Abstract
We generated four HIV-1 cultures that are resistant to a peptide fusion inhibitor corresponding to the first heptad repeat of gp41 in order to study mechanisms of resistance and gain insights into envelope glycoprotein-mediated membrane fusion. Two genetic pathways emerged that were defined by acquisition of a specific mutation in either the first or second heptad repeat region of gp41 (HR1 or the HR2, respectively). Each pathway was enriched in mutations that clustered in either HR2 and V3 or in HR1 and residues in or near CD4 contact sites. The gp41 mutations in both pathways not only accounted for resistance to the selecting HR1 peptide but also conferred cross-resistance to HR2 peptide fusion inhibitors and enhanced the stability of the six-helix bundle formed by the self-assembly of HR1 and HR2. The gp120 mutations alone enhanced fusion but did not appear to directly contribute to resistance. The implications of these findings for resistance mechanisms and regulation of envelope-mediated fusion are discussed.
INTRODUCTION
Human immunodeficiency virus (HIV) entry into cells is mediated by the envelope (Env) protein, which consists of the gp120 surface subunit and the noncovalently associated gp41 transmembrane subunit. gp120 binding to cellular CD4 and chemokine receptors induces conformational changes in Env that lead to insertion of the gp41 fusion peptide into the target membrane (reviewed in references 19 and 21) and subsequent folding of two heptad-repeat regions (HR1 and HR2, also referred to as N-HR and C-HR) in gp41 into a thermostable six-helix bundle (6HB). The 6HB structure is composed of a trimeric HR1 coiled-coil core surrounded by three HR2 helices, which pack in an antiparallel fashion into the hydrophobic grooves of the coiled coil (9, 41, 70, 76, 82). Formation of the 6HB drives viral and cellular membrane fusion, which is needed for virus entry (45).
Fusion inhibitors constitute a relatively new class of antiretrovirals that prevent virus entry into cells by interfering with HR1 and HR2 interactions to form the 6HB. Peptide fusion inhibitors corresponding to HR2 sequences, for example, enfuvirtide (also referred to as T20 or DP-178), have proven to be potent inhibitors of HIV infection both in vitro and in vivo (32, 79). From genetic studies, biochemical studies with peptides and recombinant proteins, and structural studies of HR1 and HR2 peptides that self-assemble into a thermostable 6HB (10, 11, 20, 33, 36, 46, 50, 62, 63, 74, 75), it is believed that T20 binds to HR1 along the coiled-coil HR1 grooves during conformational changes to form a peptide-gp41 6HB-like structure that interferes with formation of the viral (endogenous) gp41 6HB in a dominant negative manner. However, there are also data indicating that T20 potentially interacts with other regions of Env, for example, regions of gp41 that are near or within the membrane (35, 40, 48), and the coreceptor binding site on gp120 possibly through electrostatic interactions (3, 83). Similar to other antiretrovirals, T20 unfortunately faces the problem of emerging viral resistance. A large database of viruses resistant to T20 has been generated from clinical and laboratory studies. It will therefore be important to develop fusion inhibitors that bind to gp41 in different ways to offset the potential for cross-resistance among agents in the fusion inhibitor class.
Peptide fusion inhibitors corresponding to HR1, for example DP-107 (78), N36 (18), and 5-helix (64), also inhibit HIV fusion. It is likely that HR1 peptides, in an analogous manner to HR2 peptides, interact with HR2 to form a peptide-gp41 6HB-like structure that interferes with formation of the endogenous 6HB (18, 26, 64). HR1 peptides additionally may interact with the HR1 of gp41 in a dominant negative mechanism to form a heterologous peptide-gp41 coiled coil that interferes with the endogenous coiled coil and prevents formation of the gp41 6HB (7, 77, 78). Since HR1 and HR2 peptides can target different sites and residues in gp41, HR1 peptides potentially represent different subclasses of fusion inhibitors with different resistance profiles.
In studies aimed at understanding the mechanism of HR1 peptide inhibition and resistance, we (16) along with others (17, 30) found that viruses resistant to HR1 peptide inhibitors are associated with the mutations in HR1 and HR2. Surprisingly, some of these initial reports also showed that these mutations conferred cross-resistance to HR2 peptide inhibitors (16) and, in some cases, increased in 6HB stability (16, 30). These findings suggest an indirect mechanism of resistance that does not depend on mutation of contact residues to reduce inhibitor binding. To further investigate resistance mechanisms for HR1 peptide inhibitors and structure-function relationships in Env that control refolding of the HR1 and HR2, we studied HR1 peptide resistance in multiple virus cultures to discern patterns of escape. These studies identified two genetic pathways defined by key mutations in either HR1 or HR2 that were associated with mutations in either the CD4 binding or V3 region of gp120, respectively. In both pathways, gp41 mutations enhanced 6HB stability and conferred resistance to not only the selecting peptides but also other peptide fusion inhibitors, while the gp120 mutations improve fusion. Implications of these findings for Env entry and resistance mechanisms are discussed.
MATERIALS AND METHODS
Cells and plasmids.
PM-1 lymphoid cells (42), which endogenously express CD4, CXCR4, and CCR5 receptors (39), were obtained from Michael Norcross and Hana Golding (U.S. Food and Drug Administration [FDA], Bethesda, MD). 293T cells and U87 cells expressing CD4 and CCR5 (U87-T4-CCR5) or CCR5 were kindly provided by Dan Littman (New York University, New York, NY). HeLa cells expressing various levels of CD4 and CCR5 (JC55, JC53, RC4, and RC49) (55) were a gift from David Kabat (Oregon Health and Science University, Portland, OR). The Rev expression plasmid (pRev) (28) was provided by Tom Hope (Northwestern University Feinberg School of Medicine, Chicago, IL). Env-deficient HIV-1 genome (Gag/Pol) plasmid pCMVΔR8.2 (where CMV is cytomegalovirus) and luciferase (Luc) reporter plasmid pHR′CMV-Luc were described previously (49, 84) and obtained from Gary J. Nabel (NIH, Bethesda, MD). To express JR-CSF Env proteins, the env open reading frame (ORF), including signal peptide sequence, of wild-type (WT) or N44-selected mutants was amplified by PCR with the high-fidelity Taq polymerase (Invitrogen, Carlsbad, CA) and primer pairs JRenv-5 (ACGATCCGATATCGCCGCCACCATGAGAGTGAAGGAGAAATATC) and JRenv-3 (TCTAGAGCGGCCGCTTATAGCAAAGCCCTTTCCAAGC) and then placed into the sites of EcoRV and NotI in pCMV/R plasmid that was obtained from Gary J. Nabel (NIH, Bethesda, MD) to create pCMV/R-Env. The proviral plasmids pLAI(JR-CSF) expressing the LAI genome except with the env gene replaced with JR-CSF env, including wild-type and N44-selected mutants, were engineered as described previously (16). β-Galactosidase (β-Gal) α-subunit and ω-subunit expression plasmids (27) were generously provided by Nathaniel Landau (New York University, New York, NY).
Peptides.
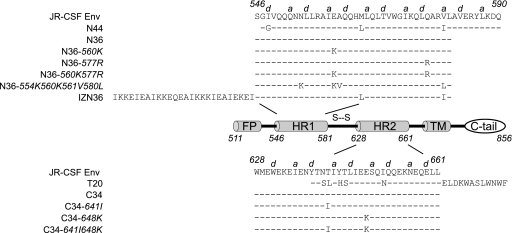
The HR1 peptide inhibitors N44, N36, IZN36, and N36 mutants (N36 with the 560K mutation [N36-560K], N36-577R, N36 with the 560K 577R double mutation [N36-560K/577R], and N36-554K/560K/561V/580L) (Fig. 1) and the HR2 peptide inhibitors T20, C34, and C34 mutants (C34-641I, C34-648K, and C34-641I/648K) (Fig. 1) were synthesized using standard 9-fluorenylmethoxy carbonyl chemistry and purified by high-pressure liquid chromatography (CBER Facility for Biotechnology Resources, Bethesda, MD). N44 has the identical sequence to the bacterially synthesized HR1 peptide used in our prior study (16). All peptides were confirmed to have the expected molecular weights using matrix-assisted laser desorption ionization–time of flight mass spectroscopy and found to be >93% pure by gel electrophoresis.
Fig. 1.
Diagram of domains in gp41 and amino acid sequences of corresponding peptides. HR1 and HR2 correspond to the first and second heptad repeat motifs in gp41, respectively. FP, fusion peptide; TM transmembrane domain; C-tail, cytoplasmic tail. Numbers denote amino acids starting at gp160 according to the nomenclature of the HXB2 reference clone. The sequence of gp41 corresponds to the JR-CSF molecular clone used for these studies a and d indicate positions in the heptad repeats.
Selection of N44-resistant virus.
Viral stocks of HIV-1 LAI(JR-CSF) were generated by transfecting 293T cells with the plasmid pLAI(JR-CSF) using Fugene 6 (Roche, Indianapolis, IN) and collecting filtered culture supernatants 48 h posttransfection, as described previously (16). Viruses were quantified by HIV-1 p24 Gag enzyme-linked immunosorbent assay (ELISA) (AIDS Vaccine Program, NCI-Frederick Cancer Research and Development Center, Frederick, MD) according to the manufacturer's instructions and stored at −80°C. N44-resistant viruses were generated as described previously (16). Briefly, 1 × 106 PM-1 cells were infected overnight with LAI(JR-CSF) virus stock containing 50 ng of p24 Gag in 4 ml of RPMI 1640 medium containing 10% fetal bovine serum in the presence of 500 nM N44. The cells were washed twice the next day and resuspended in 4 ml of RPMI 1640 medium containing the same concentration of N44. Half of the cells and supernatant were collected every 3 to 4 days and replaced with an equal volume of N44-containing RPMI 1640 medium. The supernatants were monitored for virus production by HIV-1 p24 Gag ELISA. Virus and cell samples of each passage of each independent culture were stored from several time points. Supernatants containing the peak level of p24 Gag production were then used for subsequent rounds of resistance selection by infecting with virus supernatants containing 25 ng of p24 Gag and following the same protocol as described above except with increasing N44 concentrations.
Reconstruction of resistant Envs.
Genomic DNA from the cells corresponding to the supernatants containing a peak level of p24 Gag was extracted by using a DNeasy kit (Qiagen, Valencia, CA) according to the manufacturer's instructions. The proviral env gene was then amplified by PCR with the high-fidelity Taq polymerase (Invitrogen, Carlsbad, CA) and primer pairs Envf (AAGCAAAGATCATTAGGGATTATGG) and Envr (AAAACCCACTTCCTCATCCTCA). PCR products of the entire env gene were sequenced to identify mutations. The PCR products with mutations were further engineered in the env gene from the plasmid pLAI(JR-CSF) as described previously (16), and the mutated env gene was also inserted into the pCMV/R expression plasmid, as described above. All constructs were verified to have the mutations that were present in the proviral DNA. After studies were completed, we discovered that the Env from culture 2 had an unintentional mutation of isoleucine to threonine at position 161, which was clearly introduced during a subcloning step. Further analysis of this mutation showed that it did not affect resistance or fusion phenotypes (data not shown).
Pseudovirus assay.
Pseudovirus stocks with wild-type or mutant Env proteins, carrying a Luc reporter gene, were produced by 293T cells in 10-cm-diameter dishes transfected with 5 μg of pCMVΔR8.2, 5 μg of pHR′CMV-Luc, and 2 μg of pCMV/R-Env constructs (wild type or mutants) using FuGene 6 (Roche, Indianapolis, IN) according to the manufacturers protocol. Supernatants were collected 48 h posttransfection, filtered through a 0.45-μm-pore-size low protein-binding filter, quantified by HIV-1 p24 Gag ELISA, and stored at −80°C. Pseudovirus infectivity was determined by combining three independent pseudovirus stocks and calculating the 50% tissue culture infectivity dose (TCID50) by the Reed and Muench method (58). To evaluate resistance, 2 × 104 cells/well were seeded in 96-well plates in Dulbecco's modified Eagle medium (DMEM) containing 10% fetal calf serum, 2 mM glutamine, 1× penicillin, 1× streptomycin, 1 mM sodium pyruvate, and 1× nonessential amino acids (DMEM+) 1 day before infection. U87 CD4+ CCR5+ cells were then infected with equivalent TCID50 inocula of pseudovirus stocks in the presence of peptide inhibitors in 100 μl of DMEM+ supplemented with 8 μg of Polybrene (Sigma, St. Louis, MO) per ml. After 16 h of incubation, the cells were fed with 100 μl of fresh DMEM+ alone and cultured for an additional 24 h at 37°C. The cells were then lysed and assayed for luciferase activity using luciferase kit reagents (Promega, Madison, WI) according to the manufacturer's instructions. Pseudovirus infectivity was expressed as relative luminescence units (RLU/ml).
Cell-cell fusion assay.
Env function was assessed in a cell-cell fusion assay based on β-galactoside (β-Gal) complementation (66). Briefly, 293T cells were transfected with the wild-type or mutant Env expression plasmids (pCMV/R-Env) along with Rev and β-Gal α-subunit expression plasmids using Fugene 6 reagent. Target HeLa cells expressing various levels of CD4 and CCR5 receptors were transfected with the β-Gal ω-subunit expression plasmid. Thirty hours after transfection 3 × 104 target cells were seeded in 96-well plates. Forty-eight hours after transfection, 4.5 × 104 293T cells were added to target cells that had been seeded overnight. After 4 h of incubation of cocultures in DMEM, which was determined to be in the linear range of the fusion curves, cell-cell fusion was scored by β-Gal activity in coculture cell lysates using a Galacto-Star kit (Applied Biosystems, Carlsbad, CA) according to the manufacturer's instructions. β-Gal activity was expressed as relative luminescence units (RLU/ml).
CD spectroscopy.
Stocks of HR1 peptides (N36 and its mutants) (Fig. 1) in 50 mM sodium formate (pH 3.5) and HR2 peptides (C34 and its mutants) (Fig. 1) in 20 mM sodium phosphate (pH 7.0) were mixed (10 μM each) in 20 mM sodium phosphate (pH 7.0) containing 0.2 M NaCl, as described previously (16). The isolated HR1 and HR2 peptides were also diluted to 10 μM in 20 mM sodium phosphate (pH 7.0) containing 0.2 M NaCl for testing. The peptide samples were heated for 30 min at 37°C before circular dichroism (CD) analysis. CD spectra of the peptide samples were recorded at room temperature with a Jasco J810 spectropolarimeter using a 1-nm bandwidth, 0.1-nm resolution, 0.1-cm path length, 1-s response time, and 5-nm/min scanning speed. The spectra were corrected by subtraction of a blank corresponding to the solvent. The α-helical content was calculated from the CD signal by dividing the mean residue ellipticity at 222 nm by the value expected for 100% helix formation (−33,000 degrees cm2 dmol−1) (13, 67). Thermal denaturation of HR1 and HR2 peptide mixtures were acquired at 222 nm by applying a thermal gradient of 1°C/min with a temperature slope of 10 to 90°C, 1-nm bandwidth, 1-°C step resolution, 0.1-cm path length, 1-s response time, and standard 100-millidegree (mdeg) sensitivity. Reverse melting from 90 to 10°C for analyzing reversibility of the folding of the HR1 and HR2 peptide mixtures was also recorded. The melting curve was smoothed, and the temperature at the transition midpoint (melting temperature [Tm]) was estimated from first-derivative plots of the curves by using Jasco software utilities. The fraction of unfolded molecules was analyzed according to a two-state, N ⇋ U mechanism (2).
Statistics.
At least three independent dose-response experiments using multiple batches of each pseudovirus were performed for each inhibitor. In independent experiments, each infection and luciferase measurement was run in duplicate. Uninfected cells were used for the background of luciferase activity. The 50% inhibitory concentrations (IC50) relative to each individual pseudovirus infection without inhibitor were calculated by nonlinear regression analysis with GraphPad Prism software. The geometric mean (GM) IC50 for each inhibitor was determined for the wild-type pseudovirus. To compare across independent experiments, the IC50 of each pseudovirus was normalized to the GM IC50 of the wild-type pseudovirus for each inhibitor and is represented as the fold change (mean ± standard deviation [SD]) in resistance relative to the wild-type pseudovirus. To compare wild-type and mutant Env-induced cell fusion, the fusion index (β-galactosidase activity) of each Env was normalized to the GM of the fusion index of the wild-type Env. The Student t test was performed for comparing mutants to the wild type. P values of <0.01 were considered statistically significant.
RESULTS
Generating escape mutant viruses to N44.
A molecular clone consisting of the LAI proviral genome with the env gene replaced by the JR-CSF env gene [pLAI(JR-CSF)] was used to generate the parental virus for selecting resistance to the HR1 peptide N44. N44-resistant isolates (Table 1) were selected by repeated passage of three independent cultures (cultures 1, 2, and 3) of LAI(JR-CSF) on CD4+ CXCR4+ CCR5+ lymphoblast cells (PM-1) in the presence of increasing concentrations of N44, starting at 500 nM. In addition, we continued to culture the N44-resistant isolate NC mutant (culture 4) that we reported previously (16) with a higher concentration of N44, starting at 1,500 nM. In parallel, four independent cultures without N44 served as controls (controls 1 to 4). Viral replication was monitored by the production of p24 Gag in culture supernatants. After 11 passages with increasing N44 concentrations, cultures 1, 2, and 4 obtained resistance to 5 μM N44 (10-fold increase), while culture 3 reached only 3 μM N44 (6-fold increase). Higher levels of resistance (>10-fold) were not achieved by further increasing the concentrations of N44 peptide. The relatively slow emergence of N44 resistance in our cultures contrasts with the high-level resistance that rapidly emerges with T20 selection (24, 62; also unpublished data). Control cultures without N44 selection pressure did not acquire resistance to N44 (data not shown).
Table 1.
Emerging mutations in the envelope glycoprotein after the indicated passage
| Culture and passagea | gp120 mutation(s) | gp41 mutation(s)b |
|||
|---|---|---|---|---|---|
| HR1 | Hinge | HR2 | C tail | ||
| Cltr 1 | |||||
| P3 | L125F | ||||
| P5 | L125F, I165K | E560K | |||
| P6 | L125F, I165K | E560K, V580L | |||
| P7 | L125F, I165K | N554K, E560K, V580L | T641I | ||
| P11 | L125F, I165K | N554K, E560K, A561V, V580L | T641I | ||
| Cltr 2 | |||||
| P2 | L125F | ||||
| P4 | L125F, E429K | ||||
| P5 | L125F, E429K | E560K | |||
| P7 | L125F, E429K | E560K, Q577R | |||
| P11 | L125F, E429K | E560K, Q577R | |||
| Cltr 3 | |||||
| P2 | T319I | ||||
| P4 | T319I | E648K | |||
| P11 | T319I | E648K | |||
| Cltr 4 | |||||
| P4 | Q577R | E648K | |||
| P5 | S306G, T319I | Q577R | E648K | V833I | |
| P8 | S306G, T319I, E322D | Q577R | D620N | E648K | V833I |
| P14 | S306G, T319I, E322D | Q577R | D620N | T641I, E648K | V833I |
| P15 | S306G, T319I, E322D | Q577R | D620N | T641I, E648K | V833I |
| Con 1 | |||||
| P7 | E172G | ||||
| P11 | E172G | ||||
| Con 2 | |||||
| P5 | Q170R | ||||
| P7 | S115R, Q170R | ||||
| P11 | S115R, Q170R | ||||
| Con 3 | |||||
| P5 | F317Y | ||||
| P11 | F317Y | ||||
| Con 4 | |||||
| P5 | D167N | ||||
| P11 | L125F, D167N | ||||
Mutations arising during 11 passages (P0 to P11) of HIV-1 cultures propagated in the presence (cultures 1 to 4) or absence (controls 1 to 4) of the N44 peptide are shown. Culture 4 was a continuation of the culture that we reported previously (16), which acquired the Q577R and E648K mutations after four passages (P4). Culture 4 in this report was selected for another 11 passages (to P15). Cltr, selection culture; Con, control culture; P, passage.
HR1, heptad repeat 1; HR2, heptad repeat 2; C-tail, cytoplasmic tail.
N44 resistance pathways.
To identify the mutations acquired during N44 selection, the entire env genes from integrated proviral DNA were amplified by PCR, sequenced, and then compared with those from control cultures grown without N44 selection. As shown in Table 1, all viruses from N44 resistance cultures acquired mutations in both gp120 and gp41 subunits. Interestingly, viruses from both cultures 1 and 2 first acquired a leucine-to-phenylalanine mutation at residue 125 (HXB2, gp160 numbering) (L125F) in gp120, followed by a glutamic acid-to-lysine mutation at position 560 (E560K) in gp41, while viruses from both cultures 3 and 4 acquired a glutamic acid-to-lysine mutation at residue 648 (E648K) in gp41 together with a threonine-to-isoleucine mutation at position 319 (T319I) in gp120. However, viruses from culture 1 acquired additional mutations of isoleucine to lysine at residue 165 (I165K) in gp120 and additional HR1 mutations of asparagine to lysine at residue 554 (N554K), alanine to valine at position 561 (A561V), and valine to leucine at residue 580 (V580L), along with a single HR2 mutation of threonine to isoleucine at residue 641 (T641I) that arose in later passages. Culture 2 viruses also acquired a glutamic acid-to-lysine mutation at residue 429 (E429K) in gp120 and a glutamine-to-arginine mutation at position 577 (Q577R) in HR1. Culture 4 viruses acquired further mutations including a serine-to-glycine mutation at residue 306 (S306G) and a glutamic acid-to-aspartic acid mutation at residue 322 (E322D) in gp120, the Q577R mutation, an aspartate-to-asparagine mutation at position 620 (D620N), the T641I mutation, and a valine-to-isoleucine mutation at residue 833 (V833I) in gp41. Viruses from control cultures showed no gp41 mutations and one or two mutations in gp120 after being subjected to a similar number of passages as the selection cultures. Mutations in viruses from control cultures, such as L125F and the aspartic acid-to-asparagine mutation at position 167 (D167N), which have been shown to increase sensitivity to CD4 (56), likely reflect adaptive changes for improved growth in tissue culture. Replicating viruses containing the gp120 and gp41 mutations found in the final passage of each of the resistance cultures were also generated from molecular clones and showed resistance to N44 on PM-1 cells (unpublished data).
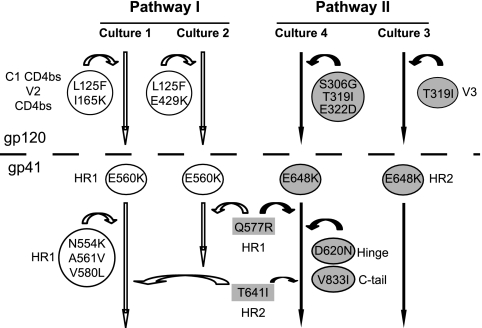
Two patterns of mutations, each occurring in half of the cultures, emerged in viruses from the four independent resistance cultures: one based on the acquisition of the E560K mutation in HR1 and the other based on the E648K mutation in HR2 (Fig. 2). Interestingly, position 648 is polymorphic (29, 37, 71, 73), and the E648K mutation was previously noted to partially compensate for the common T20 resistance mutation involving substitution of glutamic acid for asparagine at position 556 (N43D) (5, 52, 73). Using these residues to define two separate genetic pathways, we also noted that the E560K mutation (pathway I), along with other mutations in HR1 (N554K, A561V, V580L, and Q577R) emerged with the gp120 L125F mutation in both cultures while one of these cultures had an additional E429K mutation. Both L125 and E429 are in gp120 regions involved in contacts with CD4 (38). In the E648K pathway (pathway II), gp120 mutations clustered only in the V3 region. These findings suggest that there may be a direct or functional link between HR1 and regions in gp120 important for CD4 binding and between HR2 and the V3 region.
Fig. 2.
Summary of resistance pathways that emerged after selection with the N44 peptide. The mutations and their locations in specific regions of gp120 and gp41 subunits of the envelope glycoprotein are shown. E560K and E648K are common early mutations that are used to define pathways I and II, respectively. Mutations contributing to pathways I and II are shown in white and shaded circles, respectively. Mutations shown in shaded rectangles contribute to both pathways. Arrows indicate pathway, not the order of emergence of the mutations.
Mapping resistance to N44 and other peptide fusion inhibitors.
Since the mutations in the two pathways involved both gp120 and gp41 subunits, we next evaluated the roles that the mutations in each subunit play in the resistance to peptide fusion inhibitors. We constructed Env expression plasmids containing the mutations in either gp120 or gp41 for each of the four independent resistance cultures. Pseudovirus infection showed that mutations in gp41, but not gp120, accounted for almost all of acquired resistance to N44 (Fig. 3A). Mutations in gp41 of virus from culture 4 (Q577R, D620N, T641I, E648K, and V833I) showed the highest resistance to N44, while mutations in virus from culture 1 (N554K, E560K, A561V, V580L, and T641I) and culture 2 (E560K and Q577R) demonstrated slightly lower levels of resistance. The viral Env from culture 3 with only one mutation in gp41 (E648K) showed the lowest level of resistance. As seen previously (16), the fold increase in resistance seen in the pseudovirus assay using target cells with high levels of both CD4 and coreceptor, in this case U87 CD4+ CCR5+ cells, is lower than that observed in the selection cultures of replicating virus using PM-1 cells, which express relatively low levels of CD4 and CCR5 receptors. Other investigators have also noted that high levels of receptors may reduce sensitivity to entry inhibitors (44, 53, 59).
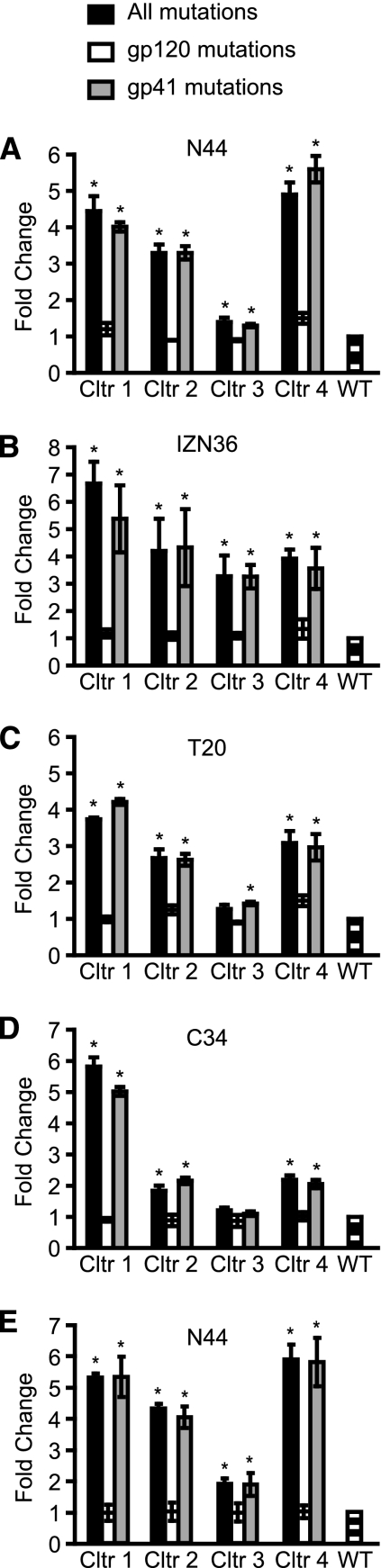
Fig. 3.
Sensitivity of the envelope glycoprotein (Env) to peptide fusion inhibitors. Wild-type Env (WT), Envs containing all mutations in both gp120 and gp41, or Envs containing all mutations in either gp120 or gp41 from viruses in the N44 resistant cultures 1, 2, 3, and 4 were incorporated into HIV pseudoviruses to assess susceptibility to the inhibition by N44 (A), IZN36 (B), T20 (C), and C34 (D) peptides on U87 CD4+ CCR5+ cells with high levels of receptor and coreceptor and by N44 on RC4 cells with lower levels of receptor and coreceptor (E). Values shown are the ratios of the IC50s of the resistant Envs compared to the IC50 of the WT Env, shown as means ± SDs from at least three independent experiments. *, P < 0.01 compared to WT (t test). Cltr, culture.
Pseudovirus infection of the PM-1 cells did not generate sufficient signal to give robust dose-response curves (data not shown), so we further investigated the contribution of the mutations to resistance in RC4 cells, which have low levels of CD4 and chemokine receptors (55). These results agreed with the U87 cells showing that the resistance could be attributed to the gp41 mutations, and there was no difference in resistance between Envs with only gp120 mutations and wild-type Env (Fig. 3E). Thus, these data clearly indicate that the acquired gp41 mutations play the major role in resistance. The gp120 mutations likely reflect adaptive changes to growth in cell culture associated with the primary gp41 resistance mutations.
We next tested the sensitivity of the N44-resistant Envs to inhibition by other peptide fusion inhibitors. Overall, the patterns of cross-resistance to other fusion inhibitors were generally similar to N44 resistance for cultures 1 to 3 though there are some differences for culture 4. While the Env from culture 4 was most resistant to N44 (Fig. 3A), it was somewhat less resistant to a trimer-stabilized HR1-peptide (IZN36) (18) (Fig. 3B). Remarkably, there was also substantial cross-resistance to two different HR2 peptide inhibitors (T20 and C34), consistent with our previous findings involving the early passage of culture 4 virus (16). The resistance patterns for T20 among the cultures were similar to those for N44, except for the Env from culture 4 (Fig. 3C), which showed slightly less resistance to T20 than N44. For the overlapping but N-terminally shifted C34 peptide, mutations from culture 1 virus again showed the highest level of cross-resistance, with less but still significant cross-resistance conferred by mutations from viruses in cultures 2 to 4 (Fig. 3D). As with N44, mutations in gp120 did not confer significant levels of resistance to any of these additional peptide fusion inhibitors. The finding of cross-resistance to different HR1 and HR2 inhibitors that bind to different regions or residues in gp41 suggests that the genetic pathways employ a common resistance mechanism that does not depend exclusively on mutation of residues that directly contact the inhibitors.
Effect of the mutations on fusion and receptor utilization.
Because the gp120 mutations clustered in regions known to be involved with CD4 or chemokine receptor use, we next evaluated the effect of the resistance mutations on Env entry. For these studies, we took advantage of stable cell lines that express various levels of CD4 and CCR5 receptors (55) in order to provide greater discrimination of fusion phenotypes that may be sensitive to receptor levels. We assessed Env function in a cell-cell fusion assay because it provided a more robust analysis than pseudovirus infection of cells with low levels of receptors, which was inefficient under the conditions tested. Cells expressing high levels of Envs with the resistance mutations along with the N-terminal fragment of β-galactosidase (α subunit) were cocultivated with target cells expressing the C-terminal fragment of β-galactosidase (ω subunit) (27) along with various levels of CD4 and CCR5 receptors. Fusion allows complementation of the subunits, providing β-galactosidase activity as readout (66). Cell numbers and surface expression levels of the Envs were controlled and similar within each set of experiments (data not shown).
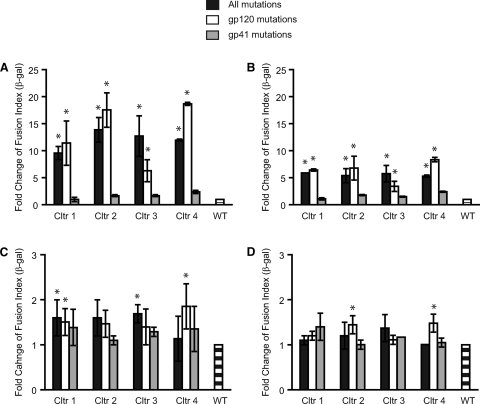
As expected, receptor levels greatly influenced fusion of the mutant Envs relative to the wild type. When cells expressing low levels of CD4 and CCR5 were used as targets, mutations from all four virus cultures enhanced fusion at least 10-fold relative to the wild type (Fig. 4A). However, the magnitude of the enhancement was reduced to 5-fold when CD4 levels remained low but CCR5 levels were increased (Fig. 4B). These findings suggested that the resistant Envs utilized CCR5 better than the wild type but that the difference was less pronounced when CCR5 levels were high. Similarly, the differences between the resistant and wild-type Envs were greatly reduced when CD4 expression levels were high (Fig. 4C and D), indicating that high-level expression of CD4 could compensate for less efficient use of CD4 in the wild-type Envs. Significantly, the enhancement of Env fusion seen in cells with reduced receptor levels mapped predominantly to mutations in gp120. Thus, the resistant Envs also appear to use receptors more efficiently.
Fig. 4.
Effect of resistance mutations on envelope glycoprotein (Env) fusion and receptor use. Wild-type Env (WT) and Envs containing resistance mutations in both gp120 and gp41 or in gp120 or gp41 alone were tested for Env-mediated cell-cell fusion using target cells expressing various levels of CD4 and CCR5: RC4 cells, low levels of CD4 and CCR5 (A); RC49 cells, low level of CD4 and high level of CCR5 (B); JC55 cells, high level of CD4 and low level of CCR5 (C), and JC53 cells, high levels of CD4 and CCR5 (D). Values are the ratios of the GM of fusion (β-galactosidase activity) of Envs with resistance mutations compared to WT Env, shown as means ± SDs from three independent experiments. *, P < 0.01 compared to WT (t test); Cltr, culture.
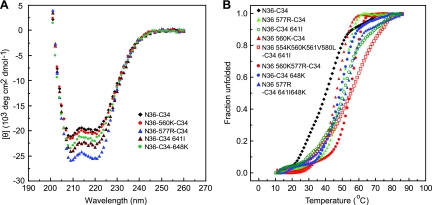
Effect of resistance mutations on 6HB thermal stability.
We next investigated whether the gp41 resistance mutations could directly interfere with inhibitor binding to gp41 by using mixtures of wild-type and mutant HR1 and HR2 peptides in thermal denaturation studies to model 6HB interactions. Since E560K and E648K are key residues that define the pathways and since Q577R and T641I contribute to both pathways (Fig. 2), we first analyzed those mutations individually. All mutations imparted higher α-helical contents to N36-C34 mixtures than the α-helical content of the wild type, with Q577R having the greatest effect (Fig. 5A). Next, we analyzed the combinations of mutant HR1 and HR2 peptides, including the N36-C34 combinations containing gp41 mutations from culture 1 (N554K, E560K, A561V, and V580L in N36, and T641I in C34), culture 2 (E560K and Q577R in N36), and culture 4 (Q577R in N36 and E648K and T641I in C34), and showed that all combinations had high α-helical contents (data not shown).
Fig. 5.
Thermal denaturation studies of equimolar mixtures of the HR1 (N36) and HR2 (C34) peptides containing wild-type residues or mutations, as indicated in Fig. 1. (A) The α-helical conformation of the complex formed by N36-C34 mixtures (10 μM each in phosphate-buffered saline) was analyzed by CD spectroscopy. (B) The fraction of unfolded N36-C34 mixtures was calculated from CD measurements at 222 nm as a function of temperature. Two independent experiments gave identical results. The N36 and C34 mixtures are as indicated.
Thermal denaturation studies showed that the helical bundles formed by all N36-C34 mixtures exhibited a simple two-state unfolding. While wild-type N36-C34 showed a transition midpoint temperature (Tm) of 44°C, N36-560K–C34, N36-577R–C34, N36-560K/577R–C34, N36–C34-641I, N36–C34-648K, N36-577R–C34-641I/648K, and N36-554K/560K/561V/580L–C34-641I all exhibited higher Tms, with values of 47°C, 50°C, 56°C, 53°C, 49°C, 54°C, and 59°C, respectively, indicating that increased 6HB thermal stability is a characteristic feature associated with the resistant Envs (Fig. 5B). The increase in Tm for each resistant Env extends our prior observation on the initial resistance mutations in culture 4 (E648K and Q577R) and is in agreement with a recent report showing that a mutation in residue 648 that emerged in an X4 virus after selection with the HR1 N36 peptide increases 6HB stability through local rearrangement of the hydrogen bond network in gp41 (30). The improved thermal stability of peptide mixtures exhibiting the resistance mutations further suggests a resistance mechanism that involves enhancement of endogenous bundle stability. While mutations in the HR2 might affect 6HB interactions in both the endogenous and inhibitor bundles, mutations in HR1 would be preferentially advantageous to the virus because the sequence of HR1 peptide inhibitor does not change. Previously, it has been noted that 6HB stability can play a role in sensitivity to T20 though the propensity of T20 to form a stable 6HB with gp41 may not always be predictive of peptide potency (23).
DISCUSSION
We generated four independent HIV-1 cultures that are resistant to an HR1 peptide inhibitor in order to identify genetic pathways of escape and elucidate potential mechanisms of resistance. Dominant negative inhibitors, such as N44, likely impose highly restricted solutions for escape because mutations that impair inhibitor binding would also likely impair the endogenous 6HB bundle formation and, consequently, Env function. Resistance to dominant negative fusion inhibitors may therefore reveal compensatory mutations that restore Env function and highlight determinants in Env that regulate conformational changes leading to membrane fusion.
Two patterns of resistance mutations defined by early acquisition of either the E560K mutation in HR1 (pathway I) or E648K mutation in HR2 (pathway II) emerged and were equally distributed among the four cultures. The two cultures exhibiting the pathway II pattern also acquired mutations in V3. In one of these cultures, mutations arose in residues 319 and 306, which are located on antiparallel β-hairpin strands in the V3 loop and possibly influence each other in a compensatory manner (65). The two cultures in pathway I were characterized by the E560K mutation in HR1 together with mutations that may affect CD4 interactions, including L125 and E429 (38), and by the Q577R mutation (16). Additional HR1 mutations were also present in one culture, while Q577R and T641I contributed to both pathways. A conservative valine-to-isoleucine mutation at position 833 (V833I) in the cytoplasmic tail was also seen in one culture though it did not contribute to resistance when examined as a single mutation (data not shown). Significantly, the same two genetic pathways defined by either mutations in HR1 and gp120 residues involving or near CD4 contact residues or mutations in HR2 and V3 were also seen in multiple additional resistance cultures that were selected with another HR1 peptide, N36 (unpublished data). N36 is contained in N44 and corresponds to HR1 sequences in the atomic structure of the six-helix bundle (9). Izumi et al. also found that N36 selected for a resistance mutation in residue 648 of an X4 virus, but in this case the resistance mutation was the substitution of a glutamine residue for glutamic acid (30).
Our findings agree with a recent report indicating that elements in the V3 loop may be involved in stabilizing gp120 interactions with gp41 in the native Env trimer (81). Other reports have also noted potential interactions between gp120 and residues in the gp41 heptad repeat regions that affect fusogenicity (12, 43, 54, 68) and sensitivity to antibody neutralization or peptide inhibition (4, 8, 14, 15, 34, 51, 69, 72, 80). Additional studies have pointed to a direct interaction between T20 and regions in gp120 important for coreceptor binding (3, 83). Along with these reports, our findings that the resistance mutations segregated into two distinct pathways suggest that there may be direct or allosteric interactions between (i) HR1 and residues involved with CD4 binding and (ii) HR2 and V3 regions.
To explore potential communication between mutations in gp120 and gp41, we analyzed Env function and sensitivity to several fusion inhibitors in Envs containing the gp120 or gp41 mutations alone or together. These studies showed that the gp41 mutations accounted for almost all of the resistance, not only to the selecting peptide and another HR1 peptide fusion inhibitor but also to two HR2 peptide fusion inhibitors (Fig. 3C and D). In contrast, the mutations in gp120 accounted for the enhanced fusion seen in cells expressing lower levels of CD4 or CCR5 receptors, but the most pronounced effect was in cells having low levels of both CD4 and CCR5 receptors. High levels of receptors apparently compensated for less efficient receptor use and diminished the differences between the resistant and wild-type Envs.
Some of the gp120 mutations, in particular, the L125F mutation, which was also present in one of the control cultures, and possibly I165K, as well as some of the V3 mutations, likely reflect Env adaptation to growth in culture. All resistance and control cultures, however, retained CCR5 tropism (data not shown). Adaptive mutations in gp120 increase virus replication, which may have facilitated acquisition of resistance mutations, though the gp120 mutations alone did not directly contribute to resistance in the cells used to generate the IC50 curves (Fig. 3). We could not assess resistance in the PM-1 cells that were used for selection due to the poor infectivity of the wild-type pseudovirus in these cells under the conditions tested, so we cannot rule out the possibility that gp120 mutations alone or in combination with specific gp41 mutations contributed to resistance in the cultures. Previously, others have reported that V3 mutations in gp120 can modulate sensitivity to T20 (14, 15, 53, 83) and that increased T20 potency can be correlated with reduced receptor levels (53, 59) and entry kinetics (1, 22, 53, 59, 61). Nonetheless, it has been suggested that overall fusion kinetics may not always predict susceptibility to inhibition by peptide because the time for escape from inhibition by an HR2 peptide did not always correlate with its potency (47). In fact, it has been shown that some T20 resistance mutations reduce Env fusion kinetics (60), presumably because the mutations simultaneously impair inhibitor binding and endogenous 6HB stability. Subsequent compensatory mutations may help to restore 6HB stability and Env function (5, 57).
The presence of two pathways dominated by early mutations in either HR1 or HR2 also raises questions about the target sites for the N44 inhibitor. According to the dominant negative mechanism of inhibition, N44 as a monomer or oligomer could interact with HR1 to interfere with formation of the endogenous HR1 coiled coil needed for 6HB formation. In addition, N44 as a trimer could bind to HR2 and interfere with formation of the 6HB. Currently, there is some evidence for both models of inhibition (7, 26). Although it may be tempting to speculate that the occurrence of two different genetic pathways for resistance might be pointing to these two different modes of inhibition, our data do not clarify this issue because the resistance mutations do not obviously directly impair inhibitor binding. Rather, the observation that both pathways provide cross-resistance not only to other HR1 peptide inhibitors but also to HR2 peptide inhibitors argues for a more general mechanism of resistance that appears to be largely independent of mutation of the residues that directly contact the inhibitors. This mechanism differs from the one generally used in T20 resistance, which often starts with mutation of residues in the N-terminal region of HR1 that directly impact peptide binding (6, 24, 62).
Our biophysical data provide insights into a potential general mechanism of resistance to peptide fusion inhibitors. HR1 and HR2 peptides with or without the resistance mutations form stable helical bundles when mixed in equimolar ratios (Fig. 5). Remarkably, all gp41 resistance mutations occurring in the heptad repeats enhanced 6HB thermostability. Bundle-stabilizing mutations in HR1 would be especially advantageous to the virus because they would favor the virus over the HR1-peptide inhibitor that does not have the mutation. Mutations that preferentially stabilize the viral 6HB compared to the peptide-gp41 6HB would also help to explain the broad cross-resistance seen among all the peptide fusion inhibitors tested.
While pathways I and II also lead to resistance to T20, these patterns of mutations are not seen after T20 selection. Presumably, mutation of residues in the N-terminal region of HR1, which imparts a high level of resistance to T20, is less costly to the virus than mutation of the residues seen in our resistance pathways. Further, the inhibitory activity of the HR peptides appears to depend on many kinetic parameters affecting inactivation of gp41, as well as the peptide-Env context (23, 31, 47, 53), suggesting that the virus may have several potential pathways to gain resistance. Efforts are under way to understand how the increase in 6HB stability observed in our resistant Env relates to the kinetics of fusion and availability of the gp41 fusion intermediate to N44. In any case, mutation of a few contact residues appears to be a less viable option for resistance to N44 and other HR1 peptide fusion inhibitors, perhaps because such contact residues may be critical to one or more conformations needed for Env entry. In this regard, we find it especially intriguing that the resistance mutations in HR1 or HR2 that enhance 6HB stability occur with mutations in specific regions of gp120 that affect receptor use. Conceivably, these gp120 mutations that affect receptor use balance gp41 resistance mutations that favor 6HB formation to better coordinate receptor activation with bundle formation. Premature refolding of Env into the 6HB prior to receptor engagement would inactivate Env fusion potential. Interestingly, a recent report has also found that changes in gp41 contribute to the reactivity of Env to ligands and temperature, further underscoring the importance of global, long-range intramolecular interactions throughout Env (25).
In summary, selection with an HR1 peptide fusion inhibitor identified two genetic pathways for resistance, which were enriched in mutations in either HR1 and residues in gp120 that are involved in CD4 interactions or in HR2 and residues in the V3 region. Both pathways involve gp41 mutations that enhance endogenous 6HB stability and play a major role in conferring cross-resistance to several different peptide fusion inhibitors. Specific patterns of gp120 mutations associated with mutations in either HR1 or HR2 provide potential clues to the regulation of receptor activation with 6HB bundle formation.
ACKNOWLEDGMENTS
We thank Keith Peden and Michael Norcross (U.S. Food and Drug Administration, Bethesda, MD) for critical reading of the manuscript.
This work was supported in part by institutional funds from the FDA and the Intramural AIDS Targeted Antiviral Program from the National Institutes of Health.
We have no competing interests.
Footnotes
Published ahead of print on 12 October 2011.
REFERENCES
- 1. Abrahamyan L. G., et al. 2005. The cytoplasmic tail slows the folding of human immunodeficiency virus type 1 Env from a late prebundle configuration into the six-helix bundle. J. Virol. 79:106–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Agashe V. R., Udgaonkar J. B. 1995. Thermodynamics of denaturation of barstar: evidence for cold denaturation and evaluation of the interaction with guanidine hydrochloride. Biochemistry 34:3286–3299 [DOI] [PubMed] [Google Scholar]
- 3. Alam S. M., et al. 2004. An inducible HIV type 1 gp41 HR-2 peptide-binding site on HIV type 1 envelope gp120. AIDS Res. Hum. Retroviruses 20:836–845 [DOI] [PubMed] [Google Scholar]
- 4. Back N. K., et al. 1993. Mutations in human immunodeficiency virus type 1 gp41 affect sensitivity to neutralization by gp120 antibodies. J. Virol. 67:6897–6902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bai X., et al. 2008. Impact of the enfuvirtide resistance mutation N43D and the associated baseline polymorphism E137K on peptide sensitivity and six-helix bundle structure. Biochemistry 47:6662–6670 [DOI] [PubMed] [Google Scholar]
- 6. Baldwin C. E., et al. 2004. Emergence of a drug-dependent human immunodeficiency virus type 1 variant during therapy with the T20 fusion inhibitor. J. Virol. 78:12428–12437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bewley C. A., Louis J. M., Ghirlando R., Clore G. M. 2002. Design of a novel peptide inhibitor of HIV fusion that disrupts the internal trimeric coiled-coil of gp41. J. Biol. Chem. 277:14238–14245 [DOI] [PubMed] [Google Scholar]
- 8. Blish C. A., Nguyen M. A., Overbaugh J. 2008. Enhancing exposure of HIV-1 neutralization epitopes through mutations in gp41. PLoS Med. 5:e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chan D. C., Fass D., Berger J. M., Kim P. S. 1997. Core structure of gp41 from the HIV envelope glycoprotein. Cell 89:263–273 [DOI] [PubMed] [Google Scholar]
- 10. Chan D. C., Kim P. S. 1998. HIV entry and its inhibition. Cell 93:681–684 [DOI] [PubMed] [Google Scholar]
- 11. Chen C.-H., Matthews T. J., McDanal C. B., Bolognesi D. P., Greenberg M. L. 1995. A molecular clasp in the human immunodeficiency virus (HIV) type 1 TM protein determines the anti-HIV activity of gp41 derivatives: implication for viral fusion. J. Virol. 69:3771–3777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chen S. S. 1994. Functional role of the zipper motif region of human immunodeficiency virus type 1 transmembrane protein gp41. J. Virol. 68:2002–2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen Y. H., Yang J. T., Chau K. H. 1974. Determination of the helix and beta form of proteins in aqueous solution by circular dichroism. Biochemistry 13:3350–3359 [DOI] [PubMed] [Google Scholar]
- 14. Derdeyn C. A., et al. 2000. Sensitivity of human immunodeficiency virus type 1 to the fusion inhibitor T-20 is modulated by coreceptor specificity defined by the V3 loop of gp120. J. Virol. 74:8358–8367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Derdeyn C. A., et al. 2001. Sensitivity of human immunodeficiency virus type 1 to fusion inhibitors targeted to the gp41 first heptad repeat involves distinct regions of gp41 and is consistently modulated by gp120 interactions with the coreceptor. J. Virol. 75:8605–8614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Desmezieres E., et al. 2005. HIV gp41 escape mutants: cross-resistance to peptide inhibitors of HIV fusion and altered receptor activation of gp120. J. Virol. 79:4774–4781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dwyer J. J., et al. 2008. Design of an engineered N-terminal HIV-1 gp41 trimer with enhanced stability and potency. Protein Sci. 17:633–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Eckert D. M., Kim P. S. 2001. Design of potent inhibitors of HIV-1 entry from the gp41 N-peptide region. Proc. Natl. Acad. Sci. U. S. A. 98:11187–11192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Eckert D. M., Kim P. S. 2001. Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 70:777–810 [DOI] [PubMed] [Google Scholar]
- 20. Furuta R. A., Wild C. T., Weng Y., Weiss C. D. 1998. Capture of an early fusion-active conformation of HIV-1 gp41. Nat. Struct. Biol. 5:276–279 [DOI] [PubMed] [Google Scholar]
- 21. Gallo S. A., et al. 2003. The HIV Env-mediated fusion reaction. Biochim. Biophys. Acta 1614:36–50 [DOI] [PubMed] [Google Scholar]
- 22. Gallo S. A., Puri A., Blumenthal R. 2001. HIV-1 gp41 six-helix bundle formation occurs rapidly after the engagement of gp120 by CXCR4 in the HIV-1 Env-mediated fusion process. Biochemistry 40:12231–12236 [DOI] [PubMed] [Google Scholar]
- 23. Gallo S. A., Sackett K., Rawat S. S., Shai Y., Blumenthal R. 2004. The stability of the intact envelope glycoproteins is a major determinant of sensitivity of HIV/SIV to peptidic fusion inhibitors. J. Mol. Biol. 340:9–14 [DOI] [PubMed] [Google Scholar]
- 24. Greenberg M. L., Cammack N. 2004. Resistance to enfuvirtide, the first HIV fusion inhibitor. J. Antimicrob. Chemother. 54:333–340 [DOI] [PubMed] [Google Scholar]
- 25. Haim H., et al. 2011. Contribution of intrinsic reactivity of the HIV-1 envelope glycoproteins to CD4-independent infection and global inhibitor sensitivity. PLoS Pathog. 7:e1002101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. He Y., et al. 2003. Peptides trap the human immunodeficiency virus type 1 envelope glycoprotein fusion intermediate at two sites. J. Virol. 77:1666–1671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Holland A. U., Munk C., Lucero G. R., Nguyen L. D., Landau N. R. 2004. Alpha-complementation assay for HIV envelope glycoprotein-mediated fusion. Virology 319:343–352 [DOI] [PubMed] [Google Scholar]
- 28. Hope T. J., McDonald D., Huang X. J., Low J., Parslow T. G. 1990. Mutational analysis of the human immunodeficiency virus type 1 Rev transactivator: essential residues near the amino terminus. J. Virol. 64:5360–5366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hudelson S. E., et al. 2009. Analysis of HIV type 1 gp41 and enfuvirtide susceptibility among men in the United States who were HIV infected prior to availability of HIV entry inhibitors. AIDS Res. Hum. Retroviruses 25:701–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Izumi K., et al. 2010. Characterization of HIV-1 resistance to a fusion inhibitor, N36, derived from the gp41 amino-terminal heptad repeat. Antiviral Res. 87:179–186 [DOI] [PubMed] [Google Scholar]
- 31. Kahle K. M., Steger H. K., Root M. J. 2009. Asymmetric deactivation of HIV-1 gp41 following fusion inhibitor binding. PLoS Pathog. 5:e1000674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kilby J. M., et al. 1998. Potent suppression of HIV-1 replication in humans by T-20, a peptide inhibitor of gp41-mediated virus entry. Nat. Med. 4:1302–1307 [DOI] [PubMed] [Google Scholar]
- 33. Kilgore N. R., Salzwedel K., Reddick M., Allaway G. P., Wild C. T. 2003. Direct evidence that C-peptide inhibitors of human immunodeficiency virus type 1 entry bind to the gp41 N-helical domain in receptor-activated viral envelope. J. Virol. 77:7669–7672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Klasse P. J., McKeating J. A., Schutten M., Reitz M. S., Jr., Robert-Guroff M. 1993. An immune-selected point mutation in the transmembrane protein of human immunodeficiency virus type 1 (HXB2-Env:Ala 582(→Thr)) decreases viral neutralization by monoclonal antibodies to the CD4-binding site. Virology 196:332–337 [DOI] [PubMed] [Google Scholar]
- 35. Kliger Y., et al. 2001. Mode of action of an antiviral peptide from HIV-1. Inhibition at a post-lipid mixing stage. J. Biol. Chem. 276:1391–1397 [DOI] [PubMed] [Google Scholar]
- 36. Koshiba T., Chan D. C. 2003. The prefusogenic intermediate of HIV-1 gp41 contains exposed C-peptide regions. J. Biol. Chem. 278:7573–7579 [DOI] [PubMed] [Google Scholar]
- 37. Kuiken C., et al. 2009. HIV sequence compendium 2009. Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, Los Alamos, NM [Google Scholar]
- 38. Kwong P. D., et al. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lee B., Sharron M., Montaner L. J., Weissman D., Doms R. W. 1999. Quantification of CD4, CCR5, and CXCR4 levels on lymphocyte subsets, dendritic cells, and differentially conditioned monocyte-derived macrophages. Proc. Natl. Acad. Sci. U. S. A. 96:5215–5220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Liu S., et al. 2005. Different from the HIV fusion inhibitor C34, the anti-HIV drug Fuzeon (T-20) inhibits HIV-1 entry by targeting multiple sites in gp41 and gp120. J. Biol. Chem. 280:11259–11273 [DOI] [PubMed] [Google Scholar]
- 41. Lu M., Blacklow S. C., Kim P. S. 1995. A trimeric structural domain of the HIV-1 transmembrane glycoprotein. Nat. Struct. Biol. 2:1075–1082 [DOI] [PubMed] [Google Scholar]
- 42. Lusso P., et al. 1995. Growth of macrophage-tropic and primary human immunodeficiency virus type 1 (HIV-1) isolates in a unique CD4+ T-cell clone (PM1): failure to downregulate CD4 and to interfere with cell-line-tropic HIV-1. J. Virol. 69:3712–3720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Maerz A. L., Drummer H. E., Wilson K. A., Poumbourios P. 2001. Functional analysis of the disulfide-bonded loop/chain reversal region of human immunodeficiency virus type 1 gp41 reveals a critical role in gp120-gp41 association. J. Virol. 75:6635–6644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Markosyan R. M., Cohen F. S., Melikyan G. B. 2003. HIV-1 envelope proteins complete their folding into six-helix bundles immediately after fusion pore formation. Mol. Biol. Cell 14:926–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Melikyan G. B., et al. 2000. Evidence that the transition of HIV-1 gp41 into a six-helix bundle, not the bundle configuration, induces membrane fusion. J. Cell Biol. 151:413–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mink M., et al. 2005. Impact of human immunodeficiency virus type 1 gp41 amino acid substitutions selected during enfuvirtide treatment on gp41 binding and antiviral potency of enfuvirtide in vitro. J. Virol. 79:12447–12454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Miyauchi K., Kozlov M. M., Melikyan G. B. 2009. Early steps of HIV-1 fusion define the sensitivity to inhibitory peptides that block 6-helix bundle formation. PLoS Pathog. 5:e1000585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Munoz-Barroso I., Durell S., Sakaguchi K., Appella E., Blumenthal R. 1998. Dilation of the human immunodeficiency virus-1 envelope glycoprotein fusion pore revealed by the inhibitory action of a synthetic peptide from gp41. J. Cell Biol. 140:315–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Naldini L., et al. 1996. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science 272:263–267 [DOI] [PubMed] [Google Scholar]
- 50. Nameki D., et al. 2005. Mutations conferring resistance to human immunodeficiency virus type 1 fusion inhibitors are restricted by gp41 and Rev-responsive element functions. J. Virol. 79:764–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Park E. J., Vujcic L. K., Anand R., Theodore T. S., Quinnan G. V., Jr 1998. Mutations in both gp120 and gp41 are responsible for the broad neutralization resistance of variant human immunodeficiency virus type 1 MN to antibodies directed at V3 and non-V3 epitopes. J. Virol. 72:7099–7107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Perez-Alvarez L., et al. 2006. Long-term monitoring of genotypic and phenotypic resistance to T20 in treated patients infected with HIV-1. J. Med. Virol. 78:141–147 [DOI] [PubMed] [Google Scholar]
- 53. Platt E. J., Durnin J. P., Kabat D. 2005. Kinetic factors control efficiencies of cell entry, efficacies of entry inhibitors, and mechanisms of adaptation of human immunodeficiency virus. J. Virol. 79:4347–4356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Platt E. J., Durnin J. P., Shinde U., Kabat D. 2007. An allosteric rheostat in HIV-1 gp120 reduces CCR5 stoichiometry required for membrane fusion and overcomes diverse entry limitations. J. Mol. Biol. 374:64–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Platt E. J., Wehrly K., Kuhmann S. E., Chesebro B., Kabat D. 1998. Effects of CCR5 and CD4 cell surface concentrations on infections by macrophagetropic isolates of human immunodeficiency virus type 1. J. Virol. 72:2855–2864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Pugach P., et al. 2004. The prolonged culture of human immunodeficiency virus type 1 in primary lymphocytes increases its sensitivity to neutralization by soluble CD4. Virology 321:8–22 [DOI] [PubMed] [Google Scholar]
- 57. Ray N., Blackburn L. A., Doms R. W. 2009. HR-2 mutations in human immunodeficiency virus type 1 gp41 restore fusion kinetics delayed by HR-1 mutations that cause clinical resistance to enfuvirtide. J. Virol. 83:2989–2995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Reed L. J., Muench H. 1938. A simple method of calculating fifty per cent endpoints. Am. J. Hyg. (Lond.) 27:493–497 [Google Scholar]
- 59. Reeves J. D., et al. 2002. Sensitivity of HIV-1 to entry inhibitors correlates with envelope/coreceptor affinity, receptor density, and fusion kinetics. Proc. Natl. Acad. Sci. U. S. A. 99:16249–16254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Reeves J. D., et al. 2005. Enfuvirtide resistance mutations: impact on human immunodeficiency virus envelope function, entry inhibitor sensitivity, and virus neutralization. J. Virol. 79:4991–4999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Reeves J. D., et al. 2004. Impact of mutations in the coreceptor binding site on human immunodeficiency virus type 1 fusion, infection, and entry inhibitor sensitivity. J. Virol. 78:5476–5485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Rimsky L. T., Shugars D. C., Matthews T. J. 1998. Determinants of human immunodeficiency virus type 1 resistance to gp41-derived inhibitory peptides. J. Virol. 72:986–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Root M. J., Hamer D. H. 2003. Targeting therapeutics to an exposed and conserved binding element of the HIV-1 fusion protein. Proc. Natl. Acad. Sci. U. S. A. 100:5016–5021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Root M. J., Kay M. S., Kim P. S. 2001. Protein design of an HIV-1 entry inhibitor. Science 291:884–888 [DOI] [PubMed] [Google Scholar]
- 65. Rosen O., Sharon M., Quadt-Akabayov S. R., Anglister J. 2006. Molecular switch for alternative conformations of the HIV-1 V3 region: implications for phenotype conversion. Proc. Natl. Acad. Sci. U. S. A. 103:13950–13955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Rossi F., Charlton C. A., Blau H. M. 1997. Monitoring protein-protein interactions in intact eukaryotic cells by beta-galactosidase complementation. Proc. Natl. Acad. Sci. U. S. A. 94:8405–8410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Shu W., et al. 2000. Helical interactions in the HIV-1 gp41 core reveal structural basis for the inhibitory activity of gp41 peptides. Biochemistry 39:1634–1642 [DOI] [PubMed] [Google Scholar]
- 68. Stern T. L., Reitz M. S., Jr., Robert-Guroff M. 1995. Spontaneous reversion of human immunodeficiency virus type 1 neutralization-resistant variant HXB2thr582: in vitro selection against cytopathicity highlights gp120-gp41 interactive regions. J. Virol. 69:1860–1867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Sullivan N., Sun Y., Li J., Hofmann W., Sodroski J. 1995. Replicative function and neutralization sensitivity of envelope glycoproteins from primary and T-cell line-passaged human immunodeficiency virus type 1 isolates. J. Virol. 69:4413–4422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Tan K., Liu J., Wang J., Shen S., Lu M. 1997. Atomic structure of a thermostable subdomain of HIV-1 gp41. Proc. Natl. Acad. Sci. U. S. A. 94:12303–12308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Teixeira C., et al. 2010. Short communication: high polymorphism rates in the HR1 and HR2 gp41 and presence of primary resistance-related mutations in HIV type 1 circulating in Brazil: possible impact on enfuvirtide efficacy. AIDS Res. Hum. Retroviruses 26:307–311 [DOI] [PubMed] [Google Scholar]
- 72. Thali M., et al. 1994. Resistance to neutralization by broadly reactive antibodies to the human immunodeficiency virus type 1 gp120 glycoprotein conferred by a gp41 amino acid change. J. Virol. 68:674–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Tolstrup M., et al. 2007. Full fusion competence rescue of the enfuvirtide resistant HIV-1 gp41 genotype (43D) by a prevalent polymorphism (137K). AIDS 21:519–521 [DOI] [PubMed] [Google Scholar]
- 74. Trivedi V. D., et al. 2003. The LLSGIV stretch of the N-terminal region of HIV-1 gp41 is critical for binding to a model peptide, T20. Protein Eng. 16:311–317 [DOI] [PubMed] [Google Scholar]
- 75. Wei X., et al. 2002. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob. Agents Chemother. 46:1896–1905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Weissenhorn W., Dessen A., Harrison S. C., Skehel J. J., Wiley D. C. 1997. Atomic structure of the ectodomain from HIV-1 gp41. Nature 387:426–430 [DOI] [PubMed] [Google Scholar]
- 77. Weng Y., Weiss C. D. 1998. Mutational analysis of residues in the coiled coil domain of the human immunodeficiency virus type-1 (HIV-1) gp41. J. Virol. 72:9676–9682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Wild C., Oas T., McDanal C., Bolognesi D., Matthews T. 1992. A synthetic peptide inhibitor of human immunodeficiency virus replication: correlation between solution structure and viral inhibition. Proc. Natl. Acad. Sci. U. S. A. 89:10537–10541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Wild C. T., Shugars D. C., Greenwell T. K., McDanal C. B., Matthews T. J. 1994. Peptides corresponding to a predictive alpha-helical domain of human immunodeficiency virus type 1 are potent inhibitors of virus infection. Proc. Natl. Acad. Sci. U. S. A. 91:9770–9774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Wyatt R., et al. 1997. Analysis of the interaction of the human immunodeficiency virus type 1 gp120 envelope glycoprotein with the gp41 transmembrane glycoprotein. J. Virol. 71:9722–9731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Xiang S. H., et al. 2010. A V3 loop-dependent gp120 element disrupted by CD4 binding stabilizes the human immunodeficiency virus envelope glycoprotein trimer. J. Virol. 84:3147–3161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Yang Z. N., et al. 1999. The crystal structure of the SIV gp41 ectodomain at 1.47 Å resolution. J. Struct. Biol. 126:131–144 [DOI] [PubMed] [Google Scholar]
- 83. Yuan W., Craig S., Si Z., Farzan M., Sodroski J. 2004. CD4-induced T-20 binding to human immunodeficiency virus type 1 gp120 blocks interaction with the CXCR4 coreceptor. J. Virol. 78:5448–5457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Zufferey R., Nagy D., Mandel R. J., Naldini L., Trono D. 1997. Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nat. Biotechnol. 15:871–875 [DOI] [PubMed] [Google Scholar]