 81.5 ms) and an intercycle delay of 1.5 s. Time points were collected in an interwoven manner to ameliorate instrumentation instabilities (13). All spectra were processed with NMRPipe and the peak intensities were quantified using Sparky (14, 15). The backbone NH peak assignments have been previously reported and assignments for the hUNG-DNA complex were obtained by tracking the chemical shift perturbations upon stepwise addition of DNA to the enzyme solution (16)
The PRE measurements were performed using samples containing 500 μL of 250 μM hUNG in a standard NMR tube. The hUNG complex with non-target DNA contained ~5 fold excess of DNA to ensure saturation of the enzyme. The PREs for each amide proton (Γ2) are defined as the difference shown in eq 1, where R2,para and R2,dia are the transverse relaxation rates of the paramagnetic and diamagnetic samples. Practically, this difference is measured using a two time point relation (Ta and Tb in eq 1), where Ta,b are two relaxation delays (Ta = 3 ms and Tb = 12 ms) during which amide protons are left transverse prior to detection of the resultant HSQC peak intensities (I)(17) in both the diamagnetic and paramagnetic samples.
| (1) |
Uncertainty in Γ2 was estimated by propagating unsigned differences in the measured peak intensities of duplicate data points as errors through equation 1. To confirm the assumption of exponential decay, a total of five points were measured for one sample of the hUNG-DNA complex using [Gd(HP-DO3A)] = 10 mM. The R2 obtained from the exponential fit to the five-time point data set and from the two-time point method were in excellent agreement for all resonances analyzed.
Calculation of Molar Relaxivity and Predicted PRE
For both free hUNG and its complex with non-target DNA, Gd(HP-DO3A) was titrated into the solution at four concentrations: 1 mM, 2.5 mM, 5 mM and 10 mM. At all concentrations of the paramagnet, the 15N1H chemical shifts were unchanged from the diamagnetic sample, indicating that the PREs result from a collisional mechanism rather than transient binding interactions between the spin label and specific sites on the enzyme (18). Additional support for a collisional relaxation mechanism was provided by the observation that the PREs were linear with respect to spin label concentration. Since in general it is not possible to find a single concentration of a paramagnetic agent that provides an optimal PRE measurement for all residues, we instead measured the molar relaxivity, which was calculated as the linear slope of a plot of the observed PRE (Γ2, eq 1) for each backbone amide proton as a function of Gd(HP-DO3A) concentration. The measured molar relaxivity was compared to the predicted PRE values obtained from theoretical calculations. For the PRE predictions we employed the Otting-LeMaster approach (18, 19), with modifications by Iwahara et al (3), where 3D lattice-based integration was replaced by shell-based integration. For these calculations, the crystal structures for free hUNG (pdb code 1AKZ) and the hUNG-nonspecific DNA complex (pdb code 2OXM) were used. The theoretical values were scaled to the same order of the measured molar relaxivities using an optimized scaling factor (3, 17). The theoretical PREs were found to correlate very well with the calculated depth index of each amide proton (see below).
Calculations of Depth Index
The crystal structures 1AKZ and 2OXM were also used for calculation of the depth index (Di,r) for each amide proton using the program SADIC(20). The depth index is defined as shown in eq 2, where Vi,r is the
| (2) |
exposed volume of a sample sphere of radius r centered on amide proton i, and V0,r is the volume of the same sphere centered on the isolated atom. Thus, the depth index is a measure of the relative exposed volume of an atom, and can take on a value between 0 (no exposure) and 2 (complete exposure). The exposed volume is the space external to the enzyme surface evaluated at a distance r in all directions around a given amide proton, and takes into account surface shape rather than just linear distances. In these calculations, the surface probe size was set at the radius of the Gd(HP-DO3A) cosolute (3.5 Å), and the optimized radius (r) for the sample sphere centered on each amide proton (i) was 10 Å.
Normal Mode Analysis
A course-grained normal mode analysis was done using elNemo webserver (http://www.igs.cnrs-mrs.fr/elnemo/) using the crystal structure of free hUNG (1AKZ). Default parameters were used for the calculation of the five lowest frequency normal modes of the structure, where the protein backbone was represented as rigid blocks containing two residues.
RESULTS
PRE Measurements of Free and DNA-Bound hUNG
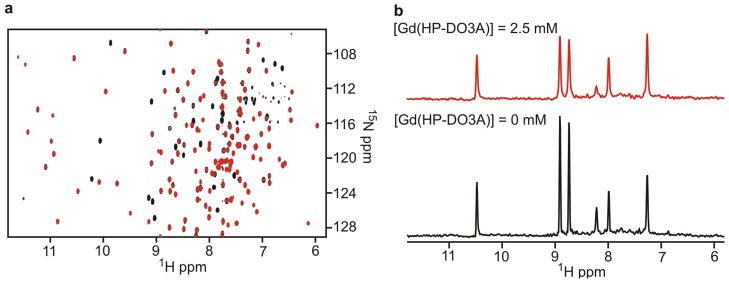
The backbone NMR assignments of hUNG have been previously reported (16), and were used in the present study since identical solution conditions were used. To minimize the complicating effects of spin diffusion on the interpretation of the PREs, and to increase the signal-to-noise of the measurements, the enzyme was prepared with uniform deuteration of all carbon-bound protons. The 1H-15N TROSY-HSQC spectra of free hUNG in the absence and presence of 2.5 mM Gd(HP-DO3A) are shown in Figure 1, and display excellent dispersion and signal-to-noise. The corresponding spectra for the enzyme-DNA complex are of comparable quality and are shown in Supplemental Figure S1. To form the complex with DNA, we titrated in the 10 mer DNA duplex while following the incremental chemical shift changes in the HSQC spectrum after each addition, which is characteristic of fast exchange on the NMR chemical shift timescale. As previously reported (16), selected residues in the DNA binding cleft and active site showed the largest chemical shift perturbations, which plateaued in the presence of a 5-fold excess of DNA over hUNG (not shown). Saturation of DNA binding by the enzyme is consistent with the Kd of hUNG for the 10 mer nonspecific DNA (previously measured to be 29 μM under these solution conditions) (16). We observed no chemical shift changes in the backbone amide resonances at any concentration of Gd(HP-DO3A), nor did addition of this reagent compete with DNA binding as judged by the invariant chemical shifts of the complex as this spin reagent was added in the range 1 to 10 mM.
Figure 1.
The 1H-15N TROSY spectra of free hUNG and the effect of the Gd(HP-DO3A) paramagnetic cosolute. (a) The spectra were obtained with isotropically 2H, 15N-enriched hUNG samples with no Gd(HP-DO3A) (black) or [Gd(HP-DO3A)] = 2.5 mM (red). (b) Representative 1D slices (f1 = 123.8 ppm) of the spectra in (a).
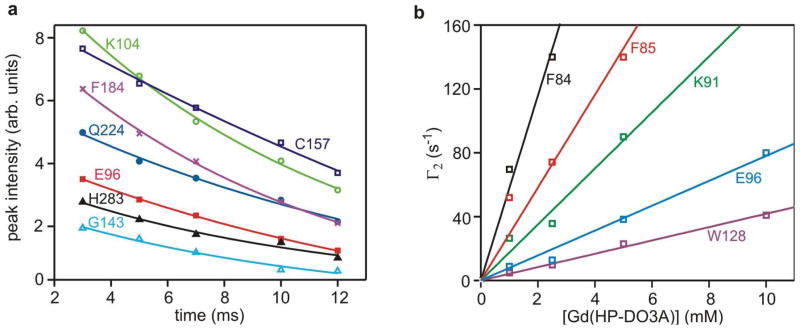
The proton transverse relaxation rates (R2) for each observed backbone amide were measured using the two-time point method (eq 1), or in the case of 10 mM Gd(HP-DO3A), five delay times to assess the errors in the two-point method (Fig. 2a). Comparison of the two methods showed that the R2 values for twenty representative residues were in agreement to within ± 8%. The R2 values for the diamagnetic and paramagnetic samples were used to calculate the PRE (Γ2, eq 1) at each concentration of Gd(HP-DO3A), and the observed PREs were plotted against [Gd(HP-DO3A)] to obtain the molar relaxivities (Γ2′) from the linear regression best-fit slopes (Fig. 2b). The observation that Γ2′ is linear over the entire concentration range of Gd(HP-DO3A), for all observed residues, is consistent with a collisional encounter mechanism. In addition, the linear slopes (Γ2′) provide a robust parameter for comparison of protons that are strongly and weakly relaxed by the paramagnet. We also note that the PRE correlation time is dominated by translational diffusion of the paramagnet cosolute, and would not be expected to be affected by the mass difference between the free enzyme and its DNA complex (17).
Figure 2.
Determination of amide proton transverse relaxation rates and molar relaxivities. (a) Exponential fits of the relaxation dispersion profiles of selected residues of the hUNG-DNA complex with [Gd(HP-DO3A)] = 10 mM. The nonlinear regression best-fit curves are shown. (b) Plot of Γ2 values obtained from eq 1 as a function of Gd(HP-DO3A) concentration for representative residues of free hUNG. The linear slope for each residue represents the spin-probe concentration-weighted PRE (Γ2′, or molar relaxivity).
Comparison of Predicted and Experimental PREs
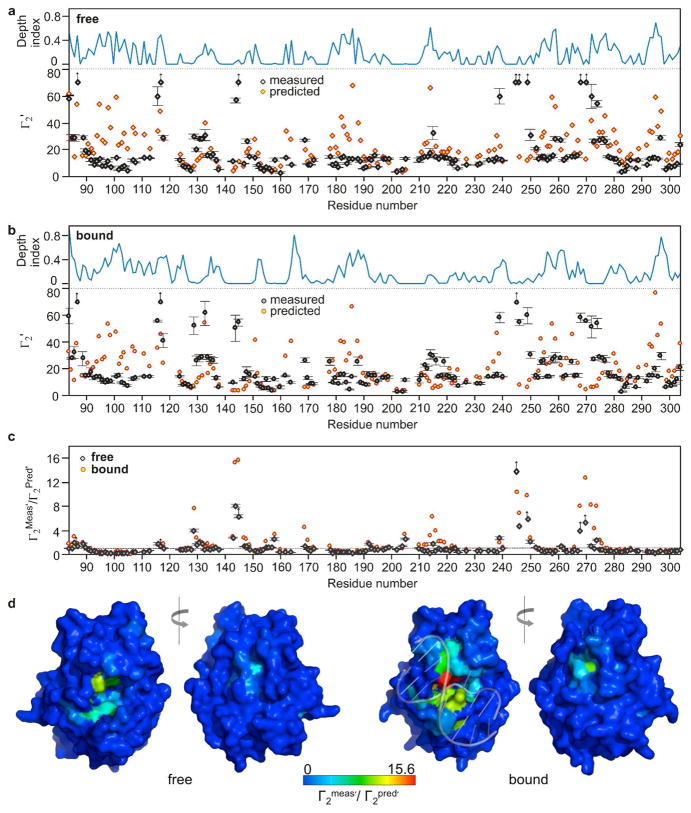
The PRE arising from a paramagnetic cosolute depends on the depth of the affected hydrogen atom in the protein structure, the translational and rotational diffusion constants, and the electron relaxation time, which is about 4 ns for Gd3+ (18). However, explicit calculation of a cosolute PRE effect using these fundamental parameters of the system is extremely complicated, and instead, an empirical approach is often used (18, 19, 21). This approach uses the coordinates obtained from the three-dimensional structure of the macromolecule, which are typically embedded in a cubic lattice. The distances of each hydrogen atom of the macromolecule to all external grid points (i.e. grid points that are not inside the volume defined by the macromolecular van der Waals surface) are summed and the resulting values are scaled to the experimental PRE values using a uniform scaling factor. This distance-based approach for calculating atom exposure is computationally facile, and has been shown to give reasonable agreement with experimental measurements (21). We have also employed a volume-based approach to calculate atom exposure (the depth index, Di,r), which tends to give higher detail for atoms that are located near the protein surface (see Methods) (20). We found that the predicted PREs are very well correlated with the depth index of each amide proton (R2 > 0.9, Supplemental Fig. S2)(Fig. 3a, b)
Figure 3.
Measured and predicted values for the amide proton molar relaxivities (Γ2′) of free hUNG and its complex with non-target DNA and the calculated depth indices for each amide proton (Di,r). (a) Measured and predicted Γ2′ values for each amide proton of free hUNG and its corresponding depth index value (eq 2). (b) Measured and predicted Γ2′ values for each amide proton of DNA-bound hUNG and its corresponding depth index value (eq 2). The predicted PREs were obtained using the grid-based approach using the atom coordinates obtained from the crystal structures of the free enzyme (1AKZ) and its non-specific DNA complex (2OXM) with water molecules omitted (3, 18, 19). The predicted values were scaled to the measured PRE values using a uniform scaling factor. (c) The ratios between the measured and predicted PRE values across the protein sequence for free hUNG and its complex with DNA. (d)) Crystal structures of free hUNG and its exosite hUNG-DNA complex with the ratios of Γ2Meas′/Γ2Pred′ color coded to the structural surfaces. In panels a, b and c, the missing points are either due to proline residues or residues with no or very weak signals. Errors of the data from the hUNG-DNA complex were omitted for clarity but are similar to those shown for the free enzyme. Supplementary Table S1 lists all of the PREs and their error values.
A comparison of the predicted and measured PRE values for free hUNG (Γ2meas′/Γ2pred′) shows very good agreement for a large majority of the backbone positions (Fig. 3a, 3c and 3d and Supplemental Table S1). However, it is also true that discrete regions of the structure show substantial deviations between the predicted and measured values, indicating that the static structure alone is not sufficient to account for all of the solution measurements. Three such regions include residues 95–108, 182–188 and 292–297 where the experimental PREs are much smaller than the predicted values. For other regions in the free enzyme, the measured PREs are significantly larger than the predicted values (Fig. 3c, black diamonds), and these regions are uniformly implicated in DNA recognition and catalysis. Hyper-reactive regions include the uracil binding site (144–145)(22), a DNA phosphate binding strand (246–250) (6), and an active site loop involved in DNA recognition (268–274) (16, 23). Highly localized, non-random deviations from the predictions indicate that the paramagnet is specifically excluded or allowed access to specific regions in a manner that does not correlate with atom exposure as indicated in the static structures used to make the PRE predictions. This conclusion follows because the predicted PREs are well-correlated with the calculated atom depths and accessible surface areas for each amide hydrogen (Supplemental Fig. S2). We considered whether the sites with large measured PREs that did not agree with prediction might arise from rapid amide proton exchange with solvent protons that had been relaxed by the paramagnet. However, this mechanism was excluded because these sites did not show significant exchange peaks with water in 15N-edited 3D-NOESY data (not shown).
Similar anomalies between predicted and measured PREs (Γ2meas′/Γ2pred′) are found for the hUNG-DNA complex (Fig. 3b, 3c, 3d, and Supplemental Table S1). The finding that large PREs persist in the active site and DNA recognition motifs within the DNA complex is striking because bound DNA would be expected to protect these regions. In addition to the regions where Γ2meas′/Γ2pred′ is large for both the free enzyme and its DNA complex, the DNA complex shows three additional regions where this ratio is large (129–132, 169–170, 214–216)(Fig. 3c, red circles). It is of interest that residues 129–132 comprise a remote “hinge” region implicated in dynamic clamping of hUNG (see Discussion and Supplementary Video S2) (16), residues 169–170 are in the region that makes contact with the deoxyuridine nucleotide in the specific hUNG-DNA complex, and residues 214–216 are part of an extended strand that interacts with the extrahelical base in the nonspecific DNA complex (23).
Free and DNA-Bound hUNG are More Similar than Predicted from Static Structures
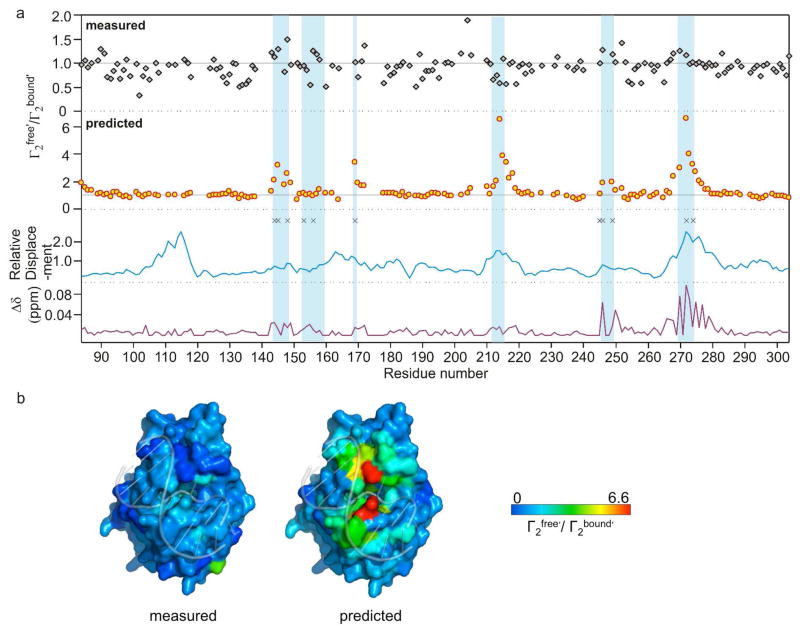
To further explore the basis for the unanticipated large experimental PREs in the DNA binding and active site regions of hUNG, we compared the ratio Γ2free′/Γ2bound′ using both the measured and predicted values (Fig. 4a). Across the entire protein sequence, the measured ratios of Γ2free′/Γ2bound′ deviate from unity by only modest amounts, even for regions that show chemical shift changes upon DNA binding (Fig. 4a), and for regions that are occluded by bound DNA in the static structure (Fig. 4b). In contrast, the predicted ratios of Γ2free′/Γ2bound′ increase markedly in regions where DNA occludes the site, as would be expected based on simple intuition (Fig. 4a, b). For comparison in Fig. 4a, we have also indicated the residues of hUNG that show millisecond time-scale dynamics in previous NMR studies (asterisks), the active site and DNA binding regions (shaded), and the relative amide nitrogen atom displacements in the lowest frequency normal mode calculated using course grain methods (16). In general, the active site and DNA binding regions exhibit millisecond time scale dynamics, display some of the largest displacements in the normal mode analysis, and show the largest deviations from the predicted PREs.
Figure 4.
Measured and predicted ratios of Γ2free′/Γ2bound′ across the hUNG linear sequence and comparison with dynamic properties, DNA induced chemical shift changes, and relative backbone amide nitrogen atom displacements obtained in a course-grain normal mode analysis. (a) From top to bottom: measured and predicted ratios of Γ2free′/Γ2bound′, amide nitrogen displacements in the lowest frequency normal mode analysis of free hUNG (1AKZ), and weighted chemical shift (1H and 15N) perturbations brought about by DNA binding. The black asterisks indicate amide positions with kex values in millisecond range (16). Vertical blue shaded bars indicate the six regions of the primary sequence that are important for DNA binding or enzyme catalytic activity. Supplementary Table S1 lists all of the PREs, chemical shift perturbations, relative atom displacements in the normal mode analysis, and residues with chemical exchange contributions to their linewidths. (b) Crystal structures of exosite hUNG-DNA complex (2OXM) with the measured (left) and predicted (right) Γ2free′/Γ2bound′ ratios color coded to the surface.
DISCUSSION
It is expected that the signal attenuations brought about by a paramagnet cosolute would reflect the frequency at which the cosolute visits various sites on the surface of a macromolecular target and the exposure of these sites. In the present case, we discover that although many of the sites behave as predicted, discrete regions of hUNG are protected from the expected effects of the paramagnet, while other regions are hyper-reactive (Fig. 3d). In addition, it is intriguing that the hyper-reactive regions are associated with DNA recognition or uracil binding, suggesting that the unanticipated exposure of these regions may be of functional significance. In the following analysis, we consider the two distinct findings of protection and hyper-reactivity, taking into account structural, electrostatic and dynamic properties of the system. The selected residues we consider below are found in clusters in the linear protein sequence (see structural representation in Fig. 5), and all show a discrepancy between the predicted and measured PREs that greatly exceeds the average deviation observed across the entire protein (Supplemental Table S2 and S3). We note that the conclusions discussed below are not dependent on whether the predicted PREs are calculated using a model that includes or excludes crystallographically observed waters, nor are they dependent on the size of probe used in the PRE predictions (i.e. r = 1.4 Å or 3.5 Å). In addition, the PREs do not correlate with crystallographic B factors.
Figure 5.
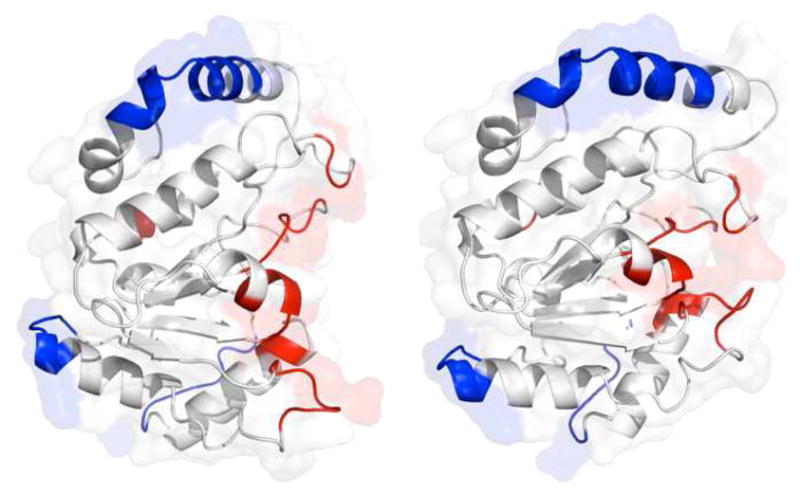
The two extrema of a course-grain normal mode analysis performed using the crystallographic coordinates of free hUNG (1AKZ). Sites that are hyper-reactive and protected from the paramagnetic probe are colored in red and blue, respectively. The left panel shows the open state and the right panel shows the closed conformation.
Protected Sites: Residues 95–108, 182–188, and 292–297
These three regions where the mean ratio Γ2meas′/Γ2pred′ is ~ 0.3 ± 0.1 (Supplemental Table S2), are not implicated directly in DNA binding or catalysis and the linear amino acid sequences of these regions are not conserved in UNGs from diverse organisms. All residues in this protected group have either coil or helical structural features, and diverse side chain composition. There is no obvious unifying structural or physical property of this protected group, other than the contiguous nature of the protected residues, that would explain the observed protection from the spin label (Fig. 5, blue shaded residues). For example, residues 95–108 have a helix-turn-helix structure, where the turn forms at residue 99, while residues 182–188 are all located in a loop region. In contrast, residues 292–297 are found at the end of a helix and then continue into a coil region. The residues in all three regions are composed of a mix of polar, charged and hydrophobic side chains making it unlikely that they possess unique properties that would exclude access by the paramagnetic probe (Supplemental Table S2). It has been previously reported that the presence of structural water molecules can prevent approach of a spin probe to nearby sites and result in lower than expected PREs (24). However, examination of the crystal structure to locate bound water molecules revealed no obvious correlation between the presence of crystallographic water and the reduced PREs, and inclusion of bound waters during the calculation of the predicted PREs makes no difference in the trends we observe in these data (not shown). Of course, we cannot exclude the possibility that differential solvation of these sites in solution could hinder close approach of a spin probe, but there is no basis for such speculation. On the basis that the above protected regions are located in flexible turns, coiled regions or helix termini, we surmise that conformations distinct from the crystal structure may be dynamically sampled in solution, giving rise to the discrepancies between the PRE predictions and measurements. The structure and relative populations of these putative alternative conformations is not known, except that they must on average protect the backbone amide protons from the cosolute as compared to the crystal conformation.
Hyper-reactive Sites: Residues 129–132, 144–145, 169–170, 214–216, 246–250 and 268–274
These regions, where the ratioΓ2meas′/Γ2pred′ can be as large as 16 (Figure 3c and 3d), are associated with DNA recognition and catalysis and are highly conserved between all UNGs (Supplemental Table S3). Similar to the protected sites, most of the residues in these regions have loop or coil structures, or short helices that may be dynamic (Fig. 5, red shaded residues). As the protected sites, these hyper-reactive regions have an unremarkable mixture of polar and hydrophobic residues, and the only charged residue in this group is Asp145, an essential active site base (Supplemental Table S3)(25). It is significant that a single unifying feature of these hyper-reactive residues is their association with DNA recognition or catalysis, and their previous identification as regions that exhibit μs to ms time scale dynamics in the presence of nonspecific DNA (16).
Residues 144–145, 169–170, 214–216, 246–250 and 268–274 are located in regions that should be directly protected by DNA binding as indicated by the high ratios of Γ2,freepred/Γ2,boundpred in Fig. 4a, yet these regions show similar elevated measured PREs in both the free and DNA bound states (Fig. 4b). Such an unexpected result virtually requires that the nonspecific enzyme-DNA complex experiences dynamic fluctuations, with the DNA and enzyme transiently losing the interactions that are observed in the crystal structure. These motions presumably expose otherwise occluded amide sites and allow access of the paramagnet cosolute. We note that the lower than anticipated protection observed in the DNA complex cannot be attributed to a significant presence of unbound enzyme because the equilibrium population of free enzyme is less than 1% of the total enzyme concentration based on the Kd of 29 μM. If such a scenario were occurring, it would be expected that the PREs of the complex would be a population-weighted average of the bound and unbound forms, yet the ratio Γ2free/Γ2bound bound for residues that would be expected to be protected by DNA are near unity. Therefore, the results more likely reflect the cosolute sampling loosely-bound exposed states that exist within the binary complex with nonspecific DNA.
These results support our previous interpretation of the dynamic behavior of these same regions observed in NMR relaxation dispersion experiments (16). Although the observation of dynamics does not provide any information on the nature of the motions involved, a previous course-grained normal mode analysis of hUNG suggests that these motions arise from an open-to-closed transition in the DNA-bound enzyme (16) (see red regions in Fig. 5 and Supplemental Video S1). A possible functional significance for this transition is to promote DNA sliding in a loose open state, and base pair interrogation in the closed state as observed in the crystal structure of the closed nonspecific complex (6). Although hyper-reactive residues 129–132 are found in a remote region located almost 180° from the active site of the enzyme, inspection of the normal mode trajectory in Supplemental Video S2 suggests that this region forms a molecular “hinge” for the open-to-closed conformational transition of hUNG. Thus, the hyper-reactivity of this region is possibly related to the same conformational dynamics that give rise to the effects in the DNA binding and active site regions. Because this course-grain normal mode analysis only reveals large-scale conformational changes in the protein, we cannot exclude the possibility that additional localized conformational dynamics in the hyper-reactive regions also contribute to the observed PRE effects.
Conclusions
The most salient finding from this work is that cosolute PRE effects correlate with the dynamic behavior of hUNG. Although correlations between cosolute PRE effects and dynamical averaging over the acquisition time of the NMR experiment have been suggested in other systems(26, 27, 28), we are unaware of any previous reports where PRE effects are unchanged, or even enhanced, upon binding of a large ligand such as DNA. Thus, the PRE gives a unique vantage point on the nature of the nonspecific complex between hUNG and DNA, which may be envisioned as the state of the enzyme during its search for uracil damaged sites in DNA. A picture emerges of the search complex as a very flexible species that can assume multiple discrete binding modes. The importance of these multiple modes may be related to the requirement that a DNA sliding enzyme must not only be able to slide, but also, switch to an damage interrogation mode (16, 29). Thus, views of DNA sliding as 1D diffusion of an enzyme on a DNA rod with a small frictional drag coefficient might be better viewed as occurring through dynamic oscillations between loose and more tightly bound forms of the enzyme. The loosely bound form(s) may allow microscopic dissociation or short range sliding, and the tightly bound form may be tuned for capture of spontaneously emerging DNA bases. Finally, it would be potentially advantageous if a theoretical and computational framework could be further developed to quantitatively account for observed discrepancies between predicted and calculated solute PREs, and derive useful structural information from these measurements (30).
Supplementary Material
Acknowledgments
We thank Junji Iwahara for providing the computer program for calculating cosolute PREs from structural coordinates.
Footnotes
SUPPORTING INFORMATION. 2D 1H-15N NMR spectra of DNA-bound hUNG, correlation plots of predicted PREs and depth indices, and three summary tables, and two videos showing the lowest frequency normal mode of hUNG. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Harel M, Spaar A, Schreiber G. Fruitful and Futile Encounters Along the Association Reaction between Proteins. Biophys J. 2009;96:4237–4248. doi: 10.1016/j.bpj.2009.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Iwahara J, Clore GM. Detecting Transient Intermediates in Macromolecular Binding by Paramagnetic NMR. Nature. 2006;440:1227–1230. doi: 10.1038/nature04673. [DOI] [PubMed] [Google Scholar]
- 3.Iwahara J, Zweckstetter M, Clore GM. NMR Structural and Kinetic Characterization of a Homeodomain Diffusing and Hopping on Nonspecific DNA. Proc Natl Acad Sci U S A. 2006;103:15062–15067. doi: 10.1073/pnas.0605868103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krosky DJ, Song F, Stivers JT. The Origins of High-Affinity Enzyme Binding to an Extrahelical DNA Base. Biochemistry. 2005;44:5949–5959. doi: 10.1021/bi050084u. [DOI] [PubMed] [Google Scholar]
- 5.Porecha RH, Stivers JT. Uracil DNA Glycosylase Uses DNA Hopping and Short-Range Sliding to Trap Extrahelical Uracils. Proc Natl Acad Sci U S A. 2008;105:10791–10796. doi: 10.1073/pnas.0801612105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parker JB, Bianchet MA, Krosky DJ, Friedman JI, Amzel LM, Stivers JT. Enzymatic Capture of an Extrahelical Thymine in the Search for Uracil in DNA. Nature. 2007;449:433–437. doi: 10.1038/nature06131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Friedman JI, Majumdar A, Stivers JT. Nontarget DNA Binding Shapes the Dynamic Landscape for Enzymatic Recognition of DNA Damage. Nucleic Acids Res. 2009 doi: 10.1093/nar/gkp161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Friedman JI, Stivers JT. Detection of Damaged DNA Bases by DNA Glycosylase Enzymes. Biochemistry. 2010;49:4957–4967. doi: 10.1021/bi100593a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parker JB, Stivers JT. Dynamics of Uracil and 5-Fluorouracil in DNA. Biochemistry. 2011;50:612–617. doi: 10.1021/bi101536k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mol CD, Arvai AS, Slupphaug G, Kavli B, Alseth I, Krokan HE, Tainer JA. Crystal Structure and Mutational Analysis of Human Uracil-DNA Glycosylase: Structural Basis for Specificity and Catalysis [See Comments] Cell. 1995;80:869–878. doi: 10.1016/0092-8674(95)90290-2. [DOI] [PubMed] [Google Scholar]
- 11.Donaldson LW, Skrynnikov NR, Choy WY, Muhandiram DR, Sarkar B, Forman-Kay JD, Kay LE. Structural Characterization of Proteins with an Attached ATCUN Motif by Paramagnetic Relaxation Enhancement NMR Spectroscopy. J Am Chem Soc. 2001;123:9843–9847. doi: 10.1021/ja011241p. [DOI] [PubMed] [Google Scholar]
- 12.Iwahara J, Tang C, Clore GM. Practical Aspects of (1)H Transverse Paramagnetic Relaxation Enhancement Measurements on Macromolecules RID A-3511-2008. Journal of Magnetic Resonance. 2007;184:185–195. doi: 10.1016/j.jmr.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pervushin K, Riek R, Wider G, Wuthrich K. Attenuated T2 Relaxation by Mutual Cancellation of Dipole-Dipole Coupling and Chemical Shift Anisotropy Indicates an Avenue to NMR Structures of very Large Biological Macromolecules in Solution. Proc Natl Acad Sci U S A. 1997;94:12366–12371. doi: 10.1073/pnas.94.23.12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: A Multidimensional Spectral Processing System Based on UNIX Pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 15.Goddard TD, Kneller DG. SPARKY. University of California; San Francisco: [Google Scholar]
- 16.Friedman JI, Majumdar A, Stivers JT. Nontarget DNA Binding Shapes the Dynamic Landscape for Enzymatic Recognition of DNA Damage. Nucleic Acids Res. 2009;37:3493–3500. doi: 10.1093/nar/gkp161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clore GM, Iwahara J. Theory, Practice, and Applications of Paramagnetic Relaxation Enhancement for the Characterization of Transient Low-Population States of Biological Macromolecules and their Complexes. Chem Rev. 2009;109:4108–4139. doi: 10.1021/cr900033p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pintacuda G, Otting G. Identification of Protein Surfaces by NMR Measurements with a Pramagnetic Gd(III) Chelate. J Am Chem Soc. 2002;124:372–373. doi: 10.1021/ja016985h. [DOI] [PubMed] [Google Scholar]
- 19.Hernandez G, Teng CL, Bryant RG, LeMaster DM. O2 Penetration and Proton Burial Depth in Proteins: Applicability to Fold Family Recognition. J Am Chem Soc. 2002;124:4463–4472. doi: 10.1021/ja017340k. [DOI] [PubMed] [Google Scholar]
- 20.Varrazzo D, Bernini A, Spiga O, Ciutti A, Chiellini S, Venditti V, Bracci L, Niccolai N. Three-Dimensional Computation of Atom Depth in Complex Molecular Structures. Bioinformatics. 2005;21:2856–2860. doi: 10.1093/bioinformatics/bti444. [DOI] [PubMed] [Google Scholar]
- 21.Iwahara J, Clore GM. Direct Observation of Enhanced Translocation of a Homeodomain between DNA Cognate Sites by NMR Exchange Spectroscopy. J Am Chem Soc. 2006;128:404–405. doi: 10.1021/ja056786o. [DOI] [PubMed] [Google Scholar]
- 22.Parikh SS, Walcher G, Jones GD, Slupphaug G, Krokan HE, Blackburn GM, Tainer JA. Uracil-DNA Glycosylase-DNA Substrate and Product Structures: Conformational Strain Promotes Catalytic Efficiency by Coupled Stereoelectronic Effects. Proc Natl Acad Sci USA. 2000;97:5083–5088. doi: 10.1073/pnas.97.10.5083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parker JB, Stivers JT. Uracil DNA Glycosylase: Revisiting Substrate-Assisted Catalysis by DNA Phosphate Anions. Biochemistry. 2008;47:8614–8622. doi: 10.1021/bi800854g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Niccolai N, Ciutti A, Spiga O, Scarselli M, Bernini A, Bracci L, Di Maro D, Dalvit C, Molinari H, Esposito G, Temussi PA. NMR Studies of Protein Surface Accessibility. J Biol Chem. 2001;276:42455–42461. doi: 10.1074/jbc.M107387200. [DOI] [PubMed] [Google Scholar]
- 25.Drohat AC, Jagadeesh J, Ferguson E, Stivers JT. The Role of Electrophilic and Base Catalysis in the Mechanism of Escherichia Coli Uracil DNA Glycosylase. Biochemistry. 1999;38:11866–11875. doi: 10.1021/bi9910878. [DOI] [PubMed] [Google Scholar]
- 26.Teng CL, Bryant RG. Mapping Oxygen Accessibility to Ribonuclease a using High-Resolution NMR Relaxation Spectroscopy. Biophys J. 2004;86:1713–1725. doi: 10.1016/S0006-3495(04)74240-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Teng CL, Hinderliter B, Bryant RG. Oxygen Accessibility to Ribonuclease a: Quantitative Interpretation of Nuclear Spin Relaxation Induced by a Freely Diffusing Paramagnet. J Phys Chem A. 2006;110:580–588. doi: 10.1021/jp0526593. [DOI] [PubMed] [Google Scholar]
- 28.Teng CL, Bryant RG. Experimental Measurement of Nonuniform Dioxygen Accessibility to Ribonuclease A Surface and Interior. J Am Chem Soc. 2000;122:2667–2668. [Google Scholar]
- 29.Slutsky M, Mirny LA. Kinetics of Protein-DNA Interaction: Facilitated Target Location in Sequence-Dependent Potential. Biophys J. 2004;87:4021–4035. doi: 10.1529/biophysj.104.050765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Madl T, Bermel W, Zangger K. Use of Relaxation Enhancements in a Paramagnetic Environment for the Structure Determination of Proteins using NMR Spectroscopy. Angew Chem Int Ed Engl. 2009;48:8259–8262. doi: 10.1002/anie.200902561. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.