Abstract
Antibodies against the CD4 binding site (CD4bs) on the HIV-1 spike protein gp120 can show exceptional potency and breadth. We determined structures of NIH45-46, a more potent clonal variant of VRC01, alone and bound to gp120. Comparisons with VRC01–gp120 revealed that a four-residue insertion in CDRH3 contributed to increased interaction between NIH45-46 and the gp120 inner domain, which correlated with enhanced neutralization. We used structure-based design to create NIH45-46G54W, a single substitution in CDRH2 that increases contact with the gp120 bridging sheet and improves breadth and potency, critical properties for potential clinical use, by an order of magnitude. Together with the NIH45-46–gp120 structure, these results indicate that gp120 inner domain/bridging sheet residues should be included in immunogens to elicit CD4bs antibodies.
Three decades after the emergence of HIV there is no vaccine, and AIDS remains a threat to global public health (1-5). However, some HIV-infected individuals eventually develop broadly neutralizing antibodies (bNAbs), i.e., antibodies that neutralize a large panel of HIV viruses (6-11) and can delay viral rebound in HIV patients (12). Such antibodies are relevant to vaccine development because passive transfer into macaques can prevent infection (13-15). Antibodies obtained by new cloning methods (7,16,17) target several epitopes on the viral spike (7,8,16,18-20). The broadest and most potent are highly active agonistic anti-CD4 binding site antibodies (HAADs) that mimic binding of the host receptor CD4 (21) by exposing the co-receptor binding site on gp120 (8,16,22-24). Despite isolation from different donors, HAADs are derived from two closely-related Ig VH genes that share gp120 contact residues (16,25). HAADs are typically members of large expanded clones (16) with variable levels of neutralizing activity despite intraclonal sequence similarities (16,25,26). Structures of gp120 complexes with VRC01, a highly potent and broad HAAD (25), and VRC03 and VRC-PG04, two new CD4-binding site (CD4bs) antibodies sharing the VRC01 germline VH gene, revealed convergence of gp120 recognition despite low sequence identities (48-57% in VH; 62-65% in VL) (26). However, sequence differences between these clonally-unrelated antibodies make it difficult to determine structural features that correlate with neutralization potency and breadth.
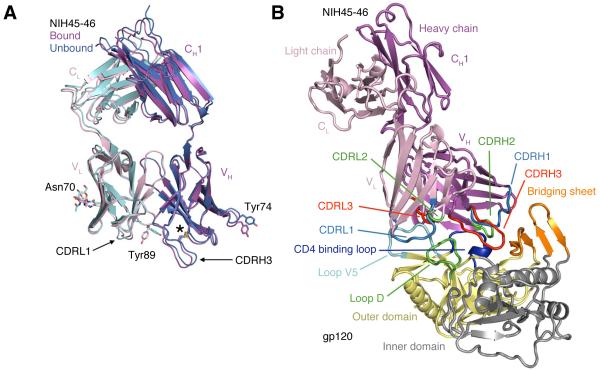
To determine structural correlates of high potency and breadth in HAADs, we solved structures of NIH45-46, alone and bound to the clade A/E 93TH057 gp120 core (27) (Table S1, Fig. 1A,1B). NIH45-46 is a more potent clonal variant of VRC01 that was isolated from the same donor using a YU2 trimer (16) instead of a resurfaced gp120 core (RSC3) as a bait (8). Comparisons of NIH45-46 Fab in its free versus gp120-bound states demonstrate that gp120 binding does not require major conformational changes (Fig. 1A). However, gp120 binding induced minor conformational changes in CDRL1, CDRH3, and in heavy chain framework region 3 (FWR3). As predicted by high sequence identity (85% in VH; 96% in VL) (Fig. S1), NIH45-46 resembles VRC01 (Fig. S2A,B). However, relative to VRC01, NIH45-46 includes a four-residue insertion within CDRH3 (Fig. 2A) that was acquired by somatic hypermutation (16).
Fig. 1.
Crystal structures of NIH45-46 Fab alone and bound to gp120. (A) Superimposition of the structures of the free (blue heavy chain and cyan light chain) and bound (magenta heavy chain and pink light chain) NIH45-46 Fab. RMSDs for free and bound VH-VH and VL-VL superimpositions (123 and 99 Cα atoms, respectively) are each 0.5 Å. The location of an extra disulfide bond that joins Cys32–Cys98 in VH is marked with an asterisk and N-linked carbohydrate attached to Asn70 of VL is shown as sticks. Arrows point to slightly different conformations in CDRL1 and CDRH3 in bound and free NIH45-46. Highlighted sidechains, Tyr89CDRL3 and Tyr74FWRH3, adopt notably different conformations upon binding gp120. (B) Structure of the NIH45-46–gp120 complex. Ribbon diagram of the NIH45-46 Fab (magenta and pink for the heavy and light chains, respectively) complexed with the inner (yellow) and outer (gray) domains of the 93TH053 gp120 core, which lacks three variable loops (V1-V2 and V3) and has N- and C-terminal truncations (25). Lines point to structural features of the gp120 core discussed in the text.
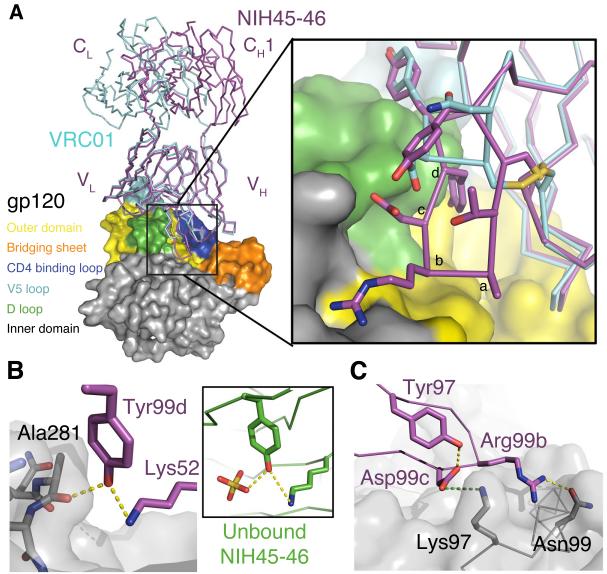
Fig. 2.
Interactions of the NIH45-46 insertion with gp120. (A) Superimposition of the gp120 portions of VRC01–93TH053 (PDB 3NGB) and NIH45-46–93TH053 structures. The Fabs are shown as magenta (NIH45-46) or cyan (VRC01) wire, and gp120 is shown as a surface with the color scheme used in Fig. 1B. The region surrounding the four-residue insertion in the NIH45-46 CDRH3 (residues 99a – 99d) is boxed. Inset: Close-up of the boxed region in which the NIH45-46 insertion residues are labeled alphabetically to correspond with residues 99a – 99d. The sidechains of relevant CDRH3 residues are shown as sticks. (B) Hydrogen bond network between the mainchain carbonyl oxygen of Ala281gp120, Tyr99dNIH45-46 in CDRH3, and Lys52NIH45-46 in CDRH2. Yellow dots represent hydrogen bonds. The conformation of Tyr99dNIH45-46 is also stabilized by a hydrogen bond with Lys52NIH45-46 in unbound NIH45-46 (inset) in which a sulfate ion (yellow) from the crystallization solution substitutes for the interaction with Ala281gp120. (C) Electrostatic interaction between Asp99cNIH45-46 and Lys97gp120 (green dots) is shown with Asp99cNIH45-46-Tyr97NIH45-46 and Arg99bNIH45-46–Asn99gp120 hydrogen bonds (yellow dots).
The crystal structure of the NIH45-46–93TH057 gp120 complex verified that NIH45-46 targets the CD4bs on gp120 (Fig. 1B,2A). The primary binding surface is the outer domain, including the CD4 binding loop (Fig. S3A), loop D and loop V5, but CDRH3NIH45-46 reaches toward the gp120 inner domain (Fig. 1B,2A-C). Important interactions in the VRC01–93TH057 structure (25) are conserved in NIH45-46 (Fig. S2B); e.g., residues C-terminal to CDRH2 of VRC01 and NIH45-46 mimic the interaction of mainchain atoms in the C” β-strand of CD4 domain, which hydrogen bond with the CD4-binding loop of gp120 (Fig. S3). In both NIH45-46 and VRC01, hydrogen bonds between CDRH2 and gp120 are water-mediated (25,26) (except for the Gly54NIH45-46/Gly54VRC01 carbonyl oxygen–Asp368gp120 backbone nitrogen H-bond (Fig. S3)), and Arg71VRC01/Arg71NIH45-46 preserves the Arg59CD4 interaction with Asp368gp120. However, the Phe43CD4 interaction with a hydrophobic pocket between α-helix 3gp120 (CD4 binding loop) and β-strand 21gp120 (bridging sheet) (Fig. 3A,B) is not mimicked by either antibody. Differences between VRC01 and NIH45-46 include the conformation of heavy chain residue Tyr74, a FWR3 residue that was substituted during somatic hypermutation (16), and a tyrosine to serine substitution in CDRL1 (Fig. S4,S5, Supplementary Discussion).
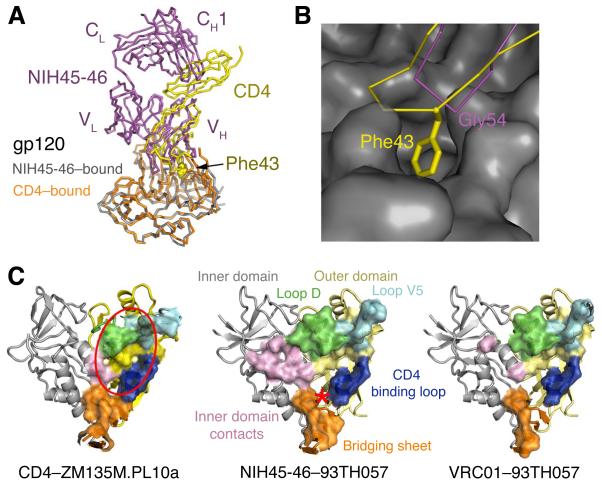
Fig. 3.
NIH45-46 mimicry of CD4 binding. (A) Superimposition of NIH45-46–gp120 (magenta and gray, respectively) and CD4–gp120 (yellow and orange, respectively) (31) calculated based on gp120 Cα positions. Phe43CD4 is shown using spheres. (B) Close-up of the CDRH2 loop of NIH45-46 (magenta) and CDR2-like loop of CD4 (yellow) interacting with gp120 (gray surface). Phe43CD4 (labeled) inserts into a hydrophobic pocket on gp120. The closest corresponding residue on NIH45-46 (Gly54NIH45-46) is labeled. (C) Comparison of contacts on gp120 made by CD4, NIH45-46, and VRC01 (left, middle and right respectively). Residues at each contact interface are highlighted on the gp120 structure (colored as in Fig. 1B) as a surface enclosing the contact residues. The location of the Phe43 cavity targeted by the tryptophan substitution in NIH45-46G54W is indicated on the middle panel with a red asterisk. The approximate location of the gp120 region defined as the initial site of CD4 attachment (25, 26) is indicated with a red oval in the left panel.
The most notable difference between VRC01 and NIH45-46 is the four-residue insertion (residues 99a–99d) in CDRH3. Three inserted residues contribute to binding to gp120 (Fig. 2A-inset), consistent with deletion of the insertion resulting in ~10-fold reduced neutralization potencies (Table S2). First, the Tyr99dNIH45-46 sidechain hydrogen bonds with the loop D Ala281gp120 carbonyl oxygen (Fig. 2B), a mainchain atom, thus preventing escape through mutation. Indeed, NIH45-46-sensitive strains accommodate different sidechains at position 281gp120 (Table S3). The importance of Tyr99dNIH45-46 for potency is demonstrated by alanine substitution (NIH45-46 Y99dA), which reduces the neutralization potency of NIH45-46 to values intermediate between wild-type NIH45-46 and the deletion mutant (Table S2). Second, Asp99cNIH45-46 interacts electrostatically with Lys97gp120 at the base of α-helix 1gp120, and third, Arg99bNIH45-46 hydrogen bonds with Asn99gp120 (Fig. 2C). The conformation of the insertion is stabilized by two intramolecular hydrogen bonds. In one, the Tyr99dNIH45-46 sidechain hydrogen bonds with the ε-amino group of Lys52NIH45-46 within CDRH2 (Fig. 2B), also seen in the unbound structure of NIH45-46 (Fig. 2B-inset), thus the Tyr99dNIH45-46 hydroxyl is poised for interacting with Ala281gp120. A second hydrogen bond between Tyr97NIH45-46 and Asp99cNIH45-46 in the gp120-bound Fab positions the negatively-charged aspartic acid for interaction with Lys97gp120 (Fig. 2C). The region of gp120 with which CDRH3NIH45-46 interacts was not included in the previously-defined vulnerable site of initial CD4 attachment on the gp120 outer domain (25) (Fig. 3C). Thus, gp120 residues that contact CDRH3NIH45-46 residues required for potent neutralization (Table S2), e.g., Lys97gp120, were mutated in RSC3 (Fig. S6), the resurfaced gp120 used for isolating bNAbs (8) and as a candidate HIV immunogen (28).
The insertion in CDRH3 contributes to a higher total buried surface area between the NIH45-46 heavy chain and gp120 compared with VRC01 (Table S4). The extra contacts with gp120 created by the CDRH3 insertion allow the NIH45-46 footprint on gp120 to more closely resemble the CD4 footprint on gp120 than does the VRC01 footprint (Fig. 3C, Table S4). The observation that NIH45-46 shows more extensive contacts relative to VRC01 with the inner domain and bridging sheet of gp120 (Fig. 3C), yet exhibits higher potency and breadth (16), is inconsistent with the suggestion that increased contact area with regions outside of the outer domain of gp120 correlate with decreased neutralization potency and/or breadth (25,26). Indeed, the observed CDRH3 contacts with the inner domain imply that the crystallographically-observed conformation of this region, whether pre-existing or induced, actively played a role in the affinity maturation events that resulted in the four-residue insertion with CDRH3.
Although NIH45-46 increases its contacts with the inner domain/bridging sheet area of gp120, like VRC01, it lacks a critical CD4 contact to a hydrophobic pocket at the boundary between the gp120 bridging sheet and outer domain made by burying Phe43CD4. This residue alone accounts for 23% of the interatomic contacts between CD4 and gp120, serving as a “linchpin” that welds CD4 to gp120 (29). On gp120, the Phe43 binding cavity is a binding site of small-molecule CD4 mimics (30) and a desirable target for compounds to disrupt CD4–gp120 interactions (29), yet it remains unfilled in the 93TH057 complexes with VRC01 (25) and NIH45-46. In a superimposition of a CD4–gp120 structure (31) and NIH45-46–gp120 (Fig. 3B), the Cα atom of heavy chain residue Gly54NIH45-46 is only ~1.4 Å from the Phe43CD4 Cα, suggesting that this important interaction might be mimicked by substituting Gly54NIH45-46 with a large hydrophobic residue. Indeed, residue 54 of VRC03 is a tryptophan, and Trp54VRC03 is accommodated within gp120’s Phe43 binding cavity to mimic Phe43CD4, while still maintaining its mainchain hydrogen bond with Asp368gp120 (PDB 3SE8) (Fig. S3). If increasing contacts with the inner domain/bridging sheet enhances antibody activity, as suggested by analysis of the NIH45-46–gp120 structure, then substituting Gly54NIH45-46 with a large hydrophobic residue should increase the potency and breadth of NIH45-46.
We constructed a series of NIH45-46 mutants to test the possibility that a hydrophobic sidechain at position 54 in NIH45-46 would improve activity. We first verified that substitutions at residue 54 did not interfere with antigen binding by assessing the ability of one mutant, NIH45-46G54W, to bind core gp120s. Surface plasmon resonance (SPR) binding analyses demonstrated that NIH45-46G54W Fab bound core gp120s with slightly higher affinities than did NIH45-46 Fab, with differences largely due to slower dissociation rates (Fig. S7). We next evaluated mutant IgGs in neutralization assays using a panel of six viruses chosen to include NIH45-46–sensitive and resistant strains (Fig. S8A). NIH45-46G54W and NIH45-46G54F showed increased potencies and NIH45-46G54W increased breadth by neutralizing three strains that are resistant to >50 μg/mL of NIH45-46. The apparent increase in breadth is likely due to increased potency as evidenced by the extrapolated IC50 for NIH45-46 against strain DU172.17 (Fig. S8B).
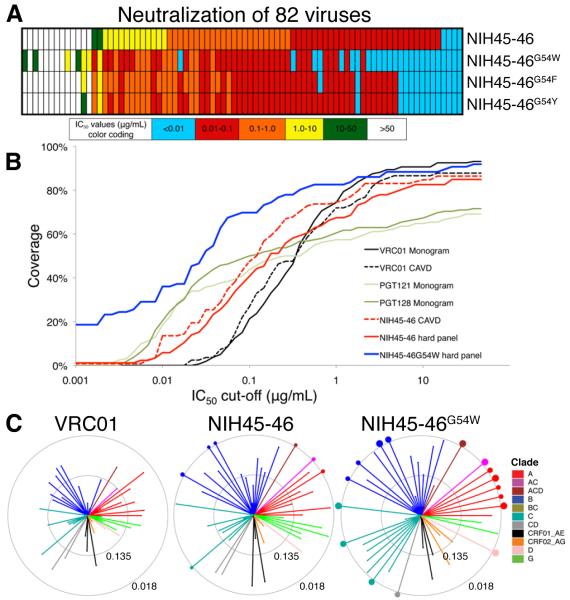
We further tested 82 viruses including 13 NIH45-46–resistant, 14 weakly-neutralized, and 55 sensitive strains representing all clades, of which 12 are transmitted founder viruses (Fig 4A, Tables S5,S6). This panel is more difficult for NIH45-46 to neutralize than a recently-published panel (16) (Fig. 4B). NIH45-46G54W showed increases in potency and breadth compared to NIH45-46 and VRC01: geometric mean IC50s (32) of 0.04 μg/mL for NIH45-46G54W, 0.41 μg/mL for NIH45-46, and 0.92 μg/mL for VRC01 (calculated for 65 viruses against which VRC01 was previously evaluated (16)) (Tables S5,S6, Fig. 4C). Notably, NIH45-46G54W gained de novo neutralization activity against six NIH45-46 resistant strains, including the only three that were sensitive to VRC01 but resistant to NIH45-46 in the panel tested in (16). Remarkably, for some strains that NIH45-46 neutralizes poorly, NIH45-46G54W was significantly more potent (e.g., improvements of >700-fold for T255-34 and 2000-fold for 3718.v3.c11). The enhanced neutralization activity of NIH45-46G54W implies that Trp54 forms a favorable hydrophobic interaction with Phe43 cavity of gp120 as seen in VRC03–gp120 (PDB 3SE8). NIH45-46G54F showed some increased activity, but other substitutions were less favorable (Fig. S8A, Tables S5,S6), including serine (25), which is found in the NIH45-46/VRC01 germline precursor. Substituting Gly54 with tryptophan adds ~140 Å2 of buried surface area on VH when complexed with gp120, consistent with the reduced dissociation rates observed in SPR experiments (Fig. S7), and provide a critical contact to a region of gp120 that cannot change without loss of CD4 tropism. By providing a tryptophan in the Phe43 cavity of gp120, NIH45-46G54W may use higher affinities and/or slower dissociation rates to overcome incompatible surface variations that render some viruses less sensitive or resistant to its effects. If so, resistance to NIH45-46G54W might require restricted access to the binding site on a native trimeric spike. Inspection of sequences of strains resistant to NIH45-46G54W and/or NIH45-46 suggests that variations at contact sites near V5 and in loop D may be responsible (Table S7) (25), consistent with mutagenesis studies of neutralization by VRC01 (33).
Fig. 4.
Increased neutralization potency of NIH45-46G54W. (A) Schematic comparing neutralization potencies for NIH45-46 and three position 54 mutants, including NIH45-46G54W. IC50 values are color-coded as shown. Numerical values for the same 82 strains are shown in Table S5 (IC50 values) and Table S6 (IC80 values). (B) Graphical comparisons of neutralization coverage and potency. The y-axis shows the cumulative frequency of IC50 values up to the concentration shown on the x-axis. The curves for VRC01, the only antibody tested in both the CAVD panel (a panel of 118 viral strains (16)) and the Monogram panel (a panel of 162 viral strains (18)), are similar, although the CAVD panel includes a few more resistant strains (compare the solid and dotted black lines). NIH45-46 (red dotted line) was tested on the CAVD panel and is more potent than VRC01 and broader than the PGT Abs. NIH45-46G54W (solid blue line) was tested with NIH45-46 (solid red line) on a panel of 82 viral strains chosen to include many NIH45-46 resistant and poorly neutralized strains (hard panel; Tables S5,S6), thus resulting in lower coverage and lower potency for NIH45-46 when evaluated using the hard panel versus using the CAVD panel (compare the solid and dotted red lines). The improved potency and breadth of NIH45-46G54W versus NIH45-46 against the hard panel are apparent. Even when evaluated using a difficult to neutralize panel of viruses, NIH45-46G54W is the most potent and broadly neutralizing antibody described to date. (C) Neutralization summary spider graphs comparing IC50 values for VRC01, NIH45-46, and NIH45-46G54W for 65 common viruses that were tested in this work or previously (16). Each color represents a different HIV clade. Length of lines and size of circles are inversely proportional to IC50. The distance between the outer and inner circle and the distance from the inner circle to the center of a spider graph each span two natural logs in IC50 concentration (concentrations in μg/mL are indicated on each circle). Dots on the outer circle indicate strains with IC50 values less than 0.018 μg/mL whose lines were truncated in the graph. The size of each dot is inversely proportional to the IC50 value.
Heavy chain residue 54 is not conserved in HAADs; in addition to glycine (NIH45-46 and VRC01), residue 54 can be threonine (3BNC60, 3BNC117, 3BNC115; VRC-PG04), tyrosine (12A12), phenylalanine (12A21), or arginine (1B2530 and 1NC9) (16, 26). Tryptophan substitution did not improve all HAADs that were tested (Table S8), thus increased potency requires a precise geometry.
Passive immunization and/or gene therapy to deliver HIV antibodies is increasingly being considered an option for prevention of HIV infection. To reduce the concentrations and numbers of antibodies required for protection to realistic and affordable levels, highly potent and broadly neutralizing antibodies are the reagents of choice for passive delivery. Although it is difficult to compare the potencies and breadth of antibodies characterized using different virus panels, the natural form of NIH45-46 exhibits superior potency to VRC01 when compared against a panel of 82 Tier 2 and 3 viruses representing all known HIV clades (16). A new set of HIV antibodies, the PGT antibodies that recognize the gp120 V3 loop and associated carbohydrates, exhibited median IC50s up to 10-fold lower than VRC01 (18), but are less potent and broad than NIH45-46G54W (Fig. 4B, Table S9). The successful improvement of the potency and breadth of NIH45-46, already one of the most potent and broad of the currently-available bNAbs (16), suggests that NIH45-46G54W should be among a small number of characterized HIV antibodies considered for clinical testing. Since escape from even the best bNAbs is possible, optimal results may be achieved using NIH45-46G54W in combination with one or more other bNAbs.
Our results also have important implications for vaccine development, i.e., the findings that contacts to the gp120 inner domain by NIH45-46 and increased interactions with the bridging sheet by NIH45-46G54W improve the breadth and potency of a HAAD antibody suggest that immunogens used in vaccine strategies should include native inner domain/bridging sheet residues. Although this sort of immunogen may elicit some non-neutralizing CD4bs antibodies, raising the most potent and broad CD4bs antibodies will require contact with regions within the gp120 inner domain and bridging sheet in addition to the initial site of CD4 attachment on the gp120 outer domain.
Supplementary Material
Acknowledgments
We thank B. Hahn, J. Baalwa, F. McCutchan, G. Shaw, D. Montefiori, M. Thomson, J. Overbaugh, R. Swanstrom, L. Morris, J. Kim, L. Zhang, D. Ellenberger, and C. Williamson for contributing the HIV-1 envelope plasmids used in our neutralization panel. We thank T. Oliveira and A. Halper-Stromberg for calculations and helpful discussions, and members of the Bjorkman and Nussenzweig laboratories for critical reading of the manuscript. R.D., J.F.S., M.C.N, and P.J.B. have pending patent applications with the U.S. Patent and Trademark Office, patent numbers U.S. 61/486,960 and 61/523,244, entitled “Human Immunodeficiency Virus Neutralizing Antibodies and Methods of Use Thereof” and “Anti-HIV Antibodies and Related Methods And Compositions,” respectively. The reagents are available with a Materials Transfer Agreement. This work was supported by Collaboration for AIDS Vaccine Discovery (CAVD) grants with support from the Bill & Melinda Gates Foundation (grant 38660 (P.J.B.), grant 38619 (M.S.S.) and grant 38619s (M.C.N.)), NIH grants P01 AI081677-01 (M.C.N.), RR00862, RR022220, and the Molecular Observatory at Caltech supported by the Gordon and Betty Moore Foundation. FK was supported by the German Research Foundation (DFG, KL 2389/1-1). Operations at SSRL are supported by the US-DOE and NIH. The data reported in this paper are tabulated in the main text and the supporting online material, and coordinates and X-ray crystallographic data for the NIH45-46–gp120 complex and NIH45-46 Fab have been deposited in the Protein Data Bank (accession codes 3U7Y and 3U7W respectively).
Footnotes
This manuscript has been accepted for publication in Science. This version has not undergone final editing. Please refer to the complete version of record at http://www.sciencemag.org/. The manuscript may not be reproduced or used in any manner that does not fall within the fair use provisions of the Copyright Act without the prior, written permission of AAAS.
Supporting Online Material□www.sciencemag.org□
References and Notes
- 1.Fauci AS, et al. Science. 2008 Jul 25;321:530. doi: 10.1126/science.1161000. [DOI] [PubMed] [Google Scholar]
- 2.McElrath MJ, Haynes BF. Immunity. 2010 Oct 29;33:542. doi: 10.1016/j.immuni.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Karlsson Hedestam GB, et al. Nat Rev Microbiol. 2008 Feb;6:143. doi: 10.1038/nrmicro1819. [DOI] [PubMed] [Google Scholar]
- 4.McMichael AJ, Borrow P, Tomaras GD, Goonetilleke N, Haynes BF. Nat Rev Immunol. 2010 Jan;10:11. doi: 10.1038/nri2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zolla Pazner S, Cardozo T. Nature reviews. Immunology. 2010;10:527. doi: 10.1038/nri2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walker LM, et al. Science. 2009 Sep 3;326:285. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scheid JF, et al. Nature. 2009 Apr 2;458:636. doi: 10.1038/nature07930. [DOI] [PubMed] [Google Scholar]
- 8.Wu X, et al. Science. 2010 Aug 13;329:856. [Google Scholar]
- 9.Stamatatos L, Morris L, Burton DR, Mascola JR. Nat Med. 2009 Aug;15:866. doi: 10.1038/nm.1949. [DOI] [PubMed] [Google Scholar]
- 10.Gray ES, et al. J Virol. 2011 May;85:4828. doi: 10.1128/JVI.00198-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mikell I, et al. PLoS Pathog. 2011;7:e1001251. doi: 10.1371/journal.ppat.1001251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trkola A, et al. Nat Med. 2005 Jun;11:615. doi: 10.1038/nm1244. [DOI] [PubMed] [Google Scholar]
- 13.Mascola JR, et al. Nat Med. 2000 Feb;6:207. doi: 10.1038/72318. [DOI] [PubMed] [Google Scholar]
- 14.Hessell AJ, et al. PLoS Pathog. 2009 May;5:e1000433. doi: 10.1371/journal.ppat.1000433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hessell AJ, et al. Nature. 2007 Sep 6;449:101. doi: 10.1038/nature06106. [DOI] [PubMed] [Google Scholar]
- 16.Scheid JF, et al. Science. 2011 Aug 11;333:1633. doi: 10.1126/science.1207227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scheid JF, et al. J Immunol Methods. 2009 Apr 15;343:65. doi: 10.1016/j.jim.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walker LM, et al. Nature. 2011 Aug 17;477:466. [Google Scholar]
- 19.Corti D, et al. PLoS One. 2010;5:e8805. doi: 10.1371/journal.pone.0008805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bonsignori M, et al. J Virol. 2011 Jul 27; [Google Scholar]
- 21.Sattentau QJ, Weiss RA. Cell. 1988 Mar 11;52:631. doi: 10.1016/0092-8674(88)90397-2. [DOI] [PubMed] [Google Scholar]
- 22.Thali M, et al. J Virol. 1993 Jul;67:3978. doi: 10.1128/jvi.67.7.3978-3988.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen B, et al. Nature. 2005 Feb 24;433:834. doi: 10.1038/nature03327. [DOI] [PubMed] [Google Scholar]
- 24.Decker JM, et al. J Exp Med. 2005 May 2;201:1407. doi: 10.1084/jem.20042510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou T, et al. Science. 2010 Aug 13;329:811. [Google Scholar]
- 26.Wu X, et al. Science. 2011 Aug 11;333:1593. [Google Scholar]
- 27.Materials and methods are available as supporting material on Science Online.
- 28.Nabel G, Kwong P, Mascola J. Philosophical transactions - Royal Society. Biological sciences. 2011;366:2759. doi: 10.1098/rstb.2011.0096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kwong PD, et al. Nature. 1998 Jun 18;393:648. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Madani N, et al. Structure. 2008 Nov 12;16:1689. doi: 10.1016/j.str.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taken from a crystal structure of Fab 21c plus CD4 domains 1 and 2 bound to the ZM135M.PL10a gp120 core at 2.45Å (to be submitted to the PDB).
- 32.Geometric mean IC50 calculations were done without excluding resistant strains by entering values of 50 μg/mL for strains with IC50 values >50 μg/mL.
- 33.Li Y, et al. J Virol. 2011 Sep;85:8954. doi: 10.1128/JVI.00754-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.