Abstract
Many bacteria spread over surfaces by “swarming” in groups. A problem for scientists who study swarming is the acquisition of statistically significant data that distinguish two observations or detail the temporal patterns and two-dimensional heterogeneities that occur. It is currently difficult to quantify differences between observed swarm phenotypes. Here, we present a method for acquisition of temporal surface motility data using time-lapse fluorescence and bioluminescence imaging. We specifically demonstrate three applications of our technique with the bacterium Pseudomonas aeruginosa. First, we quantify the temporal distribution of P. aeruginosa cells tagged with green fluorescent protein (GFP) and the surfactant rhamnolipid stained with the lipid dye Nile red. Second, we distinguish swarming of P. aeruginosa and Salmonella enterica serovar Typhimurium in a coswarming experiment. Lastly, we quantify differences in swarming and rhamnolipid production of several P. aeruginosa strains. While the best swarming strains produced the most rhamnolipid on surfaces, planktonic culture rhamnolipid production did not correlate with surface growth rhamnolipid production.
INTRODUCTION
Many bacteria utilize motility as a step in surface colonization. During one type of surface exploration, described as swarming, flagellated bacteria coordinate to spread across surfaces within a thin liquid film (26). Many bacteria swarm, including species found in diverse soil and water environments (e.g., Serratia marcescens, Salmonella enterica serovar Typhimurium, Bacillus subtilis, Vibrio cholerae, Proteus mirabilis, and Pseudomonas aeruginosa) (24, 26, 50). A critical problem during the assessment of swarming is the acquisition of statistically significant data that describe the differences (for coverage area, density, gene expression, etc.) that are often present.
In the laboratory, swarming is commonly studied by using plate motility assays. For example, swarming is assessed by observing the spread of cells on top of specific concentrations of agar (0.4% to 1.5%) (26). Reports for bacterial swarming are often absolute in nature. In most cases, an agar plate is observed at a single time point 24 or 48 h postinoculation, and a given strain or genetic mutant is judged to either swarm or not swarm. Other studies amplify this basic protocol to describe observational variations in swarm patterns. The current methods have not been extended to include imaging for the purpose of quantifying gene expression, cell density, heterogeneous patterns, rates of spreading, or the onset of other pertinent parameters.
Pseudomonas aeruginosa is a ubiquitous environmental organism that acts as an opportunistic human pathogen to cause skin, eye, lung, and blood infections (27). Numerous studies have been published in recent years that address some aspect of P. aeruginosa swarming. P. aeruginosa is generally reported to swarm agar of between 0.4 and 0.7%, and the swarming of this bacterium is greatly influenced by the production of the surfactant rhamnolipid (9, 15, 29, 34, 41). Several recent studies have addressed the importance of swarm motility to the biofilm formation and antibiotic resistance of P. aeruginosa (8, 32, 38, 41). (Comparable associations that relate swarming with antibiotic resistance, virulence, and biofilm formation have also been made for other swarming bacteria, such as Proteus mirabilis, S. enterica serovar Typhimurium, and Serratia species [3, 6, 52].) Optimal swarming and the interesting tendril patterns that are often observed during P. aeruginosa swarming are known to be dependent upon the self-produced biosurfactant rhamnolipid (9, 15, 29). Rhamnolipid production is controlled by the rhl quorum-sensing cascade of P. aeruginosa; activation of the rhl system requires a sufficient population of P. aeruginosa cells (35, 36). The influence of Pseudomonas rhamnolipids has been researched by many groups; these glycolipid surfactants show cytotoxicity to eukaryotic cells, decrease liquid surface tension and aid motility, and help maintain access to nutrients within biofilms (1, 4, 19, 29, 36, 40). Rhamnolipid production is influenced by both carbon source and surface growth conditions (25, 41, 42).
Here, we present a method to track, analyze, and quantify bacterial swarming. This procedure utilizes bacterial expression of fluorescent or bioluminescent genetic reporters like green fluorescent protein (GFP) and luciferase (lux operon) or cellular stains to enable time-lapse optical imaging. We specifically examine swarming of P. aeruginosa and the presence of P. aeruginosa rhamnolipid during swarming by staining with and fluorescence imaging of Nile red. Another feature of this method is that multiple bacteria can be inoculated and detected on the same petri dish using orthogonal genetic reporters. This ability to separate multiple optical reporters facilitates the direct observation of microbial interaction and/or gene expression during the motility process. We demonstrate and distinguish temporal interaction of dual swarming species P. aeruginosa and S. enterica serovar Typhimurium and also show the acquisition of bioluminescence data in addition to fluorescence in the same experiment. Lastly, our investigation of five common laboratory strains and three clinical isolates shows that rhamnolipid production is indeed highly variable among strains. Interestingly, the production of rhamnolipid by P. aeruginosa strains growing in planktonic cultures was not useful to predict rhamnolipid production of cultures during swarming. In summary, this technique enables the imaging and quantitative assessment of bacterial surface movement to allow for detailed analysis of spatiotemporal patterns.
MATERIALS AND METHODS
Bacterial swarm assays.
Swarming was studied for the strains listed in Table 1 using plate assays containing 0.45% noble agar and FAB medium with 12 mM glucose (P. aeruginosa) (25) or 0.6% noble agar and LB with 0.5% glucose (S. enterica serovar Typhimurium with P. aeruginosa) (47). Some plate assays contained dye mixtures that were added immediately prior to pouring the melted agar medium into plates. Approximately 6 h after pouring the plates, they were inoculated using a sterilized platinum wire with log-phase cells (optical density at 600 nm [OD600] ≈ 0.7) grown in their respective media used for the swarm experiments. Swarm plates that were imaged only for their comparative endpoint swarm development (i.e., analysis of different P. aeruginosa strains) were incubated at 30°C for 48 h prior to imaging.
Table 1.
Bacterial strains used in this study
| Strain | Select characteristics | Reference(s) |
|---|---|---|
| Pseudomonas aeruginosa strains | ||
| PAO1C | PAO laboratory strain, ATCC collection strain 15692 | 25, 41 |
| PAO1C-GFP | PAO1C harboring mini-Tn7gfpmut2 for fluorescence | 10, 18, 41 |
| PA14 | Laboratory strain, sequenced, used in many swarming studies | 8, 16, 37, 49 |
| PAO1 | Most common PAO laboratory strain, first to be sequenced, used in numerous studies | 12, 28, 44 |
| MPA01 | PAO laboratory strain, mutagenesis library—Seattle collection, often denoted also as PAO1 | 2, 23, 28 |
| PAK | Laboratory strain expressing type-a flagellin, used in several flagellar studies | 11, 16, 48 |
| SMC1587 | Mucoid phenotype clinical isolate | 7, 31, 33 |
| CF39 | Rugose (RSCV) phenotype clinical isolate | 5, 43 |
| FRD1 | Mucoid phenotype clinical isolate | 17, 30, 39, 45, 53 |
| PAO1C-ΔrhlAB | Rhamnolipid-deficient mutant of PAO1C | 41 |
| Salmonella enterica serovar Typhimurium FL6 | Harbors luxCDABE for bioluminescence | 51 |
Cell- and rhamnolipid-imaging dyes.
Swarm assays conducted with strains that were not fluorescent or bioluminescent contained Syto 24 (Invitrogen) at a concentration of 4 μl/100 ml to stain bacterial cells green.
Some plate assays were amended to include the lipid stain Nile red (20) as a visual indicator of spatial rhamnolipid distribution. These rhamnolipid indicator plates included a 1-in-100 dilution of filter-sterile stock containing 1 mg ml−1 Nile red (MP Biomedicals) dissolved in 85% propylene glycol (prepared the day of use to limit photoinactivation). Plates were cured and incubated in the dark.
Whole-plate imaging.
Imaging experiments were performed at room temperature (∼23°C) on a Carestream multispectral FX (MSFX) (Carestream Health, Woodbridge, CT) image station equipped with a 16-bit charge-coupled device (CCD) sensor and 300-W light source. Up to four plates were placed on a tray inside the MSFX. Since the camera for the MSFX captures from beneath the imaging platen, the plates were inverted so that the agar would not obstruct the optical path. The lids of the petri dishes were placed open side up on top of their counterparts that held the inoculated agar. The lids of the petri dishes were filled with water, and another tray was used to enclose the petri dishes in order to maintain humidity throughout the experiment. Five types of images were collected as follows.
Green fluorescence: excitation, 480 ± 10 nm; emission, 535 ± 17.5 nm; acquisition time, 30.0 s; f-stop, 4.0; field of view (FOV), 190 mm; focal plane, 27.5 mm; binning, none. These settings were used to capture fluorescence from the green fluorescent protein expressed in cells.
Red fluorescence: excitation, 540 ± 10 nm; emission, 600 ± 17.5 nm was used to collect the data; duration, 60.0 s; f-stop, 4.0; FOV, 190 mm; focal plane, 27.5 mm; binning, 2 × 2. These settings were used to capture fluorescence from Nile red staining of rhamnolipid.
Luminescence I: excitation, off; emission, no filter; acquisition, 10 s; f-stop, 2.5; FOV, 190 mm; focal plane, 27.5 mm; binning, 16 × 16. This luminescent image served to turn off the excitation lamp during intervals between fluorescent images to prevent any excitation light interference with bacterial growth.
Luminescence II: excitation, off; emission, no filter; acquisition, 300 s; f-stop, 2.5; FOV, 190 mm; focal plane, 27.5 mm; binning, 8 × 8. These settings were used to capture emissions from bioluminescent bacteria.
Image preparation and analysis.
Carestream Multispectral Software was used to batch export each image as a 16-bit TIFF file. The images were taken and stacked according to category (GFP, Nile red, etc.) using ImageJ editing software. Image processing proceeded as follows: (i) intensity signals were inverted, (ii) the look-up table was inverted, and (iii) background was subtracted using a rolling-ball radius with a pixel radius equal to half of one dimension of the image size (e.g., 1,024 for a 2,048 × 2,048 image).
Only GFP and Nile red images were used for quantitative image analysis. Images in which the P. aeruginosa bacteria had reached the perimeter of the plate were omitted from further analysis by deleting them from the stack. The ICA (intensity correlation analysis) look-up table was applied to the stack to better visualize the borders of the bacterial colony. Next, the threshold utility (with “dark background” checked) was used to adjust the threshold to isolate the bacterial colony. The wand (tracing tool) was then used to automatically select the area superimposed over the bacteria from the threshold utility. After selecting the area with a region of interest (ROI), the threshold function was reduced to visually observe and confirm that the ROI had accurately captured the true bacterial area. This process must be performed for each image analyzed, as the bacterial colony will swarm and its characteristics will change. It is important to note that as the bacteria swarm, the threshold will change, i.e., increase, as the other characteristics change. Additionally, different thresholds are used among different data sets (e.g., GFP and Nile red). In order for the ROI to accurately represent the bacterial growth, it is recommended that the threshold be increased in small increments between consecutive images. For the time-lapse intervals used for these 2-to-3 day experiments, analyzing every 5th or 10th image tended to create the best data set.
The wand has now created an ROI that accurately captures the current bacterial colony. Using the “Measure” utility, one can acquire the data of the ROI (the bacterial colony). Various data can be gathered, including area, perimeters, and integrated density, to quantify these parameters of swarming.
Confocal microscopy.
Images of single cells were obtained using a Nikon A1 confocal microscope equipped with a 100× LU Plan Fluor objective with simultaneous excitation at 488 nm and 561 nm with emission capture using settings of 525 ± 50 nm and 595 ± 50 nm.
LC-MS analyses of rhamnolipid.
Supernatants from FAB-glucose-grown cultures were analyzed using liquid chromatography-mass spectrometry (LC-MS) (13, 14). Supernatants were prepared by harvesting cultures after 48 h of growth at 37°C by centrifugation at 7,500 rpm for 10 min and filtration through a 0.22-μm filter. Acetonitrile (960 μl) and an internal standard (40 μl) of 16-hydroxyhexadecanoic acid dissolved in acetonitrile (concentration, 1,000 ppm) were added to 1 ml of each sample. Liquid chromatography separation was achieved by using a Dionex Acclaim rapid-separation LC (RSLC) 120 C18 column (2.2-μm particle size, 120-Å pore size, 2.1 by 100 mm) with an eluent gradient of 50/50 (water/acetonitrile) to 10/90 (water/acetonitrile) containing 0.1% formic acid using a flow of 0.5 ml/min at 50°C. MS was performed on the samples by using a Bruker micrOTOF II in negative-ion mode. Chromatograms were extracted from the total ion chromatograph using pseudomolecular ion molecular masses.
RESULTS AND DISCUSSION
Quantification of swarm images.
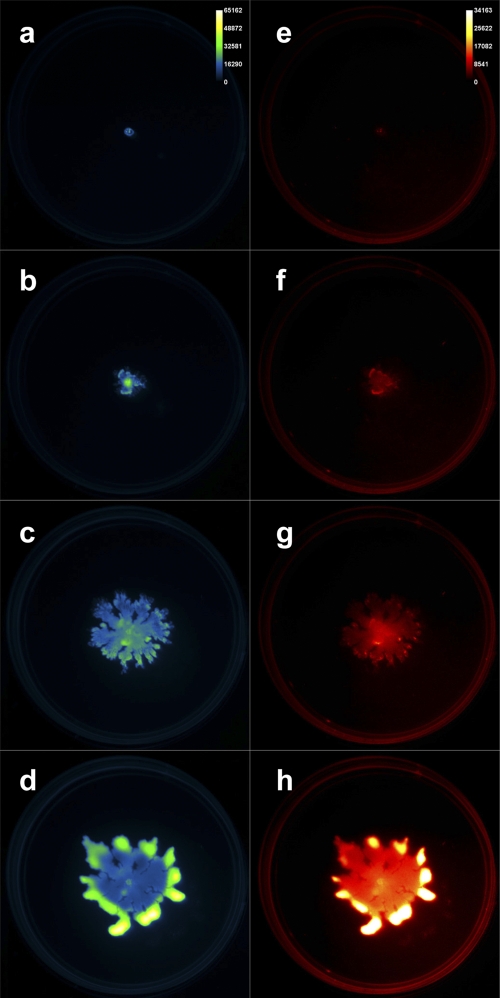
Time-lapse fluorescence imaging was performed simultaneously on three Pseudomonas aeruginosa swarm plates. A GFP-expressing P. aeruginosa strain (PAO1C-GFP) was inoculated at the center of each petri dish, which was subsequently imaged (excitation, 480 nm, and emission, 535 nm). We also captured red fluorescence (excitation, 540 nm, and emission, 600 nm) emanating from Nile red included in the agar medium to report the presence of the P. aeruginosa biosurfactant rhamnolipid, which aids swarming. The MSFX was used to acquire each of these images in an automated and sequential fashion at 22-min intervals for 2 days. Figure 1 presents four images from this time in both the green channel (left column, false-colored “green fire blue”) and red channel (right column, false-colored “red hot”). The full 48-h data set is included as movie S1 in the supplemental material.
Fig. 1.
Time-lapse images of cells and rhamnolipid during Pseudomonas aeruginosa swarming. (a to d) GFP-expressing P. aeruginosa cells. (e to h) Nile red-stained rhamnolipid. Images collected at 0-h (a, e), 10-h (b, f), 30-h (c, g), and 48-h (d, h) time points.
The raw images can be used to analyze spatial and temporal aspects of swarming. We used region-of-interest (ROI) analysis to quantify specific parameters of the acquired swarm data. Figure 2a presents the measured swarm area versus time in each fluorescent channel of the given imaging data. The total area of red fluorescence lagged that of green signal from the cells for the first 40 h of the experiment, after which it led the growth of the cells. This trend was apparent in all three experimental plates tested. Control plates were also examined with P. aeruginosa cells grown under nonswarming (hard agar) conditions, and these data are also included in Fig. 2a. Under nonswarming conditions, very little radial movement was observed compared to that in the optimal swarming experiments. Figure 2b shows the change in mean fluorescence intensity of the total swarm area over time, normalized to the initial time point. The ability to obtain such digitized fluorescence data of the entire swarm area allows for the quantification of temporal patterns and the distinction of trends such as the relative increase of rhamnolipid production with a minimal increase in cell density. Previously, such patterns have been predicted but not measured.
Fig. 2.
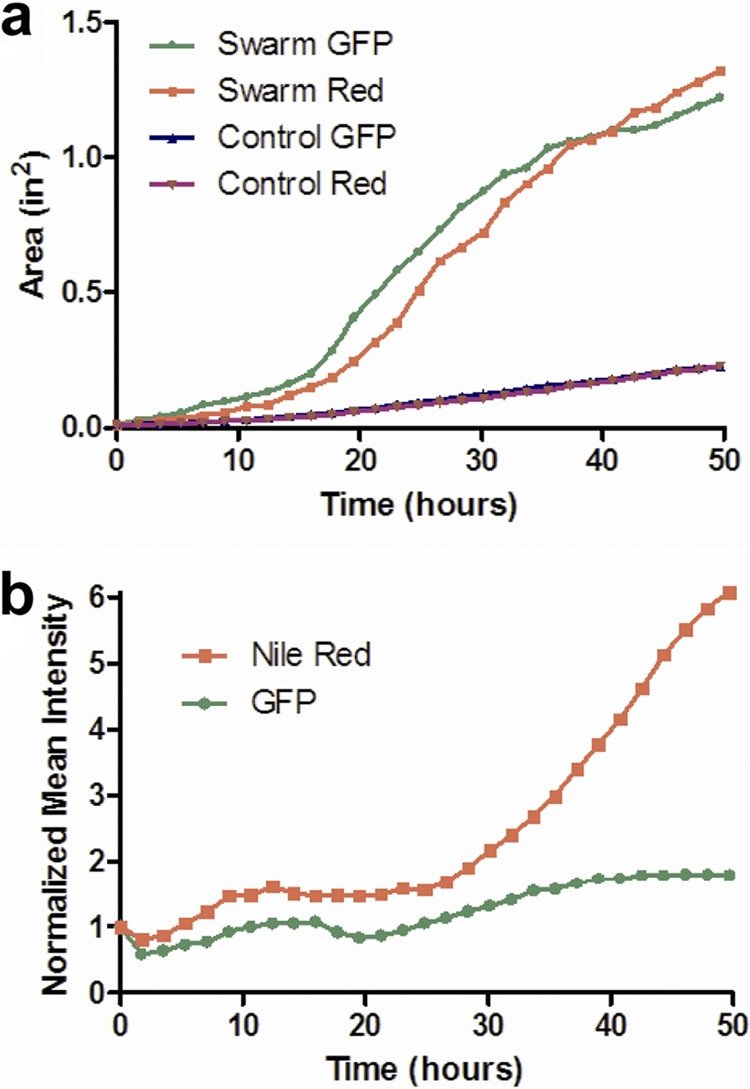
Coverage areas and fluorescence intensities of cells and rhamnolipid during Pseudomonas aeruginosa swarming. (a) Area over time for GFP-expressing P. aeruginosa cells and Nile red-stained rhamnolipid under optimal swarm conditions (0.45% agar) and control conditions with limited swarming (0.6% agar). (b) Normalized mean intensity over time for GFP-expressing P. aeruginosa cells and Nile red-stained rhamnolipid for optimal swarm conditions (0.45% agar).
Calibration of fluorescence from swarm plate assays.
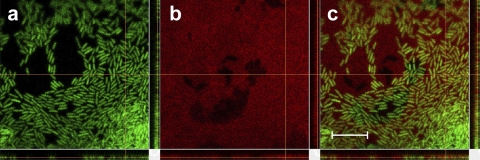
We validated a linear relationship between fluorescence and concentration for these plate assay images. For GFP-expressing P. aeruginosa cells, known amounts of cells were spotted and imaged on swarm plates. Fluorescence was measured and analyzed as for the other images, and the correlation between fluorescence and cell population was unambiguous (see Fig. S1 in the supplemental material). The same analysis was performed using known masses of rhamnolipid upon these plates containing Nile red. The correspondence between red fluorescence and rhamnolipid concentration is linear for concentrations up to 11% rhamnolipid, as shown in Fig. S2 in the supplemental material. As the rhamnolipid was not autofluorescent but required binding with Nile red, maximum fluorescence took approximately 6 h to develop (see Fig. S2b in the supplemental material). Lastly, we confirmed preferential binding of Nile red to rhamnolipid in these plate assays. Figure 3 shows P. aeruginosa cells near the edge of a swarm and Nile red-stained rhamnolipid 24 h postinoculation. While some colocalization of red with the green cells occurs, the red fluorescence is abundant over most of the field of view and is consistent with dissolved rhamnolipid within a thin liquid swarm layer that contains many P. aeruginosa cells. Thus, while the binding of Nile red to the P. aeruginosa cell wall is not negligible, the fluorescence is far greater for the stained rhamnolipid.
Fig. 3.
Close-up of P. aeruginosa swarming bacteria producing rhamnolipid upon a swarm plate containing Nile red. (a) P. aeruginosa cells expressing GFP in the 525 ± 50 emission channel. (b) Nile red-stained rhamnolipid in the 595 ± 50 emission channel. (c) Merged image of channels shown in panels a and b. Scale bar = 10 μm.
Coswarm imaging experiments.
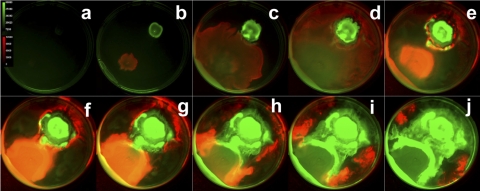
This swarm plate imaging method can be applied to study competition among two surface-spreading strains (or perhaps mutants) that possess distinguishable genetic reporters. We demonstrate such an application using a coswarming experiment with two bacterial species expressing differing photoactive molecules. We tracked GFP-expressing P. aeruginosa swarming in competition with a bioluminescent strain of Salmonella enterica serovar Typhimurium when the two cell types were inoculated on opposite sides of a petri dish. Green fluorescence and bioluminescence images were taken sequentially at 20-min intervals in order to capture the interplay between cell types over a 60-h period. Images from this time-lapse experiment are presented in Fig. 4, and the entire data set is presented in movie S2 in the supplemental material. Here, the GFP-expressing P. aeruginosa cells are false-colored green, while the Lux-expressing S. enterica serovar Typhimurium cells are colored in red. This montage and its corresponding data set capture a remarkable interplay between two bacteria as they swarm. The S. enterica serovar Typhimurium cells swarm rapidly and surround the P. aeruginosa culture, as noted in Fig. 4d. However, the P. aeruginosa cells eventually surround the competing microbes during the remainder of the time course (Fig. 4j). The biology taking place in this series is certainly complex and beyond the scope of this report. Nevertheless, this time-lapse optical imaging technique may be readily applied to yield dynamic insight into these types of microbial interactions.
Fig. 4.
Swarming of P. aeruginosa (green) and S. enterica serovar Typhimurium (red) over time. Frames show swarming at 6-h intervals.
Quantifying swarming and rhamnolipid production for several P. aeruginosa strains.
We investigated the motility of eight strains of P. aeruginosa (Table 1). Five of these strains are common laboratory strains that have been studied for decades but were each originally clinical isolates. (Three of these strains, PAO1, MPAO1, and PAO1C, are all derived from the common PAO isolate described by Holloway [21, 22]. PAO1C-GFP, used in the experiments described above, has always swarmed identically to untagged PAO1C.) In addition to these laboratory strains, three recent clinical isolates were studied for swarming: SMC1587 (7, 31, 33), FRD1 (30, 39, 45, 53), and CF39 (5, 43) are each cystic fibrosis clinical isolates that have been characterized by other research groups. The growth rates of these strains in planktonic culture were statistically equivalent under our tested culturing conditions, except for strain PA14, which doubled more slowly than the other strains (Table 2).
Table 2.
Relative swarm areas and levels of surface rhamnolipid production and of predominant liquid culture rhamnolipid congeners produced by strains of P. aeruginosad
| Strain | Swarminga | Surface rhamnolipid (Nile red intensity)a | Liquid culture rhamnolipid congenera |
Liquid culture doubling time (min) | |
|---|---|---|---|---|---|
| R-R-C10-C10 | R-C10-C10 | ||||
| PAO1C | 26.3 ± 5.1 | 32.1 ± 10.4 | 100.0 ± 5.5 | 74.1 ± 15.8 | 38.2 ± 9.8 |
| PA14 | 13.7 ± 2.7 | 22.3 ± 6.9 | 56.3 ± 6.1 | 73.0 ± 16.9 | 65.0 ± 9.1c |
| PA01 | 11.5 ± 2.6 | 2.8 ± 0.8 | 42.0 ± 9.1 | 9.6 ± 8.6 | 35.4 ± 6.1 |
| MPA01 | 15.5 ± 1.4 | 4.2 ± 0.7 | 53.2 ± 7.5 | 22.3 ± 12.2 | 35.6 ± 2.4 |
| PAK | 4.1 ± 0.4 | 1.1 ± 0.6 | 10.3 ± 2.6b | 75.7 ± 22.1b | 35.5 ± 3.1 |
| SMC1587 | 2.2 ± 0.4 | 0.2 ± 0.1 | 0.0 ± 0.0b | 0.5 ± 0.3b | 42.7 ± 9.2 |
| CF39 | 100 ± 14.4 | 100 ± 58.7 | 78.8 ± 1.9 | 100.0 ± 27.7 | 40.1 ± 11.8 |
| FRD1 | 18.1 ± 1.7 | 9.4 ± 1.7 | 13.5 ± 7.2 | 0.3 ± 0.1 | 43.4 ± 2.4 |
| PAO1C ΔrhlAB | 8.9 ± 0.8 | 1.7 ± 0.3 | ND | ND | NM |
All values except doubling time are the mean percentage ± standard deviation of the maximum level for the category.
Average and standard deviation are based upon only two measurements for these samples.
Statistically different from all other values (0.012 < P < 0.047).
ND, not detected; NM, not measured.
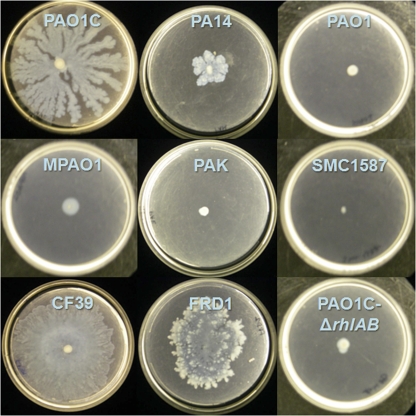
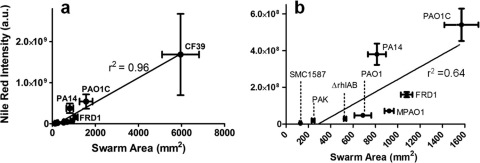
Swarming was examined for these eight strains (and a rhamnolipid-deficient, ΔrhlAB mutant of strain PAO1C) on soft-agar plates containing minimal medium with glucose as the sole carbon source (25). Many differences in swarming behavior were observed between strains, not only in the size of the swarm zone but also in tendril formation and other growth patterns at the swarm zone edges (Fig. 5). Representative images of the fluorescent patterns for stained P. aeruginosa cells and rhamnolipid are shown in Fig. S3 in the supplemental material. When considering all strains, the overall swarm area correlated well with rhamnolipid production during swarming (Fig. 6a). Strain CF39 showed significantly more surface coverage and rhamnolipid intensity than the other strains. If CF39 is excluded from consideration, the link between coverage and rhamnolipid intensity is less convincing (Fig. 6b). This poor correlation (r2 = 0.64) between rhamnolipid and swarm area suggests that while rhamnolipid aids swarming (in agreement with many previous studies), there are additional uncharacterized factors that influence the swarm differences between these P. aeruginosa strains. Table 2 includes the relative swarm area and surface rhamnolipid production for all strains.
Fig. 5.
Swarming of different Pseudomonas aeruginosa strains after 48 h on FAB-glucose medium with 0.45% noble agar.
Fig. 6.
Swarm area versus surface rhamnolipid production for strains of Pseudomonas aeruginosa. (a) All strains. (b) Blow-up of panel a that excludes data for strain CF39. Swarm area was determined by determining pixel area of cells stained with Syto 24 dye, while rhamnolipid was determined by total intensity of Nile red-stained rhamnolipid. Each data point shows the standard deviations of four measurements. a.u., arbitrary (instrument) unit.
Clinical isolate CF39 and strain PAO1C exhibited the most overall swarming (Fig. 5). Strain CF39 clearly swarmed the fastest by developing tendrils over an entire 100-mm plate within 24 h and effectively covering the entire plate within 48 h. Strain PA14 swarmed modestly, which we found curious given its well-characterized swarming ability (8, 16, 37, 49); we subsequently confirmed the ability of PA14 to robustly form tendrils when swarming on soft-agar plates containing LB medium (not shown). Strains PAO1 and MPAO1 swarmed minimally, but most replicates showed considerable “swimming” motility within these soft-agar swarm plates. Strain PAK also swarmed minimally within 48 h, but plates resting on the laboratory bench for 5 days showed evidence of swarming behavior, including tendril formation. Clinical isolate SMC1587 showed no sign of motility on these soft-agar swarm plates.
Lastly, clinical isolate FRD1 also showed substantial swarming, and its patterns were very distinct. The swarming of FRD1 is notable, as previous studies have shown that the mucoid phenotype leads to a loss of motility (45, 46, 53). (The mucoid-related loss of motility is due to AlgT, an alternative sigma factor that promotes the expression of amrZ, which represses the expression of the flagellar regulator fleQ [17, 45, 46]). Retention of the mucoid phenotype was confirmed by growth of mucoid colonies on standard LB agar plates from restreaks of swarming bacteria (however, during the swarm assays on soft agar, the strain does not appear mucoid). Therefore, our results suggest that algT or amrZ may be downregulated when P. aeruginosa grows on soft (agar) surfaces that are more hydrophobic.
Considerable variation was also present in the character of the swarm zone edge. Strains MPAO1 and PAO1 did not form tendrils on plates, but at the edge of the swarm zone, “microtendrils” formed, giving the swarm edge a rough and serrated appearance. The edges of PAO1C swarm zones did not show microtendrils but instead consisted of multiple fronts appearing as successive waves of cells that were radiating outward. Two strains, PA14 and PAK, showed a strong tendency to grow upwards (i.e., off the plate in the z-axis direction), producing swarm zones with greater depth than other strains but less plate coverage. The swarm zone edge of strain CF39 was smooth, lacking both the waves of PAO1C and the microtendrils of MPAO1 and PAO1. Even at the microscopic level, FRD1 showed extensive tendril formation. The edges of FRD1 swarm zones show deep furrows that are not as regular as microtendrils; these furrows remained connected with the main swarm zone.
In addition to swarming, all strains were probed more broadly for motility. Swim and twitching motility plates were made using 0.3% and 1.0% agar, respectively, to test for functional flagella and type IV pili (29, 41). During the swimming assays, certain strains showed a strong tendency to exhibit swarming behavior. Conversely to strains PAO1, MPAO1, and PAK that preferred to swim on swarm agar, strains PAO1C, PA14, CF39, and FRD1 swarmed on swim agar. Behavior varied between a combination of both swarming on the surface and swimming within the agar, as seen in strains PAO1C, PA14, and CF39, and only swarming on these 0.3% agar surfaces, as seen in strain FRD1. We conducted additional assays using 0.25% agar; this was sufficient to promote swimming of PAO1C, PA14, and CF39, but strain FRD1 still showed a strong preference for swarming over swimming. Wet mounts of planktonic cultures were also investigated using light microscopy; all strains were clearly motile except for SMC1587, which showed no movement by any cell.
Strains PAO1C, PAO1, MPAO1, and PA14 all readily twitched at the agar-plate interface in 1.0% agar assays. Strains PAK and FRD1 showed only minimal twitching, while CF39 and SMC1587 showed no evidence of twitching behavior.
Liquid culture rhamnolipid production did not correlate with rhamnolipid production during surface growth.
Several studies have measured rhamnolipid produced by P. aeruginosa growing in planktonic culture (13, 14, 41). We hypothesized that trends of rhamnolipid production during surface growth could not be predicted from planktonic measurements, and we compared surface rhamnolipid production with planktonic rhamnolipid production for each of these strains. The two primary rhamnolipids produced by P. aeruginosa are l-rhamnosyl-β-hydroxydecanoyl-β-hydroxydecanoate (a monorhamnolipid, commonly called R-C10-C10) and l-rhamnosyl l-rhamnosyl-β-hydroxydecanoyl-β-hydroxydecanoate (a dirhamnolipid, commonly called R-R-C10-C10) (14, 54). We investigated for these and additional rhamnolipid congeners, as substitutions have been observed with C8-hydroxyoctanoyl and C12-hydroxyduodecanoyl chains in place of the hydroxydecanoyl groups (14, 42, 54). While the two best swarming strains, CF39 and PAO1C, produced the most rhamnolipid under planktonic conditions, there was no clear correlation between planktonic and surface rhamnolipid production overall (Table 2). Strains PAO1, MPAO1, and PAK, which had poor rhamnolipid production during surface growth, produced significant amounts of rhamnolipid in planktonic culture. The strain that showed the least amount of swarming, SMC1587, produced only barely detectable amounts of rhamnolipid in liquid culture. FRD1 produced little rhamnolipid in liquid culture. Overall, surface production of rhamnolipid could not be predicted by planktonic production of rhamnolipid—Fig. S4 in the supplemental material displays the relative production of the predominant rhamnolipid congeners and surface rhamnolipid produced during swarming. While the C10 chain congeners were the most prevalent constituents overall, there was not consistency among the production of these rhamnolipids for the different strains (see Table S1 in the supplemental material).
Swarming is a highly variable trait for P. aeruginosa.
Our results suggest that strain variation accounts for some of the dissimilarity in swarming shown in many recent studies. Some differences in twitching motility were also observed among these strains, but only strain SMC1587 was nonmotile for all assays (swarming, swimming, twitching, and inspection for motile cells in planktonic culture). One significant component of the observed swarm differences was surface production of rhamnolipid by these strains—the best swarming strains produced the most rhamnolipid when growing on swarm assay plates. Some strains that produced only minimal rhamnolipid on surfaces, however, produced substantial quantities of rhamnolipid in planktonic culture. This difference between planktonic and surface rhamnolipid production is not understood and highlights some current limits of our understanding in regard to surface growth of P. aeruginosa and the overall regulation of swarming.
This fluorescent acquisition method enables the direct visualization of patterns for swarming bacteria. We additionally show the ability to visualize P. aeruginosa rhamnolipid, which is an important factor in P. aeruginosa swarming. This technique is easily modifiable to track patterns resulting from distinguishable expression of multiple fluors and luminescence, as shown in a coswarming experiment with GFP-expressing P. aeruginosa and bioluminescent S. enterica serovar Typhimurium. We anticipate that this technique can be widely applied to examine spatiotemporal patterns important to swarming to better understand the complex motions and interactions of bacteria.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Indiana Clinical and Translational Science Institute under grant no. NIH UL1RR025761 to J.D.S. W.M.L. was supported by Carestream Health, Inc., Woodbridge, CT.
We thank the David Piwnica-Worms laboratory, Washington University, St. Louis, MO, for providing S. enterica FL6 cells. Purified rhamnolipid (JB-515) was kindly provided by Pradeep Singh, University of Washington, Seattle, WA.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 7 October 2011.
REFERENCES
- 1. Abdel-Mawgoud A. M., Lepine F., Déziel E. 2010. Rhamnolipids: diversity of structures, microbial origins and roles. Appl. Microbiol. Biotechnol. 86: 1323–1336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Alvarez-Ortega C., Harwood C. S. 2007. Identification of a malate chemoreceptor in Pseudomonas aeruginosa by screening for chemotaxis defects in an energy taxis-deficient mutant. Appl. Environ. Microbiol. 73: 7793–7795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Belas R., Suvanasuthi R. 2005. The ability of Proteus mirabilis to sense surfaces and regulate virulence gene expression involves FliL, a flagellar basal body protein. J. Bacteriol. 187: 6789–6803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Boles B. R., Thoendel M., Singh P. K. 2005. Rhamnolipids mediate detachment of Pseudomonas aeruginosa from biofilms. Mol. Microbiol. 57: 1210–1223 [DOI] [PubMed] [Google Scholar]
- 5. Burns J. L., et al. 2001. Longitudinal assessment of Pseudomonas aeruginosa in young children with cystic fibrosis. J. Infect. Dis. 183: 444–452 [DOI] [PubMed] [Google Scholar]
- 6. Butler M. T., Wang Q., Harshey R. M. 2010. Cell density and mobility protect swarming bacteria against antibiotics. Proc. Natl. Acad. Sci. U. S. A. 107: 3776–3781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cady K. C., et al. 2011. Prevalence, conservation and functional analysis of Yersinia and Escherichia CRISPR regions in clinical Pseudomonas aeruginosa isolates. Microbiology 157: 430–437 [DOI] [PubMed] [Google Scholar]
- 8. Caiazza N. C., Merritt J. H., Brothers K. M., O'Toole G. A. 2007. Inverse regulation of biofilm formation and swarming motility by Pseudomonas aeruginosa PA14. J. Bacteriol. 189: 3603–3612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Caiazza N. C., Shanks R. M., O'Toole G. A. 2005. Rhamnolipids modulate swarming motility patterns of Pseudomonas aeruginosa. J. Bacteriol. 187: 7351–7361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Conrad J. C., et al. 2011. Flagella and pili-mediated near-surface single-cell motility mechanisms in P. aeruginosa. Biophys. J. 100: 1608–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dasgupta N., Ramphal R. 2001. Interaction of the antiactivator FleN with the transcriptional activator FleQ regulates flagellar number in Pseudomonas aeruginosa. J. Bacteriol. 183: 6636–6644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Davies D. G., et al. 1998. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280: 295–298 [DOI] [PubMed] [Google Scholar]
- 13. Déziel E., et al. 1999. Liquid chromatography/mass spectrometry analysis of mixtures of rhamnolipids produced by Pseudomonas aeruginosa strain 57RP grown on mannitol or naphthalene. Biochim. Biophys. Acta 1440: 244–252 [DOI] [PubMed] [Google Scholar]
- 14. Déziel E., Lépine F., Milot S., Villemur R. 2000. Mass spectrometry monitoring of rhamnolipids from a growing culture of Pseudomonas aeruginosa strain 57RP. Biochim. Biophys. Acta 1485: 145–152 [DOI] [PubMed] [Google Scholar]
- 15. Déziel E., Lépine F., Milot S., Villemur R. 2003. rhlA is required for the production of a novel biosurfactant promoting swarming motility in Pseudomonas aeruginosa: 3-(3-hydroxyalkanoyloxy)alkanoic acids (HAAs), the precursors of rhamnolipids. Microbiology 149: 2005–2013 [DOI] [PubMed] [Google Scholar]
- 16. Doyle T. B., Hawkins A. C., McCarter L. L. 2004. The complex flagellar torque generator of Pseudomonas aeruginosa. J. Bacteriol. 186: 6341–6350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Garrett E. S., Perlegas D., Wozniak D. J. 1999. Negative control of flagellum synthesis in Pseudomonas aeruginosa is modulated by the alternative sigma factor AlgT (AlgU). J. Bacteriol. 181: 7401–7404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gibiansky M. L., et al. 2010. Bacteria use type IV pili to walk upright and detach from surfaces. Science 330: 197. [DOI] [PubMed] [Google Scholar]
- 19. Glick R., et al. 2010. Increase in rhamnolipid synthesis under iron-limiting conditions influences surface motility and biofilm formation in Pseudomonas aeruginosa. J. Bacteriol. 192: 2973–2980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Greenspan P., Mayer E. P., Fowler S. D. 1985. Nile red: a selective fluorescent stain for intracellular lipid droplets. J. Cell Biol. 100: 965–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Holloway B. W. 1955. Genetic recombination in Pseudomonas aeruginosa. J. Gen. Microbiol. 13: 572–581 [DOI] [PubMed] [Google Scholar]
- 22. Holloway B. W., Krishnapillai V., Morgan A. F. 1979. Chromosomal genetics of Pseudomonas. Microbiol. Rev. 43: 73–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jacobs M. A., et al. 2003. Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 100: 14339–14344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jarrell K. F., McBride M. J. 2008. The surprisingly diverse ways that prokaryotes move. Nat. Rev. Microbiol. 6: 466–476 [DOI] [PubMed] [Google Scholar]
- 25. Kamatkar N. G., Shrout J. D. 2011. Surface hardness impairment of quorum sensing and swarming for Pseudomonas aeruginosa. PLoS One 6: e20888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kearns D. B. 2010. A field guide to bacterial swarming motility. Nat. Rev. Microbiol. 8: 634–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kerr K. G., Snelling A. M. 2009. Pseudomonas aeruginosa: a formidable and ever-present adversary. J. Hosp. Infect. 73: 338–344 [DOI] [PubMed] [Google Scholar]
- 28. Klockgether J., et al. 2010. Genome diversity of Pseudomonas aeruginosa PAO1 laboratory strains. J. Bacteriol. 192: 1113–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Köhler T., Curty L. K., Barja F., van Delden C., Pechére J. C. 2000. Swarming of Pseudomonas aeruginosa is dependent on cell-to-cell signaling and requires flagella and pili. J. Bacteriol. 182: 5990–5996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Leid J. G., et al. 2005. The exopolysaccharide alginate protects Pseudomonas aeruginosa biofilm bacteria from IFN-gamma-mediated macrophage killing. J. Immunol. 175: 7512–7518 [DOI] [PubMed] [Google Scholar]
- 31. MacEachran D. P., et al. 2007. The Pseudomonas aeruginosa secreted protein PA2934 decreases apical membrane expression of the cystic fibrosis transmembrane conductance regulator. Infect. Immun. 75: 3902–3912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Merritt J. H., Brothers K. M., Kuchma S. L., O'Toole G. A. 2007. SadC reciprocally influences biofilm formation and swarming motility via modulation of exopolysaccharide production and flagellar function. J. Bacteriol. 189: 8154–8164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Moreau-Marquis S., et al. 2008. The ΔF508-CFTR mutation results in increased biofilm formation by Pseudomonas aeruginosa by increasing iron availability. Am. J. Physiol. Lung Cell. Mol. Physiol. 295: L25–L37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nozawa T., et al. 2007. Rhamnolipid-dependent spreading growth of Pseudomonas aeruginosa on a high-agar medium: marked enhancement under CO2-rich anaerobic conditions. Microbiol. Immun. 51: 703–712 [DOI] [PubMed] [Google Scholar]
- 35. Ochsner U. A., Fiechter A., Reiser J. 1994. Isolation, characterization, and expression in Escherichia coli of the Pseudomonas aeruginosa rhlAB genes encoding a rhamnosyltransferase involved in rhamnolipid biosurfactant synthesis. J. Biol. Chem. 269: 19787–19795 [PubMed] [Google Scholar]
- 36. Ochsner U. A., Koch A. K., Fiechter A., Reiser J. 1994. Isolation and characterization of a regulatory gene affecting rhamnolipid biosurfactant synthesis in Pseudomonas aeruginosa. J. Bacteriol. 176: 2044–2054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. O'Toole G. A., Kolter R. 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30: 295–304 [DOI] [PubMed] [Google Scholar]
- 38. Overhage J., Bains M., Brazas M. D., Hancock R. E. 2008. Swarming of Pseudomonas aeruginosa is a complex adaptation leading to increased production of virulence factors and antibiotic resistance. J. Bacteriol. 190: 2671–2679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pham T. H., Webb J. S., Rehm B. H. 2004. The role of polyhydroxyalkanoate biosynthesis by Pseudomonas aeruginosa in rhamnolipid and alginate production as well as stress tolerance and biofilm formation. Microbiology 150: 3405–3413 [DOI] [PubMed] [Google Scholar]
- 40. Read R. C., et al. 1992. Effect of Pseudomonas aeruginosa rhamnolipids on mucociliary transport and ciliary beating. J. Appl. Physiol. 72: 2271–2277 [DOI] [PubMed] [Google Scholar]
- 41. Shrout J. D., et al. 2006. The impact of quorum sensing and swarming motility on Pseudomonas aeruginosa biofilm formation is nutritionally conditional. Mol. Microbiol. 62: 1264–1277 [DOI] [PubMed] [Google Scholar]
- 42. Soberón-Chávez G., Lépine F., Déziel E. 2005. Production of rhamnolipids by Pseudomonas aeruginosa. Appl. Microbiol. Biotechnol. 68: 718–725 [DOI] [PubMed] [Google Scholar]
- 43. Starkey M., et al. 2009. Pseudomonas aeruginosa rugose small-colony variants have adaptations that likely promote persistence in the cystic fibrosis lung. J. Bacteriol. 191: 3492–3503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Stover C. K., et al. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406: 959–964 [DOI] [PubMed] [Google Scholar]
- 45. Tart A. H., Blanks M. J., Wozniak D. J. 2006. The AlgT-dependent transcriptional regulator AmrZ (AlgZ) inhibits flagellum biosynthesis in mucoid, nonmotile Pseudomonas aeruginosa cystic fibrosis isolates. J. Bacteriol. 188: 6483–6489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tart A. H., Wolfgang M. C., Wozniak D. J. 2005. The alternative sigma factor AlgT represses Pseudomonas aeruginosa flagellum biosynthesis by inhibiting expression of fleQ. J. Bacteriol. 187: 7955–7962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Toguchi A., Siano M., Burkart M., Harshey R. M. 2000. Genetics of swarming motility in Salmonella enterica serovar typhimurium: critical role for lipopolysaccharide. J. Bacteriol. 182: 6308–6321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Totten P. A., Lory S. 1990. Characterization of the type a flagellin gene from Pseudomonas aeruginosa PAK. J. Bacteriol. 172: 7188–7199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tremblay J., Déziel E. 2010. Gene expression in Pseudomonas aeruginosa swarming motility. BMC Genomics 11: 587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Verstraeten N., et al. 2008. Living on a surface: swarming and biofilm formation. Trends Microbiol. 16: 496–506 [DOI] [PubMed] [Google Scholar]
- 51. White A. G., et al. 2010. Optical imaging of bacterial infection in living mice using deep-red fluorescent squaraine rotaxane probes. Bioconjug. Chem. 21: 1297–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Williamson N. R., Fineran P. C., Ogawa W., Woodley L. R., Salmond G. P. 2008. Integrated regulation involving quorum sensing, a two-component system, a GGDEF/EAL domain protein and a post-transcriptional regulator controls swarming and RhlA-dependent surfactant biosynthesis in Serratia. Environ. Microbiol. 10: 1202–1217 [DOI] [PubMed] [Google Scholar]
- 53. Wyckoff T. J., Thomas B., Hassett D. J., Wozniak D. J. 2002. Static growth of mucoid Pseudomonas aeruginosa selects for non-mucoid variants that have acquired flagellum-dependent motility. Microbiology 148: 3423–3430 [DOI] [PubMed] [Google Scholar]
- 54. Zhu K., Rock C. O. 2008. RhlA converts beta-hydroxyacyl-acyl carrier protein intermediates in fatty acid synthesis to the beta-hydroxydecanoyl-beta-hydroxydecanoate component of rhamnolipids in Pseudomonas aeruginosa. J. Bacteriol. 190: 3147–3154 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.