Abstract
The aconitase gene acn of Corynebacterium glutamicum is regulated by four transcriptional regulators, indicating that the synthesis of this enzyme is carefully controlled. To understand the causes for this elaborate regulation, the properties of the Δacn-1 deletion mutant were analyzed in detail. The mutant was glutamate auxotrophic in glucose minimal medium, showed a strong growth defect, and secreted large amounts of acetate. None of these phenotypes could be complemented by plasmid-encoded aconitase, suggesting the presence of a secondary mutation. In fact, a point mutation within the gltA gene encoding citrate synthase was identified that caused the instability of the protein and an almost complete lack of its enzymatic activity. Subsequently, 27 further, independent Δacn clones were isolated, and 15 of them were found to contain distinct mutations in gltA, causing the loss of citrate synthase activity. A similar result was observed for mutants lacking the isocitrate dehydrogenase gene icd. In this case, 8 of 24 Δicd clones contained additional mutations in gltA. Indirect evidence was obtained that elevated intracellular citrate concentrations could be the cause of this selection pressure. Accordingly, the careful control of aconitase synthesis might have evolved due to the necessity to avoid inhibitory cytoplasmic citrate levels on the one hand and to prevent the excessive synthesis of an oxygen-sensitive protein requiring both iron and sulfur on the other hand.
INTRODUCTION
For most organisms, the tricarboxylic acid (TCA) cycle, also known as the Krebs cycle, is a key metabolic pathway. In its catabolic function, acetyl-coenzyme A (CoA), a product of the degradation of sugars, fatty acids, amino acids, and other carbon sources, is oxidized to CO2, thereby generating reducing equivalents (NADH and reduced quinones) for respiration and ATP by substrate-level phosphorylation. In its anabolic function, the TCA cycle produces 2-oxoglutarate and oxaloacetate as precursors of the glutamate family and the aspartate family of amino acids, respectively, in addition to other important intermediates, such as succinyl-CoA. Moreover, in many bacteria, NADPH is produced for biosynthetic purposes by the isocitrate dehydrogenase reaction.
The TCA cycle has been studied quite intensively in Corynebacterium glutamicum. This Gram-positive soil bacterium is used for large-scale industrial amino acid production, with the major products being the flavor enhancer l-glutamate (as monosodium salt) and the feed additive l-lysine, both of which are derived from intermediates of the TCA cycle. Many enzymes have been characterized, for example, citrate synthase (13), aconitase (4), isocitrate dehydrogenase (12), the 2-oxoglutarate dehydrogenase complex (25, 43, 59), fumarase (19), succinate dehydrogenase (34), malate-menaquinone oxidoreductase, and malate dehydrogenase (18, 40, 41). In addition, a complex regulatory network for the control of TCA cycle activity has been elucidated (for a review, see reference 7) that includes a variety of transcriptional regulators and a novel mechanism for the control of 2-oxoglutarate dehydrogenase activity involving the inhibitor protein OdhI, which acts as a phosphorylation-dependent molecular switch (43).
The relevance of individual TCA cycle enzymes in C. glutamicum has been explored with the help of mutants in which the corresponding genes were disrupted or deleted, such as strains lacking citrate synthase (13, 47), isocitrate dehydrogenase (12), 2-oxoglutarate dehydrogenase (25), malate-quinone oxidoreductase (41), and malate dehydrogenase (40, 41). In the case of citrate synthase, which catalyzes the first, irreversible step of the TCA cycle, this approach revealed the presence of alternative enzymes which can at least partially take over the function of the deleted protein. Initially it was shown that an inactivation of gltA (cg0949) leads to citrate or glutamate auxotrophy, suggesting that gltA codes for the only citrate synthase in C. glutamicum (13). Later, two methylcitrate synthases were identified, PrpC1 (cg0798) and PrpC2 (cg0762), which also can catalyze the formation of citrate from oxaloacetate and acetyl-CoA (47). Their activity usually is not sufficient to substitute for the one of GltA, but enhanced prp gene expression due to the inactivation of the repressor PrpR allows for the partial complementation of a ΔgltA mutant (47).
In the present work, we have analyzed the properties of aconitase deletion mutants. According to the genome sequence, C. glutamicum contains only a single aconitase gene, named acn (cg1737). No other enzyme that can metabolize citrate, such as citrate lyase or ATP-citrate lyase, currently is known in this species. Aconitase of C. glutamicum recently has been characterized biochemically and was found to have properties similar to those of aconitases from other sources (4). It contains a 4Fe-4S cluster and is highly oxygen sensitive. The expression of the acn gene is extensively regulated by at least four different transcriptional regulators, AcnR, RipA, RamA, and GlxR. Whereas the physiological role of acn repression by AcnR is not yet clear, repression by RipA is part of iron homeostasis and serves to reduce the iron demand under iron limitation (15, 33, 65). The activation of acn transcription by RamA serves to allow the increased carbon flux through the TCA cycle during growth on acetate (14). GlxR is a global regulator that directly influences the expression of about 14% of the annotated genes of C. glutamicum (31). It binds to the acn promoter and presumably represses its transcription in the presence of cyclic AMP (cAMP) (22).
In eukaryotes and some prokaryotes, aconitase also has a regulatory function by binding to certain mRNAs and inhibiting or increasing their translation (1, 3, 5, 24, 54–56). This function is exhibited only when the Fe-S cluster is absent (5, 64). In Bacillus subtilis, for example, aconitase is required for the late sporulation phase (52). The Escherichia coli enzymes AcnA and AcnB have been shown to positively autoregulate their synthesis under oxidative stress by stabilizing their own mRNA (54). In addition, both proteins were found to be involved in the control of superoxide dismutase (56). Whether C. glutamicum aconitase also exhibits a regulatory function by binding to certain mRNAs is not known yet.
In this work, a detailed analysis of aconitase deletion mutants of C. glutamicum was performed with the aim of understanding the reasons for the elaborate transcriptional regulation of the acn gene and to test for a possible regulatory function of aconitase. In the course of our studies, it became evident that the deletion of the acn gene causes a strong selection pressure for secondary mutations within the gltA gene, which might be caused by a growth-inhibitory level of cytoplasmic citrate.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth media.
The bacterial strains and plasmids used in this study are listed in Table 1. The C. glutamicum type strain ATCC 13032 was used as the wild type. The Δacn and Δicd mutants are derivatives of the wild type containing an in-frame deletion of either the aconitase gene acn or the isocitrate dehydrogenase gene icd. For growth experiments, 20 ml of brain heart infusion broth (BHI; Difco Laboratories, Detroit, MI) supplemented with 4% (wt/vol) glucose (final concentration of 4.2% [wt/vol] glucose) was inoculated with a colony from a fresh BHI agar plate and incubated overnight at 30°C and 120 rpm. Cells of this preculture were washed once in phosphate-buffered saline (PBS; 137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4, 1.4 mM KH2PO4, pH 7.3) and used to inoculate the main culture to an optical density at 600 nm (OD600) of about 1. Main cultures were incubated at 30°C and 120 rpm in 500-ml baffled Erlenmeyer flasks containing 50 ml CGXII minimal medium (29) supplemented with 3,4-dihydroxybenzoate (30 mg liter−1) as an iron chelator and glucose, l-glutamate, l-glutamine, or sodium citrate as a carbon source at the concentrations specified throughout the text. For high-performance liquid chromatography (HPLC) analyses, citrate determination, DNA microarrays, and two-dimension (2D) gel analysis, a second preculture was included using the same medium as that for the main culture. For plasmid-harboring strains, the medium was supplemented with 25 μg ml−1 kanamycin. For the induction of the expression of the target genes cloned into the expression plasmid pEKEx2 or pAN6, isopropyl β-d-1-thiogalactopyranoside (IPTG) was used at the concentrations indicated. For cloning purposes, Escherichia coli DH5α was used and cultivated at 37°C in lysogeny broth (LB) (50).
Table 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| E. coli | ||
| DH5α | F− φ80dlacΔ(lacZ)M15 Δ(lacZYA-argF)U169 endA1 recA1 hsdR17 (rK− mK+) deoR thi-1 phoA supE44 λ−gyrA96 relA1; used for cloning procedures | 23 |
| C. glutamicum | ||
| ATCC 13032 | Biotin-auxotrophic wild type | 30 |
| ATCC 13032 Δacn-1, Δacn-2, . . . , Δacn-30 (inclusive) | Independently isolated clones with an in-frame deletion of the aconitase gene acn | This work |
| ATCC 13032 Δicd-1, Δicd-2, . . . , Δicd-39 (inclusive) | Independently isolated clones with an in-frame deletion of the isocitrate dehydrogenase gene icd | This work |
| Plasmids | ||
| pK19mobsacB | Kanr; vector for allelic exchange in C. glutamicum; pK18 oriVE. coli, sacB, lacZα | 51 |
| pK19mobsacB-Δacn | Kanr; pK19mobsacB derivative containing a PCR product covering the up- and downstream regions of the acn gene | This work |
| pK19mobsacB-Δicd | Kanr; pK19mobsacB derivative containing a PCR product covering the up- and downstream regions of the icd gene | This work |
| pEKEx2 | Kanr; C. glutamicum/E. coli shuttle vector for regulated gene expression; Ptac, lacIq, pBL1 oriVC. glutamicum., pUC18 oriVE. coli | 11 |
| pEKEx2-acn | Kanr; pEKEx2 derivative containing the acn gene of C. glutamicum under the control of the tac promoter | This work |
| pAN6 | Kanr; C. glutamicum/E. coli shuttle vector for regulated gene expression, derivative of pEKEx2 | 16 |
| pAN6-acn | Kanr; pAN6 derivative containing the acn gene of C. glutamicum under the control of the tac promoter; expression without Strep tag | This work |
| pAN6-acn-gltA | Kanr; pAN6 derivative containing the acn and the gltA gene of C. glutamicum under the control of the tac promoter | This work |
| pAN6-icd | Kanr; pAN6 derivative containing the icd gene of C. glutamicum under the control of the tac promoter | This work |
| pAN6-icd-gltA | Kanr; pAN6 derivative containing the icd and the gltA gene of C. glutamicum under the control of the tac promoter | This work |
| pAN6-citH | Kanr; pAN6 derivative containing the citH gene of C. glutamicum under the control of the tac promoter | This work |
Recombinant DNA work.
The enzymes for recombinant DNA work were obtained from New England BioLabs (NEB; Frankfurt, Germany) or Fermentas (St. Leon-Rot, Germany). The oligonucleotides used in this study were obtained from Operon (Cologne, Germany) and are listed in Table S1 in the supplemental material. Routine methods, such as PCR, DNA restriction, and ligation, were performed using standard protocols (50). Chromosomal DNA of C. glutamicum was prepared as described previously (13). E. coli plasmids were isolated using the QIAprep spin miniprep kit (Qiagen, Hilden, Germany). E. coli was transformed using the RbCl method (23). C. glutamicum was transformed by electroporation (60). DNA sequencing was performed by AGOWA (Berlin, Germany).
The in-frame acn and icd deletion mutants of C. glutamicum were constructed via a two-step homologous recombination protocol as described previously (42). The up- and downstream regions (∼500 bp) of the genes were amplified using the oligonucleotide pairs acn-A-fw-EcoRI/acn-B-rev, acn-C-fw/acn-D-BamHI, icd-A-fw-EcoRI/icd-B-rev, and icd-C-fw/icd-D-BamHI, respectively. The resulting PCR products served as templates for overlap extension PCR using the oligonucleotide pairs acn-A-fw-EcoRI/acn-D-BamHI and icd-A-fw-EcoRI/icd-D-BamHI. The resulting DNA fragments were digested with EcoRI and BamHI and cloned into pK19mobsacB. The plasmids were checked by sequencing to exclude errors. The transfer of the resulting plasmids pK19mobsacB-Δacn and pK19mobsacB-Δicd into C. glutamicum and screening for the first and second recombination event were performed as described previously (42). Kanamycin-sensitive and saccharose-resistant clones were tested by colony PCR analysis with the oligonucleotide pairs acn-upstream/acn-D-BamHI and icd-upstream/icd-D-BamHI for the deletion of acn and icd, respectively.
For the construction of plasmid pEKEx2-acn, the acn coding region of the C. glutamicum wild type was amplified with an artificial ribosome binding site using the oligonucleotide pair acn-RBS-BamHI-fw/acn-BamHI-rv and cloned into the BamHI site of pEKEx2. The plasmid was sequenced to exclude errors. For complementation experiments, the plasmids pAN6-acn and pAN6-acn-gltA were constructed as follows. The acn coding region was amplified using the primer pair acn-NdeI-fw/acn-NheI-STOP-rv and cloned into pAN6 via the NdeI and NheI sites. The plasmid was sequenced to exclude errors. The gltA coding region was amplified using the primer pair gltA-RBS-NheI-fw/gltA-NheI-rv, which introduces a ribosome binding site (GAGATA) 5 bp upstream of the start codon of gltA. The product was cloned into the NheI site of pAN6-acn. The sequence and the orientation of gltA in pAN6-acn-gltA were checked by sequencing. The plasmids pAN6-icd and pAN6-icd-gltA were constructed analogously. The icd coding region was amplified using the primer pair icd-NheI-fw/icd and cloned into pAN6 via the NdeI and NheI sites. The cloning of the gltA gene was performed as described above.
Determination of specific activities of aconitase and citrate synthase.
Aconitase activity was measured in cell extracts as described previously (33). For citrate synthase activity measurements, the Δacn strains were grown for 16 h in 50 ml BHI medium supplemented with 4% (wt/vol) glucose, harvested, washed once in 50 mM Tris-HCl buffer (pH 7.5), and resuspended in a buffer (the volume was equal to the OD of the culture multiplied by 0.2 ml) containing 50 mM Tris-HCl, pH 7.5, and 200 mM potassium glutamate. One ml of the cell suspension was mixed with 250 mg zirconia/silica beads (0.1-mm diameter; Biospec, Bartlesville, OK) in a 2-ml Eppendorf tube, and the cells were mechanically disrupted by three 30-s shakings in a Silamat S5 (Ivoclar Vivadent, Ellwangen, Germany). For the determination of citrate synthase activity in Δicd mutants, 1 ml of the cell suspension was mixed with 400 mg zirconia/silica beads in a 2-ml screw-cap tube, and the cells were mechanically disrupted by 3 min of shaking in a SpeedMill P12 homogenizer (Analytik Jena AG, Jena, Germany). Cell debris and unbroken cells were separated by two 15-min centrifugations at 16,000 × g at 4°C. Between the two centrifugation steps the supernatant was transferred to a new tube. The resulting cell extract was kept on ice until it was used for the assay, which was based on the reaction of coenzyme A with 5,5′-dithiobis(2-nitrobenzoic acid). The absorbance of the resulting yellow 2-nitro-5-thiobenzoate dianion was measured spectrophotometrically at 412 nm as described previously (47). Protein concentrations were determined using the bicinchoninic acid (BCA) protein assay kit (Pierce, Bonn, Germany) with bovine serum albumin as the standard.
Determination of glucose and organic acids.
Glucose and organic acid concentrations in supernatants were determined by ion-exchange chromatography using an Agilent 1100 LC system (Agilent Technologies, Waldbronn, Germany) equipped with a cation exchange column (organic acid Refil column; 300 by 8 mm; CS-Chromatography GmbH, Langerwehe, Germany). Substances were eluted with 100 mM H2SO4 for 42 min at 40°C using a flow rate of 0.4 ml min−1. Eluted organic acids and glucose were detected by a diode array detector at a wavelength of 215 nm or by a refraction index detector, respectively. The substances were quantified by comparing the signal areas with calibration curves based on external standards. Citrate concentrations also were determined enzymatically using a citrate assay kit (Megazyme, Bray, Ireland) according to the manufacturer's instructions.
Western blot analysis.
For the detection of citrate synthase, aconitase, or isocitrate dehydrogenase in cell extracts of C. glutamicum strains, Western blot analysis was performed. Cell extract corresponding to 10 μg protein was separated on sodium dodecyl sulfate (SDS)-polyacrylamide gels (10% separating gel) and then electroblotted onto a nitrocellulose membrane (Hybond-C extra; GE Healthcare, Munich, Germany) using a Transblot semidry transfer cell (Bio-Rad, München, Germany) and buffer containing 25 mM Tris, 192 mM glycine, and 20% methanol (58). The blocking of nonspecific binding was performed with 5% (wt/vol) skim-milk powder in PBST buffer (137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4, 1.4 mM KH2PO4, 500 μl l−1 Tween 20). Polyclonal rabbit antibodies were prepared against purified citrate synthase, aconitase, and isocitrate dehydrogenase of C. glutamicum by BioGenes (Berlin, Germany). The primary antibodies were used in a 1:10,000 dilution. The secondary antibody was a goat anti-rabbit immunoglobulin G alkaline phosphatase conjugate (Bio-Rad) and used at a 1:10,000 dilution. Detection was performed in alkaline phosphatase buffer (100 mM Tris-HCl, pH 9.5, 100 mM NaCl, 10 mM MgCl2) containing 0.3 g liter−1 nitroblue tetrazolium chloride and 0.2 g liter−1 5-bromo-4-chloro-3-indolylphosphate.
Sequencing of the gltA coding region in selected clones of C. glutamicum.
To sequence the gltA gene of selected C. glutamicum strains, the gltA coding region, including the promoters, was amplified by colony PCR using the primers gltA-fw and gltA-rv. The PCR product was purified and sequenced using the primers gltA-seq-f and gltA-seq-b, which anneal roughly in the middle of the gltA gene facing in opposite directions.
RESULTS
Phenotypic properties of an aconitase deletion mutant.
To study the role of aconitase in Corynebacterium glutamicum, acn (cg1737) in-frame deletion mutants were constructed and verified by colony PCR as described in Materials and Methods. Four independent clones were isolated (Δacn-1 to Δacn-4 clones), and the Δacn-1 clone was chosen for further characterization. As expected, the mutant was not able to grow in CGXII glucose minimal medium without the supplementation of l-glutamate or l-glutamine. As shown in Fig. S1 in the supplemental material, the final optical density (OD600) depended on the glutamate or glutamine concentration. The maximal optical densities achieved were ∼5 for 5 mM l-glutamine, ∼10 for 20 mM l-glutamine, ∼17 for 40 mM l-glutamine, and ∼30 for 100 mM l-glutamine or l-glutamate. The growth rates were about 0.15 h−1 for 5, 20, and 40 mM l-glutamine and about 0.18 h−1 for 100 mM l-glutamine or l-glutamate. Under the given conditions, the C. glutamicum wild type showed a growth rate of about 0.35 h−1 and a final OD600 of about 60 to 70.
The deletion of the acn gene also was confirmed by measuring the specific activity of aconitase in cell extracts. Whereas the wild type showed a specific activity of 0.20 ± 0.05 U mg−1, which corresponds to previously published values (33), only background activity (<0.01 U mg−1) was found for the Δacn-1 strain. Aconitase activity could be restored (2.44 ± 0.37 U mg−1) by introducing a plasmid encoding aconitase (pEKEx2-acn) into C. glutamicum Δacn-1 and the induction of expression with 1 mM IPTG. Surprisingly, neither the growth defect nor the glutamate auxotrophy could be complemented.
Organic acid formation in the wild type and Δacn mutant.
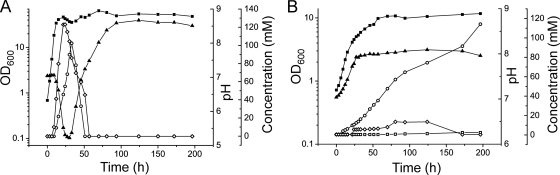
Transcriptome and proteome analyses of the Δacn-1 mutant in the exponential growth phase at an OD600 of about 5 revealed strongly increased mRNA and protein levels for enzymes involved in acetate metabolism, in particular for isocitrate lyase and malate synthase (see sections SD and SE in the supplemental material). As these enzymes are known to be induced in the presence of acetate (48, 49), the excretion of organic acids and the pH profile of the mutant and the wild type were measured during cultivation in CGXII minimal medium containing 222 mM glucose and 100 mM l-glutamate. In the exponential growth phase, the pH of the wild-type cultures dropped significantly to about 5.2 due to the accumulation of lactate (up to 120 mM) and acetate (up to 100 mM) in the medium (Fig. 1A). When the cells reached the stationary growth phase, lactate and acetate were consumed again, causing a rise of the pH, which finally stabilized at about 8.0. The accumulation of lactate is a consequence of oxygen limitation and is typical for the cultivation conditions used. In contrast, the strong acetate formation is not observed in CGXII medium containing only glucose as a carbon source. Perhaps the flux into the TCA cycle is partially controlled by the availability of glutamate and becomes reduced when high concentrations of glutamate are present, leading to increased acetate secretion. Both lactate and acetate formation started when the wild-type cultures had reached an OD600 of about 18. The profiles of the Δacn-1 strain were very different from that of the wild type (Fig. 1B). Besides the lower growth rate and the lower final OD, the pH never dropped but started to increase right from the beginning of the cultivation and stabilized at around pH 8.0. Lactate formation was strongly reduced and reached concentrations of only 15 mM. In contrast, acetate was excreted immediately after the start of the cultivation and reached final concentrations of up to 120 mM. The acetate was not consumed again. Minor concentrations of citrate (at most 2 mM) were detected in the supernatant of stationary-phase cultures of the Δacn-1 mutant, whereas citrate was not detectable by HPLC in the supernatant of the wild-type cultures. When an enzymatic assay was used, citrate concentrations at a maximum of 0.5 mM were detected in the wild-type culture supernatants (data not shown).
Fig. 1.
Growth, pH profile, and organic acid production of the C. glutamicum wild type (A) and the Δacn-1 mutant (B). Cells were cultivated in CGXII medium with 4% (wt/vol) glucose and 100 mM l-glutamate. At the indicated time points, 1-ml samples were taken and used for the determination of the optical density (OD600). Subsequently, the samples were centrifuged and the supernatant was used for pH and HPLC analysis. ▪, OD600; ▴, pH; ○, acetate; ⋄, lactate; □, citrate. Representative results from three independent experiments are shown. In the supernatant of the wild-type culture, citrate could not be detected by HPLC, therefore the corresponding curve was omitted.
The observation that the Δacn-1 mutant formed acetate right from the beginning of the cultivation explains the strongly increased mRNA levels of the isocitrate lyase (aceA) and malate synthase (aceB) genes compared to those of the wild type (see Table S4 in the supplemental material). The transcription of aceA and aceB is known to be strongly activated in the presence of acetate by the transcriptional regulator RamA (10). The RNA used for the DNA microarray experiments was isolated from cells at an OD600 of about 5, at which point the wild type had not yet started forming acetate.
Activity and content of citrate synthase in selected C. glutamicum strains.
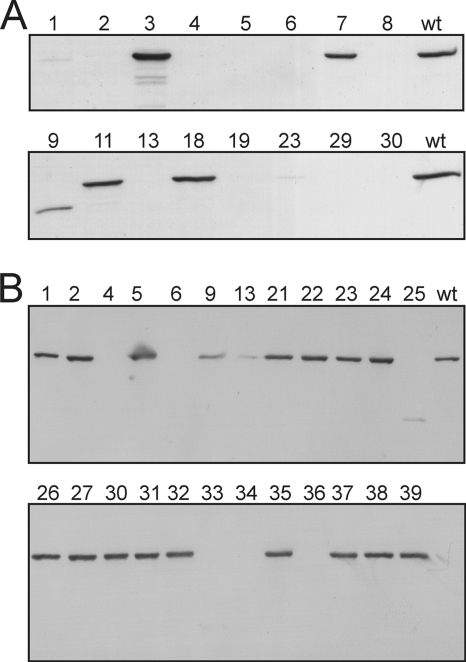
In view of the fact that the citrate synthase reaction is irreversible under physiological conditions and no other citrate-converting enzymes besides aconitase are known in C. glutamicum, higher concentrations of citrate had been expected in the Δacn-1 mutant. The oxaloacetate required for citrate synthesis can be formed by phosphoenolpyruvate (PEP) carboxylase, pyruvate carboxylase, or from glutamate via the TCA cycle. One explanation for the low citrate accumulation could be low citrate synthase activity. To test this hypothesis, both the enzymatic activity and the protein level of citrate synthase were determined. With 0.016 U mg−1, citrate synthase activity of cell extracts of the C. glutamicum Δacn-1 and Δacn-3 strains was about 50 times decreased compared to that of the wild type (0.756 U mg−1). The residual activity of the Δacn-1 strain is similar to that measured in a gltA insertion mutant (13). To probe whether citrate synthase is present (but inactive) in or absent from C. glutamicum Δacn, a Western blot analysis with citrate synthase-specific antibodies was performed. It revealed close to wild-type levels of citrate synthase in the Δacn-3 strain but an almost complete absence of citrate synthase protein from the Δacn-1 strain (Fig. 2A).
Fig. 2.
Western blot analysis of selected Δacn and Δicd clones with citrate synthase-specific antibodies. (A) Western blot with citrate synthase-specific antibodies of the Δacn clones carrying a mutation in the chromosomal gltA gene. (B) Western blot analysis of all Δicd clones analyzed in this work. Cells were cultivated in BHI medium supplemented with 4% (wt/vol) glucose for 16 h, harvested, and washed once with PBS buffer. The cells were disrupted, and cell extract corresponding to 10 μg protein (A) or 8 μg protein (B) was used for SDS-PAGE. The proteins then were blotted onto a nitrocellulose membrane. The membranes were probed with polyclonal citrate synthase-specific rabbit antibodies and an anti-rabbit alkaline phosphatase-conjugated secondary antibody. The numbers above the lanes are those given to the individual Δacn or Δicd clones tested. wt, C. glutamicum wild type.
Identification of secondary mutations in Δacn strains causing the loss of citrate synthase activity.
The results described above suggested the presence of mutations in the citrate synthase genes of the aconitase deletion strains. Therefore, the gltA coding region in Δacn-1 and Δacn-3 strains was sequenced. In the Δacn-1 strain, a mutation causing a Pro399→Leu exchange that presumably leads to the misfolding and subsequent degradation of the mutated protein was detected. The gltA gene of the Δacn-3 strain contained a mutation leading to an Asn241→Lys exchange, which probably caused an inactivation of citrate synthase. With the knowledge of these secondary mutations in gltA, another approach was used to complement the glutamate auxotrophy of the Δacn-1 strain with an expression plasmid containing not only acn but also gltA (pAN6-acn-gltA). In this case, both the glutamate auxotrophy and the growth defect could be abolished (see Fig. S2 in the supplemental material).
To test whether the absence of aconitase always causes suppressor mutations within the gltA gene, new Δacn clones were isolated and analyzed by the sequencing of the gltA gene, citrate synthase activity assays, and Western blot analysis with citrate synthase-specific antibodies. Table 2 shows an overview of the results. Of the 28 Δacn clones in total, 16 contained an altered gltA gene, and remarkably all 16 gltA mutations were different. The following mutations were observed: five single amino acid exchanges, four insertion sequence (IS) element insertions, three frameshifts, two nonsense mutations, one 200-bp deletion, and one partial duplication. Whenever a mutation was observed in the gltA coding region, there was no citrate synthase activity measurable in the cell extract of the respective clone (Table 2). Figure 2A shows the Western blot analysis of the Δacn clones with a mutated gltA gene. The majority of the clones (no. 1, 2, 4, 5, 6, 8, 13, 19, 23, 29, and 30) lacked detectable citrate synthase protein. The clones 3, 7, 11, and 18 contained citrate synthase protein in amounts close to the wild-type level, and clone 9 contains a shortened version of citrate synthase caused by the partial deletion.
Table 2.
Overview of the Δacn clones analyzed in this study
| Wild type or Δacn mutant no. | Change in: |
Presence of citrate synthase: |
||
|---|---|---|---|---|
| gltAa | GltAb | Proteinc | Activityd | |
| Wild type | + | + | ||
| 1 | C1196→T | Pro399→Leu | − | − |
| 2 | C385→T | Gln129→STOP | − | − |
| 3 | C723→A | Asn24→Lys | + | − |
| 4 | ISCg1d (cg2600) insertion before G160, Gly54 | − | − | |
| 5 | Insertion of A at A899 | Frameshift | − | − |
| 6 | ISCg13b (cg1782) insertion 50 bp upstream of start codon | − | − | |
| 7 | C1255→T | Pro419→Ser | + | − |
| 8 | ISCg5 (cg0824 or cg2915) insertion before A124, Met42 | − | − | |
| 9 | C1017-A1223 missing | 69 amino acids missing | + (smaller)e | − |
| 10 | + | + | ||
| 11 | C188→T | Ser63→Leu | + | − |
| 12 | + | + | ||
| 13 | G1127-T1172 doubled | Frameshift | − | − |
| 14 | + | + | ||
| 15 | + | + | ||
| 16 | + | + | ||
| 18 | A947→T | His316→Leu | + | − |
| 19 | C493→T | Gln165→STOP | − | − |
| 20 | + | + | ||
| 21 | + | + | ||
| 22 | + | + | ||
| 23 | ISCg1a (cg1213) insertion in the −10 region of promoter P1 | (+)f | − | |
| 24a | + | + | ||
| 24b | + | + | ||
| 27 | + | + | ||
| 28 | + | + | ||
| 29 | +C after C538 | Frameshift | − | − |
| 30 | T941 missing | Frameshift | − | − |
Mutation on the DNA level.
Consequences on the protein level.
Plus and minus indicate that the protein was detectable or undetectable, respectively, by Western blotting.
A plus indicates activity of >0.5 U (mg protein)−1; a minus indicates background activity of <0.03 U (mg protein)−1. Measurements were performed in triplicate with cell extract from one culture of each mutant.
Due to the partial deletion of 207 bp, the resulting protein is 69 amino acids smaller than the native protein (41.0 kDa and 48.9 kDa, respectively).
Due to the insertion of the IS element into the promoter of gltA, the strain shows an intermediate phenotype with a significantly reduced amount of citrate synthase protein present in the cell [indicated by (+)].
Analysis of Δicd strains.
During the studies of the Δacn strains, a Δicd strain was tested and showed no citrate synthase activity. The sequencing of the gltA gene of this strain revealed a Gly54→Val exchange (Table 3, Δicd_old). The deletion of icd therefore seemed to have an effect similar to that of the acn deletion. To further test this hypothesis, 23 additional Δicd clones were isolated and analyzed by the sequencing of the gltA gene, citrate synthase activity assays, and Western blot analysis with citrate synthase-specific antibodies (Fig. 2B and Table 3). The Western blot also was probed with isocitrate dehydrogenase-specific antibodies to confirm the absence of this enzyme. Eight of 24 Δicd clones contained a mutated gltA gene. Interestingly, two of the eight clones carried an IS element between the two promoters of gltA, which in the case of the Δicd-9 strain was located further upstream of the transcriptional start site than in the case of the Δicd-13 mutant. The Δicd-13 clone showed a strong decrease of citrate synthase activity and protein content, whereas the Δicd-9 clone showed an intermediate phenotype.
Table 3.
Overview of the Δicd clones analyzed in this study
| Wild type or Δicd mutant no. | Change in: |
Presencef of citrate synthase: |
||
|---|---|---|---|---|
| gltAa | GltAb | Proteinc | Activityd | |
| Wild type | + | + | ||
| 1 | + | + | ||
| 2 | + | + | ||
| 4 | ISCg1d (cg2600) insertion before G160, Gly54 | − | − | |
| 5 | + | + | ||
| 6 | ISCg1a (cg1213) insertion before G160, Gly54 | − | − | |
| 9 | ISCg1 between P1 and P2, 292 bp upstream of the translational start | (+) | (+)e | |
| 13 | ISCg1b (cg2725) between P1 and P2, 146 bp upstream of the translational start | (±) | (±)e | |
| 21 | + | + | ||
| 22 | + | + | ||
| 23 | + | + | ||
| 24 | + | + | ||
| 25 | C966 missing | Frameshift | (+) (smaller)g | − |
| 26 | + | + | ||
| 27 | + | + | ||
| 30 | + | + | ||
| 31 | + | + | ||
| 32 | + | + | ||
| 34 | +A after A972 | Frameshift | − | − |
| 35 | + | + | ||
| 36 | A552-C564 missing | − | − | |
| 37 | + | + | ||
| 38 | + | + | ||
| 39 | + | + | ||
| Δicd_old | G161→T | Gly54→Val | + | − |
Mutation on the DNA level.
Consequences on the protein level.
Plus and minus indicate that the protein was detectable or undetectable, respectively, by Western blotting.
A plus indicates activity of >0.25 U (mg protein)−1; a minus indicates background activity of <0.03 U (mg protein)−1. Measurements were performed in triplicate with cell extract from one culture of each mutant.
Δicd-9 strain, 0.18 U (mg protein)−1; Δicd-3 strain, 0.04 (mg protein)−1.
Symbols indicate amounts of protein in the following order: + > (+) > (±) > −.
Due to a frameshift caused by the absence of C966, the resulting protein is 115 amino acids smaller than the native protein (35.6 kDa and 48.9 kDa, respectively).
Complementation studies were performed with the Δicd-2 (active citrate synthase) and Δicd-4 (inactive citrate synthase) strains using the plasmids pAN6, pAN6-icd, and pAN6-icd-gltA. The glutamate auxotrophy of the Δicd-2 strain containing an intact gltA gene could be complemented with pAN6-icd, whereas for the complementation of the Δicd-4 mutant with a mutated gltA gene the plasmid encoding both isocitrate dehydrogenase and citrate synthase was necessary (data not shown). The results suggest that the selection pressure for citrate synthase inactivation is less strong in Δicd mutants than in Δacn mutants, which might be related to the possibility that Δicd mutants metabolize isocitrate via the glyoxylate shunt.
Evidence for growth inhibition by cytoplasmic citrate accumulation.
The results reported above show that the deletion of the aconitase gene or the isocitrate dehydrogenase gene in C. glutamicum triggered a selection pressure for secondary mutations in the citrate synthase gene, causing the loss of citrate synthase activity. A possible reason for this phenomenon could be a growth-inhibitory effect of elevated intracellular citrate levels. By gas chromatography-mass spectrometry (GC-MS) analysis, it was shown qualitatively that the Δacn-1 strain in fact contained significantly more citrate within the cells than the wild type cultivated under the same conditions (data not shown). To get some evidence for the growth-inhibitory effect of citrate, an experiment was performed in which C. glutamicum faces a sudden import of citrate into the cell. Citrate utilization and its regulation have been studied recently in C. glutamicum (8, 44). This species possesses two citrate uptake systems, CitH and TctABC. Citrate transport by CitH is dependent on Ca2+ or Sr2+, whereas TctCBA requires Mg2+ or Ca2+. The transcription of citH and tctCBA is activated by a two-component regulatory system composed of the sensor kinase CitA and the response regulator CitB when citrate is present in the medium. Cells adapted to citrate consumed it in parallel with glucose.
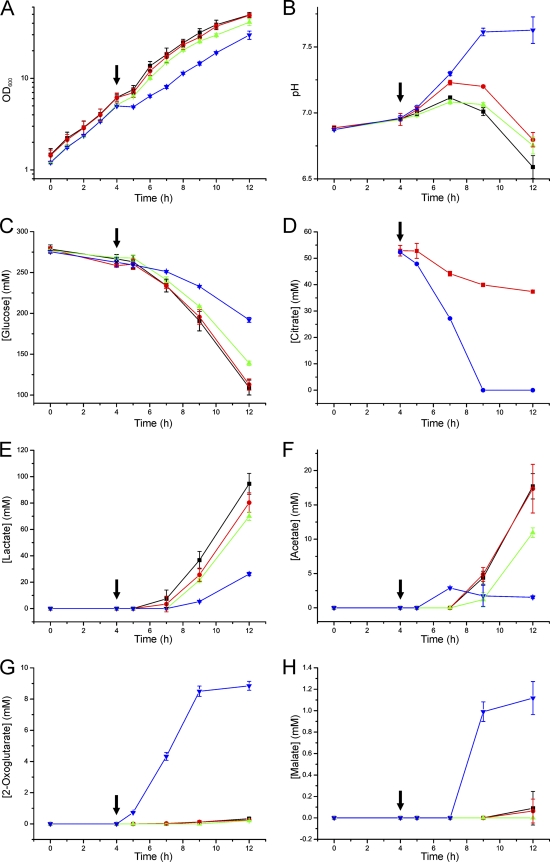
To analyze the response to a sudden high intracellular citrate concentration, the C. glutamicum wild type was transformed with the expression plasmid pAN6-citH, which carries the citH gene under the control of the tac promoter. The experimental setup is presented in Fig. S4 in the supplemental material. Two parallel cultures of the wild type carrying pAN6-citH and of the wild type carrying the control plasmid pAN6 were grown in CGXII glucose medium with 1 mM IPTG. After 4 h, 50 mM sodium citrate and 5 mM CaCl2 were added to one set, whereas no addition was made to the second set of cultures. Samples were taken during cultivation and analyzed for OD600, pH, substrate consumption, and organic acid production (Fig. 3). Before the addition of citrate, all strains showed the same growth rate. After the addition of citrate, a weak transient growth defect was observed for the control strain carrying pAN6 and for the cultures that were not supplemented with citrate. This was due to the multiple samplings at this time point and was not observed in experiments where only a single sample was taken (data not shown). Strikingly, citrate addition to the strain overexpressing citH caused a complete growth stop, which lasted for about 1 h. Thereafter the culture resumed growth and reached a similar growth rate and the same final OD600 as the control cultures. Regarding the pH profile, glucose consumption, lactate formation, and acetate formation, the cultures with and without citrate of the strain carrying pAN6 and the cultures without citrate of the strain carrying pAN6-citH behaved similarly (Fig. 3). In contrast, the cultures with citrate of the strain carrying pAN-citH showed very different behavior. The pH significantly increased after citrate addition and remained in the alkaline region. Glucose consumption was significantly retarded, whereas citrate was rapidly consumed. In the control strain carrying pAN6, measurable citrate utilization started after a lag phase of about 1 h, which presumably was required for the activation of citH and tctCBA expression by the CitAB two-component system. Lactate and acetate formation of the citrate-containing cultures of the strain with pAN6-citH was much lower than that of the other cultures, presumably due to the reduced glucose consumption. Instead, immediately after citrate addition this strain started to excrete 2-oxoglutarate (up to 10 mM) and, later, small concentrations (1.5 mM) of malate. The formation of 2-oxoglutarate indicates a metabolic bottleneck at the 2-oxoglutarate dehydrogenase complex.
Fig. 3.
Influence of citrate addition on growth, pH profile, substrate consumption, and product formation of the C. glutamicum wild type carrying either pAN6-citH (green and blue symbols) or the control plasmid pAN6 (black and red symbols). After the first preculture in BHI medium during the day, a second preculture was performed in CGXII minimal medium with 4% (wt/vol) glucose, 25 μg/ml kanamycin, and 100 μM IPTG overnight. The main culture was performed in CGXII minimal medium with 4% (wt/vol) glucose, 25 μg/ml kanamycin, and 1 mM IPTG. After 4 h of cultivation (OD600 ∼5), two of the cultures (indicated by the blue and red symbols) were supplemented with 50 mM trisodium citrate and 5 mM CaCl2 (black arrows), whereas the two other cultures did not receive a supplement (green and black symbols). Samples were taken at the indicated time points and analyzed for OD600 (A), pH of the supernatant (B), glucose consumption (C), citrate consumption (D), lactate formation (E), acetate formation (F), 2-oxoglutarate formation (G), and malate formation (H). The data shown are average values and standard deviations from three independent cultivations.
To get clues to the mechanism of citrate toxicity, the transcriptomes of the wild type carrying pAN6-citH 20 min after citrate addition and those before citrate addition were compared (see Table S7 in the supplemental material). In total, 864 genes showed a more than 2-fold altered mRNA ratio. A similar comparison was done for the control strain carrying pAN6, and in this case 218 genes had an mRNA level altered 2-fold (see Table S8 in the supplemental material). Despite these many changes, it was not possible to get obvious hints on the mechanisms of citrate toxicity. However, the fact that a sudden rapid uptake of citrate leads to a complete, transient growth stop supports the assumption that high intracellular citrate concentrations are harmful to C. glutamicum cells.
DISCUSSION
The results described in this work show that the deletion of the aconitase gene or of the isocitrate dehydrogenase gene in C. glutamicum causes selection pressure to lose citrate synthase activity (Tables 2 and 3). In four of the Δacn and four of the Δicd mutants, gltA (or its promoter region) was disrupted by the transposition of IS elements. The genome of C. glutamicum ATCC 13032 contains 24 IS elements (27), and so far only two of them have been analyzed further, ISCg1 (62) and ISCg2 (46). Our results show that ISCg1, ISCg5, and ISCg13 are induced in C. glutamicum under citrate stress conditions.
The strong selection pressure in Δacn and Δicd mutants for secondary mutations in the gltA gene was assumed to be due to a toxic effect of citrate accumulation in the cells. In the food industry, sodium citrate is used as an antibacterial agent (36), confirming that this metabolite can inhibit growth when present at unphysiologically high concentrations. C. glutamicum presumably possesses no alternative enzymes or pathways for citrate catabolism (6) except for aconitase and the TCA cycle. The mechanism(s) of the assumed toxic effect of elevated citrate concentrations still is not clear, but it seems to be important for C. glutamicum to keep the intracellular citrate level below a certain threshold. This also can explain why the expression of aconitase and citrate synthase (61) is extensively controlled at the transcriptional level by three or four different regulators and perhaps also by additional, still unknown mechanisms.
Studies on the phenotype of aconitase or isocitrate deletion mutants have been performed with a number of different organisms. In the Gram-positive soil bacterium Bacillus subtilis, the mutation of citB coding for an aconitase leads to glutamate auxotrophy and to an early sporulation defect (9). Aconitase deletion strains accumulated up to 2 mM citrate in the culture supernatant. The supplementation of the growth medium with a high excess of Mn or Fe ions could partially complement the sporulation defect, indicating that it partially results from a restricted availability of these cations due to their chelation by citrate. A triple mutant lacking citB, citA, and citZ (the latter two genes code for citrate synthases) had a 100-fold higher sporulation frequency than the citB single mutant and did not accumulate citrate. Comparable results were observed for Δicd mutants of B. subtilis (26, 38).
The Gram-negative bacterium E. coli possesses two genes coding for aconitases (acnA and acnB). An acnA single deletion mutant did not show a growth defect, probably because the aconitase activity was taken over by AcnB (21). The growth of an acnB single mutant was impaired but not abolished in glucose minimal medium (20). An acnAB mutant did not grow in glutamate-supplemented glucose minimal medium under aerobic conditions until it became rapidly overgrown by mutants that also lacked citrate synthase activity (20). In 1976 it was reported that E. coli strains lacking both icd and gltA grew faster than icd single mutants (35). For an isocitrate dehydrogenase deletion mutant of E. coli BL21(DE3), it could be shown that this strain accumulates large amounts of citrate and isocitrate intracellularly (up to 11.8 and 0.5 mM, respectively) as well as in the medium (up to 3.5 and 0.4 mM, respectively) (2).
Another organism which was subject to aconitase deletion studies is Streptomyces coelicolor (63). As in C. glutamicum and B. subtilis, S. coelicolor has just one aconitase gene (acoA), and its deletion leads to glutamate auxotrophy in glucose minimal medium and to defects in growth, antibiotic biosynthesis, and aerial hypha formation (63). The mutant secreted citrate (up to 14 mM), acetate (up to 11 mM), and pyruvate (up to 0.45 mM), causing an acidification of the growth medium to a pH of about 3.5. The growth defect of the acoA single mutant could be partially suppressed by an additional mutation of the citA gene coding for citrate synthase. For the nitrogen-fixing bacterium Sinorhizobium meliloti 1021, it was shown that the deletion of citrate synthase can restore the growth of an aconitase deletion mutant (32). For Δicd mutants of this bacterium, it was observed that there appeared to be spontaneous mutations in the gene coding for citrate synthase (39). For the human pathogen Staphylococcus aureus, an aconitase deletion mutant has been described (53), but its growth properties in glucose-based minimal medium have not been described. In Bradyrhizobium japonicum, the deletion of acnA did not lead to a clear glutamate auxotrophy, but growth was severely inhibited and could not be restored by the supplementation of the medium with glutamate (57). As only a single aconitase gene was found in the B. japonicum genome sequence (28), the acnA disruptant may possess some residual aconitase activity.
The effect of aconitase and isocitrate dehydrogenase deletions on citrate synthase is not restricted to prokaryotes, as a similar phenomenon also was found in yeast. It was shown that Δicd and Δaco1 mutants of Saccharomyces cerevisiae share several growth phenotypes, for example, poor growth on glycerol, an inability to grow with acetate as the sole carbon source, or a propensity to generate petite segregants, which can be complemented or at least moderated by the codisruption of cit1 coding for mitochondrial citrate synthase (17, 37, 45).
From this literature survey, it is obvious that several microorganisms lacking aconitase or isocitrate dehydrogenase showed a less severe growth phenotype when citrate synthase activity was absent simultaneously. This study is the first one, however, in which secondary mutations in the citrate synthase gene caused by the deletion of acn or icd were analyzed in detail and in a set of more than 50 independent mutants. Our results demonstrate that it is impossible to characterize a definitive Δacn mutant, because different clones can vary significantly.
An important result was the finding that the inactivation of citrate synthase is not the only solution to cope with the stress caused by the absence of aconitase or isocitrate dehydrogenase, as 12 Δacn and 16 Δicd mutants were found to have intact citrate synthases.
Further studies, including genome sequence analysis, are required to understand how these mutants handle the stress situation.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Bundesministerium für Bildung und Forschung (BMBF) and by Evonik Degussa GmbH, as well as by a grant from the Division Health and Nutrition, within the cluster project “SysMAP,” to M.B.
We thank Abigail Koch-Koerfges for the development of the HPLC analysis method.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 7 October 2011.
REFERENCES
- 1. Alén C., Sonenshein A. L. 1999. Bacillus subtilis aconitase is an RNA-binding protein. Proc. Natl. Acad. Sci. U. S. A. 96: 10412–10417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Aoshima M., Ishii M., Yamagishi A., Oshima T., Igarashi Y. 2003. Metabolic characteristics of an isocitrate dehydrogenase defective derivative of Escherichia coli BL21(DE3). Biotechnol. Bioeng. 84: 732–737 [DOI] [PubMed] [Google Scholar]
- 3. Banerjee S., Nandyala A. K., Raviprasad P., Ahmed N., Hasnain S. E. 2007. Iron-dependent RNA-binding activity of Mycobacterium tuberculosis aconitase. J. Bacteriol. 189: 4046–4052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Baumgart M., Bott M. 2011. Biochemical characterisation of aconitase from Corynebacterium glutamicum J. Biotechnol. 154: 163–170 [DOI] [PubMed] [Google Scholar]
- 5. Beinert H., Kennedy M. C., Stout C. D. 1996. Aconitase as iron-sulfur protein, enzyme, and iron-regulatory protein. Chem. Rev. 96: 2335–2373 [DOI] [PubMed] [Google Scholar]
- 6. Bott M. 1997. Anaerobic citrate metabolism and its regulation in enterobacteria. Arch. Microbiol. 167: 78–88 [PubMed] [Google Scholar]
- 7. Bott M. 2007. Offering surprises: TCA cycle regulation in Corynebacterium glutamicum. Trends Microbiol. 15: 417–425 [DOI] [PubMed] [Google Scholar]
- 8. Brocker M., Schaffer S., Mack C., Bott M. 2009. Citrate utilization by Corynebacterium glutamicum is controlled by the CitAB two-component system through positive regulation of the citrate transport genes citH and tctCBA. J. Bacteriol. 191: 3869–3880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Craig J. E., Ford M. J., Blaydon D. C., Sonenshein A. L. 1997. A null mutation in the Bacillus subtilis aconitase gene causes a block in SpoOA-phosphate-dependent gene expression. J. Bacteriol. 179: 7351–7359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cramer A., Gerstmeir R., Schaffer S., Bott M., Eikmanns B. J. 2006. Identification of RamA, a novel LuxR-type transcriptional regulator of genes involved in acetate metabolism of Corynebacterium glutamicum. J. Bacteriol. 188: 2554–2567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Eikmanns B. J., Kleinertz E., Liebl W., Sahm H. 1991. A family of Corynebacterium glutamicum/Escherichia coli shuttle vectors for cloning, controlled gene expression, and promoter probing. Gene 102: 93–98 [DOI] [PubMed] [Google Scholar]
- 12. Eikmanns B. J., Rittmann D., Sahm H. 1995. Cloning, sequence analysis, expression, and inactivation of the Corynebacterium glutamicum icd gene encoding isocitrate dehydrogenase and biochemical characterization of the enzyme. J. Bacteriol. 177: 774–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Eikmanns B. J., Thum-Schmitz N., Eggeling L., Lüdtke K. U., Sahm H. 1994. Nucleotide sequence, expression and transcriptional analysis of the Corynebacterium glutamicum gltA gene encoding citrate synthase. Microbiology 140: 1817–1828 [DOI] [PubMed] [Google Scholar]
- 14. Emer D., Krug A., Eikmanns B. J., Bott M. 2009. Complex expression control of the Corynebacterium glutamicum aconitase gene: identification of RamA as a third transcriptional regulator besides AcnR and RipA. J. Biotechnol. 140: 92–98 [DOI] [PubMed] [Google Scholar]
- 15. Frunzke J., Bott M. 2008. Regulation of iron homeostasis in Corynebacterium glutamicum, p. 241–266 In Burkovski A. (ed.), Corynebacteria: genomics and molecular biology. Caister Academic Press, Norfolk, United Kingdom [Google Scholar]
- 16. Frunzke J., Engels V., Hasenbein S., Gätgens C., Bott M. 2008. Co-ordinated regulation of gluconate catabolism and glucose uptake in Corynebacterium glutamicum by two functionally equivalent transcriptional regulators, GntR1 and GntR2. Mol. Microbiol. 67: 305–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gadde D. M., McCammon M. T. 1997. Mutations in the idh2 gene encoding the catalytic subunit of the yeast NAD+-dependent isocitrate dehydrogenase can be suppressed by mutations in the cit1 gene encoding citrate synthase and other genes of oxidative metabolism. Arch. Biochem. Biophys. 344: 139–149 [DOI] [PubMed] [Google Scholar]
- 18. Genda T., Nakamatsu T., Ozaki I. 2003. Purification and characterization of malate dehydrogenase from Corynebacterium glutamicum. J. Biosci. Bioeng. 95: 562–566 [PubMed] [Google Scholar]
- 19. Genda T., Watabe S., Ozaki H. 2006. Purification and characterization of fumarase from Corynebacterium glutamicum. Biosci. Biotechnol. Biochem. 70: 1102–1109 [DOI] [PubMed] [Google Scholar]
- 20. Gruer M. J., Bradbury A. J., Guest J. R. 1997. Construction and properties of aconitase mutants of Escherichia coli. Microbiology 143: 1837–1846 [DOI] [PubMed] [Google Scholar]
- 21. Gruer M. J., Guest J. R. 1994. Two genetically distinct and differentially regulated aconitases (AcnA and AcnB) in Escherichia coli. Microbiology 140: 2531–2541 [DOI] [PubMed] [Google Scholar]
- 22. Han S. O., Inui M., Yukawa H. 2008. Effect of carbon source availability and growth phase on expression of Corynebacterium glutamicum genes involved in the tricarboxylic acid cycle and glyoxylate bypass. Microbiology 154: 3073–3083 [DOI] [PubMed] [Google Scholar]
- 23. Hanahan D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166: 557–580 [DOI] [PubMed] [Google Scholar]
- 24. Hentze M. W., Kuhn L. C. 1996. Molecular control of vertebrate iron metabolism: mRNA-based regulatory circuits operated by iron, nitric oxide, and oxidative stress. Proc. Natl. Acad. Sci. U. S. A. 93: 8175–8182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hoffelder M., Raasch K., van Ooyen J., Eggeling L. 2010. The E2 domain of OdhA of Corynebacterium glutamicum has succinyltransferase activity dependent on lipoyl residues of the acetyltransferase AceF. J. Bacteriol. 192: 5203–5211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jin S. F., Levin P. A., Matsuno K., Grossman A. D., Sonenshein A. L. 1997. Deletion of the Bacillus subtilis isocitrate dehydrogenase gene causes a block at stage I of sporulation. J. Bacteriol. 179: 4725–4732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kalinowski J., et al. 2003. The complete Corynebacterium glutamicum ATCC 13032 genome sequence and its impact on the production of L-aspartate-derived amino acids and vitamins. J. Biotechnol. 104: 5–25 [DOI] [PubMed] [Google Scholar]
- 28. Kaneko T., et al. 2002. Complete genomic sequence of nitrogen-fixing symbiotic bacterium Bradyrhizobium japonicum USDA110. DNA Res. 9: 189–197 [DOI] [PubMed] [Google Scholar]
- 29. Keilhauer C., Eggeling L., Sahm H. 1993. Isoleucine synthesis in Corynebacterium glutamicum: molecular analysis of the ilvB-ilvN-ilvC operon. J. Bacteriol. 175: 5595–5603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kinoshita S., Udaka S., Shimono M. 1957. Studies on amino acid fermentation. Part I. Production of L-glutamic acid by various microorganisms. J. Gen. Appl. Microbiol. 3: 193–205 [PubMed] [Google Scholar]
- 31. Kohl T. A., Tauch A. 2009. The GlxR regulon of the amino acid producer Corynebacterium glutamicum: detection of the corynebacterial core regulon and integration into the transcriptional regulatory network model. J. Biotechnol. 143: 239–246 [DOI] [PubMed] [Google Scholar]
- 32. Koziol U., et al. 2009. Deletion of citrate synthase restores growth of Sinorhizobium meliloti 1021 aconitase mutants. J. Bacteriol. 191: 7581–7586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Krug A., Wendisch V. F., Bott M. 2005. Identification of AcnR, a TetR-type repressor of the aconitase gene acn in Corynebacterium glutamicum. J. Biol. Chem. 280: 585–595 [DOI] [PubMed] [Google Scholar]
- 34. Kurokawa T., Sakamoto J. 2005. Purification and characterization of succinate:menaquinone oxidoreductase from Corynebacterium glutamicum. Arch. Microbiol. 183: 317–324 [DOI] [PubMed] [Google Scholar]
- 35. Lakshmi T. M., Helling R. B. 1976. Selection for citrate synthase deficiency in icd mutants of Escherichia coli. J. Bacteriol. 127: 76–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lee Y. L., Cesario T., Owens J., Shanbrom E., Thrupp L. D. 2002. Antibacterial activity of citrate and acetate. Nutrition 18: 665–666 [DOI] [PubMed] [Google Scholar]
- 37. Lin A. P., Hakala K. W., Weintraub S. T., McAlister-Henn L. 2008. Suppression of metabolic defects of yeast isocitrate dehydrogenase and aconitase mutants by loss of citrate synthase. Arch. Biochem. Biophys. 474: 205–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Matsuno K., et al. 1999. Metabolic imbalance and sporulation in an isocitrate dehydrogenase mutant of Bacillus subtilis. J. Bacteriol. 181: 3382–3391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. McDermott T. R., Kahn M. L. 1992. Cloning and mutagenesis of the Rhizobium meliloti isocitrate dehydrogenase gene. J. Bacteriol. 174: 4790–4797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Molenaar D., van der Rest M. E., Drysch A., Yücel R. 2000. Functions of the membrane-associated and cytoplasmic malate dehydrogenases in the citric acid cycle of Corynebacterium glutamicum. J. Bacteriol. 182: 6884–6891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Molenaar D., van der Rest M. E., Petrovic S. 1998. Biochemical and genetic characterization of the membrane-associated malate dehydrogenase (acceptor) from Corynebacterium glutamicum. Eur. J. Biochem. 254: 395–403 [DOI] [PubMed] [Google Scholar]
- 42. Niebisch A., Bott M. 2001. Molecular analysis of the cytochrome bc1-aa3 branch of the Corynebacterium glutamicum respiratory chain containing an unusual diheme cytochrome c1. Arch. Microbiol. 175: 282–294 [DOI] [PubMed] [Google Scholar]
- 43. Niebisch A., Kabus A., Schultz C., Weil B., Bott M. 2006. Corynebacterial protein kinase G controls 2-oxoglutarate dehydrogenase activity via the phosphorylation status of the OdhI protein. J. Biol. Chem. 281: 12300–12307 [DOI] [PubMed] [Google Scholar]
- 44. Polen T., Schluesener D., Poetsch A., Bott M., Wendisch V. F. 2007. Characterization of citrate utilization in Corynebacterium glutamicum by transcriptome and proteome analysis. FEMS Microbiol. Lett. 273: 109–119 [DOI] [PubMed] [Google Scholar]
- 45. Przybyla-Zawislak B., Gadde D. M., Ducharme K., McCammon M. T. 1999. Genetic and biochemical interactions involving tricarboxylic acid cycle (TCA) function using a collection of mutants defective in all TCA cycle genes. Genetics 152: 153–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Quast K., Bathe B., Pühler A., Kalinowski J. 1999. The Corynebacterium glutamicum insertion sequence ISCg2 prefers conserved target sequences located adjacent to genes involved in aspartate and glutamate metabolism. Mol. Gen. Genet. 262: 568–578 [DOI] [PubMed] [Google Scholar]
- 47. Radmacher E., Eggeling L. 2007. The three tricarboxylate synthase activities of Corynebacterium glutamicum and increase of L-lysine synthesis. Appl. Microbiol. Biotechnol. 76: 587–595 [DOI] [PubMed] [Google Scholar]
- 48. Reinscheid D. J., Eikmanns B. J., Sahm H. 1994. Characterization of the isocitrate lyase gene from Corynebacterium glutamicum and biochemical analysis of the enzyme. J. Bacteriol. 176: 3474–3483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Reinscheid D. J., Eikmanns B. J., Sahm H. 1994. Malate synthase from Corynebacterium glutamicum: sequence analysis of the gene and biochemical characterization of the enzyme. Microbiology 140: 3099–3108 [DOI] [PubMed] [Google Scholar]
- 50. Sambrook J., Fritsch E. F., Maniatis T. 1989. Molecular cloning, a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 51. Schäfer A., Tauch A., Jäger W., Kalinowski J., Thierbach G., Pühler A. 1994. Small mobilizable multipurpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19-selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145: 69–73 [DOI] [PubMed] [Google Scholar]
- 52. Serio A. W., Pechter M. B., Sonenshein A. L. 2006. Bacillus subtilis aconitase is required for efficient late-sporulation gene expression. J. Bacteriol. 188: 6396–6405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Somerville G. A., et al. 2002. Staphylococcus aureus aconitase inactivation unexpectedly inhibits post-exponential-phase growth and enhances stationary-phase survival. Infect. Immun. 70: 6373–6382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Tang Y., Guest J. R. 1999. Direct evidence for mRNA binding and post-transcriptional regulation by Escherichia coli aconitases. Microbiology 145: 3069–3079 [DOI] [PubMed] [Google Scholar]
- 55. Tang Y., Guest J. R., Artymiuk P. J., Read R. C., Green J. 2004. Post-transcriptional regulation of bacterial motility by aconitase proteins. Mol. Microbiol. 51: 1817–1826 [DOI] [PubMed] [Google Scholar]
- 56. Tang Y., Quail M. A., Artymiuk P. J., Guest J. R., Green J. 2002. Escherichia coli aconitases and oxidative stress: post-transcriptional regulation of sodA expression. Microbiology 148: 1027–1037 [DOI] [PubMed] [Google Scholar]
- 57. Thöny-Meyer L., Künzler P. 1996. The Bradyrhizobium japonicum aconitase gene (acnA) is important for free-living growth but not for an effective root nodule symbiosis. J. Bacteriol. 178: 6166–6172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Towbin H., Staehelin T., Gordon J. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets-procedure and some applications. Proc. Natl. Acad. Sci. U. S. A. 76: 4350–4354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Usuda Y., et al. 1996. Molecular cloning of the Corynebacterium glutamicum (“Brevibacterium lactofermentum” AJ12036) odhA gene encoding a novel type of 2-oxoglutarate dehydrogenase. Microbiology 142: 3347–3354 [DOI] [PubMed] [Google Scholar]
- 60. van der Rest M. E., Lange C., Molenaar D. 1999. A heat shock following electroporation induces highly efficient transformation of Corynebacterium glutamicum with xenogeneic plasmid DNA. Appl. Microbiol. Biotechnol. 52: 541–545 [DOI] [PubMed] [Google Scholar]
- 61. van Ooyen J., et al. 2011. Citrate synthase in Corynebacterium glutamicum is encoded by two gltA transcripts which are controlled by RamA, RamB, and GlxR. J. Biotechnol. 154: 140–148 [DOI] [PubMed] [Google Scholar]
- 62. Vertès A. A., Inui M., Kobayashi M., Kurusu Y., Yukawa H. 1994. Isolation and characterization of IS31831, a transposable element from Corynebacterium glutamicum. Mol. Microbiol. 11: 739–746 [DOI] [PubMed] [Google Scholar]
- 63. Viollier P. H., et al. 2001. Roles of aconitase in growth, metabolism, and morphological differentiation of Streptomyces coelicolor. J. Bacteriol. 183: 3193–3203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Walden W. E., et al. 2006. Structure of dual function iron regulatory protein 1 complexed with ferritin IRE-RNA. Science 314: 1903–1908 [DOI] [PubMed] [Google Scholar]
- 65. Wennerhold J., Krug A., Bott M. 2005. The AraC-type regulator RipA represses aconitase and other iron proteins from Corynebacterium under iron limitation and is itself repressed by DtxR. J. Biol. Chem. 280: 40500–40508 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.