Abstract
Acetyl coenzyme A carboxylase (ACC) is a key enzyme providing a substrate for mycolic acid biosynthesis. Although in vitro studies have demonstrated that the protein encoded by accD6 (Rv2247) may be a functional carboxyltransferase subunit of ACC in Mycobacterium tuberculosis, the in vivo function and regulation of accD6 in slow- and fast-growing mycobacteria remain elusive. Here, directed mutagenesis demonstrated that although accD6 is essential for M. tuberculosis, it can be deleted in Mycobacterium smegmatis without affecting its cell envelope integrity. Moreover, we showed that although it is part of the type II fatty acid synthase operon, the accD6 gene of M. tuberculosis, but not that of M. smegmatis, possesses its own additional promoter (Pacc). The expression level of accD6Mtb placed only under the control of Pacc is 10-fold lower than that in wild-type M. tuberculosis but is sufficient to sustain cell viability. Importantly, this limited expression level affects growth, mycolic acid content, and cell morphology. These results provide the first in vivo evidence for AccD6 as a key player in the mycolate biosynthesis of M. tuberculosis, implicating AccD6 as the essential ACC subunit in pathogenic mycobacteria and an excellent target for new antitubercular compounds. Our findings also highlight important differences in the mechanism of acetyl carboxylation between pathogenic and nonpathogenic mycobacterial species.
INTRODUCTION
One of the most interesting features of all mycobacteria is their thick and highly impermeable cell envelope (45). Besides peptidoglycan surrounding the cytoplasmic membrane, this complex lipid-rich structure consists of an outer layer of mycolic acids covalently linked to arabinogalactan. Mycolic acids are long-chain (C60-90), high-molecular-weight β-hydroxy fatty acids with a short alkyl branch (C24-26) in the α position (2, 9, 17, 39). The tightly packed layer of mycolate chains in the cell wall of Mycobacterium tuberculosis is responsible for its important characteristics, including resistance to chemical injury and dehydration, low permeability to hydrophobic antibiotics, and ability to form biofilms (19, 23, 24, 52). Furthermore, mycolic acids are considered major virulence effectors, allowing M. tuberculosis to persist within the host (6, 17, 24, 83).
The biosynthesis of mycolic acid is linked to the unusual presence of two fatty acid synthases in mycobacteria (see Fig. S1 in the supplemental material): the mammalian-type multienzyme fatty acid synthase I (FAS-I) and fatty acid synthase II (FAS-II), which is a set of autonomic enzymes similar to those found in other bacteria (8, 59, 68, 82). FAS-I catalyzes the de novo synthesis of short fatty acyl primers (typically C16-26), and the FAS-II system subsequently elongates these short chains into long-chain fatty acids (C48-56). Mycolic acid biosynthesis is also the target of powerful antitubercular drugs, such as isoniazid (INH), ethionamide, and thiolactomycin (3, 27, 31, 44, 53, 70, 72), and an emerging target for future inhibitors. Although we know the functions of the most important enzymes involved in the de novo synthesis and elongation of fatty acyl chains (7, 10, 14, 22, 35, 41, 42, 60, 66, 67, 79, 84), little is known about the initial steps within this pathway, such as the synthesis of malonyl coenzyme A (malonyl-CoA).
Malonyl-CoA, the universal substrate for the synthesis of mycolic and other fatty acids, is incorporated into the growing acyl chain during the repetitive cycle of FAS-I/FAS-II reactions (see Fig. S1 in the supplemental material). It is generated by the carboxylation of acetyl-CoA in a reaction catalyzed by acetyl-CoA carboxylase (ACC) (80). This irreversible, biotin- and ATP-dependent reaction consists of two catalytic steps: (i) the carboxylation of biotin to form carboxybiotin and (ii) the transfer of a carboxyl group from the biotin to a specific substrate, such as acetyl-CoA, to generate malonyl-CoA. Each half-reaction is catalyzed by a specific ACC subunit: the first step by biotin carboxylase (BC) and the second step by carboxyltransferase (CT). In mycobacteria and other bacteria, each catalytic subunit is encoded by a separate gene (16, 37).
Three genes potentially encoding biotin carboxylase (α-subunit), accA1 to accA3, and six genes believed to encode carboxyltransferase (β-subunit), accD1 to accD6, have been identified in the M. tuberculosis genome (15). Since the β-subunits confer the substrate specificity of ACC, the large number of accD genes in mycobacterial genomes may reflect the ability of mycobacteria to carboxylate not only acetyl-CoA but also several other distinct substrates, including the short acyl chains that serve as intermediates in the biosynthesis of complex mycobacterial (glyco)lipids. Transposon site hybridization (TraSH) analysis has shown that among the six carboxyltransferase genes in M. tuberculosis, accD4, accD5, and accD6 are essential for cell survival (63, 64, 65). To date, the roles of accD4 and accD5 in mycolic acid biosynthesis have been studied (see Fig. S1 in the supplemental material) (21, 28, 38, 51, 58), and the expression profiles of all accD family members in M. tuberculosis are known (18).
In the context of malonyl-CoA synthesis, we have focused our work on accD6, which remains the least characterized carboxyltransferase gene, despite its presumable role in mycolic acid biosynthesis. The role of accD6 (Rv2247) in M. tuberculosis mycolic acid biosynthesis has been predicted from its location in the FAS-II gene cluster (15). Daniel et al. confirmed that M. tuberculosis accD6 (accD6Mtb) is highly expressed during intensive mycolate biosynthesis and showed that the AccD6 and AccA3 proteins could together reconstitute an enzyme that is able to carboxylate acetyl-CoA in vitro (see Fig. S1 in the supplemental material) (18). However, due to the essential nature of accD6, the possible involvement of AccD6 in mycolic acid biosynthesis has never been addressed in vivo. Until now, only one study concerning the probable function of accD6 in the fast-growing nonpathogenic species Mycobacterium smegmatis has been conducted (36).
Here, genetic studies were used to investigate and compare accD6 gene essentiality in M. tuberculosis and M. smegmatis. We also demonstrated that M. tuberculosis accD6 is controlled both by the FAS-II promoter and by an internal promoter upstream of the gene. This unexpected finding allowed us to generate an M. tuberculosis mutant strain in which accD6 expression was driven only by its own promoter independently of the FAS-II promoter. This strain appeared to be a valuable tool for dissecting the role of AccD6 expression with respect to growth, mycolic acid biosynthesis, and M. tuberculosis cell morphology.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
M. tuberculosis H37Rv, M. smegmatis mc2155 (73), Escherichia coli Top-10 (Invitrogen) and E. coli BL21 pLysS (Invitrogen) were used in the present study. Strains based on M. tuberculosis H37Rv were maintained on Middlebrook 7H10 agar or 7H9 broth (Becton Dickinson) with 10% OADC (oleic acid, albumin, dextrose, catalase) enrichment (Becton Dickinson). M. smegmatis derivative strains were cultured either in Nutrient Broth (Becton Dickinson) supplemented with 10.0 g liter−1 glucose or in Sauton medium. For selection, we used kanamycin (25 μg ml−1), hygromycin (50 μg ml−1), gentamicin (7.5 μg ml−1), 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal; 50 μg ml−1), or sucrose (2%, wt/vol) as appropriate. E. coli Top-10 was used as the host for cloning, whereas E. coli BL21 pLysS was used as the host for expressing recombinant AccD6Mtb. Both E. coli strains were grown in LB medium. Plasmid selection and maintenance were performed using ampicillin (10 μg ml−1), chloramphenicol (34 μg ml−1), hygromycin (200 μg ml−1), and kanamycin (50 μg ml−1). The plasmids used in this study are listed and described in Table S1 in the supplemental material. Cell densities were determined by using an Ultrospec 2000 spectrophotometer (Pharmacia Biotech); the results presented reflect the values remaining after subtraction of the optical density at 600 nm (OD600) of the culture medium.
Gene cloning strategies.
Standard molecular biology protocols were used for all cloning procedures (62). All PCR products were obtained using thermostable Pfu DNA polymerase (Fermentas). They were initially cloned into a pJET1.2/blunt vector (Fermentas), followed by sequencing and digestion with the appropriate restriction enzymes. They were then cloned into the final vectors. To facilitate subcloning, some restriction enzyme recognition sites were incorporated into the primer sequences (see Table S2 in the supplemental material), while in other cases, natural restriction sites were used.
Construction of accD6 gene replacement vectors.
All PCR primers utilized in this study are listed in Table S2 in the supplemental material. To create an unmarked deletion of the accD6 gene in M. tuberculosis and M. smegmatis, a suicidal recombination delivery vector based on p2NIL was used (54). In both cases, the recombination vector carried the region upstream of accD6 together with the 5′ end of the gene (the GR1-GR2 PCR fragment; 1,595 bp for M. tuberculosis and 1,762 bp for M. smegmatis) cloned next to the 3′ end of the gene and its downstream region (the GR3-GR4 PCR fragment; 1,197 bp for M. tuberculosis and 1,237 bp for M. smegmatis) (see Fig. S2A and S3A in the supplemental material). The 5′ and 3′ accD6 PCR fragments were ligated into p2NIL so that the resulting ΔaccD6 gene was devoid of an internal sequence (624 bp for M. smegmatis and 853 bp for M. tuberculosis). Since the ΔaccD6 gene was cloned out of frame, it encoded a nonfunctional protein.
Finally, the PacI screening cassette from pGOAL17 (54) was inserted into the prepared constructs, yielding the suicide delivery vectors pJPD6Ms and pJPD6Tb.
Testing the essentiality of the accD6 gene from M. tuberculosis.
The two-step recombination protocol of Parish and Stoker (54) was used to disrupt the gene of interest at its native locus. The plasmid DNA of suicide delivery vector pJPD6Tb was treated with NaOH (0.2 mM) and was electroporated into M. tuberculosis competent cells, where it was integrated into the chromosome by homologous recombination. The resulting single-crossover (SCO) recombinant mutant colonies were blue, Kanr, and sensitive to sucrose (2%). The recombination site was confirmed by PCR and Southern blot hybridization. A single SCO colony was then picked, resuspended in fresh 7H9 medium with OADC, poured onto solid 7H10 medium with OADC without any selective markers, and incubated at 37°C for 7 days to allow the second crossover to occur. Serial dilutions were plated onto medium containing sucrose and X-Gal to select for double crossovers (DCO). Potential double-crossover colonies (white, sucrose resistant) carrying either wild-type (wt) accD6 (wt-DCO) or the mutated ΔaccD6 gene (mut-DCO) were screened for kanamycin sensitivity and were confirmed by PCR and Southern blot hybridization. The identification of mut-DCO strains would be possible only if accD6 is dispensable for the viability of M. tuberculosis.
The PCR analysis used to distinguish among SCO, wt-DCO, and mut-DCO strains was performed on XhoI- and PvuII-digested chromosomal template DNA using primers TBaccD6-XbaIs and TBaccD6-HindIIIrev. The probe for Southern blot hybridization was generated by PCR using the same primers, with pJPD6Tb as the template. Probe labeling, hybridization, and signal detection were performed using the AlkPhos Direct labeling and detection system (GE Healthcare) according to the manufacturer's instructions.
Disruption of the M. smegmatis accD6 gene by homologous recombination.
To perform unmarked deletion of the accD6 gene from M. smegmatis, we used the two-step recombination protocol described above. The pJPD6Ms suicide delivery vector was electroporated into M. smegmatis competent cells, and the resulting blue, Kanr, sucrose-sensitive (2%) SCO recombinant mutant colonies were streaked onto solid medium without antibiotics to allow the second crossover to occur. Potential double-crossover colonies (white, sucrose resistant) carrying either wt accD6 (wt-DCO) or the mutated ΔaccD6 gene (mut-DCO) were screened for kanamycin sensitivity and were confirmed by PCR and Southern blot hybridization. The PCR analysis used to distinguish among the SCO, wt-DCO, and mut-DCO strains was performed using BamHI-digested chromosomal DNA as the template along with primers MsaccD6Xs and MsaccD6HXr. The probe for Southern blot hybridization was generated by PCR using the same primers with pJPD6Ms as the template. Probe labeling, hybridization, and signal detection were performed as described above.
Construction of complementation plasmids.
For complementation of the M. tuberculosis accD6 SCO strain, we constructed the pFASTb2, pFD6Tb1, and pPD6Tb vectors. For pFASTb2, a PCR fragment (1,028 bp after NotI/XbaI digestion) carrying the FAS-II operon promoter and the first 114 bp of the fabD gene (see Fig. 2A) was amplified from M. tuberculosis chromosomal DNA using primers TbP-fas2-Not-nat and TbP-fas2-Xba-nat, and the resulting fragment was cloned into the NotI/XbaI site of the pMV306Gm integrative vector to yield pFASTb. Then another PCR fragment (2,754 bp after XbaI/EcoRI digestion) carrying the remaining 795 bp of the fabD gene, the full sequences of acpM and kasA, and the first 259 bp of kasB, amplified using primers Tb-fas2-sense-Xb and Tb-fas2-reve-Ec, was cloned into pFASTb using the XbaI/EcoRI sites, yielding pFASTb1. Finally, a PCR fragment (2,542 bp after EcoRI digestion) carrying the remaining 1,058 bp of kasB and the full sequence of accD6 (amplified using primers Tb-fas2-senEcoRI and Tb-fas2-revEcoRI) was cloned into the EcoRI site of pFASTb1 to yield pFASTb2, which contained a reconstituted version of the entire FAS-II gene cluster (PfasII-FASIIMtb). For the construction of pFD6Tb1, the entire accD6 gene (1,486 bp) was amplified from M. tuberculosis chromosomal DNA using primers TBaccD6-XbaIs and TBaccD6-HindIIIrev and was subsequently cloned into the XbaI/HindIII site of the pMV306Hyg integrative vector to yield pFD6Tb. The latter was then used as a host for a fragment carrying the FAS-II promoter (1,028 bp after NotI/XbaI digestion), amplified using primers TbP-fas2-Not-nat and TbP-fas2-Xba-nat, and cloned into the NotI/XbaI site upstream of the accD6 gene sequence to yield the final pFD6Tb1 construct (PfasII-accD6Mtb) (see Fig. 2A).
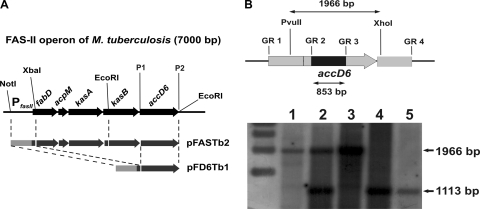
Fig. 2.
Complementation of the M. tuberculosis accD6 SCO strain. (A) Schematic demonstrating the construction of the complementation vectors that allowed us to replace the chromosomal copy of M. tuberculosis accD6 with its mutated copy. The dashed lines indicate the cloning steps, and the restriction sites are shown. P1 and P2 represent primers TBaccD6-XbaIs and TBaccD6-HindIIIrev, used to amplify the M. tuberculosis accD6 gene. (B) (Top) Map showing the length of the restriction DNA fragment (1,966 bp) and the internal deletion in the mutated gene (853 bp). The chromosomal localization of accD6 is represented by the gray arrow, while the internal deletion is marked by a black rectangle. (Bottom) Southern blot analysis confirming the deletion of the chromosomal copy of accD6 from complemented M. tuberculosis strains. Lanes represent genomic DNA from wild-type M. tuberculosis (lane 1), an SCO strain (lane 2), a DCO strain carrying the wild-type accD6 gene (wt-DCO) (lane 3), the ΔaccD6Mtb-PfasII-acc D6Mtb mutant (lane 4), and the ΔaccD6Mtb-PfasII-FASIIMtb mutant (lane 5).
For the construction of pPD6Tb (Pacc-accD6Mtb), a PCR fragment (2,542 bp after EcoRI digestion) carrying the entire accD6 (Rv2247) gene together with 1,088 bp of upstream sequence (the 30-bp intergenic region and 1,058 bp of kasB) (see Fig. 5B) was amplified using primers Tb-fas2-senEcoRI and Tb-fas2-revEcoRI and was then cloned into the EcoRI site of pMV306Gm.
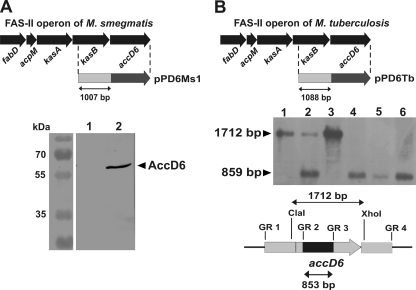
Fig. 5.
M. tuberculosis but not M. smegmatis accD6 possesses its own promoter. (A) (Top) Schematic showing the construction of the genetic construct for the complementation of the M. smegmatis ΔaccD6 mutant in order to examine whether accD6 possesses its own promoter (Pacc). The dashed lines indicate the fragment cloned into the integrative vector to produce the pPD6Ms1 construct. (Bottom) Western blot analysis of total crude lysates from the ΔaccD6Msm-Pacc-accD6Msm mutant (lane 1) and wild-type M. smegmatis (lane 2) strains with rabbit anti-AccD6 antibodies. Note the lack of a detectable signal for AccD6 protein expression driven by the 1,007-bp region upstream of the gene. (B) (Top) Schematic showing the construction of the genetic construct for complementation of the M. tuberculosis accD6 SCO mutant in order to examine whether accD6 possesses its own promoter (Pacc). The dashed lines indicate the fragment cloned into the integrative vector to produce the pPD6Tb construct. (Center) Southern blot confirming the deletion of the chromosomal copy of M. tuberculosis accD6 in the ΔaccD6Mtb-Pacc-accD6Mtb mutant, which expressed the accD6 gene exclusively from its own promoter (Pacc). The accD6Mtb gene sequence was used as the probe. Lanes: 1, wild-type M. tuberculosis; 2, single-crossover mutant (SCO); 3, double-crossover mutant carrying the wild-type accD6 gene (wt-DCO); 4 to 6, three independent ΔaccD6Mtb-Pacc-accD6Mtb mutant strains. (Bottom) Map showing the restriction DNA fragment and the size of the internal deletion in the mutated gene. The chromosomal localization of accD6 is represented by the gray arrow and the internal deletion by a black rectangle.
The three constructs were separately introduced by electroporation into the attB site of the M. tuberculosis accD6 SCO mutant. The strains obtained were selected for DCO mutants in which the chromosomal copy of the gene had been replaced by the plasmid-delivered version to generate the ΔaccD6Mtb-PfasII-FASIIMtb, ΔaccD6Mtb-PfasII-accD6Mtb, and ΔaccD6Mtb-Pacc-accD6Mtb strains.
For complementation of the M. smegmatis ΔaccD6 DCO mutant, we constructed vectors pAceD6Ms, pAceD6Tb, and pPD6Ms1. To construct pAceD6Ms (Pami-accD6Msm), a PCR fragment (1,425 bp) carrying the entire M. smegmatis accD6 (MSMEG_4329) gene was first amplified using primers MsaccD6Xs and MsaccD6Xr and then cloned into the XbaI site of the pJam2 shuttle vector (75) under the control of the acetamidase promoter (Pami), induced following addition of 4 g liter−1 of acetamide. To construct pAceD6Tb (Pami-accD6Mtb), a PCR fragment (1,422 bp) carrying the entire M. tuberculosis accD6 (Rv2247) gene was first amplified using primers TBaccD6B and TBaccD6X and then cloned into the BamHI/XbaI site of the pJam2 shuttle vector. To construct vector pPD6Ms1 (Pacc-accD6Msm), a PCR fragment (1,637 bp after ClaI/EcoRI digestion) carrying the entire M. smegmatis accD6 (MSMEG_4329) gene and 180 bp of sequence upstream of the accD6 start codon was first amplified using primers MsD6Cns and MsD6prEr and then cloned into the ClaI/EcoRI site of pMV306Km, to generate the pPD6Ms construct. Then another PCR fragment, carrying an additional 827 bp (after ClaI digestion) upstream of the 180-bp sequence described above was amplified using primers MsD6PCls and MsD6nCr and was cloned into the ClaI site of pPD6Ms to generate pPD6Ms1, in which accD6 is placed under the control of its 1,007-bp upstream sequence (see Fig. 5A).
The three constructs were separately electroporated into the M. smegmatis ΔaccD6 DCO mutant to generate the ΔaccD6Msm-Pami-accD6Msm, ΔaccD6Msm-Pami-accD6Mtb, and ΔaccD6Msm-Pacc-accD6Msm strains.
Expression and purification of recombinant KasAMtb and AccD6Mtb.
The cloning and purification of recombinant KasAMtb was described previously (32). For the generation of recombinant AccD6Mtb, the accD6Mtb (Rv2247) gene was PCR amplified from M. tuberculosis genomic DNA using primers TBaccD6s and TBaccD6r and was cloned into the BamHI/HindIII site of the pHIS.Parallel1 expression vector (69). The resulting plasmid, pHD6Tb, was verified by sequencing and was introduced into E. coli BL21 pLysS cells. The cells were grown in 1 liter LB medium at 37°C until the OD600 reached 0.4 to 0.6, whereupon expression of the His-tagged AccD6Mtb fusion protein was induced with 0.4 mM isopropyl-β-d-thiogalactopyranoside (IPTG). After 4 h of incubation at 37°C, the cells were harvested by centrifugation, resuspended in binding buffer (Novagen), and lysed by sonication. The AccD6Mtb fusion protein was purified from cell lysates with nickel affinity chromatography on a His-Bind column (Novagen) and was subsequently used to raise a rabbit polyclonal primary antibody (see below).
Preparation of antisera against KasAMtb and AccD6Mtb.
The preparation of rat anti-KasAMtb antibodies was described previously (34). An antiserum against AccD6Mtb was obtained by subcutaneous immunization of a New Zealand laboratory rabbit with three doses of the purified M. tuberculosis AccD6 antigen (150 μg, 100 μg, and 100 μg), emulsified with incomplete Freund's adjuvant (Sigma), at 3-week intervals. The levels of anti-AccD6 antibodies in serum samples from the immunized rabbit and preimmune serum samples (negative controls) were screened by an enzyme-linked immunosorbent assay (ELISA) using purified AccD6 as the coating antigen and rabbit serum samples diluted from 1:100 to 1:51,200 as the primary antibody. The immunoenzymatic reaction was developed using horseradish peroxidase-conjugated goat anti-rabbit secondary antibodies (Jackson ImmunoResearch), with 2,2′-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid diammonium salt (ABTS) (Sigma) as the chromogen. The absorbance values were measured at a λ of 405 nm. The optimal working dilution of the polyclonal anti-AccD6 serum for Western blotting was determined in preliminary titration experiments using purified AccD6 as a standard antigen, anti-AccD6 rabbit serum in dilutions from 1:100 to 1:51,200 as the primary antibodies, horseradish peroxidase-conjugated goat anti-rabbit IgG (Jackson ImmunoResearch) as the secondary antibodies, and 4-chloro-1-naphthol (Sigma) as the chromogen.
Cell wall permeability test.
Tritiated rifampin (4-methylpiperazine-3H; specific activity, 10 Ci mmol−1; Moravek Biochemicals) was used to examine the cell wall permeability of wild-type M. smegmatis and ΔaccD6 DCO mutant cells according to the modified protocol of Piddock et al. (57). In brief, mycobacterial cells were grown to mid-logarithmic phase (OD600, 0.6) in nutrient broth (Becton Dickinson) supplemented with 10.0 g liter −1 glucose. Fifty milliliters of the culture was centrifuged at 3,000 × g for 20 min at 37°C, resuspended in the same medium to an optical density of 8.0, and placed in a 37°C water bath for 10 min to equilibrate. The [3H]rifampin was added at a final concentration of 0.272 μg ml−1 (3.33 μCi ml−1), and 500-μl samples were removed at various time intervals. Each sample was mixed with 1 ml of 50 mM sodium phosphate buffer (pH 7) on ice and was centrifuged at 16,000 × g for 20 min at 4°C. The resulting cell pellets were washed again in the same buffer, recentrifuged, and mixed with Ultima Gold scintillation fluid (Perkin-Elmer). The cell-associated radioactivity was determined by liquid scintillation counting. Passive adsorption of rifampin to the cell wall (background) was estimated by performing the experiments at 0°C; the results from these experiments were subtracted from the values obtained at 37°C to determine the activity of rifampin that had actively accumulated in the cells.
RNA extraction and reverse transcription.
For quantitative real-time PCR (qRT-PCR) and FAS-II operon analysis, RNA was extracted from wild-type M. tuberculosis and M. smegmatis strains and from the ΔaccD6Mtb-PfasII-accD6Mtb and ΔaccD6Mtb-Pacc-accD6Mtb mutants using the TRIzol LS reagent (Invitrogen). Briefly, 10 ml of the aerated culture at logarithmic phase (OD600, 0.6) was centrifuged at 3,000 × g for 20 min at 4°C and was resuspended in 300 μl of water. The culture was transferred to screw-cap tubes containing 0.5 ml of 0.1-mm-diameter zirconia-silica beads (BioSpec Products) and 900 ml of TRIzol LS reagent. The bacteria were lysed using a Mini-BeadBeater-8 cell disruptor (BioSpec Products) for 3 min and were then incubated for 5 min at room temperature. Following incubation, the insoluble material was removed by centrifugation at 16,000 × g for 15 min at 4°C, and RNA was purified and treated with DNase I (Fermentas) according to the manufacturer's instructions. Finally, the RNA samples were eluted in RNase-free water and were quantified using an ND-1000 spectrophotometer (NanoDrop Technologies). Each time, the RNA samples were PCR verified in order to identify possible DNA contamination. For reverse transcription, we used a RevertAid H Minus First Strand cDNA synthesis kit (Fermentas) and performed the reactions in total volumes of 20 μl containing 1 μg of total RNA. Subsequently, 1 μl of cDNA (equivalent to 50 ng of RNA) was used in the qRT-PCR experiments (see below).
qRT-PCR.
qRT-PCR for the analysis of accD gene expression was performed using the Maxima SYBR green qPCR master mix (Fermentas) and a 7900HT real-time PCR system (Applied Biosystems). Each reaction (final volume, 25 μl) was mixed on ice and contained 1× Maxima SYBR green qPCR master mix, 50 ng of cDNA, and 0.3 μM each primer (see Table S2 in the supplemental material for primer sequences). For expression analysis of the M. tuberculosis accD genes, we used a two-step cycling protocol in which the reaction mixtures were first heated to 95°C for 10 min and then subjected to 40 cycles of 95°C for 20 s (denaturation) and 60°C for 60 s (annealing/extension). Data were acquired during the annealing/extension step. For expression analysis of the M. smegmatis accD genes, we used a three-step cycling protocol in which the reaction mixtures were first heated to 95°C for 10 min and then subjected to 40 cycles of 95°C for 20 s (denaturation), 63°C for 30 s (annealing), and 72°C for 30 s (extension). Data were acquired during the extension step. To verify the specificity and identity of the PCR products generated, melting curve analysis was performed at the end of each PCR, and the PCR products were analyzed by agarose gel electrophoresis. Each experiment was performed in triplicate, and the results are presented as means and standard errors. Our comparisons of the expression levels of various accD family members between M. tuberculosis and M. smegmatis are presented as cycle threshold (CT) values, normalized with respect to the expression of sigA (ΔCT) and converted to a linear form (2−ΔCT). For the other qRT-PCR experiments, the results reflect the fold change in the expression of a given gene in the mutant strain versus the wild-type strain, as calculated using the double delta method (2−ΔΔCT).
Total-protein isolation and Western blotting.
Ten-milliliter aliquots of bacterial culture were centrifuged, and the bacteria were resuspended in Tris-EDTA (TE) buffer and were disrupted by bead beating with 0.1-mm-diameter zirconia-silica beads. The resulting cell lysates were clarified by centrifugation. The total protein concentration in each cell lysate was determined using a bicinchoninic acid (BCA) protein assay reagent kit (Pierce). Equal amounts of proteins (20 μg) were separated on 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels and were transferred to a nitrocellulose membrane (Thermo Scientific). The membrane was saturated with 5% skim milk in phosphate-buffered saline (PBS), probed with rat anti-KasA antibodies (dilution, 1:500) or rabbit anti-AccD6 antibodies (dilution, 1:1,000), and then washed and incubated with horseradish peroxidase-conjugated anti-rat or anti-rabbit secondary antibodies (dilution, 1:1,000). The protein signals were visualized directly on the membrane using 4-chloro-1-naphthol (Sigma) as a chromogen.
Lipid and mycolic acid extraction and analysis.
Mycobacterial cultures at the mid-logarithmic growth phase were first mixed with [2-14C]acetate (specific activity, 45 to 60 mCi mmol−1; Perkin-Elmer) at 1 μCi ml−1 and then further incubated at 37°C for 4 h (for M. smegmatis) or 1, 6, and 24 h (for M. tuberculosis). The 14C-labeled cells were harvested by centrifugation at 2,000 × g, subjected to alkaline hydrolysis by incubation in 2 ml 15% tetrabutylammonium hydroxide (TBAH) (Sigma) at 100°C overnight, and then mixed with 4 ml CH2Cl2, 300 μl CH3I (Sigma), and 2 ml H2O. After 1 h, the upper, aqueous phase was discarded, and the lower, organic phase was washed twice with water and dried. The lipids were extracted using diethyl ether, dried, and then resuspended in 200 μl CH2Cl2. Equal counts of the fatty acid methyl esters (FAMES) and mycolic acid methyl esters (MAMES) from wild-type and mutant strains were applied to thin-layer chromatography (TLC) silica gel 60F254 plates (Merck) and developed in petroleum ether-acetone (19:1, vol/vol).
For complex lipid analysis, the [2-14C]acetate-labeled cells were extracted with 2 ml CH3OH-0.3% NaCl (10:1, vol/vol) and 2 ml petroleum ether. The mixture was centrifuged; the upper petroleum ether layer was removed; and an additional 2 ml petroleum ether was mixed with the lower fraction. The combined petroleum ether extracts were then evaporated under nitrogen to yield apolar lipids that were resuspended in CH2Cl2 prior to TLC analysis. For the extraction of polar lipids, the methanolic saline extract obtained after extraction of the apolar lipids was first heated at 65°C for 5 min and then mixed with 2.3 ml of CHCl3-CH3OH-0.3% NaCl (9:10:3, vol/vol). The solvent extract was then separated from the biomass by centrifugation, and the supernatant was retained. The pellet was further extracted with 0.75 ml CHCl3-CH3OH-0.3% NaCl (5:10:4, vol/vol). The combined solvent extracts were mixed with 1.3 ml CHCl3 and 1.3 ml 0.3% NaCl. After centrifugation, the lower organic layer was collected and evaporated to dryness to yield the polar lipids, which were resuspended in CHCl3-CH3OH-H2O (10:10:3, vol/vol) prior to TLC analysis.
For 2-dimensional TLC analysis of the complex lipids, five solvent systems were used to cover the polarity range of both polar and apolar mycobacterial lipids according to the work of Besra (4). Equal counts of the extracts were subjected to TLC, resolved using the appropriate solvent system, dried, and exposed overnight to X-Omat film (Kodak).
SEM.
The N-acetyl-l-cysteine-sodium citrate-NaOH (NALC-NaOH) procedure was used to prepare samples for scanning electron microscopy (SEM) (49). In brief, 10 ml of 0.05 g N-acetyl-l-cysteine in 5 ml of 2.9% citric acid and 5 ml 4% NaOH was mixed with 10 ml of mycobacterial liquid culture, and the mixture was vortexed and incubated at 37°C for 20 min. Then 20 ml of water was added, and the sample was centrifuged at 3,000 × g. The supernatant was carefully discarded, and the cells were fixed with 1.5 ml of 4% glutaraldehyde in 0.1 M phosphate buffer (pH 7.0) for 6 h (76), followed by centrifugation and resuspension in phosphate buffer. The cells were then treated with OsO4, dehydrated stepwise in a graded acetone series, dried, and sputter coated with gold using a K550X sputter coater (Quorum Technologies). The samples were examined using a Vega 3 scanning electron microscope (Tescan) with ×30,000 magnification and an accelerating voltage of 30 kV.
RESULTS
The expression profiles of the acyl-CoA carboxylase β subunits differ for pathogenic and nonpathogenic mycobacteria.
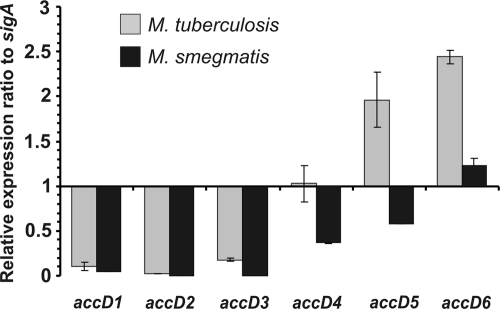
The expression profiles of all nine genes predicted to encode the carboxylase subunits of M. tuberculosis have been evaluated previously (18). Quantitative real-time PCR (qRT-PCR) analysis revealed that only the genes encoding the AccA3, AccD4, AccD5, and AccD6 carboxylase subunits were expressed at high levels during M. tuberculosis exponential growth. However, the expression profiles of their orthologs in fast-growing species have not been reported yet. Therefore, we compared the expression levels of various members of the accD family in pathogenic and nonpathogenic mycobacteria. RNA isolated from exponentially growing cells was subjected to qRT-PCR, and the relative expression level of each accD gene was calculated from the CT value, normalized with respect to the expression level of the endogenous control gene, sigA. Comparison of values obtained for M. tuberculosis and M. smegmatis indicated significant differences in the expression level, limited to three specific accD genes that are expressed and regulated mostly during the exponential growth of pathogenic mycobacteria (Fig. 1). The expression levels of accD1 to accD3 were similar in the two species. In contrast, the expression level of accD5 in M. smegmatis was found to be three times lower than that in M. tuberculosis, whereas the expression levels of accD4 and accD6 were found to be two times lower. The significant difference in the expression levels of genes encoding major carboxyltransferase subunits may involve differences in the carboxylation process during mycolate biosynthesis in these species.
Fig. 1.
Quantitative real-time PCR analysis of the expression levels of the carboxyltransferase β-subunit genes in exponentially growing cultures of M. tuberculosis and M. smegmatis. The transcript level of each subunit is indicated relative to the expression level of sigA (internal control). Values are means ± standard errors.
The accD6 (Rv2247) gene is essential for the viability of Mycobacterium tuberculosis.
The in vivo function and regulation of accD6 expression in slow- and fast-growing mycobacteria remain largely unknown. Furthermore, the belief that the accD6 gene is essential for M. tuberculosis is based solely on predictive data from high-density mutagenesis studies (63, 64, 65). To carefully evaluate whether accD6 is essential for M. tuberculosis, we used a two-step homologous recombination protocol (54) to generate single-crossover (SCO) strains carrying both an endogenous wild-type accD6 gene and an additional accD6 allele carrying an internal deletion (ΔaccD6). After the second crossover, PCR analysis of more than 50 individual DCO colonies generated from two independent SCO strains identified wt-DCO exclusively, thereby strongly suggesting that deletion of accD6 is lethal for M. tuberculosis (see Fig. S2A in the supplemental material). To further confirm the essentiality of accD6 in M. tuberculosis and to exclude potential failure in the knockout procedure, the screening was repeated with another, intact copy of the gene introduced into the attB site of the SCO strain. Two distinct constructs based on the mycobacterial integrative pMV306 vector were created and introduced into the host chromosome. In the first construct, accD6 was cloned under the control of a putative FAS-II operon promoter (PfasII-acc D6Mtb). The second construct consisted of the whole FAS-II operon sequence (PfasII-FASIIMtb) (Fig. 2A). The resulting strains were then subjected to a second crossover to generate DCO mutants (the ΔaccD6Mtb-PfasII-acc D6Mtb and ΔaccD6Mtb-PfasII-FASIIMtb mutants) that were subsequently identified by PCR (see Fig. S2B in the supplemental material) and Southern blot hybridization (Fig. 2B). The successful engineering of such strains confirmed the essentiality of accD6 in M. tuberculosis.
The accD6 (MSMEG_4329) gene is dispensable in Mycobacterium smegmatis.
The observed difference in the requirement for AccD6 between the pathogenic and the nonpathogenic strain prompted us to revisit the question of whether the accD6 gene is essential for M. smegmatis. SCO strains carrying both wild-type accD6 and a mutated copy of accD6 (ΔaccD6) were constructed and subjected to a second crossover. This resulted in the generation both of wt-DCO strains and of strains carrying only the mutated (ΔaccD6) copy of the gene (mut-DCO). The deletion of accD6 in M. smegmatis showed that, in contrast to its ortholog in M. tuberculosis, this gene is not essential for viability. The genotypes of three selected mutants identified within 25 analyzed DCO mutants were verified by PCR (see Fig. S3 in the supplemental material) and Southern blotting (Fig. 3A). The loss of the AccD6 protein in the M. smegmatis ΔaccD6 mutant was confirmed by Western blotting with a polyclonal rabbit antiserum raised against the M. tuberculosis AccD6 protein (Fig. 3B). To ensure that deletion of the accD6 gene did not affect the expression of the neighboring kasB gene, which is also dispensable in M. smegmatis, we subjected total-protein extracts from the ΔaccD6 mutant to Western blotting with a rat antiserum raised against KasA, which can also cross-react with KasB (7). Our results revealed that deletion of accD6 did not affect the expression of either of the β-ketoacyl-AcpM synthases KasA and KasB (Fig. 3C).
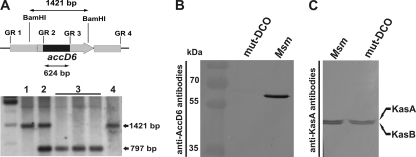
Fig. 3.
Confirmation of the loss of a functional accD6 gene in the M. smegmatis ΔaccD6 DCO mutant. (A) (Top) Schematic showing the restriction-digested DNA fragment (1,421 bp) and the size of the internal deletion in the mutated gene (624 bp). The accD6 gene is represented by the gray arrow and the internal deletion by a black rectangle. (Bottom) Southern blot confirming the deletion of accD6 in mutated M. smegmatis. Lanes: 1, wild-type M. smegmatis; 2, single-crossover strain; 3, double-crossover ΔaccD6 mutant; 4, wild-type DCO strain. (B) Western blot of total crude lysates from M. smegmatis (Msm) and a ΔaccD6 DCO mutant (mut-DCO) strain confirming the loss of AccD6 protein expression in the mutant strain, as assessed using rabbit anti-AccD6 antibodies. (C) Western blot of total crude lysates from M. smegmatis and mut-DCO strains confirming that the protein expression levels of KasA and KasB were similar in the wild-type and mutant strains, as assessed using rat anti-KasA antibodies capable of cross-reacting with KasB.
The permeability and lipid composition of the M. smegmatis ΔaccD6 cell envelope remains unaltered.
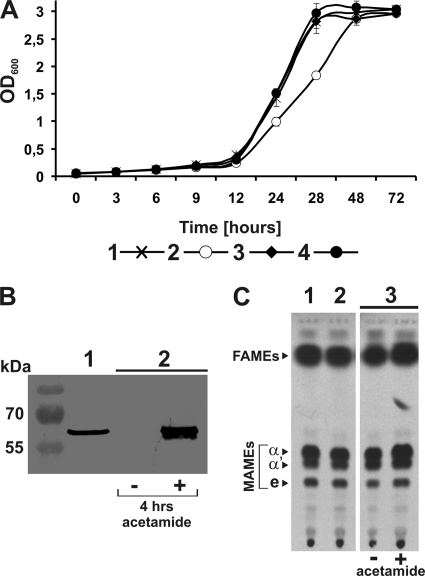
Studies on conditional depletion of enzymes that are essential for mycolic acid synthesis in M. smegmatis have demonstrated that a loss of active protein leads to the cessation of mycolate synthesis, a drop in the OD, a decrease in numbers of CFU, and finally mycobacterial cell lysis (7, 79). Thus, we reasoned that inactivation of a probable acetyl-CoA carboxylase active in the FAS-II system should also introduce changes into the mycolic acid biosynthetic pathway and the whole envelope lipid organization that in turn should affect the cell viability. OD measurements of wild-type and ΔaccD6 DCO mutant cultures growing in Sauton medium revealed only a slight decrease in mutant growth dynamics during the logarithmic phase, but in the end, the mutant and wild-type M. smegmatis strains entered the stationary phase with the same OD value. This growth rate delay was not seen in mutant strains complemented with intact copies of accD6 from M. smegmatis or M. tuberculosis (Fig. 4A). To prepare such strains, M. smegmatis and M. tuberculosis accD6 were cloned under the control of the acetamidase (Pami) promoter, and the resulting constructs (Pami-accD6Msm and Pami-accD6Mtb) were then introduced into the M. smegmatis ΔaccD6 strain to generate the ΔaccD6Msm-Pami-accD6Msm and ΔaccD6Msm-Pami-accD6Mtb mutants.
Fig. 4.
Phenotypic analysis of the M. smegmatis ΔaccD6 mutant. (A) Growth rate analysis of wild-type M. smegmatis (curve 1), the ΔaccD6 mutant (curve 2), and strains complemented with intact copies of accD6Msm and accD6Mtb expressed under the control of the acetamidase promoter: the ΔaccD6Msm-Pami-accD6Msm (curve 3) and ΔaccD6Msm-Pami-accD6Mtb (curve 4) strains. Growth rate analysis was performed on Sauton medium, and OD values are means ± standard errors from three independent experiments. (B) Western blot of total crude lysates from the M. smegmatis wild-type strain (lane 1) and the ΔaccD6Msm-Pami-accD6Mtb mutant grown in the absence (−) or presence (+) of acetamide (lane 2). (C) Thin-layer chromatography of 14C-labeled FAMEs and MAMEs extracted from wild-type M. smegmatis (lane 1), the ΔaccD6 mutant (lane 2), and the uninduced (−) and induced (+) ΔaccD6Msm-Pami-accD6Mtb mutant (lane 3). Equal counts (100,000 cpm) were loaded onto a TLC plate and were separated as described in Materials and Methods. The symbols α, α′, and e correspond to α-mycolates, α′-mycolates, and epoxy-mycolates, respectively.
To study the effect of accD6 deletion on the composition of M. smegmatis envelope lipids, the M. smegmatis wild-type and ΔaccD6 DCO mutant strains were labeled with [2-14C]acetate. The fatty and mycolic acids were extracted from labeled cells and methylated. Extracts of total fatty acid methyl esters (FAMEs) and mycolic acid methyl esters (MAMEs) were analyzed by TLC-autoradiography. As shown in Fig. 4C, disruption of the accD6 gene did not cause any detectable changes in the composition or quantity of mycolic acids. We next addressed whether complementation of the ΔaccD6 mutant with an intact M. tuberculosis accD6 gene could affect the cell wall mycolic acid profile. The ΔaccD6Msm-Pami-accD6Mtb mutant was cultured on rich medium with or without acetamide (Fig. 4B) and was labeled with [2-14C]acetate. FAMEs and MAMEs were then isolated and analyzed as described above. As shown in Fig. 4C, no significant changes in the mycolate profile were observed between the wild-type and mutant strains.
To obtain the full picture of cell envelope lipids in the ΔaccD6 mutant strain, we investigated the extractable (polar and apolar) lipid profile by 2-dimensional TLC with the use of different solvent systems. This systematic analysis did not provide significant changes in the quantity and/or quality of complex lipids (see Fig. S4A in the supplemental material). In search of more subtle changes in the structure of cell envelope as a consequence of accD6 disruption, we determined and compared the cell wall permeability in the mutant and parental strains. This was achieved using a large, hydrophobic, tritium-labeled rifampin molecule, according to the modified protocol of Piddock et al. (57). Figure S4B in the supplemental material clearly indicates that both the wild-type strain and the ΔaccD6 mutant exhibit the same rate of radiolabeled rifampin uptake. The absence of any significant differences in the permeability of the cell wall between the mutant and wild-type strains was finally confirmed by comparison of the estimated MIC99 (MIC at which 99% of bacilli are inhibited) values for rifampin and crystal violet. For both the wild-type and mutant strains, the MIC values were 20 μg ml−1 for rifampin and 6 μg ml−1 for crystal violet (data not shown).
M. tuberculosis accD6 possesses its own promoter.
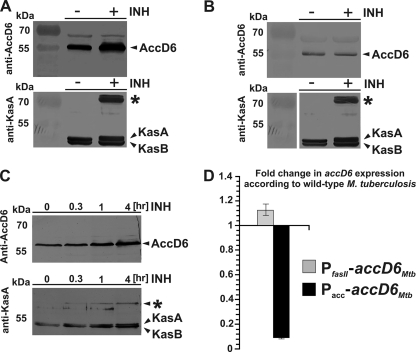
Our study is the first report of a FAS-II gene whose essentiality differs between pathogenic and nonpathogenic species. Interestingly, we also observed differences in the expression levels of this gene between the strains analyzed. In light of these results, we reasoned that accD6 may possibly be regulated separately from the other members of the FAS-II gene cluster. Therefore, using specific genetic constructs for complementation of the M. smegmatis ΔaccD6 and M. tuberculosis accD6 SCO mutants, we investigated whether accD6 possesses an endogenous and specific promoter (Pacc). In the case of M. smegmatis, the accD6 gene was cloned together with 1,007 bp of upstream sequence into the pMV306 integrative vector, and the resulting pPD6Ms1 construct was introduced into the mut-DCO (ΔaccD6) strain to generate the ΔaccD6Msm-Pacc-accD6Msm mutant. A total-protein extract of the ΔaccD6Msm-Pacc-accD6Msm mutant was assayed by Western blotting and probed with a rabbit antiserum against AccD6. As shown in Fig. 5A, the ΔaccD6Msm-Pacc-accD6Msm mutant failed to produce AccD6, demonstrating that the 1,007-bp region upstream of accD6 in M. smegmatis did not confer promoter activity under our experimental conditions. Similar experiments were undertaken with the accD6 promoter region from M. tuberculosis. The accD6 gene together with its 1,088-bp upstream sequence was cloned into pMV306, thus generating pPD6Tb, which was subsequently introduced into the M. tuberculosis SCO strain containing both the native and the disrupted (ΔaccD6) form of the gene. If it is possible for the pPD6Tb construct to complement the loss of the essential M. tuberculosis accD6 gene, one would expect, after the second crossover, to obtain a DCO strain with a deletion in the chromosomal copy of the gene. Such a strain would contain only an intact accD6 gene expressed from its own promoter (Pacc-accD6Mtb). Following completion of all selection steps, we found that the 1,088-bp DNA fragment upstream of accD6Mtb carries a promoter sequence able to drive the expression of accD6 on a level sufficient to ensure cell survival. This mutant, designated the ΔaccD6Mtb-Pacc-accD6Mtb strain, containing accD6 under the control of its own promoter, was subsequently confirmed by Southern blotting (Fig. 5B) and Western blot analysis (see Fig. 7B and 8A).
Fig. 7.
(A to C) Western blot analyses of total crude lysates of M. tuberculosis (A) and the ΔaccD6Mtb-Pacc-accD6Mtb mutant (B) grown in the presence (+) or absence (−) of INH and of total crude lysates of M. smegmatis grown in the presence of INH for 0.3, 1, and 4 h (C), probed with rabbit anti-AccD6 and rat anti-KasA antibodies. The asterisks represent an 80-kDa KasA-containing complex. (D) qRT-PCR analysis showing the fold change in the expression of accD6 in exponentially growing cultures of the ΔaccD6Mtb-PfasII-accD6Mtb (PfasII-accD6Mtb) and ΔaccD6Mtb-Pacc-accD6Mtb (Pacc-accD6Mtb) mutants relative to the expression level in wild-type M. tuberculosis, with all results normalized to the expression level of sigA. Values are means ± standard errors.
Fig. 8.
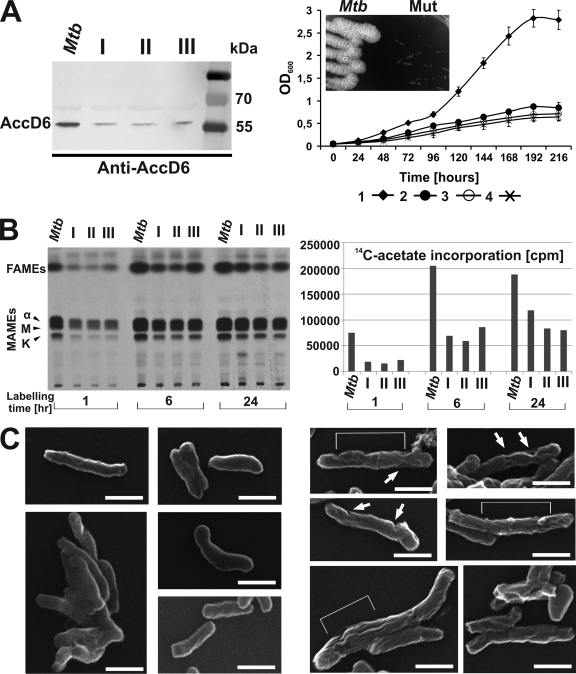
Phenotypic analysis of the ΔaccD6Mtb-Pacc-accD6Mtb mutants. (A) (Left) Western blot analysis of total crude lysates from M. tuberculosis (Mtb) and three independently obtained ΔaccD6Mtb-Pacc-accD6Mtb mutants (I, II, and III), probed with rabbit anti-AccD6 antibodies. (Right) Analysis of the growth rates of three ΔaccD6Mtb-Pacc-accD6Mtb strains (curves 2, 3, and 4) and wild-type M. tuberculosis (curve 1) grown under aeration in 7H9 medium supplemented with OADC. All values are means ± standard errors from three independent experiments. (Inset) Growth of the ΔaccD6Mtb-Pacc-accD6Mtb mutant (Mut) with respect to the growth of wild-type M. tuberculosis in 7H10-OADC medium. (B) Thin-layer chromatography (left) and scintillographic analysis (right) of 14C-labeled mycolic and fatty acid methyl esters extracted from exponentially growing wild-type M. tuberculosis and the three ΔaccD6Mtb-Pacc-accD6Mtb mutants (I, II, and III). Samples were withdrawn after 1, 6, and 24 h of labeling with [2-14C]acetate. The symbols α, M, and K correspond to α-mycolates, methoxy-mycolates, and keto-mycolates, respectively. (C) Scanning electron micrographs of wild-type M. tuberculosis (left) and the ΔaccD6Mtb-Pacc-accD6Mtb mutant (right). Several fields were examined, and representative samples are shown in both panels. Surface changes of the mutant cells are marked with white arrows and brackets. Bars, 1.0 μm.
The accD6 gene belongs to the FAS-II transcriptional unit.
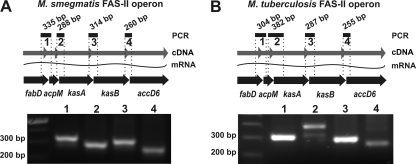
Our data showed the presence of an independent promoter sequence (Pacc) that can exclusively control accD6 expression in M. tuberculosis. Additionally, the same gene in M. smegmatis was nonessential for normal mycolate biosynthesis. These observations render questionable the theory that accD6 belongs to the FAS-II transcriptional unit. Although the accD6 open reading frame (ORF) is located within a cluster of five genes that have been shown to be involved in mycolic acid biosynthesis (15), there is no any genetic evidence that they constitute a single operon. This prompted us to analyze whether expression of all five FAS-II genes is of the monocistronic or the polycistronic type. If all five genes are expressed as a single multigenic transcript, we would expect that four PCRs performed on a total-cDNA matrix by the use of four sets of primers covering four intergenic sequences between FAS-II genes should yield four distinct PCR products. A similar procedure was used to analyze the expression mode of FAS-II genes in both M. smegmatis (Fig. 6A) and M. tuberculosis (Fig. 6B). These results indicate that, in both species, all four intergenic sequences are present. This genetic evidence conclusively demonstrates that accD6, together with other FAS-II genes, constitutes an operon that can be transcribed from a single, continuous mRNA particle.
Fig. 6.
PCR analysis of the FAS-II gene cluster structure. Four PCRs were prepared using four sets of primers covering intergenic sequences and total-cDNA matrices from M. smegmatis (A) and M. tuberculosis (B). The lane numbers (bottom) correspond to the numbered PCR products shown in the schematic (top). The expected product sizes are given. In the schematics, the genes of the FAS-II cluster are marked as thick black arrows, while the wavy lines represent isolated mRNA, and the gray arrows represent the total cDNA obtained by reverse transcription. See Table S2 in the supplemental material for a description of the primers utilized.
Expression of accD6Mtb under the control of the PfasII promoter, but not the Pacc promoter, is upregulated by INH treatment.
The data presented above indicated that, despite possessing its own independent promoter sequence, the accD6 gene of M. tuberculosis is also a member of the FAS-II transcriptional unit. The sequence and location of the FAS-II operon promoter (PfasII) are known (26). Interestingly, the first-line antitubercular drug isoniazid (INH), together with other drugs inhibiting the FAS-II pathway, can stimulate expression from the FAS-II operon promoter (5, 26, 71, 81). Here we used INH to further verify that although accD6Mtb can be expressed independently from Pacc, it can be also expressed under the control of PfasII, together with the rest of the FAS-II genes. One can assume that if accD6 is transcribed together with the other members of the operon, INH-mediated induction of the PfasII promoter should result in increased AccD6 expression levels. Moreover, the availability of the M. tuberculosis mutant expressing accD6 from the Pacc promoter allowed us to investigate whether the internal regulatory sequence is also sensitive to INH induction. Thus, wild-type M. tuberculosis and the ΔaccD6Mtb-Pacc-accD6Mtb mutant were first cultured to mid-log phase and then treated with INH (2 μg ml−1). After 24 h of incubation, samples were collected from control cultures (without INH) and INH-treated cultures. The corresponding bacterial lysates were electrophoresed, blotted, and probed with antibodies specific to AccD6. Figure 7A clearly shows an increased level of AccD6 after INH treatment in wild-type M. tuberculosis. In contrast, the ΔaccD6Mtb-Pacc-accD6Mtb mutant failed to overexpress AccD6 (Fig. 7B). As a positive control for INH-mediated induction, samples were also probed with an antiserum raised against KasA. The visualization of 80-kDa KasA-containing complexes formed as a result of KasA protein overexpression (33, 43) confirmed that the FAS-II genes were properly induced through INH-mediated action on the PfasII promoter.
Similar experiments were performed using wild-type M. smegmatis. The M. smegmatis strain was first cultured to mid-log phase and then treated with INH (15 μg ml−1). Aliquots were withdrawn at several time points and were disrupted in order to obtain total-protein extracts, and the lysates were blotted with an antiserum raised against AccD6 (Fig. 7C). Visible induction of AccD6 protein expression after 4 h of incubation with INH indicated that accD6Msm expression is strongly controlled by the PfasII promoter. The positive control for INH induction was prepared as described above for M. tuberculosis.
Decreased expression of accD6 in M. tuberculosis affects growth, cell wall lipid content, and cell morphology.
The generation of the ΔaccD6Mtb-Pacc-accD6Mtb mutant indicates that the expression level of accD6 under the control of the Pacc promoter is sufficient to sustain the function of this essential gene in M. tuberculosis and allow for cell survival. In addition, accD6 placed only under the control of the PfasII promoter successfully replaced the chromosomal copy of this gene in the ΔaccD6Mtb-PfasII-accD6Mtb mutant. These facts led to the question: what is the accD6 expression level under the control of each promoter separately? Thus, qRT-PCR analysis was performed on total cDNA from the ΔaccD6Mtb-Pacc-accD6Mtb and ΔaccD6Mtb-PfasII-accD6Mtb mutants, and the data were compared with those obtained for wild-type M. tuberculosis (Fig. 7D). The results indicated that the expression level of accD6 under the control of the Pacc promoter in the ΔaccD6Mtb-Pacc-accD6Mtb mutant was at least 10 times lower than that in wild-type M. tuberculosis. This low expression of accD6 did not significantly alter the expression of the other accD genes (data not shown). Interestingly, we did not observe any significant difference in accD6 expression between the M. tuberculosis wild-type and ΔaccD6Mtb-PfasII-accD6Mtb strains. Our qRT-PCR data showing very low accD6 expression in the ΔaccD6Mtb-Pacc-accD6Mtb mutant were consistent with the results from Western blot analysis of AccD6 protein expression in wild-type and ΔaccD6Mtb-Pacc-accD6Mtb mutant M. tuberculosis (Fig. 7A and B and 8 A). The massive decrease in the expression of this essential gene prompted us to analyze the growth dynamics of the mutant strain. OD measurements of three independently obtained mutants grown in rich medium revealed that the cultures terminated their dynamic growth after 96 to 120 h of incubation, and they entered a turbidimetric plateau after 168 h of incubation, at an OD600 that never exceeded 1.0 (Fig. 8A). It is noteworthy that at the plateau point, >60% of the cells were still viable, as confirmed by a fluorescent live/dead test (data not shown). The growth of the ΔaccD6Mtb-Pacc-accD6Mtb strain was also particularly evident when the strain was plated onto solid 7H10-OADC medium, but as for liquid cultures, a significant growth rate defect could be observed (Fig. 8A, inset). Given the results from the prior in vitro studies on the probable function of AccD6 in M. tuberculosis (18) and the indisputably defective growth of the ΔaccD6Mtb-Pacc-accD6Mtb mutants, we next analyzed their lipid content. The three independently obtained ΔaccD6Mtb-Pacc-accD6Mtb mutants and the wild-type strain were labeled with [2-14C]acetate for 1, 6, or 24 h, and equal volumes (10% of the total counts) of the extracted FAMEs and MAMEs were analyzed by TLC-autoradiography. As shown in Fig. 8B, regardless of the labeling time, the total counts of extracted lipids were typically 3- to 4-fold lower in the ΔaccD6Mtb-Pacc-accD6Mtb mutant strains than in the wild type. The difference in lipid contents was most obvious when the labeling period was short (1 h). Additionally, it is important that the [2-14C]acetate pulse-labeling studies, as well as the accD6Mtb expression analyses, were conducted in exponentially growing cultures, before the mutant cells terminated their dynamic growth.
Phenotypic analyses of the ΔaccD6Mtb-Pacc-accD6Mtb mutant offered us a unique opportunity to observe the direct in vivo effects of very low AccD6 expression. TLC analysis showed overall decreases in the quantities of both fatty acids (the FAS-I end products) and mycolic acids (the FAS-II end products), suggesting that fatty acid synthesis was inhibited at a very early stage in the mutant strain (Fig. 8B). Previous studies on genes essential for mycolic acid biosynthesis demonstrated that their inactivation or depletion in M. smegmatis led to the cells having an irregular surface (7, 79). Similarly, our SEM analyses of the ΔaccD6Mtb-Pacc-accD6Mtb strain revealed extensive changes to the surface of the mutant cell wall (Fig. 8C). Of the bacteria observed in a total of 20 fields (586 cells analyzed in total), 84.5% had a characteristic “wrinkled” appearance of grooves and dimples. This phenotype was similar to that of M. smegmatis cells following the depletion of KasA, which is also an essential member of the FAS-II biosynthetic pathway (7). Taken together, these results are the first in vivo demonstration of the role of AccD6 in the mycolic acid biosynthesis pathway of M. tuberculosis. Early-stage inhibition of fatty acid biosynthesis in the ΔaccD6Mtb-Pacc-accD6Mtb mutant implicates AccD6 as the essential, dedicated acetyl-CoA carboxylase subunit in pathogenic mycobacteria.
DISCUSSION
The mechanism of acetyl-CoA carboxylation—an essential reaction in fatty acid and mycolic biosynthesis—still requires elucidation in mycobacteria. Since AccD4 and AccD5 were the only carboxyltransferase subunits purified from mycobacterial cell extracts, they were initially considered to be major constituents of ACC complexes in tubercle bacilli (21, 51, 58). However, in vitro analysis showed that neither of them can be considered the subunit dedicated exclusively to acetyl-CoA carboxylation. In the context of malonyl-CoA synthesis in mycobacteria, the work of Daniel et al. (18) focused our attention on the third essential carboxyltransferase gene—accD6. In vitro studies showed that AccD6 protein, together with AccA3, reconstitutes an enzyme that preferentially carboxylates acetyl-CoA over propionyl-CoA (18). accD6 is the only CT subunit-encoding gene that is a member of the FAS-II gene locus (15), and it is highly expressed during intensive mycolate biosynthesis in M. tuberculosis (18).
Here we report the first detailed genetic analysis of accD6 as the gene encoding the carboxyltransferase subunit of acetyl-CoA carboxylase in mycobacteria. qRT-PCR analysis of all accD members in M. tuberculosis and M. smegmatis showed that the three accD genes highly expressed and regulated during mycolate biosynthesis in pathogenic mycobacteria were expressed at significantly lower levels in M. smegmatis. The lower expression level of M. smegmatis accD6, suggesting that there could be a species-specific difference in the requirement for the AccD6 protein, prompted us to reconsider the essential nature of accD6. Because the transposon mutagenesis method of Sassetti et al. (63, 64, 65) only predicts essentiality and may give some false results (55), we opted to use a two-step homologous-recombination method (54) to examine whether we could disrupt accD6 in the chromosomes of M. tuberculosis and M. smegmatis. This efficient method has been widely used in our laboratory for testing the essentiality of M. tuberculosis and M. smegmatis genes (11, 12, 13, 20, 29, 30). In contrast to procedures based on delivery vectors harboring a thermosensitive origin of replication (25, 56), this method does not require growth temperature shifts (54). Using this technique, we provided compelling evidence that accD6 is indeed an essential gene in M. tuberculosis under the culture conditions described, thereby confirming the previous predictive data.
However, in contrast to the previous report by Kurth et al. (36), we were able to remove the functional accD6 gene from the chromosome of M. smegmatis. Deletion of accD6 in M. smegmatis was performed in two independent experiments and was confirmed by all typical techniques, as described in Results. The basic difference between our findings concerning accD6 in M. smegmatis and the previous work of Kurth et al. (36) may result from the use of different gene knockout methodologies in the two studies. Differences in the efficiency of the integration and/or allelic exchange processes, as well as a screening procedure that requires changing the culture growth conditions, may be possible reasons for the failure to isolate the ΔaccD6Msm mutant in previous studies.
This is the first report showing a difference in the essentiality of a particular gene that belongs to the FAS-II gene locus between pathogenic and nonpathogenic mycobacteria. Similar differences between slow- and fast-growing strains have been noted previously among the genes responsible for controlling cell envelope biosynthesis. Amin et al. demonstrated the essentiality of the arabinosyltransferase-encoding gene embA in M. tuberculosis but not M. smegmatis (1). accD6 is the second member of the FAS-II gene locus found to be dispensable for the viability of M. smegmatis, since kasB was also shown to be nonessential for the in vitro growth of this bacteria (7). Interestingly, we were able to disrupt kasB and accD6 simultaneously in M. smegmatis (our unpublished data). The double mutant was still viable, but its cell envelope was significantly more permeable, in agreement with the previously described phenotype of the ΔkasB mutant of Mycobacterium marinum (22). To date, only two of the five genes in the FAS-II locus have been shown to be essential for cell viability in M. smegmatis: kasA (7) and acpM (our unpublished data).
Phenotypic analysis of the ΔaccD6Msm mutant showed that the absence of functional AccD6 in M. smegmatis is not associated with changes in the cell wall lipid content and/or permeability, indicating that the process of acetyl carboxylation was not affected in the mutant cells. This finding, which contradicts those of Kurth et al. (36), suggests that there may be another AccD subunit capable of fulfilling the function of AccD6 in M. smegmatis, or perhaps that AccD6 is not the functional subunit of acetyl-CoA carboxylase in this species. In vitro studies on M. tuberculosis AccD5 substrate specificity have suggested that despite its predominant activity as a propionyl-CoA carboxyltransferase, it is also able to transfer the carboxyl group on the acetyl-CoA (21, 51). A previous study showed that accD4 was essential for M. smegmatis (58). We tested the essentiality of all accD family members in M. smegmatis and found that only accD4 and accD5 are essential for cell survival in this bacterium (data not shown). Furthermore, qRT-PCR analysis revealed that the expression levels of the accD4 and accD5 genes were increased in the ΔaccD6Msm mutant (data not shown), prompting us to speculate that one or both of these genes could encode the putative subunit(s) that can compensate for the loss of AccD6 in M. smegmatis.
Our complementation studies on the M. smegmatis ΔaccD6Msm and M. tuberculosis accD6 SCO mutants revealed another important between-species difference in accD6 expression. Although in both cases accD6 is a member of the FAS-II transcriptional unit and its expression is controlled by the PfasII promoter, we found that accD6Mtb possesses its own, additional promoter (Pacc), located within the first 1,088 bp of its upstream sequence. In contrast to previous findings by Kurth et al. (36), no such sequence is present in the case of M. smegmatis accD6, expression of which is controlled exclusively by the PfasII promoter. Our results suggest that in the pathogenic strain M. tuberculosis, accD6 is under the influence of two (PfasII and Pacc) regulatory sequences. qRT-PCR analysis revealed that PfasII plays the dominant role in driving the expression of accD6Mtb under standard in vitro growth conditions. Although the additional Pacc promoter seems not to participate in supporting the physiological expression level of accD6Mtb under standard growth conditions, it is able to sustain the expression of this gene on a level allowing for cell survival in the absence of PfasII.
Recently, Salzman et al. (61) identified a transcriptional factor (MabR) regulating FAS-II operon expression through its action on the PfasII promoter. Also, typical inhibitors of mycolic acid biosynthesis, such as isoniazid (INH), are able to induce a transcriptional response by genes placed under the control of PfasII (5, 26, 71, 81). The identification of the additional promoter of accD6Mtb, Pacc, is the first demonstration that a particular gene of the FAS-II operon in M. tuberculosis can be regulated independently from the other operon members. We speculate that this might be possible under specific growth conditions through alternative (PfasII/Pacc) promoter usage and the activities of different transcriptional regulators, as was reported for mammalian ACC 1 and 2 (40, 50). Our studies on the ΔaccD6Mtb-Pacc-accD6Mtb mutant treated with INH suggest that Pacc is regulated independently from PfasII, since Pacc is insensitive to INH treatment. The findings of Salzman et al. (61) support this hypothesis, showing that a palindromic motif recognized by MabR is localized uniquely in PfasII, thus suggesting that this transcriptional repressor is very unlikely to influence accD6Mtb expression driven from the Pacc promoter. However, this does not exclude the possibility that AccD6 activity is regulated following posttranslational modifications. Indeed, recent studies reported that several FAS-II components, including KasA, KasB, FabH, MabA, and InhA, are regulated at the level of enzyme activity through phosphorylation (46, 47, 77, 78). The fact that all FAS-II components investigated so far are regulated by mycobacterial Ser/Thr kinases (48) leads to the hypothesis that AccD6 may also be regulated by phosphorylation, although this remains to be experimentally demonstrated.
The expression level of accD6Mtb placed only under the control of Pacc was more than 10 times lower than that in wild-type M. tuberculosis. Our ability to successfully obtain the ΔaccD6Mtb-Pacc-accD6Mtb mutant allowed us to analyze the direct phenotypic effect of low accD6 expression in tubercle bacilli. We found that this decreased expression of AccD6 protein arrested the growth of the mutant strain and inhibited proper fatty and mycolic acid biosynthesis. Inhibition of fatty acid synthesis occurred at a very early stage, likely reflecting the impaired activity of acetyl-CoA carboxylase, which provides the essential building blocks for both the FAS-I and FAS-II pathways. As shown by SEM analysis, low-level expression of AccD6 in M. tuberculosis generated cells with an irregular, “wrinkled” surface similar to that reported for INH-treated M. tuberculosis (74). Similar changes were also observed prior to lysis of M. smegmatis depleted of KasA or InhA (7, 79). These morphological changes, considered to be effects of reduced mycolic acid biosynthesis, support the hypothesis of direct in vivo involvement of AccD6Mtb in this metabolic pathway.
Along with the in vitro studies on AccD6 (18), our results demonstrate and confirm the key role of AccD6 in mycolic acid biosynthesis by M. tuberculosis. The essentiality of AccD6 to M. tuberculosis only may indicate the importance of this protein in the pathogenesis of mycobacterial infection and makes it an excellent target for the development of new antimycobacterial compounds.
Supplementary Material
ACKNOWLEDGMENTS
This research was cofinanced by the European Regional Development Fund under the Operational Programme Innovative Economy, grant POIG.01.01.02-10-107/09, and the State Committee for Scientific Research (contract N302 035 31/3172).
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 7 October 2011.
REFERENCES
- 1. Amin A. G., et al. 2008. EmbA is an essential arabinosyltransferase in Mycobacterium tuberculosis. Microbiology 154: 240–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Asselineau J., Lederer E. 1950. Structure of the mycolic acids of mycobacteria. Nature 166: 782–783 [DOI] [PubMed] [Google Scholar]
- 3. Banerjee A., et al. 1994. inhA, a gene encoding a target for isoniazid and ethionamide in Mycobacterium tuberculosis. Science 263: 227–230 [DOI] [PubMed] [Google Scholar]
- 4. Besra G. S. 1998. Preparation of cell-wall fractions from mycobacteria. Methods Mol. Biol. 101: 91–107 [DOI] [PubMed] [Google Scholar]
- 5. Betts J. C., et al. 2003. Signature gene expression profiles discriminate between isoniazid-, thiolactomycin-, and triclosan-treated Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 47: 2903–2913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bhatt A., et al. 2007. Deletion of kasB in Mycobacterium tuberculosis causes loss of acid-fastness and subclinical latent tuberculosis in immunocompetent mice. Proc. Natl. Acad. Sci. U. S. A. 104: 5157–5162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bhatt A., Kremer L., Dai A. Z., Sacchettini J. C., Jacobs W. R., Jr 2005. Conditional depletion of KasA, a key enzyme of mycolic acid biosynthesis, leads to mycobacterial cell lysis. J. Bacteriol. 187: 7596–7606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bloch K., Vance D. 1977. Control mechanisms in the synthesis of saturated fatty acids. Annu. Rev. Biochem. 46: 263–298 [DOI] [PubMed] [Google Scholar]
- 9. Brennan P. J., Nikaido H. 1995. The envelope of mycobacteria. Annu. Rev. Biochem. 64: 29–63 [DOI] [PubMed] [Google Scholar]
- 10. Brown A. K., et al. 2007. Identification of the dehydratase component of the mycobacterial mycolic acid-synthesizing fatty acid synthase-II complex. Microbiology 153: 4166–4173 [DOI] [PubMed] [Google Scholar]
- 11. Brzostek A., Dziadek B., Rumijowska-Galewicz A., Pawelczyk J., Dziadek J. 2007. Cholesterol oxidase is required for virulence of Mycobacterium tuberculosis. FEMS Microbiol. Lett. 275: 106–112 [DOI] [PubMed] [Google Scholar]
- 12. Brzostek A., Pawelczyk J., Rumijowska-Galewicz A., Dziadek B., Dziadek J. 2009. Mycobacterium tuberculosis is able to accumulate and utilize cholesterol. J. Bacteriol. 191: 6584–6591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Brzostek A., Sliwiński T., Rumijowska-Galewicz A., Korycka-Machała M., Dziadek J. 2005. Identification and targeted disruption of the gene encoding the main 3-ketosteroid dehydrogenase in Mycobacterium smegmatis. Microbiology 151: 2393–2402 [DOI] [PubMed] [Google Scholar]
- 14. Choi K. H., Kremer L., Besra G. S., Rock C. O. 2000. Identification and substrate specificity of beta-ketoacyl (acyl carrier protein) synthase III (mtFabH) from Mycobacterium tuberculosis. J. Biol. Chem. 275: 28201–28207 [DOI] [PubMed] [Google Scholar]
- 15. Cole S. T., et al. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393: 537–544 [DOI] [PubMed] [Google Scholar]
- 16. Cronan J. E., Jr., Waldrop G. L. 2002. Multi-subunit acetyl-CoA carboxylases. Prog. Lipid Res. 41: 407–435 [DOI] [PubMed] [Google Scholar]
- 17. Daffe M., Draper P. 1998. The envelope layers of mycobacteria with reference to their pathogenicity. Adv. Microb. Physiol. 39: 131–203 [DOI] [PubMed] [Google Scholar]
- 18. Daniel J., Oh T. J., Lee C. M., Kolattukudy P. E. 2007. AccD6, a member of the Fas II locus, is a functional carboxyltransferase subunit of the acyl-coenzyme A carboxylase in Mycobacterium tuberculosis. J. Bacteriol. 189: 911–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dubnau E., et al. 2000. Oxygenated mycolic acids are necessary for virulence of Mycobacterium tuberculosis in mice. Mol. Microbiol. 36: 630–637 [DOI] [PubMed] [Google Scholar]
- 20. Dziadek J., Rutherford S. A., Madiraju M. V., Atkinson M. A., Rajagopalan M. 2003. Conditional expression of Mycobacterium smegmatis ftsZ, an essential cell division gene. Microbiology 149: 1593–1603 [DOI] [PubMed] [Google Scholar]
- 21. Gago G., Kurth D., Diacovich L., Tsai S. C., Gramajo H. 2006. Biochemical and structural characterization of an essential acyl coenzyme A carboxylase from Mycobacterium tuberculosis. J. Bacteriol. 188: 477–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gao L. Y., et al. 2003. Requirement for kasB in Mycobacterium mycolic acid biosynthesis, cell wall impermeability and intracellular survival: implications for therapy. Mol. Microbiol. 49: 1547–1563 [DOI] [PubMed] [Google Scholar]
- 23. Glickman M. S., Jacobs W. R., Jr 2001. Microbial pathogenesis of Mycobacterium tuberculosis: dawn of a discipline. Cell 104: 477–485 [DOI] [PubMed] [Google Scholar]
- 24. Glickman M. S., Cox J. S., Jacobs W. R., Jr 2000. A novel mycolic acid cyclopropane synthetase is required for cording, persistence, and virulence of Mycobacterium tuberculosis. Mol. Cell 5: 717–727 [DOI] [PubMed] [Google Scholar]
- 25. Guilhot C., Gicquel B., Martin C. 1992. Temperature-sensitive mutants of the Mycobacterium plasmid pAL5000. FEMS Microbiol. Lett. 77: 181–186 [DOI] [PubMed] [Google Scholar]
- 26. Gupta N., Singh B. N. 2008. Deciphering kas operon locus in Mycobacterium aurum and genesis of a recombinant strain for rational-based drug screening. J. Appl. Microbiol. 105: 1703–1710 [DOI] [PubMed] [Google Scholar]
- 27. Hayashi T., Yamamoto O., Sasaki H., Okazaki H., Kawaguchi A. 1984. Inhibition of fatty acid synthesis by the antibiotic thiolactomycin. J. Antibiot. (Tokyo) 37: 1456–1461 [DOI] [PubMed] [Google Scholar]
- 28. Holton S. J., King-Scott S., Nasser Eddine A., Kaufmann S. H., Wilmanns M. 2006. Structural diversity in the six-fold redundant set of acyl-CoA carboxyltransferases in Mycobacterium tuberculosis. FEBS Lett. 580: 6898–6902 [DOI] [PubMed] [Google Scholar]
- 29. Korycka-Machala M., et al. 2006. Distinct DNA repair pathways involving RecA and nonhomologous end joining in Mycobacterium smegmatis. FEMS Microbiol. Lett. 258: 83–91 [DOI] [PubMed] [Google Scholar]
- 30. Korycka-Machala M., et al. 2007. Evaluation of NAD+-dependent DNA ligase of mycobacteria as a potential target for antibiotics. Antimicrob. Agents Chemother. 51: 2888–2897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kremer L., et al. 2000. Thiolactomycin and related analogues as novel anti-mycobacterial agents targeting KasA and KasB condensing enzymes in Mycobacterium tuberculosis. J. Biol. Chem. 275: 16857–16864 [DOI] [PubMed] [Google Scholar]
- 32. Kremer L., et al. 2002. Mycolic acid biosynthesis and enzymic characterization of the beta-ketoacyl-ACP synthase A-condensing enzyme from Mycobacterium tuberculosis. Biochem. J. 364: 423–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kremer L., et al. 2003. Inhibition of InhA activity, but not KasA activity, induces formation of a KasA containing complex in mycobacteria. J. Biol. Chem. 278: 20547–20554 [DOI] [PubMed] [Google Scholar]
- 34. Kremer L., Guérardel Y., Gurcha S. S., Locht C., Besra G. S. 2002. Temperature-induced changes in the cell-wall components of Mycobacterium thermoresistibile. Microbiology 148: 3145–3154 [DOI] [PubMed] [Google Scholar]
- 35. Kremer L., et al. 2001. Biochemical characterization of acyl carrier protein (AcpM) and malonyl-CoA:AcpM transacylase (mtFabD), two major components of Mycobacterium tuberculosis fatty acid synthase II. J. Biol. Chem. 276: 27967–27974 [DOI] [PubMed] [Google Scholar]
- 36. Kurth D. G., et al. 2009. ACCase 6 is the essential acetyl-CoA carboxylase involved in fatty acid and mycolic acid biosynthesis in mycobacteria. Microbiology 155: 2664–2675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lane M. D., Moss J., Polakis S. E. 1974. Acetyl coenzyme A carboxylase. Curr. Top. Cell. Regul. 8: 139–195 [PubMed] [Google Scholar]
- 38. Lin T. W., et al. 2006. Structure-based inhibitor design of AccD5, an essential acyl-CoA carboxylase carboxyltransferase domain of Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U. S. A. 103: 3072–3077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Liu J., Barry C. E., III, Besra G. S., Nikaido H. 1996. Mycolic acid structure determines the fluidity of the mycobacterial cell wall. J. Biol. Chem. 271: 29545–29551 [DOI] [PubMed] [Google Scholar]
- 40. Mao J., Chirala S. S., Wakil S. J. 2003. Human acetyl-CoA carboxylase 1 gene: presence of three promoters and heterogeneity at the 5′-untranslated mRNA region. Proc. Natl. Acad. Sci. U. S. A. 100: 7515–7520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Marrakchi H., et al. 2002. MabA (FabG1), a Mycobacterium tuberculosis protein involved in the long chain fatty acid elongation system FAS-II. Microbiology 148: 951–960 [DOI] [PubMed] [Google Scholar]
- 42. Marrakchi H., Laneelle G., Quemard A. 2000. InhA, a target of the antituberculous drug isoniazid, is involved in a mycobacterial fatty acid elongation system, FAS-II. Microbiology 146: 289–296 [DOI] [PubMed] [Google Scholar]
- 43. Mdluli K., et al. 1996. Biochemical and genetic data suggest that InhA is not the primary target for activated isoniazid in Mycobacterium tuberculosis. J. Infect. Dis. 174: 1085–1090 [DOI] [PubMed] [Google Scholar]
- 44. Mdluli K., et al. 1998. Inhibition of a Mycobacterium tuberculosis beta-ketoacyl ACP synthase by isoniazid. Science 280: 1607–1610 [DOI] [PubMed] [Google Scholar]
- 45. Minnikin D. E. 1982. Lipids: complex lipids, their chemistry, biosynthesis and roles, 95–184 In Ratledge C., Stanford J. (ed.), The biology of the mycobacteria, vol. 1. Physiology, identification and classification. Academic Press, New York, NY [Google Scholar]
- 46. Molle V., Brown A. K., Besra G. S., Cozzone A. J., Kremer L. 2006. The condensing activities of the Mycobacterium tuberculosis type II fatty acid synthase are differentially regulated by phosphorylation. J. Biol. Chem. 281: 30094–30103 [DOI] [PubMed] [Google Scholar]
- 47. Molle V., et al. 2010. Phosphorylation of InhA inhibits mycolic acid biosynthesis and growth of Mycobacterium tuberculosis. Mol. Microbiol. 78: 1591–1605 [DOI] [PubMed] [Google Scholar]
- 48. Molle V., Kremer L. 2010. Division and cell envelope regulation by Ser/Thr phosphorylation: Mycobacterium shows the way. Mol. Microbiol. 75: 1064–1077 [DOI] [PubMed] [Google Scholar]
- 49. Morcillo N., Imperiale B., Palomino J. C. 2008. New simple decontamination method improves microscopic detection and culture of mycobacteria in clinical practice. Infect. Drug Resist. 1: 21–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Oh S. Y., et al. 2005. Alternative usages of multiple promoters of the acetyl-CoA carboxylase beta gene are related to differential transcriptional regulation in human and rodent tissues. J. Biol. Chem. 280: 5909–5916 [DOI] [PubMed] [Google Scholar]
- 51. Oh T. J., Daniel J., Kim H. J., Sirakova T. D., Kolattukudy P. E. 2006. Identification and characterization of Rv3281 as a novel subunit of a biotin-dependent acyl-CoA carboxylase in Mycobacterium tuberculosis H37Rv. J. Biol. Chem. 281: 3899–3908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ojha A., et al. 2005. GroEL1: a dedicated chaperone involved in mycolic acid biosynthesis during biofilm formation in mycobacteria. Cell 123: 861–873 [DOI] [PubMed] [Google Scholar]
- 53. Parikh S. L., Xiao G., Tonge P. J. 2000. Inhibition of InhA, the enoyl reductase from Mycobacterium tuberculosis, by triclosan and isoniazid. Biochemistry 39: 7645–7650 [DOI] [PubMed] [Google Scholar]
- 54. Parish T., Stoker N. G. 2000. Use of a flexible cassette method to generate a double unmarked Mycobacterium tuberculosis tlyA plcABC mutant by gene replacement. Microbiology 146: 1969–1975 [DOI] [PubMed] [Google Scholar]
- 55. Parish T., et al. 2007. Functional complementation of the essential gene fabG1 of Mycobacterium tuberculosis by Mycobacterium smegmatis fabG but not Escherichia coli fabG. J. Bacteriol. 189: 3721–3728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Pelicic V., et al. 1997. Efficient allelic exchange and transposon mutagenesis in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U. S. A. 94: 10955–10960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Piddock L. J., Williams K. J., Ricci V. 2000. Accumulation of rifampicin by Mycobacterium aurum, Mycobacterium smegmatis and Mycobacterium tuberculosis. J. Antimicrob. Chemother. 45: 159–165 [DOI] [PubMed] [Google Scholar]
- 58. Portevin D., et al. 2005. The acyl-AMP ligase FadD32 and AccD4-containing acyl-CoA carboxylase are required for the synthesis of mycolic acids and essential for mycobacterial growth: identification of the carboxylation product and determination of the acyl-CoA carboxylase components. J. Biol. Chem. 280: 8862–8874 [DOI] [PubMed] [Google Scholar]
- 59. Rock C. O., Cronan J. E. 1996. Escherichia coli as a model for the regulation of dissociable (type II) fatty acid biosynthesis. Biochim. Biophys. Acta 1302: 1–16 [DOI] [PubMed] [Google Scholar]
- 60. Sacco E., et al. 2007. The missing piece of the type II fatty acid synthase system from Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U. S. A. 104: 14628–14633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Salzman V., et al. 2010. Transcriptional regulation of lipid homeostasis in mycobacteria. Mol. Microbiol. 78: 64–77 [DOI] [PubMed] [Google Scholar]
- 62. Sambrook J., Russell D. W. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 63. Sassetti C. M., Boyd D. H., Rubin E. J. 2001. Comprehensive identification of conditionally essential genes in mycobacteria. Proc. Natl. Acad. Sci. U. S. A. 98: 12712–12717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Sassetti C. M., Boyd D. H., Rubin E. J. 2003. Genes required for mycobacterial growth defined by high density mutagenesis. Mol. Microbiol. 48: 77–84 [DOI] [PubMed] [Google Scholar]
- 65. Sassetti C. M., Rubin E. J. 2003. Genetic requirements for mycobacterial survival during infection. Proc. Natl. Acad. Sci. U. S. A. 100: 12989–12994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Scarsdale J. N., Kazanina G., He X., Reynolds K. A., Wright H. T. 2001. Crystal structure of the Mycobacterium tuberculosis beta-ketoacyl-acyl carrier protein synthase III. J. Biol. Chem. 276: 20516–20522 [DOI] [PubMed] [Google Scholar]
- 67. Schaeffer M. L., et al. 2001. Purification and biochemical characterization of the Mycobacterium tuberculosis beta-ketoacyl-acyl carrier protein synthases KasA and KasB. J. Biol. Chem. 276: 47029–47037 [DOI] [PubMed] [Google Scholar]
- 68. Schweizer E., Hofmann J. 2004. Microbial type I fatty acid synthases (FAS): major players in a network of cellular FAS systems. Microbiol. Mol. Biol. Rev. 68: 501–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Sheffield P., Garrard S., Derewenda Z. 1999. Overcoming expression and purification problems of RhoGDI using a family of “parallel” expression vectors. Protein Expr. Purif. 15: 34–39 [DOI] [PubMed] [Google Scholar]
- 70. Slayden R. A., Barry C. E., III 2002. The role of KasA and KasB in the biosynthesis of meromycolic acids and isoniazid resistance in Mycobacterium tuberculosis. Tuberculosis (Edinb.) 82: 149–160 [DOI] [PubMed] [Google Scholar]
- 71. Slayden R. A., Lee R. E., Barry C. E., III 2000. Isoniazid affects multiple components of the type II fatty acid synthase system of Mycobacterium tuberculosis. Mol. Microbiol. 38: 514–525 [DOI] [PubMed] [Google Scholar]
- 72. Slayden R. A., et al. 1996. Antimycobacterial action of thiolactomycin: an inhibitor of fatty acid and mycolic acid synthesis. Antimicrob. Agents Chemother. 40: 2813–2819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Snapper S. B., Melton R. E., Mustafa S., Kieser T., Jacobs W. R., Jr 1990. Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol. Microbiol. 4: 1911–1919 [DOI] [PubMed] [Google Scholar]
- 74. Takayama K., Wang L., Merkal R. S. 1973. Scanning electron microscopy of the H37Ra strain of Mycobacterium tuberculosis exposed to isoniazid. Antimicrob. Agents Chemother. 4: 62–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Triccas J. A., Parish T., Britton W. J., Grequel B. 1998. An inducible expression system permitting the efficient purification of a recombinant antigen from Mycobacterium smegmatis. FEMS Microbiol. Lett. 167: 151–156 [DOI] [PubMed] [Google Scholar]
- 76. Verma I., Rohilla A., Khuller G. K. 1999. Alterations in macromolecular composition and cell wall integrity by ciprofloxacin in Mycobacterium smegmatis. Lett. Appl. Microbiol. 29: 113–117 [DOI] [PubMed] [Google Scholar]
- 77. Veyron-Churlet R., et al. 2009. The Mycobacterium tuberculosis beta-ketoacyl-acyl carrier protein synthase III activity is inhibited by phosphorylation on a single threonine residue. J. Biol. Chem. 284: 6414–6424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Veyron-Churlet R., Zanella-Cléon I., Cohen-Gonsaud M., Molle V., Kremer L. 2010. Phosphorylation of the Mycobacterium tuberculosis beta-ketoacyl-acyl carrier protein reductase MabA regulates mycolic acid biosynthesis. J. Biol. Chem. 285: 12714–12725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Vilcheze C., et al. 2000. Inactivation of the inhA-encoded fatty acid synthase II (FASII) enoyl-acyl carrier protein reductase induces accumulation of the FASI end products and cell lysis of Mycobacterium smegmatis. J. Bacteriol. 182: 4059–4067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Wakil S. J., Stoops J. K., Joshi V. C. 1983. Fatty acid synthesis and its regulation. Annu. Rev. Biochem. 52: 537–579 [DOI] [PubMed] [Google Scholar]
- 81. Wilson M., et al. 1999. Exploring drug induced alterations in gene expression in Mycobacterium tuberculosis by microarray hybridization. Proc. Natl. Acad. Sci. U. S. A. 96: 12833–12838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Wood W. I., Peterson D. O., Bloch K. 1978. Subunit structure of Mycobacterium smegmatis fatty acid synthetase. Evidence for identical multifunctional polypeptide chains. J. Biol. Chem. 253: 2650–2656 [PubMed] [Google Scholar]
- 83. Yuan Y., Zhu Y., Crane D. D., Barry C. E., III 1998. The effect of oxygenated mycolic acid composition on cell wall function and macrophage growth in Mycobacterium tuberculosis. Mol. Microbiol. 29: 1449–1458 [DOI] [PubMed] [Google Scholar]
- 84. Zimhony O., Vilcheze C., Jacobs W. R., Jr 2004. Characterization of Mycobacterium smegmatis expressing the Mycobacterium tuberculosis fatty acid synthase I (fas1) gene. J. Bacteriol. 186: 4051–4055 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.