Abstract
Coevolution of virus and host is a process that emerges in persistent virus infections. Here we studied the coevolutionary development of coxsackievirus B3 (CVB3) and cardiac myocytes representing the major target cells of CVB3 in the heart in a newly established persistently CVB3-infected murine cardiac myocyte cell line, HL-1CVB3. CVB3 persistence in HL-1CVB3 cells represented a typical carrier-state infection with high levels (106 to 108 PFU/ml) of infectious virus produced from only a small proportion (approximately 10%) of infected cells. CVB3 persistence was characterized by the evolution of a CVB3 variant (CVB3-HL1) that displayed strongly increased cytotoxicity in the naive HL-1 cell line and showed increased replication rates in cultured primary cardiac myocytes of mouse, rat, and naive HL-1 cells in vitro, whereas it was unable to establish murine cardiac infection in vivo. Resistance of HL-1CVB3 cells to CVB3-HL1 was associated with reduction of coxsackievirus and adenovirus receptor (CAR) expression. Decreasing host cell CAR expression was partially overcome by the CVB3-HL1 variant through CAR-independent entry into resistant cells. Moreover, CVB3-HL1 conserved the ability to infect cells via CAR. The employment of a soluble CAR variant resulted in the complete cure of HL-1CVB3 cells with respect to the adapted virus. In conclusion, this is the first report of a CVB3 carrier-state infection in a cardiomyocyte cell line, revealing natural coevolution of CAR downregulation with CAR-independent viral entry in resistant host cells as an important mechanism of induction of CVB3 persistence.
INTRODUCTION
The understanding of viral diseases has been drastically improved in the last decades, and it is becoming evident that sustained virus-host altercations can lead to the establishment of persistent infections in humans and animals by many cytolytic viruses. The mechanisms involved in the development of virus persistence, however, are often not well understood. It has been proposed that the in vivo innate and humoral immune response represents the most important factor involved in the development of virus persistence. Immunological pressure may result in selection of attenuated or defective virus mutants that escape immunological clearance, leading to persistent virus infection (46). In vitro models are much simpler than in vivo models and have therefore facilitated the study of both the cellular and the viral components of persistent viral infections.
Certain cytolytic viruses can establish persistent infections in vitro as well as in vivo (4, 5, 8, 28, 39). Persistent in vitro infections can be divided into two major groups. One group involves steady-state infections, which are characterized by virus infection of all cells. The virus, however, is unable to accomplish the typical lytic replication cycle. The other group includes carrier-state virus infections. These are characterized by a cytolytic infection (yielding high progeny numbers) of a small proportion of cells, which spares the majority of cells in culture from cytolysis (21–24, 39, 40).
Persistent viral infection occurring in vitro seems to result from coevolution of host cell resistance and virus virulence and develops over a prolonged period of interaction of virus with cell (1, 13, 24, 50, 68). For several viruses and virus families, such as foot and mouth disease virus (62), reoviruses (1), enteroviruses (23, 24, 28), coronaviruses (6), hepatitis C virus (68), and autonomous parvovirus (54), coevolution of cells and viruses following in vitro infection has been demonstrated. Molecular analysis revealed some key mechanisms, including mutations of the receptor and reduction of virus receptor expression (7, 24, 50), obstacles in post receptor events during the viral uptake process (14), and intracellular blocking of virus replication (13), that seem to be involved in establishing carrier-state infections in vitro. On the other hand, development of virus mutants that exhibit increased avidity to downregulated cellular receptors (6, 7), acquisition of epitope variants that enable alternative virus receptor usage (6), emergence of replication-defective virus mutants (23), and generation of small-plaque virus variants with new replication intermediates and requirements (29) during cell virus coevolution in vitro have also been reported.
The genus Enterovirus belongs to the family Picornaviridae, and the species that infect humans are further divided into human rhinoviruses A, B, and C and human enteroviruses A, B (including type B coxsackievirus), C (including poliovirus), and D. Enteroviruses are characterized by a single-stranded (positive) RNA genome packaged in a capsid containing four capsid proteins (VP1 to VP4) (42, 47). In particular, coxsackieviruses of group B (CVB) (serotypes 1 to 6) have been established as highly prevalent human pathogens and are known to be associated with a variety of acute and chronic forms of diseases, including myocarditis, meningitis, and pancreatitis (31, 41, 42, 58). Cardiac infection with coxsackievirus B3 (CVB3) can result in acute myocarditis that spontaneously resolves or chronic myocarditis with prolonged viral persistence. Long-term sequelae of chronic cardiac CVB3 infection include heart failure and development of dilated cardiomyopathy (DCM) with an adverse prognosis (48, 49).
The molecular mechanisms leading to induction and maintenance of CVB3 persistence in the human heart as well as in experimental murine models are poorly understood. Moreover, adaptation of the immune system resulting from alterations in the immunological pressure adds complexity to the in vivo situation, making it difficult to separate immune evasion from such alterations leading to modified replication and viral entry.
In vitro cell systems with carrier-state virus infections have been shown to provide a useful approach for identifying factors regulating viral persistence (23, 51). To investigate mechanisms of CVB3 persistence in cardiac cells, a CVB3 carrier-state infection of primary human myocardial fibroblasts (HMF) was established several years ago (27, 28). Unfortunately, cardiomyocytes, not fibroblasts, represent the major target cells of CVB3 in a healthy human heart, thus limiting the suitability of persistently CVB3-infected HMF cells as a model (35).
We have established a persistently CVB3-infected murine cardiac cell line, HL-1CVB3, as a more relevant model. The persistently infected HL-1CVB3 cell line showed a typical carrier-state infection, with continuous delivery of high titers of CVB3 from a low proportion of infected cells. The expression of the coxsackievirus and adenovirus receptor (CAR) was investigated as a key factor associated with resistance of HL-1CVB3 cells to infection, and the entry, replication rate, and receptor usage of the resulting CVB3-HL1 progeny virus were examined to evaluate coevolutionary viral adaptations that emerged during virus persistence.
MATERIALS AND METHODS
Viruses.
CVB-3 (Nancy strain; VR-30) was obtained from the American Type Culture Collection (ATCC) and propagated in HeLa cells. CVB3-HL1 is the variant of the CVB3 Nancy strain that emerged during persistent infection in HL-1CVB3 cells. CVB3 was collected from the supernatant of HeLa cells, and CVB3-HL1 was collected from the supernatant of HL-1CVB3 cells (passages 9 to 11). Viruses were concentrated by ultracentrifugation via a sucrose gradient procedure. Both virus strains were quantified by standard plaque assays using HeLa cells, as the genome-to-PFU ratios for the two virus strains were found to be similar by real-time reverse transcription-PCR (RT-PCR) (data not shown).
CVB3 variant CVB3-PD was kindly provided by Michaela Schmidtke (Institute of Virology and Antiviral Therapy, Friedrich Schiller University, Jena, Germany).
Cell cultures.
HeLa, C2C12, and CHO-K1 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) (Gibco BRL, Karlsruhe, Germany) supplemented with 5% fetal calf serum (FCS) and 1% penicillin-streptomycin. The HL-1 cell line, a cardiac muscle cell line established from an AT-1 mouse atrial cardiomyocyte tumor lineage, was a kind gift from William C. Claycomb (LSU Health Science Center, Department of Biochemistry and Molecular Biology, New Orleans, LA). HL-1, HL-1CVB3, and HL-1cure cells were maintained in Claycomb medium supplemented with 10% FCS (JRH Bioscience, Lenexa, KS), 1% (each) penicillin-streptomycin, 0.1 mM norepinephrine (Sigma, Munich, Germany), and 2 mM l-glutamine (Invitrogen, Karlsruhe, Germany). Embryonal mouse cardiomyocytes (EMCMs) were isolated from fetal C57BL/6 mice. Between days 13 and 14 after fertilization, embryonic mouse hearts were isolated and incubated in EDTA-trypsin at 4°C overnight. After additional incubation for 15 min in a Thermo shaker (Eppendorf, Hamburg, Germany) at 37°C and 450 rpm, 500 μl of growth medium (DMEM without glutamine, 10% fetal bovine serum [FBS], 1% penicillin-streptomycin) was added to each heart. The digested heart tissue was transferred to a 75-ml cell culture flask. In order to separate the fibroblasts from cardiomyocytes, the cells were incubated for 1 h at 37°C and 5% CO2; after fibroblasts adhered, the supernatant containing cardiomyocytes was transferred to 12-well plates and cultured for 5 days by changing growth medium containing 5-bromo-2-deoxyuridine (BrdU) (BD Pharmingen, Heidelberg, Germany) (0.1 mmol/liter) every second day. Neonatal rat cardiomyocytes (NRCM) were generated and maintained as described previously (18).
Plasmid transfection.
CHO-K1 cells were transfected with pZS2-mCAR1 (16) or pZS2 (19) by the use of Lipofectamine LTX with Plus reagent (Invitrogen GmbH, Karlsruhe, Germany) according to the recommendations of the supplier. Transfections were performed using 24-well plates with a plasmid quantity of 0.5 μg/well.
Production of sCAR-Fc.
HeLa cells were transduced with an adenoviral vector (AdG12) expressing the fusion protein of the extracellular domain of CAR and the carboxyl terminus of the human IgG1 Fc domain (sCAR-Fc) in the presence of doxycycline (Dox) (52). After transduction, cells were cultured for 3 days and Dox was added daily at a concentration of 1 μg/ml (Sigma). At days 2 and 3 after transduction, the cell culture supernatants were collected and fresh medium containing Dox was added. The collected supernatants containing sCAR-Fc were pooled, and 2-ml aliquots were stored at −30°C. The sCAR-Fc concentration in the cell culture supernatant was measured with a human IgG enzyme-linked immunosorbent assay (ELISA) (Bethyl Laboratories Inc., Montgomery, TX) according to the supplier's instructions.
Virus plaque assay.
Virus plaque assays were carried out as described previously (17). Briefly, HeLa cells were cultured in six-well culture plates as confluent monolayers at a density of 1 × 106 cells/well. After 24 h, medium was removed and cells were overlaid with 1 ml of diluted supernatant harvested from HL-1 cell culture or from homogenized mouse hearts and then incubated at 37°C for 30 min and, after removal of the supernatant, overlaid with 2 ml of agar containing Eagle's minimal essential medium (MEM). Three days later, the cells were stained with 0.025% neutral red in phosphate-buffered saline (PBS). Virus titers were determined by plaque counting at 3 h after staining. Data shown represent the results of two plaque assays, each performed in duplicate.
Real-time RT-PCR.
Total RNA was isolated with Trizol reagent (Invitrogen, Karlsruhe, Germany) according to the company's recommendations followed by DNase I digestion (Peqlab, Erlangen, Germany). RNA was then reverse transcribed using a High-Capacity cDNA reverse transcription kit (Applied Biosystems Inc., Foster City, CA). For quantification of gene expression, real-time PCR was performed using a Mastercycler ep Gradient S system (Eppendorf) and TaqMan Gene Expression Master Mix (Applied Biosystems Inc.) under the standard conditions determined by the supplier. After AmpliTaq Gold activation for 4 min at 94°C, 40 cycles were run at a denaturing temperature of 94°C (for 15 s) and an annealing and extension temperature of 60°C (for 1 min). Expression levels of CAR, and of GAPDH (glyceraldehyde-3-phosphate dehydrogenase) as a control housekeeping gene for normalization, were determined by real-time PCR with minor groove binder (MGB) probes (Applied Biosystems). PCRs were performed in triplicate, and analysis was carried out using the 2−ΔΔCT threshold cycle method (37).
CVB3-RNA quantification.
For quantitative analysis of CVB3 RNA in tissue or cells, total RNA was isolated with a High Pure viral nucleic acid kit (Roche) according to the suggestions of the supplier and CVB3 RNA levels were measured by light-cycle PCR using an artus Enterovirus LC RT-PCR kit (Qiagen, Hilden, Germany).
Cell viability assay.
The cell viability was carried out by the use of a XTT cell proliferation kit (Roche, Mannheim, Germany) following the manufacturer's instructions. The absorbance measured at 492 nm is proportional to the number of metabolically active cells and therefore to cell viability.
In situ hybridization.
CVB3 plus-strand and minus-strand genomic RNA was detected by in situ hybridization using strand-specific single-stranded 35S-labeled RNA probes as described previously (33).
Immunofluorescence.
HL-1 cells were fixed with 1 ml of Tris-buffered saline (TBS) solution containing 4% formaldehyde and permeabilized with 0.5% Triton X-100. After being blocked in TBS buffer with 5% FCS and 0.1% Triton X-100, cells were incubated with NCL-entero anti-enterovirus (VP1) mouse monoclonal antibody (Novocastra, Newcastle, United Kingdom) at a dilution of 1:10 in blocking solution for 1 h. After extensive rinsing was performed, the cells were incubated with goat anti-mouse secondary antibody conjugated with Alexa-Fluor 488 or Alexa-Fluor 594 (Molecular Probes, Karlsruhe, Germany) at a dilution of 1:400 for 1 h. Cell nuclei were stained with DAPI (4′,6-diamidino-2-phenylindole) (Sigma) (0.5 μg/ml). To stain the actin cytoskeleton, cells were incubated with Alexa-Fluor 488-conjugated phalloidin (Molecular Probes) at a dilution of 1:200 for 20 min. For propidium iodide (PI) staining, cells were washed with PBS and incubated with PBS–0.5% PI (Molecular Probes) at 37°C for 30 min.
Western blot analysis.
Cells were scraped from a confluent T25 flask and lysed with lysis buffer (20 mM Tris [ph 8], 10 mM NaCl, 0.5% [vol/vol] Triton X-100, 5 mM EDTA, 3 mM MgCl2). Protein (20 μg) was separated on NuPage Bis-Tris gels (Invitrogen) (4% to 12%) under reducing conditions and transferred onto a polyvinylidene difluoride (PVDF) membrane (Bio-Rad Laboratories, Hercules, CA). For detection of CAR, SERCA2a, and GAPDH, after the membranes were blocked with 5% dry milk–TBS, they were incubated with H-300 anti-CAR antibody (Santa Cruz, Santa Cruz, CA) at a dilution of 1:200, anti-SERCA2a antibody (Abcam, Cambridge, MA) at a dilution of 1:20,000, and anti-GAPDH antibody (Millipore, Billerica, MA) at a dilution of 1:7,500. After washing was performed three times with TBS, the membranes were incubated with species-specific horseradish peroxidase (HRP)-conjugated secondary antibodies (Dako, Hamburg, Germany) in a dilution of 1:10,000 in 5% dry milk–TBS at room temperature for 1 h. After extensive washing, detection by chemiluminescence was achieved using Rodeo ECL Western blotting reagent (USB Corporation, Santa Clara, CA).
Murine viral myocarditis model.
Male BALB/c mice (6 weeks old) were infected intraperitoneally (i.p.) with CVB3 and CVB3-HL1 at 5 × 105 PFU. Seven days after CVB3 infection, the animals were killed and hearts were analyzed for the presence of infectious CVB3 and CVB3 genomes in the ventricles. For validation of inflammation, the basal parts of the hearts were fixed in 4% formalin, embedded in paraffin, sectioned, and stained with hematoxylin and eosin. Animal experiments were performed in accordance with the principles of laboratory animal care and the German law on animal protection.
RESULTS
Characterization of a persistently CVB3-infected cardiomyocyte cell line.
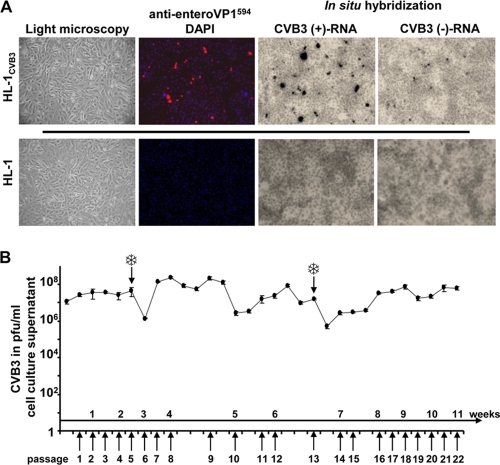
The HL-1 mouse atrial cardiomyocyte cell line was infected with CVB3 at different multiplicities of infection (MOI) (10, 1, and 0.1) and grown for 2 days at 37°C in Claycomb medium supplemented with 2% FCS. After trypsinization and reseeding, only the HL-1 cells infected at an MOI of 0.1 were able to adhere and grow to a confluent monolayer. The new cell culture, referred to as HL-1CVB3, could be maintained and passaged using the same procedures used with the naive parental HL-1 cell line. Although morphologically indistinguishable from the parental HL-1 cell line by light microscopy (Fig. 1 A), the HL-1CVB3 cells consistently released large amounts of progeny virus (106 to 108 PFU/ml) over a period of 11 weeks. During that period, aliquots of cells were frozen at subculture passages 5 and 13, and thawed cells showed no decline in virus production or cell growth compared to the original HL-1CVB3 cells (Fig. 1B). Analysis of CVB3 VP1 expression in HL-1CVB3 cells as determined by immunofluorescence revealed that only approximately 10% of the HL-1CVB3 cells expressed the viral capsid protein VP1; that finding was confirmed by in situ hybridization with CVB3 positive- and negative-strand genomic probes (Fig. 1A). We did consistently note, however, that the number of cells that were found to contain positive CVB3 strands was greater than the number that were found to contain the negative strand in the infected subpopulation of HL-1CVB3 cells (Fig. 1A). Based on these observations, CVB3 infection of HL-1CVB3 cells displays typical persistent carrier-state infection characteristics.
Fig. 1.
Characterization of persistently CVB3-infected HL-1CVB3 cardiac mouse cell line. (A) Cell morphology, virus capsid protein (VP1) expression, and determination of CVB3-RNA in HL-1CVB3 cells. Light microscopy showed no difference between HL-1CVB3 and uninfected HL-1 cells in cell morphology. CVB3 VP1 expression was determined by indirect immunofluorescence after incubation of cells with an anti-enterovirus VP1 antibody and a secondary antibody conjugated with Alexa-Fluor594 (red). Cell nuclei were costained with DAPI (blue). The presence of CVB3 positive- and negative-strand RNA was determined by in situ hybridization using specific 35S-labeled enterovirus positive- or negative-strand RNA hybridization probes. Magnification, 100-fold. (B) CVB3 titer in HL-1CVB3 cells. Levels of infectious CVB3 in the supernatant of HL-1CVB3 cells were determined by plaque assays after virus persistence was established. During the investigation period of 11 weeks, cells were passaged every 3 to 4 days. The culture supernatant of HL-1CVB3 cells showed continuous high levels of CVB3 ranging between 106 and 108 PFU/ml. At indicated time points (snowflakes), cells were frozen and reseeded.
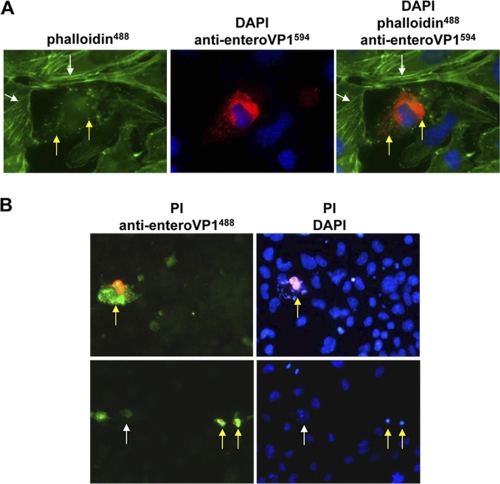
CVB3 induces a typical cytopathic effect (CPE), including disruption of the cytoskeleton, in infected cells. The cytoskeleton of CVB3-infected HL-1CVB3 cells was first stained with fluorescent phalloidin, which binds directly to the cellular F-actin filaments, and then costained for enterovirus VP1. VP1-negative cells showed intact actin filaments in the HL-1CVB3 cell culture, whereas the VP1-positive subpopulation showed a depolymerized actin cytoskeleton with disrupted actin filaments and accumulations of actin conglomerates (Fig. 2 A). Loss of membrane integrity induced by the presence of CVB3 in HL-1CVB3 cells was measured by nuclear propidium iodide (PI) staining (Fig. 2B); when cells were fixed and costained for VP1, PI was observed only inside VP1-positive cells. Moreover, late-stage destruction of CVB3-infected HL-1CVB3 cells could be confirmed by detection of karyopyknosis and fragmentation of the nucleus following DAPI staining (Fig. 2B, right upper and lower panels). Lack of PI staining in individual CVB3-infected HL-1CVB3 cells (Fig. 2B, left lower panel), however, may reflect an earlier phase of CVB3 infection, when cell death is not yet induced. CPE was detected during establishment of the persistently infected cell culture and could be observed in the cell culture as a rare event during the whole investigation period.
Fig. 2.
Pathogenic alterations caused by CVB3 infection in the persistently infected HL-1CVB3 cell line. (A) Disruption of cytoskeleton in CVB3-infected HL-1CVB3 cells. Cells were stained for detection of CVB3 VP1 expression as described for Fig. 1A. Staining of F-actin filaments was carried out with Alexa-Fluor488 (green)-conjugated phalloidin. CVB3-infected cells in the culture showed disruption of actin filaments and accumulation of actin conglomerates (green spots and yellow arrows), whereas uninfected cells showed intact filamentous actin fibers (white arrows). Magnification, 1,000-fold. (B) Detection of cytopathic effect in CVB3-infected HL-1CVB3 cells. CVB3-infected cells were incubated with propidium iodide (PI) (red) and then stained with enterovirus-specific VP1 antibody and a secondary antibody conjugated with Alexa-Fluor488 (green) (left panels) and then with DAPI (blue) (right panels). Red-green and magenta-blue costaining reveals CVB3-infected dead cells (yellow arrows), whereas cells stained solely green (white arrow, lower left panel) may represent CVB3-infected cells at an early replication stage without signs of cell death. Magnification, 400-fold.
CVB3-HL1 cells show increased cardiomyocyte-specific replication in vitro but lack cardiopathogenesis in vivo.
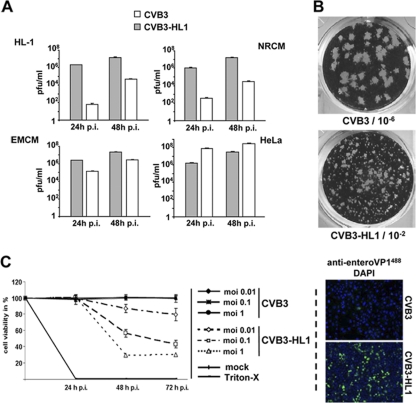
An altered replication rate is often observed during cell culture adaptation of viral strains. To study cell-type-specific CVB3 adaptation, we next investigated the replication efficiency and virulence of CVB3 progeny from HL-1CVB3 cells. HL-1CVB3 cell culture supernatants from passages 9 to 11 were collected, purified, and concentrated by ultracentrifugation using sucrose banding. The progeny virus (CVB3-HL1) showed a 10- to 100-fold reduction in virus replication (Fig. 3 A) and produced smaller virus plaques relative to those of the parental CVB3 in HeLa cells (Fig. 3B). Infection of HL-1 cells and neonatal rat cardiomyocytes (NRCM) with CVB3-HL1 yielded 100- to 10,000-fold more viral progeny at 24 and 48 h postinfection relative to the results seen with cells infected with the parental CVB3 strain (Fig. 3A). CVB3-HL1 infection of embryonic mouse cardiomyocytes (EMCM) also showed a consistently higher yield of progeny relative to parental CVB3 infection (albeit the difference was smaller than that seen with the other two cardiac cell lines) but yielded lower progeny numbers than parental CVB3 infection of HeLa cells (Fig. 3A). Moreover, 30% to 40% of HL-1 cells infected with CVB3-HL1 (MOI of 1) showed VP1 expression at 24 h postinfection compared to approximately 1% of VP1-positive HL-1 cells infected with CVB3 (Fig. 3C, right panel). Cell viability assays also revealed much greater cytotoxicity of CVB3-HL1 in HL-1 cells, while parental CVB3 cytotoxicity appeared negligible (Fig. 3C, left panel). These data indicate that CVB3-HL1 represents a new CVB3 variant that exhibits increased replication rates and virulence for cardiomyocytes in vitro.
Fig. 3.
Alteration of infectivity and replication activity of CVB3-HL1. (A) Replication of CVB3-HL1 in cardiac and HeLa cells in vitro. HeLa cells, native HL-1 cells, NRCM, and EMCM were infected with CVB3 and CVB3-HL1 at an MOI of 0.1. Plaque assays were performed 24 and 48 h later. In comparison to CVB3, CVB3-HL1 showed distinctly increased replication in the cardiac cell lines but lower replication in HeLa cells. Values are given as means ± standard deviations (SD) of the results of two independent experiments, each performed in duplicate. p.i., postinfection. (B) Representative plaque assay of CVB3 (dilution, 10−6) and CVB3-HL1 (dilution, 10−2) in HeLa cells. The CVB3-HL1 variant showed a smaller plaque size than CVB3. (C) Infectivity of CVB3-HL1 versus CVB3 in HL-1 cells. (Left panel) HL-1 cells were seeded in 96-well plates and infected with different doses of CVB3 and CVB3-HL1. Cell viability was measured by cell proliferation assays performed with uninfected cells (mock) as a control (set as 100%). CVB3-HL1, but not CVB3, induced cell lysis in HL-1 cells. Values are given as means ± SD of the results of two experiments, each performed in quadruplicate. (Right panel) HL-1 cells were infected with CVB3 and CVB3-HL1 at an MOI of 1. VP1 capsid protein was detected by immunofluorescence using an anti-enterovirus VP1 antibody (green) 24 h later. Nuclei were stained with DAPI (blue). Magnification, 100-fold. Compared to CVB3 infection, CVB3-HL1 infection showed a drastic increase in the number of VP1-positive HL-1 cells.
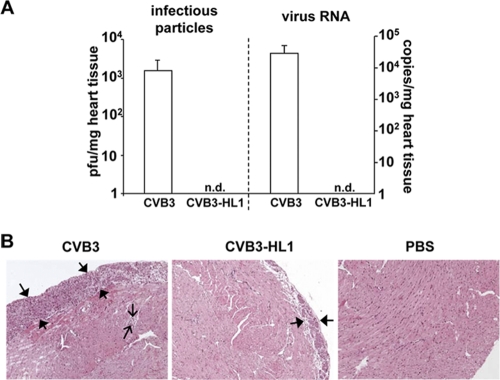
To compare the cardiopathogenic potential and cardiotropism of CVB3-HL1 to those of parental CVB3 in vivo, BALB/c mice were infected with 5 × 105 PFU CVB3-HL1 or CVB3 and investigated 7 days later for virus replication and inflammation. Infectious CVB3 particles (approximately 103/mg of heart tissue) were detected in CVB3-infected mice, confirming cardiotropism and successful viral replication of parental CVB3 in the murine heart. Surprisingly, infectious CVB3-HL1 particles were undetectable in the heart of CVB3-HL1-infected animals. Real-time RT-PCR confirmed the absence of CVB3-HL1 genomes in mice infected with the persistent strain but confirmed the presence of abundant amounts of CVB3 RNA in animals infected with the parental strain (Fig. 4 A). To document whether the absence of CVB3-HL1 in the heart correlated with a lack of pathological alterations, histological analysis of heart tissues of CVB3- and CVB3-HL1-infected animals was carried out. All 9 CVB3-infected animals showed strong pericardial inflammation and some overt inflammatory foci in the myocardium. In contrast, only 4 of the 9 animals infected with CVB3-HL1 showed inflammation in the heart. Furthermore, the area of leukocyte infiltration for those 4 animals was distinctly smaller than the area seen in hearts from parental CVB3-infected animals (Fig. 4B). Interestingly, histological examinations and in situ hybridization assays for detection of the CVB3 genome revealed strong tissue destruction and the presence of large amounts of CVB3 in the pancreas (results not shown) in both CVB3- and CVB-HL1-infected animals, thus indicating similar levels of in vivo infectivity of CVB3 and CVB3-HL1 in noncardiac CVB3 target organs. In summary, these data demonstrate that increased cardiospecific replication of CVB3-HL1 in vitro did not result in enhanced cardiotropic virulence in vivo.
Fig. 4.
Cardiotropism and cardiopathogenicity of CVB3-HL1 in vivo. (A) CVB3-HL1 lacks cardiotropism in vivo. Nine BALB/c mice were infected with 5 × 105 PFU of CVB3 or CVB-HL1. As a control, 6 mice were treated with PBS. Seven days after CVB3 infection, organs were harvested for virus detection and histopathological analysis. CVB3 positive-strand RNA and infectious virus numbers in the heart tissue were quantified by real-time RT-PCR and a standard plaque assay, respectively. Quantitative analysis showed a complete absence of CVB3-HL1 in the heart, whereas CVB3 RNA (right) and infectious CVB3 particles (left) were abundantly detected. n.d., not detectable. (B) Heart sections of CVB3- and CVB3-HL1-infected mice were stained with hematoxylin and eosin (magnification, 100-fold). CVB3-infected animals showed an expanded inflammation area at the pericardium (thick arrows) and some inflammatory spots with mononuclear cells in the myocardium (thin arrows). Heart tissues of CVB3-HL1-infected animals showed faint pericardial infiltrations with mononuclear cells.
A decrease of cellular CAR expression during CVB3 persistence in HL-1CVB3 cells induces CVB3-HL1 receptor shift.
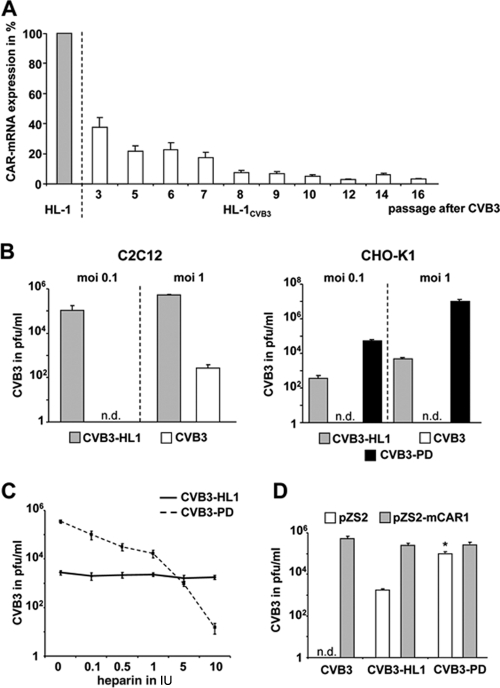
CVB3 uses cellular-decay-accelerating factor (DAF) and CAR as receptors to infect cells. However, murine DAF is not broadly expressed in mice (36), and murine DAF and related homologues are unable to bind to CVB3 (61), leaving murine CAR as the only known potential viable receptor in mice. As virus uptake is an important mechanism in the virus replication cycle, we hypothesized that changes in cellular CAR expression may be involved in the adaptation of HL-1CVB3 cells to CVB3 infection. Quantification of CAR-mRNA expression by real-time RT-PCR revealed a 60% decrease in CAR-mRNA expression at passage 3 and a decrease of more than 80% at passage 7 after infection of HL-1 cells with CVB3 relative to the results seen with the parental noninfected HL-1 cell line. A further and lasting reduction of more than 90% was seen after passage 7 (Fig. 5 A). As CAR is essential for CVB3 uptake into target cells, the continuous production of high CVB3 titers in HL-1CVB3 cells (Fig. 1B) suggested that the CAR-independent uptake mechanisms had been acquired by the CVB3-HL1-adapted strain in HL-1CVB3 cells. To test this assumption, we investigated the ability of CVB3-HL1 to infect and replicate in mouse myoblast C2C12 cells and CHO-K1 cells. CHO-K1 cells do not express CAR (57), whereas a low level of CAR expression in C2C12 cells was reported previously (32, 44). Real-time RT-PCR analysis, however, demonstrated an absence of CAR expression in the C2C12 cells used in this study (results not shown). Both cell lines were incubated with the parental CVB3 strain and with CVB3-HL1 at MOIs of 0.1 and 1. As a control for determining CAR-independent CVB3 uptake, CHO-K1 cells were also infected with CVB3-PD, a CVB3 variant that uses heparan sulfate (HS) as a receptor to infect CAR-negative cells (67). Measurement of progeny virus titers at 24 h after infection demonstrated that CVB3-HL1, but not the parental CVB3, efficiently infected C2C12 cells. CHO-K1 cells were efficiently infected with CVB3-HL1 and CVB3-PD but were completely resistant to CVB3 (Fig. 5B). These data demonstrate that CVB3-HL1 is able to infect cells via CAR-independent uptake mechanisms.
Fig. 5.
CVB3-HL1 persistence in HL-1CVB3 cells is associated with CAR downregulation and virus receptor shift. (A) CAR expression in persistently infected HL-1CVB3 cells. Total RNA was isolated from HL-1CVB3 cells at the indicated passages after CVB3 infection, and CAR mRNA was quantified by real-time RT-PCR. CAR-mRNA expression of noninfected HL-1 cells was set as 100%. (B) CVB3-HL1 utilizes a CAR-independent method to infect cells. C2C12 and CHO-K1 cells were infected with CVB3-HL1 and CVB3 at MOIs of 0.1 and 1. Replication efficiencies were measured by plaque assays 24 h later. As a control for CAR-independent virus uptake determinations, CHO-K1 cells were infected with strain CVB3-PD, which is known to infect CAR-negative cells via binding to heparan sulfate. The CVB3-HL1 variant, but not parental CVB3, replicated efficiently in CAR-negative cells. n.d., not detectable. (C) Lack of CVB3-HL1 inhibition by heparin. CVB3-PD and CVB3-HL1 (each at an MOI of 1) were preincubated with heparin at different concentrations for 1 h. Subsequently, CHO-K1 cells were infected, and replication efficiencies were measured by plaque assays 24 h later. Whereas infection of CVB3-HL1 was unaffected by heparin treatment, CVB3-PD infection was inhibited by heparin in a dose-dependent manner. (D) In spite of a CVB3-HL1 receptor shift during virus persistence in HL-1 cells, CVB3-HL1 can use CAR as a receptor to infect cells. CHO-K1 cells were transfected with mouse CAR1 (pZS2-mCAR1)-expressing plasmid or control plasmid (pZS2) and infected 48 h later with the indicated CVB3 subtypes (MOI, 1). At 8 h later, the amounts of infectious virus particles were determined by plaque assays. Note that there was a significant (P < 0.005) difference between pZS2 and pZS2-mCAR1 results for the CVB3-PD group.
To examine whether HS may act as a receptor of the CVB3-HL1 variant, we pretreated CVB3-HL1 and CVB3-PD with heparin, a soluble HS receptor analogue, before incubating CHO-K1 cells. As expected, CVB3-PD infection was strongly inhibited by heparin treatment. CVB3-HL1 infection, however, was unaffected (Fig. 5C), indicating that CVB-HL1 is able to infect cells via an unknown CAR- and HS-independent uptake mechanism.
Next, we investigated whether a CVB3-HL1 receptor shift was accompanied by an inability of CVB3-HL1 to infect cells via CAR. Therefore, CHO-K1 cells were transfected with mouse CAR-expressing plasmids and then infected with CVB3-HL1, CVB3, and CVB3-PD. The progeny virus titers were determined 8 h later. All analyzed CVB3 subtypes replicated more efficiently in CAR-expressing than in CAR negative-control-transfected CHO-K1 cells (Fig. 5D), revealing that the CVB3-HL1 variant was still capable of using CAR as a receptor for infection.
Complete cure of persistently infected HL-1 cells by sCAR-Fc treatment.
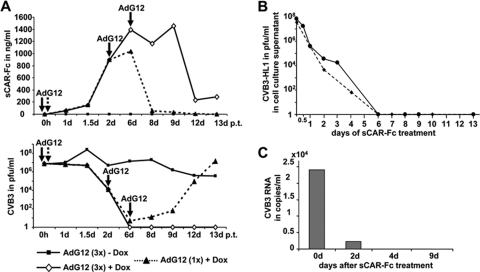
We recently showed that treatment of a persistently CVB3-infected human myocardial fibroblast (HMF) cell line with AdG12, a recombinant adenovirus vector expressing sCAR-Fc, decreased persistent CVB3 infection; however, sCAR-Fc was not capable of clearing the CVB3 infection (64). To examine whether sCAR-Fc was able to bind and inhibit the CVB3-HL1 variant, we transduced HL-1CVB3 cells with AdG12 (MOI, 30) and induced the expression of sCAR-Fc with doxycycline (Dox). A single transduction resulted in strong sCAR-Fc expression and reduced CVB3-HL1 titers by about a millionfold within 6 days. Thereafter, sCAR-Fc expression rapidly declined, becoming undetectable at day 12 after vector transduction. In parallel, CVB3-HL1 titers were found to have recovered and climbed to almost pretreatment levels by day 12. Repeated transduction of HL-1CVB3 cells with AdG12, in the presence of Dox, at days 2 and 6 after the first AdG12 transduction resulted in extended production of high-level sCAR-Fc expression and in complete clearance of CVB3 from HL-1CVB3 cells at day 6 (Fig. 6 A). External repeated addition of sCAR-Fc containing culture medium also resulted in total CVB3 clearance, indicating that intracellular delivery was not required for therapeutic efficacy (Fig. 6B). Lack of CVB3-HL1 RNA in total RNA extracts of sCAR-Fc-treated HL-1CVB3 cells at day 4 after the beginning of sCAR-Fc treatment as detected by quantitative RT-PCR (Fig. 6C) confirmed the complete clearance of HL-1CVB3 cells by recombinant sCAR-Fc. Moreover, we were unable to detect infectious CVB3-HL1 particles in the cell supernatant after “cure” of HL-1CVB3 cells (here, cured HL-1CVB3 cells are termed “HL-1cure”) over an additional period of 3 weeks (data not shown). These data demonstrate that sCAR-Fc efficiently neutralizes CVB3-HL1 and is able to clear persistently infected HL-1CVB3 cells.
Fig. 6.
Cure of HL-1CVB3 cells by sCAR-Fc. (A) Cure of HL-1CVB3 cells by sCAR-Fc expressed from an adenoviral vector. HL-1CVB3 cells on passage 16 were transduced with sCAR-Fc-expressing doxycycline (Dox)-dependent AdG12 adenovector (MOI, 30). Cells were treated once or three times at the indicated time points (arrows) and then cultured in the presence or absence of Dox as indicated. Samples of medium were taken at indicated time points to determine the amount of infectious viral particles (lower diagram) and sCAR-Fc concentration (upper diagram) in the cell culture supernatant by plaque assay and anti-human IgG ELISA, respectively. Threefold repetitive transductions, but not a single transduction, performed with AdG12 resulted in a complete elimination of CVB3-HL1 from HL-1CVB3 cells at day 6 after AdG12 transduction. d p.t., day posttransfection. (B) Cure of HL-1CVB3 cells by application of recombinant sCAR-Fc. Recombinant sCAR-Fc was produced by transduction of HL-1 cells with AdG12 in the presence of Dox. The sCAR-Fc concentration in the supernatant was quantified, and sCAR-Fc (200 ng/ml) was added to HL-1CVB3 HL cells (passage 8). The sCAR-Fc-containing medium was refreshed daily. Amounts of infectious virus particles in the cell culture supernatant were quantified by plaque assays using HeLa cells. CVB3-HL1 titers in the supernatant of HL-1CVB3 cells rapidly decreased and became undetectable at day 6 and thereafter (end of the investigation, day 13) from the beginning of sCAR-Fc treatment. Two independent experiments (dashed and solid lines) were carried out. (C) Measurement of CVB3-HL1 RNA. HL-1CVB3 cells during passage 8 were treated by daily applications of recombinant sCAR-Fc (200 ng/ml) to the cell culture medium as described for panel B. Total RNA from HL1CVB3 cells was isolated at the indicated time points, and the CVB3 RNA was quantified by real-time RT-PCR. No viral RNA was detected on day 4 or on later days after the beginning of sCAR-Fc treatment.
The HL-1cure cell line conserves resistant features of the HL-1CVB3 cell line.
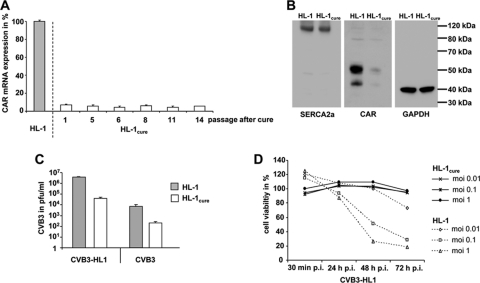
In order to elucidate whether resistant features of HL-1CVB3 cells were conserved after removing the viral pressure, HL-1cure cells were further characterized. To this end, we first determined the expression of CAR-mRNA by real-time RT-PCR and found a reduction of more than 90% in CAR mRNA expression compared to the expression seen with parental HL-1 cells (Fig. 7 A). Immunoblotting confirmed the CAR protein downregulation in HL-1cure cells (Fig. 7B), while levels of expression of SERCA2a (a cardiomyocyte-specific protein) in HL-1cure cells were similar to those seen with HL-1 cells. We next investigated the ability of CVB3 and CVB3-HL1 to infect HL-1cure cells. Both virus variants were able to infect the cell line, but the virus infection rates were about 32- and 95-fold lower, respectively, than those seen with HL-1 cells (Fig. 7C). Moreover, whereas HL-1 cells were lysed by CVB3-HL1, HL-1cure cells were resistant to CVB3-HL1-induced cell lysis (Fig. 7D). The data confirm that the HL-1cure cell line differs from the HL-1 cell line and had conserved the resistance features of the persistently infected HL1CVB3 cell line.
Fig. 7.
Characterization of HL-1cure cells. (A) Expression of CAR mRNA in HL-1cure cells. Total RNA was isolated from HL-1cure cells at the indicated passages after CVB3 infection, and CAR mRNA expression was quantified by real-time RT-PCR. CAR mRNA expression of noninfected HL-1 cells was set as 100%. (B) Western blot analysis of CAR, SERCA2a, and GAPDH expression in HL-1 and HL-1cure cells. Compared to HL-1 cells, HL-1cure cells showed decreased CAR expression but retained a similar level of expression of the SERCA2a cardiomyocyte-specific protein. (C) Resistance of HL-1cure cells to CVB3 and CVB3-HL1 infection. HL-1 and HL-1cure cells were infected with CVB3 and CVB3-HL1 at an MOI of 1. Virus titers in the cell culture supernatant were quantified by plaque assays 24 h later. Both viruses showed a significant decrease of replication in HL-1cure cells compared to HL-1 cells. Values are given as means ± SD of the results of two independent experiments, each performed in duplicate. (D) HL-1cure cells resist CVB3-HL1-induced cell lysis. HL-1 and HL-1cure cells were seeded in 96-well plates and infected with different doses of CVB3-HL1. Cell viability was measured by a cell proliferation assay (XTT), with uninfected cells (mock) as a control (set as 100%). HL-1cure cells, but not HL-1 cells, resisted CVB3-HL1-induced cell lysis. Values are given as means ± SD of the results of two experiments, each performed in quadruplicate.
DISCUSSION
Virus persistence may be viewed at three distinct levels: persistence in the population as a whole, persistence in the individual host, and persistence within a cell or group of cells (21, 33, 38). Viruses have evolved a wide variety of strategies that enable them to persist and cause diseases. A broad range of cytolytic viruses, including enteroviruses, establish persistent types of infections both in vivo and in vitro (4, 5, 8, 15, 28, 33). In most cases, this is induced by selection of virus mutants that are less cytopathic (2, 20, 53). Other mechanisms comprise coevolution of both cells and viruses (1, 6, 50). In coevolution, persistent infections are accompanied by alterations of both cells and viruses such that cellular resistance to viral replication is balanced by an enhanced capacity of the virus to infect the resistant cells.
To investigate the mechanism of CVB3 persistence in heart cells, CVB3 infection of primary human myocardial fibroblasts (HMF) or immortalized HMF was established (26–28). Unfortunately, cardiomyocytes but not fibroblasts represent the major target cells of CVB3 in the heart (35); thus, the suitability of persistently infected HMF cells for studying CVB3 infections in vitro is limited. To overcome this limitation, we established a persistently CVB3-infected cardiac cell line (HL-1CVB3) by infection of murine HL-1 cells with CVB3. Picornaviruses are able to induce two different types of persistence in vitro. The carrier-state infection is characterized by infection of a small proportion of the cell population. The infected cells release virus and are killed, but the released virus infects a small number of other cells. In a steady-state infection, all cells are infected, both virus and cell multiplication proceed without the cell being killed, and the cell culture continuously releases virus (21, 39). However, intermediate models also exist (51). HL-1CVB3 cells showed the typical signs of persistent carrier-state infection with high virus titers. The numbers of infected cells in a carrier-state infection can range from about 10% to 60% depending on the virus and host cell (6, 27, 43, 40). In line with these observations, about 10% of HL-1CVB3 cell expressed the VP1 viral capsid protein. Moreover, in situ hybridization revealed the presence of positive- and negative-strand RNA, with distinctly higher percentages of positive-strand RNAs, indicating acute, productive virus replication (35). As a further typical indication of a carrier-state infection, HL-1CVB3 cells showed virus-induced cytopathic effects (CPEs), including alterations of the cytoskeleton, loss of membrane integrity, and karyopyknosis in individual cells but not overt lytic infection of the cell culture.
The results of our study demonstrate that both the HL-1 host cell line and the CVB3 virus have evolved during persistent infection and that coevolution of target cells and virus represents the major mechanism of CVB3 persistence in HL-1 cells. Many studies have found coevolution of cells and virus during development of persistent virus infection in vitro, but elucidations of the specific mechanisms have been sparse (6, 13, 24, 68). As found here, one key feature of cellular evolution following CVB3 infection was downregulation of the CVB3 receptor CAR. Attachment to and internalization into host cells are important determinants of viral infection, and an alteration in receptor expression exerts a powerful cellular defense mechanism against viral infections. CAR acts as an essential receptor of CVB3 and mediates its internalization into the target cell (3, 12). In vitro studies have demonstrated that CAR knockdown resulted in decreased CVB3 infection, viral replication, and virus-induced cell lysis (16, 65). Moreover, heart-specific CAR ablation in vivo makes mice resistant to cardiac CVB3 infections (60).
An important aspect of development of resistance in the HL-1CVB3 cell line was the very fast downregulation of CAR in the CVB3-infected HL-1 cell culture: within 3 passages, CAR expression declined by approximately 60%, and at passage 8 and later, a consistent decrease of more than 90% in expression relative to the expression level seen with uninfected HL-1 cells was observed. Although the molecular mechanism leading to CAR downregulation was not determined in this study, it can be supposed that the initial incubation of HL-1 cells with CVB3 preferentially results in infection and subsequent disruption of cells with high levels of CAR expression. As CAR expression is variable in cell cultures, it can be supposed that a subpopulation of cells with low-level CAR expression or lacking such expression is protected from virus infection and cell lysis (19). As consequence, such cells proliferate in the culture within several passages and finally determine the level of cellular resistance to CVB3. Persistent infection of cell lines with other viruses, such as the mouse hepatitis virus (6, 55) and hepatitis C virus (68), has also been shown to be associated with cellular virus receptor downregulation. A closely similar level of poliovirus persistence was previously seen to be associated with specific mutations in the first extracellular domain of the CD155 poliovirus receptor (50). This suggests that blocking of virus binding, at the first step in infection, is a recurring mechanism in the evolution of viruses that persistently infect cell lines. Our data do not rule out the possibility that other cellular functions directly or indirectly related to the CVB3 replication cycle in persistently infected HL-1CVB3 cells are also affected, as has been reported for other cell lines with persistent virus infections (66, 68).
Based on our current knowledge, evolutionary pressure in virus-infected cell lines primarily starts from the cytopathic virus that infects and kills susceptible cells. If a cell culture is able to develop resistance, then the virus must also adapt to persist and to avoid being cleared by cell division and turnover of the host cells. In agreement with this assumption, we found that the adapted CVB3-HL1 virus was markedly different from the parental virus strain: it could infect CHO-K1 cells, which were completely resistant to parental CVB3, it had a lower capacity to infect HeLa cells, and it was much more efficient at infecting primary rodent heart-derived cells, as judged by progeny production and the presence of VP1-positive and genome-positive cells. An increased capacity to infect parental HL-1 cells was particularly notable, as CVB3 could infect only 1% of cells compared to 30% to 40% infection by the CVB3-HL1 strain. As with mouse hepatitis virus (6), yellow fever virus (63), and foot-and-mouth disease virus (59) adapted for persistent infection, we noted that CVB3-HL1 exhibited a smaller plaque size on HeLa cells than the parent strain. As reported previously from investigations performed using foot-and-mouth disease virus (59), propagation in the standard cell line (HeLa) resulted in a reversion of plaque morphology and cell tropism of CVB3-HL1 to those characteristic of the parental CVB3 strain (data not shown). Since the CVB3-HL1 strain replicates in CAR-negative cell lines, that finding suggests that the virus gained a CAR-independent entry mechanism. This, however, seems to be a crucial step in the viral coevolution process, as it helps the virus to overcome the cellular resistance acquired through CAR downregulation. Although CVB3-HL1 no longer required CAR for cell entry, it is important that CVB-HL1 could still use CAR as a cellular receptor, that CAR-based therapeutics were still capable of neutralizing the virus, and that prolonged use of such therapeutics was capable of completely clearing the adapted virus from the persistently infected cell line. This suggests only minor alterations to the capsid accompanying the altered cell tropism, as previously noted for another CVB3 variant, CVB3-PD (57).
Most unexpectedly, the enhanced ability of CVB3-HL1 to infect cardiac cells in vitro was not mirrored by an enhanced cardiotropism or cardiopathogenic capacity in vivo in BALB/c mice, demonstrating that the degree of CVB3-HL1 adaptation and virulence in vitro did not represent an indicator of the presence of in vivo cardiac infections. However, 4 of 9 CVB3-HL1-infected animals exhibited acute cardiac inflammations, suggesting that, at least in some mice, the CVB3-HL1 strain infected the heart transiently. Both host factors and viral genetic determinants influence the outcome of CVB3-induced myocarditis in genetically different inbred mouse strains (9, 30, 34). In this context, it was previously shown that different CVB3 strains (generated in vitro or in vivo) showing variations in infectivity, cell specificity, and pathology induce highly variable infections in vivo (56).
The mechanisms involved in heart resistance to CVB3-HL1 are not known, but attenuation of viruses after passages through cell culture is an often-observed phenomenon (10, 11, 25); for example, the attenuated Sabine 1 strain, the oral live polio vaccine, was derived by several passages through cell culture (45). One possible explanation for our findings is that heart resistance may have resulted from an accelerated clearance of the virus by specific immunological mechanisms directed against the CVB3-HL1 mutant; however, the additional inability of CVB3-HL1 to infect the heart efficiently, or a combination of the two mechanisms, may also be responsible for the heart resistance. The ability of CVB3-HL1 to infect other target organs in vivo in a manner typical of CVB3 was confirmed by detection of strong CVB3-HL1 infection of the pancreas. Therefore, the in vivo inability of CVB3-HL1 to infect target cells per se is rather unlikely to be the most important factor in heart resistance; antiviral immunological mechanisms are likely much more crucial.
In summary, we established and characterized a persistent CVB3 infection in a cardiomyocyte cell line for the first time. Persistent infection of cardiomyocytes was accompanied by downregulation or elimination of CAR-expressing cells in parallel to the coevolution of CAR-independent viral entry. The newly generated, persistently CVB3-infected HL-1CVB3 cell line now presents the possibility of studying virus host interaction in a more relevant cardiac in vitro system than was available before. This may help to improve understanding of the mechanisms involved in CVB3 infections of the heart. Most importantly, sCAR-Fc was found to be capable of completely eliminating the persistent adapted CVB3 strains without a requirement for intracellular delivery, supporting its candidacy to be developed as an in vivo therapeutic in the future.
ACKNOWLEDGMENTS
We thank Michaela Schmidtke for kindly providing the CVB3 mutant CVB3-PD, and we are grateful to Alice Weithäuser and Jenny Stehr for preparation of EMCM and NRCM, respectively.
This work has been supported by the Deutsche Forschungsgemeinschaft (DFG) through SFB Transregio 19 project grants to H.F., K.K., and A.D., by DFG project grants FE785/2-1 and FE785/3-1 to H.F., and by the Otto-Kuhn-Stiftung im Stifterverband für die Deutsche Wissenschaft to H.Z.
Footnotes
Published ahead of print on 5 October 2011.
REFERENCES
- 1. Ahmed R., et al. 1981. Role of the host cell in persistent viral infection: coevolution of L cells and reovoirus [sic] during persistent infection. Cell 25:325–332 [DOI] [PubMed] [Google Scholar]
- 2. Beaulieux F., et al. 2005. Cumulative mutations in the genome of Echovirus 6 during establishment of a chronic infection in precursors of glial cells. Virus Genes 30:103–112 [DOI] [PubMed] [Google Scholar]
- 3. Bergelson J. M., et al. 1997. Isolation of a common receptor for coxsackie B viruses and adenoviruses 2 and 5. Science 275:1320–1323 [DOI] [PubMed] [Google Scholar]
- 4. Borzakian S., et al. 1992. Persistent poliovirus infection: establishment and maintenance involve distinct mechanisms. Virology 186:398–408 [DOI] [PubMed] [Google Scholar]
- 5. Bowles N. E., et al. 1993. Persistence of enterovirus RNA in muscle biopsy samples suggests that some cases of chronic fatigue syndrome result from a previous, inflammatory viral myopathy. J. Med. 24:145–160 [PubMed] [Google Scholar]
- 6. Chen W., Baric R. S. 1996. Molecular anatomy of mouse hepatitis virus persistence: coevolution of increased host cell resistance and virus virulence. J. Virol. 70:3947–3960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen W., Yount B., Hensley L., Baric R. S. 1998. Receptor homologue scanning functions in the maintenance of MHV-A59 persistence in vitro. Adv. Exp. Med. Biol. 440:743–750 [DOI] [PubMed] [Google Scholar]
- 8. Chia J., Chia A., Voeller M., Lee T., Chang R. 2010. Acute enterovirus infection followed by myalgic encephalomyelitis/chronic fatigue syndrome (ME/CFS) and viral persistence. J. Clin. Pathol. 63:165–168 [DOI] [PubMed] [Google Scholar]
- 9. Chow L. H., Gauntt C. J., McManus B. M. 1991. Differential effects of myocarditic variants of coxsackievirus B3 in inbred mice. A pathologic characterization of heart tissue damage. Lab. Invest. 64:55–64 [PubMed] [Google Scholar]
- 10. Cohen J. I., Rosenblum B., Feinstone S. M., Ticehurst J., Purcell R. H. 1989. Attenuation and cell culture adaptation of hepatitis A virus (HAV): a genetic analysis with HAV cDNA. J. Virol. 63:5364–5370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Converse J. L., Kovatch R. M., Pulliam J. D., Nagle S. C., Jr., Snyder E. M. 1971. Virulence and pathogenesis of yellow fever virus serially passaged in cell culture. Appl. Microbiol. 21:1053–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Coyne C. B., Bergelson J. M. 2006. Virus-induced Abl and Fyn kinase signals permit coxsackievirus entry through epithelial tight junctions. Cell 124:119–131 [DOI] [PubMed] [Google Scholar]
- 13. de la Torre J. C., et al. 1988. Coevolution of cells and viruses in a persistent infection of foot-and-mouth disease virus in cell culture. J. Virol. 62:2050–2058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dermody T. S., Nibert M. L., Wetzel J. D., Tong X., Fields B. N. 1993. Cells and viruses with mutations affecting viral entry are selected during persistent infections of L cells with mammalian reoviruses. J. Virol. 67:2055–2063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Desailloud R., Sane F., Caloone D., Hober D. 2009. Persistent infection of a carcinoma thyroid cell line with coxsackievirus B. Thyroid 19:369–374 [DOI] [PubMed] [Google Scholar]
- 16. Fechner H., et al. 2007. Coxsackievirus B3 and adenovirus infections of cardiac cells are efficiently inhibited by vector-mediated RNA interference targeting their common receptor. Gene Ther. 14:960–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fechner H., et al. 2008. Cardiac-targeted RNA interference mediated by an AAV9 vector improves cardiac function in coxsackievirus B3 cardiomyopathy. J. Mol. Med. 86:987–997 [DOI] [PubMed] [Google Scholar]
- 18. Fechner H., et al. 2007. Highly efficient and specific modulation of cardiac calcium homeostasis by adenovector-derived short hairpin RNA targeting phospholamban. Gene Ther. 14:211–218 [DOI] [PubMed] [Google Scholar]
- 19. Fechner H., et al. 2000. Trans-complementation of vector replication versus Coxsackie-adenovirus-receptor overexpression to improve transgene expression in poorly permissive cancer cells. Gene Ther. 7:1954–1968 [DOI] [PubMed] [Google Scholar]
- 20. Frey T. K., Hemphill M. L. 1988. Generation of defective-interfering particles by rubella virus in Vero cells. Virology 164:22–29 [DOI] [PubMed] [Google Scholar]
- 21. Frisk G. 2001. Mechanisms of chronic enteroviral persistence in tissue. Curr. Opin. Infect. Dis. 14:251–256 [DOI] [PubMed] [Google Scholar]
- 22. Frisk G., Lindberg M. A., Diderholm H. 1999. Persistence of coxsackievirus B4 infection in rhabdomyosarcoma cells for 30 months. Arch. Virol. 144:2239–2245 [DOI] [PubMed] [Google Scholar]
- 23. Gibson J. P., Righthand V. F. 1985. Persistence of echovirus 6 in cloned human cells. J. Virol. 54:219–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gosselin A. S., et al. 2003. Poliovirus-induced apoptosis is reduced in cells expressing a mutant CD155 selected during persistent poliovirus infection in neuroblastoma cells. J. Virol. 77:790–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Grebennikova T. V., et al. 2004. Genomic characterization of virulent, attenuated, and revertant passages of a North American porcine reproductive and respiratory syndrome virus strain. Virology 321:383–390 [DOI] [PubMed] [Google Scholar]
- 26. Harms W., et al. 2001. Characterization of human myocardial fibroblasts immortalized by HPV16 E6-E7 genes. Exp. Cell Res. 268:252–261 [DOI] [PubMed] [Google Scholar]
- 27. Heim A., et al. 1995. Cultured human myocardial fibroblasts of pediatric origin: natural human interferon-alpha is more effective than recombinant interferon-alpha 2a in carrier-state coxsackievirus B3 replication. J. Mol. Cell. Cardiol. 27:2199–2208 [DOI] [PubMed] [Google Scholar]
- 28. Heim A., et al. 1992. Synergistic interaction of interferon-beta and interferon-gamma in coxsackievirus B3-infected carrier cultures of human myocardial fibroblasts. J. Infect. Dis. 166:958–965 [DOI] [PubMed] [Google Scholar]
- 29. Holland J., et al. 1982. Rapid evolution of RNA genomes. Science 215:1577–1585 [DOI] [PubMed] [Google Scholar]
- 30. Huber S. A. 1997. Coxsackievirus-induced myocarditis is dependent on distinct immunopathogenic responses in different strains of mice. Lab Invest. 76:691–701 [PubMed] [Google Scholar]
- 31. Kandolf R., et al. 1991. Molecular studies on enteroviral heart disease: patterns of acute and persistent infections. Eur. Heart J. 12(Suppl. D):49–55 [DOI] [PubMed] [Google Scholar]
- 32. Kimura E., et al. 2001. Efficient repetitive gene delivery to skeletal muscle using recombinant adenovirus vector containing the coxsackievirus and adenovirus receptor cDNA. Gene Ther. 8:20–27 [DOI] [PubMed] [Google Scholar]
- 33. Klingel K., et al. 1992. Ongoing enterovirus-induced myocarditis is associated with persistent heart muscle infection: quantitative analysis of virus replication, tissue damage, and inflammation. Proc. Natl. Acad. Sci. U. S. A. 89:314–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Klingel K., Kandolf R. 1993. The role of enterovirus replication in the development of acute and chronic heart muscle disease in different immunocompetent mouse strains. Scand. J. Infect. Dis. Suppl. 88:79–85 [PubMed] [Google Scholar]
- 35. Klingel K., et al. 1998. Visualization of enteroviral replication in myocardial tissue by ultrastructural in situ hybridization: identification of target cells and cytopathic effects. Lab Invest. 78:1227–1237 [PubMed] [Google Scholar]
- 36. Lin F., et al. 2001. Tissue distribution of products of the mouse decay-accelerating factor (DAF) genes. Exploitation of a Daf1 knock-out mouse and site-specific monoclonal antibodies. Immunology 104:215–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Livak K. J., Schmittgen T. D. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- 38. Mahy B. W. J., Minson A. C., Darby G. K. 1982. Virus persistence, p. 1–13 33rd symposium of the society for general microbiology. Cambridge University Press, Cambridge, United Kingdom [Google Scholar]
- 39. Mahy B. W. 1985. Strategies of virus persistence. Br. Med. Bull. 41:50–55 [DOI] [PubMed] [Google Scholar]
- 40. Matteucci D., et al. 1985. Group B coxsackieviruses readily establish persistent infections in human lymphoid cell lines. J. Virol. 56:651–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. McManus B. M., et al. 1993. Direct myocardial injury by enterovirus: a central role in the evolution of murine myocarditis. Clin. Immunol. Immunopathol. 68:159–169 [DOI] [PubMed] [Google Scholar]
- 42. Melnick J. L. 1996. Polioviruses, coxsackieviruses, echoviruses, and newer enteroviruses, p. 655–712 In Fields B. N., Knipe D. M., Howley P. M. (ed.), Fields virology, 3rd ed. Lippincott-Raven, Philadelphia, PA [Google Scholar]
- 43. Mrukowicz J. Z., et al. 1998. Viruses and cells with mutations affecting viral entry are selected during persistent rotavirus infections of MA104 cells. J. Virol. 72:3088–3097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nalbantoglu J., Pari G., Karpati G., Holland P. C. 1999. Expression of the primary coxsackie and adenovirus receptor is downregulated during skeletal muscle maturation and limits the efficacy of adenovirus-mediated gene delivery to muscle cells. Hum. Gene Ther. 10:1009–1019 [DOI] [PubMed] [Google Scholar]
- 45. Nomoto A., Iizuka N., Kohara M., Arita M. 1988. Strategy for construction of live picornavirus vaccines. Vaccine 6:134–137 [DOI] [PubMed] [Google Scholar]
- 46. Oldstone M. B. 1989. Viral persistence. Cell 56:517–520 [DOI] [PubMed] [Google Scholar]
- 47. Palacios G., Oberste M. S. 2005. Enteroviruses as agents of emerging infectious diseases. J. Neurovirol. 11:424–433 [DOI] [PubMed] [Google Scholar]
- 48. Pauschinger M., et al. 2004. Viral heart disease: molecular diagnosis, clinical prognosis, and treatment strategies. Med. Microbiol. Immunol. 193:65–69 [DOI] [PubMed] [Google Scholar]
- 49. Pauschinger M., et al. 1999. Enteroviral RNA replication in the myocardium of patients with left ventricular dysfunction and clinically suspected myocarditis. Circulation 99:889–895 [DOI] [PubMed] [Google Scholar]
- 50. Pavio N., et al. 2000. Expression of mutated poliovirus receptors in human neuroblastoma cells persistently infected with poliovirus. Virology 274:331–342 [DOI] [PubMed] [Google Scholar]
- 51. Pelletier I., Duncan G., Pavio N., Colbere-Garapin F. 1998. Molecular mechanisms of poliovirus persistence: key role of capsid determinants during the establishment phase. Cell. Mol. Life Sci. 54:1385–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Pinkert S., et al. 2009. Prevention of cardiac dysfunction in acute coxsackievirus B3 cardiomyopathy by inducible expression of a soluble coxsackievirus-adenovirus receptor. Circulation 120:2358–2366 [DOI] [PubMed] [Google Scholar]
- 53. Poidinger M., Coelen R. J., Mackenzie J. S. 1991. Persistent infection of Vero cells by the flavivirus Murray Valley encephalitis virus. J. Gen. Virol. 72(Pt. 3):573–578 [DOI] [PubMed] [Google Scholar]
- 54. Ron D., Tal J. 1985. Coevolution of cells and virus as a mechanism for the persistence of lymphotropic minute virus of mice in L-cells. J. Virol. 55:424–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sawicki S. G., Lu J. H., Holmes K. V. 1995. Persistent infection of cultured cells with mouse hepatitis virus (MHV) results from the epigenetic expression of the MHV receptor. J. Virol. 69:5535–5543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Schmidtke M., et al. 2007. The viral genetic background determines the outcome of coxsackievirus B3 infection in outbred NMRI mice. J. Med. Virol. 79:1334–1342 [DOI] [PubMed] [Google Scholar]
- 57. Schmidtke M., et al. 2000. Attachment of coxsackievirus B3 variants to various cell lines: mapping of phenotypic differences to capsid protein VP1. Virology 275:77–88 [DOI] [PubMed] [Google Scholar]
- 58. Selinka H. C., et al. 2002. Comparative analysis of two coxsackievirus B3 strains: putative influence of virus-receptor interactions on pathogenesis. J. Med. Virol. 67:224–233 [DOI] [PubMed] [Google Scholar]
- 59. Sevilla N., Domingo E. 1996. Evolution of a persistent aphthovirus in cytolytic infections: partial reversion of phenotypic traits accompanied by genetic diversification. J. Virol. 70:6617–6624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Shi Y., et al. 2009. Cardiac deletion of the coxsackievirus-adenovirus receptor abolishes coxsackievirus B3 infection and prevents myocarditis in vivo. J. Am. Coll. Cardiol. 53:1219–1226 [DOI] [PubMed] [Google Scholar]
- 61. Spiller O. B., Goodfellow I. G., Evans D. J., Almond J. W., Morgan B. P. 2000. Echoviruses and coxsackie B viruses that use human decay-accelerating factor (DAF) as a receptor do not bind the rodent analogues of DAF. J. Infect. Dis. 181:340–343 [DOI] [PubMed] [Google Scholar]
- 62. Tami C., et al. 2003. Evidence of the coevolution of antigenicity and host cell tropism of foot-and-mouth disease virus in vivo. J. Virol. 77:1219–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Vlaycheva L. A., Chambers T. J. 2002. Neuroblastoma cell-adapted yellow fever 17D virus: characterization of a viral variant associated with persistent infection and decreased virus spread. J. Virol. 76:6172–6184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Werk D., et al. 2009. Combination of soluble coxsackievirus-adenovirus receptor and anti-coxsackievirus siRNAs exerts synergistic antiviral activity against coxsackievirus B3. Antiviral Res. 83:298–306 [DOI] [PubMed] [Google Scholar]
- 65. Werk D., et al. 2005. Developing an effective RNA interference strategy against a plus-strand RNA virus: silencing of coxsackievirus B3 and its cognate coxsackievirus-adenovirus receptor. Biol. Chem. 386:857–863 [DOI] [PubMed] [Google Scholar]
- 66. Yeung M. C., Chang D. L., Camantigue R. E., Lau A. S. 1999. Inhibitory role of the host apoptogenic gene PKR in the establishment of persistent infection by encephalomyocarditis virus in U937 cells. Proc. Natl. Acad. Sci. U. S. A. 96:11860–11865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Zautner A. E., Korner U., Henke A., Badorff C., Schmidtke M. 2003. Heparan sulfates and coxsackievirus-adenovirus receptor: each one mediates coxsackievirus B3 PD infection. J. Virol. 77:10071–10077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Zhong J., et al. 2006. Persistent hepatitis C virus infection in vitro: coevolution of virus and host. J. Virol. 80:11082–11093 [DOI] [PMC free article] [PubMed] [Google Scholar]