Abstract
Polo-like kinase 1 (Plk1) plays pivotal roles in mitosis; however, little is known about its function in S phase. In this study, we show that inhibition of Plk1 impairs DNA replication and results in slow S-phase progression in cultured cancer cells. We have identified origin recognition complex 2 (Orc2), a member of the DNA replication machinery, as a Plk1 substrate and have shown that Plk1 phosphorylates Orc2 at Ser188 in vitro and in vivo. Furthermore, Orc2-S188 phosphorylation is enhanced when DNA replication is under challenge induced by ultraviolet, hydroxyurea, gemcitabine, or aphidicolin treatment. Cells expressing the unphosphorylatable mutant (S188A) of Orc2 had defects in DNA synthesis under stress, suggesting that this phosphorylation event is critical to maintain DNA replication under stress. To dissect the mechanism pertinent to this observation, we showed that Orc2-S188 phosphorylation associates with DNA replication origin and that cells expressing Orc2-S188A mutant fail to maintain the functional pre-replicative complex (pre-RC) under DNA replication stress. Furthermore, the intra-S-phase checkpoint is activated in Orc2-S188A-expressing cells to cause delay of S-phase progress. Our study suggests a novel role of Plk1 in facilitating DNA replication under conditions of stress to maintain genomic integrity.
INTRODUCTION
Two critical steps during eukaryotic cell proliferation are duplication of the genome in S phase and separation of the duplicated genome into two daughter cells in mitosis (1). Tight regulation of the cell cycle is vital in order to maintain genome stability and cell survival. Phosphorylation and proteolytic events are the two most-studied events involved in regulation of cell cycle, and Polo-like kinase 1 (Plk1), a Ser/Thr kinase, plays many important roles in this process (21). Polo kinase was first discovered in Drosophila with the loss-of-function phenotype of cells arrested in mitosis with monopolar spindles (23), and it has been extensively studied since then. Plk1 is involved in many critical aspects of mitosis, such as mitotic entry, spindle formation, and sister chromatid segregation (21). Compared to its essential functions in almost every step of mitosis, little is known about its potential functions in other stages of the cell cycle, such as DNA replication in S phase.
DNA replication is initiated by formation of the pre-replicative complex (pre-RC), whose components are recruited to origins of DNA replication in a stepwise manner beginning with the origin recognition complex (ORC) (1). Within the six-subunit ORC complex (Orc1 to Orc6), Orc2 and Orc3 form a core subcomplex with which other ORC members interact. The ORC recruits Cdc6 and Cdt1, both of which are required for subsequent loading of the minichromosome maintenance complex 2-7 (Mcm2-7), the helicase that is essential for DNA unwinding prior to replication. Formation of the pre-RC occurs during late M phase right after sister chromatid segregation and licenses the DNA for replication during the next S phase (25).
In addition to multiple mitotic functions, several recent reports support a connection between mammalian Plk1 and DNA replication. First of all, Plk1 accumulates mainly in the nucleus during S phase (11) and interacts with almost all members of Mcm2-7 protein complex (27). Second, mouse Plk1 interacts with Orc2, further indicating a potential new function for Plk1 in DNA replication (22). Most recently, lentivirus-based RNA interference (RNAi) was used to demonstrate that Plk1 is also required for proper DNA replication during undisturbed cell cycle progression in mammalian cells (30). It was shown that Plk1 depletion leads to destruction of pre-RC formation and DNA damage checkpoint activation (30). However, it is still not clear how Plk1-associated kinase activity regulates DNA replication in mammalian cells.
We report here that Plk1 phosphorylates Orc2 in vitro and in vivo. We provide evidence that Plk1 promotes DNA replication through phosphorylation of Orc2 at Ser188 on the replication origin under stressful conditions.
MATERIALS AND METHODS
Cell culture and DNA transfections.
HeLa, U2OS, HEK293T, Panc-1, and T98G cells were cultured in Dulbecco modified Eagle medium (Invitrogen) supplemented with 10% fetal bovine serum in the presence of antibiotics at 37°C in 5% CO2. RWPE-1 cells were cultured in keratinocyte/serum-free medium supplemented with 5 ng of human recombinant epithelial growth factor/ml and 0.05 mg of bovine pituitary extract/ml. Plasmid DNA was transfected with MegaTran (Origene) according to the manufacturer's protocols.
Vector construction and RNAi.
To specifically deplete endogenous Orc2, plasmid pLKO.1-Orc2 was constructed with the targeting sequence UGCUCCUCUCAUGUGGGAUCA. To specifically deplete Plk1, lentivirus was generated with the targeting sequence AGATCACCCTCCTTAAATATT (12). Lentivirus depletion was performed in the presence of Polybrene (10 μg/ml) and HEPES (10 mM).
Immunoprecipitation and Western blotting.
Cell lysates were incubated with Flag (Sigma; catalog no. F-3165), Plk1 (Santa Cruz; sc-17783), Orc2 (Calbiochem; NA73), or cyclin A (Santa Cruz; sc-239) antibodies overnight at 4°C, followed by 2 h of incubation with protein A/G Plus-Agarose beads (Santa Cruz, sc-2003). Immunocomplexes were resolved by SDS-PAGE, and coimmunoprecipitated proteins were detected by Western blotting with antibodies against T7 (Novagen; 69522-3), Flag, β-actin (Sigma; A5441), α-tubulin (Sigma; T5168), green fluorescent protein (GFP; Invitrogen; A11122), Plk1, Orc2 (BD Pharmingen; 559266), Mcm2 (Biolegend; 602201), Mcm7 (BioLegend; 602701), Orc5 (Novus Biologicals; H00005001-A01), Orc3 (Santa Cruz; sc-98930), PCNA (BD Pharmingen; 610664), Sp1 (Santa Cruz; sc-59), p53 (Oncogene; OP43-100), p21 (Santa Cruz; sc-397), or cyclin A, as indicated in specific experiments.
Recombinant protein purification.
Various domains of Orc2 were amplified by PCR, subcloned into pGEX-KG vector, expressed in Escherichia coli, and purified. Point mutations were generated with the QuikChange site-directed mutagenesis kit (Stratagene).
Kinase assay.
In vitro kinase assays were performed in TBMD buffer (50 mM Tris [pH 7.5], 10 mM MgCl2, 5 mM dithiothreitol, 2 mM EGTA, 0.5 mM sodium vanadate, 20 mM p-nitrophenyl phosphate) supplemented with 125 μM ATP and 10 μCi of [γ-32P]ATP at 30°C for 30 min in the presence of GST-Orc2 proteins. After the reaction mixtures were resolved by SDS-PAGE, the gels were stained with Coomassie brilliant blue, dried, and subjected to autoradiography. For immunoprecipitation (IP)/kinase assays, total cell extracts were immunoprecipitated with cyclin A antibodies, and the immunoprecipitated proteins were subjected to kinase assays with histone H1 (Upstate) as a substrate.
FACS analysis.
Cells were harvested by trypsinization, fixed in 75% ethanol, stained with propidium iodide solution at a final concentration of 50 μg/ml, and subjected to fluorescence-activated cell sorting (FACS) analysis.
BrdU labeling assay.
BrdU labeling assays were performed with a kit from Roche according to the manufacturer's instructions with slight modification.
ChIP.
Chromatin immunoprecipitation (ChIP) assays were performed as described previously (3) with slight modifications. In brief, HeLa cells were treated with 1% (wt/vol) formaldehyde for 10 min at room temperature with occasional swirling. Glycine was added to a final concentration of 0.2 M, followed by incubation for an additional 5 min. Cells were collected, washed with phosphate-buffered saline twice, cell lysis buffer (10 mM Tris-Cl [pH 7.5], 10 mM NaCl, 0.5% NP-40, protease, and phosphatase inhibitor cocktail) twice, and resuspended in SDS lysis buffer (10 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.5% SDS, 1 mM EDTA, protease, and phosphatase inhibitor cocktail from Thermo Scientific). After lysates were sonicated on ice to reach an average DNA fragment of 2.5 kb, the chromatin solution was clarified by centrifugation at 15,000 × g at 4°C for 30 min. The chromatin solution from an equivalent of 3 × 107 cells was diluted with IP dilution buffer (20 mM Tris-Cl [pH 8], 1 mM EDTA, 0.1% NP-40, and 150 mM NaCl,), precleared with protein G Dynabeads (Invitrogen) for 1 h and subjected to IP with adequate antibodies. After 1 h of incubation, the beads were washed with 0.1% SDS-1% Triton X-100-2 mM EDTA-20 mM Tris-HCl (pH 8.0), with 150 mM NaCl in the first wash and 500 mM NaCl in the second wash. Further washes were performed with 0.25 M LiCl-1% NP-40-1% sodium deoxycholate-1 mM EDTA-10 mM Tris-HCl (pH 8.0) and with 10 mM Tris-HCl (pH 8.0)-1 mM EDTA twice. After the immunoprecipitated material was eluted from the beads with 1% SDS-0.1 M NaHCO3-10 mM dithiothreitol, samples were incubated with 0.2 M NaCl overnight at 65°C to reverse the cross-links. The samples were then treated with DNase-free RNase A at 37°C for 30 min and digested with proteinase K at 42°C for 60 min, followed by extraction with phenol-chloroform-isoamyl alcohol and chloroform. Finally, the samples were ethanol precipitated in the presence of glycogen and resuspended in double-distilled H2O for PCR analysis.
Quantitative real-time PCR analysis.
Real-time PCR were performed in triplicate using SYBR green I with quantification as previously described (8). The primer sequences were as follows: In6 F, 5′-GACATTCTGCTTCCATAGATGTGG-3′; In6 R, 5′-GTTGGGAAAGATGTCATCATCAGG-3′; Ex9 F, 5′-ATGTCTTCCGGAGACTCCTGAAGC-3′; Ex9 R, 5′-GGCCTCCTATTCTCAGAATCATGC-3′; MCM4 F, 5′-AAACCAGAAGTAGGCCTCGCTCGG-3′; and MCM4 R, 5′-GTCTGACCTGCGGAGGTAGTTTGG-3′. The annealing temperature was 62°C. To quantify genomic DNA in the precipitated material, standard DNA samples (genomic HeLa cell DNA) were serially diluted to 50 ng, 5 ng, 500 pg, 50 pg, and 5 pg. Dilutions are based on the estimate that 50 ng of DNA correspond to 104 genomic units. Following the PCRs, the threshold cycle (CT) value for each standard sample was plotted against the logarithm of genomic unit to generate a standard curve. Genomic equivalents of DNA samples were determined by extrapolation from the standard curves.
RESULTS
Inhibition of Plk1 reduces DNA synthesis.
Using lentivirus-based RNAi to deplete Plk1 in thymidine-synchronized cells, Yim and Erikson previously showed that Plk1 depletion inhibits DNA synthesis (30). To rule out this novel finding is a secondary effect of a Plk1 depletion-induced mitotic defect, we used BI2536, a small-molecule Plk1 inhibitor (10) during various times of the cell cycle. We first synchronized U2OS cells with a double thymidine block protocol to arrest cells at the G1/S boundary. BI2536 was added to the medium 1 h prior to release to ensure the cells would enter S phase with inhibited Plk1 activity. After the second thymidine block, cells were released into BI2536-containing medium and were harvested at different times for various analyses. FACS was used to monitor cell cycle progression. As expected, BI2536-treated cells progressed into S phase more slowly than control cells (Fig. 1A, compare FACS profiles at 4 and 6 h after release). In line with the FACS profiles, DNA synthesis, indicated by 5-bromo-2′-deoxyuridine (BrdU) incorporation, was reduced in BI2536-treated cells relative to control cells (Fig. 1B). However, we might point that the DNA synthesis defect was detected only after short-term BrdU incubation (10 min) but not after longer BrdU incubation (30 min), suggesting that Plk1 activity might contribute to DNA replication but is not absolutely required for the process in undisturbed cell cycle progression.
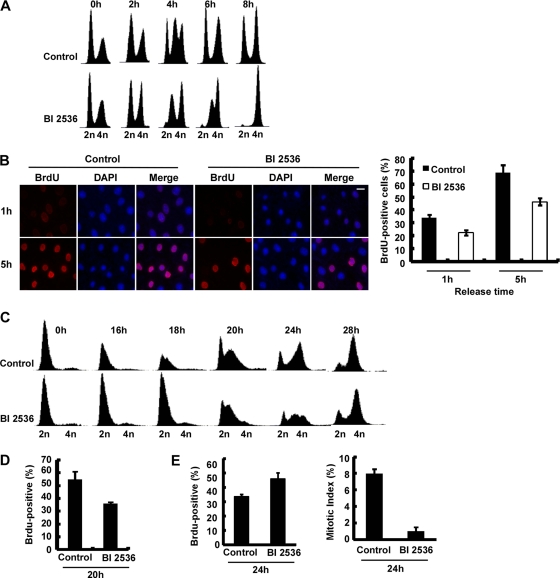
Fig. 1.
Effects of inhibition of Plk1 on DNA replication. (A) FACS analysis of cell cycle progression in U2OS cells. After U2OS cells were synchronized with a double thymidine block (DTB; 16 h treatment with thymidine, 8 h release, and a second thymidine block for 16 h), they were released into medium with or without BI2536 for different times as indicated and processed for FACS analysis. BI2536 was added into the medium 1 h prior to release. (B) Representative images of BrdU labeling in control and Plk1-inhibited cells. U20S cells were synchronized with the DTB, released into medium with or without BI2536 (added 1 h prior to release) for 1 or 5 h, labeled with BrdU for 10 min, and stained with anti-BrdU antibodies. DNA was stained with DAPI (4′,6′-diamidino-2-phenylindole). BrdU-positive cells were quantified in the right panel. (C) FACS analysis of cell cycle progression in T98G cells. After T98G cells were serum starved for 3 days to synchronize at G0, they were released into fresh medium containing 20% fetal bovine serum with the addition of nocodazole or nocodazole plus BI2536 at 12 h postrelease and harvested for FACS analysis at different times. (D) DNA synthesis of T98G cells harvested 20 h after release from serum starvation. At 20 h after release of cells prepared as in panel C, the cells were incubated with BrdU for 10 min and harvested. (E) At 24 h after release of cells prepared as in panel C, the cells were harvested for BrdU labeling, and mitotic indices were also determined based on chromosome morphology.
To further confirm this observation in a cell line that can be better synchronized, we serum starved T98G cells for 3 days to arrest cells at G0 phase (24). After release into medium containing 20% serum for 12 h, a point when the cells were enriched at G1/S, nocodazole only (control) or nocodazole-BI2536 was added, and the cells were harvested at different times for various analyses. Consistent with the data obtained with U2OS cells, BI2536-treated T98G cells showed a slower progression through S phase (Fig. 1C, compare the FACS profiles at the 18-, 20-, and 24-h points). Direct measurement of DNA synthesis supports this result. At 20 h postrelease, BrdU incorporation of the BI2536-treated cells was lower than that of control cells (Fig. 1D), since BI2536 treatment slowed down S-phase progression. In striking contrast, at 24 h postrelease, BrdU incorporation was higher in BI2536-treated cells than in control cells, since the control cells were already in G2/M phase, whereas the BI2536-treated cells were mainly in S phase at this time (Fig. 1E). Altogether, these inhibitor-based results suggest that inhibition of Plk1 reduces DNA synthesis and slows S-phase progression.
Plk1 phosphorylates Orc2 at Ser188 in vitro and in vivo.
As a component of pre-RC, Orc2 was previously suggested to be a potential Plk1 target (22). To investigate how Plk1-associated kinase activity is involved in DNA synthesis, we further characterized the relationship between Plk1 and Orc2 in detail. To determine whether Orc2 interacts with Plk1 directly, HEK293T cells were transfected with T7-Orc2, synchronized with hydroxyurea to arrest at S phase, and harvested for Plk1 IP, followed by Western blotting. Coimmunoprecipitation assays showed an interaction between overexpressed T7-Orc2 and endogenous Plk1 (Fig. 2A), indicating that Orc2 interacts with Plk1 in S phase.
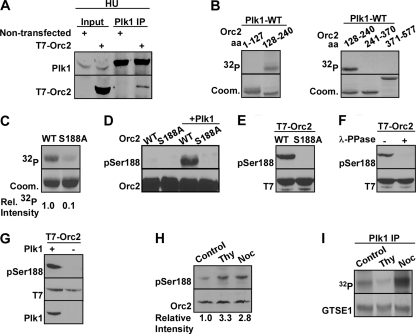
Fig. 2.
Plk1 phosphorylates Orc2 at Ser188 in vitro and in vivo. (A) Plk1 interacts with T7-Orc2 in vivo. HeLa cells were transfected with T7-Orc2, treated with hydroxyurea (HU) for 24 h, and harvested for anti-Plk1 IP, followed by Western blotting. (B) Plk1 phosphorylates recombinant Orc2 protein region (aa128-240) in vitro. Purified Plk1 was incubated with purified GST-Orc2 regions (aa1-127, aa128-240, aa241-370, and aa371-577) in the presence of [γ-32P]ATP. The reaction mixtures were resolved by SDS-PAGE, stained with Coomassie brilliant blue (Coom.), and detected by autoradiography. (C) Plk1 phosphorylates Orc2 at Ser188 in vitro. Purified Plk1 was incubated with recombinant Orc2-aa128-240 (WT or S188A mutant) and analyzed as in panel B. (D) The pS188-Orc2 antibody specifically recognizes the phosphorylated forms of Orc2. Different recombinant Orc2 proteins were incubated with or without purified Plk1 in the presence of unlabeled ATP, and immunoblotted with the pS188-Orc2 antibody. (E and F) Orc2-Ser188 is phosphorylated in vivo. (E) HEK293T cells were transfected with T7-Orc2 (WT or S188A) and harvested for Western blotting. (F) Lysates from HEK293T cells transfected with T7-Orc2-WT were treated with λ-phosphatase and subjected to Western blotting. (G) Plk1 phosphorylates Orc2 at Ser188. HeLa cells were transfected with T7-Orc2, infected with lentivirus to deplete Plk1, and immunoblotted with indicated antibodies. (H) Endogenous Orc2 is phosphorylated at S188. HeLa cells were treated with thymidine (Thy) or nocodazole (Noc) and immunoblotted with the indicated antibodies. (I) HeLa cells were treated with Thy or Noc and subjected to anti-Plk1 IP/kinase assay using GST-GTSE1 as a substrate (13).
Considering that Orc2 and Plk1 interact with each other in cells, we then sought to determine whether Orc2 is a substrate of Plk1. Accordingly, a series of in vitro kinase assays were performed with purified wild-type (WT) Plk1 and various recombinant glutathione S-transferase (GST)-Orc2 regions, purified from bacteria, in the presence of [γ-32P]ATP. As indicated, the Orc2 region from amino acids 128 to 240 (aa128-240) was phosphorylated by Plk1 in vitro (Fig. 2B). To map the phosphorylation site(s), we mutated virtually every serine/threonine to alanine within the aa128-240 region by site-directed mutagenesis and performed kinase assays with these mutants. Compared to the phosphorylation level of Orc2-WT (aa128-240), the phosphorylation level of Orc2-S188A by Plk1 was decreased 10-fold, suggesting that S188 is the major phosphorylation site by Plk1 in vitro (Fig. 2C).
In order to confirm that Orc2-S188 is indeed phosphorylated by Plk1 in vivo, a phospho-specific antibody (anti-pS188) was generated by immunization of rabbits with a 17-amino-acid peptide containing phospho-S188 and affinity purified. To characterize the specificity of the antibody, we incubated purified GST-Orc2-aa128-240 (WT or S188A) with or without WT-Plk1 in the presence of unlabeled ATP, followed by anti-pS188 Western blot. As shown in Fig. 2D, an anti-pS188 signal was detected only in the sample that was preincubated with Plk1, suggesting that the pS188 antibody does not recognize unphosphorylated Orc2. Furthermore, the S188A mutation completely abolished the anti-pS188 signal, confirming that the antibody only recognizes Orc2 phosphorylated at Ser188. Next, HEK293T cells expressing different forms of T7-Orc2 (WT or S188A) were analyzed by Western blotting with the pS188 antibody. The anti-pS188 signal was detected only in cells expressing T7-Orc2-WT, but not T7-Orc2-S188A (Fig. 2E), indicating that Ser188 of T7-Orc2 is phosphorylated in vivo. To further confirm this conclusion, we treated cell lysates of HEK293T cell expressing T7-Orc2-WT with λ-phosphatase prior to Western blotting. As expected, the anti-pS188 signal was abolished upon phosphatase treatment (Fig. 2F). Significantly, RNAi-mediated Plk1 depletion completely abolished the pS188 epitope, suggesting that Plk1 is the major kinase responsible for Orc2-S188 phosphorylation in vivo (Fig. 2G). We also sought to determine whether endogenous Orc2 is phosphorylated at Ser188. Accordingly, HeLa cells were treated with thymidine or nocodazole and harvested for Orc2 IP, followed by Western blotting. As shown in Fig. 2H, we were able to detect endogenous Orc2-S188 phosphorylation upon thymidine treatment, and the phosphorylation remains the similar level in nocodazole-treated cells. As expected, Plk1 activity is detectable in thymidine-treated cells and is elevated in nocodazole-treated cells (Fig. 2I). These cell cycle regulatory profiles of Orc2-S188 phosphorylation and Plk1 specific activity suggest that the functional significance of this phosphorylation event is likely in S phase but not in mitosis. The maintained Orc2 phosphorylation in M phase could be due to the increased Plk1 activity, consequently resulting in a slow dephosphorylation process.
Phosphorylation of Ser188 is enhanced under replication stress.
Plx1, the Xenopus homolog of Plk1, functions in DNA replication when replication forks stall (26). To rigorously test whether Plk1 phosphorylation of Orc2 contributes to DNA replication under stress in mammalian cells, we first examined the phosphorylation state of Orc2 4 h after ultraviolet (UV) irradiation. As shown in Fig. 3A, a 10-J/m2 UV treatment enhanced the phosphorylation of endogenous Orc2 at Ser188. Exposure of cells to UV irradiation at 10 J/m2 did not affect Plk1 protein expression or its kinase activity (Fig. 3B). With an increase of UV dose, the phosphorylation level of Orc2-S188 was decreased, as Plk1 is a target of DNA damage (19). The data presented in Fig. 3A suggest that at a low UV dose (10 J/m2) Plk1 kinase activity toward Orc2 might play a critical role in DNA replication. Therefore, we chose 10-J/m2 UV irradiation dose as one major way to introduce replication stress for the rest of experiments. First, UV treatment clearly enhanced Orc2-S188 phosphorylation in asynchronized HEK293T cells expressing T7-Orc2 (Fig. 3C). To assess the possible cell cycle effect on increased phosphorylation of Ser188 upon replication stress, we synchronized T98G cell in G0 phase by serum starvation, released into S phase, and treated with or without replication stress. As shown in Fig. 3D, phosphorylation of Orc2 at Ser188 increased upon UV-induced replication stress, indicating that this phosphorylation is stress-specific. Moreover, the protein levels of cyclin A (S-phase marker) from two samples were the same, suggesting that cells were in the comparable cell cycle phase, and this phosphorylation is not a simple consequence of enrichment of S-phase cells. Second, DNA replication stress is correlated with genome instability, a hallmark of cancer cells (15). To test whether this phosphorylation event is cancer cell specific, we treated RWPE-1 cells (an immortalized cell line derived from normal human adult prostate) and Panc-1 cells (a cancer cell line from human pancreatic carcinoma) with thymidine or UV. Thymidine, which represents a milder replication stress, inhibits DNA replication by a mechanism different from that of UV irradiation. The pS188 signal was barely detected in RWPE-1 cells treated with thymidine but was increased in both cell lines to a certain extent after UV treatment (Fig. 3E), suggesting that replication stress-induced Orc2-S188 phosphorylation is not cancer-cell specific. To further expand our observation, we used two additional nontransformed cell lines, and detected similar increase of Orc2-S188 phosphorylation upon replication stress (Fig. 3F and G). Third, RNAi-mediated depletion of Plk1 or Orc2 in HeLa cells after thymidine or UV treatment further confirmed that Plk1 is the kinase responsible for this phosphorylation event (Fig. 3H and I). In Fig. 3H, the reason that enough Orc2 remained after Orc2 RNAi was because we only infected cells with lentivirus targeting Orc2 for a short period due to the time limit of combining RNAi and drug treatment. Orc2 was completely knocked down after a longer incubation (Fig. 4C). Finally, to determine whether this phosphorylation event occurs under other DNA replication stress but is not restricted to UV treatment, HeLa cells were treated with other DNA replication inhibitors (hydroxyurea, gemcitabine, and aphidicolin) and harvested for anti-pS188 Western blotting. Treatment with various DNA replication inhibitors all caused an increase of the pS188 epitope (Fig. 3J), suggesting that Plk1 phosphorylation of Orc2-S188 might be a generic mechanism for cellular response to replication stress.
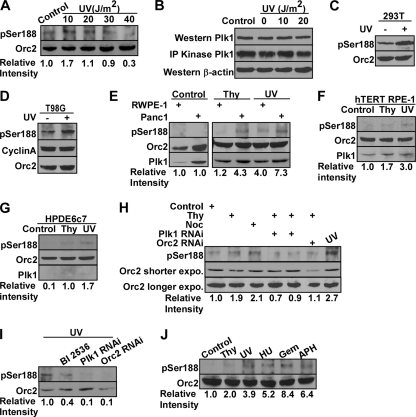
Fig. 3.
Phosphorylation of Ser188 is enhanced under replication stress. (A) HeLa cells were exposed to UV radiation with increasing doses, incubated for 4 h and harvested for Orc2 IP, followed by Western blotting. (B) HeLa cells were treated with different doses of UV, harvested 4 h post-UV treatment, and subjected to Western blot or Plk1 IP/kinase assay using GTSE1 as a substrate. (C) HEK293T cells were transfected with T7-Orc2, treated with UV (10 J/m2), incubated for 4 h, and harvested for Western blotting. (D) T98G cells were serum starved for 3 days to synchronize at G0, transfected with T7-Orc2, released into fresh medium containing 20% fetal bovine serum for 15 h, treated with or without UV (10 J/m2), and harvested 4 h post-UV treatment for various Western blot analyses. (E) RWPE-1 and Panc-1 cells were untreated or treated with thymidine or UV (10 J/m2) and subjected to anti-Orc2 IP, followed by Western blotting. (F) hTERT-RPE-1 cells were treated with thymidine or UV (10 J/m2) and subjected to anti-Orc2 IP, followed by Western blotting. (G) HPDE6c7 cells were treated with thymidine or UV (10 J/m2) and subjected to anti-Orc2 IP, followed by Western blotting. (H) HeLa cells were infected with lentiviruses to deplete Plk1 or Orc2 for 12 h, treated with thymidine, nocodazole or UV (10 J/m2), and harvested for anti-Orc2 IP, followed by Western blotting. (I) After HeLa cells were infected with lentiviruses to deplete Plk1 or Orc2 or treated with BI2536, the cells were exposed to UV (10 J/m2) and subjected to anti-Orc2 IP, followed by Western blotting. (J) Phosphorylation of Orc2 at S188 under different conditions of DNA replication stress. HeLa cells were treated with different DNA replication inhibitors and subjected to anti-Orc2 IP, followed by Western blotting. Gem, gemcitabine; APH, aphidicolin.
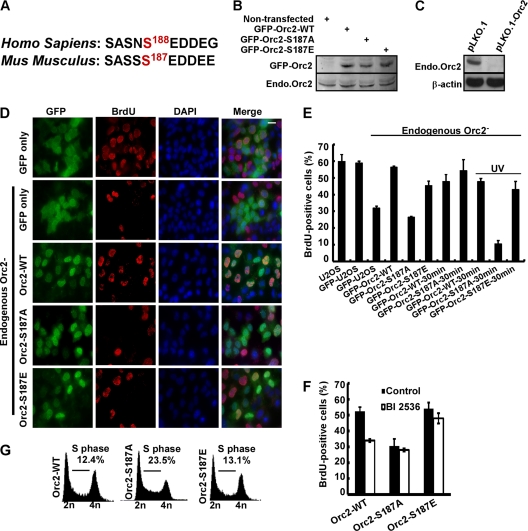
Fig. 4.
Phosphorylation of Orc2 by Plk1 promotes DNA replication. (A) Alignment of Orc2 protein sequences containing Ser188. (B) U2OS cells were transfected with RNAi-resistant murine GFP-Orc2 (WT, S187A, and S187E), selected with G418 for 2 weeks, and harvested for Western blotting. (C) U2OS cells were transfected with pLKO.1 or pLKO.1-Orc2, selected with puromycin for 48 h, and harvested for Western blotting. (D) Representative images of BrdU labeling in different cells. U2OS cells were transfected with RNAi-resistant murine GFP-Orc2 constructs (WT, S187A, and S187E) and selected with G418 for 2 weeks. Endogenous Orc2 was then depleted by transfection with pLKO.1-Orc2 and selected with puromycin for 2 days. (E) A BrdU labeling assay was performed in these cells with either 10 or 30 min of incubation or UV treatment and quantified. (F) U2OS cells stably expressing RNAi-resistant GFP-Orc2 constructs (WT, S187A, or S187E) were synchronized with the DTB, released into fresh medium with or without BI2536 (added 1 h prior to release) for 5 h, and subjected to BrdU labeling assay. (G) FACS analysis of the cells as described in panel D.
Phosphorylation of Orc2 by Plk1 promotes DNA replication.
Next, we investigated the functional significance of this phosphorylation event. Considering that inhibition of Plk1 prevents DNA synthesis and that Plk1 phosphorylates Orc2, a critical component of the DNA replication machinery under stress, we hypothesize that Plk1 phosphorylation of Orc2 at Ser188 might regulate DNA synthesis under conditions of stress. Based on protein sequence alignment, human Ser188 corresponds to murine Ser187 (Fig. 4A). A series of U2OS cell lines (a cancer cell line derived from human osteosarcoma) stably expressing murine RNAi-resistant GFP-Orc2 (WT, S187A, or S187E) were generated, and the expression levels of GFP-Orc2-WT, -S187A, and -S187E were similar to that of endogenous Orc2 (Fig. 4B). It was previously reported that knockdown of Orc2 led to reduction of DNA synthesis (17). To test whether Plk1 phosphorylation of Orc2 affects DNA replication under stress, we first compared the DNA replication in U2OS cell lines stably expressing different forms of Orc2 (WT, S187A, or S187E) after endogenous Orc2 had been depleted (Fig. 4C). As expected, Orc2 depletion led to reduction of BrdU incorporation and expression of mouse Orc2-WT and -S187E, a phospho-mimic construct, reversed the defect in DNA replication. In striking contrast, cells expressing Orc2-S187A remained replication incompetent (Fig. 4D and E). We might point out that the reduced replication in cells expressing Orc2-S187A was detected only with 10 min of BrdU labeling, but not with 30 min of BrdU labeling (Fig. 4E), suggesting that Plk1 phosphorylation of Orc2 contributes to but is not absolutely essential for DNA replication without additional stress. When UV was introduced as replication stress, we detected a reduction of BrdU incorporation in UV-treated cells expressing Orc2-S187A even after 30 min of labeling (Fig. 4E), supporting the concept that Plk1 phosphorylation of Orc2 is essential for DNA replication under stress. Furthermore, we found that Plk1 inhibition decreased DNA synthesis in cells expressing Orc2-WT but only have slight effect in cells either expressing Orc2-S187A or Orc2-S187E, suggesting that Orc2 might be a major substrate of Plk1 in terms of the regulation of DNA replication in S phase (Fig. 4F).
The significance of Plk1 phosphorylation of Orc2 in cell cycle progression was further examined by FACS analysis. As indicated in Fig. 4G, cells expressing the Plk1-unphosphorylatable mutant (Orc2-S187A) tended to accumulate at S phase, in contrast to the cells expressing Orc2-WT and -S187E. To monitor DNA synthesis in more detail, cells stably expressing different Orc2 constructs (WT, S187A, or S187E) were synchronized by the double thymidine block (DTB) protocol, released for different times, and harvested for FACS analysis. As indicated in Fig. 5A, cells expressing Orc2-S187A progressed through S phase more slowly than cells expressing Orc2-WT and -S187E, a finding consistent with the phenotype of U2OS cells after BI2536 treatment (Fig. 1A). We also directly measured DNA synthesis by BrdU labeling assays. As indicated, cells expressing Orc2-WT and -S187E started to enter S phase after 2 h of release from the DTB, reached a peak of DNA synthesis after 4 h of release, and exited S phase after 6 h of release. In contrast, a 2-h delay was observed in DNA synthesis in cells expressing Orc2-S187A (Fig. 5B), further confirming that Plk1 phosphorylation of Orc2 facilitates DNA replication, thus promoting S-phase progression under stress.
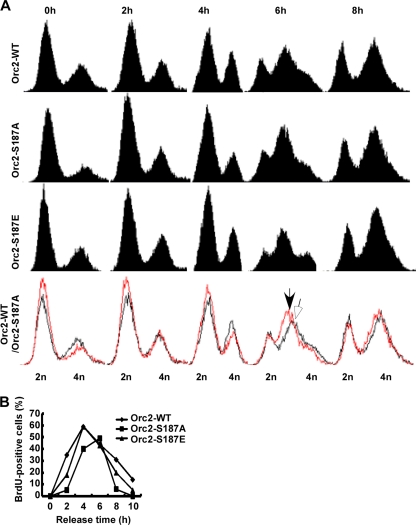
Fig. 5.
Cells expressing Orc2-S187A progress through S phase more slowly than cells expressing Orc2-WT. (A) U2OS cells stably expressing RNAi-resistant murine GFP-Orc2 (WT, S187A, or S187E) were synchronized with the DTB protocol and released for FACS analysis at different times. The bottom row shows an overlay of cell cycle profiles of U2OS cells expressing GFP-Orc2-WT (black) and -S187A (red). The peaks of FACS profiles at the 6-h point are indicated by an open arrow (WT) and a closed arrow (S187A), respectively. (B) S-phase progressions in different GFP-Orc2 cell populations (WT, S187A, and S187E). Cells as described in Fig. 4D were synchronized with the DTB, released for different times, and harvested for the BrdU labeling assay.
Plk1 phosphorylation of Orc2 regulates pre-RC maintenance under conditions of stress.
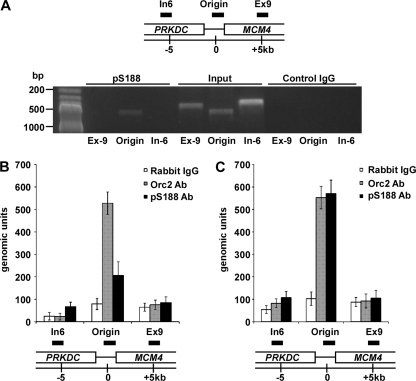
To understand the underlying mechanism of the DNA replication defect in Orc2-S187A-expressing cells, we sought to determine whether Orc2-S188 phosphorylation associates with DNA replication origin. Toward that end, we performed a series of chromatin immunoprecipitation (ChIP) experiments with antibodies against Orc2 and pS188-Orc2. By the regular PCR method, we detected a specific signal on MCM4 origin, a well-defined mammalian replication origin for Orc2 (8, 18), in ChIP against pS188 antibody but not in ChIP against control rabbit IgG after UV treatment (Fig. 6A), suggesting that the pS188 antibody could be used for ChIP assay. To further quantify the signal detected, we used a quantitative real-time PCR method (8) with Orc2 antibody as an internal control. In S-phase cells without stress, we were able to detect the signal by Orc2 antibody on MCM4 origin (∼500 genomic units) and a weak signal by pS188 antibody (∼200 genomic units) (Fig. 6B). After UV treatment, the signal by pS188 antibody increased to 500 genomic units (an ∼2.5-fold increase of nontreated cells), whereas the signal by Orc2 antibody remains the same level (Fig. 6C). These data, consistent with our observation that the phosphorylation of Ser188 was increased in response to UV treatment by Western blot analysis (Fig. 3), provide direct evidence that the phosphorylation level of Orc2 associating with the origin is increased under replication stress.
Fig. 6.
Orc2-Ser188 phosphorylation associates with DNA replication origin under stress condition. (A) HeLa cells were synchronized in G1 phase by mimosine (400 μM) treatment and released into S phase. After UV (10 J/m2) treatment, the cells were incubated for 4 h and subjected to ChIP analysis. Regular PCR was performed with DNA extracted from chromatin precipitated with antibodies against pS188-Orc2 or rabbit IgG as control. In6 is the region 6 kb upstream of MCM4 origin, Ex9 is the region 5 kb downstream of MCM4 replication origin. The PCR products of In6, MCM4, and Ex9 are 346, 483, and 387 bp, respectively. (B) HeLa cells were synchronized in G1 phase by mimosine (400 μM) treatment, released into S phase, and subjected to ChIP analysis. Quantitative real-time PCR was performed with DNA extracted from chromatin precipitated with antibodies against Orc2, pS188, or rabbit IgG as a control. In6 is the region that is 6 kb upstream of the MCM4 replication origin, and Ex9 is the region that is 5 kb downstream of the MCM4 origin (8). (C) HeLa cells were synchronized in G1 phase by mimosine treatment, released into S phase, and treated with UV (10 J/m2). After incubation for 4 h, the cells were subjected to ChIP analysis as described in panel B.
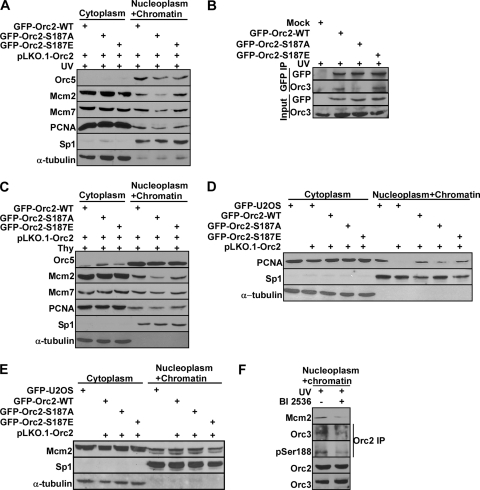
To evaluate the significance of this phosphorylation event on pre-RC complex that binds on DNA replication origin, we then tested the integrity of the pre-RC complex in cell lines expressing different Orc2 constructs. Cells expressing Orc2-S187A showed a decrease in chromatin/nucleoplasm Mcm2 protein level and a moderate decrease in chromatin/nucleoplasm Orc5, Mcm7, and PCNA protein levels in comparison to cells expressing Orc2-WT and -S187E upon UV treatment (Fig. 7A). Next, we further tested the binding affinity between endogenous Orc3 and different forms of Orc2 (WT, S187A, and S187E), since Orc3 was reported as a direct Orc2-binding partner to form the core subcomplex of ORC (2). Introduction of the S187A mutation in Orc2 decreased its binding affinity to Orc3 upon UV treatment (Fig. 7B), providing an explanation for the decreased levels of other pre-RC components (Fig. 7A). Because the Orc2 phosphorylation signal is enhanced by DNA replication stress in S phase (Fig. 3) and because pre-RC formation occurs in late mitosis and early G1, we believe that Plk1-mediated Orc2 phosphorylation is unlikely to affect the loading of the pre-RC but is required for the maintenance of the pre-RC under stress conditions in S phase. To determine whether the requirement of Plk1 phosphorylation of Orc2 in the maintenance of pre-RC is UV specific, we also analyzed the integrity of the pre-RC of thymidine-treated cells stably expressing Orc2. Similar to what we observed in UV-treated cells, cells expressing Orc2-S187A also had a reduced level of chromatin/nucleoplasm Orc5, Mcm2, Mcm7, and PCNA compared to cells expressing Orc2-WT or Orc2-S187E (Fig. 7C). The effect of thymidine treatment was not as dramatic as that of UV irradiation, probably because of different stress mechanisms of these two agents (a milder replication stress by thymidine). Finally, we also examined the integrity of the pre-RC in the absence of additional stress. In randomly growing cells expressing different forms of Orc2, introduction of the Orc2-S187A mutation slightly affected the chromatin/nucleoplasm protein level of PCNA (Fig. 7D), but not Mcm2 protein (Fig. 7E), further suggesting that Plk1 phosphorylation of Orc2 is a critical event for the maintenance of pre-RC under conditions of stress. To confirm this conclusion with endogenous proteins, we monitored pre-RC complex in HeLa cells with UV-induced replication stress in the presence or absence of Plk1 inhibitor BI2536. As shown in Fig. 7F, along with reduced Orc2-S188 phosphorylation, inhibition of Plk1 decreased nucleoplasm and chromatin fraction of Mcm2, and decreased the binding affinity between Orc2 and Orc3. Altogether, we concluded that the kinase activity of Plk1 is required for maintaining of pre-RC under conditions of stress.
Fig. 7.
Plk1 phosphorylation of Orc2 regulates pre-RC maintenance under conditions of stress. (A) Distribution of DNA replication proteins in cells expressing different Orc2 constructs after UV irradiation. U2OS cells stably expressing RNAi-resistant GFP-Orc2 constructs (WT, S187A, or S187E) were infected with lentivirus to deplete the endogenous Orc2, treated with UV (10 J/m2), and incubated for 4 h. Cells were then harvested, fractionated into cytoplasmic and nucleoplasm/chromatin fractions, and analyzed by Western blot analysis. (B) Binding between different forms of Orc2 and endogenous Orc3 after UV treatment. HEK293T cells were transfected with GFP-Orc2 constructs (WT, S187A or S187E), treated with UV (10 J/m2), incubated for 4 h, and harvested, and nucleoplasm/chromatin extracts were subjected to anti-GFP IP, followed by Western blotting. (C) Distribution of DNA replication factors in cells expressing different Orc2 constructs after thymidine treatment. U2OS cells stably expressing GFP-Orc2 constructs (WT, S187A, or S187E) were depleted of Orc2 by RNAi and subjected to the DTB treatment. The cells were then harvested, fractionated into cytoplasmic and nucleoplasm/chromatin fractions, and analyzed by Western blotting. (D and E) Relative abundance of PCNA (D) and Mcm2 (E) in cell populations expressing different forms of Orc2. Randomly growing U2OS cells stably expressing GFP-Orc2 constructs (WT, S187A, and S187E) were depleted of endogenous Orc2, harvested, fractionated into cytoplasmic and nucleoplasm/chromatin fractions, and analyzed by Western blotting. (F) HeLa cells were exposed to UV (10 J/m2), treated with or without BI2536 (added 1 h prior to UV) for 6 h, and harvested. After nucleoplasm/chromatin fractions were extracted, samples were analyzed by Orc2 IP, followed by Western blotting.
Slow S-phase progression is due to intra-S-phase checkpoint activation.
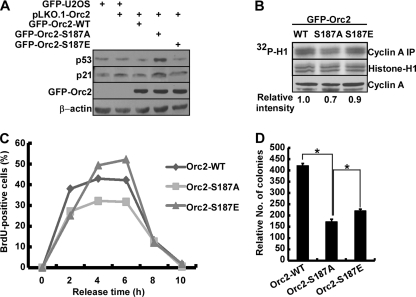
Having established the molecular mechanism of the DNA replication defect in cells expressing Orc2-187A, we then sought to determine why we observed a slower S-phase progression in these cells (Fig. 5B). Because Plk1 depletion induces S-phase checkpoint activation (30), an apparent increased S-phase population of randomly growing cells expressing Orc2-S187A might be due to activation of the intra-S-phase checkpoint (Fig. 4G). Given the fact that the p53 pathway plays an important role in arresting cells in response to checkpoint activation (16), p53 levels were analyzed in cells stably expressing Orc2 constructs (WT, S187A, or S187E) in the absence of endogenous protein. As expected, the levels of p53 and its major downstream target p21 were elevated in cells expressing Orc2-S187A (Fig. 8A), indicating activation of the intra-S-phase checkpoint in the Orc2-S187A cells. Optimal Cdk2/cyclin A activity is also critical for S-phase progression (31). Considering that p21, the inhibitor of Cdk2/cyclin A (14), was elevated in cells expressing Orc2-S187A, we performed anti-cyclin A IP/kinase assays to test whether Cdk2/cyclin A kinase activity was affected in these cells. As expected, cells expressing Orc2-S187A indeed had lower Cdk2/cyclin A activity than cells expressing Orc2-WT and -S187E (Fig. 8B), suggesting that the reduced Cdk2/cyclin A activity might contribute to the slow S-phase progression in Orc2-S187A-expressing cells. To evaluate the significance of p53 on slow S-phase progression, we transfected GFP-Orc2-WT, -S187A, or -S187E into p53-deficient cells (H1299) and monitored S-phase progression by measurement of BrdU incorporation. As shown in Fig. 8C, H1299 cells expressing Orc2-S187A showed a similar pattern of S-phase progression as cells expressing Orc2-WT. The 2-h delay of S-phase progression in p53 wild-type cells (U2OS) (Fig. 5B) was not observed in p53-deficient cells (H1299), suggesting that p53 plays a major role in activation of intra S-phase checkpoint that contributes to slow S-phase progression in S187A-expressing U2OS cells.
Fig. 8.
Slow S-phase progression is due to intra-S-phase checkpoint activation. (A) U2OS cells stably expressing GFP-Orc2 constructs (WT, S187A, or S187E) were depleted of endogenous Orc2 and subjected to anti-p53 and anti-p21 Western blots. (B) Cells as in panel A were subjected to anti-cyclin A IP/kinase assays using histone H1 as a substrate. (C) H1299 cells were synchronized with the DTB, transfected with murine RNAi-resistant GFP-Orc2 constructs (WT, S187A, or S187E), released for different times, and harvested for BrdU labeling assay. (D) U2OS cells stably expressing murine RNAi-resistant GFP-Orc2 constructs (WT, S187A, or S187E) were grown on soft agar and treated with hydroxyurea (50 μM) for 5 days, and the relative colony numbers were determined by a CellTiter-Glo luminescent cell viability assay (*, P < 0.05).
Since Plk1 phosphorylation of Orc2 promotes the maintenance of pre-RC to ensure smooth S-phase progression under replication stress, we further sought to determine whether S187A-expressing cells are more sensitive to replication stress. We performed a soft agar quantitative assay by treating U2OS cells stably expressing Orc2-WT, -S187A, or -S187E with low-dose hydroxyurea for 5 days. As shown in Fig. 8D, cells expressing Orc2-S187A form fewer colonies than cells expressing Orc2-WT or Orc2-S187E in the presence of replication stress, suggesting that phosphorylation of Ser187 did protect cells from replication stress. Cells expressing Orc2-S187E mutant showed more colonies than cells expressing S187A but did not fully rescue the phenotype, indicating that phosphorylation of Ser187 alone is not sufficient to maintain cell viability under replication stress.
DISCUSSION
Plk1 in DNA replication.
Plk1, the best-characterized member of a family of Ser/Thr protein kinases, is an important regulator of the cell cycle. The protein expression level of Plk1 is tightly regulated during the cell cycle; it is detected at the S phase, continues to increase at the G2 phase, and reaches a peak during mitosis (6). Genetic and biochemical experiments have shown that Plk1 is involved in many aspects of mitosis (21). However, little is known about the function of Plk1 beyond mitosis. We show here that Orc2, an essential component of DNA replication machinery, is a Plk1 substrate in vivo. Under conditions of stress, Plk1 phosphorylation of Orc2 at Ser188 on DNA replication origin promotes maintenance of the pre-RC, thus ensuring smooth S-phase progression. These findings reveal a novel function of mammalian Plk1 and provide a mechanism to understand how Plk1 facilitates DNA replication under stress.
BI2536, a Plk1 inhibitor, instantly and reversibly inhibits Plk1 kinase activity (10). Since most of the functions of Plk1 occur during mitosis, we used BI2536 to inhibit Plk1 activity at times other than mitosis, thus excluding possible secondary effects generated by Plk1 depletion-induced mitotic defects. We show that inhibition of Plk1 slowed S-phase progression in different cancer cell lines (Fig. 1). We acknowledge that we cannot exclude the possibility that the observed phenotype is due to partial inhibition of Plk2 and Plk3, two polo kinases with interphase functions (20). The BI2536-associated specificity issue can be partially addressed by the RNAi approach, which was used to show slow S-phase progression upon Plk1 depletion in HeLa cells (30). In addition, we previously reported that Plk1 phosphorylation of Hbo1 (histone acetyltransferase binding to Orc1), another component of the DNA replication machinery, in late mitosis is essential for pre-RC formation (29). The experiments described here are designed to address the question whether Plk1 is directly involved in DNA replication during S-phase progression, as we analyzed cells that were released from block in either late G1 or G0 phase. We conclude that Plk1 activity contributes to S-phase progression in cancer cells but is not absolutely required for DNA replication in the absence of additional stress. Due to perturbed regulation and genomic instability, cancer cells consistently encounter DNA replication stress in comparison with normal cells. Since our central hypothesis is that Plk1 is particularly critical for DNA replication under stress, we would predict that normal cells are less dependent on Plk1 for their replication. In support, our results showed that the anti-pS188-Orc2 signal is more robust in cancer cells (Panc-1, HeLa, and T98G cells) than in nontransformed cells (RWPE-1, HPDE6c7, and hTERT-RPE1 cells) (Fig. 3). Moreover, the pS188-Orc2 epitope increased in both cancer cells and normal cells after low-dose UV treatment, suggesting that this phosphorylation event is not cancer-cell specific. Therefore, Plk1-mediated phosphorylation of Orc2 represents a universal mechanism of cells to maintain genome integrity. We are aware that even normal cells encounter replication errors at some point. Although we could not detect the pS188-Orc2 epitope in normal cells when DNA replication is not disturbed, others showed that long-term Plk1 depletion leads to slow S-phase progression, likely due to checkpoint activation in normal cells (9).
Orc2 in DNA replication.
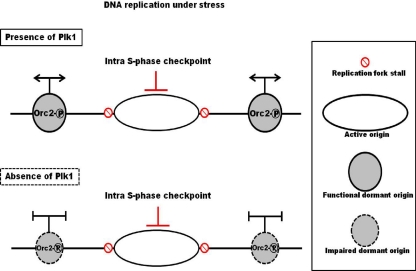
Orc2 is a component of core subcomplex (Orc2/3) that interacts with other ORC members. Depletion of Orc2 reduces the protein levels of other ORC subunits, such as Orc1 and Orc3-6, supporting a very critical role of Orc2 in DNA replication (14). In HeLa cells, it was reported that a fraction of Orc2 protein releases from the chromatin with Orc1 protein after DNA replication initiation in S phase. However, a large amount of Orc2 still binds to the chromatin with unknown functions (7). Two recent publications indicate that there is actually excess loading of Mcm proteins on the chromatin to form dormant origins. Once the DNA replication fork stalls under stress conditions, these dormant origins will fire to ensure genome integrity (5, 28). However, how these excess Mcm proteins are regulated is still not well understood. We showed that Orc2-S188 phosphorylation associates with DNA replication origin under replication stress during S phase (Fig. 6) and that the chromatin/nucleoplasm portions of Mcm proteins are reduced in cells expressing Orc2-S187A upon UV treatment (Fig. 7A). Furthermore, we found that Orc2-S187A had a low affinity to bind to Orc3 under stress (Fig. 7B). These results suggest a potential role of this phosphorylation event in pre-RC function. However, in randomly growing cells expressing Orc2-S187A, no Mcm protein defect was detected (Fig. 7E), and a replication defect in these cells could be detected only under the condition of 10 min of BrdU labeling. Considering that Mcm2-7 is continuing to be loaded onto origins in these cycling cells, we propose that Plk1 phosphorylation of Orc2 is critical for Mcm maintenance rather than Mcm loading onto the origins (Fig. 9). The temporal regulation of this phosphorylation event also supports this notion, since the phosphorylation occurs long after pre-RC loading and is enhanced upon DNA replication stress. In cells expressing the Plk1-unphosphorylatable mutant of Orc2, the excess Mcm2-7 failed to be maintained on chromatin, thus impairing the firing of dormant origins in the presence of replication stress. Therefore, our study suggests a novel mechanism in which Plk1 phosphorylation of Orc2 at Ser188 maintains functional dormant origins to promote DNA replication under stress (Fig. 9). Because Plx1 is recruited to the stalled replication fork by Mcm2 and is required for ATM/ATR-induced checkpoint adaptation for unknown reasons (26), we speculate that mammalian Plk1 might play an important role in promoting DNA replication under stress via phosphorylation of Orc2 at Ser188 to stabilize the dormant pre-RC.
Fig. 9.
Model illustrating how Plk1 phosphorylation of Orc2 regulates DNA replication in cells under conditions of stress. Cells under DNA replication stress activate the intra-S-phase checkpoint to induce stalled replication forks. In the presence of Plk1, Plk1 phosphorylation of Orc2 at Ser188 on the origin of DNA replication helps maintain the pre-RC to assure firing of the dormant origins under stress. In the absence of Plk1, Orc2 is not phosphorylated at Ser188 and the pre-RC is impaired, leading to DNA replication defects and a slowed S-phase progression.
In addition to an S-phase defect, Orc2 depletion in human cells causes destruction of heterochromatin protein 1 (HP1) and abnormally condensed chromosomes, indicating that Orc2 has functions other than pre-RC formation (17). Reduced levels of HP1 members have been associated with increased DNA repair (4). Whether Plk1 phosphorylation of Orc2 also affects other functions of Orc2 during the cell cycle needs further experimentation.
Plk1 depletion causes intra-S-phase checkpoint activation (30), likely due to lack of Orc2-Ser188 phosphorylation (Fig. 8). It is well documented that Plk1 elevation is correlated with proliferation and transformation and Plk1 depletion causes cell death in cancer cells but not in normal cells (12), arguing that cancer cells rely more heavily on Plk1 than normal cells. Genome instability associated with tumorigenesis will consistently put cancer cells under stress. Since Plk1 phosphorylation of Orc2 provides a unique mechanism for cancer cells to continue DNA replication in the presence of stress, gradual elevation of Plk1 in normal cells under chronic stress will likely contribute to transformation, consequently cancer progression.
ACKNOWLEDGMENTS
We thank Kevin Struhl from Harvard Medical School for sharing primers in ChIP assay and Benoit Miotto for discussions with ChIP techniques. We thank Scott Briggs, Paul South, and Hai-Ning Du from Purdue University for discussions of ChIP techniques. We thank Bruce Stillman for the T7-Orc2 construct. We appreciate Eleanor Erikson for critical reading of the manuscript.
X.L. is a recipient of the Howard Temin Award from the National Cancer Institute (K01CA114401 ). Support from the Purdue University Center for Cancer Research Small Grants Program is gratefully acknowledged. This project was also supported by National Science Foundation (MCB-1049693), the Elsa U. Pardee Foundation (grant 204937), and Uniting against Lung Cancer (grant 09107892).
Footnotes
Published ahead of print on 26 September 2011.
REFERENCES
- 1. Bell S. P., Dutta A. 2002. DNA replication in eukaryotic cells. Annu. Rev. Biochem. 71: 333–374 [DOI] [PubMed] [Google Scholar]
- 2. Brand N., Faul T., Grummt F. 2007. Interactions and subcellular distribution of DNA replication initiation proteins in eukaryotic cells. Mol. Genet. Genomics 278: 623–632 [DOI] [PubMed] [Google Scholar]
- 3. Cawley S., et al. 2004. Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell 116: 499–509 [DOI] [PubMed] [Google Scholar]
- 4. Dinant C., Luijsterburg M. S. 2009. The emerging role of HP1 in the DNA damage response. Mol. Cell. Biol. 29: 6335–6340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ge X. Q., Jackson D. A., Blow J. J. 2007. Dormant origins licensed by excess Mcm2-7 are required for human cells to survive replicative stress. Genes Dev. 21: 3331–3341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Golsteyn R. M., Mundt K. E., Fry A. M., Nigg E. A. 1995. Cell cycle regulation of the activity and subcellular localization of Plk1, a human protein kinase implicated in mitotic spindle function. J. Cell Biol. 129: 1617–1628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kreitz S., Ritzi M., Baack M., Knippers R. 2001. The human origin recognition complex protein 1 dissociates from chromatin during S phase in HeLa cells. J. Biol. Chem. 276: 6337–6342 [DOI] [PubMed] [Google Scholar]
- 8. Ladenburger E. M., Keller C., Knippers R. 2002. Identification of a binding region for human origin recognition complex proteins 1 and 2 that coincides with an origin of DNA replication. Mol. Cell. Biol. 22: 1036–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lei M., Erikson R. L. 2008. Plk1 depletion in nontransformed diploid cells activates the DNA-damage checkpoint. Oncogene 27: 3935–3943 [DOI] [PubMed] [Google Scholar]
- 10. Lenart P., et al. 2007. The small-molecule inhibitor BI 2536 reveals novel insights into mitotic roles of polo-like kinase 1. Curr. Biol. 17: 304–315 [DOI] [PubMed] [Google Scholar]
- 11. Li H., Wang Y., Liu X. 2008. Plk1-dependent phosphorylation regulates functions of DNA topoisomerase IIα in cell cycle progression. J. Biol. Chem. 283: 6209–6221 [DOI] [PubMed] [Google Scholar]
- 12. Liu X., Lei M., Erikson R. L. 2006. Normal cells, but not cancer cells, survive severe Plk1 depletion. Mol. Cell. Biol. 26: 2093–2108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liu X. S., Li H., Song B., Liu X. 2010. Polo-like kinase 1 phosphorylation of G2 and S-phase-expressed 1 protein is essential for p53 inactivation during G2 checkpoint recovery. EMBO Rep. 11: 626–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Machida Y. J., Teer J. K., Dutta A. 2005. Acute reduction of an origin recognition complex (ORC) subunit in human cells reveals a requirement of ORC for Cdk2 activation. J. Biol. Chem. 280: 27624–27630 [DOI] [PubMed] [Google Scholar]
- 15. Negrini S., Gorgoulis V. G., Halazonetis T. D. 2010. Genomic instability: an evolving hallmark of cancer. Nat. Rev. Mol. Cell. Biol. 11: 220–228 [DOI] [PubMed] [Google Scholar]
- 16. Petermann E., Caldecott K. W. 2006. Evidence that the ATR/Chk1 pathway maintains normal replication fork progression during unperturbed S phase. Cell Cycle 5: 2203–2209 [DOI] [PubMed] [Google Scholar]
- 17. Prasanth S. G., Prasanth K. V., Siddiqui K., Spector D. L., Stillman B. 2004. Human Orc2 localizes to centrosomes, centromeres and heterochromatin during chromosome inheritance. EMBO J. 23: 2651–2663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Schaarschmidt D., Ladenburger E. M., Keller C., Knippers R. 2002. Human Mcm proteins at a replication origin during the G1 to S phase transition. Nucleic Acids Res. 30: 4176–4185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Smits V. A., et al. 2000. Polo-like kinase-1 is a target of the DNA damage checkpoint. Nat. Cell Biol. 2: 672–676 [DOI] [PubMed] [Google Scholar]
- 20. Steegmaier M., et al. 2007. BI 2536, a potent and selective inhibitor of polo-like kinase 1, inhibits tumor growth in vivo. Curr. Biol. 17: 316–322 [DOI] [PubMed] [Google Scholar]
- 21. Strebhardt K. 2010. Multifaceted polo-like kinases: drug targets and anti-targets for cancer therapy. Nat. Rev. Drug Discov. 9: 643–660 [DOI] [PubMed] [Google Scholar]
- 22. Stuermer A., et al. 2007. Mouse pre-replicative complex proteins colocalize and interact with the centrosome. Eur. J. Cell Biol. 86: 37–50 [DOI] [PubMed] [Google Scholar]
- 23. Sunkel C. E., Glover D. M. 1988. polo, a mitotic mutant of Drosophila displaying abnormal spindle poles. J. Cell Sci. 89 (Pt. 1): 25–38 [DOI] [PubMed] [Google Scholar]
- 24. Takahashi Y., Rayman J. B., Dynlacht B. D. 2000. Analysis of promoter binding by the E2F and pRB families in vivo: distinct E2F proteins mediate activation and repression. Genes Dev. 14: 804–816 [PMC free article] [PubMed] [Google Scholar]
- 25. Takeda D. Y., Dutta A. 2005. DNA replication and progression through S phase. Oncogene 24: 2827–2843 [DOI] [PubMed] [Google Scholar]
- 26. Trenz K., Errico A., Costanzo V. 2008. Plx1 is required for chromosomal DNA replication under stressful conditions. EMBO J. 27: 876–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tsvetkov L., Stern D. F. 2005. Interaction of chromatin-associated Plk1 and Mcm7. J. Biol. Chem. 280: 11943–11947 [DOI] [PubMed] [Google Scholar]
- 28. Woodward A. M., et al. 2006. Excess Mcm2-7 license dormant origins of replication that can be used under conditions of replicative stress. J. Cell Biol. 173: 673–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wu Z. Q., Liu X. 2008. Role for Plk1 phosphorylation of Hbo1 in regulation of replication licensing. Proc. Natl. Acad. Sci. U. S. A. 105: 1919–1924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yim H., Erikson R. L. 2009. Polo-like kinase 1 depletion induces DNA damage in early S prior to caspase activation. Mol. Cell. Biol. 29: 2609–2621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhu Y., et al. 2004. Intra-S-phase checkpoint activation by direct CDK2 inhibition. Mol. Cell. Biol. 24: 6268–6277 [DOI] [PMC free article] [PubMed] [Google Scholar]