Abstract
Certain antibodies from HIV-infected humans bind conserved transition state (CD4 induced [CD4i]) domains on the HIV envelope glycoprotein, gp120, and demonstrate extreme dependence on the formation of a gp120-human CD4 receptor complex. The epitopes recognized by these antibodies remain undefined although recent crystallographic studies of the anti-CD4i monoclonal antibody (MAb) 21c suggest that contacts with CD4 as well as gp120 might occur. Here, we explore the possibility of hybrid epitopes that demand the collaboration of both gp120 and CD4 residues to enable antibody reactivity. Analyses with a panel of human anti-CD4i MAbs and gp120-CD4 antigens with specific mutations in predicted binding domains revealed one putative hybrid epitope, defined by the human anti-CD4i MAb 19e. In virological and immunological tests, MAb 19e did not bind native or constrained gp120 except in the presence of CD4. This contrasted with other anti-CD4i MAbs, including MAb 21c, which bound unliganded, full-length gp120 held in a constrained conformation. Conversely, MAb 19e exhibited no specific reactivity with free human CD4. Computational modeling of MAb 19e interactions with gp120-CD4 complexes suggested a distinct binding profile involving antibody heavy chain interactions with CD4 and light chain interactions with gp120. In accordance, targeted mutations in CD4 based on this model specifically reduced MAb 19e interactions with stable gp120-CD4 complexes that retained reactivity with other anti-CD4i MAbs. These data represent a rare instance of an antibody response that is specific to a pathogen-host cell protein interaction and underscore the diversity of immunogenic CD4i epitope structures that exist during natural infection.
INTRODUCTION
The HIV surface glycoprotein, gp120, comprises an array of conserved domains within the context of a trimeric envelope spike that is otherwise highly variable with respect to conformation, sequence, and structure. A subset of such domains is displayed when gp120 binds to the HIV surface receptor, CD4 (8, 22, 36, 39, 44, 48, 54), and converts from a flexible structure to one that is relatively constrained and functionally essential. This conversion in part stabilizes a critical domain, termed the bridging sheet (5, 27, 56). This action, along with concurrent repositioning of the V1V2 and V3 loops (5, 6, 27, 55), forms a coreceptor binding site that facilitates critical gp120 interactions with chemokine receptors that trigger viral membrane fusion and entry.
The epitopes located in and around the coreceptor binding site are generically termed “CD4 induced” (CD4i) in accordance with their enhanced exposure and/or stabilization within gp120 after CD4 engagement. The immunogenic and antigenic nature of these epitopes has been subject to ongoing investigation and interpretation. Based on computational models, it has been argued that CD4i epitopes are immunologically silent (3, 4, 29, 37) because they are either buried in the free trimer structure or occluded from immunoglobulin interactions postattachment by the restrictive orientation of the CD4 ectodomain (28, 29). Nevertheless, repeated observations that CD4i epitopes are typically immunogenic in HIV infection and elicit cognate humoral immune responses (7, 9, 20, 21, 32, 40, 47) strongly indicate that certain CD4i epitopes are available to surface immunoglobulin during the course of HIV infection (32, 47). In agreement with this idea, it was recently reported that immunization of macaques with soluble envelope trimers raised low-titer, transient CD4i responses as a consequence of CD4 binding (12, 15). Two reports have linked broadly cross-reactive neutralizing responses in part to signature sequences within the CD4i coreceptor binding domain (19, 25).
Independent of their origin, CD4i epitopes and their cognate antibodies display a unique array of highly conserved characteristics and specificities. Recently, it was reported that an Fab fragment of the human monoclonal antibody (MAb) 21c, isolated from an HIV-infected person, binds an epitope comprising gp120 and CD4 residues (11). Crystallographic data indicate that the light chain variable domain contacts CD4, whereas the heavy chain variable domain (VH) binds gp120. Such interactions are apparently distinct from the canonical definition of CD4i specificity and introduce the possibility of hybrid (or bimolecular) epitopes that are entirely dependent upon the juxtaposition of proximal gp120 and CD4 sequences within an attachment complex.
In this study, we used immunochemical assays and targeted mutagenesis to examine this possibility. We report that one human anti-gp120 antibody, MAb 19e, recognizes a unique hybrid epitope with antigenic features that are entirely dependent upon the collaboration of gp120 and CD4 sequences. This feature is distinct from other known CD4i epitopes including MAb 21c, which can bind gp120 in the absence of CD4.
MATERIALS AND METHODS
Cell lines, reagents, and antibodies.
The 293 cells and TZM-bl cell lines were obtained from the NIH AIDS Reagent Repository (Bethesda, MD). Monoclonal antibodies 17b, 21c (34, 38, 46, 57), 19e, ED47 (9), C11 (34, 38, 46, 57), and A32 (38, 46) and/or their respective Epstein-Barr virus (EBV)-transformed human B-cell lines were generously provided by James Robinson (Tulane University, New Orleans, LA). Antibody 2G12 (34, 49, 50) was provided by Hermann Katinger (Polymun Scientific Immunobiologische Forschung GmbH, Vienna, Austria). Soluble four-domain human CD4 (sCD4) was provided by Werner Meier (Biogen, Cambridge, MA). For some experiments, antibodies were produced by transient transfection of 293 cells with plasmids encoding published heavy and light chain immunoglobulin sequences, as previously reported (21). Antibodies were purified from culture supernatants by protein A affinity chromatography, dialyzed against phosphate-buffered saline (PBS), and quantified by bicinchoninic acid (BCA) assay (Bio-Rad, Hercules, CA) according to the manufacturer's protocol. Expression plasmids encoding HIV BaL gp120, FLSC (full-length single-chain gp120-CD4 complex), and SCBaL/M9 (FLSC containing the CD4M9 peptide sequence) were constructed as previously reported (16, 17). Mutations at codons for amino acid position 199 in gp120 in the FLSC antigen or for amino acid positions 58 and 60 in CD4 were introduced by site-directed mutagenesis using targeted primers and a Quickchange II kit (Stratagene, LaJolla, CA) using the manufacturer's protocols. Changes were verified by nucleotide sequencing of the modified expression plasmids. HIV envelope-based antigens were expressed by transiently transfecting plasmids into 293 cells using Fugene (Boeringer Mannheim, Indianapolis, IN) according to the manufacturer's protocol. After 48 h, supernatants were collected for protein purification via lectin affinity chromatography using Galanthus nivalis lectin coupled to 4% agarose beads (Sigma, St. Louis, MO) (18). Bound protein was eluted with 1 M methyl α-d-mannopyranoside and dialyzed against PBS. Protein concentrations were determined by BCA assay (Bio-Rad, Hercules, CA) according to the manufacturer's protocol.
HIV envelope ELISA.
HIV envelope capture enzyme-linked immunosorbent assays (ELISAs) were performed as previously described (17) with various single-chain complexes (as indicated in the text) or HIV BaL gp120 as target antigens that were captured by antibody D7324 adsorbed to the solid phase. Captured antigens were incubated with serial concentrations of human MAbs (as indicated in the text and figure legends), followed by washing. Bound antibodies were reacted with horseradish peroxidase (HRP)-labeled goat anti-human antibody (KPL, Gaithersburg, MD), followed by washing. Tetramethylbenzidine (TMB) HRP substrate (KPL, Gaithersburg, MD) was added, and bound antibody was measured as a function of absorbance at 450 nm. Background assays were carried out in the absence of antigen; values were subtracted from all test absorbance readings. Corrected antibody concentration versus absorbance curves were generated and analyzed to derive one-half maximum binding concentrations for various antibodies using SigmaPlot (Systat GmbH, Germany).
Neutralization assay.
TZM-bl cells (4.5 × 103) expressing CD4 and CCR5 were added to wells in a 96-well microtiter plate and incubated overnight at 37°C in 100 μl of complete medium. Viruses (HIV BaL or HIV BaL 6133) were diluted in 100 μl of culture medium to achieve tissue culture infective doses (TCIDs) of approximately 250,000 relative light unit (RLU) equivalents and incubated with 100 μl of culture medium containing 3-fold serially diluted concentrations of test antibodies starting at 50 μg/ml for 1 h at 37°C. Culture medium was then removed from the microtiter plates and replaced with 200 μl of medium containing virus and various concentrations of antibody. Following incubation at 37°C for 48 h, the cells were washed in PBS, and luminescence was detected using a Bright-Glo Luciferase Assay System (Promega, Madison, WI) according to the manufacturer's protocol. Infection was quantified using a Victor2 (EG&G Wallac, Gaithersburg, MD) fluorescence plate reader. Values were corrected by subtracting background measurements in the absence of virus. The percent infection was calculated by dividing the corrected number of relative light units for each experimental well by the corrected number of relative light units for control wells containing only cells and virus. All assays were carried out in triplicate.
SPR analyses.
Surface plasmon resonance (SPR) analyses of human MAb binding to HIV BaL gp120 or single-chain complexes was accomplished following the methods of Steckbeck et al. (43) using a Biacore T100 instrument (GE Healthcare, Uppsala, Sweden). Protein A (Pierce, Rockford, IL) was immobilized using amine coupling chemistry to consecutive flow cells of a CM5 sensor chip (yielding immobilization levels of approximately 4,000 response units [RU]) to serve as a capture surface for MAbs 19e and 21c. This orientation ensured that the MAb-envelope binding occurred as a homogenous 1:1 Langmuir interaction. MAb capture was standardized to yield approximately 100 RU of captured protein (this was typically achieved with 2 μg/ml antibody). These conditions were used in kinetic binding experiments with both FLSC and FLSC carrying the mutations R to A at position 58 and S to A at position 60 (FLSC RASA). An increasing concentration series of 0, 6.25, 12.5, 25, 50, 100, and 200 nM was injected in duplicate and flowed over the immobilized antibodies at 30 μl/min, with a dissociation phase of 10 min. A buffer injection served as a background control. Upon completion of each association and dissociation cycle, surfaces were pulsed with regeneration solution (100 mM HCl) at a flow rate of 100 μl/min. MAb association rates (ka), dissociation rates (kd), and affinity constants (KD) were calculated from the binding isotherms using Biacore T100 evaluation software, version 1.1.1 (GE Healthcare, Uppsala, Sweden). Binding curves were fit to a 1:1 binding model. Immobilized MAbs C11, 21c, and 19e were analyzed with His-tagged sCD4 in the absence of gp120 using a CM4 chip, which minimizes nonspecific substrate interactions.
Flow cytometry.
CD4+ CEM-NKr-CCR5 cells (5 × 106) were suspended in 1 ml of PBS containing 2% fetal bovine serum and 0.2% sodium azide with or without 50 μg of HIV BaL gp120. After incubation for 1 h at ambient temperature, cells were washed to remove unbound gp120, suspended in the same buffer, and treated with serial concentrations (4-fold, beginning at 40 μg/ml) of test MAb. After 30 min at 4°C, cells were washed and treated with phycoerythrin-conjugated mouse anti-human IgG (Becton Dickinson, Franklin Lakes NJ). After 45 min at 4°C, cells were washed, fixed with paraformaldehyde, and analyzed with a FACSCalibur (Becton Dickinson, Franklin Lakes, NJ) flow cytometer. Data were processed and analyzed using FlowJo software (Tree Star, Ashland, OR).
Antibody 19e docking model.
Based on sequence data, a model of the 19e antibody fragment containing the variable region (Fv) was developed using the AUTOALIGN option for the Web antibody modeling (52, 53) (WAM) server at the University of Bath. The Fv model was examined for the absence of conflicts and exhibited none. From there, the Fv structure was then used to develop a docking model with the YU2 gp120-CD4 complex based on published coordinates (26). The PatchDock algorithm (41, 42) was used in order to derive models interpretable in the context of our chemical data. This algorithm delivered 100 statistically ranked docking solutions of the 19e Fv binding to the YU2 gp120-CD4 complex that were not influenced by any preselected bias for molecular positioning. The highest-scoring model was chosen for further study, given its interpretability in the context of our chemical and mutagenesis data. The model was not refined further and is used as a heuristic guide to interpret the experimental data.
RESULTS
Anti-CD4i antibody binding to constrained envelope antigens.
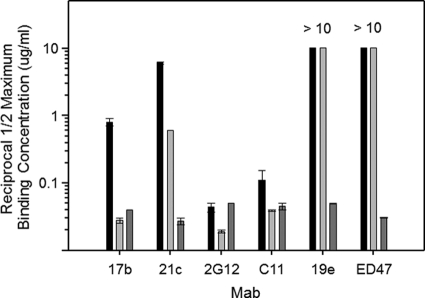
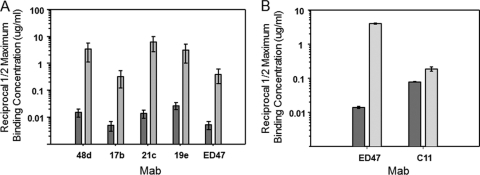
The behavior of anti-CD4i epitope antibodies might be explained by their dependence on CD4-constrained gp120 structures, or, alternatively, on the distinct composition of both gp120 and CD4 sequences aligned within a gp120-CD4 complex (i.e., a hybrid epitope). To differentiate these possibilities, we used ELISAs (16, 17) to characterize the binding of several human anti-envelope MAbs to either single-chain HIV BaL gp120-CD4 (17) or HIV BaL gp120-CD4 mimetic complexes (16), which display the same constrained gp120 structures in the presence or absence of CD4, respectively. The single-chain CD4 mimetic complexes (designated SCBaL/M9) contain modified scorpion toxin (51) scaffolds (CD4M9) that interact with the CD4 binding site on gp120 and stabilize a CD4-induced structure. The single-chain gp120-CD4 complexes (designated FLSC) contain human CD4 domains 1 and 2 and form a natural gp120-CD4 configuration (17). Representative MAb binding to the two antigens versus free gp120 is shown in Fig. 1. As expected, MAbs to non-CD4i epitopes (C11 and 2G12) bound equally well to all antigens. In comparison, MAb 17b exhibited more efficient binding (i.e., lower one-half maximum binding concentrations) to both FLSC and SCBaL/M9 than free gp120 (10). Anti-CD4i MAb 21c bound strongly to FLSC and exhibited lower but significant binding to SCBaL/M9. In contrast, MAb ED47 and MAb 19e bound only to FLSC but did not bind either gp120 or SCBaL/M9, indicating that they have an unusual dependence on constrained gp120 structures that involve CD4.
Fig. 1.
ELISAs of human anti-gp120 MAb binding to gp120 versus constrained gp120 with or without CD4. Monomeric HIV BaL gp120 (black bars), SCBaL/M9 (light gray bars), which presents a constrained gp120 structure in the absence of CD4, and FLSC (dark gray bars), which presents a single-chain gp120-CD4 complex, were captured on the substrate as described in Materials and Methods. The captured antigens were reacted with 3-fold serial concentrations of the indicated human MAbs starting at 10 μg/ml; bound antibody was detected as described in Materials and Methods. Reciprocal one-half maximum binding titers were calculated from the concentration versus absorbance curves after subtraction of background signal. As MAbs 19e and ED47 did not achieve a one-half maximum signal with either gp120 or SCBaL/M9, titers are given as >10. All assays were performed in duplicate; average values are shown. Error bars indicate highest and lowest values.
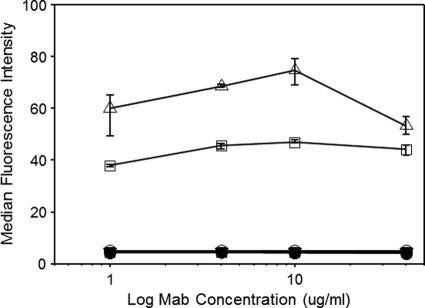
In flow cytometric binding assays with CD4+ CEM-NKr-CCR5 cells, MAb 19e failed to produce binding signals at antibody concentrations up to 40 μg/ml (Fig. 2). In comparison, strong binding was seen using cells coated with HIV BaL gp120. Similar results were obtained with MAb 21c, which was reported to have a certain sCD4 binding capacity (11). As expected, the control MAb b12, directed against the CD4 binding site on gp120, did not bind to uncoated cells or to coated cells, in the latter case because the CD4 binding site was competitively occupied by CD4. To further evaluate the capacities of MAbs 19e and 21c to interact with CD4, SPR analyses were carried out with high concentrations of sCD4 (beginning at 130 μM; 6 mg/ml) in the absence of gp120. The anti-gp120 MAb C11, which recognizes a conserved epitope in the C1-C5 domain, distal to the CD4 binding site (35), was tested as a control. As shown in Fig. 3, higher serial concentrations of sCD4 produced nonsaturating binding signals when flowed over MAb 21c, similar to results of previous studies (11), and MAb 19e. However, the same binding pattern was also seen with MAb C11.
Fig. 2.
Flow cytometric analyses of MAb 19e on CD4+ cells. The indicated concentrations of MAbs 19e (squares), 21c (triangles), and b12 (circles) were tested against CD4+ CEM-NKr-CCR5 cells that were either treated with HIV BaL gp120 (open symbols) or left untreated (closed symbols). Bound antibody was detected as described in Materials and Methods. All assays were performed in triplicate; average values are shown. Error bars indicate standard deviations.
Fig. 3.
SPR analyses of sCD4 interactions with human anti-gp120 MAbs. Threefold serial concentrations of sCD4 were flowed over human anti-gp120 MAbs that were immobilized on a protein A-coated CM4 chip. MAb 19e (filled circles), MAb 21c (open circles), and MAb C11 (filled triangles) were tested in duplicate experiments. Average values are shown; error bars indicate highest and lowest values. The inset shows sensorgrams for each MAb tested at 130 μM: MAb 19e, black line; MAb 21c, red line; MAb C11, blue line.
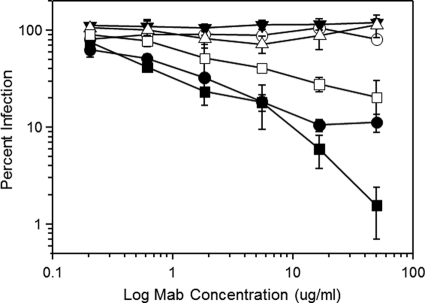
To examine MAb reactivity with virion trimers, we employed an assay design that compares neutralization potency against HIV BaL versus an engineered clone (HIV BaL 6133) that constitutively expresses a CD4-bound envelope structure as a consequence of replacing the gp41 cytoplasmic domain sequence with that of the HXB2 isolate (45). This alteration confers a capacity to infect susceptible target cells expressing low levels of CCR5 as well as CD4-negative cells and an increased reactivity with anti-CD4i epitope MAbs (45). This format specifically assesses the impact of conformational constraints on HIV envelope immunoreactivity without requiring sCD4, alterations in the ectodomain sequence, and/or epitope scaffolding within non-HIV constructs (45). In this case, more potent neutralization of HIV BaL 6133 than HIV BaL indicates that a constrained gp120 structure promotes antibody binding to a functional trimer. As expected, preincubation of the two HIV variants with the canonical anti-CD4i MAb 17b, which recognizes an epitope in the coreceptor binding site, produced more potent neutralization of HIV BaL 6133 than of HIV BaL (Fig. 4). A greater difference was observed with MAb 21c, which potently neutralized HIV BaL 6133 but poorly inhibited HIV BaL. In contrast, MAb 19e failed to neutralize either virus at concentrations up to 100 μg/ml. These results indicate that a constrained gp120 structure on the virion surface satisfies the requirements for MAb 21c binding but is insufficient to support MAb 19e binding.
Fig. 4.
Anti-CD4i epitope MAb-mediated neutralization of viruses presenting native versus constitutively constrained HIV BaL envelope. Viruses (native HIV BaL envelope, open symbols; constrained HIV BaL 6133 envelope; filled symbols) were preincubated with the indicated concentrations of MAb 21c (circles), MAb 19e (triangles), and MAb 17b (squares). Infectivity was measured using TZM-bl cells as described in Materials and Methods. As a control, irrelevant human IgG was tested in an identical manner and caused no reduction in infectivity at any concentration tested (data not shown). All assays were performed in triplicate; average values are shown. Error bars indicate standard deviations. The experiment was repeated with similar results.
Definition of a hybrid epitope by site-directed mutagenesis.
ELISA analyses with a variety of mutant HIV BaL gp120 antigens revealed that a serine-to-leucine modification at position 199 (S199L; residue numbering based on the prototypic HxBc2 gp120 sequence) in the V2 stem extensively reduced the binding of anti-CD4i MAbs 17b, 48d, 21c 19e, and ED47 to gp120-sCD4 complexes (Fig. 5 A). In general, the binding of these MAbs to complexes formed with the S199L gp120 mutant was at least 2 orders of magnitude less efficient (higher one-half maximum binding concentrations) than to complexes containing native gp120. Such differences in reactivities did not extend to the conserved gp120 epitope C11 (Fig. 5B). These data suggested that many anti-CD4i antibodies including MAb 19e share a binding domain that includes residue 199 as one key contact on gp120.
Fig. 5.
Effect of S199L substitution on the binding of anti-CD4i epitope MAbs in ELISA. (A) Native HIV BaL gp120 (black bars) or HIV BaL gp120 S199L (gray bars) antigens were captured on microtiter plate wells as described in Materials and Methods and reacted with sCD4 (2.5 μg/ml). The resulting complexes were then treated with 3-fold serial concentrations of the indicated MAbs starting at 5 μg/ml. Bound antibody was detected as described in Materials and Methods. Reciprocal one-half maximum binding titers were calculated from the concentration versus absorbance curves after subtraction of background signal. (B) In separate experiments, the binding of anti-CD4i MAb ED47 was compared to that of human MAb C11, which recognizes an unrelated epitope. Assays were performed as described for panel A. All assays were performed twice; average values are shown. Error bars indicate highest and lowest values.
Using position 199 as a point of reference, we developed a framework in silico model for estimating MAb 19e reactivity based on published coordinates of a gp120-CD4 complex (27). We first placed residue 199 in the theoretical epicenter of a gp120 epitope and then examined the proximal residues that fell within a 12- to 15-Å radius, which circumscribes an area that might be covered by a typical immunoglobulin paratope. According to this modeling exercise, a portion of the CD4 sequence (58RRSLWDQA64) had the potential to provide the CD4 component of a putative hybrid 19e epitope.
To examine this possibility, site-directed mutagenesis was employed to produce a modified FLSC construct that contained mutations R to A at position 58 (FLSC RA) or S to A at position 60 (FLSC SA) in the CD4 sequence. A third construct was designed to contain both mutations (FLSC RASA). These mutations were selected based on previous evidence (1, 2, 33) that such changes do not significantly impact gp120-CD4 interactions. We first verified that the mutant FLSC complexes were functionally intact using flow cytometry, which determined that they retained their ability to bind cell surface CCR5 compared to the parental construct. Specifically, the RASA double mutant exhibited a half-maximal surface binding titer (0.6 μg/ml) that was not significantly different from that of FLSC (0.3 μg/ml). Thus, the introduction of the CD4 mutations did not alter the presentation of the coreceptor binding site. In accordance, SPR analyses of FLSC or FLSC RASA showed no detectable binding of sCD4 (data not shown) to either protein. This apparent absence of free CD4 binding sites on the two constructs further indicated that each maintained stable intramolecular interactions between their respective gp120 and CD4 moieties.
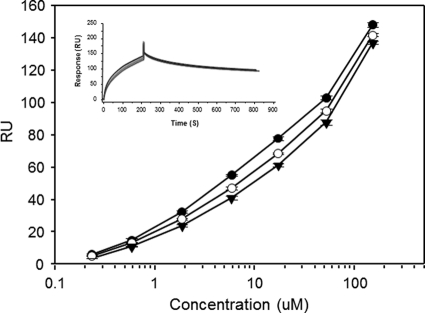
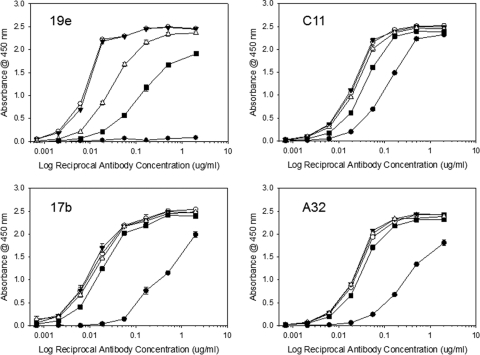
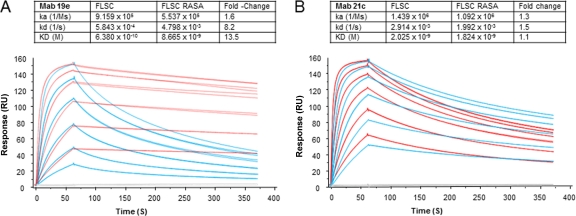
The mutant FLSC antigens were then compared with FLSC for reactivity with MAb 19e versus MAb C11 and two other antibodies to CD4i epitopes: MAb 17b and MAb A32. The latter antibody recognizes a conformational domain distal to the coreceptor binding site (14). As shown in Fig. 6, MAbs A32 and 17b exhibited very similar binding curves with FLSC and all three mutant complexes. As expected, MAbs 17b and A32 also showed detectable but lower binding (i.e., higher one-half maximum binding concentrations) to free gp120. These patterns strongly indicated that the mutant complexes present similarly constrained gp120 antigens. MAb C11 also exhibited equivalent reactivities with all single-chain complex antigens but slightly lower binding to monomeric gp120. In contrast, the one-half maximum binding concentration for MAb 19 reactivity with FLSC RASA was roughly 20-fold higher than binding to FLSC. MAb 19e binding to the FLSC RA single mutant was nearly identical to that seen with FLSC although its reactivity with FLSC SA was slightly less efficient. Concordant results were obtained by SPR analyses of MAb 19e binding to FLSC and FLSC RASA antigens. In these experiments, MAb 21c was tested in parallel for comparison. As shown in Fig. 7, there were no significant differences in MAb 21c binding to FLSC versus FLSC RASA with respect to any kinetic parameter. In contrast, MAb 19e binding clearly differed between antigens, exhibiting an off-rate (kd) more than 8-fold faster and an affinity constant (KD) more than 10-fold higher with FLSC RASA than with FLSC. Collectively, these data indicated that MAb 19 binding is uniquely dependent upon the identity of residues within positions 58 to 64 in CD4.
Fig. 6.
Effects of CD4 mutations R58A and S60A on the binding of human anti-gp120 MAbs to single-chain complexes in ELISA. Single-chain complexes containing the R58A mutation (FLSC RA; filled triangles), the S60A mutation (FLSC SA; open triangles), or both mutations (FLSC RASA; filled squares) were captured on microtiter plate wells as described in Materials and Methods. Unmodified FLSC (open circles) and monomeric gp120 (filled circles) were tested for comparison. The captured antigens were then reacted with 4-fold dilutions of the indicated MAbs starting at 2 μg/ml. Bound antibody was detected as described in Materials and Methods. All assays were performed in duplicate; average values are shown. Error bars indicate highest and lowest values.
Fig. 7.
Measurements of MAb 19e binding kinetics with native versus mutant single-chain gp120-CD4 complexes. Sensorgrams represent SPR experiments (described in Materials and Methods) in which a concentration series of FLSC (red traces) or FLSC RASA (blue traces) was flowed over immobilized MAb 19e (A). For comparison, experiments with MAb 21c (B) were carried out in parallel. All binding conditions were tested in duplicate. Replicate sensorgram plots are shown. Blank experiments in which no antigen was used are shown as gray traces. Binding constants derived from the sensorgrams are tabulated (insets) along with calculated differences between FLSC and FLSC RASA binding parameters. The experiment was repeated with similar results.
Computational modeling of a hybrid epitope.
To interpret these data in a structural context, we evaluated MAb 19e binding to a gp120-CD4 complex using in silico modeling based on known protein structures. This approach was taken as we were unable to obtain useful MAb crystals. We first determined the MAb 19e heavy and light chain gene sequences by reverse transcription-PCR (RT-PCR) of the producer cell line as previously described (21). The heavy chain CDR3 loop sequence was of particular interest as this domain is considered to be critical for interactions between anti-CD4i epitope antibodies and cognate epitopes in and around the coreceptor binding site (24). Sequencing of the MAb 19 heavy and light chains revealed that the 19e heavy chain is derived from the VH1-24 gene, which contrasts with most other known anti-CD4i antibodies that are derived from VH1-69 (21, 24). The 19e heavy chain contains a relatively long CDRH3 loop (∼18 residues), as is typically the case for anti-CD4i epitope antibodies; however, compared to the other known anti-CD4i antibodies, the 19e CDR3 loop (sequence, KPLRNFDWSRRGDYGMDV) was predicted to be slightly more basic overall.
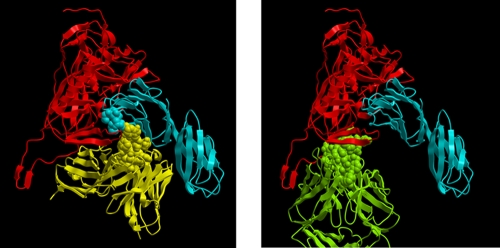
Based on the sequence data, we generated a computational model of the 19e Fv (see Materials and Methods), which was then used to develop a docking model with the YU2 gp120-CD4 complex (containing the V3 loop) based on published coordinates (23, 26). The PatchDock algorithm (41, 42) was used in order to derive models interpretable in the context of our chemical data. The highest-scoring (most likely) model indicated that MAb 19e binds a gp120-CD4 complex in an orientation that differs from what has been reported for other anti-CD4i epitope antibodies such as MAb X5 (Fig. 8). Overall, this model predicts that the MAb 19e heavy chain is situated in proximity to CD4; the light chain is in proximity to gp120. This orientation allows one reasonable interpretation of the mutagenesis data as contacts are predicted to involve CD4 residues within the 58RRSLWDQA64 sequence and elements of MAb 19e. For example, CD4 residue L61 is predicted to contact residue D215 in the 19 heavy chain; CD4 position R58 is predicted to contact Y31 in the 19e light chain. gp120 contacts are predicted to exclusively involve the light chain, within the limits of the modeling exercise. Notably, gp120 residues D197, T198, and V200 (in accordance with the S199L mutagenesis data) are predicted to be particularly important for the light chain interactions.
Fig. 8.
Molecular model of 19e Fv interactions with a gp120-CD4 complex. The predicted 19e Fv model (see Materials and Methods) is shown at left. For comparison, an image from the published crystal structure of anti-CD4i MAb X5 bound to a gp120-CD4 complex (23) is shown at right. In both panels, gp120 is shown in red, and CD4 is shown in blue. 19e Fv is shown in yellow; the heavy chain CDR3 loop proximal to CD4 is shown as a Corey-Pauling-Koltun (CPK) rendering. The SLWDQ sequence in CD4 is shown as a blue CPK rendering. Fab X5 is shown in green with the heavy chain CDR3 loop shown as a CPK rendering.
DISCUSSION
The family of so-called CD4i epitopes on the HIV gp120-CD4 complex comprises immunogenic, highly conserved and functional domains that are logically viewed as potential vaccine and therapeutic targets (10, 13). Since the gp120-CD4 complex positions heterologous sequences within extremely stable and immunogenic proximities, it creates a potential environment for the presentation of hybrid epitopes comprised of residues supplied by two proteins. Spatial considerations for epitope formation and paratope positioning around the coreceptor binding domain suggest that an array of overlapping hybrid specificities is possible (10, 11, 27).
One potential hybrid epitope was recently revealed by crystallography of an Fab fragment of MAb 21c in a complex with a clade C gp120 and CD4 (11). Our analyses indicate that this MAb is capable of significant reactivity with constrained forms of monomeric clade B gp120 in the absence of CD4 (Fig. 1). The neutralization assays (Fig. 4) further indicate that MAb 21c can also bind unliganded envelope trimers, provided they are presented in a constrained conformation. Specifically, MAb 21c neutralized HIV BaL 6133, which expressed a constitutively constrained envelope, but did not affect native HIV BaL in an assay with CD4-positive target cells. Taken together, these properties suggest that MAb 21c does not have an absolute requirement for CD4 in order to establish interactions with the HIV envelope. Given the existing structural information (11), it seems possible that MAb 21c establishes its main contacts with gp120 and engages CD4 as a consequence of surrounding spatial constraints under certain circumstances. This may represent a case where gp120 specificity evolved during B cell ontogeny toward a capacity for accommodating and exploiting the proximity of CD4. However, we note that our studies are limited to a full-length clade B envelope and do not exclude the possibility that MAb 21c exhibits stronger CD4 dependence with certain envelopes from other clades, particularly when they are expressed as truncated core structures (11).
In comparison, MAb 19e is exquisitely dependent on the formation of gp120-CD4 complex as it did not react with native or constrained gp120 in the absence of CD4 (Fig. 1) and did not neutralize HIV BaL 6133 or HIV BaL virus (Fig. 4). The inability of this MAb to neutralize either virus underscores its inability to recognize unliganded envelope regardless of conformational state. Further, the absence of neutralizing activity against either virus indicates that the 19e epitope is highly restricted on HIV BaL trimer-CD4 complexes, in contrast to 17b. Notably, MAb 19e mediates neutralization of HIV-2 pseudotyped virions pretreated with sCD4 (9). However, MAb 19e failed to neutralize HIV BaL in the presence of subinhibitory amounts of sCD4 (data not shown). This contrasts with the ability of MAb 19e to recognize CD4+ cells coated with gp120 (Fig. 2). Thus, there may be important variability in hybrid epitope occlusion, depending on the disposition of the envelope antigen being tested.
In SPR analyses, MAb 19e and MAb 21c bound very poorly to free sCD4 (Fig. 3). Although nonsaturation binding was observed with high sCD4 concentrations, the same pattern was evident in control tests with MAb C11. The latter result indicates that much of the observed binding is due to intrinsic interactions between high concentrations of sCD4 and nonspecific immunoglobulin similar to those already described (30). A previous study showed that an artificially glycosylated form of sCD4 failed to bind MAb 21c (11). This effect was attributed to occlusion of the putative autoreactive epitope; however, it is possible that the glycan moiety served to abrogate nonspecific interactions with immunoglobulin. In any case, the extrapolation of sCD4 binding in SPR experiments to predictions of autoreactivity under conditions of natural infection seems problematic, particularly since MAbs 19e and 21c did not exhibit detectable binding to CD4+ cells in the absence of gp120 (Fig. 2). It seems more likely that the immunoreactivity of MAb 21c in vivo is determined by the conformational status of gp120.
Antigen-antibody docking models generated in silico provide a useful means for obtaining positional information, provided that interpretations are made in the context of immunochemical and biochemical data. The unbiased 19e Fv model (Fig. 8) was in good agreement with our mutagenesis data in that contacts were predicted to involve residues that were identical, or immediately adjacent, to ones that impacted MAb 19e binding to gp120-CD4 complexes. Specifically, our findings that MAb 19e binding was strongly reduced by the S199L mutation in gp120 (Fig. 5) agrees with predicted contacts between residues 197 to 200 and the antibody light chain. Further, the negative impact of the R58A and S60A CD4 mutations on MAb 19e (Fig. 6) binding agree with predicted contacts between CD4 and the antibody light and heavy chains, respectively. At the same time, additional contacts between the MAb 19e heavy chain and CD4 residues distal to the SLWDQ region were also possible. This could explain why one or both of the CD4 mutations did not totally eliminate MAb 19e binding to a single-chain complex whereas replacing CD4 with a mimetic peptide (missing an SLWDQ sequence) abolishes immunoreactivity (Fig. 1).
Notably, MAb 21c binding was reported to involve CD4 residues within the 58RRSLWDQA64 sequence (11). An interesting feature is that CD4 residues R59 and S60 form a pocket that captures L105 in the MAb 21c heavy chain CDR3 domain (11). In MAb 19e, adjacent CD4 residue L61 is predicted to contact D215, which is situated in the same CDR3 position as the analogous L105 in MAb 21c. Nevertheless, MAb 21c orients such that its light chain is closest to CD4 and thereby establishes contacts. In the case of MAb 19e, the docking model predicts that the heavy chain is oriented toward CD4. In accordance with this model, the two MAbs were distinguished by SPR analyses, which indicated that the combined R58A and S60A CD4 mutations in FLSC RASA impacted kinetic parameters of MAb 19e binding but did not significantly impact MAb 21c binding compared to FLSC (Fig. 7).
Overall, our data indicate that MAb 19e binds a unique hybrid epitope that depends absolutely on the juxtaposition of gp120 and CD4 sequences within a gp120-CD4 complex for immunoreactivity with cognate antibody. As such, this epitope may represent a relatively pure form of hybrid specificity that depends on CD4 not only for structural perturbation of gp120 but also for epitope completion. Notably, the anti-CD4i epitope MAb ED47 also failed to bind a gp120-CD4 mimetic peptide complex (Fig. 1). This suggests that antibodies against hybrid epitopes such as 19e may not be rare. The frequency of such responses remains largely unexplored as most efforts to census the anti-gp120 reactive memory B cell pools of HIV-positive subjects typically do not employ gp120-CD4 complexes for screening (31, 40).
The provenance of responses to hybrid epitopes such as 19e also remains unknown, but by definition this must involve some form of stable interaction between cell surface CD4 and either HIV gp120, free envelope trimers, or virions. However, this scenario disagrees with molecular models of gp120-CD4 complexes, which predict that epitopes surrounding the coreceptor binding site on gp120 are always occluded from immunoglobulin-B cell receptor interactions as a consequence of the restrictive “height” of the CD4 receptor (29). According to the standing models, such constraints should apply to complexes formed by either envelope monomers or trimers. Nevertheless, the presence of antibodies such as MAb 19e in natural HIV infection points toward situations where immunogenic CD4i epitope exposure occurs within the envelope-receptor complexes that arise during natural HIV infection. It will be important to design new screening tools and structural models that allow a more comprehensive understanding of how HIV manages to elicit immune responses to virus-host antigen complexes.
ACKNOWLEDGMENTS
This work was supported by RO1 A160481-01A1-02 to A.L.D. S.I. was supported by a grant from the Fogarty International Center (5-D43 TW 01041-05).
Footnotes
Published ahead of print on 12 October 2011.
REFERENCES
- 1. Ashkenazi A., et al. 1990. Mapping the CD4 binding site for human immunodeficiency virus by alanine-scanning mutagenesis. Proc. Natl. Acad. Sci. U. S. A. 87:7150–7154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Brodsky M. H., Warton M., Myers R. M., Littman D. R. 1990. Analysis of the site in CD4 that binds to the HIV envelope glycoprotein. J. Immunol. 144:3078–3086 [PubMed] [Google Scholar]
- 3. Burton D. R., et al. 2004. HIV vaccine design and the neutralizing antibody problem. Nat. Immunol. 5:233–236 [DOI] [PubMed] [Google Scholar]
- 4. Burton D. R., Stanfield R. L., Wilson I. A. 2005. Antibody vs. HIV in a clash of evolutionary titans.. Proc. Natl. Acad. Sci. U. S. A. 102:14943–14948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chen B., et al. 2005. Structure of an unliganded simian immunodeficiency virus gp120 core. Nature 433:834–841 [DOI] [PubMed] [Google Scholar]
- 6. Cocchi F., et al. 1996. The V3 domain of the HIV-1 gp120 envelope glycoprotein is critical for chemokine-mediated blockade of infection. Nat. Med. 2:1244–1247 [DOI] [PubMed] [Google Scholar]
- 7. Crooks E. T., et al. 2005. Characterizing anti-HIV monoclonal antibodies and immune sera by defining the mechanism of neutralization. Hum. Antibodies 14:101–113 [PMC free article] [PubMed] [Google Scholar]
- 8. Dalgleish A. G., et al. 1984. The CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature 312:763–767 [DOI] [PubMed] [Google Scholar]
- 9. Decker J. M., et al. 2005. Antigenic conservation and immunogenicity of the HIV coreceptor binding site. J. Exp. Med. 201:1407–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. DeVico A. L. 2007. CD4-induced epitopes in the HIV envelope glycoprotein, gp120. Curr. HIV Res. 5:561–571 [DOI] [PubMed] [Google Scholar]
- 11. Diskin R., Marcovecchio P. M., Bjorkman P. J. 2010. Structure of a clade C HIV-1 gp120 bound to CD4 and CD4-induced antibody reveals anti-CD4 polyreactivity. Nat. Struct. Mol. Biol. 17:608–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Douagi I., et al. 2010. Influence of novel CD4 binding-defective HIV-1 envelope glycoprotein immunogens on neutralizing antibody and T-cell responses in non-human primates. J. Virol. 84:1683–1695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ferrari G., et al. 2011. An HIV-1 gp120 envelope human monoclonal antibody that recognizes a C1 conformational epitope mediates potent antibody-dependent cellular cytotoxicity (ADCC) activity and defines a common ADCC epitope in human HIV-1 serum. J. Virol. 85:7029–7036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Finnegan C. M., Berg W., Lewis G. K., DeVico A. L. 2001. Antigenic properties of the human immunodeficiency virus envelope during cell-cell fusion. J. Virol. 75:11096–11105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Forsell M. N., et al. 2008. B cell recognition of the conserved HIV-1 co-receptor binding site is altered by endogenous primate CD4. PLoS Pathog. 4:e1000171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fouts T., et al. 2002. Crosslinked HIV-1 envelope-CD4 receptor complexes elicit broadly cross-reactive neutralizing antibodies in rhesus macaques. Proc. Natl. Acad. Sci. U. S. A. 99:11842–11847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fouts T. R., et al. 2000. Expression and characterization of a single-chain polypeptide analogue of the human immunodeficiency virus type 1 gp120-CD4 receptor complex. J. Virol. 74:11427–11436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gilljam G. 1993. Envelope glycoproteins of HIV-1, HIV-2, and SIV purified with Galanthus nivalis agglutinin induce strong immune responses. AIDS Res. Hum. Retroviruses 9:431–438 [DOI] [PubMed] [Google Scholar]
- 19. Gnanakaran S., et al. 2010. Genetic signatures in the envelope glycoproteins of HIV-1 that associate with broadly neutralizing antibodies. PLoS Comput. Biol. 6:e1000955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gray E. S., et al. 2007. Neutralizing antibody responses in acute human immunodeficiency virus type 1 subtype C infection. J. Virol. 81:6187–6196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Guan Y., et al. 2009. Discordant memory B cell and circulating anti-Env antibody responses in HIV-1 infection. Proc. Natl. Acad. Sci. U. S. A. 106:3952–3957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hart T. K., et al. 1991. Binding of soluble CD4 proteins to human immunodeficiency virus type 1 and infected cells induces release of envelope glycoprotein gp120. Proc. Natl. Acad. Sci. U. S. A. 88:2189–2193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Huang C. C., et al. 2005. Structure of a V3-containing HIV-1 gp120 core. Science 310:1025–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Huang C. C., et al. 2004. Structural basis of tyrosine sulfation and VH-gene usage in antibodies that recognize the HIV type 1 coreceptor-binding site on gp120. Proc. Natl. Acad. Sci. U. S. A. 101:2706–2711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kirchherr J. L., et al. 2011. Identification of amino acid substitutions associated with neutralization phenotype in the human immunodeficiency virus type-1 subtype C gp120. Virology 409:163–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kwong P. D., et al. 2000. Structures of HIV-1 gp120 envelope glycoproteins from laboratory-adapted and primary isolates. Structure 8:1329–1339 [DOI] [PubMed] [Google Scholar]
- 27. Kwong P. D., et al. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kwong P. D., Wyatt R., Sattentau Q. J., Sodroski J., Hendrickson W. A. 2000. Oligomeric modeling and electrostatic analysis of the gp120 envelope glycoprotein of human immunodeficiency virus. J. Virol. 74:1961–1972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Labrijn A. F., et al. 2003. Access of antibody molecules to the conserved coreceptor binding site on glycoprotein gp120 is sterically restricted on primary human immunodeficiency virus type 1. J. Virol. 77:10557–10565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lenert P., Kroon D., Spiegelberg H., Golub E. S., Zanetti M. 1990. Human CD4 binds immunoglobulins. Science 248:1639–1643 [DOI] [PubMed] [Google Scholar]
- 31. Li Y., et al. 2009. Analysis of neutralization specificities in polyclonal sera derived from human immunodeficiency virus type 1-infected individuals. J. Virol. 83:1045–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. McMichael A. J., Borrow P., Tomaras G. D., Goonetilleke N., Haynes B. F. 2010. The immune response during acute HIV-1 infection: clues for vaccine development. Nat. Rev. Immunol. 10:11–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Moebius U., Clayton L. K., Abraham S., Harrison S. C., Reinherz E. L. 1992. The human immunodeficiency virus gp120 binding site on CD4: delineation by quantitative equilibrium and kinetic binding studies of mutants in conjunction with a high-resolution CD4 atomic structure. J. Exp. Med. 176:507–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Moore J. P., Sodroski J. 1996. Antibody cross-competition analysis of the human immunodeficiency virus type 1 gp120 exterior envelope glycoprotein. J. Virol. 70:1863–1872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Moore J. P., Willey R. L., Lewis G. K., Robinson J., Sodroski J. 1994. Immunological evidence for interactions between the first, second, and fifth conserved domains of the gp120 surface glycoprotein of human immunodeficiency virus type 1. J. Virol. 68:6836–6847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pal R., DeVico A., Rittenhouse S., Sarngadharan M. G. 1993. Conformational perturbation of the envelope glycoprotein gp120 of human immunodeficiency virus type 1 by soluble CD4 and the lectin succinyl Con A. Virology 194:833–837 [DOI] [PubMed] [Google Scholar]
- 37. Poignard P., Saphire E. O., Parren P. W., Burton D. R. 2001. Gp120: biologic aspects of structural features. Annu. Rev. Immunol. 19:253–274 [DOI] [PubMed] [Google Scholar]
- 38. Robinson J. E., Yoshiyama H., Holton D., Elliott S., Ho D. D. 1992. Distinct antigenic sites on HIV gp120 identified by a panel of human monoclonal antibodies. J. Cell. Biochem. 50(Suppl. 16E):71 [Google Scholar]
- 39. Sattentau Q. J., Moore J. P., Vignaux F., Traincard F., Poignard P. 1993. Conformational changes induced in the envelope glycoproteins of the human and simian immunodeficiency viruses by soluble receptor binding. J. Virol. 67:7383–7393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Scheid J. F., et al. 2009. Broad diversity of neutralizing antibodies isolated from memory B cells in HIV-infected individuals. Nature 458:636–640 [DOI] [PubMed] [Google Scholar]
- 41. Schneidman-Duhovny D., Inbar Y., Nussinov R., Wolfson H. J. 2005. PatchDock and SymmDock: servers for rigid and symmetric docking. Nucleic Acids Res. 33:W363–W367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schneidman-Duhovny D., et al. 2003. Taking geometry to its edge: fast unbound rigid (and hinge-bent) docking. Proteins 52:107–112 [DOI] [PubMed] [Google Scholar]
- 43. Steckbeck J. D., et al. 2005. Kinetic rates of antibody binding correlate with neutralization sensitivity of variant simian immunodeficiency virus strains. J. Virol. 79:12311–12320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sullivan N., et al. 1998. CD4-induced conformational changes in the human immunodeficiency virus type 1 gp120 glycoprotein: consequences for virus entry and neutralization. J. Virol. 72:4694–4703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Taylor B. M., et al. 2008. An alteration of HIV gp41 leading to reduced CCR5 dependence and CD4 independence. J. Virol. 82:5460–5471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Thali M., et al. 1993. Characterization of conserved human immunodeficiency virus type 1 gp120 neutralization epitopes exposed upon gp120-CD4 binding. J. Virol. 67:3978–3986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tomaras G. D., et al. 2008. Initial B-cell responses to transmitted human immunodeficiency virus type 1: virion-binding immunoglobulin M (IgM) and IgG antibodies followed by plasma anti-gp41 antibodies with ineffective control of initial viremia. J. Virol. 82:12449–12463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Trkola A., et al. 1996. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR-5. Nature 384:184–186 [DOI] [PubMed] [Google Scholar]
- 49. Trkola A., et al. 1995. Cross-clade neutralization of primary isolates of human immunodeficiency virus type 1 by human monoclonal antibodies and tetrameric CD4-IgG. J. Virol. 69:6609–6617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Trkola A., et al. 1996. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J. Virol. 70:1100–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Vita C., et al. 1999. Rational engineering of a miniprotein that reproduces the core of the CD4 site interacting with HIV-1 envelope glycoprotein. Proc. Natl. Acad. Sci. U. S. A. 96:13091–13096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Whitelegg N., Rees A. R. 2004. Antibody variable regions: toward a unified modeling method. Methods Mol. Biol. 248:51–91 [DOI] [PubMed] [Google Scholar]
- 53. Whitelegg N. R., Rees A. R. 2000. WAM: an improved algorithm for modelling antibodies on the WEB. Protein Eng. 13:819–824 [DOI] [PubMed] [Google Scholar]
- 54. Wu L., et al. 1996. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature 384:179–183 [DOI] [PubMed] [Google Scholar]
- 55. Wyatt R., et al. 1998. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature 393:705–711 [DOI] [PubMed] [Google Scholar]
- 56. Wyatt R., Sodroski J. 1998. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science 280:1884–1888 [DOI] [PubMed] [Google Scholar]
- 57. Xiang S. H., Doka N., Choudhary R. K., Sodroski J., Robinson J. E. 2002. Characterization of CD4-induced epitopes on the HIV type 1 gp120 envelope glycoprotein recognized by neutralizing human monoclonal antibodies. AIDS Res. Hum. Retroviruses 18:1207–1217 [DOI] [PubMed] [Google Scholar]