Abstract
a) Objectives
MMP-2, MMP-9, their complexes and ADAM12 are detected in the urine of breast cancer patients and predict disease status. We assessed the use of FRET-based substrates in an assay to distinguish breast cancer patients from controls.
b) Design and Methods
Substrates with varying specificities for MMP-9 and MMP-2 and several ADAMs were screened. Flsub21 and Flsub13, substrates for ADAM12 and ADAM8 respectively, were studied.
c) Results
Flsub21 and Flsub13 cleavage activity was detected in the urine of patients with invasive and metastatic breast cancer at significantly higher frequencies compared to controls. Our model predicted probabilities of 90% when both Flsub21 and Flsub13 were positive, 65% when Flsub21 alone was positive, 55% when Flsub13 alone was positive and 20% when both substrates were negative.
d) Conclusions
These data suggest the potential utility of FRET substrates to non-invasively identify invasive and/or metastatic breast cancer.
Keywords: MMPs, ADAMs, FRET substrates, urine, enzyme activity, breast cancer, invasive, metastatic
Introduction
Matrix metalloproteases (MMPs) are a multigene family of zinc-dependent endopeptidases that have been implicated in tumor growth, invasion and metastasis in experimental cancer models and in human tumors [1–8]. The characteristic domain structure of MMPs includes (1) the signal peptide domain, which guides the enzyme into the rough endoplasmic reticulum during synthesis, (2) the propeptide domain, which sustains the latency of these enzymes until it is removed or disrupted, (3) the catalytic domain, which houses the highly conserved Zn2+ binding region (HExGHxxGxxHS/T) and is responsible for enzyme activity, (4) the hemopexin domain, which determines the substrate specificity of MMPs and (5) a small hinge region, which enables the hemopexin region to present substrate to the active core of the catalytic domain. A subfamily of membrane-type MMPs (MT-MMPs) possesses an additional transmembrane domain. Two members of this family in particular, MMP-2 and MMP-9, degrade, among other substrates, typeIV collagen, fibronectin and laminin, major components of the basement membrane and are commonly used as markers of the malignant phenotype. MMP activity is regulated by a group of four endogenous tissue inhibitors of metalloproteases (TIMPs) [1,5, 9].
More recently, a related family of disintegrin metalloproteases, the ADAMs (A Disintegrin And Metalloprotease) have also been implicated in tumor growth and metastasis. Most ADAMs possess the conserved Zn-binding catalytic domain similar to MMPs and are proteolytically active. ADAM family members are typically membrane-bound, however, some can have alternatively spliced secreted isoforms as well [10,11]. ADAM substrates include cell-surface-associated type I or II integral membrane proteins and a number of them also possess matrix-degrading activity similar to the MMPs. Based on their metalloprotease function and substrate specificity, MMPs and ADAMs have been shown to be involved in normal developmental processes such as cardiac and neuronal development [12], mammary involution [13] and bone turnover [14] and when dysregulated, their activity can lead to diseases including cancer, inflammation, obesity and cardiac hypertrophy [15–17].
Overexpression of MMPs and/or ADAMs in tumor tissue and/or stroma can result in increased levels of MMP activity in various body fluids. Evidence is emerging that members of the MMP and/or ADAM family can serve not only as potential markers for diagnosis and prognosis, early detection and risk assessment but also as indicators of tumor recurrence, metastatic spread and response to primary and adjuvant therapy for breast cancer [18–21]. MMP-9 has been detected in the serum and plasma of tumor bearing rats and in humans with malignant tumors [22–25]. We have previously reported that MMPs can be detected in urine from patients with a variety of cancers and are independent predictors of disease status [19,20,26–29]. MMP-9 levels in tumor tissue as well as serum, plasma and urine are significantly elevated in breast cancer patients [24,26,27,30]. ADAM12 can be detected in the urine of breast [31] and bladder [32] cancer patients and its levels have been shown to correlate with disease status, stage and cancer risk [18,31].
In addition to zymography and other biochemical approaches to measuring enzyme activity, fluorescent and colorimetric substrates can be used easily to assess enzyme activities. Fluorescence resonance energy transfer (FRET) substrates have been used, for example, to detect stromelysin activity in synovial fluid [33]. In this study, we asked whether FRET substrates might be utilized as the basis of a non-invasive test for cancer. This would require only the addition of the substrate to urine and measurement of an increase in fluorescence over a short period of time using a fluorescence plate reader. Fluorescent substrates for MMPs and ADAMs have been designed [34–36] based on either previously known physiological substrates of these enzymes or results from substrate mapping experiments. Here, we have used fluorescent substrates described in the literature to determine whether they are suitable to indicate the presence of breast cancer.
Materials and methods
Study Population
Eighty-nine samples were analyzed in this study, including samples from patients diagnosed with ductal carcinoma in situ (n=24), invasive breast cancer (n=22) and advanced metastatic breast cancer (n=18) cancer and age-matched controls (n=21). All diagnoses were confirmed by biopsy. Specimens were obtained prior to surgical or other therapeutic intervention. Institutional Review Board approval for the study was obtained.
Urine sample collection and processing
For a pilot study, five normal and five metastatic age-matched urines were purchased from Bioreclammation, Inc. All of the remaining urine samples were collected according to the institutional bioethical guidelines pertaining to discarded clinical material as previously reported by us [27]. Samples were collected in sterile containers and immediately frozen at −20°C. Urine was tested for presence of blood and leukocytes using Multistix 9 Urinalysis Strips (Bayer, Elkhart IN) and samples containing blood or leukocytes were excluded. Protein concentration of urine was determined by the Bradford method using bovine serum albumin as the standard.
Screening of urines with fluorescence substrates
Initially, a pilot study composed of five normal and five metastatic age-matched urines was conducted. Flsub8, DabPChaGC(Me)HAK(Fam)-NH2, Flsub10, Dabcyl-SPLAQAVRSSK(Fam)-NH2, Flsub11, Dabcyl-GPLGMRGK(Fam)-NH2, Flsub13, Dabcyl-HGDQMAQKSK(Fam)-NH2, Flsub21, Dabcyl-LAQAHomopheRSK(Fam)-NH2, and Flsub 63, Dabcyl-SNLAYYTAK(Fam)K-NH2 were prepared as 10 mM stock solutions in dimethyl sulfoxide (DMSO). Substrates were diluted to 20 µM in buffer containing 50 mM Tris, pH8, 10 mM CaCl2, and 0.01% Brij for assays measuring ADAM activity and 50 mM Tris, pH 7.5, 150 mM NaCl, 5 mM CaCl2, 1 µM ZnSO4, 0.01% Brij for assays measuring MMP activity. For the assay, 75 µl of urine was added to a Grenier 96 well black coated plate (Sigma-Aldrich, St. Louis, MO). To start the reaction, 25 µl of substrate was added via multipipettor. Fluorescence values were measured in a Fluroskan II at excitation and emission wavelengths of 485 and 530nm respectively. Control wells contained urine with substrate buffer alone or 25 µl of substrate buffer and 75 µl of phosphate buffered saline. The total running time for each assay was 2.5 hr, readings were taken every 2 min. Ninety points were used to determine slope values which were linear. Fluorescence units (FU) versus time were plotted and slopes of the initial velocities were obtained by linear fit. Protein concentrations were determined using the Pierce protein assay. Absorbance was measured at 595 nm. Specific activities (U/mg) were determined by dividing the slopes from the FU versus time graphs by total protein present in the urine for each assay.
Measurement of Flsub21 and Flsub13 cleavage activities
Stock substrate solutions of Flsub21 and Flsub13 were prepared at 5 mM concentration in DMSO and stored at −80 °C. Substrates were thawed and diluted into assay buffer (50 mM Tris, pH 7.5, 5 mM CaCl2, 1 µM ZnSO4) to obtain a working concentration of 20 µM for the assays.
Reactions were conducted in 96-well white polystyrene flat bottom plates (Whatman, GE Healthcare, Piscataway, NJ) at room temperature. All assays were conducted in duplicate. The assay mixture consisted of 80 µl urine sample and 20 µl substrate (final substrate concentration 4 µM). To determine background fluorescent levels, control wells containing assay buffer only, substrate alone or urine sample alone were used and background fluorescent levels subtracted before activity calculation. The enzyme, assay buffer or urine samples were added to the wells first and subsequently the reaction was started by adding the substrate using a multichannel pipettor. The reaction was monitored using a Wallace Victor2 1420 Multilabel counter (Perkin Elmer, Waltham, MA). Excitation and emission filters were set to 485 and 530 nm respectively. For all assays fluorescence measurements were recorded every 15 min over a period of 3 h. Experimental data from the fluorimeter was imported into Excel for specific activity calculations. Net fluorescence was obtained by the subtraction of background fluorescence from each well. Slope was calculated using net fluorescence increase (using twelve data points) in the linear range versus time curves. Slope values were initially divided by an arbitrary number 1000. To obtain substrate cleavage activity (U/ml), slope values were multiplied by 12.5. Finally, specific activity (U/mg) was calculated using protein concentration values of each urine sample.
Statistical Analysis
Cleavage of FRET substrates Flsub21 and Flsub13 were compared between breast cancer urine samples and control urine samples using the nonparametric Mann-Whitney U-test since these substrates displayed skewness as assessed by the Kolmogorov-Smirnov test of normality [37]. Data are presented as medians and interquartile ranges. Using an activity value of greater than 0 U/mg for Flsub21 and Flsub13 to identify possible breast cancer, sensitivity and specificity were calculated for breast cancer patients versus controls, and for invasive breast cancer (IBC)/metastatic disease versus controls using Fisher’s exact test. Multivariable logistic regression was used to test whether Flsub21 and Flsub13 (based on a positive or negative test results) were independently predictive of 1) cancer versus control, and 2) IBC/metastatic disease compared to controls with the odds ratio and 95% confidence interval (CI) for determining risk [38,39]. Receiver operating characteristic (ROC) curve analysis was applied to determine the diagnostic accuracy of each urinary substrate in differentiating cancer patients from controls using area under the curve (AUC) with a 95% CI as the measure of prediction [40]. Statistical analysis was performed using the SPSS software package (version 18.0, SPSS Inc./IBM, Chicago, IL). Two-tailed values of P < 0.05 were considered statistically significant.
Results
Screening of fluorescent substrates for MMP/ADAM activity
Several different MMPs and ADAMs were screened for their ability to cleave a range of fluorescent substrates including Flsub8, −10, −11, −13, −21 and −63 (Table 1). Substrates were chosen based on their ability to distinguish between MMP-9, MMP-2, ADAM12 and ADAM8. For example, Flsub8 and −11 were chosen because of their selectivity for MMP-9 over MMP-2. In addition, these two substrates were not very reactive towards other MMPs with the exceptions of MMP-13, and ADAM family members, ADAM8, −9, −10, −12, and −17. Since the presence of MMP-9 and not MMP-2 in urine was shown to correlate with breast cancer, we reasoned that these two substrates could potentially be used successfully to predict disease status. Flsub63 is a substrate selective for MMP-2 over MMP-9 and therefore served as a negative control. Finally, Flsub10, −13 and −21 are useful fluorescent substrates to measure ADAM activity. Flsub10 is not a selective substrate but is very sensitive for ADAM17. Flsub21 is the best substrate known to date for ADAM12 [35] although it can be cleaved by ADAM17 and ADAM10 as well. Flsub13 is used typically to detect ADAM8 activity. Since a secreted isoform of ADAM8 has been reported and since ADAM8 levels have been shown to correlate with head and neck cancer [41], we chose to use Flsub13 in the event that ADAM8 is also present in the urine samples. In addition, Flsub13 is selective for ADAM8 over most of the MMPs.
Table 1.
Screening of Fluorescent Substrate Cleavage Activity by MMPs and ADAMs
| Flsub10c | Flsub21b | Flsub13b | Flsub63a | Flsub8b | Flsub11a | |
|---|---|---|---|---|---|---|
| ADAM8 | 3.3 × 104 | 1.0 × 105 | 5.3 × 104 | 2.5 × 103 | 1.7 × 103 | 2.4 × 103 |
| ADAM9 | 1.1 × 105 | 8.3 × 103 | 1.6 × 103 | 2.2 × 104 | 4.5 × 103 | 2.3 × 102 |
| ADAM10 | 3.6 × 103 | 6.2 × 103 | 2.7 × 102 | 1.4 × 104 | 1.9 × 102 | <1 × 101 |
| ADAM12 | 3.8 × 104 | 2.8 × 105 | 4.0 × 101 | NAd | 3.0 × 103 | NAd |
| ADAM17 | 9.6 × 105 | 4.3 × 105 | NDe | 1.1 × 103 | 3.8 × 104 | 3.7 × 103 |
| MMP-1 | 5.1 × 104 | 2.8 × 104 | NDe | 3.2 × 103 | 7.6 × 104 | 4.5 × 104 |
| MMP-2 | 1.7 × 105 | 3.2 × 105 | 2.4 × 103 | 1.3 × 106 | 2.9 × 104 | 4.8 × 105 |
| MMP-3 | 3.9 × 104 | 4.0 × 103 | NDe | 2.2 × 104 | 5.2 × 101 | 1.6 × 103 |
| MMP-8 | 2.6 × 104 | 1.4 × 105 | NDe | 3.4 × 105 | 2.4 × 104 | 3.6 × 104 |
| MMP-9 | 6.0 × 105 | 2.2 × 105 | NDe | 1.4 × 105 | 8.5 × 105 | 1.4 × 106 |
| MMP-13 | 1.7 × 106 | 4.6 × 105 | NDe | 1.6 × 106 | 2.1 × 106 | 3.3 × 106 |
Specificity values at 10 µM substrate concentration were measured by back calculation of enzyme concentrations after determination of kcat/Km for known substrates. Enzyme concentrations were determined previously by active site titration as described in Rasmussen et. al. [34]
Values were taken from Moss et. al. [35]
Values were taken from Moss et. al. [42]
Not attempted
No activity detected
To ascertain whether these fluorescent substrates could be used to predict disease status, urine samples from patients with metastatic breast cancer and age- and sex-matched controls were initially screened with several substrates that are known to be efficiently cleaved by MMP-2, MMP-9, ADAM8 and ADAM12.
Table 2 indicates the specific activity (fluorescent units (FU)/min.mg) calculated using the fluorescence units versus time plots for each of the urine samples using six distinct substrates. The MMP-9-specific substrates Flsub8 and Flsub11 were only partially selective, with specific activity values for the slopes (mean FU/min.mg of 5 vs. 28 and 13.8 vs. 19) for the normal and metastatic groups respectively. Mean activity values for the metastatic and control groups using the MMP-2 substrate Flsub63 were very similar (Table 2) and all the specific activities were above 1 with the exception of 3N. Similar findings were obtained with the general ADAM substrate, Flsub10, as all the values were positive, even though there was a 33-fold mean difference for the normal and metastatic groups. Interestingly, fluorescent substrates Flsub13 and −21, with high specificity for ADAM8 and ADAM12 respectively, showed the most discrimination between the two test groups. The activity values for Flsub13 and Flsub21 were either negative or approached zero for all urine samples from normal controls with the exception of sample 1N (Table 2). In contrast, urine samples from patients with metastatic disease all tested positive for Flsub21 cleavage activity while 4 out of 5 samples tested were positive for Flsub13 cleavage activity.
Table 2.
Initial Screening of Urine Samples for Fluorescent Substrate Cleavage Activity
| Substratea | Flsub11 | Flsub63 | Flsub8 | Flsub10 | Flsub21 | Flsub13 | |
|---|---|---|---|---|---|---|---|
| Urines | |||||||
| Normal | |||||||
| 1N | 69b | 24 | 45 | 39 | 44 | 29 | |
| 2N | 0 | 0.8 | 0 | 15 | 0 | 0 | |
| 3N | 0 | 0 | 0 | 76 | 0 | 0 | |
| 4N | 0 | 15 | 0 | 87 | 0 | 0 | |
| 5N | 0 | 36 | 0 | 1.3 | 0 | 0 | |
| Average N | 13.8 | 15 | 5 | 43.6 | 8.8 | 5.8 | |
| Metastatic | |||||||
| 1M | 0 | 46 | 17 | 1000 | 4.7 | 4.8 | |
| 2M | 33 | 15 | 3.8 | 190 | 16 | 5.4 | |
| 3M | 47 | 13 | 100 | 1300 | 17 | 0 | |
| 4M | 15 | 7.9 | 21 | 3600 | 25 | 29 | |
| 5M | 0 | 10 | 0 | 1100 | 22 | 7.4 | |
| Average M | 19 | 18 | 28 | 1438 | 17 | 9 | |
Reaction conditions provided in the Materials and methods section
Specific activity (FU/min.mg)
Our initial screening studies indicated that Flsub21 and Flsub13 could prove to be useful substrates for screening urine samples from breast cancer patients. Purified recombinant human ADAM12 has been previously reported to cleave Flsub21 efficiently but not Flsub13 [35] whereas ADAM8 could selectively cleave Flsub13 [35]. Next, we assessed the effect of metalloprotease inhibitors on Flsub21 and Flsub13 cleavage activity in urine samples. The general MMP inhibitor, GM6001 (500nM) had no effect on Flsub21 and Flsub13 cleavage activity in urine samples. However, when the broad spectrum ADAM inhibitor, TMI (5 µM) was used, urine from cancer patients displayed inhibition for Flsub13- (20–100%) and Flsub21-cleavage activity (0–45%), whereas no inhibitable activity was present in any of the normal control urines regardless of the substrate used (data not shown). These findings prompted us to test urine samples using the fluorescent substrates Flsub21 and Flsub13 to determine whether there was a correlation between activity and disease status. A total of 85 urine samples were tested including 64 from breast cancer patients and 21 from age- and sex-matched controls. The breast cancer cohort included samples from across the disease spectrum, ductal carcinoma in-situ (DCIS, n=24), locally invasive breast cancer (IBC, n=22) and advanced metastatic disease (n=18) (Fig. 1). Urine samples were tested in triplicate. Urinary Flsub21 and Flsub13 cleavage activity for breast cancer patients and normal controls is presented in Table 3. Using Flsub21, only 19% of urines from normal controls displayed cleavage activity whereas 34%, 68% and 89% samples from DCIS, IBC and metastatic patients respectively displayed Flsub21 cleavage activity. For this substrate, the median specific activity values for samples from patients with DCIS were similar to normal controls, however, median specific activity levels were significantly higher in urine samples from invasive (P<0.001) and metastatic breast cancer (P<0.001) (Table 3). Similar trends were observed for Flsub13, while only 28% of samples from normal controls displayed any activity 37%, 77% and 89% samples from DCIS, IBC and metastatic patients respectively had positive Flsub13 activity. In addition, the median specific activity values using Flsub13 were not very different for normal controls or DCIS samples and significantly higher median specific activities were observed for urines from patients with IBC (P<0.001) and metastatic breast cancer (P<0.001).
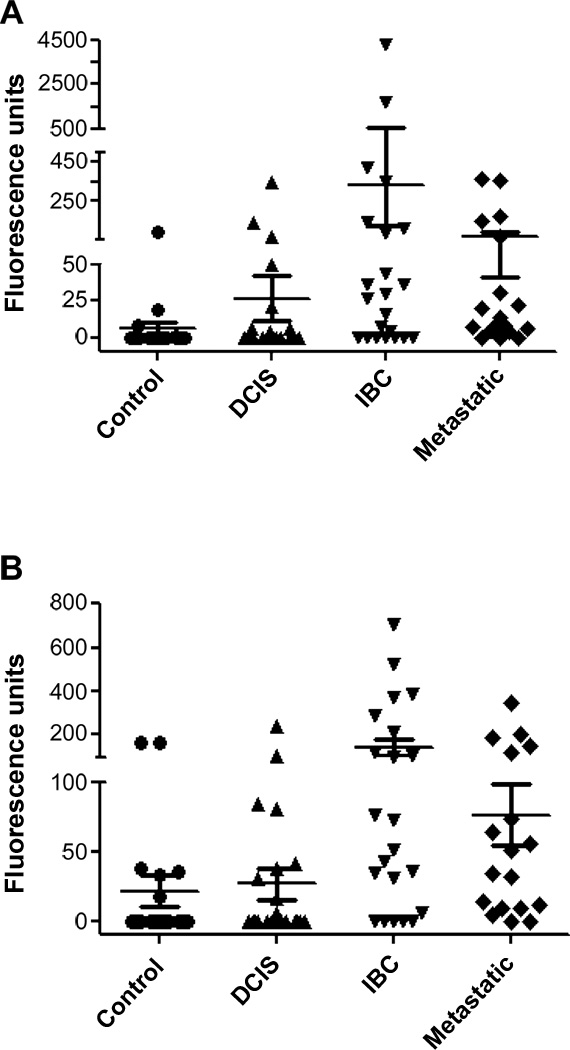
Figure 1.
Flsub21 and Flsub13 cleavage activity is significantly higher in urine of patients with invasive and metastatic breast cancer compared to controls. Scatter plots representing urinary Flsub21 (A) and Flsub13 (B) cleavage activity from breast cancer groups; DCIS (triangle), IBC (inverted triangle), metastatic disease (diamonds) and normal controls (circles). Each assay was conducted in triplicate and the results are expressed as the mean (±SEM). Using Flsub21, specific activity levels were significantly higher in urine samples from invasive (p<0.001) and metastatic breast cancer (p<0.001) compared to controls. Similarly for Flsub13, significantly higher specific activities were observed for urines from patients with IBC (p<0.001) and metastatic breast cancer (p<0.001) compared to controls.
Table 3.
Urinary Flsub21 and Flsub13 Activity in Breast Cancer Patients and Controls
| Substrate (U/mg) |
Controls (n = 21) |
Breast Cancer (n = 64) |
DCIS (n = 24) |
IBC (n = 22) |
Metastatic (n = 18) |
|---|---|---|---|---|---|
| Flsub21 | |||||
| Median | 0 | 5.7 | 0 | 27.5 | 11.0 |
| IQR | 0-0 | 0–48 | 0–6 | 0–114 | 5–88 |
| Range | 0–94 | 0–4319 | 0–347 | 0–4319 | 0–362 |
| P value | − | <0.001* | 0.24 | <0.001* | <0.001* |
| Flsub13 | |||||
| Median | 0 | 35.0 | 0 | 73.7 | 43.0 |
| IQR | 0–25 | 0–101 | 0–40 | 32–270 | 9–127 |
| Range | 0–165 | 0–3702 | 0–242 | 0–3702 | 0–347 |
| P value | − | 0.002* | 0.31 | <0.001* | <0.001* |
Statistically significant, P values vs. controls, Mann-Whitney U-test. IQR = interquartile range, DCIS = ductal carcinoma in situ, IBC = invasive breast cancer
This data was analyzed by univariate and multivariate statistics (Table 3 and 4). Multivariable logistic regression using binary cut-off values for the two substrates indicated that Flsub21 provided significant predictive information in differentiating breast cancer patients from controls (Table 4; odds ratio 7.7, 95% CI: 2.3 – 25.8, P < 0.001). Flsub13 was not found to provide additional predictive information (P = 0.12). In a subgroup analysis considering the 40 IBC/metastatic disease patients and the 21 controls, multivariable logistic regression indicated that using binary cut-off values, Flsub21 (P = 0.009) and Flsub13 (P = 0.028) cleavage activities were significant independent biomarkers. The odds of IBC/metastatic disease based on a value greater than 0 U/mg for Flsub21 was almost 8 times higher (odds ratio 7.9, 95% CI: 2.0 – 37.4), whereas the odds of IBC/metastatic disease were over 5 times higher in individuals having a positive Flsub13 activity (odds ratio 5.7, 95% CI: 1.3 – 27.2).
Table 4.
Diagnostic Performance Characteristics of the Substrates
| Breast Cancer vs. Controls | IBC/Metastatic vs. Controls | |||||
|---|---|---|---|---|---|---|
| AUC | 95% CI | P value | AUC | 95% CI | P value | |
| Flsub21 | 0.745 | 0.633–0.857 | 0.001 | 0.842 | 0.735–0.946 | <0.001 |
| Flsub13 | 0.724 | 0.601–0.646 | 0.002 | 0.811 | 0.692–0.929 | <0.001 |
| Sensitivity | Specificity | P value | Sensitivity | Specificity | P value | |
| Flsub21 | 41/64=64% | 17/21=81% | 0.001 | 31/40=78% | 17/21=81% | <0.001 |
| Flsub13 | 44/62=71% | 15/21=71% | 0.002 | 33/38=87% | 15/21=71% | <0.001 |
IBC = invasive breast cancer. AUC = area under the curve based on ROC curve analysis. CI = confidence interval.
Overall, there were significant differences in the median specific activity using both Flsub21 and Flsub13 between all 64 cancer patients and 21 controls (Table 3). Examining each group separately, it was obvious that the substrates show differences although these differences are statistically significant only for the IBC and metastatic group compared to controls and did not reach significance for the DCIS groups. Medians, interquartile ranges (25th – 75th percentiles), and full ranges were used with the nonparametric Mann-Whitney U-test (due to skewness of the substrate data) for comparing groups. Multivariable analysis using logistic regression indicated that while both Flsub21 and Flsub13 displayed some value in differentiating cancer versus controls, this was largely due to higher Flsub21 and Flsub13 values in the more advanced cancer subgroups (IBC or metastatic disease). Urinary Flsub21 and Flsub13 activities were highly correlated and logistic regression confirmed that they are not independent markers. Therefore, one of these two substrates would probably be sufficient from the perspective of predicting the presence of any cancer. The utility of these substrates is limited for distinguishing cancer from non-cancer when considering the DCIS cancer subgroup, since there were too many DCIS urine samples with non-measurable Flsub21 and/or Flsub13 activity.
Based on multivariable regression modeling, using the two fluorescent substrates for predicting IBC and/or advanced metastatic disease, the model would predict a 20% probability if both substrates were negative, 55% probability if Flsub13 was positive and Flsub21 was negative, 65% probability if Flsub21 was positive and Flsub13 was negative, and a 90% probability of advanced breast cancer (IBC or metastatic disease) if urinary Flsub21 and Flsub13 cleavage activities were both positive. In general, the value of these substrates increases substantially in differentiating the more advanced breast cancer from normal controls or from the DCIS subgroup.
Table 4 summarizes results of ROC analysis and shows that while the AUC was significant for substrates in differentiating all cancers (n=64) compared to normal controls (n=21), the diagnostic performance based on AUC values was better when differentiating invasive and/or metastatic breast cancer from normal controls as opposed to differentiating between controls and all cancers.
Using any positive urinary FRET substrate cleavage activity measurement, the table summarizes sensitivity and specificity for Flsub21 and Flsub13 for all breast cancer groups versus control and for IBC/metastatic versus controls (the sensitivity for each substrate is much higher). In conclusion, the substrates Flsub21 and Flsub13 offer useful diagnostic characteristics as predictive biomarkers, however the data suggest that this value lies mainly in differentiating more advanced breast cancer disease from normal controls.
Discussion
A number of the MMP and ADAM family members are found in biological fluids and are now being appreciated as potential biomarkers for cancer. In the current study, we have used fluorescent substrates to determine whether they have the potential to be used in predicting the presence of disease using urine samples from patients with breast cancer and normal controls. Fluorescent substrates designed to be specific for ADAM family members were predictive of disease status. Some ADAM substrates proved not to be useful, such as the best TACE substrate Flsub10 (data not shown) and broad spectrum substrates described in Moss et. al. [42]. During the initial screening process, of all the substrates tested, those more selective for MMP-9 proved to be the most useful. The best MMP-9 substrate tested was Flsub11 which is slightly more specific than Flsub8 for MMP-9 compared to the ADAMs and other MMPs. Flsub11 is based on a previous substrate that was demonstrated to be useful for detection of MMP-13 activity [34]. However, its specificity constant is greater than 106 for MMP-9 and as a result appears to be very useful in detecting MMP-9 activity. In contrast, Flsub63, which is a MMP-2 specific substrate, was not indicative of disease status.
The substrates that proved to be the best for large scale screening of urines were Flsub13 and Flsub21. Flsub13, an ADAM8 substrate that is based on a cleavage sequence of CD23, is most promising since only activities in urines from cancer patients were inhibited by the broad spectrum ADAM inhibitor, TMI-1 (data not shown). Flsub13 is unique in that it is not processed well by any of the MMPs tested. Flsub21 is based on the processed site in TNF-alpha but has homophenylalanyl in place of valine at S1’. This substitution was shown previously to be helpful in increasing TACE activity [43] although it is cleaved by a number of MMP and ADAM family members. The closely related Flsub10, with the native TNF-alpha substrate sequence, is not as useful as Flsub21, suggesting that the unnatural amino acid substitution is beneficial. Another ADAM selective substrate, PEPDAB014, which is based on the TGF-alpha cleavage site [42], was not a useful substrate, suggesting that TACE activity may not be predictive of the presence of metastatic breast cancer. Therefore, substrates based on physiological cleavage sites of proteins that are shed by ADAMs appear to be useful in detecting ADAM family members in human urine samples. In fact, combinations of semi-selective substrates along with the use of inhibitors as in the multiple enzyme reagent assay system (MEMRAS) technique [34] has been modified recently by Miller et. al. to quantify ADAM17 and -10 levels in cellular assays, even though there were non-specific proteinase activities present [44].
More importantly, however, is the finding that a combination of Flsub21 and Flsub13 activity determined, with 90% certainty, that the individual has either invasive breast cancer or metastatic disease. While the substrates studied here were not useful in detecting the presence of other stages of the disease, combinations of semi-selective substrates may ultimately be useful in doing so [44]. Traditional techniques to measure metalloproteinase activities have relied on ELISA assays and zymography. While ELISAs can be quite useful, they do not measure how catalytically active an enzyme is in a biological fluid. The use of fluorescence substrates offers a unique, quantifiable and sensitive way to determine active enzyme concentrations. Furthermore, unlike zymography which is gel-based, these fluorescence assays can be high throughput, completed within a shorter time frame and automated if necessary.
In this report, we have presented a proof-of-concept study in which fluorescent substrate cleavage activity was used to predict disease status in patients with breast cancer. Using a cohort of samples from across the disease spectrum and comparing these to normal controls, we found that Flsub21 and Flsub13 cleavage activity may be used to distinguish between the presence of invasive and/or metastatic breast cancer from normal controls.
Mosesetal _Highlights.
We analyzed proteolysis of FRET substrates in urine of breast cancer patients and controls.
Flsub21 and Flsub13 cleavage is detected at significantly higher frequencies in urine of breast patients.
FRET substrates may be used to non-invasively identify invasive and/or metastatic breast cancer.
Acknowledgements
This work was supported by The Breast Cancer Research Foundation, NIH PO1 CA45548 and The Fortin Foundation.
Abbreviations
- FRET
fluorescence resonance energy transfer
- MMP
matrix metalloprotease
- ADAM
a disintegrin and metalloprotease
- IBC
invasive breast cancer
- DCIS
ductal carcinoma in situ
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Roy R, Zhang B, Moses MA. Making the cut: protease-mediated regulation of angiogenesis. Exp Cell Res. 2006;312:608–622. doi: 10.1016/j.yexcr.2005.11.022. [DOI] [PubMed] [Google Scholar]
- 2.Chambers AF, Matrisian LM. Changing views of the role of matrix metalloproteinases in metastasis. J Natl Cancer Inst. 1997;89:1260–1270. doi: 10.1093/jnci/89.17.1260. [DOI] [PubMed] [Google Scholar]
- 3.Kleiner DE, Stetler-Stevenson WG. Matrix metalloproteinases and metastasis. Cancer Chemother Pharmacol. 1999;43 Suppl:S42–S51. doi: 10.1007/s002800051097. [DOI] [PubMed] [Google Scholar]
- 4.Curran S, Murray GI. Matrix metalloproteinases in tumour invasion and metastasis. J Pathol. 1999;189:300–308. doi: 10.1002/(SICI)1096-9896(199911)189:3<300::AID-PATH456>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 5.Nagase H, Woessner JF., Jr Matrix metalloproteinases. J Biol Chem. 1999;274:21491–21494. doi: 10.1074/jbc.274.31.21491. [DOI] [PubMed] [Google Scholar]
- 6.Fang J, Shing Y, Wiederschain D, Yan L, Butterfield C, Jackson G, Harper J, Tamvakopoulos G, Moses MA. Matrix metalloproteinase-2 is required for the switch to the angiogenic phenotype in a tumor model. Proc Natl Acad Sci U S A. 2000;97:3884–3889. doi: 10.1073/pnas.97.8.3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bergers G, Javaherian K, Lo KM, Folkman J, Hanahan D. Effects of angiogenesis inhibitors on multistage carcinogenesis in mice. Science. 1999;284:808–812. doi: 10.1126/science.284.5415.808. [DOI] [PubMed] [Google Scholar]
- 8.Egeblad M, Werb Z. New functions for the matrix metalloproteinases in cancer progression. Nat Rev Cancer. 2002;2:161–174. doi: 10.1038/nrc745. [DOI] [PubMed] [Google Scholar]
- 9.Bode W. Structural basis of matrix metalloproteinase function. Biochem Soc Symp. 2003:1–14. doi: 10.1042/bss0700001. [DOI] [PubMed] [Google Scholar]
- 10.Seals DF, Courtneidge SA. The ADAMs family of metalloproteases: multidomain proteins with multiple functions. Genes Dev. 2003;17:7–30. doi: 10.1101/gad.1039703. [DOI] [PubMed] [Google Scholar]
- 11.Edwards DR, Handsley MM, Pennington CJ. The ADAM metalloproteinases. Mol Aspects Med. 2008;29:258–289. doi: 10.1016/j.mam.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maretzky T, Schulte M, Ludwig A, Rose-John S, Blobel C, Hartmann D, Altevogt P, Saftig P, Reiss K. L1 is sequentially processed by two differently activated metalloproteases and presenilin/gamma-secretase and regulates neural cell adhesion, cell migration, and neurite outgrowth. Mol Cell Biol. 2005;25:9040–9053. doi: 10.1128/MCB.25.20.9040-9053.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khokha R, Werb Z. Mammary gland reprogramming: metalloproteinases couple form with function. Cold Spring Harb Perspect Biol. 3 doi: 10.1101/cshperspect.a004333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cawston TE, Wilson AJ. Understanding the role of tissue degrading enzymes and their inhibitors in development and disease. Best Pract Res Clin Rheumatol. 2006;20:983–1002. doi: 10.1016/j.berh.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 15.Turner SL, Blair-Zajdel ME, Bunning RA. ADAMs and ADAMTSs in cancer. Br J Biomed Sci. 2009;66:117–128. doi: 10.1080/09674845.2009.11730257. [DOI] [PubMed] [Google Scholar]
- 16.Moali C, Hulmes DJ. Extracellular and cell surface proteases in wound healing: new players are still emerging. Eur J Dermatol. 2009;19:552–564. doi: 10.1684/ejd.2009.0770. [DOI] [PubMed] [Google Scholar]
- 17.Aiken A, Khokha R. Unraveling metalloproteinase function in skeletal biology and disease using genetically altered mice. Biochim Biophys Acta. 2010;1803:121–132. doi: 10.1016/j.bbamcr.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 18.Pories SE, Zurakowski D, Roy R, Lamb CC, Raza S, Exarhopoulos A, Scheib RG, Schumer S, Lenahan C, Borges V, Louis GW, Anand A, Isakovich N, Hirshfield-Bartek J, Wewer U, Lotz MM, Moses MA. Urinary metalloproteinases: noninvasive biomarkers for breast cancer risk assessment. Cancer Epidemiol Biomarkers Prev. 2008;17:1034–1042. doi: 10.1158/1055-9965.EPI-07-0365. [DOI] [PubMed] [Google Scholar]
- 19.Smith ER, Zurakowski D, Saad A, Scott RM, Moses MA. Urinary biomarkers predict brain tumor presence and response to therapy. Clin Cancer Res. 2008;14:2378–2386. doi: 10.1158/1078-0432.CCR-07-1253. [DOI] [PubMed] [Google Scholar]
- 20.Roy R, Louis G, Loughlin KR, Wiederschain D, Kilroy SM, Lamb CC, Zurakowski D, Moses MA. Tumor-specific urinary matrix metalloproteinase fingerprinting: identification of high molecular weight urinary matrix metalloproteinase species. Clin Cancer Res. 2008;14:6610–6617. doi: 10.1158/1078-0432.CCR-08-1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chan LW, Moses MA, Goley E, Sproull M, Muanza T, Coleman CN, Figg WD, Albert PS, Menard C, Camphausen K. Urinary VEGF and MMP levels as predictive markers of 1-year progression-free survival in cancer patients treated with radiation therapy: a longitudinal study of protein kinetics throughout tumor progression and therapy. J Clin Oncol. 2004;22:499–506. doi: 10.1200/JCO.2004.07.022. [DOI] [PubMed] [Google Scholar]
- 22.Jung K, Krell HW, Ortel B, Hasan T, Romer A, Schnorr D, Loening SA, Lein M. Plasma matrix metalloproteinase 9 as biomarker of prostate cancer progression in Dunning (Copenhagen) rats. Prostate. 2003;54:206–211. doi: 10.1002/pros.10183. [DOI] [PubMed] [Google Scholar]
- 23.Davies B, Waxman J, Wasan H, Abel P, Williams G, Krausz T, Neal D, Thomas D, Hanby A, Balkwill F. Levels of matrix metalloproteases in bladder cancer correlate with tumor grade and invasion. Cancer Res. 1993;53:5365–5369. [PubMed] [Google Scholar]
- 24.Wu ZS, Wu Q, Yang JH, Wang HQ, Ding XD, Yang F, Xu XC. Prognostic significance of MMP-9 and TIMP-1 serum and tissue expression in breast cancer. Int J Cancer. 2008;122:2050–2056. doi: 10.1002/ijc.23337. [DOI] [PubMed] [Google Scholar]
- 25.Vasala K, Paakko P, Turpeenniemi-Hujanen T. Matrix metalloproteinase-9 (MMP-9) immunoreactive protein in urinary bladder cancer: a marker of favorable prognosis. Anticancer Res. 2008;28:1757–1761. [PubMed] [Google Scholar]
- 26.Fernandez CA, Yan L, Louis G, Yang J, Kutok JL, Moses MA. The matrix metalloproteinase-9/neutrophil gelatinase-associated lipocalin complex plays a role in breast tumor growth and is present in the urine of breast cancer patients. Clin Cancer Res. 2005;11:5390–5395. doi: 10.1158/1078-0432.CCR-04-2391. [DOI] [PubMed] [Google Scholar]
- 27.Moses MA, Wiederschain D, Loughlin KR, Zurakowski D, Lamb CC, Freeman MR. Increased incidence of matrix metalloproteinases in urine of cancer patients. Cancer Res. 1998;58:1395–1399. [PubMed] [Google Scholar]
- 28.Yan L, Borregaard N, Kjeldsen L, Moses MA. The high molecular weight urinary matrix metalloproteinase (MMP) activity is a complex of gelatinase B/MMP-9 and neutrophil gelatinase-associated lipocalin (NGAL). Modulation of MMP-9 activity by NGAL. J Biol Chem. 2001;276:37258–37265. doi: 10.1074/jbc.M106089200. [DOI] [PubMed] [Google Scholar]
- 29.Smith ER, Manfredi M, Scott RM, Black PM, Moses MA. A recurrent craniopharyngioma illustrates the potential usefulness of urinary matrix metalloproteinases as noninvasive biomarkers: case report. Neurosurgery. 2007;60:E1148–E1149. doi: 10.1227/01.NEU.0000255464.37634.3C. discussion E1149. [DOI] [PubMed] [Google Scholar]
- 30.La Rocca G, Pucci-Minafra I, Marrazzo A, Taormina P, Minafra S. Zymographic detection and clinical correlations of MMP-2 and MMP-9 in breast cancer sera. Br J Cancer. 2004;90:1414–1421. doi: 10.1038/sj.bjc.6601725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roy R, Wewer UM, Zurakowski D, Pories SE, Moses MA. ADAM 12 cleaves extracellular matrix proteins and correlates with cancer status and stage. J Biol Chem. 2004;279:51323–51330. doi: 10.1074/jbc.M409565200. [DOI] [PubMed] [Google Scholar]
- 32.Frohlich C, Albrechtsen R, Dyrskjot L, Rudkjaer L, Orntoft TF, Wewer UM. Molecular profiling of ADAM12 in human bladder cancer. Clin Cancer Res. 2006;12:7359–7368. doi: 10.1158/1078-0432.CCR-06-1066. [DOI] [PubMed] [Google Scholar]
- 33.Beekman B, van El B, Drijfhout JW, Ronday HK, TeKoppele JM. Highly increased levels of active stromelysin in rheumatoid synovial fluid determined by a selective fluorogenic assay. FEBS Lett. 1997;418:305–309. doi: 10.1016/s0014-5793(97)01371-9. [DOI] [PubMed] [Google Scholar]
- 34.Rasmussen FH, Yeung N, Kiefer L, Murphy G, Lopez-Otin C, Vitek MP, Moss ML. Use of a multiple-enzyme/multiple-reagent assay system to quantify activity levels in samples containing mixtures of matrix metalloproteinases. Biochemistry. 2004;43:2987–2995. doi: 10.1021/bi036063m. [DOI] [PubMed] [Google Scholar]
- 35.Moss ML, Rasmussen FH. Fluorescent substrates for the proteinases ADAM17, ADAM10, ADAM8, and ADAM12 useful for high-throughput inhibitor screening. Anal Biochem. 2007;366:144–148. doi: 10.1016/j.ab.2007.04.043. [DOI] [PubMed] [Google Scholar]
- 36.Moss ML, Sklair-Tavron L, Nudelman R. Drug insight: tumor necrosis factor-converting enzyme as a pharmaceutical target for rheumatoid arthritis. Nat Clin Pract Rheumatol. 2008;4:300–309. doi: 10.1038/ncprheum0797. [DOI] [PubMed] [Google Scholar]
- 37.Altman D. Practical statistics for medical research. New York: Chapman & Hall; 1991. [Google Scholar]
- 38.Katz M. Multivariable analysis: a practical guide for clinicians. New York: Cambridge University Press; 2006. [Google Scholar]
- 39.Harrell F. Regression modeling strategies. With applications to linear models, logistic regression, and survival analysis. New York: Springer; 2001. [Google Scholar]
- 40.Pepe M. The statistical evaluation of medical tests for classification and prediction. New York: Oxford University Press; 2004. [Google Scholar]
- 41.Stokes A, Joutsa J, Ala-Aho R, Pitchers M, Pennington CJ, Martin C, Premachandra DJ, Okada Y, Peltonen J, Grenman R, James HA, Edwards DR, Kahari VM. Expression profiles and clinical correlations of degradome components in the tumor microenvironment of head and neck squamous cell carcinoma. Clin Cancer Res. 2010;16:2022–2035. doi: 10.1158/1078-0432.CCR-09-2525. [DOI] [PubMed] [Google Scholar]
- 42.Moss ML, Rasmussen FH, Nudelman R, Dempsey PJ, Williams J. Fluorescent substrates useful as high-throughput screening tools for ADAM9. Comb Chem High Throughput Screen. 2010;13:358–365. doi: 10.2174/138620710791054259. [DOI] [PubMed] [Google Scholar]
- 43.Lambert MH, Blackburn RK, Seaton TD, Kassel DB, Kinder DS, Leesnitzer MA, Bickett DM, Warner JR, Andersen MW, Badiang JG, Cowan DJ, Gaul MD, Petrov KG, Rabinowitz MH, Wiethe RW, Becherer JD, McDougald DL, Musso DL, Andrews RC, Moss ML. Substrate specificity and novel selective inhibitors of TNF-alpha converting enzyme (TACE) from two-dimensional substrate mapping. Comb Chem High Throughput Screen. 2005;8:327–339. doi: 10.2174/1386207054020840. [DOI] [PubMed] [Google Scholar]
- 44.Miller MA, Barkal L, Jeng K, Herrlich A, Moss M, Griffith LG, Lauffenburger DA. Proteolytic Activity Matrix Analysis (PrAMA) for simultaneous determination of multiple protease activities. Integr Biol (Camb) doi: 10.1039/c0ib00083c. [DOI] [PMC free article] [PubMed] [Google Scholar]