Abstract
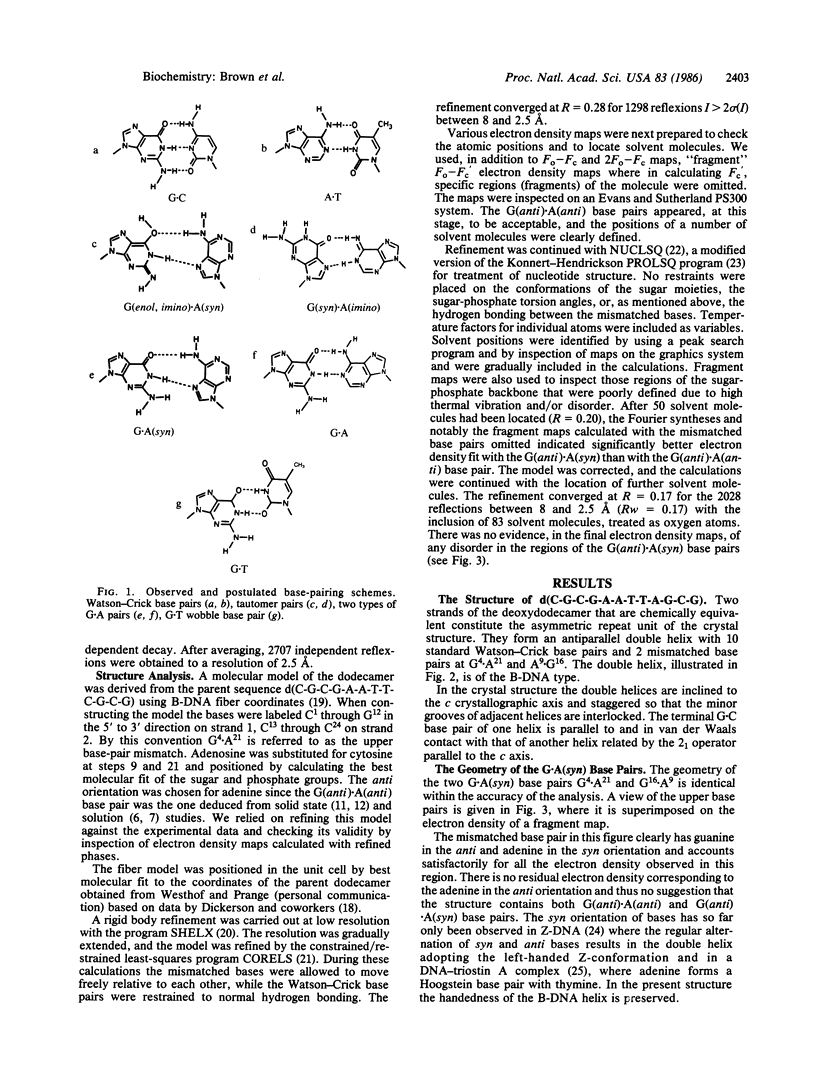
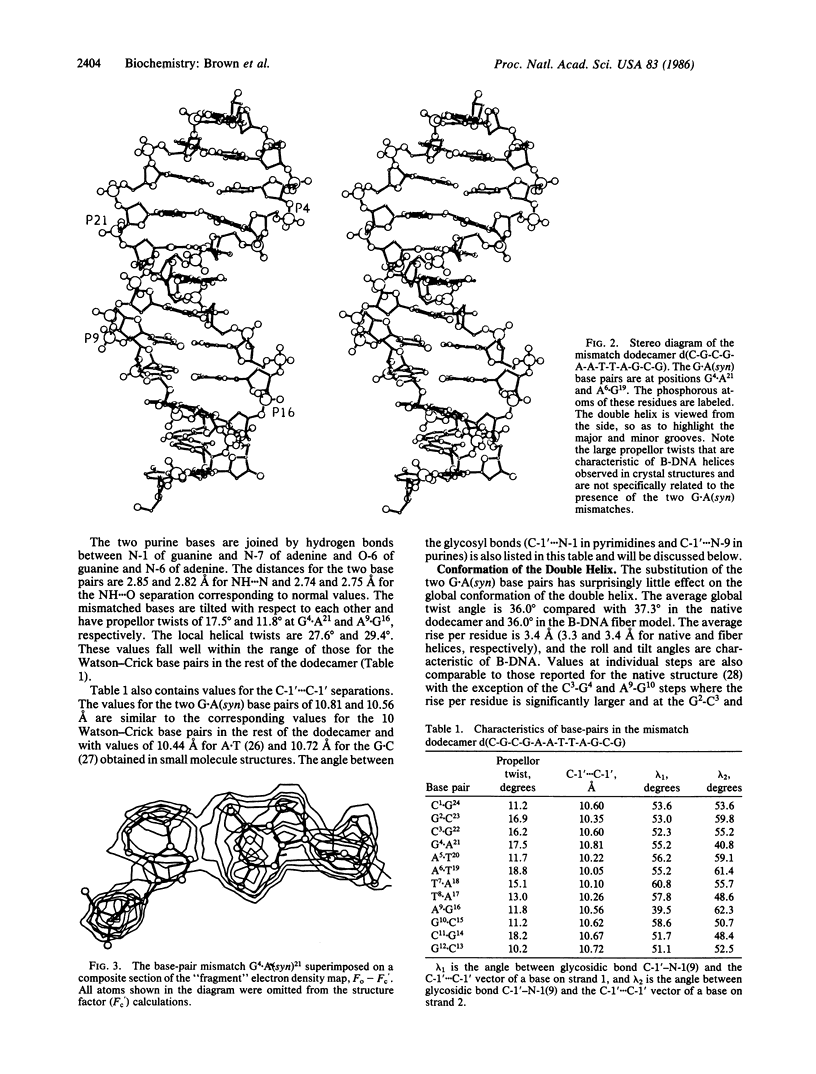
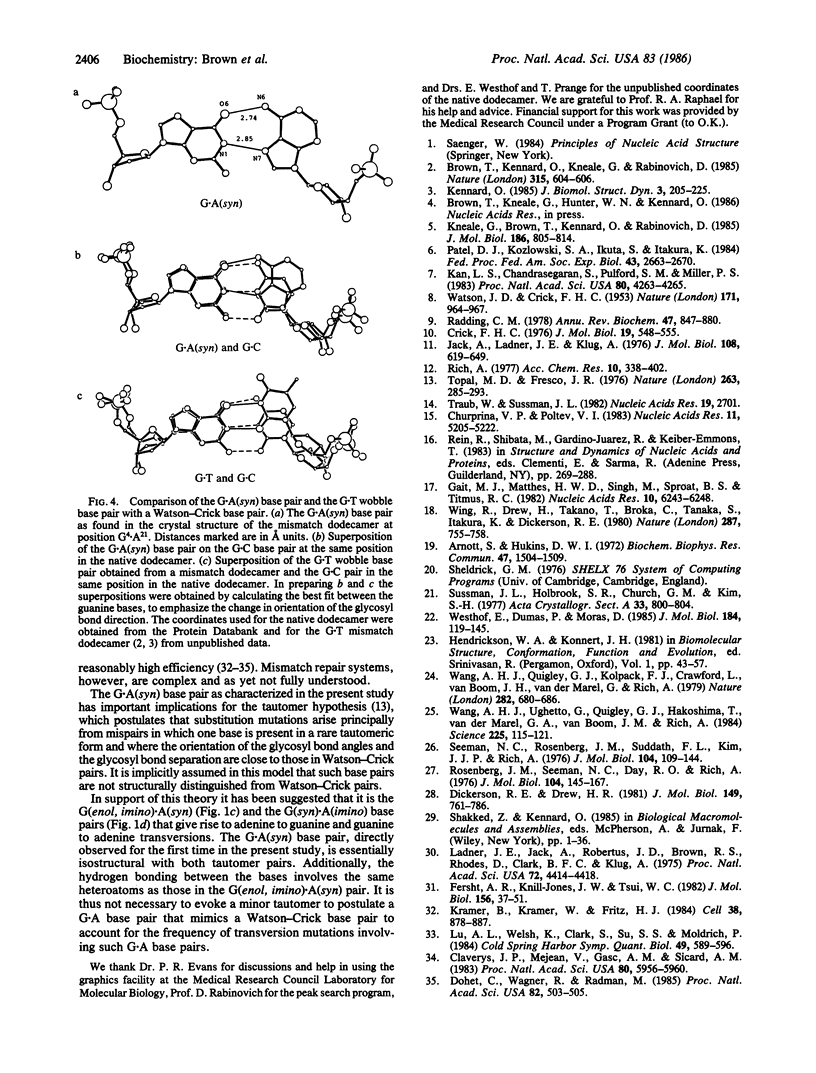
The synthetic deoxydodecamer d(C-G-C-G-A-A-T-T-A-G-C-G) was analyzed by x-ray diffraction methods, and the structure was refined to a residual error of R = 0.17 at 2.5-A resolution (2 sigma data) with 83 water molecules located. The sequence crystallizes as a full turn of a B-DNA helix and contains 2 purine X purine (G.A) base pairs and 10 Watson-Crick base pairs. The analysis shows conclusively that adenine is in the syn orientation with respect to the sugar moiety whereas guanine adopts the usual trans orientation. Nitrogen atoms of both bases are involved in hydrogen bonding with the N-1 of guanine 2.84 A from the N-7 of adenine and the N-6 of adenine within 2.74 A of the O-6 of guanine. The C-1'...C-1' separation is 10.7 A close to that for standard Watson-Crick base pairs. The incorporation of the purine.purine base pairs at two steps in the dodecamer causes little perturbation of either the local or the global conformation of the double helix. Comparison of the structural features with those of the G.T wobble pair and the standard G.C pair suggests a rationale for the differential enzymatic repair of the two types of base-pair mismatches.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arnott S., Hukins D. W. Optimised parameters for A-DNA and B-DNA. Biochem Biophys Res Commun. 1972 Jun 28;47(6):1504–1509. doi: 10.1016/0006-291x(72)90243-4. [DOI] [PubMed] [Google Scholar]
- Brown T., Kennard O., Kneale G., Rabinovich D. High-resolution structure of a DNA helix containing mismatched base pairs. Nature. 1985 Jun 13;315(6020):604–606. doi: 10.1038/315604a0. [DOI] [PubMed] [Google Scholar]
- Chuprina V. P., Poltev V. I. Possible conformations of double-helical polynucleotides containing incorrect base pairs. Nucleic Acids Res. 1983 Aug 11;11(15):5205–5222. doi: 10.1093/nar/11.15.5205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claverys J. P., Méjean V., Gasc A. M., Sicard A. M. Mismatch repair in Streptococcus pneumoniae: relationship between base mismatches and transformation efficiencies. Proc Natl Acad Sci U S A. 1983 Oct;80(19):5956–5960. doi: 10.1073/pnas.80.19.5956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crick F. H. Codon--anticodon pairing: the wobble hypothesis. J Mol Biol. 1966 Aug;19(2):548–555. doi: 10.1016/s0022-2836(66)80022-0. [DOI] [PubMed] [Google Scholar]
- Dickerson R. E., Drew H. R. Structure of a B-DNA dodecamer. II. Influence of base sequence on helix structure. J Mol Biol. 1981 Jul 15;149(4):761–786. doi: 10.1016/0022-2836(81)90357-0. [DOI] [PubMed] [Google Scholar]
- Dohet C., Wagner R., Radman M. Repair of defined single base-pair mismatches in Escherichia coli. Proc Natl Acad Sci U S A. 1985 Jan;82(2):503–505. doi: 10.1073/pnas.82.2.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fersht A. R., Knill-Jones J. W., Tsui W. C. Kinetic basis of spontaneous mutation. Misinsertion frequencies, proofreading specificities and cost of proofreading by DNA polymerases of Escherichia coli. J Mol Biol. 1982 Mar 25;156(1):37–51. doi: 10.1016/0022-2836(82)90457-0. [DOI] [PubMed] [Google Scholar]
- Gait M. J., Matthes H. W., Singh M., Sproat B. S., Titmas R. C. Rapid synthesis of oligodeoxyribonucleotides. VII. Solid phase synthesis of oligodeoxyribonucleotides by a continuous flow phosphotriester method on a kieselguhr-polyamide support. Nucleic Acids Res. 1982 Oct 25;10(20):6243–6254. doi: 10.1093/nar/10.20.6243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack A., Ladner J. E., Klug A. Crystallographic refinement of yeast phenylalanine transfer RNA at 2-5A resolution. J Mol Biol. 1976 Dec 25;108(4):619–649. doi: 10.1016/s0022-2836(76)80109-x. [DOI] [PubMed] [Google Scholar]
- Kan L. S., Chandrasegaran S., Pulford S. M., Miller P. S. Detection of a guanine X adenine base pair in a decadeoxyribonucleotide by proton magnetic resonance spectroscopy. Proc Natl Acad Sci U S A. 1983 Jul;80(14):4263–4265. doi: 10.1073/pnas.80.14.4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennard O. Structural studies of DNA fragments: the G.T wobble base pair in A, B and Z DNA; the G.A base pair in B-DNA. J Biomol Struct Dyn. 1985 Oct;3(2):205–226. doi: 10.1080/07391102.1985.10508412. [DOI] [PubMed] [Google Scholar]
- Kneale G., Brown T., Kennard O., Rabinovich D. G . T base-pairs in a DNA helix: the crystal structure of d(G-G-G-G-T-C-C-C). J Mol Biol. 1985 Dec 20;186(4):805–814. doi: 10.1016/0022-2836(85)90398-5. [DOI] [PubMed] [Google Scholar]
- Kramer B., Kramer W., Fritz H. J. Different base/base mismatches are corrected with different efficiencies by the methyl-directed DNA mismatch-repair system of E. coli. Cell. 1984 Oct;38(3):879–887. doi: 10.1016/0092-8674(84)90283-6. [DOI] [PubMed] [Google Scholar]
- Ladner J. E., Jack A., Robertus J. D., Brown R. S., Rhodes D., Clark B. F., Klug A. Structure of yeast phenylalanine transfer RNA at 2.5 A resolution. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4414–4418. doi: 10.1073/pnas.72.11.4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu A. L., Welsh K., Clark S., Su S. S., Modrich P. Repair of DNA base-pair mismatches in extracts of Escherichia coli. Cold Spring Harb Symp Quant Biol. 1984;49:589–596. doi: 10.1101/sqb.1984.049.01.066. [DOI] [PubMed] [Google Scholar]
- Patel D. J., Kozlowski S. A., Ikuta S., Itakura K. Dynamics of DNA duplexes containing internal G.T, G.A, A.C, and T.C pairs: hydrogen exchange at and adjacent to mismatch sites. Fed Proc. 1984 Aug;43(11):2663–2670. [PubMed] [Google Scholar]
- Radding C. M. Genetic recombination: strand transfer and mismatch repair. Annu Rev Biochem. 1978;47:847–880. doi: 10.1146/annurev.bi.47.070178.004215. [DOI] [PubMed] [Google Scholar]
- Rosenberg J. M., Seeman N. C., Day R. O., Rich A. RNA double-helical fragments at atomic resolution. II. The crystal structure of sodium guanylyl-3',5'-cytidine nonahydrate. J Mol Biol. 1976 Jun 14;104(1):145–167. doi: 10.1016/0022-2836(76)90006-1. [DOI] [PubMed] [Google Scholar]
- Seeman N. C., Rosenberg J. M., Suddath F. L., Kim J. J., Rich A. RNA double-helical fragments at atomic resolution. I. The crystal and molecular structure of sodium adenylyl-3',5'-uridine hexahydrate. J Mol Biol. 1976 Jun 14;104(1):109–144. doi: 10.1016/0022-2836(76)90005-x. [DOI] [PubMed] [Google Scholar]
- Topal M. D., Fresco J. R. Base pairing and fidelity in codon-anticodon interaction. Nature. 1976 Sep 23;263(5575):289–293. doi: 10.1038/263289a0. [DOI] [PubMed] [Google Scholar]
- Traub W., Sussman J. L. Adenine-guanine base pairing ribosomal RNA. Nucleic Acids Res. 1982 Apr 24;10(8):2701–2708. doi: 10.1093/nar/10.8.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WATSON J. D., CRICK F. H. Genetical implications of the structure of deoxyribonucleic acid. Nature. 1953 May 30;171(4361):964–967. doi: 10.1038/171964b0. [DOI] [PubMed] [Google Scholar]
- Wang A. H., Quigley G. J., Kolpak F. J., Crawford J. L., van Boom J. H., van der Marel G., Rich A. Molecular structure of a left-handed double helical DNA fragment at atomic resolution. Nature. 1979 Dec 13;282(5740):680–686. doi: 10.1038/282680a0. [DOI] [PubMed] [Google Scholar]
- Westhof E., Dumas P., Moras D. Crystallographic refinement of yeast aspartic acid transfer RNA. J Mol Biol. 1985 Jul 5;184(1):119–145. doi: 10.1016/0022-2836(85)90048-8. [DOI] [PubMed] [Google Scholar]
- Wing R., Drew H., Takano T., Broka C., Tanaka S., Itakura K., Dickerson R. E. Crystal structure analysis of a complete turn of B-DNA. Nature. 1980 Oct 23;287(5784):755–758. doi: 10.1038/287755a0. [DOI] [PubMed] [Google Scholar]


