Abstract
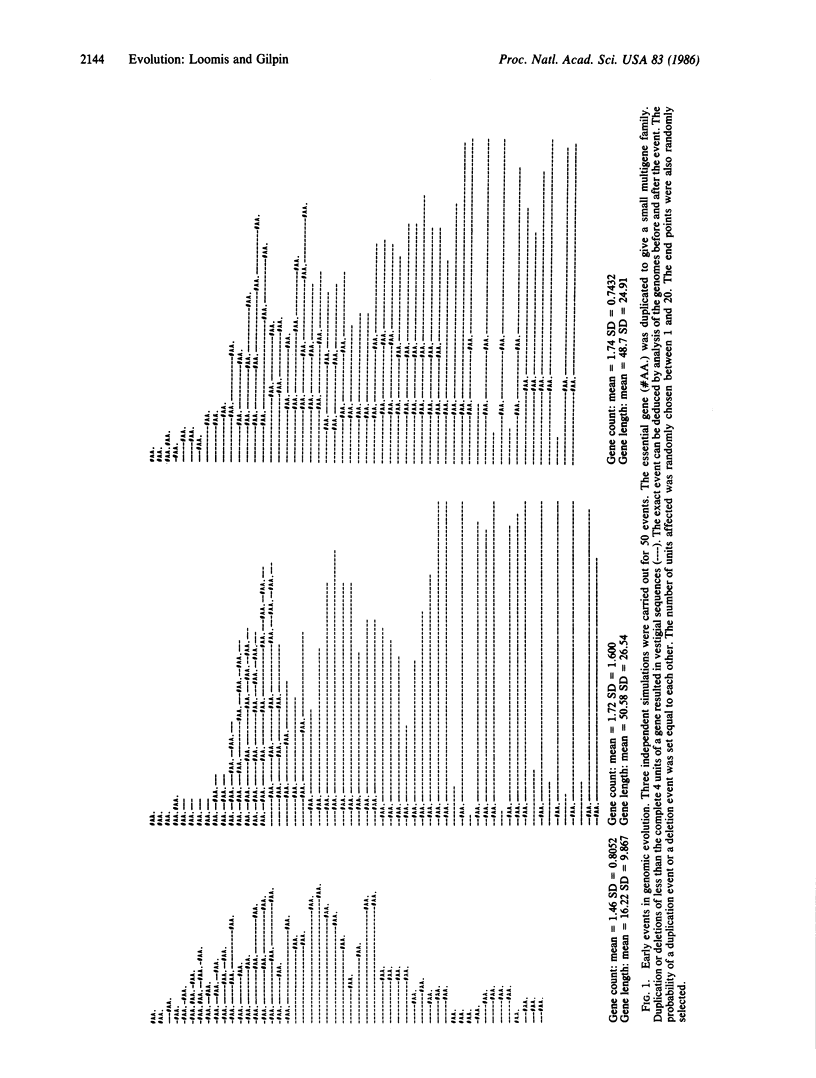
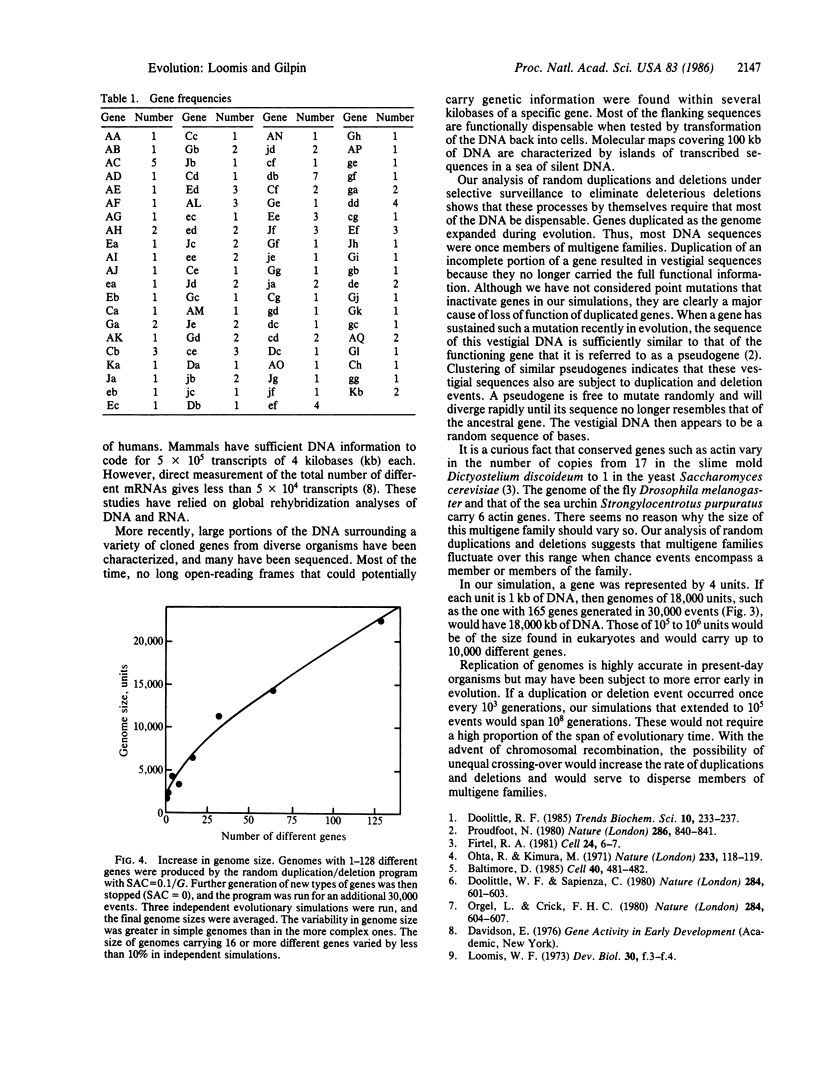
Random duplication and deletion events generate complex genomes carrying a large amount of dispensable sequences. We have simulated such events in a computer model. We followed the evolution of a genome carrying at least one copy of each type of gene. Partial duplications and deletions of genes generated nonfunctional vestigial sequences that were dispensable. The size of the genome stabilized only when the amount of dispensable sequences had increased to the point that most deletions did not affect vital genes. Within such genomes, the number of copies of specific genes fluctuated, thereby generating small multigene families. The parameters of the model were tested over 100,000 events in both simple and complex genomes. The results indicate that when the size of the genome is not critical to survival, as appears to be the case within limits in most eukaryotic organisms, the genome carries vestigial sequences that are no longer functional and that many genes are present in multigene families by chance.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baltimore D. Retroviruses and retrotransposons: the role of reverse transcription in shaping the eukaryotic genome. Cell. 1985 Mar;40(3):481–482. doi: 10.1016/0092-8674(85)90190-4. [DOI] [PubMed] [Google Scholar]
- Doolittle W. F., Sapienza C. Selfish genes, the phenotype paradigm and genome evolution. Nature. 1980 Apr 17;284(5757):601–603. doi: 10.1038/284601a0. [DOI] [PubMed] [Google Scholar]
- Firtel R. A. Multigene families encoding actin and tubulin. Cell. 1981 Apr;24(1):6–7. doi: 10.1016/0092-8674(81)90494-3. [DOI] [PubMed] [Google Scholar]
- Ohta T., Kimura M. Functional organization of genetic material as a product of molecular evolution. Nature. 1971 Sep 10;233(5315):118–119. doi: 10.1038/233118a0. [DOI] [PubMed] [Google Scholar]
- Orgel L. E., Crick F. H. Selfish DNA: the ultimate parasite. Nature. 1980 Apr 17;284(5757):604–607. doi: 10.1038/284604a0. [DOI] [PubMed] [Google Scholar]
- Proudfoot N. Pseudogenes. Nature. 1980 Aug 28;286(5776):840–841. doi: 10.1038/286840a0. [DOI] [PubMed] [Google Scholar]


