Abstract
Summary: Bacteria and fungi can form a range of physical associations that depend on various modes of molecular communication for their development and functioning. These bacterial-fungal interactions often result in changes to the pathogenicity or the nutritional influence of one or both partners toward plants or animals (including humans). They can also result in unique contributions to biogeochemical cycles and biotechnological processes. Thus, the interactions between bacteria and fungi are of central importance to numerous biological questions in agriculture, forestry, environmental science, food production, and medicine. Here we present a structured review of bacterial-fungal interactions, illustrated by examples sourced from many diverse scientific fields. We consider the general and specific properties of these interactions, providing a global perspective across this emerging multidisciplinary research area. We show that in many cases, parallels can be drawn between different scenarios in which bacterial-fungal interactions are important. Finally, we discuss how new avenues of investigation may enhance our ability to combat, manipulate, or exploit bacterial-fungal complexes for the economic and practical benefit of humanity as well as reshape our current understanding of bacterial and fungal ecology.
INTRODUCTION
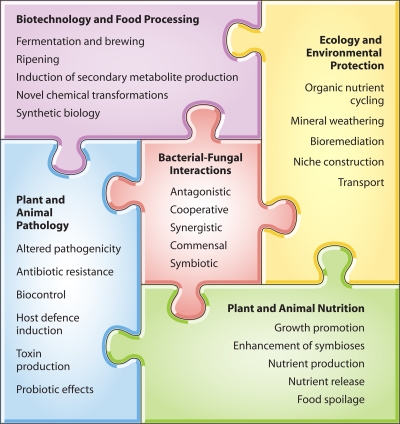
Historically, the classical separation of microbiological research between bacteriologists and mycologists has led to the study of bacteria and fungi in axenic settings. This compartmentalization has overlooked the fact that in many environments, bacteria and fungi coexist and interact. Furthermore, these bacterial-fungal interactions (BFIs) often have important ramifications for the biology of the interacting partners. In recent years, research in this area has developed significantly in both breadth and depth. Contemporary studies have revealed that fungi and bacteria often form physically and metabolically interdependent consortia that harbor properties distinct from those of their single components (379). These reports have also highlighted the multiple practical relevancies of these interactions (Fig. 1) to an exceptionally diverse variety of fields, including agriculture, forestry, environmental protection, food processing, biotechnology, medicine, and dentistry (119, 120, 200, 397).
Fig. 1.
Relevance of BFIs to different areas of scientific study.
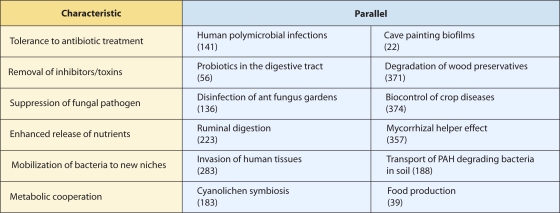
In each of the disciplines for which BFIs are important, research has progressed somewhat differently. This is likely a reflection of their distinct contexts but also a reflection of a lack of interactions among researchers working in these different areas. However, many commonalities exist among the BFIs in these different settings, and a greater appreciation of them would help scientists to identify potentially relevant studies outside their normal specialty, a step that is often important when searching for new ideas, hypotheses, methods, or collaborators. Here we attempt to bridge this gap by integrating advances in research from all these fields into a single source. By not focusing exclusively on one area of application, we seek to achieve a novel unifying perspective on BFIs that enables the identification of fundamental themes, mechanisms, and areas of mutual interest. Given the burgeoning research field, it is an ideal time to attempt such an appraisal, as it will soon become impossible to integrate these many facets into a single work.
In this review, we will consider a BFI as a scenario in which the fungus or bacterium has a direct effect on the other microorganism, thus excluding situations of mere physical proximity. We will also exclude situations in which the effect of one partner on the other is mediated solely through a third organism, such as the systemic induction of plant immunity (151). We will occasionally draw upon examples involving oomycetes, which, while not true fungi in a phylogenetic sense, often share similar general morphologies and ecological niches. The BFI theme will be considered at three levels: the diversity of interactions between bacteria and fungi, the effects of bacterial-fungal consortia on other organisms and the environment, and, finally, their use for (or in) biotechnological applications. Thus, our objective is not to discuss in great depth very-well-known model systems but to attempt to give a wider perspective, revealing commonalities and differences that exist in different BFI contexts. Today, humankind faces many practical challenges relevant to BFIs that are important for health, food security, and sustainable ecosystem management. At the same time, technological developments look to be set to transform our ability to address these problems through science. So far, soil, plant, food, and animal bacteriologists and mycologists have neglected each other's research fields; we hope that this review will contribute to closer collaborations between them.
INTERACTIONS BETWEEN FUNGI AND BACTERIA
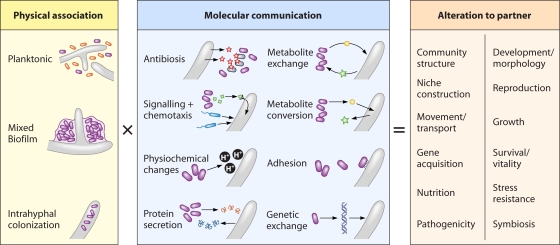
Associations between bacteria and fungi exist in many different contexts and can be considered from different perspectives (Fig. 2). In this section, we begin by describing the general characteristics of BFIs, reviewing the degrees of intimacy that are exhibited between the two partners in terms of their different physical associations. We then assess the various forms of molecular communication that occur in BFIs, which range from antibiosis and signaling to genetic exchange. Finally, we discuss the consequences that BFIs can have on the development and life cycles of bacteria and fungi. While not exhaustive, our examples have deliberately been drawn from a wide range of different contexts, both to emphasize the diversity of BFIs and to stimulate readers to consider other systems in which parallels to their own field may exist.
Fig. 2.
The BFI equation. The combination of physical associations and molecular interactions between bacteria and fungi can result in a variety of different outcomes for each partner. In turn, these changes may affect the influence of the bacterial-fungal complex on their biotic and abiotic environment.
Physical Complexes between Bacteria and Fungi
Complexes containing bacteria and fungi are found in many distinct environments, such as the lungs of cystic fibrosis patients (31); the human oral cavity (16); the production of foods such as cheese, wine, tempeh, and sourdough (408); and agricultural and forest environments (119, 402). The physical associations between them can range from seemingly disordered polymicrobial communities to highly specific symbiotic associations of fungal hyphae and bacterial cells. The description of the taxonomic diversity of polymicrobial communities has gained fresh momentum from newly available DNA sequencing technologies that can resolve community structures to a high level. These methods are likely to provide new insights into bacterial and fungal community compositions, their associations, and also their responses to each other and the environment; for example, Rousk et al. recently demonstrated contrasting influences of soil pH on the bacterial and fungal communities (329). Mixed biofilms containing both fungi (filamentous or nonfilamentous) and bacteria can be considered to be a second, more intimate level of bacterial-fungal association. This arrangement differs from mixed communities, as in a biofilm, the microorganisms form structured communities held together by an extracellular matrix of microorganism-derived macromolecules that have physical and physiological properties distinct from those of free-living cells (99). Bacterial-fungal biofilms can exist as mixed complexes of the two, or fungi may provide biotic support for the establishment of a bacterial biofilm (166, 364). Bacterial-fungal contact and adhesion are likely to be important early events in the process of the formation of mixed bacterial-fungal biofilms. Some specificity may exist in these interactions, depending on the cellular developmental and physiological status; for example, Pseudomonas aeruginosa is able to colonize the hyphal but not the yeast form of Candida albicans. Bacterial-fungal biofilm structures are an area of considerable interest in a clinical context owing to their prevalence in certain infections (166) and in medical devices such as catheters, prostheses, and mechanical ventilators (18, 309). Microorganisms occupying such structures typically show enhanced resistance to antibiotic therapies; for example, the presence of C. albicans has been shown to significantly enhance Staphylococcus aureus biofilm formation and its resistance to vancomycin in serum (153, 154). Mixed bacterial-fungal biofilms have also been reported in other contexts, such as mycorrhizal root systems (283, 334) and rice wine production (187), and are implicated in the degradation of historic artifacts such as mural paintings (136, 388).
The most intimate BFIs occur when the two partners establish a symbiosis. These symbioses can be classified as either an ectosymbiotic relationship, in which bacteria remain external to the fungal plasma membrane, or an endosymbiotic relationship, in which bacteria are located inside the fungal cell. A recently described example of the former is the association of certain Klebsiella and Pantoea species with the fungus gardens of leaf cutter ants (310). Cyanolichens, symbioses formed between fungi (typically ascomycetes, e.g., Geosiphon pyriformis) and photosynthetic cyanobacteria (typically belonging to Nostoc spp.), are also typically ectosymbiotic (196, 325). However, a clear example of a cyanolichen endosymbiotic relationship can also be found in the symbiosis between Geosiphon pyriformis and Nostoc punctiforme (196, 199). Nonphotosynthetic endobacteria and ectosymbiotic bacterial partners are also associated with cyanolichens, but their diversity is only beginning to be explored (29, 43, 61, 140, 141, 268, 272, 325, 355). While cyanobacteria are intracellular in the association between Geosiphon pyriformis and Nostoc punctiforme, they are enclosed in a specialized swollen multinucleate fungal “bladder” that is morphologically distinct from the rest of the hyphae, and within this bladder, the cyanobacteria are surrounded by a host-derived symbiosome membrane (199, 227). This arrangement differs from those of other BFIs involving endobacteria, such as those that occur between Burkholderia and Rhizopus hyphae (209), Burkholderia and Mortierella elongata (337), as well as a variety of other endobacteria associated with ecto- and arbuscular mycorrhizal fungi (37, 40, 48, 272, 365). In these pairings, no specialized hyphal structure is present; the bacteria occupy the cytoplasm of hyphae within the fungal mycelium and, in some cases, also fungal spores (227). Indeed, it has been hypothesized that some endobacteria, such as “Candidatus Glomeribacter gigasporarum,” are obligately dependent on the fungus, as after their isolation from their fungal host, it has not been possible to cultivate them independently in laboratory media (41, 175). A recent report describing over 400 phylogenetically diverse associations of endophytic bacteria with fungi isolated from foliar tissues indicated that such associations may be far more common than was previously appreciated (163).
Bacterial-Fungal Molecular Interactions and Communication
Central to BFIs is the communication or “dialogue” between the bacterium and the fungus, which we outline here to provide a context for a later discussion of the impact of BFIs. Further discussions of these processes, particularly in a clinical context, where Candida albicans-Pseudomonas aeruginosa interactions are an intensively studied model system, can be found in several recent reviews (301, 379, 397).
Interactions via antibiosis.
Probably the best-known and most extensively studied category of bacterial-fungal communication is antibiosis, a chemical warfare that is typified by the diffusion of deleterious and often chemically complex molecules from one partner to the other. Research investigating antibiosis has led to the development of numerous important antibiotics to combat microbial infections, the most famous of which is the beta-lactam antibiotic penicillin, which was developed based on the antibiosis of a Penicillium mold contaminating a Staphylococcus culture (109). The mechanisms of antibiotic action in BFIs have been of particular interest, especially in the context of the increasing incidence of antibiotic resistance in a clinical setting. A wide variety of mechanisms have been identified, including the inhibition of key cellular functions such as cellular respiration (e.g., hydrogen cyanide and fusaric acid), cell wall synthesis (e.g., penicillin and butyric acid), and transport systems (e.g., β-phenylethanol), while others impair the integrity of cell membranes (e.g., hydrolytic enzymes, cyclic lipopeptides, and polymyxin B). Environments in which antibiotics are present exert a strong selective pressure favoring fungi and bacteria that are either insensitive or that possess effective antibiotic resistance mechanisms. For example, exposure to phenazines and phloroglucinols produced by certain Pseudomonas isolates induces the expression of several ABC transporters in the fungal phytopathogen Botrytis cinerea, which are thought to prevent the intrahyphal accumulation of antifungal metabolites (348, 349). In addition, a B. cinerea laccase was found to be responsible for the production of reactive species that detoxify 2,4-diacetylphloroglucinol (349). Interestingly, isolates of another phytopathogen, Fusarium oxysporum, produce fusaric acid in response to 2,4-diacetylphloroglucinol. Fusaric acid was also shown to repress phenazine biosynthesis in Pseudomonas chlororaphis PCL 1391 as well as a virulence-associated quorum-sensing system (350, 390). The interaction between the mycorrhizal fungus Amanita muscaria and Streptomyces sp. strain AcH505 also leads to the suppression of bacterial antibiotic production. In this case, the fungus represses the biosynthesis of the antibiotics WS-5995 B and WS-5995 C by organic acid production (323).
Signaling-based interactions.
Other molecules have more subtle effects than antibiotics during BFIs by acting as signaling molecules. Some bacterial metabolites stimulate hyphal growth; for instance, during its interaction with Amanita muscaria, Streptomyces sp. AcH505 shows an enhanced production of the secondary metabolite auxofuran, which promotes the extension of the fungal mycelium. Unidentified volatile substances produced by some bark beetle-associated bacteria stimulate the growth of their symbiotic fungi (1). Bacterial peptidoglycans have been shown to induce Candida albicans hyphal growth, while the presence of the C. albicans metabolite farnesol can modulate the expression of virulence genes in Pseudomonas aeruginosa by influencing bacterial quorum sensing (82, 83). Interestingly, in the case of the in vitro C. albicans-P. aeruginosa BFI, the reverse situation can also occur, in which a bacterial quorum-sensing molecule, 3-oxo-C12 homoserine lactone, inhibits yeast filamentation (165). The reciprocal effect of farnesol and 3-oxo-C12 homoserine lactone is considered to be due to the presence of a 12-carbon chain within their chemical structures, since other chemically similar molecules with different carbon chain lengths do not cause similar signaling effects (165). Rasmussen et al. (318) showed that Penicillium sp. can also produce inhibitors of bacterial quorum sensing. This supports a more general role for quorum-sensing molecules in bacterial-fungal signaling, as does the discovery that another structurally unrelated bacterial quorum-sensing molecule, the 22-amino-acid competence-stimulating peptide of the Gram-positive organism Streptococcus mutans, also inhibits the yeast-hypha transition in C. albicans (176). As yet, the role of quorum-sensing signaling in nonmedical BFIs is largely unexplored, although quorum-sensing systems are present in many environmental bacteria, and there is some evidence that mycorrhizal fungi can degrade quorum-sensing molecules (387). Interestingly, low concentrations of some antibiotics that do not induce bacterial stress responses can have signaling effects on bacterial biofilm formation and motility (105, 218). This concept has not been extensively studied in the field of BFIs but may be of significant relevance to them, particularly during the early stages of the formation of bacterial-fungal complexes, when antibiotic metabolites may be present in only small quantities.
Interaction via modulation of the physiochemical environment.
As well as bacterial-fungal communication that is mediated by a specific molecule and a target/receptor, communication in BFIs may occur via modifications of the physiochemical properties of their environment. A common effect is an alteration of the pH, since although some microorganisms (e.g., streptococci, lactobacilli, and Candida) can occupy environments under a broad range of pH conditions, most are susceptible to acidic pHs below 4 (288). Thus, changes in pH can affect microbial community structure by either promoting or inhibiting the growth of acid-sensitive organisms, as demonstrated in the phyllosphere, the human gut, and cheese and wine production (5, 13, 78, 113, 289). On cheese surfaces, for example, yeast lactate metabolism and the production of alkaline metabolites such as ammonia cause deacidification that favors the growth of less-acid-tolerant bacterial strains that are essential for cheese ripening (79). Similarly, the presence of the alkalinizing yeast Geotrichum candidum enhances the growth of Salmonella on tomato fruit surfaces (395). Coinoculation with Saccharomyces cerevisiae was found to significantly improve the viability of Pseudomonas putida in grape juice and in a synthetic glucose-rich medium (328). This was attributed to yeast glucose metabolism, the action of which results in a reduced concentration of deleterious gluconic acid produced by bacteria under these nutrient conditions (328). In addition to its effects on microbial growth, environmental pH can also influence other microbial processes; for example, the rate of synthesis of the secondary metabolite aflatoxin by Aspergillus parasiticus is higher under acidic growth conditions, while alkaline medium increases the production of penicillin by Aspergillus nidulans (59, 302).
Interactions via chemotaxis and cellular contacts.
While diffusible molecules play a significant role in many BFIs, migration and physical contact are also important processes in the establishment of BFIs. Chemotaxis (directed movement) of bacteria toward fungi and fungally derived molecules has been demonstrated in several instances; for example, both detrimental and beneficial Pseudomonas species exhibit taxis toward fungal mycelial exudates (93, 139). In the case of the chemotactic response of the biocontrol strain Pseudomonas fluorescens WCS365, Fusarium oxysporum fusaric acid has been identified as an important fungally derived chemotactic signal (95). Cell-cell contact between fungi and bacteria can result in important changes to their physiology and interactions. These interactions may also be modulated by the environment; for example, nutritional conditions have been shown to modulate coadherence between C. albicans and oral bacteria (278). The molecular nature of bacterial-fungal contact has been examined in only a few systems, and these studies have, perhaps unsurprisingly, highlighted important roles for membrane proteins. For example, the attachment of Acinetobacter baumannii to C. albicans (122) is mediated by the bacterial major outer membrane protein OmpA, while the contact-based signaling of Streptococcus gordonii toward C. albicans is mediated partly via the bacterial cell wall-anchored proteins SspA and SspB (19) and the hyphal wall protein Als3 (369).
Adhesive interactions mediated by polysaccharide for the attachment by Pseudomonas bacteria with antifungal activity onto the hyphae of the button mushroom Agaricus bisporus have been reported (315). A role for extracellular polysaccharides in the attachment of bacterial species to arbuscular mycorrhizal fungi has also been reported (39), while in the brewing industry, the coflocculation of the fission yeast Schizosaccharomyces pombe with the Gram-positive lactic acid bacterium Pediococcus damnosus appears to be mediated in part by yeast cell surface mannose and galactose residues, with the latter shielding the former from P. damnosus lectin receptors (303). Bacterial lipopolysaccharides are also involved in BFIs. Indeed, Burkholderia mutants impaired in poly-d-galactofuranose O-antigen synthesis are not able to establish a successful symbiosis with Rhizopus (215). In this case, the bacterial lipopolysaccharide was suggested to promote the evasion of host defense systems, since galactofuranose is a common component of the hyphae of filamentous fungi (215).
Bacterial-fungal contact-based interactions may not be solely adhesive in nature; for example, a lack of O-linked glycans in C. albicans confers hypersensitivity to contact-dependent hyphal death caused by P. aeruginosa (51). Another interesting example of cell-cell recognition is found in cyanolichens, where an arginase secreted by the fungal partner was recently reported to act as a lectin (97, 392). The binding of this arginase to a polygalactosylated Nostoc receptor is paralleled in lichen symbioses formed between algae and fungi and thus may be a key early signaling event in the establishment of lichen symbioses in general (97, 392). A potential role for lectin proteins in BFIs was reported for the truffle fungus Tuber borchii (66). The main soluble protein found in its fruiting bodies was shown to bind to exopolysaccharides from truffle-associated Rhizobium isolates, suggesting that lectin-mediated interactions may contribute to the selection imposed by the truffle on its associated bacterial community (66).
Trophic interactions.
Nutritional interactions between fungi and bacteria are important to many BFIs. Trophic competition between fungi and bacteria is well documented in the plant root environment (rhizosphere), where bacterial competition for nutrients such as carbon (104, 106, 213, 371), nitrogen (239), or iron (104, 213, 214, 402) can be an effective biocontrol mechanism against fungal root pathogens. Examples of bacterial-fungal trophic competition in other environments include competition for carbon substrates during the decomposition of leaves (256); the uptake and release of nutrients by yeast during wine fermentation, which greatly affects growth of malolactic bacteria (5); and competition between the feed additive S. cerevisiae CNCM I-1077 and rumen bacteria in an in vitro rumen system (71). Protocooperative behavior, which is advantageous but not essential to the two partners involved, is thought to be important for many BFIs and has been of particular interest to scientists investigating the functioning of complex bacterial-fungal communities in the natural environment (177, 391) and in food processing (223, 274).
Some bacteria, such as the phytopathogen Pseudomonas syringae B728a, the mushroom pathogen Pseudomonas tolaasii, and some collimonads isolated from sand dunes, have been described as being mycophagous; i.e., they are able to acquire all the nutrients that they need for growth from fungi (90, 216, 374, 403). Since nonphotosynthetic endobacteria are completely enclosed within the fungal hypha, they must also obtain their nutrients solely from the fungal cytoplasm. Little is known of the trophic exchanges that occur during these interactions; however, while fungal endobacteria have been observed in dead hyphae (37, 255), they are generally not considered to cause hyphal damage like other mycophagous bacteria. The flow of nutrients in a BFI may not always be directed to the bacterial partner (Fig. 3). Indeed, in cyanolichens, the intrahyphal cyanobacteria probably represent a significant source of carbon and possibly nitrogen for the fungi (80, 197, 198, 356). Some evidence even suggests an enhancement of cyanobacterial photosynthesis when it is engaged in the symbiosis (42). Nitrogen-fixing bacteria have also been isolated from mycorrhizal (including truffle) fungi and in the fungus gardens of leaf cutter ants, further suggesting positive inputs of endobacteria to fungal nutrition (21, 252, 310, 380).
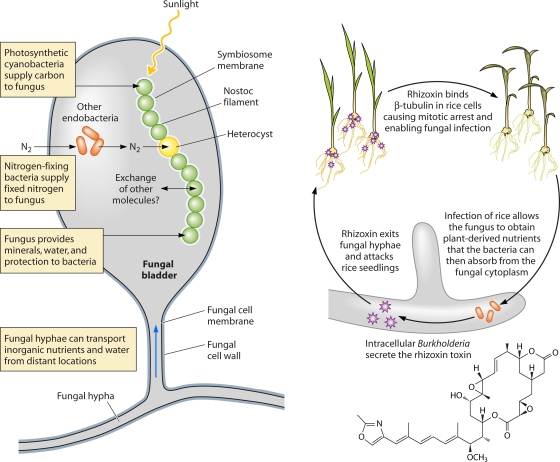
Fig. 3.
Role of autotrophic versus heterotrophic endobacteria in promoting fungal nutrition. (Left) Cartoon illustrating the nutritional relationship between Nostoc cyanobacteria and the lichenous fungus Geosiphon pyriforme during their BFI. Fungal cell structures such as vacuoles, nuclei, lipid bodies, and mitochondria are omitted for clarity. Carbon fixation occurs in photosynthetic cyanobacterial cells, while Nostoc heterocysts are able to fix atmospheric nitrogen (197, 198). Bacteria benefit from micronutrients supplied by the fungus, such as phosphate (356). Other nonphotosynthetic endobacteria living within the bladder may also supply fixed nitrogen to the fungus (355). (Right) Cartoon illustrating the role of Burkholderia rhizoxinica in the nutrition of Rhizopus microcarpus. In contrast to the cyanolichen example, the bacterium is not a primary producer of organic carbon or nitrogen for the fungus (208). However, the bacterial biosynthesis of the toxin rhizoxin is crucial for fungal pathogenicity toward rice seedlings and therefore to the fungal exploitation of plant-derived carbon and nitrogen (298). R. microcarpus also requires B. rhizoxinica for vegetative reproduction (299).
As well as purely trophic interactions, bacteria or fungi can benefit from specific compounds that are produced by the other partner if they cannot produce it themselves or if they can produce it in only growth-limiting quantities. Several mycorrhizal helper bacteria secrete citric and malic acids that are metabolized by Laccaria bicolor, promoting its growth (100). The essential micronutrient thiamine (vitamin B1) has been implicated in the growth promotion exhibited by Bacillus sp. strain TB-1 toward the thermophilic yeast Debaryomyces vanriji (Schwanniomyces vanrijiae) (324) and in the growth-promoting effect of the mycorrhizal helper strain Pseudomonas fluorescens BBc6R8 on the ectomycorrhizal fungus L. bicolor S238N (93). Conversely, ectomycorrhizal fungi may produce organic acids or sugars, such as trehalose, that can affect the composition and growth of associated bacterial communities (116, 287). In the interaction between S. cerevisiae and several Acinetobacter species, ethanol secreted by the former was shown to stimulate the growth of the bacterial species and also to act as a signaling molecule, altering cell physiology to increase salt tolerance and, in the case of A. baumannii, enhancing in vitro pathogenicity toward the nematode Caenorhabditis elegans (372).
Interactions via cooperative metabolism.
An interesting extension to the concept of the utilization of specific metabolites from one BFI partner to aid the general growth requirements of the other is that of metabolite conversion. In this scenario, metabolite exchange in the BFI results in the formation or degradation of a molecule that neither partner can produce alone. In the case of several complex food products that require BFIs for their production, like wine and cheese, each partner contributes to the synthesis and organoleptic qualities of the final end product. For example, in cheese ripening, the interaction between Kluyveromyces lactis and Brevibacterium linens results in an altered profile of aromatic volatile sulfur compounds, which is believed to be due to the supply of an important precursor molecule, methanethiol, by the bacterium to the yeast (10). Another example of such anabolism is the synthesis of melanin pigments during the Klebsiella aerogenes-Cryptococcus neoformans BFI (114, 115). C. neoformans is able to use homogentisic acid produced by K. aerogenes as a precursor for the synthesis of melanin pigments that enhance the virulence of the fungus and afford it protection against UV and other environmental stresses (114, 115). A less beneficial conversion of a bacterial metabolite occurs upon coculturing of P. aeruginosa and C. albicans, since a pyocyanin precursor produced by the former is converted to a toxic red pigment within the fungus (131, 257). Cooperation to achieve novel catabolic reactions is also known in some instances; for example, mixed bacterial-fungal consortia are able to biodegrade and mineralize high-molecular-weight polycyclic aromatic hydrocarbons (49, 203). The degradation of fungal self-inhibitory molecules by bacteria was hypothesized to play an important role in the growth-promoting effects of bacteria on certain fungi, including the metabolism of toxic polyphenols secreted by the ectomycorrhizal fungus Paxillus involutus (100) and the induction of mushroom formation in Agaricus bisporus by Pseudomonas putida (314). Conversely, fungi or bacteria may benefit from the production of toxins by their BFI partner. A particularly interesting example of this is found with Rhizopus, a phytopathogenic fungus that causes rice seedling blight (298). Rhizoxin, the antimitotic toxin responsible for the initiation of the disease, was shown to be synthesized not by the fungus but by Burkholderia rhizoxinica and Burkholderia endofungorum, bacteria that live within the fungal hyphae (297, 298).
Interactions via protein secretion and gene transfer.
In addition to the transfer of nutritive metabolites, antibiotics, and signaling molecules, the exchange of other biomolecules between bacteria and fungi can also occur. Many bacteria rely on secretion systems to translocate molecules, such as proteins and DNA, into neighboring cells and the extracellular milieu (46). In Gram-negative bacteria, these secretion systems can range from simple transporters to multicomponent protein complexes and have been classified into six categories (type I secretion system [T1SS] to T6SS). T1SS and T2SS substrates, such as lipases, proteases, and beta-glucanases, are implicated in the antifungal activities of several different bacterial species (11, 38, 172, 229, 330, 374) and in some cases may function synergistically with bacterial secondary metabolites (110). Other secretion systems, such as the T3SS and T4SS, are responsible for the direct delivery of bacterial proteins or DNA into the host cytoplasm and have been widely studied in the context of bacterial virulence toward higher eukaryotes (7, 129, 312). The heterologous expression of T3SS effector proteins from a range of plant- and animal-pathogenic bacteria in S. cerevisiae has been used successfully to identify their potential cellular functions (368). Surprisingly, evidence for the translocation of T3SS effectors into the fungal cytoplasm does not exist; however, two recent studies employing T3SS mutants suggested that these systems may play a role in BFIs (87, 206). Cusano et al. (87) demonstrated that a disruption of the T3SS of Pseudomonas fluorescens BBc6R8 abolished its mycorrhizal helper effect, although whether the plant of the fungus was the T3SS target is unknown. Lackner et al. (206) found that the disruption of a T3SS gene cluster in the endobacterium Burkholderia rhizoxinica resulted in a reduced intrahyphal survival of the bacterium and a failure to elicit the sporulation of the Rhizopus microsporus host.
Scientists have employed the T4SS of bacteria to transform fungi with foreign DNA for over 2 decades. This was first achieved by using Escherichia coli to transform S. cerevisiae (158, 279) but is becoming more widely adopted as a method of fungal transformation, since Agrobacterium tumefaciens, a natural plant genetic engineer, is capable of transforming both S. cerevisiae (55) and a range of filamentous fungi (92, 188, 294). Interestingly, evidence for natural horizontal gene transfer between bacteria and fungi is also mounting as more genome sequences become available (56, 107, 322, 343, 401, 403). Mallet et al. (232) found that among all putative lateral gene transfers in Aspergillus fumigatus, the main proportion (40%) were of bacterial origin. Another recent study identified 713 genes of probable prokaryotic origin in fungal genomes, putatively involved in amino acid isomerization, arsenate detoxification, and peptidoglycan synthesis (237). One might speculate that these genes were acquired from bacteria that were previously part of a BFI with the fungi in question and that may have fulfilled the same functional role as that of the horizontally acquired fungal gene.
Consequences of Bacterial-Fungal Interactions for Participating Organisms
The successful establishment of an association between bacteria and fungi has profound consequences for both organisms. In this section, we describe the main outcomes of BFIs in relation to changes in the bacterial and fungal partners' physiology, life cycles, and survival.
Effects on fungal development.
Extracellular bacteria can affect fungal development and spore production, to the benefit or the detriment of the fungus. Bacteria stimulate spore germination in several fungi, including the plant-pathogenic oomycete Phytophthora alni (68), the saprophytic cheese-associated fungus Penicillium roqueforti (152), several bark beetle fungal symbionts (1), and the arbuscular mycorrhizal fungus Glomus intraradices Sy167 (161, 162). Interestingly, antibiotic treatment to “cure” Rhizopus of the Burkholderia endobacteria living within its hyphae results in a fungus that no longer produces reproductive sporangia or spores (299). This is believed to be a mechanism that ensures the presence of bacteria within fungal spores during vegetative reproduction, guaranteeing the vertical transmission of the bacteria (299), and thus is likely to have been essential for spreading the symbiosis globally (205). As such, it promotes a permanent association with the fungus, which is distinct from the cyclical associations seen for cyanolichens (227). Another notable effect on fungal development is seen in the life cycle of the edible button mushroom Agaricus bisporus. The commercial production of mushrooms occurs via the initial colonization of mushroom compost by the fungal mycelium followed by casing with a layer of a peat/limestone mix that stimulates fruiting body initiation (280). The presence of bacteria, notably Pseudomonas putida, in this casing layer is highly beneficial for the induction of mushroom production by A. bisporus (103, 157, 316). Pseudomonads are also strongly associated with the fruiting bodies of the ascomycete truffle fungus Tuber borchii and the oyster mushroom Pleurotus ostreatus and may play a role in their development (73, 75, 338, 339). A stimulatory effect on Pleurotus mushroom formation caused by Bradyrhizobium has also been observed (307). Effects of bacteria on fungal growth, measured by biomass, hyphal branching patterns, elongation, morphology, and density, have been documented in a number of settings, including mushroom formation (316), in vitro studies of mycorrhizal fungi (94, 230), rice wine fermentation (187), and clinically relevant BFIs (165, 189, 282). The mycorrhizal helper strain Streptomyces sp. AcH505 was shown to have a pronounced effect on the organization of the cytoskeleton of the ectomycorrhizal fungus Amanita muscaria (351). The same strain also produces auxofuran, a metabolite that contributes to its growth-promoting effect, the synthesis of which is promoted in bacterial-fungal cocultures and at acidic pH values that typify the growth conditions of ectomycorrhizal fungi (323).
Effects on fungal pathogenicity.
Associations with bacteria can have a considerable influence on the pathogenicity of fungi; for example, the presence of bacteria associated with a strain of Fusarium oxysporum appears to be important for allowing it to adopt invasive\pathogenic growth (253). Indeed, effects of bacteria on fungal life cycles often influence fungal pathogenicity. An early observation of a negative effect on fungal pathogenicity is the inhibition of the spore germination of the phytopathogen Botrytis cinerea by an antagonistic bacterial community on Chrysanthemum leaves (44). Another study examining the interaction of B. cinerea with soil bacteria provided evidence that the bacterial degradation of oxalic acid produced by the fungus can reduce B. cinerea pathogenicity (347). In a clinical setting, various bacterial small molecules have been shown to affect the morphological transition of fungi from the yeast form to the filamentous form, which is critical in the context of fungal pathogenicity. These include short-chain fatty acids from lactic acid bacteria (282) and mutanobactin A from Streptococcus mutans (182), which inhibit this transition in C. albicans. Bacterial effects on fungal biofilm formation have also been documented; for example, a recent study indicated that diffusible molecules from P. aeruginosa suppress Aspergillus fumigatus biofilm formation in vitro, which may explain why the fungus causes only low mortality in the lungs of cystic fibrosis patients in which P. aeruginosa is a common inhabitant (262).
Effects on bacterial and fungal physiology.
Observation of the effects of fungi on bacterial development is difficult due to the small size and single-cell nature of prokaryotes. However, if we consider bacterial-fungal biofilms, it is clear that fungi can promote distinct differences in bacterial development by contributing to a distinctive ecological niche, within which bacteria exhibit physiological differences, such as resistance to antibiotics, stress, and an altered expression of virulence genes, compared to free-living bacteria (153, 154, 306). There is also body of literature that points to the influence of fungi on bacterial community structure at both the taxonomic and functional levels, notably in the mycorrhizosphere and pathorhizosphere (119, 332, 370) but also in other contexts, such as cheese and wine production (2, 58, 173, 261, 386). However, it is a significant challenge to make the link between cell-cell communication in BFIs and community-level organization.
The effects of fungal interactions on bacterial physiology (and vice versa) can be assessed by using global techniques such as proteomics and transcriptomics, which may reveal changes that are undetectable by other means. Indeed, various studies have used “omics” techniques to probe the changes that occur during BFIs at the subcellular level (25, 94, 167, 231, 235, 259, 281, 352); for example, a recent study of the arbuscular mycorrhizal fungus Gigaspora margarita using combined proteomic and lipid metabolite profiling revealed that the presence of endobacteria had a significant impact on fungal physiology (331). Targeted approaches assessing the proteomic or transcriptomic responses of one BFI partner to molecules produced by the other also allow the testing of specific hypotheses relating to BFIs, such as responses to antibiotics (245, 246). However, many mechanistic investigations of BFIs are performed in controlled microcosms with a very limited number of interacting microorganisms, whereas natural interactions often involve a complex community of microorganisms whose combined properties may be unpredictable. A good illustration of this was reported by de Boer et al. (91), who observed that the consortia of soil bacteria reduce the expansion of pathogenic fungi, whereas the individual species had no effect on the growth of the fungi. New sequencing technologies are now beginning to allow us to address more complex microbial communities but have yet to be applied to bacterial-fungal consortia.
Effects on survival, dispersal, and colonization.
Fungi and bacteria also play important roles in promoting the survival of their interacting partners. This effect can be reciprocal; for example, P. fluorescens BBc6R8 promotes the viability of the mycorrhizal fungus Laccaria bicolor under unfavorable growth conditions in soil (54), while the fungus can also promote the survival of the bacterium (93). Filamentous fungi can also provide a vector for bacteria by transporting them to new locations where they may access new niches or substrates, as was observed for the degradation of polycyclic aromatic hydrocarbon pollutants (202). This may have relevance in other contexts as well; for example, it has been postulated that the association of Staphylococcus aureus with Candida hyphae provides a mechanism for the bacterial invasion of otherwise inaccessible tissues, such as epithelial layers (306). BFIs may also favor the colonization of surfaces that are otherwise inaccessible to some microorganisms. C. albicans was shown to strongly enhance biofilm formation by Staphylococcus aureus in an in vitro polystyrene-serum system, with the bacteria associating with the fungal hyphae rather than the plastic substrate as part of a polymicrobial biofilm (154). Similar effects may be important for the colonization of medical devices by a range of microbes (99).
Evidence for heritable changes.
The evolutionary consequences of BFIs are generally poorly understood, although in certain circumstances, such as intrahyphal bacteria or horizontal gene transfer between bacteria and fungi, a clear heritable component to the interaction can be discerned. The first complete genome sequence of an intrahyphal bacterium, the rhizoxin-producing bacterium Burkholderia rhizoxinica (207), has revealed a genome size reduction compared to the genome sizes of other Burkholderia bacteria, a characteristic that is common in the genomes of many bacteria that have adapted to obligate or symbiotic associations with eukaryotes (258), although this reduction is less extreme than those of some bacterial symbionts of insects, and those authors suggested that B. rhizoxinica has a “genome in transition” to adaptation to the intrahyphal niche (208). The B. rhizoxinica genome also suggests some metabolic adaptation to an intrahyphal existence by the bacterium and possesses a surprisingly large number of genes that are predicted to be involved in the biosynthesis of nonribosomal peptides (208). As more intrahyphal bacterial genomes become available, such as that of “Candidatus Glomeribacter gigasporarum” (175), it will be interesting to see if there are common features that are indicative of strains that can live within fungi. On the fungal side of the symbiosis, recent studies of Rhizopus microsporus β-tubulin sequences indicate that its resistance to the endobacterial rhizoxin toxin is likely to predate the establishment of the symbiosis, since other Zygomycota that do not harbor the endobacterium are also rhizoxin resistant (344). Two studies suggested that the horizontal gene transfer of bacterial genes into Saccharomyces may contribute significantly to its metabolic potential. In S. cerevisiae and Saccharomyces bayanus, an alphaproteobacterium is thought to have been the source of a gene encoding a sulfatase, which catalyzes the hydrolysis of sulfate esters (150). Those authors and Gojkovic et al. (135) also identified a second gene in S. cerevisiae, encoding a dihydroorotate dehydrogenase involved in de novo pyrimidine biosynthesis, which is likely to have been acquired from the Lactobacillales. Interestingly, the ancestral eukaryotic gene, which encodes a mitochondrial rather than a cytoplasmic form of the enzyme, has been lost in S. cerevisiae. Analysis of the role of dihydroorotate dehydrogenases in Saccharomyces kluyveri (which possesses both forms of the enzyme) suggests that only the prokaryotic form functions in the absence of oxygen, suggesting that this innovation may have contributed to the adaptation of S. cerevisiae to anaerobic growth (135).
Complexity in life cycles.
As well as studying evolutionary questions, it is also important to appreciate the temporal aspect of BFIs over shorter time periods, since interactions may occur transiently or have different outcomes depending upon the physiological or developmental stages in which the two partners meet. A good example of this is seen for the biology of mushroom production by Agaricus bisporus (Fig. 4), which, as described above, is partly dependent on the presence of Pseudomonas putida in the casing layer (103). However, pseudomonads also cause disease on mushrooms, resulting in symptoms such as brown blotch, ginger blotch, and drippy gill (381, 383, 407). It appears that the tolaasin toxin used by P. tolaasii to cause brown blotch symptoms is either not expressed or ineffective against hyphae during their initial colonization of the mushroom bed, since mushroom beds are initially symptomless (317). Furthermore, other pseudomonads and the products of their metabolism have been shown to antagonize the pathogenicity of P. tolaasii (270, 373). Thus, within a single genus, we can see growth- or development-promoting, deleterious, and biocontrol activities of bacteria toward fungi, highlighting the complexity and specificity of the interactions that can exist in BFIs through time. This also illustrates the notion of a dynamic relationship between bacteria and fungi, with the potential for both beneficial and detrimental effects depending on the environment and phenology of the organisms. Identification of the key mechanistic determinants for these different behaviors, both genetic and environmental, therefore represents the first step in controlling or manipulating them.
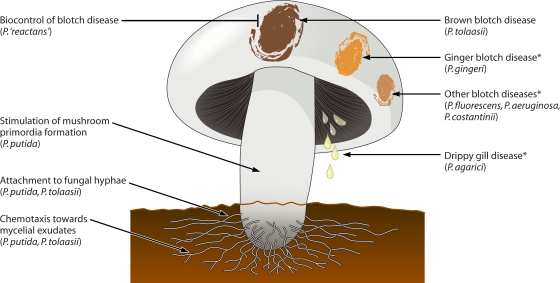
Fig. 4.
Interactions of pseudomonads with Agaricus bisporus lead to both positive and negative outcomes for the fungus, depending on the bacterial isolate and the developmental stage of the fungus. The toxin tolaasin is the primary factor responsible for brown blotch disease caused by P. tolaasii; other contributing factors are a secreted protease, lipase, and exopolysaccharide. *, mechanism unknown.
IMPACT ON OTHER ORGANISMS AND THE ENVIRONMENT
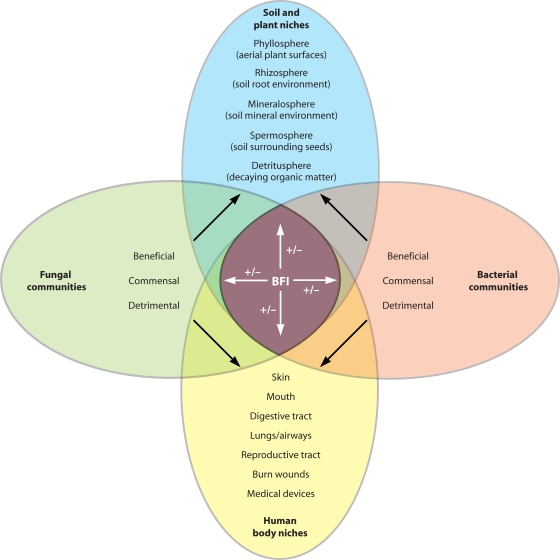
The impact of BFIs on the nutrition and health of the host organisms is often significant, as both plants and animals host consortia of bacteria and fungi in a variety of niches (Fig. 5). In the soil environment, BFIs play a role in processes such as mineralization and the degradation of toxic compounds. In this section, we describe the relevance of BFIs in these different contexts, highlighting common or contrasting themes.
Fig. 5.
Coexistence and impact of bacteria and fungi in contrasting microbial ecosystems. Bacterial and fungal communities occupy overlapping niches in soil or when associated with plants and humans or other animals. Disturbing the communities that occupy these niches, for example, by the introduction or removal of key members, may alter the balance that exists between them. This can cause changes to the influences of bacteria and fungi on their niche, with consequences for the functioning of the ecosystem. BFIs may also result in novel effects in niches that do not occur in their absence.
Influence on Host Nutrition
Effects on plants.
Plants photosynthesize to obtain the carbon that they require, but the other nutrients that are essential for their growth and development must be obtained from the soil. In terrestrial ecosystems, the root systems of most plant species interact with mycorrhizal fungi to form symbiotic structures called mycorrhizas (178, 241). Mycorrhiza formation enhances root access to water and poorly available mineral nutrients in the soil, while heterotrophic fungi benefit from plant carbohydrates exuded by roots. Under natural conditions, mycorrhizas are surrounded by a wide diversity of bacterial communities that interact with the symbiosis at physical, metabolic, and functional levels, forming a multitrophic mycorrhizal complex (118). Individual strains have been isolated from these and other soil bacterial communities that behave as mycorrhiza helper bacteria (124), since they promote mycorrhiza establishment or functionality (23, 119, 180). Additive or synergistic effects on plant growth and nutrition between helper or mycorrhiza-associated bacteria and mycorrhizal fungi are now well documented. The nature of the nutritional benefit provided by a BFI for plants has also been demonstrated in some instances. A Bacillus subtilis helper strain with a phosphate-solubilizing ability significantly increased the biomass and nitrogen and phosphorus accumulation in onion tissues when inoculated with the mycorrhizal fungus Glomus intraradices in a soil with low phosphorus bioavailability (382). In another study, Requena et al. (321) identified Rhizobium strains that were able to enhance the Anthyllis cytisoides-G. intraradices symbiosis and improve the plant nitrogen status. Therefore, it is clear that mycorrhiza-associated bacteria and mycorrhizal fungi can positively interact to promote a sustainable nutrient supply to plants.
Various hypotheses have been proposed to explain the mechanistic basis for the bacterial mycorrhiza-helper effect (119), some of which may be mediated through the host, such as the suppression of plant defense responses (212). Interestingly, it appears that there may be some overlap between strains that can suppress fungal diseases of plants and strains that can act as helper bacteria; for example, recently, Pivato et al. (311) revealed that Pseudomonas fluorescens C7R12, which can reduce wilt caused by Fusarium, can also behave as a helper strain by specifically promoting the symbiosis between the legume Medicago truncatula and Glomus mosseae. In contrast, Lehr et al. (212) observed that the helper strain Streptomyces sp. AcH505 facilitates the colonization of plant roots by the phytopathogen Heterobasidion abietinum. Nevertheless, others have demonstrated that the mycorrhizal helper effect is not a general phenomenon among all soil bacteria (15, 186). An understanding of the basis of mycorrhizal helper bacteria specificity will aid the selection of the best strains for application in crop plants to enhance mycorrhization and may provide insight into BFIs beyond those found in the context of mycorrhizas.
Effects on humans and other animals.
In some ways, the digestive tracts of humans and other animals can be considered analogous to the zone surrounding plant root systems. Digestive tracts are ecosystems containing diverse polymicrobial microbiota that exhibit complex physical, chemical, and functional interrelationships, and their contents can be considered “external” to the body, since they have not been internalized by a membrane system (361). Human gut microbial communities are thought to play a major role in health and disease, but an exhaustive analysis of their composition and diversity has only recently become feasible with the advent of new DNA sequencing technologies (273, 313). To date, the scientific community has focused on the rich bacterial communities (101) colonizing the digestive tract, whereas the diversity of fungi inhabiting the human gut is very poorly documented (340, 354). While the level of fungal diversity in the human gut is almost certainly much lower than the level of bacterial diversity, a culture-independent rRNA oligonucleotide fingerprinting approach for the murine intestine suggested that an abundant and diverse fungal microbiota is likely to be present (358).
Fungi may play unique functional roles in the gut that are complementary to those ascribed to bacteria, such as nutrient release and exchange (375). However, the extent to which the fungi and bacteria interact in the human gut is largely undetermined. More is known about BFIs in the digestive tracts of ruminant mammals, where bacteria are believed to play a major role because of their metabolic diversity, while ruminal fungi, although less numerous, are able to weaken and degrade the more recalcitrant plant tissues thanks to the wide range of polysaccharide-hydrolyzing enzymes that they secrete. However, the precise role and overall contribution of each fungal and bacterial group to the in vivo degradation and fermentation of plant cell wall materials are still hypothetical (111). Interactions between ruminal fungi and bacteria have been studied largely in vitro, sometimes in artificial rumen ecosystems (30, 36, 179, 211, 242). These interactions range from synergism to antagonism, depending on the microbial composition and the type of plant substrate. Hydrogen-utilizing bacteria such as methanogens were reported to improve the efficacy of ruminal fungi for cellulose degradation (30, 242). In contrast, some rumen cellulolytic bacteria were observed to inhibit the ability of ruminal fungi to degrade cellulose (36). Further in vivo studies of the bacterial-fungal interactions in the rumen will be necessary to clarify their real impact on animal nutrition and will open new perspectives in the worrying context of increasing greenhouse gas production, to which the contribution of livestock is significant.
To improve animal performance and health, yeasts such as Saccharomyces cerevisiae have been used widely in recent years as probiotic additives in the diets of ruminant mammals and horses (102). In vivo investigations have demonstrated that ruminant diets supplemented with S. cerevisiae increase cellulolytic bacterial densities as well as the rate of straw degradation in the rumen (70, 275). When supplied to young ruminants switching over from a milky diet to solid feed, yeast probiotics could thus be of value in stabilizing the rumen microbial ecosystem (70). Recently, live S. cerevisiae cells coincubated with three different rumen bacterial species were found to reduce bacterial proteinase activities compared to the activities in bacterial monocultures (71). In vivo, such an effect could be harnessed to reduce the proteolysis of rapidly degradable dietary proteins in the rumen and consequently to avoid the accumulation of ammonia. Bacteria, yeast, and also protozoa may also have beneficial effects in the rumen due to their ability to degrade mycotoxins produced by members of the fungal genera Fusarium, Penicillium, and Aspergillus present in ingested fodder (277, 384).
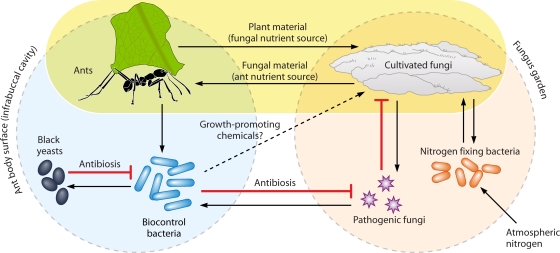
A clear beneficial role for BFIs in host nutrition is found in the mutualism between “fungus-gardening” (attine) ants and their fungal associates, which is thought to have existed for an estimated 45 to 65 million years (263). Fungus-growing ants are dependent on their fungal associates, since the cultivars serve as the sole food source for the ant larvae and queen; however, their fungus gardens may come under attack from members of the ascomycete genus Escovopsis (85). The ants keep their gardens free of microbial pathogens thanks in part to host-beneficial BFIs (Fig. 6). Attine ants support populations of actinomycete bacteria (usually Streptomyces or Pseudonocardia species) by means of cuticular crypts fed by exocrine glands (84). The bacteria produce antibiotics that antagonize the parasitic Escovopsis populations; for example, Trachymyrmex ants rear symbiotic antibiotic-producing bacteria in an infrabuccal pocket into which they transfer parasitic Escovopsis spores and hyphae, aiding in the sanitization of their fungal gardens (221). Recent studies have identified several antibiotics produced by bacteria associated with attine ants, such as the cyclic depsipeptide dentigerumycin and the polyenes mycangimycin and candicidin (149, 285, 286), that selectively target Escovopsis but not the fungal garden (24). A similar strategy of using a bacterium-derived antibiotic to control an antagonistic fungus is also utilized by the southern Pine beetle, Dendroctonus frontalis (357). The beetle is able to selectively suppress Ophiostoma minus, a competitor fungus of its beneficial symbiotic fungus Entomocorticium sp., using the polyene peroxide mycangimycin that is produced by the actinomycete bacteria that it carries in a specialized storage compartment (357).
Fig. 6.
Role of BFIs in ant-fungus mutualism. Nutrient flows and inhibitory interactions between organisms are indicated by arrows and blocked arrowheads, respectively. Major inputs of carbon (plant biomass) and nitrogen (plant biomass and nitrogen-fixing bacteria) provide the raw materials to support the web of interactions and are indicated with bold arrows. Ants transfer pathogenic Escovopsis fungi to their infrabuccal pocket during sanitization of the fungus garden.
Another fungus associated with attine ants is Phialophora, a genus of ascomycete black yeasts. Phialophora cells grow on the ants' cuticle and appear to be localized to sites where mutualistic bacteria reside (220). The basis for this localization is unknown, but the yeasts appear to obtain nutrients from the bacteria and have a negative effect on their growth, consequently reducing their antibiotic effect on Escovopsis (219). Bacteria also provide other benefits to fungi and attine ants beyond pathogen control. A recent study reported the presence of symbiotic nitrogen-fixing bacteria within the fungus gardens of several ant species (310). The presence of these bacteria is important due to the low nitrogen input available from plant leaves (310). Bacterial nitrogen fixation, thought to be performed mainly by members of the genus Klebsiella, is likely to be of substantial nutritional benefit to both the fungi and, consequently, their ant gardeners. Interestingly, the fungal cultivars of Apterostigma ants grow faster under the influence of Streptomyces culture filtrates, suggesting that the bacterium may produce fungal growth-promoting compounds, although their identities are unknown (85).
Roles in Host Health and Disease
Effects on plants.
The efforts of ants to control the parasitic fungi that attack their fungal gardens are paralleled by the attempts made by humans to control fungal diseases of plant crops. It has been recognized for a long time that many bacteria living in the root environment are able to promote plant health and growth (226) and that in some cases this is a consequence of their antagonistic impact on fungal pathogens in the soil (400). There is a large body of literature describing the use of rhizosphere bacterial isolates as potential “biocontrol” agents targeting a variety of soilborne fungal pathogens, indicating the potential of biocontrol bacteria to complement or even replace the application of chemical fungicides (147, 399, 402). Indeed, several commercial biocontrol agents targeting pre- and postharvest fungal diseases are available (34). However, despite meeting with a high degree of success in controlled environments, the widespread use of biocontrol bacterial strains is challenging, since each crop protection strategy is to some extent unique due to differences in cultivars, soil chemistries, and environmental conditions (147). Moreover, the usefulness of particular strains is not always predictable, mainly because of the variable abilities of the biocontrol bacteria to compete with resident rhizobacteria during root colonization, although they are often more efficient in disturbed natural ecosystems (77, 398). In some instances, attempts have been made to use combinations of bacterial and fungal biocontrol agents to afford plants better protection from disease (142, 228, 234). Whether interactions between the biocontrol agents has affected the outcome of these tests has generally not been investigated, except in the case of P. fluorescens CHA0 and Trichoderma atroviride, which appear to have enhanced the expression of key biocontrol factors in each other's presence (228).
Compared to the extensive literature describing the effect of biocontrol strains on soilborne phytopathogenic fungi, much less is known about the effect of their application on resident nonphytopathogenic fungi. Interestingly, a study of biocontrol inoculation of bacteria to combat the effect of phytopathogenic Rhizoctonia solani on lettuce found that the bacterial treatments had only small short-term effects on endophytic fungi and resident rhizobacteria (342). Another study reported that the application of biocontrol strains to protect lettuce from R. solani reduced the disturbance to the indigenous bacterial and fungal communities caused when the phytopathogen alone was present (3). Small but detectable effects of the antibiotic-producing biocontrol strain P. fluorescens CHA0 on fungi resident in the cucumber rhizosphere have been reported, but in this case, no target phytopathogen was included, so the effect of the biocontrol interaction on resident fungi is unknown (133). Uncontrolled side effects on symbiotic mycorrhizal fungi may partly explain the inconsistent results of some large-scale biocontrol field experiments (398). However, biocontrol strains will not necessarily be antagonistic toward mycorrhizal fungi; for example, Barea et al. (22) found that the biocontrol strain Pseudomonas fluorescens F113, which produces the antifungal metabolite 2,4-diacetylphloroglucinol, did not exhibit antibiosis toward the mycorrhizal fungus Glomus mosseae and actually promoted root colonization by the fungal hyphae. The antifungal specificity of biocontrol could therefore be one criterion for the selection and evaluation of biocontrol strains in the future. An interesting illustration of the importance of the context in which an antifungal metabolite acts is seen with rhizoxin. A gene cluster similar to that of B. rhizoxinica that is responsible for the biosynthesis of several rhizoxin analogs has been identified in the plant-associated biocontrol strain P. fluorescens Pf-5 (52, 225). However, while Pf-5 rhizoxin analogs can cause effects on rice seedling root morphology that are similar to those seen with rice seedling blight, in a biocontrol context, Pf-5 probably uses rhizoxin to attack fungal hyphae rather than to promote fungal growth in the manner seen for the Rhizopus-Burkholderia BFI (225). The need for establishing the “fungus specificity” of mycorrhizal helper bacteria has also been recognized, since some strains have been shown to promote mycorrhiza formation by a few fungal species and to inhibit other mycorrhizal fungi (124).
Bacteria and fungi can both elicit defense responses that protect plants, via the mechanisms of induced systemic resistance and systemic acquired resistance, against subsequent pathogen attack (151). Since these effects are mediated by the plant, we will not consider them further here. Plants are usually occupied by endophytic bacteria and fungi, some of which have been isolated and tested in vitro and in field tests with the view to exploit them for the biocontrol of pathogens (9, 17, 20, 35, 238, 247). Results from these studies indicate the potential for antibiotic-mediated or other BFIs among naturally occurring endophytes in planta, but a demonstration of direct BFIs in this setting is very difficult due to potential indirect plant-mediated effects. As discussed above, under natural conditions, plant roots benefit from the presence of mycorrhizal fungi; however, mycorrhizal fungi not only improve plant nutrition but also can protect plant roots against fungal disease. This results from direct effects of the mycorrhizal fungus on the plant but also from indirect effects of the mycorrhizal symbiosis on the surrounding bacterial communities. Indeed, in vitro antagonism against phytopathogenic fungi by mycorrhiza-associated bacteria has been frequently reported (12, 74, 217, 230), and there is evidence for the enrichment of bacterial communities with a high antagonistic potential toward fungal pathogens by mycorrhizal and other fungi (88, 117). Further experiments will be necessary to determine whether these observations result exclusively from fungally induced selection on the bacterial community from the soil reservoir.
As well as the plant-beneficial bacterial-fungal relationships, one should not forget that some plant-detrimental BFIs also take place. Associated bacteria were shown to enhance the pathogenicity of the foliar fungal pathogen Stagonospora nodorum when infecting wheat, even though the bacteria themselves were nonpathogenic toward the host (96). Synergistic interactions between Pectobacterium atrosepticum (Erwinia carotovora subsp. atroseptica) and the foliar pathogen Septoria tritici on wheat have also been reported (276). In the rhizosphere, different functions can coexist, with Pseudomonas bacteria increasing or decreasing the plant pathogenesis of the soilborne fungus Gaeumannomyces (335). The antimitotic toxin rhizoxin is an important component in the pathogenicity of the rice seedling blight pathogen Rhizopus microsporus; however, the synthesis of rhizoxin is performed not by the fungus but by Burkholderia bacteria living within its hyphae (298, 341).
Effects on humans and other animals.
Humans and other animals are naturally colonized by fungi and bacteria that occupy an array of niches, including the skin, the oral cavity, and the respiratory, digestive, and genital tracts (125). In healthy individuals, these microorganisms are commensal and in some cases even beneficial to human health. Pathogenic fungi and bacteria do, however, represent a serious threat to the health of individuals, especially those who are immunocompromised, since many pathogens are opportunistic rather than obligate. There are many instances where bacteria and fungi have been found together in infections of human burn wounds and keratitis (144, 146, 160, 233, 300). However, clinical therapies that have been developed to target either fungal or bacterial infections have generally failed to consider that fungi and bacteria often coinfect, forming mixed communities that have virulence and resistance properties significantly different from those of the single-species communities (366, 397). An understanding of the functioning of BFIs in human health is therefore an emerging and important challenge for medical researchers, particularly given the development of strains that are resistant to drug therapies (127, 243, 290, 292).
The lungs of immunocompromised individuals are also frequently cocolonized by bacteria, notably Pseudomonas aeruginosa, and fungi such as Candida albicans and Aspergillus fumigatus. Coinfection by these pathogens is considered to worsen lung function relative to infection by each pathogen alone (265, 301). Most studies of clinical BFIs to date have focused on Candida albicans (366, 397), a yeast which is common in the buccal, dermal, and intestinal human microbiota but which can also cause a large number of infections (308). Gupta et al. (146) revealed a significant in vivo inhibition of C. albicans growth in the presence of P. aeruginosa in burn wounds, which are highly susceptible to bacterial and fungal superinfections. A similar detrimental effect of P. aeruginosa on C. albicans has been demonstrated in vitro, specifically toward the virulent filamentous form of the fungus (164). P. aeruginosa can also grow at the expense of dermatophyte fungi associated with skin and nail infections (112). Contrastingly, Escherichia coli and Staphylococcus aureus can both behave as synergistic C. albicans copathogens, increasing mortality rates in animal models of polymicrobial peritonitis (62, 63, 194). All these examples clearly illustrate the need for a combination of clinical studies and in vitro experimentation to understand how BFIs impact human health.
The oral cavity is another site at which the potential for BFIs exists in humans. A recent study of 20 healthy individuals revealed a variety of culturable and nonculturable fungi to be present in this setting (130). In vitro studies suggested that oral bacteria may modulate the ability of the yeast Candida albicans to form biofilm structures (168, 291), while a mixed C. albicans-E. coli biofilm was found to possess enhanced resistance to an oral antiseptic. Thus, a fuller understanding of these interactions may lead to improved dental health. BFIs also contribute to the increasing number of nosocomial (hospital-acquired) infections; for example, Curvale-Fauchet et al. (86) reported that a high proportion of hospital intravascular catheters were simultaneously colonized by the pathogenic lipophilic yeast Malassezia and bacteria, including staphylococci. These mixed biofilms, which have also been evidenced with C. albicans (195, 289), can be more resistant to antibiotic treatments; for example, the presence of Staphylococcus epidermidis was shown to delay the diffusion of antifungal drugs in mixed Candida-Staphylococcus biofilms (6). Bacterial-fungal associations with potentially detrimental impacts on human health may also occur in moldy houses (170). In these damp environments, actinobacteria frequently cohabit with fungal genera associated with moisture damage, such as Stachybotrys and Aspergillus. Research evaluating the impact of such BFIs in cell lines indicates a significant potential for synergistic effects between these bacteria and fungi on cytotoxicity and mammalian inflammatory responses (170, 267, 304, 305). While mixed bacterial-fungal communities have been shown to have detrimental effects on human health, to our knowledge, there have been no reports describing the roles of intrahyphal bacteria in fungal infections of humans. Several clinical zygomycete isolates harboring intrahyphal Burkholderia bacteria that produce the rhizoxin toxin have been identified, but the presence of these bacteria is not universal in clinical zygomycoses (171, 295). In contrast to the important role of these bacteria in rice seedling blight, there is currently no evidence that intrahyphal Burkholderia promotes infections of humans by Rhizopus. Compared to parental strains, bacterium-cured R. microsporus organisms are equally destructive toward cell cultures and show no difference in virulence in animal models (171). Deleterious effects of the bacteria may, however, exist later during infection, while from the perspective of the bacteria, the fungus may provide better access to a niche that they can later exploit if released from hyphae (126, 171).
Despite the many examples of detrimental BFIs, these relationships can also have beneficial effects on animal and human health. Researchers have demonstrated that chytridiomycosis, a serious disease affecting amphibian populations worldwide which is caused by the chytrid pathogenic fungus Batrachochytrium dendrobatidis, can be successfully tackled in frogs and salamanders by exploiting antifungal-producing bacteria that naturally live on their skin (53, 155, 156). In the marine environment, a similar scenario was revealed for the shrimp Palaemon macrodactylus (132). The production of the metabolite 2,3-indolinedione by a surface-associated Alteromonas strain was shown to protect shrimp embryos from attack by the oomycete pathogen Lagenidium callinectes (132). In the insect world, the European beewolf wasp cultivates Streptomyces in antennal glands and uses the bacterium to protect its larvae from fungal infestation, again probably due to antibiotic production (185, 204).
BFI-based interventions are also of great interest in the context of mammalian health. “Probiotic” bacteria and yeasts have been used extensively within the livestock industry because of beneficial impacts on host defenses and as alternatives to the use of antibiotics in promoting animal health (123). The addition of the probiotic Saccharomyces cerevisiae CNCM I-1077 led to the elimination of Shiga toxin-producing Escherichia coli in rumen fluid (69), while in poultry, mannanoligosaccharides derived from S. cerevisiae were shown to promote the colonization of beneficial bacteria and reduce colonization by pathogenic strains (32). In human health, there is evidence for a probiotic BFI effect with the use of Saccharomyces boulardii to combat antibiotic-associated diarrhea caused by the loss of indigenous microbes after antibiotic therapy and subsequent colonization by pathogens, notably Clostridium difficile (411). The effect of S. boulardii has been attributed to a variety of mechanisms (411), including some direct BFI effects: the degradation of C. difficile toxins by yeast protease activity (64, 65) and an alteration of E. coli lipopolysaccharide by a yeast phosphatase (57). Reduced bacterial adherence and colonization of intestinal epithelial cells as well as reduced expression of virulence factors were also reported for colon inflammation caused by Citrobacter rodentium in the presence of S. boulardii (409). The question of gut fungi in microbial immune homeostasis is a novel avenue for studies in this area and can now be addressed with the advent of modern techniques to profile microbes associated with the human body. Further studies will be necessary to evaluate whether BFIs within this context may be of benefit or harm to human and animal health.
Roles in Biochemical Cycles and the Environment
Terrestrial and aquatic ecosystems.
Fungi and bacteria play central roles in terrestrial ecosystems, where they participate in numerous biochemical cycles. Lignocellulolytic substrates, such as wood, that constitute very abundant but very recalcitrant organic compounds and play a central role in the carbon cycle are considered to be degraded mainly by fungi under aerobic conditions (45, 76, 138, 193). This is despite the ubiquitous presence of bacteria on these substrates that may negatively or positively interact with the fungal degraders (389). Previous studies have shown that competitive interactions between fungi and bacteria can be important during the fungal degradation of recalcitrant organic matter (89), but this does not appear to be universally true. Wood decay by the white rot fungus Heterobasidion annosum was shown to be inhibited or promoted by bacteria found in association with it, depending on the relative timing of the fungal and bacterial inoculations, suggesting a more complex ecological relationship (266). Phanerochaete chrysosporium, another white rot fungus, was also reported to live in close association with several bacterial species, even in in vitro cultures from which bacteria had been thought to be eliminated (360). Whether this coexistence is obligate and whether the fungus-associated bacteria actively participate in the degradation of lignocellulolytic substrates still remain to be determined. Certainly, bacteria can cause changes in wood chemistry, permeability, and structure, which may favor subsequent succession by fungi (76). Bacteria may also provide nutritional benefits to wood-degrading fungi, for example, by nitrogen fixation in situ or in the surrounding soil that the fungal hyphae can transport to the site of wood degradation (4, 76, 359). Bacteria may also assist in the fungal degradation of preserved woods through the sequestration or detoxification of preserving agents (76, 174, 396). In the context of wood preservation and industrial wood transformation, more studies are needed to understand better how bacteria interact with wood-degrading fungi and the practical implications of these BFIs. This will aid in the development of novel preservation strategies targeting not only wood-decaying fungi but also their associated helper bacterial communities and may improve wood transformation processes by exploiting the cooperative effect of fungi and bacteria in the breakdown of the lignocellulose complex.
BFIs also contribute to cycling on inorganic nutrients. In nutrient-poor soils such as those of forests, inorganic mineral ions derived from rocks by weathering are a key source of important tree nutrients, and both mycorrhiza-associated bacteria and mycorrhizal fungi are known to contribute to mineral weathering (385). Evidence now suggests that these microorganisms cooperate in nutrient mobilization from soil minerals (386); for example, using a budget analysis, Koele et al. (201) recently quantified the respective impacts of two ectomycorrhizal fungal strains (Laccaria bicolor and Scleroderma citrinum) and two mycorrhizosphere bacterial strains (Burkholderia glathei and Collimonas sp.) on the weathering of a phyllosilicate. The level of weathering tended to be highest for all the bacterial-fungal coinoculation treatments, supporting the hypothesis that bacterial-fungal cooperation may contribute to the cycling of inorganic nutrients in forest ecosystems and, therefore, to forest sustainability in nutrient-poor soils. The coinoculation of a phosphate-solubilizing strain of Penicillium ostreatus and a nitrogen-fixing strain of Bradyrhizobium elkanii significantly increases the quantity of phosphorus released from rock phosphate (177). Cyanolichen BFIs have also been shown to enhance the release of essential elements from minerals in the soil (192, 362). In agricultural soils, there is now an increasing need for sustainable ecofriendly fertilization strategies. Efforts should be made now to develop new biotechnological processes based on the coinoculation of bacterial-fungal consortia with weathering abilities to enable the efficient utilization of natural phosphate rocks as alternative phosphate fertilizers. To date, the roles and importance of bacterial-fungal consortia in environmental nutrient cycling have been largely overlooked. Qualitative and quantitative analysis of the contribution of these consortia to different key biogeochemical cycles (e.g., nitrogen, iron, sulfur, carbon, and phosphorus) may provide important insights into these processes that could aid in the development of ecofriendly technologies with beneficial plant and human impacts.
Despite evidence that fungi can colonize even extreme aquatic environments such as deep-sea hydrothermal vents (210), the presence and distribution of fungi in aquatic environments are poorly documented, while the co-occurrence of fungi and bacteria is even more poorly appreciated. In aquatic environments, bacteria and fungi dominate the decomposition of aquatic plant residues and live in close proximity to each other. However, only a few studies have addressed the role of BFIs during the decomposition process, and these have described mostly antagonistic relationships between fungi and bacteria (145, 251, 256, 406). Interestingly, Mille-Lindblom et al. (250) found indications of a trade-off between the fungal growth rate and the tolerance of fungi toward antagonistic and competing bacterial communities. After the cocultivation of six different fungal strains isolated from aquatic plants with a complex natural bacterial community originating from aquatic plant litter, those authors observed that the fungal strains that grew best in the absence of bacteria were most severely affected by the presence of bacteria; in contrast, those that were less inhibited in cocultures with bacteria had lower maximal growth rates in the absence of bacteria. The authors of that study hypothesized that different fungi may allocate resources differentially between their growth and mechanisms to tolerate resident microbes. Given that plant decomposition by fungi occurs in a heterogenous and nonsterile environment, clearly, account must be taken of BFIs when one tries to understand or improve such processes.
Threat of bacterial-fungal interactions to cultural heritage.
The conservation of cultural heritage, such as cave rock art, archaeological sites, historic buildings, and books, is a major challenge worldwide. Whatever the material (e.g., stone, ceramics, metals, window glasses, paintings, mortars, and adobes) and the location (indoors or outdoors) of the heritage to preserve, surfaces are often colonized by complex microbial communities. The number of studies reporting an inventory of the bacterial and fungal diversity colonizing these surfaces has increased substantially in recent years thanks to advances in molecular techniques that now allow the analysis of nonculturable microbial communities (27, 136, 249, 293). These microbial communities are often organized into biofilms that may be aggressive, producing pigments and/or mineral-weathering agents that deteriorate the colonized surfaces (169, 264, 333). These microbial communities may also threaten the health of visitors in indoor environments due to the release of bacterial and fungal toxins (293).
Many studies are under way to identify strategies for the control or elimination of deleterious biofilms associated with cultural heritage. In the view of what is already known for other environments and surfaces containing mixed bacterial-fungal communities, such as medical devices and soil minerals, such studies should consider the potential importance of BFIs occurring inside these biofilms. Bacterial antibiotics are thought to play a role in preventing the fungal colonization of cave paintings (183). Furthermore, a study of the cave of Dona Trinidad in Spain pointed to the potential of nutritional complementation between fungi and bacteria under nutrient-poor conditions; bacteria may benefit from phosphate and organic nutrients exuded by fungi whose growth can be limited by ammonium availability (378). The role of human interventions in such environments is also important (28, 378). As has been described for the human gut, antimicrobials applied to artifacts disturb the indigenous microbiota, providing an open niche for resistant microorganisms to colonize. In environments such as rock surfaces, resistant strains may also benefit significantly from the cell lysis of sensitive microbes, which can provide an additional source of nutrients to them. BFIs may influence the success of these disinfection procedures, as seems to have occurred in the prehistoric Lascaux cave. In 2001, 38 years after its closure to visitors, a bacterial-fungal invasion (Fusarium solani associated with Ralstonia sp. and Pseudomonas fluorescens) occurred in the cave (27). The first biocidal treatments resulted in an unexpected prompt development of the F. solani colonies despite the known in vitro efficiency of benzalkonium chloride against Fusarium solani. It has been hypothesized that the bacteria protected the associated F. solani that was embedded in the same biofilm against the biocidal treatment (27, 28). Moreover, Pseudomonas may also have been able to degrade the antifungal molecule (26).
APPLICATIONS OF BACTERIAL-FUNGAL INTERACTIONS
Food Processing
Fermentation and brewing.
Mixed bacterial-fungal communities play a key role in determining the taste, quality, and safety of a wide range of foods (367). The complex interactions between filamentous fungi, yeast, and bacteria that lead to wine production begin in the vineyard, on the surfaces of ripening grape berries (224, 320), and continue throughout the fermentation process until packaging (108). The biotransformation of pressed grape juice into wine results from a primary alcoholic fermentation performed by Saccharomyces cerevisiae that is frequently followed by a secondary so-called “malolactic fermentation.” Deacidification by this conversion of l-malic acid to l-lactic acid occurs either spontaneously, owing to the activity of bacteria naturally present in musts and wine, or after the inoculation of commercially available bacterial strains (184). Achieving appropriate malolactic conversion is critical for wine quality, as it contributes significantly to establishing the desired organoleptic parameters as well as wine microbiological properties and, thus, stability. The lactic acid bacterial species Oenococcus oeni has become the dominant “starter” species among those used for triggering malolactic fermentation (376). An important parameter can be the timing of the addition of the lactic acid bacterium at the end of the primary fermentation, as it must tolerate an environment that now contains yeast-derived ethanol and other toxic metabolites, such as sulfur dioxide, as well as having a low nutrient availability and a low pH (184). Interestingly, the coinoculation of yeast and a malolactic fermentor in the production of Chardonnay was shown to reduce fermentation times while producing wines that were broadly comparable to those produced through more typical two-phase inoculations (184). Dual bacterial-fungal starter cultures may also improve organoleptic properties in some instances (410). Relationships ranging from competition to inhibition to mutualism between yeast and bacteria during winemaking have been reported; for example, studies have established that yeast nitrogen metabolism can have significant and various effects on malolactic fermentation (143, 319). Further descriptions of these relationships can be found in reviews by Fleet (108) and Alexandre et al. (5). A fuller understanding of the ecology and biochemistry of this microbial ecosystem will undoubtedly have significant benefits for the wine industry and may also provide insights into the cometabolism of BFIs in other contexts.
A variety of other food products are formed due to the actions of mixed bacterial-fungal cultures (367, 408). The fermented soy product tempeh is produced by the action of Rhizopus oligosporus, which inhibits contamination by other microorganisms, including many bacteria (148). Lactic acid bacteria also contribute to the tempeh fermentation process and can similarly inhibit the growth of potentially harmful pathogenic bacteria (14). Not all Lactobacillus species are capable of coculture with Rhizopus, and the rate of fungal growth can be reduced by the bacterium, an effect which also seems to be dependent on the starting inoculum of bacteria (148). A detailed analysis of BFIs in sourdough between lactic acid bacteria and yeasts pointed strongly to key trophic relationships such as the lack of competition for preferred nitrogen sources and even the stabilization of the population of lactobacilli due to yeast-derived amino acids and peptides (134). In some cases, multiple fungi and bacteria may be involved in food production. Soy sauce fermentation involves an initial inoculation with koji (Aspergillus oryzae or A. soyae) to break down complex oligosaccharides and proteins in soybeans, followed by the transfer of the koji to brine, where it is further fermented by a mixture of bacteria (Tetragenococcus halophilus and lactic acid bacteria such as Lactobacillus delbrueckii) and yeasts (Zygosaccharomyces rouxii and Candida sp.), which are metabolically adapted for the osmotic and pH conditions of the brine (60). Similarly, a range of different yeasts and Acetobacter sp. are needed to form the fermented sweetened tea kombucha (244).
Cheese ripening.
Like wine production, cheese manufacture involves complex microbial ecosystems where BFIs play a central role, and rich mixed communities are often present (2). In bacterial smear surface-ripened cheeses such as German “Limburger,” French “Munster,” and Italian “Taleggio,” molds and yeast such as Debaryomyces hansenii and Geotrichum candidum initially dominate the cheese surface because they are acid and salt tolerant (79). They utilize lactate and thus deacidify the cheese surface, enabling the establishment of less-acid-tolerant bacterial communities, which play a significant role in the quality and safety of the final product (79). To understand the BFIs that occur during ripening, Bonaiti et al. (47) developed a microbial Livarot cheese model by selecting microbial associations contributing to cheese aroma. They succeeded in simplifying an ecosystem of 83 microbial strains from a commercial Livarot cheese to a subecosystem composed of nine species that had a strong similarity to that of the commercial cheese. In this model ecosystem, yeast appeared to play a key role in bacterial colonization and bacterial diversity; for example, the growth of some bacterial species, such as Leucobacter sp. or Brevibacterium aurantiacum, significantly relied on the presence of the yeast Geotrichum candidum (261). An understanding of the specificity of the BFIs that occur on the yeast surface will help industries that commercialize ripening microbial cultures to select the most adapted bacterial strains. Indeed, it was shown that despite massive early inoculations, commercial inocula do not always colonize the cheese surface due to the rapid development of adventitious microbiota originating from the cheese production environment (260). Unraveling the ecology of the multispecies cheese ecosystem may have benefits for food safety as well. Control of the development of the human pathogen Listeria monocytogenes in cheese is difficult due to the favorable growth conditions and its ubiquity. However, some smear-ripening bacterial communities have strong antilisterial activities (236) that might be harnessed to restrict or prevent the development of human pathogens such as L. monocytogenes during cheese ripening.
Food spoilage.
While BFIs are exploited for the production of certain foods, fungi and bacteria are also key food spoilage organisms; however, few studies have examined whether BFIs contribute to food spoilage, perhaps due to an emphasis on diagnostics/systematics and shelf life prediction within the food industry (327). Some studies have examined the potential of fungi to assist colonization by human-pathogenic bacteria; for example, spoilage fungi on tomatoes can promote Salmonella colonization, an effect which is believed to be due to their effect on pH (393, 394). The presence of rhizonin toxins produced by endobacteria inhabiting Rhizopus microsporus is a critical cause of food spoilage due to their hepatotoxicity (296). Similarly, rhizoxin-producing endobacteria present a potential threat in food processing since R. microsporus is used during the fermentative production of tempeh (209, 296, 326). Bacteria can also cause spoilage on mushroom tissues, although the effect is largely on mushroom yield and cosmetic appeal to the consumer rather than on human health (374). There has been some interest in the role of bacteria, notably lactic acid bacteria, as biopreservatives of food and animal feed (128, 336, 346). Increasing our knowledge of the antifungal activities of beneficial food-associated bacteria and also their ecology, notably their relationships with beneficial and spoilage fungi within food and animal feed, will help industries to select either strains with a wide range of applications or, conversely, strains that are well adapted to specific environments and applications.
Bioremediation of Pollutants
The microbial degradation of polycyclic aromatic hydrocarbons (PAHs) has significant potential for use in bioremediation (67, 181). PAHs are a large group of toxic environmental contaminants originating for fossil fuels and also made as a result of the incomplete combustion of various carbon-containing fuels. Most PAHs are hydrophobic and so bind to soils and sediments, thus reducing their bioavailability; they also accumulate in food chains. Due to the failure to isolate a single microorganism capable of growing on and mineralizing PAHs with five and or more benzene rings and the ubiquitous coexistence of bacteria and fungi in the environment, some authors have investigated the degradation efficacy of mixed bacterial-fungal cocultures. Boonchan et al. (49) compared the degradations of pyrene, benzo[a]pyrene, and dibenz[a,h]anthracene in monocultures of Penicillium janthinellum and Stenotrophomonas maltophilia and in cocultures of the two. Penicillium janthinellum by itself could partially degrade pyrene and benzo[a]pyrene but did not use either of them as a carbon source. Stenotrophomonas maltophilia by itself could use pyrene as a carbon source and cometabolize benzo[a]pyrene. Bacterial-fungal cocultures grew on pyrene, benzo[a]pyrene, and dibenz[a,h]anthracene and significantly improved the benzo[a]pyrene mineralization. Moreover, they significantly reduced the mutagenicity of contaminated soils with benzo[a]pyrene or dibenz[a,h]anthracene (49). Similarly, a Penicillium sp. culture and three bacterial strains originating from a hydrocarbon-contaminated soil were shown to have a synergistic effect on the removal of polluting phenanthrene when coinoculated into contaminated soil (72). Bacterial-fungal cocultures have also been found to have an enhanced ability to decolorize waste azo dyes that cause environmental pollution (137), while a mixed bacterial-fungal biofilm of Penicillium frequentans and Bacillus mycoides demonstrated a greater ability to degrade polyethylene than either partner alone (363). Similar effects of bacterial-fungal cocultures on polycaprolactone degradation have also been reported (33). These results highlight the fact that bacteria and fungi can cooperatively or synergistically mineralize soil pollutants. These results open interesting practical applications for the complete bioremediation of PAH-contaminated sites, and in view of these results, one should consider microbial interactions as well as the individual potential of isolates for pollutant degradation to improve the PAH bioremediation process.
Naturally occurring BFIs also play a key role in bioremediation in contaminated soil ecosystems. Heinonsalo et al. (159) observed decreased levels of mineral oil (nonpolar hydrocarbons) in petroleum hydrocarbon-contaminated soil colonized by pine roots and mycorrhizal fungi naturally associated with soil bacterial communities. P. fluorescens isolates that possess xylE and xylMA, plasmid-borne genes that are involved in the degradation of monoaromatics, have been found to naturally form biofilms around the hyphae of ectomycorrhizal fungi symbiotically associated with Scots pine seedlings, providing strong evidence for a bacterium-assisted breakdown of monoaromatic compounds in petroleum-contaminated soil colonized by ectomycorrhizal fungi (334). Air and a lack of moisture create barriers to the mobility of pollutant-degrading bacteria in the soil, which prevent them from spreading and delay the breakdown of pollutants. The potential of hydrophilic fungal hyphae to serve as vectors for the dispersion of several pollutant-degrading bacteria was first demonstrated by Kohlmeier et al. (202). Hyphae of the oomycete Pythium ultimum were subsequently also shown to facilitate the movement of the phenanthrene-degrading strain Pseudomonas putida PpG7 in soil (121, 404). The use of bacteria that are able to move across this “fungal highway” could help speed up the remediation of contaminated land (202). To fully exploit this potential, further research is needed to address whether all bacteria are able to use such fungal vectors and whether all fungi are able to route bacteria and to assess what impact fungal and bacterial cell walls have on this process.
Natural Product Discovery and Synthetic Biology
Humans are increasingly using BFIs, or harnessing knowledge garnered from them, for many novel applications. For instance, basic research on BFIs continues to be of importance to the development of novel agents against pathogens. This is exemplified by the discovery of the fungal defensin peptide plectasin from Pseudoplectania nigrella (345). Plectasin targets lipid II, a key component in bacterial cell wall biosynthesis, and thus provides a promising lead for a new antibiotic therapy in the face of the increased incidence of multiple-antibiotic-resistant bacteria in hospitals (269, 345). As well as the identification of secondary metabolites, recent findings suggest that BFIs can also be used to activate putative fungal secondary metabolite gene clusters that are not expressed under other culture conditions (50). Schroeckh et al. (353) demonstrated that the specific BFI between Aspergillus nidulans and Streptomyces hygroscopicus ATCC 29253 induces the expression of a fungal gene cluster responsible for the biosynthesis of several polyketide metabolites, including orsellinic acid and lecanoric acid, the latter being a typical lichen metabolite, in a contact-dependent manner. Similarly, the biosynthesis of the chlorinated benzophenone antibiotic pestalone by the marine fungus Pestalotia was triggered when the fungus was cocultivated with the marine alphaproteobacterium CNJ-328 (81). Strain CNJ-328 was also used to induce the synthesis of novel diterpenoids from the marine fungus Libertella sp. (284). Several other studies have supplemented fungal cultures with bacteria and bacterial supernatants to induce or enhance the production of fungal natural products, illustrating potential industrial applications for BFIs in this context (98, 248). An example of the fungal stimulation of a bacterial product can be seen in the enhancement of nisin (a food preservative) biosynthesis by Lactococcus lactis upon cocultivation with yeast that is able to metabolize inhibitory lactic acid that is produced during the fermentation (222). An understanding of neutral BFIs may also be of benefit in some circumstances; for example, the selection of vitamin-producing bacterial strains that can be cocultured with Rhizopus was suggested as a method for the provision of vitamin B12 in tempeh production (190, 405).
A more reductive approach to harnessing BFIs involves the transfer of specific genes provided by one partner to the other. This may have benefits in allowing a more controlled expression of the heterologous trait, providing a more straightforward means of obtaining the same result, or allowing the expression of a trait where a “real” BFI is either undesirable or unachievable. Such approaches have been pioneered in yeast thanks to the availability of amenable genetic and cultivation techniques. In one case, an S. cerevisiae strain was engineered to produce a levanase from Bacillus subtilis that increased its ability to produce bioethanol from waste plant biomass (240). The enzyme allows the yeast to use novel carbon sources which otherwise need to be released by a separate pretreatment of the input plant material (240). In wine production, S. cerevisiae and Schizosaccharomyces pombe have been engineered to express the malolactic gene from Lactococcus lactis in order to enable them to perform both the alcoholic and malolactic fermentation stages without the need for malolactic bacteria (8). E. coli strains have been engineered with a combination of genes from S. cerevisiae and from other bacterial strains to convert dihydroxyacetone phosphate into 1,3-propanediol, which can be used as building blocks for industrial polymers (271). Bringing together subelements (e.g., protein domains) from bacterial and fungal systems into a single unit also has the potential for many novel applications; for example, protein domain sequences from a fungal nonribosomal peptide synthetase have been used to modify a similar gene in Bacillus subtilis, resulting in the production of novel secondary metabolites (377). In another example, a hybrid protein containing a fungal (Trichoderma viride HK-75) cellulose binding domain from an exoglucanase I enzyme fused to a bacterial (Bacillus subtilis BSE616) endo-beta-1,4-glucanase has improved binding and hydrolytic activities toward microcrystalline cellulose (191). Such hybrid constructions have been investigated with the purpose of producing artificial cellulosomes with enhanced abilities to biodegrade complex plant cell wall components (254). Clearly, the explosion in the amount of genome data from both the bacterial and fungal kingdoms is opening up tremendous possibilities for innovations in synthetic biology that may draw inspiration from our knowledge of BFIs.
CONCLUDING REMARKS
Everywhere, including in unexpected places and environments, bacteria and fungi have been found to live side by side and interact, and clear parallels can be drawn between some of these diverse BFIs (Fig. 7). The range of mechanisms for both antagonism and cooperation exhibited during BFIs is undoubtedly a reflection of the continuous interplay between bacteria and fungi that has occurred over the course of their evolutionary histories. It is likely that many more BFIs exist, particularly in natural environments such as the ocean, that are relatively understudied from a microbiological perspective. We are beginning to appreciate the range of effects that BFIs bring about, but there are many interesting aspects of BFIs that warrant further investigation and that will benefit from technological advances made in recent years. While many examples of bacterial-fungal communities are known, in many instances, our knowledge of the factors contributing to their initial formation (particularly, complex communities such as those in the mycorrhizosphere) and subsequent stability/homeostasis is scant. The availability of high-throughput metagenomic methodologies and data analysis techniques will enable researchers to robustly address questions such as the specificity that exists in bacterial-fungal community interactions and the changes in functionality that occur as mixed bacterial-fungal communities develop, although these sequencing-based approaches can be fully exploited only with complementary information gathered through other means, such as metabolite profiling and isotopic labeling. The ability to perform culture-independent microbial identification may also reveal previously unrecognized bacterial-fungal consortia if researchers take care to perform simultaneous prokaryotic and eukaryotic identification from environmental DNA samples.
Fig. 7.
Examples of some parallels among BFIs in different settings (22, 39, 56, 136, 141, 183, 188, 223, 283, 357, 371, 374).
The study of BFIs has led to the discovery of a great number of natural products and tools to transform fungi, but there is currently a dearth of data pertaining to non-antibiotic-mediated cell-cell communication between fungi and bacteria at the molecular level. A deeper appreciation of BFI signaling at the cellular level may lead to novel therapeutic targets for combating single- and mixed-microbial infections and conversely may also allow the enhancement of beneficial BFIs. From a biotechnology perspective, knowledge of the metabolic relationships during BFIs has significant potential for exploitation in numerous fields but has not received much attention beyond food technology research. The metabolic effect of bacterial-fungal communities on their environment will also be of great interest in the context of climate change, bioremediation, and searches for novel antimicrobials and drug therapies. Fungal endobacteria may represent an excellent approach for producing easy-to-apply “stabilized” BFIs in which modified bacteria reside within fungal hyphae, but currently, our understanding of the global ecology, evolution, trophic relationships, and functioning of these and other BFIs remains in its infancy, hampering the development of such applications. Research addressing these issues will have major implications for our view of bacterial and fungal biology and ecology.
ACKNOWLEDGMENTS
We thank Jean Garbaye for providing helpful suggestions for the manuscript.
We also gratefully acknowledge the financial backing provided by INRA to P. Burlinson and the support that has been given towards research on bacterial-fungal interactions in the Tree-Microbe Interactions Department by both INRA and the Lorraine Region for over 10 years.
Biographies

P. Frey-Klett is director of research and head of the Tree-Microbe Interaction Department at the INRA in Nancy, France. She completed her M.Sc. in plant pathology at the University of Paris XI—Orsay. She then moved to INRA-Nancy to work in the group of Dr. Jean Garbaye and was awarded her Ph.D. in 1996. Since her recruitment as a junior INRA scientist in 1997, she has been developing the bacterial-fungal interaction thematic in forest soils, focusing on mycorrhiza helper bacteria and intrafungal bacterial communities. Her strong interest in the broadness of this research area has driven her to try to induce closer collaborations between soil, plant, food, and animal bacteriologists and mycologists.

P. Burlinson obtained his undergraduate degree in molecular biology and biochemistry from Durham University in 2004. He then moved to Oxford University to work in the group of Dr. Gail Preston and was awarded his doctorate in 2008. In 2008 to 2009, he held a postdoctoral BBSRC Wain Fellowship at the University of Toronto in Canada. Subsequently, he moved to INRA-Nancy in France to work on bacterial-fungal interactions with Dr. Pascale Frey-Klett. His main research interests are in molecular microbiology and bacterial genomics, currently focused on Pseudomonas.

A. Deveau obtained her undergraduate degree in cellular biology and physiology from the Paris 7 University in 2003 and completed her M.Sc. in forest biology at the University of Nancy, Nancy, France. She pursued her Ph.D. with Pascale Frey-Klett at the University of Nancy, studying the molecular mechanisms of the effect of mycorrhiza helper bacteria. In 2008 to 2010, she held a postdoctoral Fellowship at Dartmouth Medical University, where she studied the mechanisms of intracellular signaling in Candida albicans. Since 2011, she has worked as a junior INRA scientist in Nancy, France. Her research focuses on the ecology and the molecular mechanisms of the interactions between soil bacteria and forest fungi.

M. Barret studied molecular biology and microbiology at the University of Rennes 1 (France) and obtained his Ph.D. in microbiology in 2009 at Agrocampus Ouest, studying bacterial-fungal interactions in the rhizosphere. Since 2009, he has worked as a postdoctoral researcher in the BIOMERIT Research Centre at University College Cork in Ireland. His research focuses on the role of Gram-negative secretion systems during bacterial-host interactions.

M. Tarkka is a Senior Scientist in the Department of Soil Ecology at the Helmholtz Center for Environmental Research-UFZ in Halle, Germany. He completed his M.Sc. in plant physiology at the University of Helsinki, Helsinki, Finland, and pursued his Ph.D. with Marjatta Raudaskoski at the University of Helsinki, focused on plant and fungal development in mycorrhizal symbiosis. As a postdoctoral fellow and research associate with Rüdiger Hampp at the University of Tübingen, Tübingen, Germany, he investigated how streptomycetes impact mycorrhizal symbiosis formation and affect plant disease resistance. Dr. Tarkka assumed his current position at the UFZ Halle in 2007, and his research focuses on molecular mechanisms by which streptomycetes, plants, and fungi interact with each other and on the functional properties of streptomycete communities related to disease suppression and carbon turnover.

A. Sarniguet is director of research at INRA, in the research centre of Rennes (Le Rheu, France). He received his Ph.D. from the University of Paris XI—Orsay in 1990. He was an invited researcher at the USDA-ARS in Joyce Loper's laboratory at Corvallis, OR, from 1992 to 1993. His interest is in microbial ecology in the rhizosphere, with a great emphasis on bacterial-fungal interactions and plant-pathogenic fungus populations. His main work with his current team has been focused on biocontrol bacteria, especially Pseudomonas sp., against the take-all disease of wheat.
REFERENCES
- 1. Adams A. S., Currie C. R., Cardoza Y., Klepzig K. D., Raffa K. F. 2009. Effects of symbiotic bacteria and tree chemistry on the growth and reproduction of bark beetle fungal symbionts. Can. J. Forest Res. 39:1133–1147 [Google Scholar]
- 2. Addis E., Fleet G. H., Cox J. M., Kolak D., Leung T. 2001. The growth, properties and interactions of yeasts and bacteria associated with the maturation of Camembert and blue-veined cheeses. Int. J. Food Microbiol. 69:25–36 [DOI] [PubMed] [Google Scholar]
- 3. Adesina M. F., Grosch R., Lembke A., Vatchev T. D., Smalla K. 2009. In vitro antagonists of Rhizoctonia solani tested on lettuce: rhizosphere competence, biocontrol efficiency and rhizosphere microbial community response. FEMS Microbiol. Ecol. 69:62–74 [DOI] [PubMed] [Google Scholar]
- 4. Aho P. E., Seidler R. J., Evans H. J., Raju P. N. 1974. Distribution, enumeration, and identification of nitrogen-fixing bacteria associated with decay in living white fir trees. Phytopathology 64:1413–1420 [Google Scholar]
- 5. Alexandre H., Costello P. J., Remize F., Guzzo J., Guilloux-Benatier M. 2004. Saccharomyces cerevisiae-Oenococcus oeni interactions in wine: current knowledge and perspectives. Int. J. Food Microbiol. 93:141–154 [DOI] [PubMed] [Google Scholar]
- 6. Al-Fattani M. A., Douglas L. J. 2004. Penetration of Candida biofilms by antifungal agents. Antimicrob. Agents Chemother. 48:3291–3297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Alvarez-Martinez C. E., Christie P. J. 2009. Biological diversity of prokaryotic type IV secretion systems. Microbiol. Mol. Biol. Rev. 73:775–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ansanay V., et al. 1996. Malolactic fermentation by engineered Saccharomyces cerevisiae as compared with engineered Schizosaccharomyces pombe. Yeast 12:215–225 [DOI] [PubMed] [Google Scholar]
- 9. Araujo W. L., et al. 2001. Variability and interactions between endophytic bacteria and fungi isolated from leaf tissues of citrus rootstocks. Can. J. Microbiol. 47:229–236 [DOI] [PubMed] [Google Scholar]
- 10. Arfi K., Leclercq-Perlat M. N., Spinnler H. E., Bonnarme P. 2004. Importance of curd-neutralising yeasts on the aromatic potential of Brevibacterium linens during cheese ripening. Int. Dairy J. 15:883–891 [Google Scholar]
- 11. Arora N. K., Kim M. J., Kang S. C., Maheshwari D. K. 2007. Role of chitinase and beta-1,3-glucanase activities produced by a fluorescent pseudomonad and in vitro inhibition of Phytophthora capsici and Rhizoctonia solani. Can. J. Microbiol. 53:207–212 [DOI] [PubMed] [Google Scholar]
- 12. Artursson V. 2005. Bacterial-fungal interactions highlighted using microbiomics: potential application for plant growth enhancement. Ph.D. thesis. Swedish University of Agricultural Sciences, Uppsala, Sweden [Google Scholar]
- 13. Aruscavage D., Lee K., Miller S., LeJeune J. T. 2006. Interactions affecting the proliferation and control of human pathogens on edible plants. J. Food Sci. 71:R89–R99 [Google Scholar]
- 14. Ashenafi M., Busse M. 1991. Growth of Bacillus cereus in fermenting tempeh made from various beans and its inhibition by Lactobacillus plantarum. J. Appl. Bacteriol. 70:329–333 [DOI] [PubMed] [Google Scholar]
- 15. Aspray T. J., et al. 2006. Mycorrhization helper bacteria: a case of specificity for altering ectomycorrhiza architecture but not ectomycorrhiza formation. Mycorrhiza 16:533–541 [DOI] [PubMed] [Google Scholar]
- 16. Avila M., Ojcius D. M., Yilmaz O. 2009. The oral microbiota: living with a permanent guest. DNA Cell Biol. 28:405–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Backman P. A., Sikora R. A. 2008. Endophytes: an emerging tool for biological control. Biol. Control 46:1–3 [Google Scholar]
- 18. Baena-Monroy T., et al. 2005. Candida albicans, Staphylococcus aureus and Streptococcus mutans colonization in patients wearing dental prosthesis. Med. Oral Patol. Oral Cir. Bucal 10(Suppl. 1):E27–E39 [PubMed] [Google Scholar]
- 19. Bamford C. V., et al. 2009. Streptococcus gordonii modulates Candida albicans biofilm formation through intergeneric communication. Infect. Immun. 77:3696–3704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bandara W. M., Seneviratne G., Kulasooriya S. A. 2006. Interactions among endophytic bacteria and fungi: effects and potentials. J. Biosci. 31:645–650 [DOI] [PubMed] [Google Scholar]
- 21. Barbieri E., et al. 2010. New evidence for nitrogen fixation within the Italian white truffle Tuber magnatum. Fungal Biol. 114:936–942 [DOI] [PubMed] [Google Scholar]
- 22. Barea J. M., et al. 1998. Impact on arbuscular mycorrhiza formation of Pseudomonas strains used as inoculants for biocontrol of soil-borne fungal plant pathogens. Appl. Environ. Microbiol. 64:2304–2307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Barea J. M., Azcon R., Azcon-Aguilar C. 2002. Mycorrhizosphere interactions to improve plant fitness and soil quality. Antonie Van Leeuwenhoek 81:343–351 [DOI] [PubMed] [Google Scholar]
- 24. Barke J., et al. 2010. A mixed community of actinomycetes produce multiple antibiotics for the fungus farming ant Acromyrmex octospinosus. BMC Biol. 8:109 doi:10.1186/1741-7007-8-109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Barret M., et al. 2009. The plant pathogenic fungus Gaeumannomyces graminis var. tritici improves bacterial growth and triggers early gene regulations in the biocontrol strain Pseudomonas fluorescens Pf29Arp. New Phytol. 181:435–447 [DOI] [PubMed] [Google Scholar]
- 26. Bastian F., Alabouvette C. 2009. Lights and shadows on the conservation of a rock art cave: the case of Lascaux Cave. Int. J. Speleol. 38:55–60 [Google Scholar]
- 27. Bastian F., Alabouvette C., Jurado V., Saiz-Jimenez C. 2009. Impact of biocide treatments on the bacterial communities of the Lascaux Cave. Naturwissenschaften 96:863–868 [DOI] [PubMed] [Google Scholar]
- 28. Bastian F., Jurado V., Novakova A., Alabouvette C., Saiz-Jimenez C. 2010. The microbiology of Lascaux Cave. Microbiology 156:644–652 [DOI] [PubMed] [Google Scholar]
- 29. Bates S. T., Cropsey G. W., Caporaso J. G., Knight R., Fierer N. 2011. Bacterial communities associated with the lichen symbiosis. Appl. Environ. Microbiol. 77:1309–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bauchop T., Mountfort D. O. 1981. Cellulose fermentation by a rumen anaerobic fungus in both the absence and the presence of rumen methanogens. Appl. Environ. Microbiol. 42:1103–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bauernfeind A., et al. 1987. Qualitative and quantitative microbiological analysis of sputa of 102 patients with cystic fibrosis. Infection 15:270–277 [DOI] [PubMed] [Google Scholar]
- 32. Baurhoo B., Goldflus F., Zhao X. 2009. Purified cell wall of Saccharomyces cerevisiae increases protection against intestinal pathogens in broiler chickens. Int. J. Poult. Sci. 8:133–137 [Google Scholar]
- 33. Benedict C. V., Cameron J. A., Huang S. J. 1983. Polycaprolactone degradation by mixed and pure cultures of bacteria and a yeast. J. Appl. Polym. Sci. 28:335–342 [Google Scholar]
- 34. Berg G. 2009. Plant-microbe interactions promoting plant growth and health: perspectives for controlled use of microorganisms in agriculture. Appl. Microbiol. Biotechnol. 84:11–18 [DOI] [PubMed] [Google Scholar]
- 35. Berg G., et al. 2005. Endophytic and ectophytic potato-associated bacterial communities differ in structure and antagonistic function against plant pathogenic fungi. FEMS Microbiol. Ecol. 51:215–229 [DOI] [PubMed] [Google Scholar]
- 36. Bernalier A., Fonty G., Bonnemoy F., Gouet P. 1992. Degradation and fermentation of cellulose by the rumen anaerobic fungi in axenic cultures or in association with cellulolytic bacteria. Curr. Microbiol. 25:143–148 [Google Scholar]
- 37. Bertaux J., et al. 2005. Occurrence and distribution of endobacteria in the plant-associated mycelium of the ectomycorrhizal fungus Laccaria bicolor S238N. Environ. Microbiol. 7:1786–1795 [DOI] [PubMed] [Google Scholar]
- 38. Bhattacharya D., Nagpure A., Gupta R. K. 2007. Bacterial chitinases: properties and potential. Crit. Rev. Biotechnol. 27:21–28 [DOI] [PubMed] [Google Scholar]
- 39. Bianciotto V., Andreotti S., Balestrini R., Bonfante P., Perotto S. 2001. Extracellular polysaccharides are involved in the attachment of Azospirillum brasilense and Rhizobium leguminosarum to arbuscular mycorrhizal structures. Eur. J. Histochem. 45:39–49 [DOI] [PubMed] [Google Scholar]
- 40. Bianciotto V., et al. 1996. An obligately endosymbiotic mycorrhizal fungus itself harbors obligately intracellular bacteria. Appl. Environ. Microbiol. 62:3005–3010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bianciotto V., et al. 2004. Vertical transmission of endobacteria in the arbuscular mycorrhizal fungus Gigaspora margarita through generation of vegetative spores. Appl. Environ. Microbiol. 70:3600–3608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bilger W., Büdel B., Mollenhauer R., Mollenhauer D. 1994. Photosynthetic activity of two developmental stages of a Nostoc strain (Cyanobacteria) isolated from Geosiphon pyriforme (Mycota). J. Phycol. 30:225–230 [Google Scholar]
- 43. Bjelland T., et al. 2011. Microbial metacommunities in the lichen-rock habitat. Environ. Microbiol. Rep. 4:434–442 [DOI] [PubMed] [Google Scholar]
- 44. Blakeman J. P., Fraser A. K. 1971. Inhibition of Botrytis cinerea spores by bacteria on the surface of chrysanthemum leaves. Physiol. Plant Pathol. 1:45–54 [Google Scholar]
- 45. Blanchette R. A. 1977. Associations among bacteria, yeasts, and basidiomycetes during wood decay. Phytopathology 68:631–637 [Google Scholar]
- 46. Bleves S., et al. 2010. Protein secretion systems in Pseudomonas aeruginosa: a wealth of pathogenic weapons. Int. J. Med. Microbiol. 300:534–543 [DOI] [PubMed] [Google Scholar]
- 47. Bonaiti C., Irlinger F., Spinnler H. E., Engel E. 2005. An iterative sensory procedure to select odor-active associations in complex consortia of microorganisms: application to the construction of a cheese model. J. Dairy Sci. 88:1671–1684 [DOI] [PubMed] [Google Scholar]
- 48. Bonfante P., Anca I. A. 2009. Plants, mycorrhizal fungi, and bacteria: a network of interactions. Annu. Rev. Microbiol. 63:363–383 [DOI] [PubMed] [Google Scholar]
- 49. Boonchan S., Britz M. L., Stanley G. A. 2000. Degradation and mineralization of high-molecular-weight polycyclic aromatic hydrocarbons by defined fungal-bacterial cocultures. Appl. Environ. Microbiol. 66:1007–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Brakhage A. A., Schroeckh V. 2011. Fungal secondary metabolites—strategies to activate silent gene clusters. Fungal Genet. Biol. 48:15–22 [DOI] [PubMed] [Google Scholar]
- 51. Brand A., Barnes J. D., Mackenzie K. S., Odds F. C., Gow N. A. 2008. Cell wall glycans and soluble factors determine the interactions between the hyphae of Candida albicans and Pseudomonas aeruginosa. FEMS Microbiol. Lett. 287:48–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Brendel N., Partida-Martinez L. P., Scherlach K., Hertweck C. 2007. A cryptic PKS-NRPS gene locus in the plant commensal Pseudomonas fluorescens Pf-5 codes for the biosynthesis of an antimitotic rhizoxin complex. Org. Biomol. Chem. 5:2211–2213 [DOI] [PubMed] [Google Scholar]
- 53. Brucker R. M., et al. 2008. Amphibian chemical defense: antifungal metabolites of the microsymbiont Janthinobacterium lividum on the salamander Plethodon cinereus. J. Chem. Ecol. 34:1422–1429 [DOI] [PubMed] [Google Scholar]
- 54. Brule C., et al. 2001. Survival in the soil of the ectomycorrhizal fungus Laccaria bicolor and the effects of a mycorrhiza helper Pseudomonas fluorescens. Soil Biol. Biochem. 33:1683–1694 [Google Scholar]
- 55. Bundock P., den Dulk-Ras A., Beijersbergen A., Hooykaas P. J. 1995. Trans-kingdom T-DNA transfer from Agrobacterium tumefaciens to Saccharomyces cerevisiae. EMBO J. 14:3206–3214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bushley K. E., Turgeon B. G. 2010. Phylogenomics reveals subfamilies of fungal nonribosomal peptide synthetases and their evolutionary relationships. BMC Evol. Biol. 10:26 doi:10.1186/1471-2148-10-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Buts J. P., et al. 2006. Saccharomyces boulardii produces in rat small intestine a novel protein phosphatase that inhibits Escherichia coli endotoxin by dephosphorylation. Pediatr. Res. 60:24–29 [DOI] [PubMed] [Google Scholar]
- 58. Calvaruso C., Turpault M. P., Leclerc E., Frey-Klett P. 2007. Impact of ectomycorrhizosphere on the functional diversity of soil bacterial and fungal communities from a forest stand in relation to nutrient mobilization processes. Microb. Ecol. 54:567–577 [DOI] [PubMed] [Google Scholar]
- 59. Calvo A. M., Wilson R. A., Bok J. W., Keller N. P. 2002. Relationship between secondary metabolism and fungal development. Microbiol. Mol. Biol. Rev. 66:447–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Caplice E., Fitzgerald G. F. 1999. Food fermentations: role of microorganisms in food production and preservation. Int. J. Food Microbiol. 50:131–149 [DOI] [PubMed] [Google Scholar]
- 61. Cardinale M., Vieira de Castro J., Jr., Muller H., Berg G., Grube M. 2008. In situ analysis of the bacterial community associated with the reindeer lichen Cladonia arbuscula reveals predominance of Alphaproteobacteria. FEMS Microbiol. Ecol. 66:63–71 [DOI] [PubMed] [Google Scholar]
- 62. Carlson E. 1983. Effect of strain of Staphylococcus aureus on synergism with Candida albicans resulting in mouse mortality and morbidity. Infect. Immun. 42:285–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Carlson E. 1983. Enhancement by Candida albicans of Staphylococcus aureus, Serratia marcescens, and Streptococcus faecalis in the establishment of infection in mice. Infect. Immun. 39:193–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Castagliuolo I., LaMont J. T., Nikulasson S. T., Pothoulakis C. 1996. Saccharomyces boulardii protease inhibits Clostridium difficile toxin A effects in the rat ileum. Infect. Immun. 64:5225–5232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Castagliuolo I., Riegler M. F., Valenick L., LaMont J. T., Pothoulakis C. 1999. Saccharomyces boulardii protease inhibits the effects of Clostridium difficile toxins A and B in human colonic mucosa. Infect. Immun. 67:302–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Cerigini E., Palma F., Barbieri E., Buffalini M., Stocchi V. 2008. The Tuber borchii fruiting body-specific protein TBF-1, a novel lectin which interacts with associated Rhizobium species. FEMS Microbiol. Lett. 284:197–203 [DOI] [PubMed] [Google Scholar]
- 67. Cerniglia C. E., Sutherland J. B. 2006. Relative roles of bacteria and fungi in polycyclic aromatic hydrocarbon biodegradation and bioremediation of contaminated soils, p. 182–211In Gadd G. M. (ed.), Fungi in biogeochemical cycles. Cambridge University Press, New York, NY [Google Scholar]
- 68. Chandelier A., Abras S., Laurent F., Debruxelles N., Cavelier M. 2006. Effect of temperature and bacteria on sporulation of Phytophthora alni in river water. Commun. Agric. Appl. Biol. Sci. 71:873–880 [PubMed] [Google Scholar]
- 69. Chaucheyras-Durand F., Faqir F., Ameilbonne A., Rozand C., Martin C. 2010. Fates of acid-resistant and non-acid-resistant Shiga toxin-producing Escherichia coli strains in ruminant digestive contents in the absence and presence of probiotics. Appl. Environ. Microbiol. 76:640–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Chaucheyras-Durand F., Fonty G. 2001. Establishment of cellulolytic bacteria and development of fermentative activities in the rumen of gnotobiotically-reared lambs receiving the microbial additive Saccharomyces cerevisiae CNCM I-1077. Reprod. Nutr. Dev. 41:57–68 [DOI] [PubMed] [Google Scholar]
- 71. Chaucheyras-Durand F., Masseglia S., Fonty G. 2005. Effect of the microbial feed additive Saccharomyces cerevisiae CNCM I-1077 on protein and peptide degrading activities of rumen bacteria grown in vitro. Curr. Microbiol. 50:96–101 [DOI] [PubMed] [Google Scholar]
- 72. Chavez-Gomez B., et al. 2003. Removal of phenanthrene from soil by co-cultures of bacteria and fungi pregrown on sugarcane bagasse pith. Bioresour. Technol. 89:177–183 [DOI] [PubMed] [Google Scholar]
- 73. Cho Y., Kim J., Crowley D., Cho B. 2003. Growth promotion of the edible fungus Pleurotus ostreatus by fluorescent pseudomonads. FEMS Microbiol. Lett. 218:271–276 [DOI] [PubMed] [Google Scholar]
- 74. Citernesi A. S., Fortuna P., Filippi C., Bagnoli G., Giovannetti M. 1996. The occurrence of antagonistic bacteria in Glomus mosseae pot cultures. Agronomie 16:671–677 [Google Scholar]
- 75. Citterio B., et al. 2001. Possible involvement of Pseudomonas fluorescens and Bacillaceae in structural modifications of Tuber borchii fruit bodies. Can. J. Microbiol. 47:264–268 [DOI] [PubMed] [Google Scholar]
- 76. Clausen C. A. 1996. Bacterial associations with decaying wood: a review. Int. Biodeterior. Biodegradation 37:101–107 [Google Scholar]
- 77. Compant S., Duffy B., Nowak J., Clement C., Barka E. A. 2005. Use of plant growth-promoting bacteria for biocontrol of plant diseases: principles, mechanisms of action, and future prospects. Appl. Environ. Microbiol. 71:4951–4959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Corsetti A., et al. 2001. Phenotypic and molecular identification and clustering of lactic acid bacteria and yeasts from wheat (species Triticum durum and Triticum aestivum) sourdoughs of Southern Italy. Int. J. Food Microbiol. 64:95–104 [DOI] [PubMed] [Google Scholar]
- 79. Corsetti A., Rossi J., Gobbetti M. 2001. Interactions between yeasts and bacteria in the smear surface-ripened cheeses. Int. J. Food Microbiol. 69:1–10 [DOI] [PubMed] [Google Scholar]
- 80. Crittenden P. D., Llimona X., Sancho L. G. 2007. Lichenized unicellular cyanobacteria fix nitrogen in the light. Can. J. Bot. 85:1003–1006 [Google Scholar]
- 81. Cueto M., et al. 2001. Pestalone, a new antibiotic produced by a marine fungus in response to bacterial challenge. J. Nat. Prod. 64:1444–1446 [DOI] [PubMed] [Google Scholar]
- 82. Cugini C., et al. 2007. Farnesol, a common sesquiterpene, inhibits PQS production in Pseudomonas aeruginosa. Mol. Microbiol. 65:896–906 [DOI] [PubMed] [Google Scholar]
- 83. Cugini C., Morales D. K., Hogan D. A. 2010. Candida albicans-produced farnesol stimulates Pseudomonas quinolone signal production in LasR-defective Pseudomonas aeruginosa strains. Microbiology 156:3096–3107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Currie C. R., Poulsen M., Mendenhall J., Boomsma J. J., Billen J. 2006. Coevolved crypts and exocrine glands support mutualistic bacteria in fungus-growing ants. Science 311:81–83 [DOI] [PubMed] [Google Scholar]
- 85. Currie C. R., Scott J. A., Summerbell R. C., Malloch D. 1999. Fungus-growing ants use antibiotic-producing bacteria to control garden parasites. Nature 398:701–704 [Google Scholar]
- 86. Curvale-Fauchet N., Botterel F., Legrand P., Guillot J., Bretagne S. 2004. Frequency of intravascular catheter colonization by Malassezia spp. in adult patients. Mycoses 47:491–494 [DOI] [PubMed] [Google Scholar]
- 87. Cusano A. M., et al. 2011. Pseudomonas fluorescens BBc6R8 type III secretion mutants no longer promote ectomycorrhizal symbiosis. Environ. Microbiol. Rep. 3:203–210 [DOI] [PubMed] [Google Scholar]
- 88. de Boer W., de Ridder-Duine A. S., Klein Gunnewiek P. J. A., Smant W., Van Veen J. A. 2008. Rhizosphere bacteria from sites with higher fungal densities exhibit greater levels of potential antifungal properties. Soil Biol. Biochem. 40:1542–1544 [Google Scholar]
- 89. de Boer W., Folman L. B., Summerbell R. C., Boddy L. 2005. Living in a fungal world: impact of fungi on soil bacterial niche development. FEMS Microbiol. Rev. 29:795–811 [DOI] [PubMed] [Google Scholar]
- 90. de Boer W., et al. 2004. Collimonas fungivorans gen. nov., sp. nov., a chitinolytic soil bacterium with the ability to grow on living fungal hyphae. Int. J. Syst. Evol. Microbiol. 54:857–864 [DOI] [PubMed] [Google Scholar]
- 91. de Boer W., Wagenaar A. M., Klein Gunnewiek P. J., van Veen J. A. 2007. In vitro suppression of fungi caused by combinations of apparently non-antagonistic soil bacteria. FEMS Microbiol. Ecol. 59:177–185 [DOI] [PubMed] [Google Scholar]
- 92. De Groot M. J., Bundock P., Hooykaas P. J., Beijersbergen A. G. 1998. Agrobacterium tumefaciens-mediated transformation of filamentous fungi. Nat. Biotechnol. 16:839–842 [DOI] [PubMed] [Google Scholar]
- 93. Deveau A., et al. 2010. Role of fungal trehalose and bacterial thiamine in the improved survival and growth of the ectomycorrhizal fungus Laccaria bicolor S238N and the helper bacterium Pseudomonas fluorescens BBc6R8. Environ. Microbiol. Rep. 2:560–568 [DOI] [PubMed] [Google Scholar]
- 94. Deveau A., et al. 2007. The mycorrhiza helper Pseudomonas fluorescens BBc6R8 has a specific priming effect on the growth, morphology and gene expression of the ectomycorrhizal fungus Laccaria bicolor S238N. New Phytol. 175:743–755 [DOI] [PubMed] [Google Scholar]
- 95. de Weert S., Kuiper I., Lagendijk E. L., Lamers G. E., Lugtenberg B. J. 2004. Role of chemotaxis toward fusaric acid in colonization of hyphae of Fusarium oxysporum f. sp. radicis-lycopersici by Pseudomonas fluorescens WCS365. Mol. Plant Microbe Interact. 17:1185–1191 [DOI] [PubMed] [Google Scholar]
- 96. Dewey F. M., Wong Y. L., Seery R., Hollins T. W., Gurr S. J. 1999. Bacteria associated with Stagonospora (Septoria) nodorum increase pathogenicity of the fungus. New Phytol. 144:489–497 [DOI] [PubMed] [Google Scholar]
- 97. Diaz E. M., Sacristan M., Legaz M. E., Vicente C. 2009. Isolation and characterization of a cyanobacterium-binding protein and its cell wall receptor in the lichen Peltigera canina. Plant Signal. Behav. 4:598–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Dixon E., et al. 2010. Bacteria-induced static batch fungal fermentation of the diterpenoid cyathin A(3), a small-molecule inducer of nerve growth factor. J. Ind. Microbiol. Biotechnol. 38:607–615 [DOI] [PubMed] [Google Scholar]
- 99. Donlan R. M., Costerton J. W. 2002. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 15:167–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Duponnois R., Garbaye J. 1990. Some mechanisms involved in growth stimulation of ectomycorrhizal fungi by bacteria. Can. J. Bot. 68:2148–2152 [Google Scholar]
- 101. Eckburg P. B., et al. 2005. Diversity of the human intestinal microbial flora. Science 308:1635–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. EFSA 2006. Opinion of the Scientific Panel on Additives and Products or Substances Used in Animal Feed on the safety and efficacy of the product “Biosaf Sc 47,” a preparation of Saccharomyces cerevisiae, as a feed additive for horses (question no. EFSA-Q-2005-025). EFSA J. 384:1–9 [Google Scholar]
- 103. Eger G. 1972. Experiments and comments on the action of bacteria on sporophore initiation in Agaricus bisporus. Mushroom Sci. 8:719–725 [Google Scholar]
- 104. Elad Y., Baker R. 1985. The role of competition for iron and carbon in suppression of chlamydospore germination of Fusarium spp. by Pseudomonas spp. Phytopathology 75:1053–1059 [Google Scholar]
- 105. Fajardo A., Martínez J. L. 2008. Antibiotics as signals that trigger specific bacterial responses. Curr. Opin. Microbiol. 11:161–167 [DOI] [PubMed] [Google Scholar]
- 106. Finstein M. S., Alexander M. 1962. Competition for carbon and nitrogen between Fusarium and bacteria. Soil Sci. 94:334–339 [Google Scholar]
- 107. Fitzpatrick D. A., Logue M. E., Butler G. 2008. Evidence of recent interkingdom horizontal gene transfer between bacteria and Candida parapsilosis. BMC Evol. Biol. 8:181 doi: 10.1186/1471-2148-8-181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Fleet G. H. 2003. Yeast interactions and wine flavour. Int. J. Food Microbiol. 86:11–22 [DOI] [PubMed] [Google Scholar]
- 109. Fleming A. 1929. On the antibacterial action of cultures of a Penicillium, with special reference to their use in the isolation of B. influenzae. Br. J. Exp. Pathol. 10:226–236 [Google Scholar]
- 110. Fogliano V., et al. 2002. Pseudomonas lipodepsipeptides and fungal cell wall-degrading enzymes act synergistically in biological control. Mol. Plant Microbe Interact. 15:323–333 [DOI] [PubMed] [Google Scholar]
- 111. Forano E., Chaucheyras-Durand F., Fonty G. 2007. How bacterial-fungal interactions optimise the functioning of the rumen. Biofutur 283:20–23 [Google Scholar]
- 112. Foster K. W., Thomas L., Warner J., Desmond R., Elewski B. E. 2005. A bipartite interaction between Pseudomonas aeruginosa and fungi in onychomycosis. Arch. Dermatol. 141:1467–1468 [DOI] [PubMed] [Google Scholar]
- 113. Fox P. F., Lucey J. A., Cogan T. M. 1990. Glycolysis and related reactions during cheese manufacture and ripening. Crit. Rev. Food Sci. Nutr. 29:237–253 [DOI] [PubMed] [Google Scholar]
- 114. Frases S., Chaskes S., Dadachova E., Casadevall A. 2006. Induction by Klebsiella aerogenes of a melanin-like pigment in Cryptococcus neoformans. Appl. Environ. Microbiol. 72:1542–1550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Frases S., Salazar A., Dadachova E., Casadevall A. 2007. Cryptococcus neoformans can utilize the bacterial melanin precursor homogentisic acid for fungal melanogenesis. Appl. Environ. Microbiol. 73:615–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Frey P., Frey-Klett P., Garbaye J., Berge O., Heulin T. 1997. Metabolic and genotypic fingerprinting of fluorescent pseudomonads associated with the douglas fir-Laccaria bicolor mycorrhizosphere. Appl. Environ. Microbiol. 63:1852–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Frey-Klett P., et al. 2005. Ectomycorrhizal symbiosis affects functional diversity of rhizosphere fluorescent pseudomonads. New Phytol. 165:317–328 [DOI] [PubMed] [Google Scholar]
- 118. Frey-Klett P., Garbaye J. 2005. Mycorrhiza helper bacteria: a promising model for the genomic analysis of fungal-bacterial interactions. New Phytol. 168:4–8 [DOI] [PubMed] [Google Scholar]
- 119. Frey-Klett P., Garbaye J., Tarkka M. 2007. The mycorrhiza helper bacteria revisited. New Phytol. 176:22–36 [DOI] [PubMed] [Google Scholar]
- 120. Frey-Klett P., Sarniguet A. 2008. Champignons et bactéries, les secrets de leur vie commune. Biofutur 284:19 [Google Scholar]
- 121. Furuno S., et al. 2010. Fungal mycelia allow chemotactic dispersal of polycyclic aromatic hydrocarbon-degrading bacteria in water-unsaturated systems. Environ. Microbiol. 12:1391–1398 [DOI] [PubMed] [Google Scholar]
- 122. Gaddy J. A., Tomaras A. P., Actis L. A. 2009. The Acinetobacter baumannii 19606 OmpA protein plays a role in biofilm formation on abiotic surfaces and in the interaction of this pathogen with eukaryotic cells. Infect. Immun. 77:3150–3160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Gaggia F., Mattarelli P., Biavati B. 2010. Probiotics and prebiotics in animal feeding for safe food production. Int. J. Food Microbiol. 141(Suppl. 1):S15–S28 [DOI] [PubMed] [Google Scholar]
- 124. Garbaye J. 1994. Helper bacteria—a new dimension to the mycorrhizal symbiosis. New Phytol. 128:197–210 [DOI] [PubMed] [Google Scholar]
- 125. Gastebois A., Latge J. P. 2008. Les bacteries et champignons pathogenes de l'homme, amis-ennemis. Biofutur 284:22–25 [Google Scholar]
- 126. Gee J. E., et al. 2011. Characterization of Burkholderia rhizoxinica and B. endofungorum isolated from clinical specimens. PLoS One 6:e15731 doi: 10.1371/journal.pone.0015731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Georgopapadakou N. H., Walsh T. J. 1994. Human mycoses: drugs and targets for emerging pathogens. Science 264:371–373 [DOI] [PubMed] [Google Scholar]
- 128. Gerez C. L., Carbajo M. S., Rollan G., Torres Leal G., Font de Valdez G. 2010. Inhibition of citrus fungal pathogens by using lactic acid bacteria. J. Food Sci. 75:M354–M359 [DOI] [PubMed] [Google Scholar]
- 129. Gerlach R. G., Hensel M. 2007. Protein secretion systems and adhesins: the molecular armory of Gram-negative pathogens. Int. J. Med. Microbiol. 297:401–415 [DOI] [PubMed] [Google Scholar]
- 130. Ghannoum M. A., et al. 2010. Characterization of the oral fungal microbiome (mycobiome) in healthy individuals. PLoS Pathog. 6:e1000713 doi: 10.1371/journal.ppat.1000713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Gibson J., Sood A., Hogan D. A. 2009. Pseudomonas aeruginosa-Candida albicans interactions: localization and fungal toxicity of a phenazine derivative. Appl. Environ. Microbiol. 75:504–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Gil-Turnes M. S., Hay M. E., Fenical W. 1989. Symbiotic marine bacteria chemically defend crustacean embryos from a pathogenic fungus. Science 246:116–118 [DOI] [PubMed] [Google Scholar]
- 133. Girlanda M., et al. 2001. Impact of biocontrol Pseudomonas fluorescens CHA0 and a genetically modified derivative on the diversity of culturable fungi in the cucumber rhizosphere. Appl. Environ. Microbiol. 67:1851–1864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Gobbetti M. 1998. The sourdough microflora: interactions of lactic acid bacteria and yeasts. Trends Food Sci. Technol. 9:267–274 [Google Scholar]
- 135. Gojkovic Z., et al. 2004. Horizontal gene transfer promoted evolution of the ability to propagate under anaerobic conditions in yeasts. Mol. Genet. Genomics 271:387–393 [DOI] [PubMed] [Google Scholar]
- 136. Gorbushina A. A., et al. 2004. Bacterial and fungal diversity and biodeterioration problems in mural painting environments of St. Martins church (Greene-Kreiensen, Germany). Int. Biodeterior. Biodegradation 53:13–24 [Google Scholar]
- 137. Gou M., Qu Y., Zhou J., Ma F., Tan L. 2009. Azo dye decolorization by a new fungal isolate, Penicillium sp. QQ and fungal-bacterial cocultures. J. Hazard. Mater. 170:314–319 [DOI] [PubMed] [Google Scholar]
- 138. Greaves H. 1971. The bacterial factor in wood decay. Wood Sci. Technol. 5:6–16 [Google Scholar]
- 139. Grewal S., Rainey P. 1991. Phenotypic variation of Pseudomonas putida and P. tolaasii affects the chemotactic response to Agaricus bisporus mycelial exudate. J. Gen. Microbiol. 137:2761–2768 [DOI] [PubMed] [Google Scholar]
- 140. Grube M., Berg G. 2009. Microbial consortia of bacteria and fungi with focus on the lichen symbiosis. Fungal Biol. Rev. 23:72–85 [Google Scholar]
- 141. Grube M., Cardinale M., de Castro J. V., Jr., Muller H., Berg G. 2009. Species-specific structural and functional diversity of bacterial communities in lichen symbioses. ISME J. 3:1105–1115 [DOI] [PubMed] [Google Scholar]
- 142. Guetsky R., Shtienberg D., Elad Y., Dinoor A. 2001. Combining biocontrol agents to reduce the variability of biological control. Phytopathology 91:621–627 [DOI] [PubMed] [Google Scholar]
- 143. Guilloux-Benatier M., Remize F., Gal L., Guzzo J., Alexandre H. 2006. Effects of yeast proteolytic activity on Oenococcus oeni and malolactic fermentation. FEMS Microbiol. Lett. 263:183–188 [DOI] [PubMed] [Google Scholar]
- 144. Guillouzouic A., et al. 2008. Fatal coinfection with Legionella pneumophila serogroup 8 and Aspergillus fumigatus. Diagn. Microbiol. Infect. Dis. 60:193–195 [DOI] [PubMed] [Google Scholar]
- 145. Gulis V., Suberkropp K. 2003. Interactions between stream fungi and bacteria associated with decomposing leaf litter at different levels of nutrient availability. Aquat. Microb. Ecol. 30:149–157 [Google Scholar]
- 146. Gupta N., Haque A., Mukhopadhyay G., Narayan R. P., Prasad R. 2005. Interactions between bacteria and Candida in the burn wound. Burns 31:375–378 [DOI] [PubMed] [Google Scholar]
- 147. Haas D., Defago G. 2005. Biological control of soil-borne pathogens by fluorescent pseudomonads. Nat. Rev. Microbiol. 3:307–319 [DOI] [PubMed] [Google Scholar]
- 148. Hachmeister K. A., Fung D. Y. 1993. Tempeh: a mold-modified indigenous fermented food made from soybeans and/or cereal grains. Crit. Rev. Microbiol. 19:137–188 [DOI] [PubMed] [Google Scholar]
- 149. Haeder S., Wirth R., Herz H., Spiteller D. 2009. Candicidin-producing Streptomyces support leaf-cutting ants to protect their fungus garden against the pathogenic fungus Escovopsis. Proc. Natl. Acad. Sci. U. S. A. 106:4742–4746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Hall C., Brachat S., Dietrich F. S. 2005. Contribution of horizontal gene transfer to the evolution of Saccharomyces cerevisiae. Eukaryot. Cell 4:1102–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Hammerschmidt R. 1999. Induced disease resistance: how do induced plants stop pathogens? Physiol. Mol. Plant Pathol. 55:77–84 [Google Scholar]
- 152. Hansen T. K., Jakobsen M. 1997. Possible role of microbial interactions for growth and sporulation of Penicillium roqueforti in Danablu. Lait 77:479–488 [Google Scholar]
- 153. Harriott M. M., Noverr M. C. 2010. Ability of Candida albicans mutants to induce Staphylococcus aureus vancomycin resistance during polymicrobial biofilm formation. Antimicrob. Agents Chemother. 54:3746–3755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Harriott M. M., Noverr M. C. 2009. Candida albicans and Staphylococcus aureus form polymicrobial biofilms: effects on antimicrobial resistance. Antimicrob. Agents Chemother. 53:3914–3922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Harris R. N., et al. 2009. Skin microbes on frogs prevent morbidity and mortality caused by a lethal skin fungus. ISME J. 3:818–824 [DOI] [PubMed] [Google Scholar]
- 156. Harris R. N., Lauer A., Simon M. A., Banning J. L., Alford R. A. 2009. Addition of antifungal skin bacteria to salamanders ameliorates the effects of chytridiomycosis. Dis. Aquat. Organ. 83:11–16 [DOI] [PubMed] [Google Scholar]
- 157. Hayes W. A., Randle P. E., Last F. T. 1969. The nature of the microbial stimulus affecting sporophore formation in Agaricus bisporus (Lange) Sing. Ann. Appl. Biol. 64:177–187 [Google Scholar]
- 158. Heinemann J. A., Sprague G. F., Jr 1989. Bacterial conjugative plasmids mobilize DNA transfer between bacteria and yeast. Nature 340:205–209 [DOI] [PubMed] [Google Scholar]
- 159. Heinonsalo J., Jorgensen K. S., Haahtela K., Sen R. 2000. Effects of Pinus sylvestris root growth and mycorrhizosphere development on bacterial carbon source utilization and hydrocarbon oxidation in forest and petroleum-contaminated soils. Can. J. Microbiol. 46:451–464 [DOI] [PubMed] [Google Scholar]
- 160. Hermann C., Hermann J., Munzel U., Ruchel R. 1999. Bacterial flora accompanying Candida yeasts in clinical specimens. Mycoses 42:619–627 [DOI] [PubMed] [Google Scholar]
- 161. Hildebrandt U., Janetta K., Bothe H. 2002. Towards growth of arbuscular mycorrhizal fungi independent of a plant host. Appl. Environ. Microbiol. 68:1919–1924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Hildebrandt U., Ouziad F., Marner F. J., Bothe H. 2006. The bacterium Paenibacillus validus stimulates growth of the arbuscular mycorrhizal fungus Glomus intraradices up to the formation of fertile spores. FEMS Microbiol. Lett. 254:258–267 [DOI] [PubMed] [Google Scholar]
- 163. Hoffman M. T., Arnold A. E. 2010. Diverse bacteria inhabit living hyphae of phylogenetically diverse fungal endophytes. Appl. Environ. Microbiol. 76:4063–4075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Hogan D., Kolter R. 2002. Pseudomonas-Candida interactions: an ecological role for virulence factors. Science 296:2229–2232 [DOI] [PubMed] [Google Scholar]
- 165. Hogan D., Vik A., Kolter R. 2004. A Pseudomonas aeruginosa quorum-sensing molecule influences Candida albicans morphology. Mol. Microbiol. 54:1212–1223 [DOI] [PubMed] [Google Scholar]
- 166. Hogan D. A., Wargo M. J., Beck N. 2007. Bacterial biofilms on fungal surfaces, p. 235–245In Kjelleberg S., Givskov M. (ed.), The biofilm mode of life: mechanisms and adaptations. Horizon Scientific Press, Norfolk, United Kingdom [Google Scholar]
- 167. Holcombe L. J., et al. 2010. Pseudomonas aeruginosa secreted factors impair biofilm development in Candida albicans. Microbiology 156:1476–1486 [DOI] [PubMed] [Google Scholar]
- 168. Holmes A. R., Cannon R. D., Jenkinson H. F. 1995. Interactions of Candida albicans with bacteria and salivary molecules in oral biofilms. J. Ind. Microbiol. 15:208–213 [DOI] [PubMed] [Google Scholar]
- 169. Hoppert M., König S. 2006. The succession of biofilms on building stone and its possible impact on biogenic weathering, p. 311–315In Fort R., Álvarez de Buergo M., Gomez-Heras M., Vazquez-Calvo C. (ed.), Heritage, weathering, and conservation: proceedings of the international conference, HWC-2006. Taylor & Francis, London, United Kingdom [Google Scholar]
- 170. Huttunen K., et al. 2004. Synergistic interaction in simultaneous exposure to Streptomyces californicus and Stachybotrys chartarum. Environ. Health Perspect. 112:659–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Ibrahim A. S., et al. 2008. Bacterial endosymbiosis is widely present among zygomycetes but does not contribute to the pathogenesis of mucormycosis. J. Infect. Dis. 198:1083–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Inbar J., Chet I. 1991. Evidence that chitinase produced by Aeromonas caviae is involved in the biological control of soil-borne plant pathogens by this bacterium. Soil Biol. Biochem. 23:973–978 [Google Scholar]
- 173. Izumi H., Finlay R. D. 2011. Ectomycorrhizal roots select distinctive bacterial and ascomycete communities in Swedish subarctic forests. Environ. Microbiol. 13:819–830 [DOI] [PubMed] [Google Scholar]
- 174. Jakobs-Schonwandt D., et al. 2010. Biodegradation of a biocide (Cu-N-cyclohexyldiazenium dioxide) component of a wood preservative by a defined soil bacterial community. Appl. Environ. Microbiol. 76:8076–8083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Jargeat P., et al. 2004. Isolation, free-living capacities, and genome structure of “Candidatus Glomeribacter gigasporarum,” the endocellular bacterium of the mycorrhizal fungus Gigaspora margarita. J. Bacteriol. 186:6876–6884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Jarosz L. M., Deng D. M., van der Mei H. C., Crielaard W., Krom B. P. 2009. Streptococcus mutans competence-stimulating peptide inhibits Candida albicans hypha formation. Eukaryot. Cell 8:1658–1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Jayasinghearachchi H. S., Seneviratne G. 2006. Fungal solubilization of rock phosphate is enhanced by forming fungal-rhizobial biofilms. Soil Biol. Biochem. 38:405–408 [Google Scholar]
- 178. Jeffries P., Barea J. 2001. Arbuscular mycorrhiza: a key component of sustainable plant-soil ecosystems, p. 95–113In Hock B. (ed.), The Mycota, vol. 9 Fungal associations. Springer, Berlin, Germany [Google Scholar]
- 179. Joblin K. N., Naylor G. E., Williams A. G. 1990. Effect of Methanobrevibacter smithii on xylanolytic activity of anaerobic ruminal fungi. Appl. Environ. Microbiol. 56:2287–2295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Johansson J. F., Paul L. R., Finlay R. D. 2004. Microbial interactions in the mycorrhizosphere and their significance for sustainable agriculture. FEMS Microbiol. Ecol. 48:1–13 [DOI] [PubMed] [Google Scholar]
- 181. Johnsen A. R., Wick L. Y., Harms H. 2005. Principles of microbial PAH-degradation in soil. Environ. Pollut. 133:71–84 [DOI] [PubMed] [Google Scholar]
- 182. Joyner P. M., et al. 2010. Mutanobactin A from the human oral pathogen Streptococcus mutans is a cross-kingdom regulator of the yeast-mycelium transition. Org. Biomol. Chem. 8:5486–5489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Jurado V., et al. 2009. The fungal colonisation of rock-art caves: experimental evidence. Naturwissenschaften 96:1027–1034 [DOI] [PubMed] [Google Scholar]
- 184. Jussier D., Dube Morneau A., Mira de Orduna R. 2006. Effect of simultaneous inoculation with yeast and bacteria on fermentation kinetics and key wine parameters of cool-climate Chardonnay. Appl. Environ. Microbiol. 72:221–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Kaltenpoth M., Gottler W., Herzner G., Strohm E. 2005. Symbiotic bacteria protect wasp larvae from fungal infestation. Curr. Biol. 15:475–479 [DOI] [PubMed] [Google Scholar]
- 186. Kataoka R., Taniguchi T., Futai K. 2009. Fungal selectivity of two mycorrhiza helper bacteria on five mycorrhizal fungi associated with Pinus thunbergii. World J. Microbiol. Biotechnol. 25:1815–1819 [Google Scholar]
- 187. Kawarai T., Furukawa S., Ogihara H., Yamasaki M. 2007. Mixed-species biofilm formation by lactic acid bacteria and rice wine yeasts. Appl. Environ. Microbiol. 73:4673–4676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Kemppainen M., Circosta A., Tagu D., Martin F., Pardo A. G. 2005. Agrobacterium-mediated transformation of the ectomycorrhizal symbiont Laccaria bicolor S238N. Mycorrhiza 16:19–22 [DOI] [PubMed] [Google Scholar]
- 189. Kerr J. R., et al. 1999. Pseudomonas aeruginosa pyocyanin and 1-hydroxyphenazine inhibit fungal growth. J. Clin. Pathol. 52:385–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Keuth S., Bisping B. 1993. Formation of vitamins by pure cultures of tempe moulds and bacteria during the tempe solid substrate fermentation. J. Appl. Microbiol. 75:427–434 [DOI] [PubMed] [Google Scholar]
- 191. Kim H., et al. 1998. Functional analysis of a hybrid endoglucanase of bacterial origin having a cellulose binding domain from a fungal exoglucanase. Appl. Biochem. Biotechnol. 75:193–204 [DOI] [PubMed] [Google Scholar]
- 192. Kimball T. H., Rosemary L. P. 1993. Cyanobacteria and cyanolichens: can they enhance availability of essential minerals for higher plants? Western North Am. Nat. 53:59–72 [Google Scholar]
- 193. Kirk T. K., Farrell R. L. 1987. Enzymatic “combustion”: the microbial degradation of lignin. Annu. Rev. Microbiol. 41:465–505 [DOI] [PubMed] [Google Scholar]
- 194. Klaerner H. G., et al. 1997. Candida albicans and Escherichia coli are synergistic pathogens during experimental microbial peritonitis. J. Surg. Res. 70:161–165 [DOI] [PubMed] [Google Scholar]
- 195. Klotz S. A., Chasin B. S., Powell B., Gaur N. K., Lipke P. N. 2007. Polymicrobial bloodstream infections involving Candida species: analysis of patients and review of the literature. Diagn. Microbiol. Infect. Dis. 59:401–406 [DOI] [PubMed] [Google Scholar]
- 196. Kluge M. 2002. A fungus eats a cyanobacterium: the story of the Geosiphon pyriformis endocyanosis. Biol. Environ. 102:11–14 [Google Scholar]
- 197. Kluge M., Mollenhauer D., Mollenhauer R. 1991. Photosynthetic carbon assimilation in Geosiphon pyriforme (Kützing) F. v. Wettstein, an endosymbiotic association of fungus and cyanobacterium. Planta 185:311–315 [DOI] [PubMed] [Google Scholar]
- 198. Kluge M., Mollenhauer D., Mollenhauer R., Kape R. 1992. Geosiphon pyriforme, an endosymbiotic consortium of a fungus and a cyanobacterium (Nostoc), fixes nitrogen. Bot. Acta 105:343–344 [Google Scholar]
- 199. Kluge M., Mollenhauer D., Wolf E., Schüßler A. 2002. The Nostoc-Geosiphon endocytobiosis, p. 19–30In Rai A., Bergman B., Rasmussen U. (ed.), Cyanobacteria in symbiosis. Kluwer Academic Publishers, Dordrecht, Netherlands [Google Scholar]
- 200. Kobayashi D. Y., Crouch J. A. 2009. Bacterial/fungal interactions: from pathogens to mutualistic endosymbionts. Annu. Rev. Phytopathol. 47:63–82 [DOI] [PubMed] [Google Scholar]
- 201. Koele N., Turpault M.-P., Hildebrand E. E., Uroz S., Frey-Klett P. 2009. Interactions between mycorrhizal fungi and mycorrhizosphere bacteria during mineral weathering: budget analysis and bacterial quantification. Soil Biol. Biochem. 41:1935–1942 [Google Scholar]
- 202. Kohlmeier S., et al. 2005. Taking the fungal highway: mobilization of pollutant-degrading bacteria by fungi. Environ. Sci. Technol. 39:4640–4646 [DOI] [PubMed] [Google Scholar]
- 203. Kotterman M. J., Vis E. H., Field J. A. 1998. Successive mineralization and detoxification of benzo[a]pyrene by the white rot fungus Bjerkandera sp.strain BOS55 and indigenous microflora. Appl. Environ. Microbiol. 64:2853–2858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204. Kroiss J., et al. 2010. Symbiotic streptomycetes provide antibiotic combination prophylaxis for wasp offspring. Nat. Chem. Biol. 6:261–263 [DOI] [PubMed] [Google Scholar]
- 205. Lackner G., et al. 2009. Global distribution and evolution of a toxinogenic Burkholderia-Rhizopus symbiosis. Appl. Environ. Microbiol. 75:2982–2986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206. Lackner G., Moebius N., Hertweck C. 2011. Endofungal bacterium controls its host by an hrp type III secretion system. ISME J. 5:252–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207. Lackner G., Moebius N., Partida-Martinez L., Hertweck C. 2011. Complete genome sequence of Burkholderia rhizoxinica, the endosymbiont of Rhizopus microsporus. J. Bacteriol. 193:783–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208. Lackner G., Moebius N., Partida-Martinez L. P., Boland S., Hertweck C. 2011. Evolution of an endofungal lifestyle: deductions from the Burkholderia rhizoxinica genome. BMC Genomics 12:210 doi:10.1186/1471-2164-12-210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209. Lackner G., Partida-Martinez L. P., Hertweck C. 2009. Endofungal bacteria as producers of mycotoxins. Trends Microbiol. 17:570–576 [DOI] [PubMed] [Google Scholar]
- 210. Le Calvez T., Burgaud G., Mahe S., Barbier G., Vandenkoornhuyse P. 2009. Fungal diversity in deep-sea hydrothermal ecosystems. Appl. Environ. Microbiol. 75:6415–6421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211. Lee S. S., Ha J. K., Cheng K. 2000. Relative contributions of bacteria, protozoa, and fungi to in vitro degradation of orchard grass cell walls and their interactions. Appl. Environ. Microbiol. 66:3807–3813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212. Lehr N. A., Schrey S. D., Bauer R., Hampp R., Tarkka M. T. 2007. Suppression of plant defence response by a mycorrhiza helper bacterium. New Phytol. 174:892–903 [DOI] [PubMed] [Google Scholar]
- 213. Lemanceau P., Bakker P. A., De Kogel W. J., Alabouvette C., Schippers B. 1993. Antagonistic effect of nonpathogenic Fusarium oxysporum Fo47 and pseudobactin 358 upon pathogenic Fusarium oxysporum f. sp. dianthi. Appl. Environ. Microbiol. 59:74–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214. Lemanceau P., Bakker P. A., De Kogel W. J., Alabouvette C., Schippers B. 1992. Effect of pseudobactin 358 production by Pseudomonas putida WCS358 on suppression of fusarium wilt of carnations by nonpathogenic Fusarium oxysporum Fo47. Appl. Environ. Microbiol. 58:2978–2982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215. Leone M. R., et al. 2010. An unusual galactofuranose lipopolysaccharide that ensures the intracellular survival of toxin-producing bacteria in their fungal host. Angew. Chem. Int. Ed. Engl. 49:7476–7480 [DOI] [PubMed] [Google Scholar]
- 216. Leveau J. H., Preston G. M. 2007. Bacterial mycophagy: definition and diagnosis of a unique bacterial-fungal interaction. New Phytol. 177:859–876 [DOI] [PubMed] [Google Scholar]
- 217. Li B., Ravnskov S., Xie G., Larsen J. 2007. Biocontrol of Pythium damping-off in cucumber by arbuscular mycorrhiza-associated bacteria from the genus Paenibacillus. Biocontrol 52:863–875 [Google Scholar]
- 218. Linares J. F., Gustafsson I., Baquero F., Martinez J. L. 2006. Antibiotics as intermicrobial signaling agents instead of weapons. Proc. Natl. Acad. Sci. U. S. A. 103:19484–19489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219. Little A. E., Currie C. R. 2008. Black yeast symbionts compromise the efficiency of antibiotic defenses in fungus-growing ants. Ecology 89:1216–1222 [DOI] [PubMed] [Google Scholar]
- 220. Little A. E., Currie C. R. 2007. Symbiotic complexity: discovery of a fifth symbiont in the attine ant-microbe symbiosis. Biol. Lett. 3:501–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221. Little A. E., Murakami T., Mueller U. G., Currie C. R. 2006. Defending against parasites: fungus-growing ants combine specialized behaviours and microbial symbionts to protect their fungus gardens. Biol. Lett. 2:12–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222. Liu C., Hu B., Liu Y., Chen S. 2006. Stimulation of nisin production from whey by a mixed culture of Lactococcus lactis and Saccharomyces cerevisiae. Appl. Biochem. Biotechnol. 131:751–761 [DOI] [PubMed] [Google Scholar]
- 223. Liu S.-Q., Tsao M. 2009. Enhancement of survival of probiotic and non-probiotic lactic acid bacteria by yeasts in fermented milk under non-refrigerated conditions. Int. J. Food Microbiol. 135:34–38 [DOI] [PubMed] [Google Scholar]
- 224. Lonvaud-Funel A. 2008. Du raisin au vin: l'activité d'un système microbien dynamique. Biofutur 294:26–29 [Google Scholar]
- 225. Loper J. E., Henkels M. D., Shaffer B. T., Valeriote F. A., Gross H. 2008. Isolation and identification of rhizoxin analogs from Pseudomonas fluorescens Pf-5 by using a genomic mining strategy. Appl. Environ. Microbiol. 74:3085–3093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226. Lugtenberg B., Kamilova F. 2009. Plant-growth-promoting rhizobacteria. Annu. Rev. Microbiol. 63:541–556 [DOI] [PubMed] [Google Scholar]
- 227. Lumini E., Ghignone S., Bianciotto V., Bonfante P. 2006. Endobacteria or bacterial endosymbionts? To be or not to be. New Phytol. 170:205–208 [DOI] [PubMed] [Google Scholar]
- 228. Lutz M. P., Wenger S., Maurhofer M., Defago G., Duffy B. 2004. Signaling between bacterial and fungal biocontrol agents in a strain mixture. FEMS Microbiol. Ecol. 48:447–455 [DOI] [PubMed] [Google Scholar]
- 229. Mahadevan B., Crawford D. L. 1997. Properties of the chitinase of the antifungal biocontrol agent Streptomyces lydicus WYEC108. Enzyme Microb. Technol. 20:489–493 [Google Scholar]
- 230. Maier A., Riedlinger J., Fiedler H.-P., Hampp R. 2004. Actinomycetales bacteria from a spruce stand: characterization and effects on growth of root symbiotic and plant parasitic soil fungi in dual culture. Mycol. Prog. 3:129–136 [Google Scholar]
- 231. Maligoy M., Mercade M., Cocaign-Bousquet M., Loubiere P. 2008. Transcriptome analysis of Lactococcus lactis in coculture with Saccharomyces cerevisiae. Appl. Environ. Microbiol. 74:485–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232. Mallet L. V., Becq J., Deschavanne P. 2010. Whole genome evaluation of horizontal transfers in the pathogenic fungus Aspergillus fumigatus. BMC Genomics 11:171 doi:10.1186/1471-2164-11-171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233. Mangan A. 1969. Interactions between some aural Aspergillus species and bacteria. J. Gen. Microbiol. 58:261–266 [DOI] [PubMed] [Google Scholar]
- 234. Manjula K., Krishna Kishore G., Girish A. G., Singh S. D. 2004. Combined application of Pseudomonas fluorescens and Trichoderma viride has an improved biocontrol activity against stem rot in groundnut. Plant Pathol. J. 20:75–80 [Google Scholar]
- 235. Mansour S., et al. 2009. Investigation of associations of Yarrowia lipolytica, Staphylococcus xylosus, and Lactococcus lactis in culture as a first step in microbial interaction analysis. Appl. Environ. Microbiol. 75:6422–6430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236. Maoz A., Mayr R., Scherer S. 2003. Temporal stability and biodiversity of two complex antilisterial cheese-ripening microbial consortia. Appl. Environ. Microbiol. 69:4012–4018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237. Marcet-Houben M., Gabaldon T. 2009. Acquisition of prokaryotic genes by fungal genomes. Trends Genet. 26:5–8 [DOI] [PubMed] [Google Scholar]
- 238. Mari M., Guizzardi M., Pratella G. C. 1996. Biological control of gray mold in pears by antagonistic bacteria. Biol. Control 7:30–37 [Google Scholar]
- 239. Marshall K. C., Alexander M. 1960. Competition between soil bacteria and fusarium. Plant Soil 12:143–153 [Google Scholar]
- 240. Martel C. M., et al. 2011. Expression of bacterial levanase in yeast enables simultaneous saccharification and fermentation of grass juice to bioethanol. Bioresour. Technol. 102:1503–1508 [DOI] [PubMed] [Google Scholar]
- 241. Martin F., Tunlid A. 2009. The ectomycorrhizal symbiosis: a marriage of convenience, p. 237–257In Deising H. B. (ed.), The Mycota, 2nd ed., vol. 5 Plant relationships Springer, Berlin, Germany [Google Scholar]
- 242. Marvin-Sikkema F. D., Richardson A. J., Stewart C. S., Gottschal J. C., Prins R. A. 1990. Influence of hydrogen-consuming bacteria on cellulose degradation by anaerobic fungi. Appl. Environ. Microbiol. 56:3793–3797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243. Mathew B. P., Nath M. 2009. Recent approaches to antifungal therapy for invasive mycoses. ChemMedChem 4:310–323 [DOI] [PubMed] [Google Scholar]
- 244. Mayser P., Fromme S., Leitzmann C., Grunder K. 1995. The yeast spectrum of the ‘tea fungus Kombucha’. Mycoses 38:289–295 [DOI] [PubMed] [Google Scholar]
- 245. Melin P., Schnurer J., Wagner E. G. 1999. Changes in Aspergillus nidulans gene expression induced by bafilomycin, a Streptomyces-produced antibiotic. Microbiology 145:1115–1122 [DOI] [PubMed] [Google Scholar]
- 246. Melin P., Schnurer J., Wagner E. G. 2002. Proteome analysis of Aspergillus nidulans reveals proteins associated with the response to the antibiotic concanamycin A, produced by Streptomyces species. Mol. Genet. Genomics 267:695–702 [DOI] [PubMed] [Google Scholar]
- 247. Melnick R. L., et al. 2008. Bacterial endophytes: Bacillus spp. from annual crops as potential biological control agents of black pod rot of cacao. Biol. Control 46:46–56 [Google Scholar]
- 248. Miao L., Kwong T. F., Qian P. Y. 2006. Effect of culture conditions on mycelial growth, antibacterial activity, and metabolite profiles of the marine-derived fungus Arthrinium c.f. saccharicola. Appl. Microbiol. Biotechnol. 72:1063–1073 [DOI] [PubMed] [Google Scholar]
- 249. Milanesi C., et al. 2006. Fungal deterioration of medieval wall fresco determined by analysing small fragments containing copper. Int. Biodeterior. Biodegradation 57:7–13 [Google Scholar]
- 250. Mille-Lindblom C., Fischer H., Tranvik L. J. 2006. Antagonism between bacteria and fungi: substrate competition and a possible tradeoff between fungal growth and tolerance towards bacteria. Oikos 113:233–242 [Google Scholar]
- 251. Mille-Lindblom C., Tranvik L. J. 2003. Antagonism between bacteria and fungi on decomposing aquatic plant litter. Microb. Ecol. 45:173–182 [DOI] [PubMed] [Google Scholar]
- 252. Minerdi D., Fani R., Gallo R., Boarino A., Bonfante P. 2001. Nitrogen fixation genes in an endosymbiotic Burkholderia strain. Appl. Environ. Microbiol. 67:725–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253. Minerdi D., et al. 2008. Bacterial ectosymbionts and virulence silencing in a Fusarium oxysporum strain. Environ. Microbiol. 10:1725–1741 [DOI] [PubMed] [Google Scholar]
- 254. Mingardon F., et al. 2007. Incorporation of fungal cellulases in bacterial minicellulosomes yields viable, synergistically acting cellulolytic complexes. Appl. Environ. Microbiol. 73:3822–3832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255. Mogge B., Loferer C., Agerer R., Hutzler P., Hartmann A. 2000. Bacterial community structure and colonization patterns of Fagus sylvatica L. ectomycorrhizospheres as determined by fluorescence in situ hybridization and confocal laser scanning microscopy. Mycorrhiza 9:271–278 [Google Scholar]
- 256. Moller J., Miller M., Kjoller A. 1999. Fungal-bacterial interaction on beech leaves: influence on decomposition and dissolved organic carbon quality. Soil Biol. Biochem. 31:367–374 [Google Scholar]
- 257. Morales D. K., et al. 2010. Antifungal mechanisms by which a novel Pseudomonas aeruginosa phenazine toxin kills Candida albicans in biofilms. Mol. Microbiol. 78:1379–1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258. Moran N. A. 2002. Microbial minimalism: genome reduction in bacterial pathogens. Cell 108:583–586 [DOI] [PubMed] [Google Scholar]
- 259. Moretti M., et al. 2010. A proteomics approach to study synergistic and antagonistic interactions of the fungal-bacterial consortium Fusarium oxysporum wild-type MSA 35. Proteomics 10:3292–3320 [DOI] [PubMed] [Google Scholar]
- 260. Mounier J., et al. 2006. Sources of the adventitious microflora of a smear-ripened cheese. J. Appl. Microbiol. 101:668–681 [DOI] [PubMed] [Google Scholar]
- 261. Mounier J., et al. 2008. Microbial interactions within a cheese microbial community. Appl. Environ. Microbiol. 74:172–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262. Mowat E., et al. 2010. Pseudomonas aeruginosa and their small diffusible extracellular molecules inhibit Aspergillus fumigatus biofilm formation. FEMS Microbiol. Lett. 313:96–102 [DOI] [PubMed] [Google Scholar]
- 263. Mueller U. G., Schultz T. R., Currie C. R., Adams R. M., Malloch D. 2001. The origin of the attine ant-fungus mutualism. Q. Rev. Biol. 76:169–197 [DOI] [PubMed] [Google Scholar]
- 264. Müller E., Drewello U., Drewello R., Weißmann R., Wuertz S. 2001. In situ analysis of biofilms on historic window glass using confocal laser scanning microscopy. J. Cult. Herit. 2:31–42 [Google Scholar]
- 265. Muller F. M., Seidler M. 2010. Characteristics of pathogenic fungi and antifungal therapy in cystic fibrosis. Expert Rev. Anti Infect. Ther. 8:957–964 [DOI] [PubMed] [Google Scholar]
- 266. Murray A. C., Woodward S. 2003. In vitro interactions between bacteria isolated from Sitka spruce stumps and Heterobasidion annosum. Forest Pathol. 33:53–67 [Google Scholar]
- 267. Murtoniemi T., Penttinen P., Nevalainen A., Hirvonen M. R. 2005. Effects of microbial cocultivation on inflammatory and cytotoxic potential of spores. Inhal. Toxicol. 17:681–693 [DOI] [PubMed] [Google Scholar]
- 268. Mushegian A. A., Peterson C. N., Baker C. C., Pringle A. 2011. Bacterial diversity across individual lichens. Appl. Environ. Microbiol. 77:4249–4252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269. Mygind P. H., et al. 2005. Plectasin is a peptide antibiotic with therapeutic potential from a saprophytic fungus. Nature 437:975–980 [DOI] [PubMed] [Google Scholar]
- 270. Nair N. G., Fahy P. C. 1972. Bacteria antagonistic to Pseudomonas tolaasii and their control of brown blotch of the cultivated mushroom Agaricus bisporus. J. Appl. Bacteriol. 35:439–442 [DOI] [PubMed] [Google Scholar]
- 271. Nakamura C. E., Whited G. M. 2003. Metabolic engineering for the microbial production of 1,3-propanediol. Curr. Opin. Biotechnol. 14:454–459 [DOI] [PubMed] [Google Scholar]
- 272. Naumann M., Schussler A., Bonfante P. 2010. The obligate endobacteria of arbuscular mycorrhizal fungi are ancient heritable components related to the Mollicutes. ISME J. 4:862–871 [DOI] [PubMed] [Google Scholar]
- 273. Nelson K. E., et al. 2010. A catalog of reference genomes from the human microbiome. Science 328:994–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274. Neviani E., Gatti M., Vannini L., Gardini F., Suzzi G. 2001. Contribution of Gal-lactic acid bacteria to Saccharomyces cerevisiae metabolic activity in milk. Int. J. Food Microbiol. 69:91–99 [DOI] [PubMed] [Google Scholar]
- 275. Newbold C. J., Wallace R. J., Chen X. B., McIntosh F. M. 1995. Different strains of Saccharomyces cerevisiae differ in their effects on ruminal bacterial numbers in vitro and in sheep. J. Anim. Sci. 73:1811–1818 [DOI] [PubMed] [Google Scholar]
- 276. Newton A. C., Toth I. K., Neave P., Hyman L. J. 2004. Bacterial inoculum from a previous crop affects fungal disease development on subsequent nonhost crops. New Phytol. 163:133–138 [DOI] [PubMed] [Google Scholar]
- 277. Niderkorn V., Boudra H., Morgavi D. P. 2007. Les fusariotoxines: comment limiter leur présence dans les ensilages et leur impact chez les ruminants? Fourrages 189:111–123 [Google Scholar]
- 278. Nikawa H., et al. 2001. Alteration of the coadherence of Candida albicans with oral bacteria by dietary sugars. Oral Microbiol. Immunol. 16:279–283 [DOI] [PubMed] [Google Scholar]
- 279. Nishikawa M., Suzuki K., Yoshida K. 1992. DNA integration into recipient yeast chromosomes by trans-kingdom conjugation between Escherichia coli and Saccharomyces cerevisiae. Curr. Genet. 21:101–108 [DOI] [PubMed] [Google Scholar]
- 280. Noble R., et al. 2003. Primordia initiation of mushroom (Agaricus bisporus) strains on axenic casing materials. Mycologia 95:620–629 [DOI] [PubMed] [Google Scholar]
- 281. Nouaille S., et al. 2009. Transcriptomic response of Lactococcus lactis in mixed culture with Staphylococcus aureus. Appl. Environ. Microbiol. 75:4473–4482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282. Noverr M. C., Huffnagle G. B. 2004. Regulation of Candida albicans morphogenesis by fatty acid metabolites. Infect. Immun. 72:6206–6210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283. Nurmiaho-Lassila E. L., Timonen S., Haahtela K., Sen R. 1997. Bacterial colonization patterns of intact Pinus sylvestris mycorrhizospheres in dry pine forest soil: an electron microscopy study. Can. J. Microbiol. 43:1017–1035 [Google Scholar]
- 284. Oh D. C., Jensen P. R., Kauffman C. A., Fenical W. 2005. Libertellenones A-D: induction of cytotoxic diterpenoid biosynthesis by marine microbial competition. Bioorg. Med. Chem. 13:5267–5273 [DOI] [PubMed] [Google Scholar]
- 285. Oh D. C., Poulsen M., Currie C. R., Clardy J. 2009. Dentigerumycin: a bacterial mediator of an ant-fungus symbiosis. Nat. Chem. Biol. 5:391–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286. Oh D. C., Scott J. J., Currie C. R., Clardy J. 2009. Mycangimycin, a polyene peroxide from a mutualist Streptomyces sp. Org. Lett. 11:633–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287. Olsson P. A., Wallander H. 1998. Interactions between ectomycorrhizal fungi and the bacterial community in soils amended with various primary minerals. FEMS Microbiol. Ecol. 27:195–205 [Google Scholar]
- 288. O'May G. A., Reynolds N., Macfarlane G. T. 2005. Effect of pH on an in vitro model of gastric microbiota in enteral nutrition patients. Appl. Environ. Microbiol. 71:4777–4783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289. O'May G. A., Reynolds N., Smith A. R., Kennedy A., Macfarlane G. T. 2005. Effect of pH and antibiotics on microbial overgrowth in the stomachs and duodena of patients undergoing percutaneous endoscopic gastrostomy feeding. J. Clin. Microbiol. 43:3059–3065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290. Ostrosky-Zeichner L., Casadevall A., Galgiani J. N., Odds F. C., Rex J. H. 2010. An insight into the antifungal pipeline: selected new molecules and beyond. Nat. Rev. Drug Discov. 9:719–727 [DOI] [PubMed] [Google Scholar]
- 291. O'Sullivan J. M., Jenkinson H. F., Cannon R. D. 2000. Adhesion of Candida albicans to oral streptococci is promoted by selective adsorption of salivary proteins to the streptococcal cell surface. Microbiology 146:41–48 [DOI] [PubMed] [Google Scholar]
- 292. Page M. G., Heim J. 2009. Prospects for the next anti-Pseudomonas drug. Curr. Opin. Pharmacol. 9:558–565 [DOI] [PubMed] [Google Scholar]
- 293. Palla F., Marineo S., Lombardo G., Anello L. 2006. Characterization of bacterial community in indoor environment, p. 361–365In Fort R., Buergo M. Alvarez de, Gomez-Heras M., Vazquez-Calvo C. (ed.), Heritage, weathering, and conservation: proceedings of the international conference, HWC-2006. Taylor & Francis, London, United Kingdom [Google Scholar]
- 294. Pardo A. G., et al. 2005. T-DNA transfer from Agrobacterium tumefaciens to the ectomycorrhizal fungus Pisolithus microcarpus. Rev. Argent. Microbiol. 37:69–72 [PubMed] [Google Scholar]
- 295. Partida-Martinez L. P., Bandemer S., Ruchel R., Dannaoui E., Hertweck C. 2008. Lack of evidence of endosymbiotic toxin-producing bacteria in clinical Rhizopus isolates. Mycoses 51:266–269 [DOI] [PubMed] [Google Scholar]
- 296. Partida-Martinez L. P., et al. 2007. Rhizonin, the first mycotoxin isolated from the zygomycota, is not a fungal metabolite but is produced by bacterial endosymbionts. Appl. Environ. Microbiol. 73:793–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297. Partida-Martinez L. P., et al. 2007. Burkholderia rhizoxinica sp. nov. and Burkholderia endofungorum sp. nov., bacterial endosymbionts of the plant-pathogenic fungus Rhizopus microsporus. Int. J. Syst. Evol. Microbiol. 57:2583–2590 [DOI] [PubMed] [Google Scholar]
- 298. Partida-Martinez L. P., Hertweck C. 2005. Pathogenic fungus harbours endosymbiotic bacteria for toxin production. Nature 437:884–888 [DOI] [PubMed] [Google Scholar]
- 299. Partida-Martinez L. P., Monajembashi S., Greulich K. O., Hertweck C. 2007. Endosymbiont-dependent host reproduction maintains bacterial-fungal mutualism. Curr. Biol. 17:773–777 [DOI] [PubMed] [Google Scholar]
- 300. Pate J. C., Jones D. B., Wilhelmus K. R. 2006. Prevalence and spectrum of bacterial co-infection during fungal keratitis. Br. J. Ophthalmol. 90:289–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301. Peleg A. Y., Hogan D. A., Mylonakis E. 2010. Medically important bacterial-fungal interactions. Nat. Rev. Microbiol. 8:340–349 [DOI] [PubMed] [Google Scholar]
- 302. Penalva M. A., Arst H. N., Jr 2002. Regulation of gene expression by ambient pH in filamentous fungi and yeasts. Microbiol. Mol. Biol. Rev. 66:426–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303. Peng X., Sun J., Michiels C., Iserentant D., Verachtert H. 2001. Decrease in cell surface galactose residues of Schizosaccharomyces pombe enhances its coflocculation with Pediococcus damnosus. Appl. Environ. Microbiol. 67:3413–3417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304. Penttinen P., Huttunen K., Pelkonen J., Hirvonen M. R. 2005. The proportions of Streptomyces californicus and Stachybotrys chartarum in simultaneous exposure affect inflammatory responses in mouse RAW264.7 macrophages. Inhal. Toxicol. 17:79–85 [DOI] [PubMed] [Google Scholar]
- 305. Penttinen P., Pelkonen J., Huttunen K., Toivola M., Hirvonen M. R. 2005. Interactions between Streptomyces californicus and Stachybotrys chartarum can induce apoptosis and cell cycle arrest in mouse RAW264.7 macrophages. Toxicol. Appl. Pharmacol. 202:278–288 [DOI] [PubMed] [Google Scholar]
- 306. Peters B. M., et al. 2010. Microbial interactions and differential protein expression in Staphylococcus aureus-Candida albicans dual-species biofilms. FEMS Immunol. Med. Microbiol. 59:493–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307. Peyvast G., Olfati J. A., Kariminia A., Fallah A. 2009. Rhizobia as biofertilizer for oyster mushroom cultivation. J. Pure Appl. Microbiol. 3:421–424 [Google Scholar]
- 308. Pfaller M. A., Diekema D. J. 2007. Epidemiology of invasive candidiasis: a persistent public health problem. Clin. Microbiol. Rev. 20:133–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309. Pierce G. E. 2005. Pseudomonas aeruginosa, Candida albicans, and device-related nosocomial infections: implications, trends, and potential approaches for control. J. Ind. Microbiol. Biotechnol. 32:309–318 [DOI] [PubMed] [Google Scholar]
- 310. Pinto-Tomas A. A., et al. 2009. Symbiotic nitrogen fixation in the fungus gardens of leaf-cutter ants. Science 326:1120–1123 [DOI] [PubMed] [Google Scholar]
- 311. Pivato B., et al. 2009. Bacterial effects on arbuscular mycorrhizal fungi and mycorrhiza development as influenced by the bacteria, fungi, and host plant. Mycorrhiza 19:81–90 [DOI] [PubMed] [Google Scholar]
- 312. Preston G., Studholme D., Caldelari I. 2005. Profiling the secretomes of plant pathogenic Proteobacteria. FEMS Microbiol. Rev. 29:331–360 [DOI] [PubMed] [Google Scholar]
- 313. Qin J., et al. 2010. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464:59–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314. Rainey P. 1991. Effect of Pseudomonas putida on hyphal growth of Agaricus bisporus. Mycol. Res. 95:699–704 [Google Scholar]
- 315. Rainey P. 1991. Phenotypic variation of Pseudomonas putida and P. tolaasii affects attachment to Agaricus bisporus mycelium. J. Gen. Microbiol. 137:2769–2779 [DOI] [PubMed] [Google Scholar]
- 316. Rainey P., Cole A., Fermor T., Wood D. 1990. A model system for examining involvement of bacteria in basidiodome initiation of Agaricus bisporus. Mycol. Res. 94:191–195 [Google Scholar]
- 317. Rainey P. B., Brodey C. L., Johnstone K. 1992. Biology of Pseudomonas tolaasii, cause of brown blotch disease of the cultivated mushroom. Adv. Plant Pathol. 8:95–117 [Google Scholar]
- 318. Rasmussen T. B., et al. 2005. Identity and effects of quorum-sensing inhibitors produced by Penicillium species. Microbiology 151:1325–1340 [DOI] [PubMed] [Google Scholar]
- 319. Remize F., Augagneur Y., Guilloux-Benatier M., Guzzo J. 2005. Effect of nitrogen limitation and nature of the feed upon Oenococcus oeni metabolism and extracellular protein production. J. Appl. Microbiol. 98:652–661 [DOI] [PubMed] [Google Scholar]
- 320. Renouf V., Claisse O., Lonvaud-Funel A. 2007. Inventory and monitoring of wine microbial consortia. Appl. Microbiol. Biotechnol. 75:149–164 [DOI] [PubMed] [Google Scholar]
- 321. Requena N., Jimenez I., Toro M., Barea J. M. 1997. Interactions between plant-growth-promoting rhizobacteria (PGPR), arbuscular mycorrhizal fungi and Rhizobium spp. in the rhizosphere of Anthyllis cytisoides, a model legume for revegetation in Mediterranean semi-arid ecosystems. New Phytol. 136:667–677 [DOI] [PubMed] [Google Scholar]
- 322. Richards T. A., Dacks J. B., Jenkinson J. M., Thornton C. R., Talbot N. J. 2006. Evolution of filamentous plant pathogens: gene exchange across eukaryotic kingdoms. Curr. Biol. 16:1857–1864 [DOI] [PubMed] [Google Scholar]
- 323. Riedlinger J., et al. 2006. Auxofuran, a novel metabolite that stimulates the growth of fly agaric, is produced by the mycorrhiza helper bacterium Streptomyces strain AcH 505. Appl. Environ. Microbiol. 72:3550–3557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324. Rikhvanov E. G., Varakina N. N., Sozinov D. Y., Voinikov V. K. 1999. Association of bacteria and yeasts in hot springs. Appl. Environ. Microbiol. 65:4292–4293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325. Rikkinen J., Oksanen I., Lohtander K. 2002. Lichen guilds share related cyanobacterial symbionts. Science 297:357. [DOI] [PubMed] [Google Scholar]
- 326. Rohm B., Scherlach K., Mobius N., Partida-Martinez L. P., Hertweck C. 2010. Toxin production by bacterial endosymbionts of a Rhizopus microsporus strain used for tempe/sufu processing. Int. J. Food Microbiol. 136:368–371 [DOI] [PubMed] [Google Scholar]
- 327. Roller S. 1999. Physiology of food spoilage organisms. Int. J. Food Microbiol. 50:151–153 [DOI] [PubMed] [Google Scholar]
- 328. Romano J., Kolter R. 2005. Pseudomonas-Saccharomyces interactions: influence of fungal metabolism on bacterial physiology and survival. J. Bacteriol. 187:940–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329. Rousk J., et al. 2010. Soil bacterial and fungal communities across a pH gradient in an arable soil. ISME J. 4:1340–1351 [DOI] [PubMed] [Google Scholar]
- 330. Sajben E., Manczinger L., Nagy A., Kredics L., Vagvolgyi C. 2010. Characterization of pseudomonads isolated from decaying sporocarps of oyster mushroom. Microbiol. Res. 166:255–267 [DOI] [PubMed] [Google Scholar]
- 331. Salvioli A., et al. 2010. Endobacteria affect the metabolic profile of their host Gigaspora margarita, an arbuscular mycorrhizal fungus. Environ. Microbiol. 12:2083–2095 [DOI] [PubMed] [Google Scholar]
- 332. Sanguin H., Sarniguet A., Gazengel K., Moenne-Loccoz Y., Grundmann G. L. 2009. Rhizosphere bacterial communities associated with disease suppressiveness stages of take-all decline in wheat monoculture. New Phytol. 184:694–707 [DOI] [PubMed] [Google Scholar]
- 333. Santos A., et al. 2009. Application of molecular techniques to the elucidation of the microbial community structure of antique paintings. Microb. Ecol. 58:692–702 [DOI] [PubMed] [Google Scholar]
- 334. Sarand I., et al. 1998. Microbial biofilms and catabolic plasmid harbouring degradative fluorescent pseudomonads in Scots pine mycorrhizospheres developed on petroleum contaminated soil. FEMS Microbiol. Ecol. 27:115–126 [Google Scholar]
- 335. Sarniguet A., Lucas P., Lucas M., Samson R. 1992. Soil conduciveness to take-all of wheat: influence of the nitrogen fertilizers on the structure of populations of fluorescent pseudomonads. Plant Soil 145:29–36 [Google Scholar]
- 336. Sathe S. J., Nawani N. N., Dhakephalkar P. K., Kapadnis B. P. 2007. Antifungal lactic acid bacteria with potential to prolong shelf-life of fresh vegetables. J. Appl. Microbiol. 103:2622–2628 [DOI] [PubMed] [Google Scholar]
- 337. Sato Y., et al. 2009. Detection of Betaproteobacteria inside the mycelium of the fungus Mortierella elongata. Microbes Environ. 25:321–324 [DOI] [PubMed] [Google Scholar]
- 338. Sbrana C., et al. 2002. Diversity of culturable bacterial populations associated to Tuber borchii ectomycorrhizas and their activity on T. borchii mycelial growth. FEMS Microbiol. Lett. 211:195–201 [DOI] [PubMed] [Google Scholar]
- 339. Sbrana C., et al. 2000. Adhesion to hyphal matrix and antifungal activity of Pseudomonas strains isolated from Tuber borchii ascocarps. Can. J. Microbiol. 46:259–268 [DOI] [PubMed] [Google Scholar]
- 340. Scanlan P. D., Marchesi J. R. 2008. Micro-eukaryotic diversity of the human distal gut microbiota: qualitative assessment using culture-dependent and -independent analysis of faeces. ISME J. 2:1183–1193 [DOI] [PubMed] [Google Scholar]
- 341. Scherlach K., Partida-Martinez L. P., Dahse H. M., Hertweck C. 2006. Antimitotic rhizoxin derivatives from a cultured bacterial endosymbiont of the rice pathogenic fungus Rhizopus microsporus. J. Am. Chem. Soc. 128:11529–11536 [DOI] [PubMed] [Google Scholar]
- 342. Scherwinski K., Grosch R., Berg G. 2008. Effect of bacterial antagonists on lettuce: active biocontrol of Rhizoctonia solani and negligible, short-term effects on nontarget microorganisms. FEMS Microbiol. Ecol. 64:106–116 [DOI] [PubMed] [Google Scholar]
- 343. Schmitt I., Lumbsch H. T. 2009. Ancient horizontal gene transfer from bacteria enhances biosynthetic capabilities of fungi. PLoS One 4:e4437 doi:10.1371/journal.pone.0004437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344. Schmitt I., et al. 2008. Evolution of host resistance in a toxin-producing bacterial-fungal alliance. ISME J. 2:632–641 [DOI] [PubMed] [Google Scholar]
- 345. Schneider T., et al. 2010. Plectasin, a fungal defensin, targets the bacterial cell wall precursor lipid II. Science 328:1168–1172 [DOI] [PubMed] [Google Scholar]
- 346. Schnürer J., Magnusson J. 2005. Antifungal lactic acid bacteria as biopreservatives. Trends Food Sci. Technol. 16:70–78 [Google Scholar]
- 347. Schoonbeek H. J., Jacquat-Bovet A. C., Mascher F., Metraux J. P. 2007. Oxalate-degrading bacteria can protect Arabidopsis thaliana and crop plants against Botrytis cinerea. Mol. Plant Microbe Interact. 20:1535–1544 [DOI] [PubMed] [Google Scholar]
- 348. Schoonbeek H. J., Raaijmakers J. M., De Waard M. A. 2002. Fungal ABC transporters and microbial interactions in natural environments. Mol. Plant Microbe Interact. 15:1165–1172 [DOI] [PubMed] [Google Scholar]
- 349. Schouten A., Maksimova O., Cuesta-Arenas Y., van den Berg G., Raaijmakers J. M. 2008. Involvement of the ABC transporter BcAtrB and the laccase BcLCC2 in defence of Botrytis cinerea against the broad-spectrum antibiotic 2,4-diacetylphloroglucinol. Environ. Microbiol. 10:145–157 [DOI] [PubMed] [Google Scholar]
- 350. Schouten A., et al. 2004. Defense responses of Fusarium oxysporum to 2,4-diacetylphloroglucinol, a broad-spectrum antibiotic produced by Pseudomonas fluorescens. Mol. Plant Microbe Interact. 17:1201–1211 [DOI] [PubMed] [Google Scholar]
- 351. Schrey S. D., et al. 2007. Interaction with mycorrhiza helper bacterium Streptomyces sp. AcH 505 modifies organisation of actin cytoskeleton in the ectomycorrhizal fungus Amanita muscaria (fly agaric). Curr. Genet. 52:77–85 [DOI] [PubMed] [Google Scholar]
- 352. Schrey S. D., Schellhammer M., Ecke M., Hampp R., Tarkka M. T. 2005. Mycorrhiza helper bacterium Streptomyces AcH 505 induces differential gene expression in the ectomycorrhizal fungus Amanita muscaria. New Phytol. 168:205–216 [DOI] [PubMed] [Google Scholar]
- 353. Schroeckh V., et al. 2009. Intimate bacterial-fungal interaction triggers biosynthesis of archetypal polyketides in Aspergillus nidulans. Proc. Natl. Acad. Sci. U. S. A. 106:14558–14563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354. Schulze J., Sonnenborn U. 2009. Yeasts in the gut: from commensals to infectious agents. Dtsch. Arztebl. Int. 106:837–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355. Schüßler A., Bonfante P., Schnepf E., Mollenhauer D., Kluge M. 1996. Characterization of the Geosiphon pyriforme symbiosome by affinity techniques: confocal laser scanning microscopy (CLSM) and electron microscopy. Protoplasma 190:53–67 [Google Scholar]
- 356. Schussler A., Martin H., Cohen D., Fitz M., Wipf D. 2006. Characterization of a carbohydrate transporter from symbiotic glomeromycotan fungi. Nature 444:933–936 [DOI] [PubMed] [Google Scholar]
- 357. Scott J. J., et al. 2008. Bacterial protection of beetle-fungus mutualism. Science 322:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358. Scupham A. J., et al. 2006. Abundant and diverse fungal microbiota in the murine intestine. Appl. Environ. Microbiol. 72:793–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359. Seidler R. J., Aho P. E., Raju P. N., Evans H. J. 1972. Nitrogen fixation by bacterial isolates from decay in living white fir trees [Abies concolor (Gord. and Glend.) Lindl.]. Microbiology 73:413–416 [Google Scholar]
- 360. Seigle-Murandi F., Guiraud P., Croize J., Falsen E., Eriksson K. L. 1996. Bacteria are omnipresent on Phanerochaete chrysosporium Burdsall. Appl. Environ. Microbiol. 62:2477–2481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361. Sen R. 2001. Microbial biofilms in the mycorrhizospheres of Scots pine: organization and functioning as an ‘external rumen/gut’ in boreal forest soils, abstr. MO.050, p. 121 Abstr. 9th Int. Symp. Microb. Ecol., Amsterdam, Netherlands, 26 to 31 August 2001 [Google Scholar]
- 362. Seneviratne G., Indrasena I. K. 2006. Nitrogen fixation in lichens is important for improved rock weathering. J. Biosci. 31:639–643 [DOI] [PubMed] [Google Scholar]
- 363. Seneviratne G., Tennakoon N. S., Weerasekara M. L. M. A. W., Nandasena K. A. 2006. Polyethylene biodegradation by a developed Penicillium-Bacillus biofilm. Curr. Sci. 90:20–21 [Google Scholar]
- 364. Seneviratne G., Zavahir J. S., Bandara W. M. M. S., Weerasekara M. L. M. A. W. 2008. Fungal-bacterial biofilms: their development for novel biotechnological applications. World J. Microbiol. Biotechnol. 24:739–743 [Google Scholar]
- 365. Sharma M., et al. 2008. Detection and identification of bacteria intimately associated with fungi of the order Sebacinales. Cell. Microbiol. 10:2235–2246 [DOI] [PubMed] [Google Scholar]
- 366. Shirtliff M. E., Peters B. M., Jabra-Rizk M. A. 2009. Cross-kingdom interactions: Candida albicans and bacteria. FEMS Microbiol. Lett. 299:1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367. Sieuwerts S., de Bok F. A., Hugenholtz J., van Hylckama Vlieg J. E. 2008. Unraveling microbial interactions in food fermentations: from classical to genomics approaches. Appl. Environ. Microbiol. 74:4997–5007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368. Siggers K. A., Lesser C. F. 2008. The yeast Saccharomyces cerevisiae: a versatile model system for the identification and characterization of bacterial virulence proteins. Cell Host Microbe 4:8–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369. Silverman R. J., Nobbs A. H., Vickerman M. M., Barbour M. E., Jenkinson H. F. 2010. Interaction of Candida albicans cell wall Als3 protein with Streptococcus gordonii SspB adhesin promotes development of mixed-species communities. Infect. Immun. 78:4644–4652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370. Singh B. K., et al. 2008. Relationship between assemblages of mycorrhizal fungi and bacteria on grass roots. Environ. Microbiol. 10:534–541 [DOI] [PubMed] [Google Scholar]
- 371. Sivan A., Chet I. 1989. The possible role of competition between Trichoderma harzianum and Fusarium oxysporum on rhizosphere colonization. Phytopathology 79:198–203 [Google Scholar]
- 372. Smith M. G., Des Etages S. G., Snyder M. 2004. Microbial synergy via an ethanol-triggered pathway. Mol. Cell. Biol. 24:3874–3884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373. Soler-Rivas C., Arpin N., Olivier J., Wichers H. 1999. WLIP, a lipodepsipeptide of Pseudomonas ‘reactans’, as inhibitor of the symptoms of the brown blotch disease of Agaricus bisporus. J. Appl. Microbiol. 86:635–641 [Google Scholar]
- 374. Soler-Rivas C., Jolivet S., Arpin N., Olivier J., Wichers H. 1999. Biochemical and physiological aspects of brown blotch disease of Agaricus bisporus. FEMS Microbiol. Rev. 23:591–614 [DOI] [PubMed] [Google Scholar]
- 375. Sonnenburg J. L., Angenent L. T., Gordon J. I. 2004. Getting a grip on things: how do communities of bacterial symbionts become established in our intestine? Nat. Immunol. 5:569–573 [DOI] [PubMed] [Google Scholar]
- 376. Spano G., Massa S. 2006. Environmental stress response in wine lactic acid bacteria: beyond Bacillus subtilis. Crit. Rev. Microbiol. 32:77–86 [DOI] [PubMed] [Google Scholar]
- 377. Stachelhaus T., Schneider A., Marahiel M. A. 1995. Rational design of peptide antibiotics by targeted replacement of bacterial and fungal domains. Science 269:69–72 [DOI] [PubMed] [Google Scholar]
- 378. Stomeo F., Portillo M., Gonzalez J. 2009. Assessment of bacterial and fungal growth on natural substrates: consequences for preserving caves with prehistoric paintings. Curr. Microbiol. 59:321–325 [DOI] [PubMed] [Google Scholar]
- 379. Tarkka M. T., Sarniguet A., Frey-Klett P. 2009. Inter-kingdom encounters: recent advances in molecular bacterium-fungus interactions. Curr. Genet. 55:233–243 [DOI] [PubMed] [Google Scholar]
- 380. Tilak K., Li C., Ho I. 1989. Occurrence of nitrogen-fixing Azospirillum in vesicular-arbuscular mycorrhizal fungi. Plant Soil 116:286–288 [Google Scholar]
- 381. Tolaas A. G. 1915. A bacterial disease of cultivated mushrooms. Phytopathology 5:51–54 [Google Scholar]
- 382. Toro M., Azcon R., Barea J. 1997. Improvement of arbuscular mycorrhiza development by inoculation of soil with phosphate-solubilizing rhizobacteria to improve rock phosphate bioavailability (32P) and nutrient cycling. Appl. Environ. Microbiol. 63:4408–4412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 383. Tucker C. M., Routien J. B. 1942. The mummy disease of cultivated mushrooms. Bull. Mo. Agric. Exp. Station 358:1–27 [Google Scholar]
- 384. Upadhaya S. D., Park M. A., Ha J. K. 2010. Mycotoxins and their biotransformation in the rumen: a review. Asian-Aust. J. Anim. Sci. 23:1250–1260 [Google Scholar]
- 385. Uroz S., Calvaruso C., Turpault M. P., Frey-Klett P. 2009. Mineral weathering by bacteria: ecology, actors and mechanisms. Trends Microbiol. 17:378–387 [DOI] [PubMed] [Google Scholar]
- 386. Uroz S., et al. 2007. Effect of the mycorrhizosphere on the genotypic and metabolic diversity of the bacterial communities involved in mineral weathering in a forest soil. Appl. Environ. Microbiol. 73:3019–3027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 387. Uroz S., Heinonsalo J. 2008. Degradation of N-acyl homoserine lactone quorum sensing signal molecules by forest root-associated fungi. FEMS Microbiol. Ecol. 65:271–278 [DOI] [PubMed] [Google Scholar]
- 388. Urzì C., De Leo F., Bruno L., Albertano P. 2010. Microbial diversity in paleolithic caves: a study case on the phototrophic biofilms of the cave of bats (Zuheros, Spain). Microb. Ecol. 60:116–129 [DOI] [PubMed] [Google Scholar]
- 389. Valaskova V., de Boer W., Gunnewiek P. J., Pospisek M., Baldrian P. 2009. Phylogenetic composition and properties of bacteria coexisting with the fungus Hypholoma fasciculare in decaying wood. ISME J. 3:1218–1221 [DOI] [PubMed] [Google Scholar]
- 390. van Rij E. T., Girard G., Lugtenberg B. J., Bloemberg G. V. 2005. Influence of fusaric acid on phenazine-1-carboxamide synthesis and gene expression of Pseudomonas chlororaphis strain PCL1391. Microbiology 151:2805–2814 [DOI] [PubMed] [Google Scholar]
- 391. Villegas J., Fortin J. A. 2002. Phosphorus solubilization and pH changes as a result of the interactions between soil bacteria and arbuscular mycorrhizal fungi on a medium containing NO3− as nitrogen source. Can. J. Bot. 80:571–576 [Google Scholar]
- 392. Vivas M., Sacristan M., Legaz M. E., Vicente C. 2010. The cell recognition model in chlorolichens involving a fungal lectin binding to an algal ligand can be extended to cyanolichens. Plant Biol. 12:615–621 [DOI] [PubMed] [Google Scholar]
- 393. Wade W. N., Beuchat L. R. 2003. Metabiosis of proteolytic moulds and Salmonella in raw, ripe tomatoes. J. Appl. Microbiol. 95:437–450 [DOI] [PubMed] [Google Scholar]
- 394. Wade W. N., Beuchat L. R. 2003. Proteolytic fungi isolated from decayed and damaged raw tomatoes and implications associated with changes in pericarp pH favorable for survival and growth of foodborne pathogens. J. Food Prot. 66:911–917 [DOI] [PubMed] [Google Scholar]
- 395. Wade W. N., Vasdinnyei R., Deak T., Beuchat L. R. 2003. Proteolytic yeasts isolated from raw, ripe tomatoes and metabiotic association of Geotrichum candidum with Salmonella. Int. J. Food Microbiol. 86:101–111 [DOI] [PubMed] [Google Scholar]
- 396. Wallace D. F., Cook S. R., Dickinson D. J. 2008. The role of non-decay microorganisms in the degradation of organic wood preservatives. Dev. Commer. Wood Preservatives 982:312–322 [Google Scholar]
- 397. Wargo M. J., Hogan D. A. 2006. Fungal-bacterial interactions: a mixed bag of mingling microbes. Curr. Opin. Microbiol. 9:359–364 [DOI] [PubMed] [Google Scholar]
- 398. Weller D. M. 1988. Biological control of soilborne plant pathogens in the rhizosphere with bacteria. Annu. Rev. Phytopathol. 26:379–407 [Google Scholar]
- 399. Weller D. M. 2007. Pseudomonas biocontrol agents of soilborne pathogens: looking back over 30 years. Phytopathology 97:250–256 [DOI] [PubMed] [Google Scholar]
- 400. Weller D. M., Raaijmakers J. M., Gardener B. B., Thomashow L. S. 2002. Microbial populations responsible for specific soil suppressiveness to plant pathogens. Annu. Rev. Phytopathol. 40:309–348 [DOI] [PubMed] [Google Scholar]
- 401. Wenzl P., Wong L., Kwang-won K., Jefferson R. A. 2005. A functional screen identifies lateral transfer of beta-glucuronidase (gus) from bacteria to fungi. Mol. Biol. Evol. 22:308–316 [DOI] [PubMed] [Google Scholar]
- 402. Whipps J. M. 2001. Microbial interactions and biocontrol in the rhizosphere. J. Exp. Bot. 52:487–511 [DOI] [PubMed] [Google Scholar]
- 403. Wichmann G., Sun J., Dementhon K., Glass N. L., Lindow S. E. 2008. A novel gene, phcA from Pseudomonas syringae induces programmed cell death in the filamentous fungus Neurospora crassa. Mol. Microbiol. 68:672–689 [DOI] [PubMed] [Google Scholar]
- 404. Wick L. Y., et al. 2007. Effect of fungal hyphae on the access of bacteria to phenanthrene in soil. Environ. Sci. Technol. 41:500–505 [DOI] [PubMed] [Google Scholar]
- 405. Wiesel I., Rehm H. J., Bisping B. 1997. Improvement of tempe fermentations by application of mixed cultures consisting of Rhizopus sp. and bacterial strains. Appl. Microbiol. Biotechnol. 47:218–225 [Google Scholar]
- 406. Wohl D. L., McArthur J. V. 2001. Aquatic actinomycete-fungal interactions and their effects on organic matter decomposition: a microcosm study. Microb. Ecol. 42:446–457 [DOI] [PubMed] [Google Scholar]
- 407. Wong W., Fletcher J., Unsworth B., Preece T. 1982. A note on ginger blotch, a new bacterial disease of the cultivated mushroom Agaricus bisporus. J. Appl. Bacteriol. 52:43–48 [Google Scholar]
- 408. Wood B. J. B. 1998. Microbiology of fermented foods, 2nd ed. Springer, Berlin, Germany [Google Scholar]
- 409. Wu X., et al. 2008. Saccharomyces boulardii ameliorates Citrobacter rodentium-induced colitis through actions on bacterial virulence factors. Am. J. Physiol. Gastrointest. Liver Physiol. 294:G295–G306 [DOI] [PubMed] [Google Scholar]
- 410. Yoo K. S., et al. 2010. Improvement in sensory characteristics of Campbell Early wine by adding dual starters of Saccharomyces cerevisiae and Oenococcus oeni. J. Microbiol. Biotechnol. 20:1121–1127 [DOI] [PubMed] [Google Scholar]
- 411. Zanello G., Meurens F., Berri M., Salmon H. 2009. Saccharomyces boulardii effects on gastrointestinal diseases. Curr. Issues Mol. Biol. 11:47–58 [PubMed] [Google Scholar]