Abstract
Pseudomonas aeruginosa can develop resistance to polymyxin and other cationic antimicrobial peptides. Previous work has shown that mutations in the PmrAB and PhoPQ regulatory systems can confer low to moderate levels of polymyxin resistance (MICs of 8 to 64 mg/liter) in laboratory and clinical strains of this organism. To explore the role of PhoPQ in high-level clinical polymyxin resistance, P. aeruginosa strains with colistin MICs > 512 mg/liter that had been isolated from cystic fibrosis patients treated with inhaled colistin (polymyxin E) were analyzed. Probable loss-of-function phoQ alleles found in these cystic fibrosis strains conferred resistance to polymyxin. Partial and complete suppressor mutations in phoP were identified in some cystic fibrosis strains with resistance-conferring phoQ mutations, suggesting that additional loci can be involved in polymyxin resistance in P. aeruginosa. Disruption of chromosomal phoQ in the presence of an intact phoP allele stimulated 4-amino-l-arabinose addition to lipid A and induced transcription from the promoter of the pmrH (arnB) operon, consistent with the known role of this lipid A modification in polymyxin resistance. These results indicate that phoQ loss-of-function mutations can contribute to high-level polymyxin resistance in clinical strains of P. aeruginosa.
INTRODUCTION
Pseudomonas aeruginosa is capable of causing substantial morbidity and mortality in individuals with compromised host defense mechanisms. Those with cystic fibrosis (CF) are particularly susceptible to P. aeruginosa infections of the respiratory tract (41). In this disorder, unusually viscous secretions are retained within the airways, providing an ecological niche conducive to growth of such opportunistic pathogens (38).
In many cases, P. aeruginosa isolated from the respiratory tract of a patient with CF represents a unique episode of adaptation by a strain acquired from an environmental reservoir (16). However, in some cases, epidemic strains of P. aeruginosa have been observed to spread through local, regional, national, or even international CF patient populations (1, 5, 10, 52, 55). Such CF epidemic strains often display phenotypes such as specific antibiotic resistance patterns (10, 52), increased virulence (1, 3, 5, 44, 54), and enhanced transmissibility (51). These characteristics likely promote P. aeruginosa persistence in the lungs of CF patients despite aggressive use of antipseudomonal antibiotic combinations. In the face of such selection pressure, many CF strains of P. aeruginosa eventually become multidrug resistant or even pan-drug resistant, mediated by diverse mechanisms of resistance and tolerance (4, 21, 32).
As a consequence, clinicians increasingly rely on second-line antipseudomonal agents such as polymyxin (Pm) (48). Pm represents a family of antimicrobial cyclic oligopeptides synthesized by the Gram-positive organism Bacillus polymyxa. The clinically available forms, polymyxin B (PMB) sulfate and colistimethate, a prodrug form of colistin (CST; also known as polymyxin E), are administered to CF patients intravenously or by inhalation. Pm binds to lipopolysaccharide (LPS), the major constituent of the Gram-negative outer membrane, thus promoting membrane permeabilization and diffusion of peptide across the periplasm. Pm insertion at the inner membrane disrupts cellular respiration and results in cell lysis and death (57).
The Gram-negative pathogens for which Pm is most commonly used in clinical practice are P. aeruginosa and Acinetobacter baumannii. Unfortunately, Pm-resistant (Pmr) clinical isolates of these organisms are increasingly being reported (2, 8, 15, 18, 30, 37, 40). Pmr strains are generally resistant to both PMB and CST. In P. aeruginosa, Pm resistance is associated with covalent addition of 4-amino-l-arabinose (L-Ara4N) to phosphate groups within the lipid A and core oligosaccharide moieties of LPS (7, 47, 50). Genes in the pmrHFIJKLME operon (also known as arnBCADTEFpmrE) encode enzymes responsible for the synthesis and transfer of L-Ara4N to lipid A (26, 28). This amino-sugar modification is thought to hinder charge interactions between phosphate groups within LPS and amino groups within the cyclic Pm oligopeptide.
In P. aeruginosa and some other Gram-negative organisms, the PmrAB two-component regulatory system activates transcription of the pmrHFIJKLME operon in response to antimicrobial peptide exposure or divalent cation depletion (26, 28, 45) or as a consequence of mutation (8, 47, 53). However, many Pmr CF isolates of P. aeruginosa have wild-type (WT) pmrAB alleles (8). We therefore hypothesized that mutations in additional regulatory systems stimulate the activation of the L-Ara4N operon in clinical Pm resistance. In this study, we used laboratory strains and CF isolates of P. aeruginosa to define mutation of the PhoPQ two-component regulatory system as an important mechanism of this resistance.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and genotyping.
Laboratory-adapted strains and clinical isolates of P. aeruginosa used in this study are listed in Table 1. Clinical isolates of P. aeruginosa were collected from the sputum of CF patients in Denmark. The Institutional Review Board of Seattle Children's Hospital and the Institutional Review Board of Massachusetts General Hospital reviewed and approved the use of the clinical isolates for this study. Escherichia coli strain DH5α was used as the host strain for manipulation of recombinant plasmids. P. aeruginosa and E. coli were grown at 37°C on Luria-Bertani (LB) agar plates or in LB broth with aeration. Antibiotics were used for selection at the following concentrations: kanamycin (50 mg/liter) or gentamicin (GEN) (10 mg/liter) for E. coli DH5α and GEN (100 mg/liter) for P. aeruginosa strain PAK and its derivatives. Strains were stored in a 16% glycerol-LB broth solution at −80°C. Genotypes of clinical isolates were determined by multilocus sequence typing (13); novel alleles and sequence types have been added to a P. aeruginosa database available at http://pubmlst.org/paeruginosa.
Table 1.
Laboratory-adapted strains and CF patient isolates of P. aeruginosa used in this work
| Strain no. | Designation (patient no.: yr of isolation) | Multilocus sequence type (ST) | CST MIC (mg/liter) | Strain or isolate origin or provider |
|---|---|---|---|---|
| Laboratory-adapted strains | ||||
| 1026 | PAK | 0.5 | S. Lory | |
| 1812 | PAK ΔpmrAB | 0.5 | This work | |
| 1995 | PAK ΔpmrABphoQ6 | 8 | This work | |
| 2248 | 1995 ΔphoPQ | 0.5 | This work | |
| 2244 | PAK ΔpmrAB ΔphoPQ | 2 | This work | |
| 2307 | PAK ΔpmrAB ΔphoQ | 8 | This work | |
| 2326 | PAK ΔphoQ | 32 | This work | |
| 2243 | PAK ΔphoPQ | 0.5 | This work | |
| 1555 | PAO1 | NDa | B. Iglewski | |
| 2898 | PAO1 ΔphoPQ Ω[attP::Φ(PpmrH-lacZ+)] | ND | This work | |
| Clinical isolates | ||||
| 2047 | CF isolate (patient 2: 1985) | ST387 | 1 | N. Høiby |
| 1016 | CF isolate (patient 2: 1996) | ST387 | >512 | N. Høiby |
| 1018 | CF isolate (patient 4: 1998) | ST387 | >512 | N. Høiby |
| 1019 | CF isolate (patient 5: 1998) | ST387 | >512 | N. Høiby |
| 1565 | CF isolate (patient 7: 2002) | ST387 | 8 | N. Høiby |
| 1576 | CF isolate (patient 8: 1999) | ST387 | 1 | N. Høiby |
| 1571 | CF isolate (patient 8: 2002) | ST387 | >512 | N. Høiby |
| 1579 | CF isolate (patient 9: 1999) | (ST387-related) | >512 | N. Høiby |
| 1581 | CF isolate (patient 9: 2001) | ST404 (ST387-related) | 0.5 | N. Høiby |
| 1577 | CF isolate (patient 9: 2002) | (ST387-related) | >512 | N. Høiby |
| 1585 | CF isolate (patient 10: 1995) | ST387 | 1 | N. Høiby |
| 1582 | CF isolate (patient 10: 2002) | ST387 | >512 | N. Høiby |
| 1590 | CF isolate (patient 11: 1992) | ST387 | 1 | N. Høiby |
| 1586 | CF isolate (patient 11: 2002) | ST387 | >512 | N. Høiby |
| 1597 | CF isolate (patient 12: 1999) | ST387 | >512 | N. Høiby |
| 1601 | CF isolate (patient 13: 1997) | ST387 | 0.25 | N. Høiby |
| 1021 | CF isolate (patient 13: 1998) | ST387 | >512 | N. Høiby |
| 1598 | CF isolate (patient 13: 2002) | ST387 | >512 | N. Høiby |
| 1604 | CF isolate (patient 14: 1991) | ST399 | >512 | N. Høiby |
| 1605 | CF isolate (patient 14: 1992) | ST400 (ST399 related) | 16 | N. Høiby |
| 1603 | CF isolate (patient 14: 2003) | ST398 (ST399 related) | >512 | N. Høiby |
ND, not determined.
Isolation of Pmr mutants.
A derivative of P. aeruginosa strain PAK from which the pmrAB locus had been deleted (strain 1812) was inoculated at a density of 109 CFU on 90-mm-diameter LB agar plates containing PMB (US Biochemical, Cleveland, OH) (50 to 100 mg/liter). Plates were incubated at 30°C overnight and then at ambient temperature (∼22°C) for up to 2 weeks and inspected daily for the appearance of Pmr colonies. Candidates were replated on PMB-containing LB agar to verify resistance. To assess phenotypic stability, putative PAK ΔpmrAB Pmr mutants were inoculated into LB broth without Pm, grown overnight to stationary phase, and then subjected to passage using fresh Pm-free culture medium on each of five consecutive days; after the fifth passage, each putative mutant was retested on PMB-containing LB agar.
Molecular methods.
Bacterial plasmids and chromosomal DNA were isolated using commercially available kits (QIAquick and DNeasy [Qiagen, Valencia, CA] and MasterPure [Epicentre Biotechnologies, Madison, WI]). Plasmids were introduced into P. aeruginosa by electroporation (11). Sequencing of plasmids and chromosomal DNA was performed on both strands by the use of oligonucleotide primers (see Table S1 in the supplemental material) spaced ∼400 bp apart. PCR amplification was performed with the following final concentrations or amounts: 10% dimethyl sulfoxide, 0.2 mM (each) deoxynucleotide triphosphate, 0.4 μM (each) oligonucleotide primer (see Table S2 and Table S3 in the supplemental material), approximately 10 ng of template DNA, 5 U of Pfu Turbo polymerase, and 1× Pfu reaction buffer (Stratagene, La Jolla, CA).
To construct deletions, ∼1-kb chromosomal DNA segments flanking the targeted locus were PCR amplified using chromosomal DNA and specific oligonucleotide primers (see Table S2 in the supplemental material) and joined by splicing-by-overlap-extension PCR (33); deletions were marked by inclusion of a unique restriction site in the overlapping (internal) primers. For constructs made using SS-series primers, the resulting ∼2-kb DNA fragment was inserted into the suicide plasmid pEX18Gm (34) by the use of XbaI and HinDIII restriction sites. For constructs made using SM-series primers, the DNA fragment was inserted into the Gateway entry vector pDONR201 (Gateway cloning system; Invitrogen, Carlsbad, CA) via a BP recombinase reaction and transferred into the Gateway destination vector pEXGmGW (60) via an LR recombinase reaction. Deletion construct plasmids introduced into P. aeruginosa were selected for chromosomal insertion on LB agar containing GEN and then counterselected for loss of the plasmid backbone on LB agar containing 5% sucrose (34). To confirm deletions, the region surrounding the target gene was PCR amplified from chromosomal DNA and digested with restriction endonucleases BamHI (for constructs made using SS-series primers) and HinDIII (for constructs made using SM-series primers) to detect the unique marker.
To construct expression plasmids, the target genes were PCR amplified using genomic DNA and specific oligonucleotide primers (see Table S3 in the supplemental material), and the Gateway cloning system was used to insert them into pJN105D, a version of plasmid pJN105 (49) that had been converted to a Gateway destination vector. Expression plasmids introduced into P. aeruginosa were selected on LB agar containing GEN.
Susceptibility testing.
Pm agar dilution testing was performed as described previously (12); a Nunc 96-pin replicator with 1-mm pins and an OmniTray copier were used to inoculate the surface of OmniTray plates (Nunc International, Rochester, NY) containing 2-fold serial dilutions (0.125 to 512 mg/liter) of CST sulfate salt (Sigma-Aldrich, St. Louis, MO) in Difco Mueller-Hinton agar (Becton Dickinson Diagnostic Systems, Sparks, MD). Consistent with current CLSI recommendations, a CST MIC of ≤2 mg/liter was interpreted as indicating Pm susceptibility (“S”), a CST MIC of 4 mg/liter was interpreted as indicating intermediate Pm susceptibility (“I”), and a CST MIC of ≥8 mg/liter was interpreted as indicating Pm resistance (“R”). For Pmr strains, a CST MIC of 8 to 32 mg/liter was interpreted as indicating low-level resistance, a CST MIC of 64 to 256 mg/liter was interpreted as indicating moderate-level resistance, and a CST MIC of ≥512 mg/liter was interpreted as indicating high-level resistance.
In an alternative PMB plate assay, bacterial strains were inoculated into LB broth containing 1 mM MgCl2 and grown at 37°C with aeration for 16 to 20 h to an optical density at 600 nm (OD600) of ∼3.0 to 5.0. The culture was diluted 1:50 into fresh medium and grown for 2 to 3 h at 30°C with aeration to an OD600 of ∼0.8. Dilutions containing ∼50 to 200 CFU per 0.1 ml were spread on LB agar plates containing 1 mM MgCl2 and PMB at 0 to 100 mg/liter. Susceptibility testing media used for strains with pJN105D-derived plasmids contained 0.1% l-arabinose and GEN or PMB. To estimate the PMB concentration representing a 50% lethal dose (LD50), percent mean survival at each PMB concentration was calculated in relation to the mean CFU on the PMB-free control plates. A PMB LD50 of <4 mg/liter was interpreted as indicating susceptibility (“S”), a PMB LD50 of ≥4 mg/liter but <10 was interpreted as indicating intermediate susceptibility (“I”), and a PMB LD50 of ≥10 mg/liter was interpreted as indicating resistance (“R”).
For quantitative bactericidal assays (47), strains were grown and subcultured as described above for the alternative PMB plate assay, diluted in Mueller-Hinton broth to a final density of 2 × 104 CFU per ml, exposed to 2-fold serial dilutions of PMB (0.5 to 512 mg/liter) as well as drug-free controls for 30 min at 37°C, spread on LB agar after 1:10 dilution, and incubated 16 to 20 h at 37°C for enumeration.
Lipid A isolation and analysis.
LPS was isolated after growth in LB broth supplemented with 1 mM MgCl2 (61). Lipid A was isolated from LPS by hydrolysis (9). Lipid A structure was analyzed using matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry in negative-ion mode (17). All MALDI-TOF analyses were performed using a Bruker Autoflex II mass spectrometer (Bruker Daltonics). The matrix used for lipid A analysis was 5-chloro-2-mercaptobenzothiazole (20 mg/liter in 1:1 chloroform:methanol).
Transcriptional analysis.
A P. aeruginosa reporter strain for transcriptional analysis was constructed in PAO1 (B. H. Iglewski). A lacZ fusion to the promoter of the pmrHFIJKLME operon (PA3552 to 3559 in the PAO1 genome) was constructed using a pMini-CTX::lacZ vector system (35). The region upstream of pmrHFIJKLME was amplified using specific oligonucleotide primers (see Table S4 in the supplemental material) and inserted into pMini-CTX::lacZ by the use of EcoRI and BamHI restriction sites. The fusion was integrated into the CTX site of the chromosome followed by removal of the plasmid backbone by the use of Flp recombinase. To confirm the reporter construct, the CTX region was PCR amplified from chromosomal DNA. Reporter strains carrying expression plasmids were grown for 16 to 20 h at 37°C with aeration in LB broth supplemented with 1 mM MgCl2 and GEN. Cultures were diluted 1:100 into fresh medium supplemented with 0.1% arabinose, grown for 90 min, and assayed for β-galactosidase activity (39).
Nucleotide sequence accession numbers.
The GenBank accession numbers for the new DNA sequences are JN868715 (phoPQ6 allele, strain 1995), JN868716 (phoPQ WT allele, strain 2047), JN868717 (phoPQ30 allele, strain 1016), JN868718 (phoPQ21 allele, strain 1018), JN868719 (phoP23Q23 allele, strain 1565), JN868720 (phoPQ25 allele, strain 1571), JN868721 (phoPQ22 allele, strain 1582), JN868722 (phoP24Q24 allele, strain 1604), JN868723 (pmrAB44 allele, strain 1018), and JN868724 (pmrAB34 allele, strain 1603).
RESULTS
Isolation and genetic analysis of a Pmr phoQ mutant in a ΔpmrAB strain of P. aeruginosa.
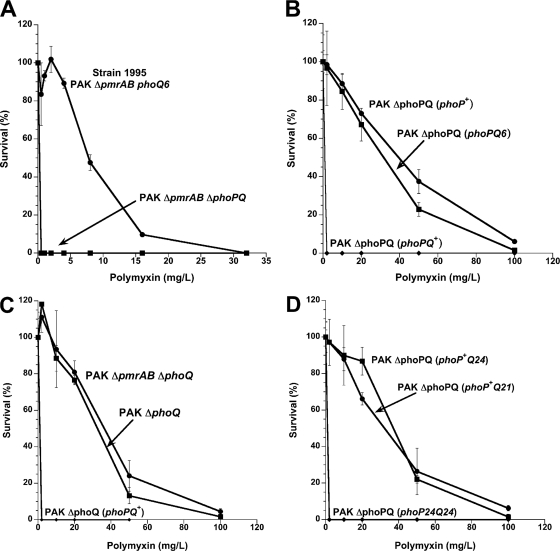
We previously showed that specific point mutations in pmrB, which encodes the sensor kinase component of the PmrAB regulatory system, conferred Pm resistance in a laboratory-adapted strain of P. aeruginosa (47). However, a survey of Pmr clinical isolates of P. aeruginosa from CF patients indicated that many do not have resistance-conferring mutations at the pmrAB locus (data not shown), suggesting that mutation of additional regulatory loci can confer clinical Pm resistance in this organism. To define such loci, we isolated spontaneous Pmr mutants of strain PAK ΔpmrAB by plating on LB agar containing PMB at 50 to 100 mg/liter. Of nine putative mutants, strain 1995 displayed the strongest and most reproducible Pm resistance (Fig. 1A). Interestingly, in contrast to Pmr pmrB mutants (47), strain 1995 had minimal cross-resistance to other cationic antimicrobial peptides (CAPs) such as protegrin-1 (56) and C18G (14) (Table 2).
Fig. 1.
Effect of phoPQ mutations on P. aeruginosa Pm resistance. (A) PMB quantitative bactericidal assay of strains 1995 (PAK ΔpmrAB phoQ6) and 2248 (PAK ΔpmrAB ΔphoPQ). (B) Alternative PMB plate assay of the PAK ΔphoPQ strain expressing the phoPQ+, phoPQ6, or phoP+ allele inserted in plasmid pJN105D. (C) Alternative PMB plate assay of chromosomal deletion mutants PAK ΔphoQ and PAK ΔpmrAB ΔphoQ as well as PAK ΔphoQ expressing the phoPQ+ alleles inserted in plasmid pJN105D. (D) Alternative PMB plate assay of the PAK ΔphoPQ strain expressing mutant phoPQ alleles from CF clinical isolates in plasmid pJN105D. The phoP+Q21 and phoP+Q24 alleles encode deletions from PhoQ of 2 and 276 amino acids, respectively, while the phoP24Q24 allele also encodes replacement in PhoP of Arg 118 with Cys.
Table 2.
Antimicrobial peptide resistance of a P. aeruginosa PAK ΔpmrAB phoQ6 mutant
| CAP | PAK ΔpmrAB phoQ+ LD50a (mg/liter) | PAK ΔpmrAB phoQ6 |
|
|---|---|---|---|
| LD50 (mg/liter) | Fold increase in Pm resistance | ||
| PMB | <0.5 | 8 | >16 |
| Protegrin-1 | 0.5 | 0.5 | 1.0 |
| C18G | 3 | 5 | 1.7 |
LD50, 50% lethal dose.
Disruption of the gene encoding the PhoQ sensor kinase (designated PA1180 in the PAO1 genome) is known to confer CAP resistance in P. aeruginosa (42, 43); in this context, PhoQ appears to act as a repressor of PhoP transcriptional activity (43). Deletion of the phoPQ locus in strain 1995 (PAK ΔpmrAB ΔphoPQ) resulted in loss of Pm resistance (Fig. 1A). The phoPQ alleles from wild-type PAK (designated phoPQ+) and strain 1995 (designated phoPQ6) were inserted into the broad-host-range expression plasmid pJN105D under the control of an arabinose-inducible promoter; expression of pJN105D::phoPQ6 in PAK ΔphoPQ conferred Pm resistance, whereas expression of pJN105D::phoPQ+ did not (Fig. 1B). Expression of pJN105D::phoPQ6 in wild-type PAK failed to confer Pm resistance (data not shown), indicating a recessive effect of phoPQ6 on Pm resistance.
Sequence analysis of the phoPQ6 allele from strain 1995 revealed a single nonsynonymous nucleotide change relative to strain PAK, namely, deletion of thymidine 1262 in the phoQ gene. This resulted in a frameshift of codons 421 to 428 and a premature stop at codon 429, with truncation of the 28 carboxy-terminal amino acids of the PhoQ protein. This truncation removes a portion of the G-box, a Gly-rich motif in the cytosolic domain of PhoQ that is important for nucleotide binding (20). The nucleotide sequences of the phoP gene in strains 1995, PAK, and PAO1 were identical (58, 59).
Effect of PhoQ truncation or deletion on polymyxin resistance.
To assess whether the frameshift associated with the phoQ6 mutant allele was functionally equivalent to carboxy-terminal truncation of PhoQ, we constructed additional phoQ mutant alleles encoding in-frame truncations (Table 3). The phoQ11 mutant allele was designed to encode conversion of Ile 421 (the first altered codon of the phoQ6 frameshift) to a stop codon (TGA), with loss of 28 carboxy-terminal residues. The phoQ12 mutant allele was similarly designed to encode conversion of Asp 433 to a stop codon (TGA), with retention of the native G-box and loss of only 16 carboxy-terminal residues, including six residues highly conserved among Salmonella enterica, E. coli, and P. aeruginosa. Expression of pJN105D::phoPQ11, pJN105D::phoPQ12, or pJN105D::phoP+ in strain PAK ΔphoPQ conferred Pm resistance at a level similar to expression of phoPQ6 (Table 3 and Fig. 1B). Strains PAK ΔpmrAB ΔphoQ (2307) and PAK ΔphoQ (2326) were created to assess the effect of single-copy chromosomal expression of PhoP in the absence of PhoQ. These strains exhibited constitutive Pm resistance that expression of pJN105D::phoPQ+ could repress (Fig. 1C). Interestingly, strain 1995 as well as the phoQ truncation mutants also displayed a small-colony morphology, reflecting a growth retardation phenotype; as with its effect in repressing Pm resistance, expression of WT phoQ from a plasmid restored these strains to the WT colony size (data not shown). However, several CF patient isolates of P. aeruginosa with small-colony morphology were found to be susceptible to Pm in the alternative PMB plate assay (data not shown), indicating that growth retardation per se does not account for the Pmr phenotype of the phoQ mutants.
Table 3.
phoPQ alleles and associated Pm susceptibility
| Allele designation | Strain no. | Amino acid change(s)a |
Pm susceptibilityb | |
|---|---|---|---|---|
| PhoP | PhoQ | |||
| phoPQ | WT | WT | S | |
| phoPQ6 | WT | frameshift at I421 (and 8 amino acids) | R | |
| phoPQ11 | WT | I421X | R | |
| phoPQ12 | WT | D433X | R | |
| phoPQ21 | 1018 (and 9 other isolates) | WT | ΔL364-G365 | R |
| phoPQ22 | 1582 | WT | ΔL364-G365; R444C | R |
| phoP23Q23 | 1565 | M175I | R6C; ΔL364-G365 | I |
| phoP24Q24 | 1604 (and 2 other isolates) | R118C | ΔV57-Q332 | S |
| phoPQ24 | WT | ΔV57-Q332 | R | |
| phoPQ25 | 1571 | WT | frameshift at V448 (and 30 amino acids) | S |
| phoPQ30 | 1016 | WT | E72G | S |
C, Cys; D, Asp; E, Glu; G, Gly; I, Ile; L, Leu; M, Met; Q, Gln; R, Arg; V, Val; X, Ter.
Alleles were inserted into pJN105D and expressed in PAK ΔphoPQ, with Pm susceptibility determined by alternative PMB plate assays and interpreted as defined in Materials and Methods. S, susceptible, I, intermediate, and R, resistant.
Analysis of phoPQ alleles in Pmr isolates of P. aeruginosa from CF patients.
To define the role of phoPQ mutations in clinical Pm resistance, we analyzed 15 Pmr and 6 Pm-susceptible (Pms) isolates of P. aeruginosa from 11 Danish CF patients who had been continuously treated with inhaled CST for up to 15 years. The phoPQ and pmrAB alleles from these isolates, which represented two distinct genotypic backgrounds (ST387 and ST399; Table 1), were PCR amplified and inserted into the pJN105D broad-host-range expression plasmid. For six of the patients, the analysis included both resistant and susceptible isolates derived from the ST387 genotypic background (Table 1).
A mutant allele with a six-nucleotide deletion (phoQ21) confers Pm resistance in an epidemic CF strain of P. aeruginosa.
The phoPQ allele from 10 Pmr isolates and 2 Pms isolates conferred resistance when expressed in strain PAK ΔphoPQ (Fig. 1D and data not shown). These 12 isolates, cultured from eight different CF patients, were ST387 derivatives with wild-type (i.e., non-resistance-conferring) pmrAB alleles (M. K. Brannon and S. M. Moskowitz, unpublished results). All 12 isolates possessed the same resistance-conferring phoQ mutation, a six-nucleotide deletion at positions 1091 to 1096 corresponding to in-frame deletion of Leu 364 and Gly 365. For eight Pm-resistant isolates (1018, 1019, 1021, 1577, 1579, 1586, 1597, and 1598) and both Pm-susceptible isolates (1581 and 1601), this was the only mutation found in the phoPQ locus; the allele was designated phoQ21 (Table 3). A plasmid bearing this allele, pJN105D::phoPQ21, conferred Pm resistance on PAK ΔphoPQ at a level similar to that conferred by pJN105D::phoPQ6. The two Pm-susceptible isolates carrying the phoQ21 allele presumably harbor unidentified secondary suppressor mutations in one or more loci required for PhoPQ-dependent Pm resistance. A third Pms isolate (1590) cultured from patient 11 10 years earlier than Pmr isolate 1586 had WT phoPQ. The pmrB gene in isolates with the phoQ21 allele (or with related alleles as described below) contained a single nucleotide mutation (Ala 211 to Val) and was designated pmrB44; a plasmid bearing the pmrAB44 allele failed to confer Pm resistance on wild-type strain PAK, indicating that this pmrB mutation represents a polymorphism.
Two resistance-conferring phoQ alleles are variants of phoQ21.
In addition to the Leu 364 and Gly 365 deletions, the phoQ gene of one Pmr isolate (1582) also contained a C-to-T transition at nucleotide 1330 of the WT sequence (designated phoQ22), corresponding to a change of Arg 444 to Cys (Table 3). A plasmid bearing this allele, pJN105D::phoPQ22, conferred Pm resistance on PAK ΔphoPQ at a level similar to that conferred by pJN105D::phoPQ21. The phoQ gene of another Pmr isolate (1565) also contained an A-to-T transversion at nucleotide 16 (designated phoQ23), corresponding to a change of Arg 6 to Cys, while the phoP gene contained a G-to-A transition at nucleotide 545 (designated phoP23), corresponding to a change of Met 175 to Ile. A plasmid bearing this allele, pJN105D::phoP23Q23, conferred Pm resistance on PAK ΔphoPQ at a lower level than the those bearing the phoQ6, phoQ21, or phoQ22 alleles (Table 3). Because pJN105D::phoP23 conferred Pm resistance on PAK ΔphoPQ at the same diminished level as that conferred by pJN105D::phoP23Q23, whereas pJN105D:: phoP+Q23 conferred Pm resistance at a level similar to that conferred by pJN105D::phoPQ21 (data not shown), the mutation of Met 175 to Ile in phoP23 represented a partial suppressor of phoQ23.
A phoP mutation completely suppresses the Pm resistance conferred by a phoQ mutant allele.
The phoQ gene of a Pmr isolate (1604) and two clonal Pmr isolates from the same patient (1603 and 1605; all ST399 derivatives) contained an in-frame deletion of nucleotides 169 through 996 (designated phoQ24), corresponding to removal of Val 57 through Gln 332. This deletion encompasses part of the periplasmic domain, the entirety of the second transmembrane domain, and the H-box motif of PhoQ. The phoP gene of these isolates had a C-to-T transition at nucleotide 354 (designated phoP24), corresponding to a change of Arg 118 to Cys. A plasmid bearing the phoQ24 allele in conjunction with wild-type phoP, pJN105D::phoP+phoQ24, conferred Pm resistance on PAK ΔphoPQ (Fig. 1D). In contrast, both pJN105D::phoP24Q24 and pJN105D::phoP24 failed to confer Pm resistance on PAK ΔphoPQ (data not shown), confirming that phoP24 is a complete suppressor of phoQ24. The pmrB gene in isolates with the phoP24Q24 allele contained a single nucleotide mutation resulting in a change of Ala 248 to Thr (designated pmrB34); a plasmid bearing the pmrAB34 allele failed to confer Pm resistance on wild-type strain PAK.
Some phoQ mutant alleles fail to confer Pm resistance on PAK ΔphoPQ.
The phoQ allele of one Pmr isolate (1571; an ST387 derivative) contained a frameshift mutation (deletion of T at nucleotide 1343; designated phoQ25) corresponding to a change of Val 448 (the carboxy-terminal residue) to Ala, loss of the normal opal termination codon, and addition of 30 amino acids to the C terminus of PhoQ before reaching an alternative termination codon; its phoP allele was WT. The phoQ allele of another Pm-resistant isolate (1016; also an ST387 derivative) contained an A-to-G transition at nucleotide 215 (designated phoQ30), corresponding to a change of Glu 72 to Gly; its phoP allele was WT. Pms isolates from the same patients (isolates 1576 and 2047, respectively) were both ST387 derivatives that contained wild-type phoQ alleles. Neither pJN105D::phoPQ25 nor pJN105D::phoPQ30 conferred Pm resistance on PAK ΔphoPQ (data not shown). Isolates 1571 and 1016 both contain resistance-conferring pmrAB alleles (S. M. Moskowitz et al., submitted for publication).
Lipid A modifications in PmrAB-independent Pmr strains of P. aeruginosa.
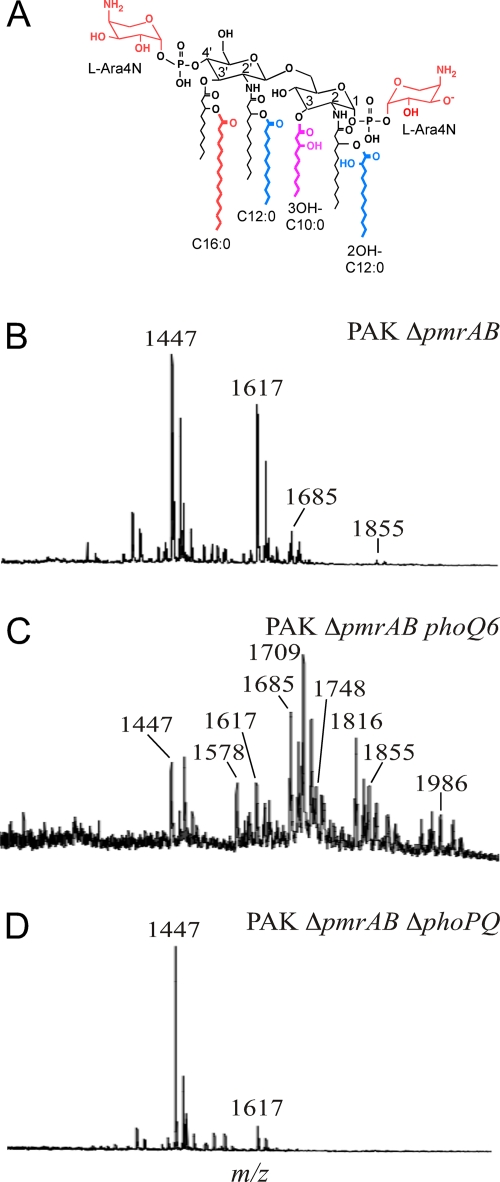
Addition of L-Ara4N to the 1 and 4′ phosphate groups of lipid A is associated with Pm resistance in many Gram-negative organisms (31). The pmrHFIJKLME operon encodes enzymes responsible for biosynthesis and attachment of L-Ara4N to lipid A (46). Other covalent modifications of P. aeruginosa lipid A (Fig. 2A) include an acyl-oxy-acyl addition of laurate (C12:0) to 3-hydroxylaurate at the 2 and 2′ positions by HtrB1 and HtrB2, 2-hydroxylation of the acyl-oxy-acyl laurates (2OH-C12:0) by LpxO1 and LpxO2, removal of 3-hydroxydecanoate (3OH-C10:0) from the 3 position by PagL, and acyl-oxy-acyl addition of palmitate (C16:0) to 3-hydroxydecanoate at the 3′ position by PagP (46). To assess the presence or absence of these modifications, lipid A was purified from Pmr and Pms P. aeruginosa strains and isolates and analyzed qualitatively by MALDI-TOF mass spectrometry.
Fig. 2.
Lipid A structures of Pms and Pmr strains. (A) Diagram of lipid A structural modifications found in P. aeruginosa. (B to D) MALDI-TOF mass spectra for (B) PAK ΔpmrAB (strain 1812), (C) PAK ΔpmrAB phoQ6 (strain 1995), and (D) PAK ΔpmrAB ΔphoPQ (strain 2244).
Lipid A from the PAK wild-type strain had major peaks at mass/charge ratios (m/z) 1,617 and 1,447 (data not shown), corresponding to hexa- and penta-acylated species that differ in the presence or absence of 3-hydroxydecanoate (Δm/z = 170). Lipid A from the PAK ΔpmrAB strain grown in LB broth with 1 mM Mg2+ had additional peaks at m/z 1,855 and 1,685 (Fig. 2B), corresponding to hepta- and hexa-acylated species of lipid A that contain palmitate (Δm/z = 238). Ernst et al. previously showed that Mg2+ depletion induces PhoPQ-dependent palmitoylation of lipid A (17); these results suggest that loss of PmrAB derepresses lipid A palmitoylation under noninducing (Mg2+-replete) conditions.
Lipid A from PAK ΔpmrAB phoQ6 had peaks corresponding to all of the species found in PAK ΔpmrAB (Fig. 2C), as well as peaks corresponding to addition of one or two L-Ara4N moieties (Δm/z = 131) to the following species: m/z 1,748 (1617 + L-Ara4N), 1,578 (1447 + L-Ara4N), 1,709 (1447 + 2[L-Ara4N]), 1,986 (1855 + L-Ara4N), and 1,816 (1685 + L-Ara4N). Lipid A from PAK ΔpmrAB ΔphoQ (strain 2307) similarly had both palmitate and L-Ara4N (Table 4). Interestingly, lipid A from PAK ΔphoQ (strain 2326) also had peaks corresponding to addition of both palmitate and L-Ara4N (data not shown), indicating that phoQ deletion promotes lipid A palmitoylation under noninducing conditions even in the presence of an intact PmrAB system. Conversely, lipid A from PAK ΔpmrAB ΔphoPQ (strain 2244) lacked both modifications (Fig. 2D), indicating that the effect of pmrAB deletion on lipid A palmitoylation is dependent on PhoP. In these laboratory strains, modification of lipid A with L-Ara4N consistently correlated with Pm resistance; in contrast, modification of lipid A with palmitate was seen in PAK ΔpmrAB, a strain that is Pm susceptible.
Table 4.
Pm susceptibility and lipid A modifications of P. aeruginosa phoPQ mutants
| Laboratory strain or clinical isolate(s) | phoPQ allele (pmrAB allele, laboratory strain) | Pm susceptibilitya | Lipid A modification |
|||
|---|---|---|---|---|---|---|
| Addition of L-Ara4N | Addition of C16:0 | Presence of 2OH-C12:0 | Removal of 3OH-C10:0 | |||
| 1026 | phoPQ+ (pmrAB+) | S | − | − | + | + |
| 1812 | phoPQ+ (ΔpmrAB) | S | − | + | + | + |
| 1995 | phoQ6 (ΔpmrAB) | R | + | + | + | + |
| 2307 | ΔphoQ (ΔpmrAB) | R | + | + | + | + |
| 2244 | ΔphoPQ (ΔpmrAB) | S | − | − | + | + |
| 2243 | ΔphoPQ (pmrAB+) | S | − | − | + | + |
| 2326 | ΔphoQ (pmrAB+) | R | + | + | + | + |
| 1018 | phoQ21 | R | + | + | + | + |
| 1597, 1021 | phoQ21 | R | + | + | − | + |
| 1019, 1586, 1598 | phoQ21 | R | + | − | − | + |
| 1581, 1601 | phoQ21 | S | − | + | − | + |
| 1582 | phoQ22 | R | + | + | − | + |
| 1565 | phoP23Q23 | R | + | + | − | + |
| 1603 | phoP24Q24 | R | + | + | − | + |
Data represent the results of measurements by colistin agar dilution. S, susceptible (CST MIC ≤ 2 mg/liter); R, resistant (CST MIC ≥ 8 mg/liter).
Among nine Pmr CF isolates of P. aeruginosa that carry phoQ21, phoQ22, or phoQ23 alleles, lipid A analysis demonstrated the addition of L-Ara4N to lipid A in all cases, and all but one of the isolates lacked 2-hydroxylaurate (Table 4). Six of the nine isolates displayed palmitate addition; all exhibited 3-hydroxydecanoate deacylase activity. Two Pms isolates that harbor the phoQ21 allele (1581 and 1601) displayed addition of palmitate but not L-Ara4N to lipid A; these isolates presumably have secondary suppressor mutations outside the phoPQ locus that interfere with this lipid A modification.
Effect of phoPQ alleles on transcriptional activation of the L-Ara4N operon.
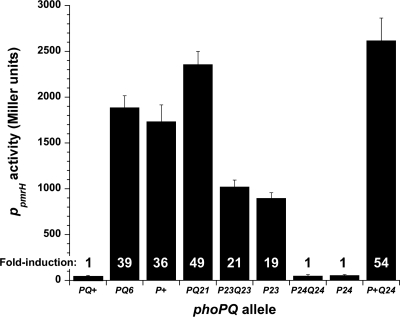
A transcriptional reporter was used to assess activation of the pmrHFIJKLME promoter by phoPQ mutant alleles. Plasmids encoding phoPQ alleles were expressed in a PAO1 ΔphoPQ strain in which a copy of the pmrH promoter region linked to a single-copy lacZ reporter had been inserted at the ΦCTX site {PAO1 ΔphoPQ Ω[attP::Φ(PpmrH-lacZ+)]}. The phoPQ6, phoP+, and phoPQ21 constructs conferred a high level of transcriptional activity, whereas the phoPQ+ construct conferred minimal transcriptional activity (Fig. 3). The phoP23Q23 and phoP23 constructs conferred an intermediate level of transcriptional activity, consistent with their intermediate level of Pm resistance. The phoP24Q24 and phoP24 constructs conferred minimal transcriptional activity, whereas the phoP+Q24 construct conferred a high level of transcriptional activity, consistent with the Pm resistance phenotypes conferred by these allelic combinations.
Fig. 3.
Transcriptional activity of phoPQ+, phoPQ6, phoP+, phoP+Q21, phoP23Q23, phoP23, phoP24Q24, phoP24, and phoP+Q24 alleles expressed from pJN105D in reporter strain PAO1 ΔphoPQ Ω[attP::Φ(PpmrH-lacZ+)]. Data shown represent the means of the results of triplicate biological experiments; error bars represent ± standard deviations. Fold induction values shown were calculated relative to the values determined for the WT phoPQ+ allele.
DISCUSSION
This work establishes phoQ disruption as a mechanism of both epidemic and sporadic Pm resistance in CF isolates of P. aeruginosa, thus complementing and extending previous reports of Pm resistance induced by phoQ mutation in non-CF and laboratory-adapted strains (6, 42, 43). Mutations disrupting the phoQ gene (e.g., small and large in-frame deletions, frameshifts, or truncations that removed as few as 16 C-terminal residues) resulted in a multifaceted phenotype that encompassed growth retardation, Pm resistance, and addition of palmitate and L-Ara4N to lipid A; phoQ mutation stimulated the latter LPS modification via induction of the pmrHFIJKLME operon, as has been previously observed (23). Mutation of phoQ in P. aeruginosa is associated with a variety of phenotypes that would seem to decrease its fitness for in vivo infection, including impaired twitching motility and decreased cytotoxicity (23). The data presented here indicate that these phenotypes do not substantially mitigate the potential for Pm resistance and epidemic spread of clinical phoQ mutants.
Mutant phoQ alleles conferred Pm resistance that was dependent on phoP. Unlike its activity in other Gram-negative pathogens (25), the PhoQ sensor kinase of P. aeruginosa appears to act as a repressor of PhoP transcriptional activity (43). In the absence of functional PhoQ, native or episomal expression of a WT phoP allele promotes addition of palmitate and L-Ara4N to lipid A and induces Pm resistance. This contrasts with S. enterica, in which phoP alleles with specific activating mutations induce Pm resistance and associated lipid A modifications but a WT phoP allele does not (24). Whether the transcriptionally active form of PhoP is phosphorylated in P. aeruginosa has not been determined; thus, it remains unclear whether PhoP repression by PhoQ is mediated by a phosphatase activity or a kinase activity.
Secondary suppressor mutations in the phoP gene were observed in some Pmr clinical isolates with phoQ mutations; these isolates presumably have resistance-conferring mutations in other regulatory loci. Conversely, in the case of CST-exposed but Pms clinical isolates with resistance-conferring phoPQ mutant alleles, we inferred the occurrence of secondary suppressor mutations in other loci, presumably components of the P. aeruginosa PhoPQ regulon (23), that appear to be required for Pm resistance and addition of L-Ara4N to lipid A.
The PhoPQ and PmrAB systems of P. aeruginosa regulate L-Ara4N addition and Pm resistance in a convergent or redundant fashion, in contrast to the hierarchical relationship in which PhoPQ regulates PmrAB in S. enterica (26–28). In P. aeruginosa, mutation of phoQ can confer Pm resistance regardless of the presence or absence of a functional PmrAB system. Our data indicate a correlation between the level of Pm resistance that phoPQ alleles confer (Fig. 1D) and the level of transcriptional activity that they induce at the L-Ara4N operon promoter (Fig. 3).
In P. aeruginosa, the PhoPQ and PmrAB systems appear to regulate palmitoylation of lipid A in an antagonistic fashion; these systems also behave divergently with respect to their ability to induce resistance to CAPs other than Pm. Specifically, PhoP activation induces acyl-oxy-acyl palmitoylation of the 3′ 3-hydroxydecanoate without appearing to promote resistance to CAPs other than Pm, whereas PmrAB activation represses palmitoylation while promoting CAP resistance (47). In contrast, PhoPQ activation in S. enterica induces acyl-oxy-acyl palmitoylation at position 2 of lipid A, a modification that promotes resistance to CAPs other than Pm (29). This suggests that the specific position of the palmitate addition within lipid A determines the effect of this modification on CAP resistance.
Our results also indicate that PhoPQ and PmrAB are not the only two-component regulatory systems influencing Pm resistance in P. aeruginosa. This was confirmed recently when a third system (ParRS) was shown to control inducible Pm resistance (19). Whether the PhoPQ or PmrAB systems modulate CAP resistance through interactions with ParRS or other regulatory systems remains to be determined.
Analysis of Pmr P. aeruginosa isolates collected from Danish CF patients revealed epidemic spread of a phoQ mutant strain as well as the occurrence of additional phoQ mutants in individual patients. A CF center in the United Kingdom has previously reported epidemic spread of a Pmr P. aeruginosa strain among four patients; two others at that center had strains with unique genotypes (15). In Denmark, continuous treatment of many CF patients with inhaled CST over 10 to 15 years starting in the early to mid-1980s likely provided selection pressure for the emergence and spread of Pmr P. aeruginosa strains (22, 36, 37).
The observation that some Danish CF patients were failing to respond clinically to continuous treatment with inhaled CST coincided with the detection of these phoQ mutant strains. CST inhalation was stopped for such patients, likely facilitating the emergence of Pms suppressor mutations in phoP and other loci. Such suppressor mutants readily outgrow phoQ strains that must divert a portion of their metabolic capacity to the modification of outer membrane components such as LPS. The subsequent resumption of CST inhalation in Danish CF patients with phoP suppressor mutants presumably provided renewed selection pressure that promoted reemergence of Pmr strains with mutations in other loci that regulate L-Ara4N modification and other resistance mechanisms.
In summary, this work has shown that mutation of the PhoPQ system is one of several regulatory mechanisms underlying clinical Pm resistance. As with mutation of the PmrAB system, addition of L-Ara4N to lipid A is a consistent biochemical consequence of such mutations and appears to be required for Pm resistance. However, the PhoPQ system itself is not required for Pm resistance, as demonstrated by clinical strains that have lost PhoPQ functionality through secondary suppressor mutations in phoP but nonetheless display a high-level resistance phenotype. Most alarmingly, this work has shown that Pmr phoQ mutant strains are capable of epidemic spread through CF clinical populations. This indicates the need for heightened infection control and clinical vigilance when CF patients are treated with inhaled CST or PMB for prolonged periods.
Supplementary Material
ACKNOWLEDGMENTS
We thank Miyuki Pier, Ulla Johansen, Pia Poss, and Helle Nordbjerg for technical assistance.
This work was supported by Public Health Service grants K08HL067903 to S.M.M. from the National Heart Lung and Blood Institute, R01AI067653 to S.M.M. from the National Institute of Allergy and Infectious Diseases, and R01AI030479 to S.I.M. from the National Institute of Allergy and Infectious Diseases. This work was also supported by grant MOSKOW01A1 to S.M.M. from the CF Foundation.
Footnotes
Supplemental material for this article may be found at http://aac.asm.org/.
Published ahead of print on 3 October 2011.
REFERENCES
- 1. Aaron S. D., et al. 2010. Infection with transmissible strains of Pseudomonas aeruginosa and clinical outcomes in adults with cystic fibrosis. JAMA 304: 2145–2153 [DOI] [PubMed] [Google Scholar]
- 2. Adams M. D., et al. 2009. Resistance to colistin in Acinetobacter baumannii associated with mutations in the PmrAB two-component system. Antimicrob. Agents Chemother. 53: 3628–3634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Al-Aloul M., et al. 2004. Increased morbidity associated with chronic infection by an epidemic Pseudomonas aeruginosa strain in CF patients. Thorax 59: 334–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Alonso A., Campanario E., Martinez J. L. 1999. Emergence of multidrug-resistant mutants is increased under antibiotic selective pressure in Pseudomonas aeruginosa. Microbiology 145(Pt. 10): 2857–2862 [DOI] [PubMed] [Google Scholar]
- 5. Armstrong D. S., et al. 2002. Detection of a widespread clone of Pseudomonas aeruginosa in a pediatric cystic fibrosis clinic. Am. J. Respir. Crit. Care Med. 166: 983–987 [DOI] [PubMed] [Google Scholar]
- 6. Barrow K., Kwon D. H. 2009. Alterations in two-component regulatory systems of phoPQ and pmrAB are associated with polymyxin B resistance in clinical isolates of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 53: 5150–5154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Boll M., Radziejewska-Lebrecht J., Warth C., Krajewska-Pietrasik D., Mayer H. 1994. 4-Amino-4-deoxy-l-arabinose in LPS of enterobacterial R-mutants and its possible role for their polymyxin reactivity. FEMS Immunol. Med. Microbiol. 8: 329–341 [DOI] [PubMed] [Google Scholar]
- 8. Brannon M. K., et al. 2005. Colistin-treated cystic fibrosis patients harbor polymyxin-resistant strains of Pseudomonas aeruginosa, some with mutation of PmrAB, a two-component regulator of lipid A structure. Abstr. Pediatr. Pulmonol. Suppl. 28, abstr. 289 [Google Scholar]
- 9. Caroff M., Tacken A., Szabo L. 1988. Detergent-accelerated hydrolysis of bacterial endotoxins and determination of the anomeric configuration of the glycosyl phosphate present in the “isolated lipid A” fragment of the Bordetella pertussis endotoxin. Carbohydr. Res. 175: 273–282 [DOI] [PubMed] [Google Scholar]
- 10. Cheng K., et al. 1996. Spread of beta-lactam-resistant Pseudomonas aeruginosa in a cystic fibrosis clinic. Lancet 348: 639–642 [DOI] [PubMed] [Google Scholar]
- 11. Choi K. H., Kumar A., Schweizer H. P. 2006. A 10-min method for preparation of highly electrocompetent Pseudomonas aeruginosa cells: application for DNA fragment transfer between chromosomes and plasmid transformation. J. Microbiol. Methods 64: 391–397 [DOI] [PubMed] [Google Scholar]
- 12. Clinical Laboratory Standards Institute 2006. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard, seventh ed. M7-A7. CLSI, Wayne, PA [Google Scholar]
- 13. Curran B., Jonas D., Grundmann H., Pitt T., Dowson C. G. 2004. Development of a multilocus sequence typing scheme for the opportunistic pathogen Pseudomonas aeruginosa. J. Clin. Microbiol. 42: 5644–5649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Darveau R. P., et al. 1992. Peptides related to the carboxyl terminus of human platelet factor IV with antibacterial activity. J. Clin. Invest. 90: 447–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Denton M., et al. 2002. Transmission of colistin-resistant Pseudomonas aeruginosa between patients attending a pediatric cystic fibrosis center. Pediatr. Pulmonol. 34: 257–261 [DOI] [PubMed] [Google Scholar]
- 16. Ernst R. K., et al. 2003. Genome mosaicism is conserved but not unique in Pseudomonas aeruginosa isolates from the airways of young children with cystic fibrosis. Environ. Microbiol. 5: 1341–1349 [DOI] [PubMed] [Google Scholar]
- 17. Ernst R. K., et al. 1999. Specific lipopolysaccharide found in cystic fibrosis airway Pseudomonas aeruginosa. Science 286: 1561–1565 [DOI] [PubMed] [Google Scholar]
- 18. Falagas M. E., Bliziotis I. A. 2007. Pandrug-resistant Gram-negative bacteria: the dawn of the post-antibiotic era? Int. J. Antimicrob. Agents 29: 630–636 [DOI] [PubMed] [Google Scholar]
- 19. Fernández L., et al. 2010. Adaptive resistance to the “last hope” antibiotics polymyxin B and colistin in Pseudomonas aeruginosa is mediated by the novel two-component regulatory system ParR-ParS. Antimicrob. Agents Chemother. 54: 3372–3382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Filippou P. S., Kasemian L. D., Panagiotidis C. A., Kyriakidis D. A. 2008. Functional characterization of the histidine kinase of the E. coli two-component signal transduction system AtoS-AtoC. Biochim. Biophys. Acta 1780: 1023–1031 [DOI] [PubMed] [Google Scholar]
- 21. Foweraker J. E., Laughton C. R., Brown D. F., Bilton D. 2009. Comparison of methods to test antibiotic combinations against heterogeneous populations of multiresistant Pseudomonas aeruginosa from patients with acute infective exacerbations in cystic fibrosis. Antimicrob. Agents Chemother. 53: 4809–4815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Frederiksen B., Koch C., Høiby N. 1997. Antibiotic treatment of initial colonization with Pseudomonas aeruginosa postpones chronic infection and prevents deterioration of pulmonary function in cystic fibrosis. Pediatr. Pulmonol. 23: 330–335 [DOI] [PubMed] [Google Scholar]
- 23. Gooderham W. J., et al. 2009. The sensor kinase PhoQ mediates virulence in Pseudomonas aeruginosa. Microbiology 155: 699–711 [DOI] [PubMed] [Google Scholar]
- 24. Gunn J. S., Ernst R. K., McCoy A. J., Miller S. I. 2000. Constitutive mutations of the Salmonella enterica serovar Typhimurium transcriptional virulence regulator phoP. Infect. Immun. 68: 3758–3762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gunn J. S., Hohmann E. L., Miller S. I. 1996. Transcriptional regulation of Salmonella virulence: a PhoQ periplasmic domain mutation results in increased net phosphotransfer to PhoP. J. Bacteriol. 178: 6369–6373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gunn J. S., et al. 1998. PmrA-PmrB-regulated genes necessary for 4-aminoarabinose lipid A modification and polymyxin resistance. Mol. Microbiol. 27: 1171–1182 [DOI] [PubMed] [Google Scholar]
- 27. Gunn J. S., Miller S. I. 1996. PhoP-PhoQ activates transcription of pmrAB, encoding a two-component regulatory system involved in Salmonella typhimurium antimicrobial peptide resistance. J. Bacteriol. 178: 6857–6864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gunn J. S., Ryan S. S., Van Velkinburgh J. C., Ernst R. K., Miller S. I. 2000. Genetic and functional analysis of a PmrA-PmrB-regulated locus necessary for lipopolysaccharide modification, antimicrobial peptide resistance, and oral virulence of Salmonella enterica serovar Typhimurium. Infect. Immun. 68: 6139–6146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Guo L., et al. 1998. Lipid A acylation and bacterial resistance against vertebrate antimicrobial peptides. Cell 95: 189–198 [DOI] [PubMed] [Google Scholar]
- 30. Hawley J. S., Murray C. K., Jorgensen J. H. 2008. Colistin heteroresistance in Acinetobacter and its association with previous colistin therapy. Antimicrob. Agents Chemother. 52: 351–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Helander I. M., Nummila K., Kilpelainen I., Vaara M. 1995. Increased substitution of phosphate groups in lipopolysaccharides and lipid A of polymyxin-resistant mutants of Salmonella typhimurium and Escherichia coli. Prog. Clin. Biol. Res. 392: 15–23 [PubMed] [Google Scholar]
- 32. Henrichfreise B., Wiegand I., Pfister W., Wiedemann B. 2007. Resistance mechanisms of multiresistant Pseudomonas aeruginosa strains from Germany and correlation with hypermutation. Antimicrob. Agents Chemother. 51: 4062–4070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ho S. N., Horton R. M. June 1991. Method for gene splicing by overlap extension using the polymerase chain reaction. U.S. patent 5023171
- 34. Hoang T. T., Karkhoff-Schweizer R. R., Kutchma A. J., Schweizer H. P. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212: 77–86 [DOI] [PubMed] [Google Scholar]
- 35. Hoang T. T., Kutchma A. J., Becher A., Schweizer H. P. 2000. Integration-proficient plasmids for Pseudomonas aeruginosa: site-specific integration and use for engineering of reporter and expression strains. Plasmid 43: 59–72 [DOI] [PubMed] [Google Scholar]
- 36. Jensen T., et al. 1987. Colistin inhalation therapy in cystic fibrosis patients with chronic Pseudomonas aeruginosa lung infection. J. Antimicrob. Chemother. 19: 831–838 [DOI] [PubMed] [Google Scholar]
- 37. Johansen H. K., Moskowitz S. M., Ciofu O., Pressler T., Høiby N. 2008. Spread of colistin resistant non-mucoid Pseudomonas aeruginosa among chronically infected Danish cystic fibrosis patients. J. Cyst. Fibros. 7: 391–397 [DOI] [PubMed] [Google Scholar]
- 38. Knowles M. R., Boucher R. C. 2002. Mucus clearance as a primary innate defense mechanism for mammalian airways. J. Clin. Invest. 109: 571–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kulasekara H. D., et al. 2005. A novel two-component system controls the expression of Pseudomonas aeruginosa fimbrial cup genes. Mol. Microbiol. 55: 368–380 [DOI] [PubMed] [Google Scholar]
- 40. Li J., et al. 2006. Heteroresistance to colistin in multidrug-resistant Acinetobacter baumannii. Antimicrob. Agents Chemother. 50: 2946–2950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lyczak J. B., Cannon C. L., Pier G. B. 2000. Establishment of Pseudomonas aeruginosa infection: lessons from a versatile opportunist. Microbes Infect. 2: 1051–1060 [DOI] [PubMed] [Google Scholar]
- 42. Macfarlane E. L., Kwasnicka A., Hancock R. E. 2000. Role of Pseudomonas aeruginosa PhoP-PhoQ in resistance to antimicrobial cationic peptides and aminoglycosides. Microbiology 146: 2543–2554 [DOI] [PubMed] [Google Scholar]
- 43. Macfarlane E. L., Kwasnicka A., Ochs M. M., Hancock R. E. 1999. PhoP-PhoQ homologues in Pseudomonas aeruginosa regulate expression of the outer-membrane protein OprH and polymyxin B resistance. Mol. Microbiol. 34: 305–316 [DOI] [PubMed] [Google Scholar]
- 44. McCallum S. J., et al. 2002. Spread of an epidemic Pseudomonas aeruginosa strain from a patient with cystic fibrosis (CF) to non-CF relatives. Thorax 57: 559–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. McPhee J. B., Lewenza S., Hancock R. E. 2003. Cationic antimicrobial peptides activate a two-component regulatory system, PmrA-PmrB, that regulates resistance to polymyxin B and cationic antimicrobial peptides in Pseudomonas aeruginosa. Mol. Microbiol. 50: 205–217 [DOI] [PubMed] [Google Scholar]
- 46. Moskowitz S. M., Ernst R. K. 2010. The role of Pseudomonas lipopolysaccharide in cystic fibrosis airway infection. Subcell. Biochem. 53: 241–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Moskowitz S. M., Ernst R. K., Miller S. I. 2004. PmrAB, a two-component regulatory system of Pseudomonas aeruginosa that modulates resistance to cationic antimicrobial peptides and addition of aminoarabinose to lipid A. J. Bacteriol. 186: 575–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Moskowitz S. M., et al. 2008. Shifting patterns of inhaled antibiotic use in cystic fibrosis. Pediatr. Pulmonol. 43: 874–881 [DOI] [PubMed] [Google Scholar]
- 49. Newman J. R., Fuqua C. 1999. Broad-host-range expression vectors that carry the L-arabinose-inducible Escherichia coli araBAD promoter and the araC regulator. Gene 227: 197–203 [DOI] [PubMed] [Google Scholar]
- 50. Nummila K., Kilpelainen I., Zahringer U., Vaara M., Helander I. M. 1995. Lipopolysaccharides of polymyxin B-resistant mutants of Escherichia coli are extensively substituted by 2-aminoethyl pyrophosphate and contain aminoarabinose in lipid A. Mol. Microbiol. 16: 271–278 [DOI] [PubMed] [Google Scholar]
- 51. Panagea S., Winstanley C., Walshaw M. J., Ledson M. J., Hart C. A. 2005. Environmental contamination with an epidemic strain of Pseudomonas aeruginosa in a Liverpool cystic fibrosis centre, and study of its survival on dry surfaces. J. Hosp. Infect. 59: 102–107 [DOI] [PubMed] [Google Scholar]
- 52. Pedersen S. S., Koch C., Høiby N., Rosendal K. 1986. An epidemic spread of multiresistant Pseudomonas aeruginosa in a cystic fibrosis centre. J. Antimicrob. Chemother. 17: 505–516 [DOI] [PubMed] [Google Scholar]
- 53. Roland K. L., Martin L. E., Esther C. R., Spitznagel J. K. 1993. Spontaneous pmrA mutants of Salmonella typhimurium LT2 define a new two-component regulatory system with a possible role in virulence. J. Bacteriol. 175: 4154–4164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Salunkhe P., et al. 2005. A cystic fibrosis epidemic strain of Pseudomonas aeruginosa displays enhanced virulence and antimicrobial resistance. J. Bacteriol. 187: 4908–4920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Scott F. W., Pitt T. L. 2004. Identification and characterization of transmissible Pseudomonas aeruginosa strains in cystic fibrosis patients in England and Wales. J. Med. Microbiol. 53: 609–615 [DOI] [PubMed] [Google Scholar]
- 56. Steinberg D. A., et al. 1997. Protegrin-1: a broad-spectrum, rapidly microbicidal peptide with in vivo activity. Antimicrob. Agents Chemother. 41: 1738–1742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Storm D. R., Rosenthal K. S., Swanson P. E. 1977. Polymyxin and related peptide antibiotics. Annu. Rev. Biochem. 46: 723–763 [DOI] [PubMed] [Google Scholar]
- 58. Stover C. K., et al. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406: 959–964 [DOI] [PubMed] [Google Scholar]
- 59. Winsor G. L., et al. 2011. Pseudomonas Genome Database: improved comparative analysis and population genomics capability for Pseudomonas genomes. Nucleic Acids Res. 39: D596–D600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wolfgang M. C., Lee V. T., Gilmore M. E., Lory S. 2003. Coordinate regulation of bacterial virulence genes by a novel adenylate cyclase-dependent signaling pathway. Dev. Cell 4: 253–263 [DOI] [PubMed] [Google Scholar]
- 61. Yi E. C., Hackett M. 2000. Rapid isolation method for lipopolysaccharide and lipid A from gram-negative bacteria. Analyst 125: 651–656 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.