Abstract
TMC310911 is a novel human immunodeficiency virus type 1 (HIV-1) protease inhibitor (PI) structurally closely related to darunavir (DRV) but with improved virological characteristics. TMC310911 has potent activity against wild-type (WT) HIV-1 (median 50% effective concentration [EC50], 14 nM) and a wide spectrum of recombinant HIV-1 clinical isolates, including multiple-PI-resistant strains with decreased susceptibility to currently approved PIs (fold change [FC] in EC50, >10). For a panel of 2,011 recombinant clinical isolates with decreased susceptibility to at least one of the currently approved PIs, the FC in TMC310911 EC50 was ≤4 for 82% of isolates and ≤10 for 96% of isolates. The FC in TMC310911 EC50 was ≤4 and ≤10 for 72% and 94% of isolates with decreased susceptibility to DRV, respectively. In vitro resistance selection (IVRS) experiments with WT virus and TMC310911 selected for mutations R41G or R41E, but selection of resistant virus required a longer time than IVRS performed with WT virus and DRV. IVRS performed with r13025, a multiple-PI-resistant recombinant clinical isolate, and TMC310911 selected for mutations L10F, I47V, and L90M (FC in TMC310911 EC50 = 16). IVRS performed with r13025 in the presence of DRV required less time and resulted in more PI resistance-associated mutations (V32I, I50V, G73S, L76V, and V82I; FC in DRV EC50 = 258). The activity against a comprehensive panel of PI-resistant mutants and the limited in vitro selection of resistant viruses under drug pressure suggest that TMC310911 represents a potential drug candidate for the management of HIV-1 infection for a broad range of patients, including those with multiple PI resistance.
INTRODUCTION
The introduction of more efficacious, convenient, safer, and better-tolerated antiretroviral (ARV) agents for the treatment of human immunodeficiency virus type-1 (HIV-1) infection over the past several years has significantly improved treatment outcomes, especially for highly treatment-experienced patients (30). Despite this remarkable success, emergence of resistance to these new ARV agents remains a critical factor in highly active antiretroviral therapy (HAART) failure. Hence, there remains a need for new, safer, more convenient ARV agents without significant drug-drug interactions and with high genetic barriers to resistance to further improve long-term treatment efficacy for patients with multidrug-resistant HIV-1 infection (36).
Darunavir (DRV) is used extensively as a first-line HIV-1 protease inhibitor (PI) for the treatment of drug-naive HIV-1-infected patients (22) and treatment-experienced patients (1, 21), including those who are resistant to multiple PIs. DRV has been reported to possess a high genetic barrier to the development of resistance (6) and to exhibit activity against HIV-1 isolates with a high number of PI resistance-associated mutations (RAMs). Analyses of the influence of baseline mutations on virological response and on in vitro susceptibility to DRV, and of mutations in patients experiencing virologic failure in the POWER 1, 2, and 3 and the DUET 1 and 2 trials, identified a set of 11 DRV RAMs (V11I, V32I, L33F, I47V, I50V, I54L and I54M, T74P, L76V, I84V, and L89V) that may reduce the susceptibility to DRV when present in combinations of 3 or more (7, 8).
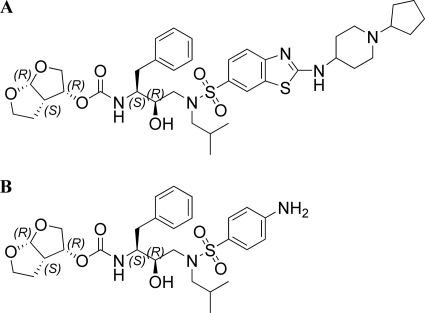
Although the population with high treatment experience is on the decline due to the increased effectiveness of treatment using recently approved ARVs, highly treatment-experienced patients with multidrug-resistant virus who fail multiple PI regimens have limited therapeutic options. Therefore, an internal HIV-1 protease research program was initiated to discover novel PIs with improved resistance profiles and limited selection of resistant virus under drug pressure. A series of fused heteroaromatic sulfonamides that showed extension into the P2′ pocket of the HIV-1 protease and exhibited exceptional activity against a panel of highly PI-cross-resistant mutants was discovered (32). The present report gives a virological characterization of TMC310911 (Fig. 1), a 2-(substituted-amino) benzothiazole sulfonamide, including details of antiviral activity, resistance profile for PI-resistant recombinant clinical isolates, and in vitro selection of resistant virus under drug pressure.
Fig. 1.
Structures of (A) TMC310911 and (B) darunavir.
MATERIALS AND METHODS
Compounds.
TMC310911 was prepared as reported previously for related structures (32). DRV was prepared as reported previously (33). Atazanavir (ATV) and tipranavir (TPV) were synthesized in-house. Amprenavir (APV), indinavir (IDV), lopinavir (LPV), and saquinavir (SQV) were purified from commercially available formulations.
Cells and viruses.
MT4 cells are human lymphoblastoic T cells that are permissive for HIV-1 infection and show a rapid and strong cytopathic effect (CPE). MT4-LTR-EGFP cells were generated by transfecting MT4 cells with a selectable construct encompassing the coding sequences for the HIV long terminal repeat (LTR) as a promoter for expression of enhanced green fluorescent protein (EGFP). Through subsequent selection, a stably transfected cell line was obtained. MT4-CMV-EGFP cells constitutively expressing the EGFP reporter under the control of a CMV promoter (the cytomegalovirus immediate-early [IE] promoter) were obtained by selection for permanently transformed MT4 cells with a CMV-EGFP reporter gene.
Fresh human peripheral blood mononuclear cells (PBMCs) were isolated from donors seronegative for HIV and hepatitis B virus (HBV) (Biological Specialty Corporation, Colmar, PA). Informed consent was not required (donors were compensated and identities were blinded). For use in drug susceptibility assays, PBMCs were purified and stimulated with phytohemagglutinin (4 μg/ml) for 48 to 72 h and further cultured in the presence of human interleukin 2 (20 U/ml) as described previously (9). Mature monocytes and macrophages (M/Ms) were isolated from PBMCs by adhesion as described previously (29). All cells were cultured in RPMI 1640 medium supplemented with fetal bovine serum combined with penicillin and streptomycin in a humidified incubator with a 5% CO2 atmosphere at 37°C. Virus stocks of HIV-1 and HIV-2 strains were harvested from MT4 cells, except for HIV-1 BaL, which was grown in M/Ms.
Recombinant clinical isolates.
Recombinant viruses, derived from clinical samples, were constructed by cotransfection of MT4 cells with patient-derived viral protease (PR) and reverse transcriptase (RT) coding sequences and an HIV-1 HXB2-derived proviral clone with deletions in the same regions as previously described (11).
Antiviral assays.
The activity of compounds against laboratory-adapted HIV strains and patient-derived recombinant viruses was evaluated using MT4-LTR-EGFP cells and a reporter gene assay (RGA). In short, various concentrations of test compounds were added to wells of a flat-bottom microtiter plate. Subsequently, virus and MT4 cells were added for final concentrations of 200 to 250 50% cell culture infectious doses (CCID50)/well and 30,000 cells/well, respectively. After 3 days of incubation (37°C and 5% CO2), the relative fluorescence of treated cultures was measured and compared with the relative fluorescence of untreated cultures. The results of antiviral assays were expressed as EC50 values, representing the concentration of a compound achieving 50% inhibition of infection compared to the drug-free control results, or as EC90 values, representing the dose achieving 90% inhibition of infection. In some cases, a fold change (FC) in susceptibility was calculated by dividing the EC50 for the tested virus by the EC50 for the wild-type (WT) virus (HIV-1/LAI) tested in parallel. The cytotoxicity of the test compound was determined in parallel with the antiviral activity. In short, compounds were added to MT4-CMV-EGFP cells and fluorescence was measured after 3 days of incubation (37°C and 5% CO2). Toxicity results were expressed as CC50 values representing the concentration of a compound that resulted in a 50% reduction in cell viability compared to the drug-free control results. The selectivity index (SI) was calculated as CC50/EC50.
The antiviral assays with PBMCs and M/Ms were carried out as previously described (9, 29). PBMCs were infected with HIV-1/LAI and HIV-1/SF2. Primary M/Ms were acutely infected with HIV-1/BaL at a multiplicity of infection (MOI) of ± 0.01. The viral replication was measured by a p24 enzyme-linked immunosorbent assay (ELISA).
Genotyping.
Genotypic analysis was performed by automated population-based sequencing. Sequence data were aligned to the WT HIV-1/HXB2CG reference (available in the GenBank database at http://www.ncbi.nlm.nih.gov/nuccore/K03455) and are reported as amino acid changes along the PR (11, 19).
In vitro resistance selection (IVRS).
HIV-1 was subjected to serial passages in MT4-LTR-EGFP cells in the presence of increasing concentrations of HIV-1 PIs. In short, cells were acutely infected with HIV-1 at an MOI of 0.001 to 0.01. Cells were resuspended in 10 ml of complete medium at a density of 1.5 × 105/ml. The concentrations of the compounds at passage 1 were 1 to 5 times the respective EC50 values. The cultures were subcultured and scored microscopically with respect to virus-induced fluorescence and cytopathicity every 3 to 4 days. Cultures were maintained in the presence of the same concentration of compound until the full virus CPE was observed and subsequently at a higher compound concentration to select for variants able to grow in the presence of the highest possible PI concentration.
Protein crystallography.
The WT HIV-1 protease used for crystallization was expressed, isolated, and purified from inclusion bodies as previously described (14). WT HIV-1 protease was further purified using a Pharmacia Superdex 75 fast-performance liquid chromatography column equilibrated with 0.05 M sodium acetate buffer at pH 5.5, containing 10% glycerol, 5% ethylene glycol, and 5 mM dithiothreitol, just before crystal trays were set. Cocrystals of the inhibitor with the WT protease were grown at room temperature by the hanging-drop vapor diffusion method. A protease concentration of 1.6 mg/ml with a 3-fold molar excess of inhibitors was used to set the crystallization drops, with the reservoir solution consisting of 126 mM phosphate buffer at pH 6.2, 63 mM sodium citrate, and 24 to 29% ammonium sulfate.
Intensity data on the protease crystals were collected at −80°C on an in-house Rigaku X-ray generator equipped with an R-axis IV image plate system. A total of 180 frames with an angular separation of 1° and without overlap between frames were collected. The data processing of the frames was carried out using the programs DENZO (23) and ScalePack (28). The crystal structure was solved and refined with the programs within the CCP4 interface (4). A structure solution for the WT protease-TMC310911 complex was obtained using the molecular replacement package AMoRe (27) and 1F7A as the starting model. After the solution was obtained, the molecular replacement phases were further improved using ARP/wARP (25) to build solvent molecules into the unaccounted regions of electron density. Model building was performed using the interactive graphics programs O (13) and Coot (10). Conjugate gradient refinement was performed using Refmac5 (26) by incorporating the Schomaker and Trueblood tensor formulation of TLS (translation, libration, screw-rotation) parameters (18). The working R (Rfactor) and its cross-validation (Rfree) were monitored throughout the refinement. The data collection and refinement statistics can be found in Table S1 in the supplemental material. The hydrogen bonding figure was made using PyMOL software (The PyMOL Molecular Graphics System, version 1.2r3pre; Schrödinger, LLC.).
Protein structure accession number.
The final coordinates and structure factors for TMC310911 in complex with WT HIV-1 protease have been deposited with the Protein Data Bank (accession number 3R4B).
RESULTS
Activity against WT HIV.
The in vitro activity of TMC310911 against laboratory HIV strains was evaluated using acutely infected MT4 cells, PBMCs, and M/M cells. Results are presented in Table 1. The TMC310911 EC50 values against WT HIV-1 ranged from 2.2 to 14 nM, and the corresponding EC90 values ranged from 5.0 to 94.7 nM. The TMC310911 CC50 for MT4 cells was 9.9 μM (interquartile ranges [IQR] = 8.57 to 12.19), with a resulting SI of 692. The TMC310911 EC50 values increased by a median factor of 7 in the presence of 50% human serum (I. Dierynck; data not shown).
Table 1.
Activity of TMC310911 against WT HIVa
| Virus/strain | Cell | Assay | EC50 (nM) |
EC90 (nM) |
||||
|---|---|---|---|---|---|---|---|---|
| n | Median | IQR | n | Median | IQR | |||
| HIV-1/LAI | MT4 | RGAb | 60 | 14.2 | 7.9–19.0 | 57 | 94.7 | 27.5–311.1 |
| HIV-1/LAI | PBMC | p24c | 1 | 2.2 | NA | 1 | 7.0 | NA |
| HIV-1/SF2 | PBMC | p24 | 1 | 2.3 | NA | 1 | 5.0 | NA |
| HIV-1/BaL | M/M | p24 | 1 | 13.9 | NA | 1 | 72.7 | NA |
| HIV-2/ROD | MT4 | RGA | 2 | 2.7 | 2.3–2.9 | 1 | 6.4 | NA |
n, number of determinations; IQR, interquartile range; NA, not applicable.
RGA, determination of inhibition of HIV LTR-driven reporter gene expression as measured in a reporter gene assay.
p24, determination of inhibition of p24 antigen accumulation.
Activity against a panel of PI-resistant recombinant clinical isolates.
To assess the activity of TMC310911 against PI-resistant HIV-1, TMC310911 was used to challenge 3,444 recombinant clinical isolates of HIV-1 submitted for routine clinical testing with various degrees of genotypic diversity and phenotypic resistance as determined by the VircoType HIV-1 and Antivirogram methods, respectively. Since DRV was extensively evaluated in highly pretreated patients with PI mutant viruses, the composition of this panel of recombinant clinical isolates was biased toward viruses with genotypic and/or phenotypic determinants of resistance to DRV. The median number (range) of major PI RAMs (12) in the panel of clinical isolates was 4 (0 to 9), with 0 major PI RAMs in 13% of isolates, 1 to 2 major PI RAMs in 14% of isolates, 3 to 6 major PI RAMs in 66% of isolates, 7 major PI RAMs in 6% of isolates, and 8 to 9 major PI RAMs in 1% of isolates. Individual major PI RAMs were present in 3% (D30N), 33% (V32I), 66% (M46I/L), 22% (I47A/V), 5.8% (G48V), 8.7% (I50L/V), 31% (I54L/M), 21% (Q58E), 11% (T74P), 6.5% (L76V), 47% (V82A/F/S/L/T), 46% (I84V), 0.8% (N88S), and 62% (L90M) of the clinical isolates.
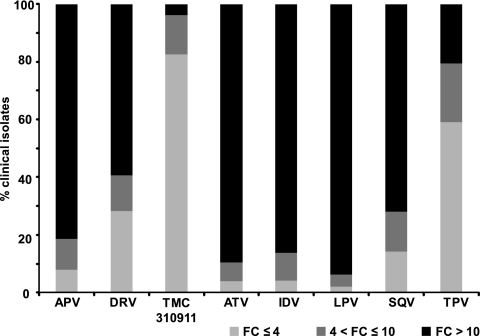
Among the analyzed samples with an Antivirogram phenotype available for all currently approved PIs (APV, ATV, DRV, IDV, LPV, SQV, and TPV; n = 2,402), 2,011 (84%) showed decreased susceptibility to at least one of these PIs, with an EC50 FC of >10. Only 3.8% (76/2,011) of these samples showed an EC50 FC of >10 for TMC310911, in comparison to 81%, 90%, 59%, 86%, 94%, 72%, and 21% for APV, ATV, DRV, IDV, LPV, SQV, and TPV, respectively (Fig. 2). The multiple-PI-resistant viruses with a TMC310911 EC50 FC > 10 contained ≥11 PI RAMs.
Fig. 2.
Activity of different PIs against a series (n = 2,011) of recombinant clinical isolates with various levels of genotypic diversity and phenotypic susceptibility and an EC50 FC > 10 for at least 1 of the currently approved PIs (APV, ATV, DRV, IDV, LPV, SQV and TPV). Antivirogram clinical cutoff (CCO): DRV FC = 10, LPV FC = 10, TPV FC = 3. Antivirogram biological cutoff (BCO): APV FC = 2.2, ATV FC = 2.1, IDV FC = 2.3, SQV FC = 1.8.
In further analyses of the antiviral activity of TMC310911, an EC50 FC > 4 and an EC50 FC > 10 were used as criteria to describe potential resistance to TMC310911. The TMC310911 EC50 FC was ≤4 for most (82%; n = 1,658) of the 2,011 isolates with decreased susceptibility (FC > 10) to at least one of the current PIs and was between 4 and 10 for 14% (n = 277) of isolates (Fig. 2). For isolates with decreased susceptibility to the most-prescribed PIs, namely, DRV (EC50 FC > 10, representing the clinical cutoff [CCO]), LPV (EC50 FC > 10; CCO), and ATV (EC50 FC > 2.2, representing the biological cutoff [BCO]), 71% (879/1,233) and 94% (1,155/1,233) had TMC310911 EC50 FC values of ≤4 and ≤10, respectively.
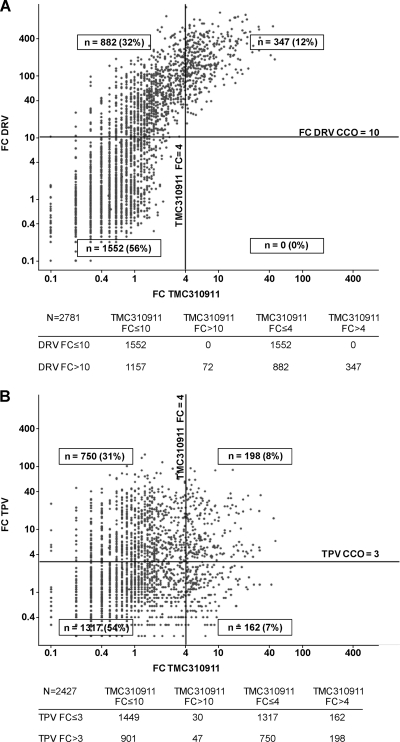
TMC310911 showed a more comprehensive antiviral inhibition of the diverse set of clinical isolates than the currently prescribed PIs, including DRV. As shown in Fig. 3A (data shown represent 2,781 samples with a DRV and TMC310911 phenotype available), TMC310911 exhibited EC50 FC values of ≤4 and ≤10 for 72% (882/1,229) and 94% (1,157/1,229), respectively, of samples with decreased susceptibility to DRV (EC50 FC > 10; CCO).
Fig. 3.
Comparison of the activity of TMC310911 against DRV (A) and TPV (B) determined with a series of recombinant clinical isolates (n = 2,781 [A] and 2,427 [B]) with various levels of genotypic diversity and phenotypic susceptibility (censored values are not shown). The Antivirogram clinical cutoff (CCO) for DRV (FC = 10) and TPV (FC = 3) are represented by horizontal lines; the TMC310911 FC value of 4 is represented by vertical lines.
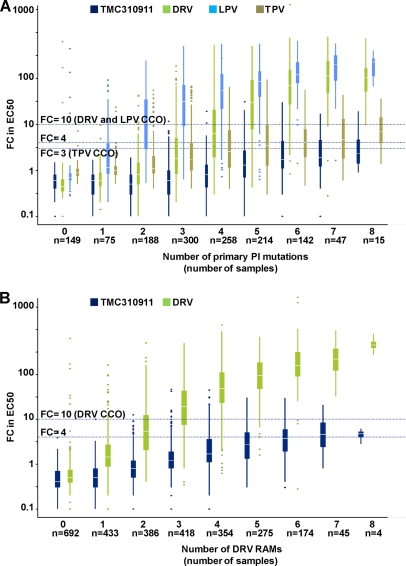
The antiviral activity of TMC310911 was affected to a small extent by the presence of an increasing number of major PI RAMs or DRV RAMs (12) compared with other PIs (Fig. 4). While the median FC value for DRV exceeded 10 (i.e., the DRV CCO) in the presence of 3 or more DRV RAMs, the median FC value for TMC310911 remained below 4 in the presence of 6 DRV RAMs and below 10 in the presence of 8 DRV RAMs (Fig. 4A). The median FC values for TMC310911 were 2.3 and 4.8 in the presence of 8 major PI RAMs and DRV RAMs, respectively. The increase in the median in the TMC310911 EC50 FC with increasing numbers of major PI RAMs was also lower than that observed for TPV (Fig. 4B), a PI that also has shown activity against many HIV-1 strains with multiple PI RAMs (20). TMC310911 had EC50 FC values ≤ 10 and ≤ 4 for 95% (901/948) and 79% (750/948), respectively, of clinical isolates with an EC50 FC value > 3 (i.e., the TPV CCO) for TPV (Fig. 3B; data represent 2,427 samples with TPV and TMC310911 phenotype available).
Fig. 4.
Activity (FC in EC50) against a series of recombinant clinical isolates with various levels of genotypic diversity and phenotypic susceptibility (censored values not shown) according to the number of (A) major PI RAMs for TMC310911, DRV, LPV and TPV (n = 1,388) and (B) DRV RAMs for TMC310911 and DRV (n = 2,781). Each box plot depicts the median value (horizontal lines), lower (Q1) and upper (Q3) quartile (squared bar), upper (largest value ≤ Q3 + 1.5 IQR) and lower (lowest value ≥ Q1 − 1.5 IQR) adjacent values (vertical lines), and outlier values (dots). The Antivirogram clinical cutoff (CCO) values for DRV and LPV (FC = 10) and TPV (FC = 3) and the FC value of 4 for TMC310911 are represented by horizontal lines. DRV RAMs: V11I, V32I, L33F, I47V, I50V, I54L/M, T74P, L76V, I84V, and L89V (12). Major PI RAMs: D30N, V32I, M46I/L, I47A/V, G48V, I50L/V, I54L/M, Q58E, T74P, L76V, V82A/F/L/S/T, I84V, N88S, and L90M (12).
IVRS performed with WT HIV-1/LAI.
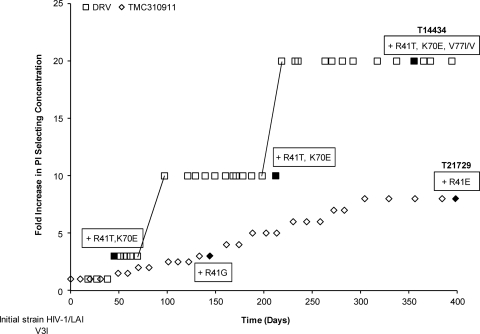
In order to study the development of resistance in the presence of TMC310911, IVRS experiments with WT HIV-1/LAI under pressure from TMC310911 were performed. As with IVRS performed with WT HIV-1 and DRV (6), selection of resistant viruses in the presence of TMC310911 was very difficult, as viruses were frequently found unable to replicate, resulting in the failure of the resistant mutant to propagate. A possible explanation might be the low replication capacity of the selected viruses. Although initial attempts to select for TMC310911-resistant virus were unsuccessful, IVRS performed with WT HIV-1/LAI subsequently resulted in selection of TMC310911-resistant virus. The results of a comparison of the data from this experiment to IVRS data from a DRV experiment are presented in Fig. 5 and Table 2. Under selective pressure from TMC310911, viruses harboring the R41G mutation and those harboring the R41E mutation were isolated at 60 and 160 nM TMC310911, respectively. The concentration of TMC310911 could not be increased above 160 nM even after prolonged culture at this concentration due to the inability of the resistant virus to propagate at higher concentrations of drug, resulting in an EC50 FC increase of 12.
Fig. 5.
In vitro selection of resistant HIV-1 starting from WT HIV-1/LAI in the presence of TMC310911 or DRV. Selection curves have been normalized, and starting selection concentrations were 20 and 10 nM for TMC310911 and DRV, respectively. Genotypes of virus strains selected at defined time points (indicated by filled symbols) list all changes from the starting strain HIV-1/LAI.
Table 2.
Phenotypic data showing PI susceptibility of the final selected virus strains starting from WT HIV-1/LAI as determined in IVRS experiments performed with TMC310911 and DRV
| Strain (compound) | Emerging protease mutation(s) | EC50 in nM (FC)a |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| TMC310911 | DRV | IDV | SQV | APV | ATV | LPV | TPV | ||
| T21729 (TMC310911) | R41E | 110 (12) | 99 (13) | 408 (8.0) | 313 (8.0) | 188 (3.8) | 85 (8.0) | 99 (10) | 2,587 (6.4) |
| T14434 (DRV) | R41T, K70E, V77I/V | 85 (22) | 62 (23) | 43 (3.2) | 24 (4.3) | 47 (3.2) | 46 (14) | 25 (6.4) | 175 (4.0) |
Median values of at least three EC50 determinations are presented. Values in parentheses represent FC in drug EC50 relative to the EC50 for HIV-1/LAI measured within the same antiviral experiment.
IVRS performed with r13025, a PI-resistant recombinant clinical isolate.
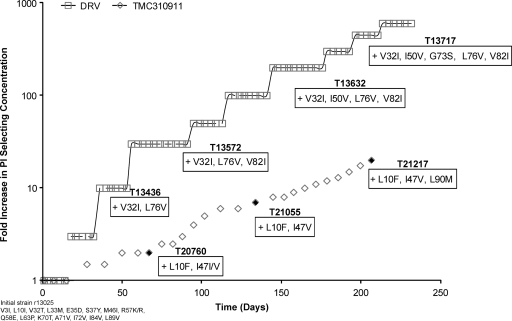
In order to determine whether strains with decreased sensitivity to TMC310911 could be generated from viruses harboring mutations conferring resistance to currently approved PIs, IVRS was performed with r13025, a multiple-PI-resistant clinical isolate (Fig. 6). The r13025 virus contained 7 PI RAMs (L10I, M46I, Q58E, L63P, A71V, I84V, and L89V) and showed decreased susceptibility to APV (FC = 20), ATV (FC = 238), IDV (FC = 98), SQV (FC = 28), and TPV (FC = 15) but not to TMC310911 (FC = 1.4), DRV (FC = 1.9) or LPV (FC = 1.4) (Table 3). The r13025 virus was passaged in increasing concentrations of TMC310911 (starting from 20 nM); after 66 days, a 2-fold increase in drug concentration was found to be associated with the emergence of mutations L10F and I47I/V (TMC310911 EC50 FC = 3.4). At the conclusion of IVRS (>200 days), the drug concentration was increased 20-fold and mutations L10F, I47V, and L90M appeared (strain T21217 in Fig. 6) and were found to be associated with a 12-fold increase in the TMC310911 EC50 (EC50 = 312 nM; FC = 16) in comparison with the value determined with the initial virus (Table 3). Strain T21217 showed a 4-fold or 7-fold decrease in susceptibility to DRV (FC = 7.4) or LPV (FC = 10), respectively, compared to the initial virus. IVRS performed with r13025 in the presence of TMC310911 was much slower than IVRS performed with r13025 and DRV (Fig. 6) or LPV (I. Dierynck; data not shown). Within a similar time frame of IVRS, the DRV concentration was increased >400-fold, resulting in the development of mutations V32I, I50V, G73S, L76V, and V82I and a 30-fold increase in the DRV EC50 (EC50 = 670 nM; FC = 258) compared with the DRV EC50 seen with the initial virus (Table 3). IVRS performed with r13025 and LPV resulted after 244 days in the selection of a virus containing the additional mutations L10F, L24I, V32I, I54V, G73S, and V82A (LPV FC = 324) and a 45-fold increase in drug concentration (I. Dierynck; data not shown).
Fig. 6.
In vitro selection of resistant HIV-1 starting from r13025, a PI-resistant recombinant clinical isolate, in the presence of TMC310911 or DRV. Selection curves have been normalized, and the starting selection concentration was 20 nM for TMC310911 and DRV. Genotypes of virus strains selected at defined time points (indicated by filled symbols) list the mutations that developed in the protease compared to the starting r13025 strain, which already contained 7 PI RAMs.
Table 3.
Phenotypic data showing PI susceptibility of the selected virus strains starting from r13205, a PI-resistant recombinant clinical isolate, in IVRS experiments performed with TMC310911 or DRV
| Strain | Emerging PR mutationsb | EC50 in nM (FC)a |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| TMC310911 | DRV | IDV | SQV | APV | ATV | LPV | TPV | ||
| r13205 | NA | 26 (1.4) | 22 (1.9) | 6,274 (98) | 818 (28) | 1,601 (20) | 4,000 (238) | 28 (1.4) | 729 (15) |
| TMC310911 expt | |||||||||
| T20760 | L10F, I47I/V | 65 (3.4) | 39 (3.4) | 3,530 (55) | 653 (23) | 1,496 (18) | 8,325 (496) | 112 (5.8) | 2,087 (4.4) |
| T21055 | L10F, I47V | 308 (16) | 122 (11) | 5,969 (94) | 1,915 (67) | 6,372 (78) | 9,601 (572) | 426 (22) | 13,903 (29) |
| T21217 | L10F, I47V, L90M | 312 (16) | 84 (7.4) | 5,406 (85) | 5,408 (188) | 3,650 (44) | 6,884 (410) | 198 (10) | 11,800 (25) |
| DRV expt | |||||||||
| T13436 | V32I, L76V | 5.1 (1.3) | 49 (21) | 28 (1.9) | 2.8 (0.5) | 2,037 (71) | 110 (33) | 263 (42) | 180 (2.9) |
| T13572 | V32I, L76V, V82I | 1.5 (3.7) | 234 (99) | 61 (4.1) | 10 (1.8) | 4,816 (169) | 317 (99) | 511 (81) | 240 (3.7) |
| T13632 | V32I, I50V, L76V, V82I | 42 (10) | 624 (265) | 107 (7.1) | 1.08 (0.2) | 12,111 (425) | 75 (20) | 1,597 (254) | 92 (1.4) |
| T13717 | V32I, I50V, G73S, L76V, V82I | 68 (17) | 670 (258) | 320 (27) | 4.2 (0.8) | 30,000 (1500) | 89 (34) | 1,900 (413) | 86 (1.3) |
Median values of at least three EC50 determinations are presented. Values in parentheses represent FC in drug EC50 relative to the EC50 for HIV-1/LAI measured within the same antiviral experiment.
Mutations that developed under selective drug pressure.
Structural basis of TMC310911 and WT HIV-1 protease interaction.
To enhance our understanding of the resistance profile of TMC310911 compared with DRV, we determined the crystal structure of TMC310911 in complex with the HIV-1 WT protease to 1.9Å. The TMC310911 complex crystallized in a P212121 space group with one protease dimer in the asymmetric unit and with a final Rfactor of 17.9 (Rfree, 22.7) (see Table S1 in supplemental material). The inhibitor was modeled in two alternate conformations in the active site, with 50% occupancy for each conformation. Each of the alternative conformations had continuous electron density for the inhibitor, except for partial density of the piperidine ring and no density of the cyclopentyl ring that was exposed to the solvent. Because of the disorder of these two rings in the crystal structure, the interactions shown by the piperidine ring and the cyclopentyl ring were not considered in analyzing the interactions of TMC310911 with the protease.
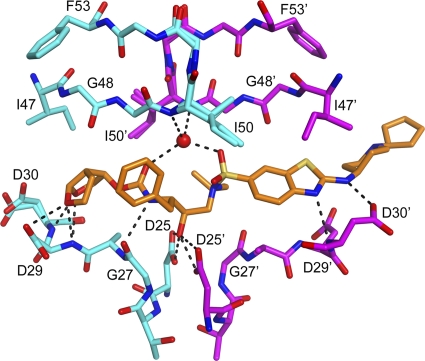
The two conformations of TMC310911 form similar hydrogen bond interactions with the protease, as shown in Fig. 7. The TMC310911-WT HIV-1 protease interactions were compared with those of DRV and WT HIV-1 protease as reported previously by us (15). Table 4 lists all the hydrogen bond interactions observed for DRV and the two conformations of TMC310911. Given the structural resemblance between DRV and TMC310911, the enzyme-inhibitor interactions were comparable. The major difference between the interactions of the WT protease with TMC310911 or DRV is that residue Asp30 supports two hydrogen bonds with the amine group of DRV whereas it forms one hydrogen bond with the nitrogen atom of benzothiazole in TMC310911 and another with the nitrogen atom of the amino group attached to the benzothiazole fragment. Although the interactions of TMC310911 are comparable to those of DRV, the additional interactions that can be formed by the piperidine ring may account for the improved resistance profile or limited selection of TMC310911-resistant virus over time.
Fig. 7.
The hydrogen bond network formed between TMC310911 and the WT HIV-1 protease. The hydrogen bonds formed by one conformation of TMC310911 are shown here. A second conformation resulted in a similar hydrogen-bonding network. The piperidine and cyclopentyl rings are not well ordered in either conformation.
Table 4.
Overview of the hydrogen bond interactions observed between the HIV-1 WT protease and DRV or two alternative TMC310911 conformers
| Protease atom or molecule | Inhibitor atom | Distance (Å) for: |
||
|---|---|---|---|---|
| DRV | TMC310911-1 | TMC310911-2 | ||
| Asp 29 N | O26 | 3.1 | 3.3 | 3.4 |
| Asp 30 N | O26 | 3.1 | 3.2 | 3.4 |
| Asp 29 N | O28 | 2.9 | 3.0 | 2.9 |
| Asp 29 OD2 | O28 | 3.3 | 3.2 | 3.5 |
| Water | O22 | 2.8 | 2.9 | 3.0 |
| Gly 27 O | N20 | 3.1 | 3.3 | 3.4 |
| Asp 25 OD1 | O18 | 2.6 | 2.6 | 3.0 |
| Asp 25 OD2 | O18 | 3.0 | 2.6 | 2.8 |
| Asp 25′ OD1 | O18 | 3.0 | 2.7 | 2.7 |
| Asp 25′ OD2 | O18 | 2.5 | 3.2 | 2.9 |
| Water | O9 | 2.9 | 2.4 | 2.5 |
| Water | N1 | 3.0 | ||
| Asp 30′ N | N1 | 3.4 | 3.2 | 3.0 |
| Asp 30′ O | N1 | 3.1 | ||
| Asp 30′ OD2 | N24 | 2.5 | 3.0 | |
DISCUSSION
The high degree of cross-resistance between most HIV-1 PIs often limits their sequential use in patients on PI-containing regimens who show virological failure. The introduction of DRV and TPV, two HIV-1 PIs with a high genetic barrier to development of resistance and activity against a wide range of PI-resistant HIV-1 strains, has significantly improved response rates in patients with a history of multiple PI failure. However, highly treatment-experienced patients failing a DRV-containing regimen have limited PI treatment options. TPV often exhibits activity against DRV-resistant isolates, and DRV often exhibits activity against TPV-resistant isolates, but in contrast to DRV, the poor tolerability of TPV has limited its use in treatment regimens (2). Hence, there is a need for well-tolerated PIs with activity against DRV- and TPV-resistant isolates that may prolong the use of this class of ARV agents.
In this study, we characterized the antiviral activity, resistance profile, and development of resistance to TMC310911, a novel PI belonging to the same chemical class as DRV. TMC310911 is a potent PI, with activity against WT HIV-1 at concentrations ranging from 2.2 to 14 nM. With few exceptions, this compound retained potent activity against a comprehensive panel of recombinant clinical isolates with decreased susceptibility to currently approved PIs, including DRV and TPV. The TMC310911 EC50 FC was >10 for only 3.8% of the multiple-PI-resistant viruses that contained ≥11 PI RAMs. The reduction in susceptibility corresponding to increasing numbers of major PI RAMs was limited and lower in magnitude for TMC310911 than for other PIs such as DRV and TPV with broad activity against multiple-PI-resistant strains. The median TMC310911 EC50 FC values were 2.3 and 4.8 in the presence of 8 major PI RAMs and DRV RAMs, respectively.
IVRS performed with WT HIV-1/LAI and the multiple-PI-resistant r13025 clinical isolate confirmed the limited emergence of resistant virus under pressure from TMC310911 over time. Starting with WT HIV-1, selection of resistant HIV strains by the use of APV, ATV, LPV, NFV, SQV, or TPV is usually easy to achieve, resulting in the emergence of strains carrying PI RAMs (6, 24), including mutations observed in the clinic with patients failing PI-containing treatment regimens. In contrast, drug concentrations could not be increased rapidly in IVRS performed with WT HIV-1 and TMC310911 or DRV, and it was not possible to observe virus replication at concentrations higher than 160 nM and 200 nM TMC310911 and DRV, respectively. Viruses containing a mutation at position 41 were selected for using both TMC310911 (R41E or R41G) and DRV (R41T) but have not been found to be associated with decreased susceptibility to PIs in clinical studies (12) and are present in only <0.05% of samples submitted for routine clinical testing (Virco BVBA; personal communication). The viruses selected under drug pressure from TMC310911 or DRV exhibited reduced susceptibility to both TMC310911 and DRV (FC > 10). It was shown using site-directed mutagenesis that the individual PR mutations selected during IVRS had no effect on susceptibility to DRV. However, several mutations accumulated in the GAG gene of the selected viruses and were found to be associated with decreased susceptibility to DRV in the corresponding GAG-PR recombinant viruses (5). These findings suggest that mutations in the GAG gene could be an implicating factor in the cross-resistance observed between DRV and TMC310911-selected viruses.
Starting from r13025, a multiple-PI-resistant clinical isolate, the development of resistant viruses in the presence of TMC310911 was reduced compared with DRV or LPV: the increase in the drug selection concentration was slower and fewer mutations appeared in the presence of TMC310911 compared with DRV or LPV. The mutations L10F, I47V, and L90M are known PI RAMs, and the resulting virus showed decreased susceptibility to all investigated PIs. Interestingly, strain T13717, selected under pressure from DRV, did not acquire resistance to IDV, ATV, SQV, or TPV but was more susceptible to those PIs than the initial r13025 strain. This could be explained by the selection of L76V, which can cause resensitization to ATV, SQV, and TPV and also to IDV, depending on the genetic mutation profile of the virus (3, 34, 35). TPV exhibits a resistance profile distinct from that of other currently available PIs (20); therefore, it was important to determine whether TMC310911 retained activity against TPV-resistant isolates. Our results showed that TMC310911 was active (FC ≤ 4) against the majority (79%) of clinical isolates with decreased susceptibility to TPV, highlighting TMC310911 as a potential drug candidate for treatment-experienced patients with resistance to TPV.
The genotypic and phenotypic determinants of DRV resistance that have been previously reported were confirmed in our experiments: phenotypic susceptibility to DRV decreased (FC > 10) in the presence of 3 or more DRV RAMs in a broad panel of recombinant clinical isolates, while most of the mutations (V32I, I50V, G73S, and L76V) that were selected when a multiple-PI-resistant clinical isolate was grown under pressure from DRV had previously been shown to be associated with reduced susceptibility to DRV (7, 8, 12). Mutation V82I does not affect susceptibility to DRV, but other mutations at this position (V82F and V82L) have been reported in a commercial database to confer resistance to DRV (31). Recently, selection of DRV-resistant variants by propagation of a mixture of HIV-1 variants isolated from patients failing a DRV-containing regimen was reported (16). Here, we showed that DRV-resistant virus could be generated using a single HIV-1 clinical isolate. The improved resistance profile or even reduced in vitro selection of viruses resistant to TMC310911 compared to DRV might be explained by an additional backbone interaction with Asp30 compared to DRV as observed in the crystal structure of the TMC310911-WT HIV-1 protease complex. Koh et al. have reported that, in addition to inhibiting the proteolytic activity of mature HIV-1 protease, DRV also inhibits the HIV-1 protease dimerization in vitro (17). This dual anti-HIV-1 function of DRV might play a role in its potent activity against multiple-PI-resistant isolates. Interactions of DRV involving Asp29, and not Asp30, were suggested to be linked with the observed dimerization inhibition. The effect of TMC310911 on HIV-1 protease dimerization has not been studied.
In addition to improved activity against multiple resistant viruses and a higher genetic barrier to the development of resistance, new HIV-1 PIs should also have an acceptable oral pharmacokinetic profile in order to lower pill burden, thereby improving convenience and adherence, as is often the case for recently approved ARVs. A good safety and tolerability profile is also desired, but this study did not address the safety and tolerability of TMC310911. Data from a proof-of-concept trial indicate the potential for once-daily dosing with TMC310911, and clinical studies evaluating this are ongoing (R. Verloes, unpublished data).
In conclusion, our findings suggest that TMC310911 exhibits broader in vitro activity against a diverse set of recombinant clinical isolates, including multiple-PI-resistant samples, and has a higher genetic barrier to the development of resistant virus in vitro than any of the currently approved PIs, including DRV. These properties support the evaluation of TMC310911 as a candidate drug suitable initially for highly treatment-experienced patients and subsequently for first-line treatment.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge the contribution of Piet Wigerinck and Herman de Kock in the discovery of TMC310911. We also gratefully acknowledge Virco BVBA for performing the antiviral analyses.
M.N.L.N. and C.A.S. were supported by the National Institutes of Health (grant P01-GM66524) and Tibotec/COSAT.
Footnotes
Supplemental material for this article may be found at http://aac.asm.org/.
Published ahead of print on 6 September 2011.
REFERENCES
- 1. Arastéh K., et al. 2009. Efficacy and safety of darunavir/ritonavir in treatment-experienced HIV type-1 patients in the POWER 1, 2 and 3 trials at week 96. Antivir. Ther. 14:859–864 [DOI] [PubMed] [Google Scholar]
- 2. Arribas J. R. 2009. Drugs in traditional drug classes (nucleoside reverse transcriptase inhibitor/nonnucleoside reverse transcriptase inhibitor/protease inhibitors) with activity against drug-resistant virus (tipranavir, darunavir, etravirine). Curr. Opin. HIV AIDS 4:507–512 [DOI] [PubMed] [Google Scholar]
- 3. Braun P., et al. 2007. Clinically relevant resensitization of protease inhibitors (PIs) saquinavir and atazanavir (ATV) by L76V mutations in multidrug-resistant HIV-1-infected patients, abstr. 129. Antivir. Ther. 12:S142 [Google Scholar]
- 4. Collaborative Computational Project, Number 4 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50:760–763 [DOI] [PubMed] [Google Scholar]
- 5. De Meyer S., et al. 2006. The pathway leading to TMC114 resistance is different for TMC114 compared with other protease inhibitors, abstr. 19. Antivir. Ther. 11:S24 [Google Scholar]
- 6. De Meyer S., et al. 2005. TMC114, a novel human immunodeficiency virus type 1 protease inhibitor active against protease inhibitor-resistant viruses, including a broad range of clinical isolates. Antimicrob. Agents Chemother. 49:2314–2321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. De Meyer S., et al. 2008. Phenotypic and genotypic determinants of resistance to darunavir: analysis of data from treatment-experienced patients in POWER 1, 2, 3 and DUET-1 and 2, abstr. 31. Antivir. Ther. 13(Suppl. 3):A33 [Google Scholar]
- 8. De Meyer S., et al. 2008. Resistance profile of darunavir: combined 24-week results from the POWER trials. AIDS Res. Hum. Retroviruses 24:379–388 [DOI] [PubMed] [Google Scholar]
- 9. Division of AIDS, National Institute of Allergy and Infectious Diseases 1997. DAIDS virology manual for HIV laboratories. Publication NIH-97-3828. U.S. Department of Health and Human Services, Washington, DC [Google Scholar]
- 10. Emsley P., Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126–2132 [DOI] [PubMed] [Google Scholar]
- 11. Hertogs K., et al. 1998. A rapid method for simultaneous detection of phenotypic resistance to inhibitors of protease and reverse transcriptase in recombinant human immunodeficiency virus type 1 isolates from patients treated with antiretroviral drugs. Antimicrob. Agents Chemother. 42:269–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Johnson V. A., et al. 2009. Update of the drug resistance mutations in HIV-1: December 2009. Top. HIV Med. 17:138–145 [PubMed] [Google Scholar]
- 13. Jones T. A., Bergdoll M., Kjeldgaard M. 1990. O: a macromolecular modeling environment, p. 189–195In Bugg C., Ealick S.(ed.), Crystallographic and modeling methods in molecular design. Springer-Verlag Press, Berlin, Germany [Google Scholar]
- 14. King N. M., et al. 2002. Lack of synergy for inhibitors targeting a multi-drug-resistant HIV-1 protease. Protein Sci. 11:418–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. King N. M., et al. 2004. Structural and thermodynamic basis for the binding of TMC114, a next-generation human immunodeficiency virus type 1 protease inhibitor. J. Virol. 78:12012–12021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Koh Y., et al. 2010. In vitro selection of highly darunavir-resistant and replication-competent HIV-1 variants by using a mixture of clinical HIV-1 isolates resistant to multiple conventional protease inhibitors. J. Virol. 84:11961–11969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Koh Y., et al. 2007. Potent inhibition of HIV-1 replication by novel non-peptidyl small molecule inhibitors of protease dimerization. J. Biol. Chem. 282:28709–28720 [DOI] [PubMed] [Google Scholar]
- 18. Kuriyan J., Weis W. I. 1991. Rigid protein motion as a model for crystallographic temperature factors. Proc. Natl. Acad. Sci. U. S. A. 88:2773–2777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Larder B. A., et al. 1993. Quantitative detection of HIV-1 drug resistance mutations by automated DNA sequencing. Nature 365:671–673 [DOI] [PubMed] [Google Scholar]
- 20. Luna B., Townsend M. U. 2007. Tipranavir: the first nonpeptidic protease inhibitor for the treatment of protease resistance. Clin. Ther. 29:2309–2318 [DOI] [PubMed] [Google Scholar]
- 21. Madruga J. V., et al. 2007. Efficacy and safety of darunavir-ritonavir compared with that of lopinavir-ritonavir at 48 weeks in treatment-experienced, HIV-infected patients in TITAN: a randomised controlled phase III trial. Lancet 370:49–58 [DOI] [PubMed] [Google Scholar]
- 22. Mills A. M., et al. 2009. Once-daily darunavir/ritonavir vs. lopinavir/ritonavir in treatment-naive, HIV-1-infected patients: 96-week analysis. AIDS 23:1679–1688 [DOI] [PubMed] [Google Scholar]
- 23. Minor W. 1993. XDISPLAYF (program). Purdue University, West Lafayette, IN [Google Scholar]
- 24. Mo H., et al. 2003. Characterization of resistant HIV variants generated by in vitro passage with lopinavir/ritonavir. Antiviral Res. 59:173–180 [DOI] [PubMed] [Google Scholar]
- 25. Morris R. J., Perrakis A., Lamzin V. S. 2002. ARP/wARP's model-building algorithms. I. The main chain. Acta Crystallogr. D Biol. Crystallogr. 58:968–975 [DOI] [PubMed] [Google Scholar]
- 26. Murshudov G. N., Vagin A. A., Dodson E. J. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53:240–255 [DOI] [PubMed] [Google Scholar]
- 27. Navaza J. 1994. AMoRe: an automated package for molecular replacement. Acta Cryst. A 50:157–163 [Google Scholar]
- 28. Otwinowski Z., Minor W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307–326 [DOI] [PubMed] [Google Scholar]
- 29. Perno C. F., Yarchoan R. 1993. Culture of HIV in monocytes and macrophages, p. 12.4.1–12.4.11 In Coligan J. E., Kruisbeek A. M., Margulies D. H., Shevach E. M., Strober W. (ed.), Current protocols in immunology, vol 3 John Wiley & Sons, New York, NY [Google Scholar]
- 30. Pomerantz R. J., Horn D. L. 2003. Twenty years of therapy for HIV-1 infection. Nat. Med. 9:867–873 [DOI] [PubMed] [Google Scholar]
- 31. Stawiski E., et al. 2010. Identification of novel mutations strongly associated with darunavir (DRV) and tipranavir (TPV) resistance and their trends in a commercial database, abstr. H-912. Abstr. 50th Intersci. Conf. Antimicrob. Agents Chemother., Boston, MA [Google Scholar]
- 32. Surleraux D. L. N. G., et al. 2005. Design of HIV-1 protease inhibitors active on multidrug-resistant virus. J. Med. Chem. 48:1965–1973 [DOI] [PubMed] [Google Scholar]
- 33. Surleraux D. L. N. G., et al. 2005. Discovery and selection of TMC114, a next generation HIV-1 protease inhibitor. J. Med. Chem. 48:1813–1822 [DOI] [PubMed] [Google Scholar]
- 34. Van Der Borght K., et al. 2006. The effects of the 76V mutation on protease inhibitor (PI) susceptibility are PI- and context-specific, abstr. THPE0038. Abstr. 16th Int. AIDS Conf., Toronto, Canada [Google Scholar]
- 35. Vermeiren H., et al. 2007. Prediction of HIV-1 drug susceptibility phenotype from the viral genotype using linear regression modeling. J. Virol. Methods 145:47–55 [DOI] [PubMed] [Google Scholar]
- 36. Wilson L. E., Gallant J. E. 2009. HIV/AIDS: the management of treatment-experienced HIV-infected patients: new drugs and drug combinations. Clin. Infect. Dis. 48:214–221 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.