Abstract
Recent evidence indicates a new role for histone deacetylases (HDACs) in the activation of genes governing the host immune response. Virus, along with other pathogenic stimuli, triggers an antiviral defense mechanism through the induction of IFN, IFN-stimulated genes, and other proinflammatory cytokines. Many of these genes have been shown to be regulated by transcription factors of the IFN regulatory factor (IRF) family. Recent studies from IRF5 knockout mice have confirmed a critical role for IRF5 in virus-induced type I IFN expression and proinflammatory cytokines IL-6, IL-12, and TNF-α; yet, little is known of the molecular mechanism of IRF5-mediated proinflammatory cytokine expression. In this study, we show that both HDACs and histone acetyltransferases (HATs) associate with IRF5, leading to alterations in its transactivation ability. Using the HDAC inhibitor trichostatin A, we demonstrate that ISRE, IFNA, and IL6 promoters require HDAC activity for transactivation and transcription, whereas TNFα does not. Mapping the interaction of corepressor proteins (HDAC1, silencing mediator of retinoid and thyroid receptor/nuclear corepressor of retinoid receptor, and Sin3a) and HATs to IRF5 revealed distinct differences, including the dependence of IRF5 phosphorylation on HAT association resulting in IRF5 acetylation. Data presented in this study support a mechanism whereby virus triggers the dynamic conversion of an IRF5-mediated silencing complex to that of an activating complex on promoters of target genes. These data provide the first evidence, to our knowledge, of a tightly controlled transcriptional mechanism whereby IRF5 regulates proinflammatory cytokine expression in conjunction with HATs and HDACs.
The histone acetylation/deacetylation modification has emerged as a major form of epigenetic mechanism that regulates the expression of genes in eukaryotic cells (1). Acetylation of nucleosomal histones in vivo is a dynamic, reversible process governed by the opposing activities of histone acetyltransferases (HATs) and histone deacetylases (HDACs) (2). Active gene transcription has been tightly associated with hyperacetylation of histones, whereas hypoacetylation has been correlated with reduced transcription or gene silencing. Recent studies indicate that gene regulation by acetylation is more dynamic and complex and that HATs can also act as repressors and HDACs can function as activators (3). Besides histones, HATs and HDACs may also act on nonhistone proteins such as transcription factors. The recruitment of HATs or HDACs by specific transcription factors enables them to regulate specific sets of target genes.
The importance of HDACs as activators of gene expression is underscored by gene expression profiling of cells treated with HDAC inhibitors (HDACi). Only a small percentage of the total genes examined responded to the treatment (~7% of mRNAs), with similar proportions of genes activated or repressed (4, 5). The inhibition of gene expression by HDACi is not well understood, but may be linked to the findings that several transcription factors are also regulated by acetylation/deacetylation (6–10). Detailed studies of the IFN-β promoter have shown that acetylation and deacetylation of histones are essential for enhanceosome function (11). Perhaps the best example of the transcription-activating role of deacetylation is in studies of cytokine-inducible gene regulation. Recent reports indicate that stimulation of gene expression downstream of IFN (either IFN-α/β or IFN-γ) fails after general blockade of HDAC activity (10, 12–14), and many HDACi reduce proinflammatory cytokine expression (15–17). Under normal conditions, the expression of genes coding for many proinflammatory cytokines remains silent. Mechanisms of transcriptional repression dominate until overcome by stimulation from extracellular stress signals, whereby transcription factors are often posttranslationally modified and other coactivators brought in to initiate de novo gene expression.
Regulation of type I IFN (IFNA and B) gene expression is controlled primarily at the transcriptional level and has been widely studied as a model to understand the mechanisms of stable repression, transient expression, and postinduction repression of genes (18). Transcription factors of the IFN regulatory factor (IRF) family have been identified as critical mediators of early inflammatory gene transcription in response to virus and TLR signaling. A less well-characterized IRF family member, IRF5, was demonstrated recently by our laboratory and others to play an intrinsic role in innate immunity. Initial studies on human IRF5 revealed a critical role in virus-induced cytokine and chemokine expression, including type I IFNs, and additional studies have demonstrated its activation in response to TLR7/8 signaling (19–22). Subsequently, animals with a targeted gene disruption of IRF5 were found to have defective proinflammatory cytokine responses (IL-6, IL-12, and TNF-α) to TLR signaling (23) and virus-induced type I IFN expression (24).
Human IRF5 exists as multiple alternatively spliced isoforms, each with distinct cell type-specific expression, regulation, cellular localization, and function (25). The identical polypeptide encoded by IRF5 variant 3 (V3; GenBank accession number AY504946; http://www.ncbi.nlm.nih.gov/genbank/) and variant 4 (V4; AY504947) cDNAs has been shown to be the most potent inducer of IFN-α in virus-infected cells (25). Indeed, ectopic IRF5 V3/V4 binds to the endogenous IFNA promoters in both uninfected and virus-infected cells; yet, levels were dramatically increased in response to Newcastle disease virus (NDV) (19, 20). These data, combined with results from promoter reporter assays (19, 20, 25) and microarray analysis of IRF5 target genes (21), suggested that IRF5 may also be involved in the repression of IFNA gene expression. Furthermore, whereas IRF5 was shown to be a critical mediator of IL-6, IL-12, and TNF-α in the transgenic knockout mouse (23), little is known of its mechanism of gene regulation.
To elucidate the mechanism(s) of IRF5-mediated proinflammatory cytokine expression, we have characterized its transactivation function in the presence or absence of HDACi, HDACs, and HATs. Unexpectedly, we find that IRF5 interacts with both HATs and HDACs, along with other corepressor proteins; however, these interactions were, in part, dependent on IRF5 post-translational modification in response to virus. We demonstrate that formation of the corepressor or coactivator complexes occurs on distinct regions of the IRF5 polypeptide and that acetylase and/or deacetylase activities are important components of IRF5-regulated proinflammatory cytokine expression. The recruitment of these complexes to the IFNA promoter, in particular, required an elaborate exchange of interacting partners that was at least partially dependent on virus-induced IRF5 phosphorylation. Together, these results highlight the importance of both HATs and HDACs in controlling IRF5-mediated proinflammatory cytokine expression.
Materials and Methods
Cells, reagents, and Abs
Human fibroblasts (2fTGH) were from G. Stark (Cleveland Clinic, Cleveland, OH), 2fTGH/Flag-tagged IRF5 V3/V4-expressing cells were previously described (19), primary human fibroblasts (HFs) were from G. Hayward (Johns Hopkins University, Baltimore, MD), Hek TLR9 cells were from Imgenex (San Diego, CA), immortalized macrophages from irf5−/− mice were from K. Fitzgerald (University of Massachusetts, Amherst, MA), and THP-1 monocytic macrophages were from American Type Culture Collection (Manassas, VA). PBMCs were freshly isolated by Ficoll-Hypaque density centrifugation (Lymphoprep, Accurate Chemical and Scientific, Westbury, NY) of heparinized blood from healthy donors. Human PBMC studies were approved by the Institutional Review Board of the New Jersey Medical School (Newark, NJ). Purified populations of human monocytes were obtained from PBMC using CD14 Microbeads (Miltenyi Biotec, Auburn, CA). All adherent cell lines were grown in DMEM supplemented with 10% FBS; all suspension cells were grown in RPMI 1640 supplemented with 10% FBS. NDV was obtained from American Type Culture Collection (VR-699), trichostatin A (TSA) from Calbiochem (San Diego, CA), R848 from Invivogen (San Diego, CA), and CpG-B from Coley Pharmaceuticals (Langenfeld, Germany). IFNA1-secreted alkaline phosphatase (SAP) promoter reporter was previously described (26); GFP-IRF-5N1 and GFP-IRF-5C1 were described (20); HDAC1 and HDAC5 expression plasmids were from S.L. Schreiber (Harvard, Cambridge, MA); silencing mediator for retinoid or thyroid-hormone receptor (SMRT) was from R. Evans (Salk Institute, San Diego, CA) and Sin3a from C. Laherty (Seattle, WA); HA–CREB-binding protein (CBP), p300, and Flag-p300/CBP-associated factor (PCAF) were from G. Nabel (University of Michigan, Ann Arbor, MI). Rabbit polyclonal Abs against actin (c-11), p300 (n-15), CBP (A-22), and Sp1 were purchased from Santa Cruz Biotechnology (Santa Cruz, CA); SMRT (PA1-842) and mSin3a (PA1-870) from Affinity Bioreagents (Golden, CO); and nuclear receptor corepressor (06-892), PCAF (07-141), HDAC1 (06-720), and acetyl-histone H3 from Upstate Biotechnology (Lake Placid, NY). IRF3 mAbs were from BD Pharmingen (San Diego, CA) and IRF5 goat polyclonal Abs from Abcam (Cambridge, MA). IRF5 phospho-specific (S427/ S430) Abs, recognizing both Ser427 and Ser430, were generated by Affinity Bioreagents. M2-Flag mAbs were from Sigma-Aldrich (St. Louis, MO); Living Colors gfp polyclonal Abs from BD Clontech (Palo Alto, CA); and myc-tag and acetylated-lysine polyclonal Abs were from Cell Signaling Technology (Beverly, MA).
Reporter assays
A total of 2 × 105 2fTGH, 2fTGH/IRF5 V3/V4, HEK TLR9, or irf5−/− macrophages were transfected with a constant amount of DNA (2 μg/six-well plate) by using the Superfect (Qiagen, Valencia, CA) or Lipofect-amine (Invitrogen, Carlsbad, CA) transfection reagent. For the SAP assay, equal amounts of the IFNA1 SAP reporter plasmid and the indicated expression plasmids were cotransfected with the β-galactosidase expression plasmid (50 ng), except where HDAC1 was transfected at increasing amounts (0.2 μg, 0.4 μg, 0.6 μg, and 1 μg). For the dual luciferase assay, equal amounts of an ISRE-containing, IL6, or TNFA promoter reporter was transfected as previously described, with the thymidine Renilla-luciferase reporter gene for normalization (22). Transfected cells were split 16 h later, incubated for an additional 6 h, and either left uninfected, infected with NDV, treated with R848 (1 μg/ml) or CpG-B (3 μg/ml) for 16 h, and/or treated with TSA for 6 h. The SAP was determined as described using β-galactosidase expression levels to normalize transfection efficiency (19, 26); luciferase activity was measured using the Dual Luciferase Assay System (Promega, Madison, WI). Each experiment was performed in triplicate and repeated three times. Data in figures are expressed as the mean ± SD. Statistical comparisons were carried out using the unpaired two-tailed Student t test.
RNA analysis
A total of 1 μg total RNA was prepared using the Qiagen RNeasy kit (Qiagen), digested with DNase I, and subjected to reverse transcriptase as previously described (19, 20). One-tenth of the resulting cDNA product was used as a template for 25 cycles of PCR using specific primers as indicated. Primers for IFNA and β-actin were previously described (27). Additional primer sequences used were as follows: IL6 forward 5′-GCC-TTCGGTCCAGTTGCCTT-3′ and reverse 5′-GCAGAATGAGATGAGT-TGTC-3′; and TNFA forward 5′-AGGCGCTCCCCAAGAAGACA-3′ and reverse 5′-TCCTTGGCAAAACTGCACCT-3′.
IFN cytopathic effect assay
2fTGH cell lines (0.5 × 105 cells/well of a 12-well plate) were transiently transfected with 100 ng indicated expression plasmids. Sixteen hours later, cells were split in duplicate to a 24-well plate, incubated for 8 h, and either left uninfected or infected with NDV for an additional 16 h. The levels of biologically active type I IFN in the cell culture supernatants were determined by the viral cytopathic effect assay (28). Vesicular stomatitis virus was used as the challenging virus, and the cytopathic effect was determined in HF.
Immunoprecipitations, immunoblot, and GST pulldown assays
Cell extracts were prepared as described (29) from control untreated or NDV-infected (6 or 8 h) 2fTGH cells and THP-1 cells stimulated with TLR ligands (1 μg/ml R848 or 3 μg/ml CpG-B) for 1 h. Extracts (300 μg protein) were incubated with the indicated Abs for 16 h at 4°C. Protein A-or G-Sepharose was then added and incubated for an additional 1 h, beads were washed, and bound proteins identified by immunoblotting (29). Signals were visualized using the ECL detection reagents (Amersham Pharmacia Biotech, Piscataway, NJ). For direct immunoblot assays, cell extracts were prepared as described above and expression assayed using 20 μg whole cell lysate. The GST pulldown assay was performed as previously described using 250 μg whole cell lysates (30). Cytoplasmic and nuclear extracts were prepared as previously described (19, 20).
Chromatin immunoprecipitation assay
The chromatin immunoprecipitation (ChIP) assay was performed as previously described (19, 30) using the ChIP assay kit from Upstate Bio-technology or ChapionChIP One-Day Kit (SA Biosciences) following the manufacturer’s instructions. For 2fTGH or THP-1 cell lines, cells were left uninfected or infected with NDV over a time course or for 3 h. Human primary monocytes (1.5 × 106/ChIP assay) were infected with NDV for 6 h. The samples were analyzed by standard semiquantitative PCR with primer pairs specific for the indicated promoter regions. For binding to endogenous IFNA promoters, the DNA template was amplified with universal primers corresponding to the regions of human endogenous IFNA genes that are conserved in all subtypes (27). For binding to the endogenous IL6 promoter, we used forward 5′-GCCTGTTAATCTGGTCACTG-3′ and reverse 5′-GCTTAGGTCGTCATTGAGGC-3′; for binding to the endogenous TNFA promoter, we used forward 5′-CAGGACCTCCAGG-TATGGAA-3′ and reverse 5′-CCGGGAATTCACAGACCACTG-3′.
Results
TSA impairs IRF5-mediated proinflammatory cytokine induction
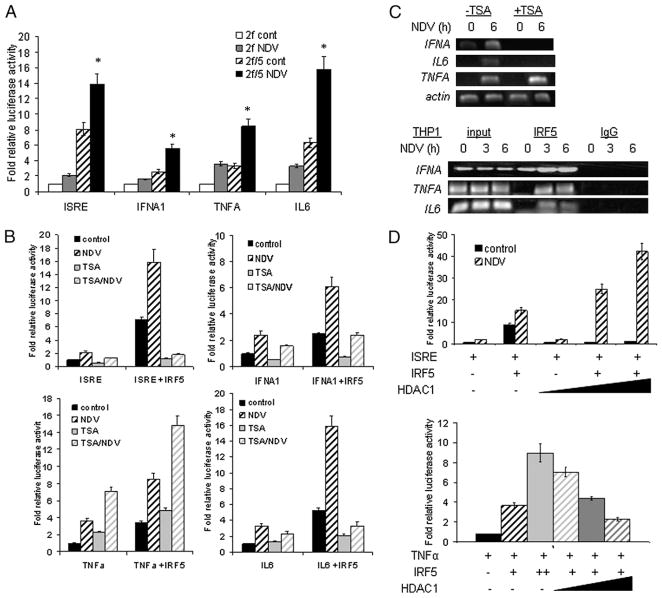
IRF family members are thought to regulate transcription by binding to IFN-stimulated response elements (ISREs) and/or IRF elements (IRF-Es) found in the promoters of target genes. We have previously demonstrated the ability of human IRF5 to bind to and transactivate IFNA/B promoters (19). To confirm the regulation of proinflammatory cytokines IL-6 and TNF-α by IRF5, we compared their transactivation potential in 2fTGH cells generated to stably overexpress IRF5 V3/V4 (19) to that of an ISRE-containing promoter reporter and the IFNA1 promoter reporter. All four luciferase promoter reporters were transactivated to differing amounts by IRF5 in uninfected cells, and promoter activity was further enhanced postinfection with NDV (Fig. 1A). Because it has previously been shown that the pharmacological HDACi TSA blocks IFN-induced ISRE transactivation (12), we used this as a control to examine the effect of TSA on other promoters. 2fTGH cell lines were transfected with the indicated promoter reporters and left untreated, treated with TSA, or infected with NDV in the presence or absence of TSA. IRF5-mediated trans-activation of the ISRE, IFNA1, and IL6 promoter reporters in the presence or absence of virus was inhibited by TSA (Fig. 1B). Surprisingly, IRF5-mediated TNFA promoter activity was enhanced by TSA. Similar results were obtained with a second structurally distinct HDACi, NaB (data not shown), ruling out any nonspecific effects of TSA. To confirm the effects of TSA on virus-induced IRF5-mediated proinflammatory cytokine expression, we analyzed endogenous cytokine transcript levels in the same cells. 2fTGH or 2fTGH/IRF5 expressing cells were left uninfected or infected with NDV in the presence or absence of TSA, and total RNA was prepared at 0 and 6 h posttreatment. After reverse transcription, PCR was performed with primers specific for each gene. In all cases, IRF5 was required for sufficient induction (Fig. 1C, top panel) because transcription of IFNA, IL6, and TNFA was not observed in parental 2fTGH cells lacking endogenous IRF5 (19 and data not shown). Coincubation with TSA inhibited the IRF5-mediated induction of IFNA and IL6 but not TNFA. Importantly, there was no inhibition of a control mRNA, β-actin. These data correlated with findings from our reporter assays. Furthermore, we demonstrate in a more biologically relevant cell line, THP-1, which expresses high levels of endogenous IRF5 (25), that IRF5 recognizes and binds to ISRE/IRF-E sites in the IFNA, IL6, and TNFA promoters (Fig. 1C, bottom panel).
FIGURE 1.
TSA selectively impairs IRF5-mediated proinflammatory cytokine expression. A, 2fTGH or 2fTGH/IRF5 V3/V4 stable expressing cells were transiently transfected with ISRE, IFNA1, IL6, or TNFA luciferase promoter reporters and left uninfected or infected with NDV. Fold relative activity is shown compared with untreated controls after normalization to protein and thymidine Renilla-luciferase activity. Results are representative of at least three independent experiments run in triplicate. B, Same as in A except cells were also treated with TSA. C, Levels of endogenous IFNA, IL6, and TNFA transcripts were detected by semiquantitative PCR in 2fTGH/IRF5-expressing cells left uninfected (0 h) or infected with NDV (6 h) in the presence or absence of TSA. Levels of β-actin are shown as a loading control and for RNA integrity. Results from the endogenous ChIP assay in THP-1 cells left uninfected (0 h) or infected with NDV over the indicated time course are shown in the bottom panels. Cell lysates were immunoprecipitated with anti-IRF5 Abs and bound DNA amplified with primers recognizing PRD/ISRE/IRF-E regions in each promoter. Results are representative of three independent experiments. D, Same as in A except increasing levels of HDAC1 were cotransfected with the indicated promoters to 2fTGH cells. *p < 0.05 compared with IRF5 expressing (2f/5) uninfected (cont) cells.
Given that TSA inhibited transactivation of the ISRE, IFNA, and IL6 promoter reporters by IRF5, yet enhanced transactivation of the TNFA promoter, combined with the fact that HDAC1 was recently identified to be essential for STAT-dependent gene expression on IFN-stimulated gene promoters (10, 12), we examined the involvement of HDAC1 in IRF5-mediated proinflammatory cytokine expression. Similar to experiments in Fig. 1A, 2fTGH cells were cotransfected with IRF5, individual promoter reporters, and increasing amounts of HDAC1. A positive regulatory role for HDAC1 was observed in IRF5-expressing NDV-infected cells with each ISRE, IFNA, and IL6 promoter reporter; data from the ISRE reporter is shown (Fig. 1D, top panel) as representative of activity from each of the three reporters (data not shown). Importantly, HDAC1 acted as a repressor to IRF5 in uninfected cells and had no effect on the promoters in the absence of IRF5. Conversely, a negative regulatory role for HDAC1 was observed on the TNFA promoter (Fig. 1D, bottom panel). To examine the specificity of HDAC1 to IRF5-mediated reporter transactivation, the class II HDAC5 was also transfected in combination with IRF5, yet no significant effect was observed (data not shown). Together, these results strongly support the conclusion that HDAC activity is required for the regulation of some IRF5-mediated proinflammatory cytokines, implicating HDAC1 as a critical positive coactivator of IFNA and IL6 gene expression.
IRF5 is part of a corepressor complex silencing IFNA gene expression in uninfected cells
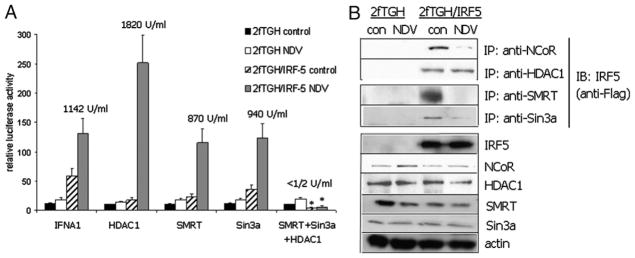
Transcriptional repression is mediated in part by non-DNA–binding corepressors. The corepressor proteins SMRT/nuclear receptor corepressor (NCoR) and Sin3a have been shown to be recruited to many classes of transcription factors and are components of multiple protein complexes containing HDACs (31, 32). SMRT/NCoR and Sin3a have been shown to interact with HDAC1, leading to the repression of gene transcription (33). Because previous data (19, 20) and data shown in Fig. 1C indicated a role for IRF5 in both the activation and repression of IFNA gene expression, likely involving HDAC1, we examined whether cotransfection of corepressor proteins with HDAC1 would have any effect on IRF5-mediated IFNA1 promoter activity or the synthesis of biologically active type I IFNs in the supernatants of 2fTGH cell lines. For these studies, 2fTGH- and 2fTGH/IRF5-expressing cells were transiently cotransfected with HDAC1, SMRT, Sin3a, or the combination of all three with the IFNA1 promoter reporter. Cells were left uninfected or infected with NDV for 16 h; media supernatants were used for the IFN cytopathic effect assay and cell lysates for luciferase activity. Exogenous corepressors had little effect on IFNA1 promoter activity in the absence of IRF5; however, in the presence of IRF5, all three repressors suppressed IRF5-mediated transactivation in the absence of virus infection (Fig. 2A). In virus-infected cells, SMRT and Sin3a alone had little effect on IRF5-mediated IFNA1 reporter activity whereas HDAC1 enhanced transactivation. When SMRT, Sin3a, and HDAC1 were cotransfected together with the IFNA1 reporter, activity was completely abolished in IRF5-expressing cells only, and this was independent of virus infection. Similar data were obtained when examining levels of endogenous, biologically active type I IFNs in the supernatants of 2fTGH/IRF5-expressing cells (Fig. 2A).
FIGURE 2.
IRF5 interacts with corepressor proteins to repress IFNA/IFN-α expression. A, 2fTGH and 2fTGH/IRF5 V3/V4-expressing cells were cotransfected with an IFNA1 luciferase reporter and equal amounts of individual corepressor proteins (HDAC1, SMRT, and Sin3a) or the combination of HDAC1, SMRT, and Sin3a. Cells were left uninfected or infected with virus for 16 h and relative luciferase activity determined after normalization to protein and thymidine Renilla-luciferase activity. Supernatants were assayed for endogenous biologically active type I IFNs by the cytopathic effect assay in HF cells. Levels of synthesized type I IFNs are shown as units/ml. B, Coimmunoprecipitations were performed in 2fTGH and 2fTGH/IRF5 expressing cells left uninfected or infected with NDV for 6 h. Endogenous proteins were immunoprecipitated as indicated and bound IRF5 detected with anti-Flag Abs. Levels of proteins expressed are shown in the bottom panels. Results are representative of three independent experiments. *p < 0.05 compared with 2fTGH/IRF5 cells lacking corepressors.
We next examined by coimmunoprecipitation whether IRF5 V3/ V4 could interact with any of these endogenous corepressor proteins. 2fTGH cell lines were left uninfected or infected with NDV for 6 h, endogenous proteins were immunoprecipitated with the indicated Abs, and interaction with ectopic IRF5 was determined by immunoblot with M2 Flag Abs. Surprisingly, interaction with NCoR, SMRT, and Sin3a occurred primarily in uninfected cell lysates (Fig. 2B); interaction with HDAC1 was observed in both uninfected and virus-infected cells. Similar results were obtained in THP-1 monocytic leukemia cells (data not shown) that express high levels of endogenous IRF5 (19, 25).
Virus triggers interaction of IRF5 with HATs
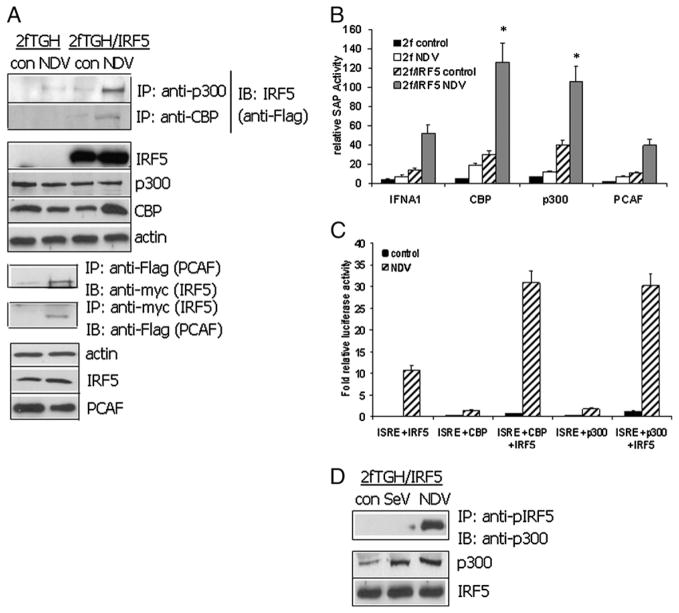
It is known that phosphorylated IRF3 forms a strong association with HATs CBP and p300 that facilitates its retention in the nucleus and transcriptional activation of type I IFNs along with other target genes (33, 34–38). To determine whether IRF5 V3/V4 could interact with HATs and whether this interaction would contribute to IRF5-mediated transcriptional activities in response to virus infection, we performed coimmunoprecipitation and promoter reporter assays (Fig. 3). Endogenous CBP and p300 were immunoprecipiated from uninfected and virus-infected 2fTGH cells transfected with empty vector control or Flag-tagged IRF5 and associated proteins identified by immunoblot with anti-Flag Ab (Fig. 3A, top panels). To examine interaction of IRF5 with another HAT, PCAF, shown to interact with IRF7 (39), we transiently transfected Flag-PCAF and myc-IRF5 to 2fTGH cells and performed reciprocal immunoprecipitations before and after virus infection (Fig. 3A, bottom panels). The results clearly indicate that all three HATs are capable of interacting with IRF5 only after virus infection.
FIGURE 3.
Virus triggers interaction of IRF5 with HAT proteins. A, Coimmunoprecipitations were performed in 2fTGH- or 2fTGH/IRF5-expressing cells left uninfected or infected with NDV for 6 h. In the top panels, endogenous proteins were immunoprecipitated as indicated and bound IRF5 detected with anti-Flag Abs. In the bottom panels, Flag-tagged PCAF and myc-tagged IRF5 were transiently transfected to 2fTGH cells and infected as described above. Reciprocal immunoprecipitations were performed as indicated. Levels of proteins expressed are shown. Results are representative of three independent experiments. B, 2fTGH and 2fTGH/IRF5 V3/V4-expressing cells were cotransfected with an IFNA1 SAP promoter reporter and equal amounts of the indicated HATs. Cells were left uninfected or infected with NDV for 16 h. Relative SAP activity was determined after normalization to protein and β-galactosidase. Experiments were performed in triplicate and repeated three times. C, Same as in B except transactivation of the ISRE luciferase reporter was determined. D, 2fTGH/IRF5-expressing cells were left uninfected or infected with the indicated virus for 6 h and immunoprecipitated with Abs recognizing phosphorylated Ser427 and Ser430 of V3/V4. Interaction of IRF5 with p300 was determined with anti-p300 Abs. Results are representative of three independent experiments. *p < 0.05 compared with 2f/IRF5 NDV-infected cells lacking HATs.
To determine whether these interactions conferred transcriptional activity to the complex, we examined IFNA1 and ISRE promoter reporter activity after transient cotransfection. Similar to data in Fig. 1A, transactivation of the IFNA1 promoter reporter by IRF5 V3/V4 occurred in both uninfected and virus-infected cells; cotransfection of V3/V4 with CBP or p300 gave significant increases in IFNA1 reporter activity (Fig. 3B). Somewhat surprising, coexpression with PCAF gave no significant change in reporter activity. This was not due to differences in expression as CBP, p300, and PCAF protein levels appeared equivalent by immunoblot analysis (data not shown). Similar effects were observed using an ISRE-containing promoter reporter (Fig. 3C), indicating that IRF5 V3/V4 and CBP/p300 form a functional coactivator complex on the IFNA1 and ISRE promoters.
Given that interaction of IRF5 with p300, CBP, or PCAF occurred in virus-infected cells only (Fig. 3A) suggests that similar to IRF3, phosphorylation of IRF5 may be a prerequisite for association. To examine this further, Abs recognizing phosphorylated serine residues Ser427 and Ser430 of the IRF5 V3/V4 polypeptide were used for immunoprecipitation, and p300 association was detected by immunoblot. These two residues were previously shown to be critical sites for NDV-induced IRF5 activation (19, 20); Sendai virus was used as a negative control as it does not induce IRF5 phosphorylation/activation. IRF5 phospho-specific Abs immunoprecipitated endogenous p300 from NDV- but not SeV-infected or uninfected cells (Fig. 3D). These data are in agreement with previous findings of IRF5 V3/V4 virus-specific activation (19, 20) and indicate that only phosphorylated IRF5 interacts with p300.
HDAC corepressor proteins and HATs interact with distinct regions of IRF5
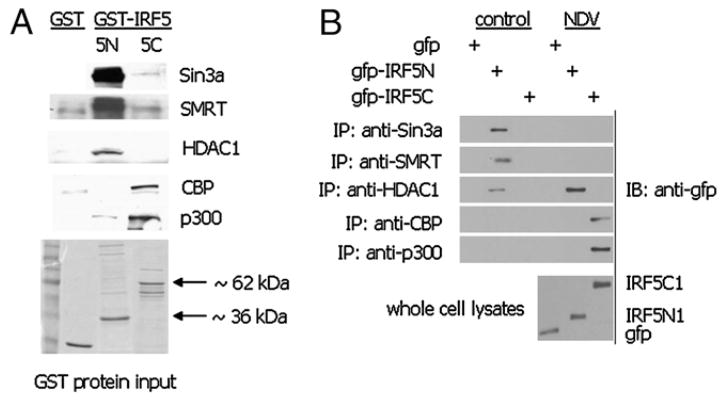
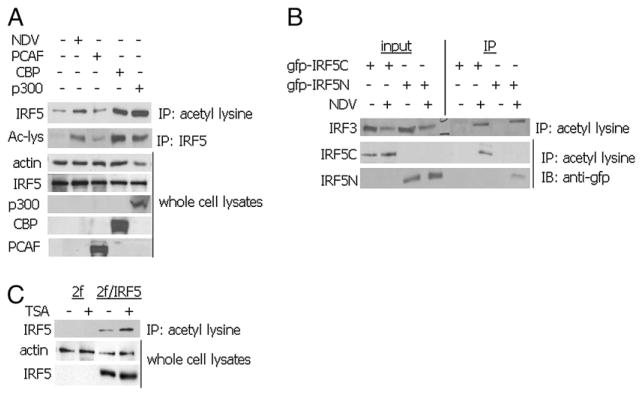
All IRF proteins share similar domain structures (i.e., a homologous DNA-binding in the N terminus and a nonhomologous IRF association domain [IAD] somewhere in the C terminus) (35, 40). Crystal structure data have revealed that CBP interacts with the IRF3 IAD (41). To determine the domain structure on IRF5 that HDAC corepressor proteins or HATs bind to, we performed the GST pulldown assay using N-terminal or C-terminal fragments of IRF5 bound to sepharose-linked GST. Overexpressed proteins pulled down by GST alone, GST-5N (amino), or GST-5C (carboxyl) were detected by immunoblot with the indicated Abs (Fig. 4A). Data clearly show a distinct binding pattern where co-repressor proteins associated primarily with the DNA binding domain of IRF5, whereas HATs CBP and p300 associated with the IAD. To confirm this exogenous data, we analyzed interaction of the endogenous proteins with gfp-tagged IRF5N or 5C after transient transfection to 2fTGH cells; cells expressing empty vector gfp were used as a negative control (Fig. 4B). 2fTGH cells were left uninfected or infected with NDV for 6 h, and endogenous proteins were immunoprecipitated from whole cell lysates. Associated proteins were identified by immunoblot with anti-GFP Ab. Similar results as those shown for the GST pulldown were obtained.
FIGURE 4.
HDAC corepressor proteins and HATs interact with distinct regions of IRF5. A, Interaction of ectopically expressed HDAC corepressor proteins or HATs with IRF5 was determined by the GST pulldown assay. 2fTGH cells were transiently transfected with the indicated expression plasmids and whole cell lysates applied to GST or GST-IRF5 Sepharose beads containing the N terminus (5 N) or the C terminus of IRF5 V3 (5C). Bound proteins were resolved by SDS-PAGE and detected with Abs recognizing Sin3a, SMRT, HDAC1, CBP, or p300. Levels of each GST fusion protein are shown after staining with Coomassie blue. Results are representative of three independent experiments. B, 2fTGH cells were transiently transfected with gfp vector control plasmid, gfp-IRF5N, or gfp-IRF5C and left uninfected or infected with NDV for 6 h. Immunoprecipitations were performed with Abs recognizing the indicated endogenous proteins and bound gfp-IRF5 was detected with anti-gfp Abs. Levels of ectopically expressed gfp fusion proteins are shown at the bottom. Results are representative of three independent experiments.
HATs p300 and CBP acetylate IRF5 in vivo
Family members IRF1 and IRF2 were shown to be acetylated by p300 and PCAF (42), whereas IRF7 was acetylated by PCAF and GCN5 (39). We tested whether IRF5 V3/V4 could be similarly acetylated by exogenous HATs in 2fTGH cells. Cell lysates were immunoprecipitated with anti-acetylated lysine Abs or those specific for IRF5, and the level of IRF5 acetylation determined by immunoblot analysis with the reciprocal Ab. We examined whether IRF5 could be acetylated in response to NDV infection as compared with overexpression with individual HATs. In both immunoprecipitation experiments, low levels of constitutive basal IRF5 acetylation were observed in uninfected cells, yet acetylation was greatly enhanced (>4-fold) by infection with NDV (Fig. 5A). Slight differences were observed in the ability of PCAF to acetylate Flag-tagged IRF5 between immunoprecipitation experiments, yet both methods gave high levels of IRF5 acetylation after cotransfection with CBP or p300. All three HAT proteins were expressed and were active in that they were able to acetylate endogenous substrates in vivo in transfected cells (data not shown).
FIGURE 5.
IRF5 is acetylated in vivo after NDV infection or coexpression with HATs. A, 2fTGH/IRF5-expressing cells were left uninfected, infected with NDV, or transiently transfected with PCAF, CBP, or p300 expression plasmids. Whole cell lysates were immunoprecipitated with either anti-acetyl lysine Abs or IRF5 Abs and acetylated IRF5 detected with the reciprocal Ab. Levels of transfected proteins are shown at the bottom. Results are representative of three independent experiments. B, NDV-induced acetylation of IRF5 occurs in both the amino- and C terminus. gfp-IRF5 fusion proteins were transiently transfected to 2fTGH cells and left uninfected or infected with NDV. Similar to A, whole cell lysates were immunoprecipitated with anti-acetyl lysine Abs and acetylated IRF5 detected with anti-gfp Abs. The same membrane was stripped and reprobed with anti-IRF3 Abs. Results are representative of three independent experiments. C, Treatment with TSA increases basal IRF5 acetylation. 2fTGH and 2fTGH/IRF5 expressing cells were left untreated or treated with TSA, immunoprecipitated with antiacetyl lysine Abs, and acetylated IRF5 detected with anti-IRF5. Results are representative of three independent experiments.
Similar to experiments performed in Fig. 4B, we used the identical GFP–IRF5 fusion constructs to gain preliminary insight into where on the IRF5 V3/V4 polypeptide HAT-associated acetylation occurs. For these studies, 2fTGH cells were transiently transfected with the indicated GFP-tagged fusion constructs and left uninfected or infected with NDV for 6 h. As shown in Fig. 5B, IRF5 acetylation occurred primarily in the C terminus of IRF5, yet low levels of acetylation were also detected in the N-terminal DNA binding domain after virus infection. As a positive control, blots were stripped and reprobed with anti-IRF3 Ab, revealing virus-induced acetylation of endogenous IRF3. These data are in agreement with previously published reports (32, 34, 43). We next examined the effect of TSA on basally acetylated IRF5. As expected, TSA increased the levels of IRF5 V3/V4 acetylation (Fig. 5C). Furthermore, overexpression of HDAC1 in NDV-infected 2fTGH/IRF5-expressing cells led to a significant decrease in acetylated IRF5 levels (data not shown), indicating that IRF5 is subjected to reversible acetylation.
Characterization of IRF5 binding to endogenous IFNA promoters
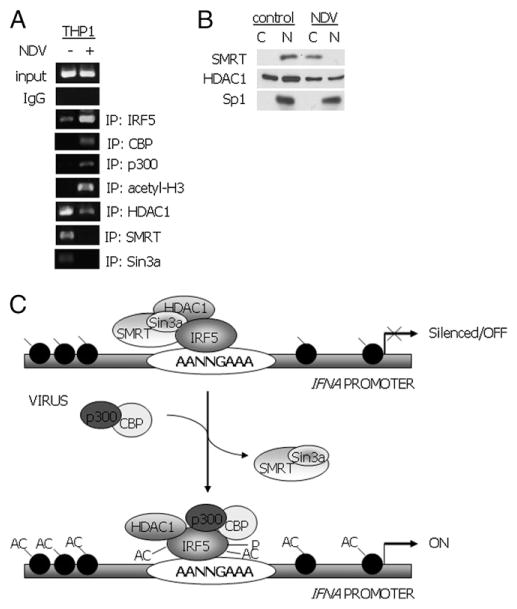
We have previously shown that ectopic IRF5 V3/V4 binds to exogenous and endogenous IFNA promoters in both uninfected and virus-infected cells (19, 20). In this study, we confirmed these findings by ChIP assay after immunoprecipitation of endogenous IRF5 from THP-1 cells, shown in Fig. 1C. THP-1 cells express high levels of IRF5 consisting primarily of the isoforms encoded by V1, V3, and V4 (25 and data not shown). We repeated this in vivo ChIP assay multiple times to analyze specific cofactors that might be involved in IRF5-mediated transcriptional repression and/or activation of the endogenous IFNA promoters. We analyzed recruitment of coactivator proteins CBP and p300, along with corepressor proteins HDAC1, SMRT/N-CoR, and Sin3a in both uninfected and virus-infected cells; at the same time, acetylation of histone H3 was determined as a marker of IFNA promoter activation (44, 45).
THP-1 cells were left uninfected or infected with NDV for 3 h, cells were cross-linked, and lysates isolated for the indicated immunoprecipitations (Fig. 6A). As shown previously, the ability of Abs specific for IRF5 to recover IFNA promoter sequences was greatly enhanced by viral infection, demonstrating that IRF5, although bound to these promoters in uninfected cells, is further recruited in response to virus infection. Similarly, p300 and CBP were recruited to the IFNA promoters in response to virus infection with a concomitant increase in acetylated histone H3. Low levels of endogenous HDAC1 were also detected as bound to the promoters in virus-infected cells, supporting data from our promoter reporter assays (Fig. 1) and coimmunoprecipitations (Fig. 2); however, significantly more HDAC1 was detected as bound to the promoters in uninfected cells. Along with HDAC1, we detected corepressor proteins SMRT/N-CoR, Sin3a, and IRF5 at the IFNA promoters. Similar data were obtained in 2fTGH cells generated to stably overexpress IRF5 V3/V4 (data not shown).
FIGURE 6.
Alterations in IRF5-mediated IFNA promoter regulation are dependent on virus. A, In vivo binding of IRF5 in THP-1 cells was determined by the ChIP assay. THP-1 cells were left uninfected or infected with NDV for 3 h. DNA recovered from ChIP after immunoprecipitation with the indicated Abs was amplified using universal primers specific for all endogenous IFNA gene promoters. Input corresponds to the amplified IFNA promoter region from DNA-protein complexes before immunoprecipitation; IgG controls are shown for IRF5 Ab specificity. Results are representative of three independent experiments. B, Cellular localization of endogenous SMRT was determined by cytoplasmic (C)/nuclear (N) extraction from THP-1 cells left uninfected or infected with NDV for 3 h. Localization of SMRT, HDAC1, and Sp1 were determined by immunoblot with Abs specific for each. C, Schematic representation of how IRF5 switches from a silencing to an activating complex in cells expressing high levels of IRF5. In uninfected cells, the IFNA promoter is silenced by chromatin remodeling through the binding of a corepressor complex comprising of HDAC1, Sin3a, and SMRT to the N terminus of IRF5 V3; histones are deacetylated. Virus infection triggers a change not only in the IRF5 polypeptide leading to enhanced nuclear localization, but it also signals the export of SMRT out of the nucleus to the cytoplasm, resulting in the association of IRF5 with coactivator proteins p300/CBP and acetylation of histones resulting in gene transcription. AC, acetylated histones; P, IRF5 phosphorylation.
To further examine mechanisms that may contribute to the observed exchange of corepressor proteins for coactivator proteins at the IFNA promoters in uninfected and virus-infected cells, we analyzed the cellular localization of SMRT. Numerous signaling pathways regulate transcription by converging on SMRT/NCoR corepressor complexes to eliminate HDAC activities through a process called derepression (46). Derepression is associated with phosphorylation and nuclear export of corepressor complexes. This has been shown to apply to SMRT after the overexpression of several serine/threonine kinases (47). To analyze SMRT localization under the context of virus infection, cytoplasmic and nuclear extracts were isolated from uninfected and virus-infected THP-1 cells after 4 h incubation and 20 μg protein was resolved by SDS-PAGE. Data in Fig. 6B demonstrate that SMRT is localized in the nucleus of uninfected cells; this finding is in agreement with previous reports (47, 48). However, in response to NDV infection, SMRT was exported from the nucleus to the cytoplasm. Localization of HDAC1 was also examined and was detected in both the cytoplasm and nucleus of uninfected and virus-infected cells, whereas Sp1, a control for the integrity of our purified extracts, resided in the nucleus of both uninfected and virus-infected cells. These data suggest that virus triggers not only IRF5 phosphorylation and translocation to the nucleus, but also the export of SMRT from the nucleus to the cytoplasm, both of which would be expected to contribute to the conversion of an IRF5-mediated silencing complex to that of an activating complex on promoters of target genes.
Discussion
IRF5 is a transcription factor that plays a crucial role in mediating both virus- and TLR-induced type I IFN and proinflammatory cytokine expression (19–24). Numerous genetic association studies have implicated IRF5 in the pathogenesis of a variety of autoimmune diseases, such as systemic lupus erythematosus (SLE), due to IRF5 polymorphisms that were found to be associated with disease susceptibility (reviewed in Refs. 49, 50). We recently demonstrated that IRF5 expression is significantly elevated in SLE patients compared with healthy donors and that upregulation of both IRF5 transcript and protein levels was associated with an SLE risk haplotype (51). Current data indicate that single nucleotide polymorphisms in the IRF5 gene may define isoform expression; for instance, the risk alleles of single nucleotide polymorphism rs10488631 were highly associated with elevated expression of IRF5 from noncoding exon 1c corresponding to IRF5 V3 (51). Support has been building around the hypothesis that polymorphisms in the IRF5 gene alter expression, regulation, and function of specific IRF5 isoforms, ultimately contributing to the dysregulation of proinflammatory cytokines, such as IFN-α, IL-6, and TNF-α, detected in the serum of SLE patients (51–53).
The ongoing type I IFN production in SLE is thought to be induced by nucleic acid containing immune complexes internalized by plasmacytoid dendritic cells via the FcγRIIa, followed by engagement of endosomal TLR7 or 9 (54, 55). A number of recent studies have provided significant evidence supporting a role for TLR7 and 9 in SLE pathology (reviewed in Refs. 56, 57). Given that TLR7/8 sense single-stranded RNA from viruses, such as NDV, and TLR9 senses unmethylated CpG motifs in viral and bacterial DNA, data presented in this study suggest similar mechanisms regulating IRF5 transcriptional activity in response to virus or TLR signaling. Indeed, IRF5 interacts with CBP after stimulation of THP-1 cells with R848 or CpG-B (Supplemental Fig. 1A), and TSA inhibits CpG-induced IRF5-mediated IL6 and ISRE promoter transactivation (Supplemental Fig. 1B–D).
Because HDAC inhibition has recently been shown to reduce inflammation in models of autoimmune and inflammatory diseases (58–63), including murine models of SLE, we sought to examine the molecular mechanism(s) by which IRF5 regulates proinflammatory cytokines associated with SLE. In most of these models, inhibition of HDACs was associated with a decrease in cytokines and disease severity. The treatment of lupus-prone mice with TSA reduced proteinuria, the infiltration of destructive inflammatory cells into the glomerulus, and spleen weight (59). The clinical benefit of TSA in these mice was associated with decreases in IL-12, IFN-γ, and IL-6. Because specific blockade of cytokines such as TNF-α, IL-1β, IL-6, or IL-12 is effective in diseases such as rheumatoid arthritis (61), SLE (59), Crohn’s disease (64), and psoriasis (65), novel therapeutic strategies to target these cytokines are an expanding field of research and clinical application. Reducing the production and activity of more than one cytokine may be an advantage because disease-associated cytokines often act synergistically. The present work expands the finding that HDACi reduce cytokine production, particularly cytokines relevant to autoimmune/inflammatory diseases, and lends significant insight into the mechanism(s) by which IRF5 regulates some of these cytokines.
Our study focused on the identical isoform encoded by IRF5 V3/V4 cDNAs because it was previously shown to be the most potent inducer of virus-mediated type I IFN expression (25) and was significantly upregulated in SLE patients with the risk haplotype (51). We began with the use of transient promoter reporter assays to determine whether IRF5 was a direct mediator of IL-6 and TNF-α expression, both of which were recently shown to be dependent on IRF5 (23). Results clearly demonstrated that IRF5 could transactivate all examined promoter reporter constructs, to different degrees, in both uninfected and virus-infected cells; virus infection gave a significant increase in promoter activity that was mediated by IRF5. Binding of IRF5 on the endogenous promoters in THP-1 cells was observed (Fig. 1C, bottom panel), and binding of IRF5 with CBP on the endogenous promoters was confirmed in primary human monocytes (Supplemental Fig. 2). Transactivation of the ISRE, IFNA1, and IL6 promoter reporters by IRF5 in response to virus (Fig. 1C) or TLR signaling (Supplemental Fig. 1B–D) was significantly inhibited by TSA, suggesting that HDACs might be involved in IRF5-mediated transcriptional regulation of these genes. The exact role of HDACs in TNFA expression are a bit controversial in the literature because differing results have been obtained depending on the HDACi used, periods of incubation, and whether the study was performed in vitro or in vivo. For instance, similar to our findings (Fig. 1B), HDAC inhibition has been shown to upregulate TNFA expression in vitro, whereas the opposite effect was observed in vivo (15, 16). It has previously been reported that IL-6 is negatively regulated by HDACi (15). The antithetic effect of TSA on IL6 and TNFA promoters is not known, but may in part be due to differential binding of HDAC1 to these promoters rather than differential binding of IRF5 or HATs (Supplemental Fig. 3). TSA works by inhibiting HDAC activity and histone acetylation plays a critical role in the regulation of TNF-α expression (66). By overexpression of a class I HDAC, HDAC1, previously shown to augment the IFN-α response (12) in a similar manner to that observed in this study for the ISRE, IFNA1, IL6and promoter reporters, we provide clear support for a role of HDAC1 in IRF5-mediated gene regulation. Whether other class I HDACs have similar effects as HDAC1 is not currently known but under investigation; however, the class II HDAC, HDAC5, had no effect on IRF5-mediated transactivation of these promoters.
We have always been intrigued by the fact that we could detect binding of ectopic IRF5 and not IRF3 or IRF7 to the promoters of IFNA genes in uninfected cells (19, 33), suggesting that IRF5 may play a unique role in gene repression. It was not until we confirmed this finding by the in vivo ChIP assay in THP-1 cells (Figs. 1C, bottom panel, 6A) that we were confident that our earlier observations were not simply due to overexpression of IRF5. To examine potential repressor functions of IRF5, we co-expressed it with other corepressor proteins SMRT/N-CoR, Sin3a, and HDAC1 and found that in the presence of all three, IRF5-mediated transactivation of the IFNA1 promoter was completely abolished, along with the synthesis of biologically active type I IFNs. Important to point out, constitutive binding of IRF5 to the endogenous IFNA promoters does not occur in every cell type and is likely dependent on the levels of IRF5 expressed and possibly the isoform expressed; we have only ever observed this phenomenon in cells that express high levels of IRF5 V3/V4 (data not shown). Coimmunoprecipitation experiments confirmed that IRF5 interacts with corepressor proteins in uninfected cells. However, binding to HDAC1 was also observed after virus infection, supporting a role for HDAC1 and IRF5 in the positive regulation of target genes as well as negative regulation. Data from coimmunoprecipitation experiments of IRF5 with HATs (Fig. 3, Supplemental Fig. 1A) further elucidated this mechanism whereby IRF5 can mediate corepressor versus coactivator functions. Based on our earlier data demonstrating virus-induced IRF5 phosphorylation (19, 20) along with recent crystallographic data on dimeric IRF5 (67), we postulate that virus triggers structural modifications in the IRF5 protein (e.g., phosphorylation and acetylation) that releases it from corepressor proteins and enhances its ability to interact with coactivator proteins (Fig. 6C). Similar results were found for IRF3 (32, 34, 40). Indeed, IRF5 cooperated with CBP/p300 to further transactivate target promoter reporters (Fig. 3B, 3C) and could be detected as bound to endogenous IL6 and TNFA promoters with IRF5 in response to virus infection (Fig. 6A, Supplemental Fig. 2).
GST pulldown assays in combination with coimmunoprecipitation experiments revealed that corepressor proteins primarily interacted with the N-terminal DNA binding domain of IRF5, whereas coactivator proteins, CBP and p300, interacted with the C terminus (Fig. 4). This was not so surprising because CBP is known to interact with the C terminus of IRF3 (40). The fact that each group of proteins interacted with distinct regions of the IRF5 polypeptide lends some insight into how HDAC1 might be able to interact with IRF5 in both uninfected and virus-infected cells without inhibiting an interaction with the HATs after virus infection. We also identified that interaction of IRF5 with the HATs was not a static event in that IRF5 acetylation was observed in response to NDV or overexpression of specific HATs. This is a newly identified posttranslational event for IRF5, now indicating that virus induces both the phosphorylation and acetylation of IRF5. Recently, IRF5 was also shown to be ubiquitinated by TNFR-associated factor 6, enhancing its IFNA promoter inducing activity (68). Although acetylation has been shown to occur at a conserved lysine residue in the N-terminal DNA binding domain of IRF1, -2, and -7 (39, 42), acetylation of IRF5 appears to be more global because it was detected in both the N and C terminus (Fig. 5B). A detailed analysis of IRF5 acetylation has yet to be performed because IRF5 V3 contains 55 lysine residues.
Recent genetic and biochemical studies have revealed that HDACs and HATs do not act independently and that their activities in some cases may be linked to one another, along with their sharing common space in the nucleus and coprecipitating with each other (69, 70). The mechanism(s) regulating transcriptional repressor or activator activity for HDACs is not known but likely involves posttranslational modifications. HDACs can be post-translationally modified through phosphorylation (71, 72), ubiquitination (73), and sumoylation (74) and have been known to associate with proteins that modulate its deacetylase activity and recruitment to genomic regions. Data presented in this study support a complex role for both HDAC1 and IRF5 in mediating the transcriptional induction of proinflammatory cytokines implicated in autoimmune diseases such as SLE.
Supplementary Material
Acknowledgments
We thank G. Stark, G. Hayward, and K. Fitzgerald for cells and S.L. Schreiber, R. Evans, C. Laherty, and G. Nabel for expression plasmids.
This work was supported in part by grants from the National Institutes of Health/National Institute of Arthritis and Musculoskeletal and Skin Diseases (5R03AR054070) and the Arthritis Foundation (to B.J.B.).
Abbreviations used in this paper
- AC
acetylated histones
- C
cytoplasmic
- CBP
CREB-binding protein
- ChIP
chromatin immunoprecipitation
- HAT
histone acetyl-transferase
- HDAC
histone deacetylase
- HDACi
histone deacetylase inhibitor
- HF
human fibroblast
- IAD
IFN regulatory factor association domain
- IRF
IFN regulatory factor
- IRF-E
IFN regulatory factor element
- ISRE
IFN-stimulated response element
- N
nuclear
- NCoR
nuclear receptor corepressor
- NDV
Newcastle disease virus
- P
IRF5 phosphorylation
- PCAF
p300/CREB-binding protein-associated factor
- SAP
secreted alkaline phosphatase
- SLE
systemic lupus erythematosus
- SMRT
silencing mediator for retinoid or thyroid-hormone receptor
- TSA
trichostatin A
- V
variant
Footnotes
The online version of this article contains supplemental material.
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 2.Cress WD, Seto E. Histone deacetylases, transcriptional control, and cancer. J Cell Physiol. 2000;184:1–16. doi: 10.1002/(SICI)1097-4652(200007)184:1<1::AID-JCP1>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 3.Nusinzon I, Horvath CM. Histone deaceylases as transcriptional activators? Role reversal in inducible gene regulation. Sci STKE. 2005;2005:re11. doi: 10.1126/stke.2962005re11. [DOI] [PubMed] [Google Scholar]
- 4.Mariadason JM, Corner GA, Augenlicht LH. Genetic reprogramming in pathways of colonic cell maturation induced by short chain fatty acids: comparison with trichostatin A, sulindac, and curcumin and implications for chemoprevention of colon cancer. Cancer Res. 2000;60:4561–4572. [PubMed] [Google Scholar]
- 5.Chambers AE, Banerjee S, Chaplin T, Dunne J, Debernardi S, Joel SP, Young BD. Histone acetylation-mediated regulation of genes in leukaemic cells. Eur J Cancer. 2003;39:1165–1175. doi: 10.1016/s0959-8049(03)00072-8. [DOI] [PubMed] [Google Scholar]
- 6.Bannister AJ, Oehler T, Wilhelm D, Angel P, Kouzarides T. Stimulation of c-Jun activity by CBP: c-Jun residues Ser63/73 are required for CBP induced stimulation in vivo and CBP binding in vitro. Oncogene. 1995;11:2509–2514. [PubMed] [Google Scholar]
- 7.Yuan W, Condorelli G, Caruso M, Felsani A, Giordano A. Human p300 protein is a coactivator for the transcription factor MyoD. J Biol Chem. 1996;271:9009–9013. doi: 10.1074/jbc.271.15.9009. [DOI] [PubMed] [Google Scholar]
- 8.Kamei Y, Xu L, Heinzel T, Torchia J, Kurokawa R, Gloss B, Lin SC, Heyman RA, Rose DW, Glass CK, Rosenfeld MG. A CBP integrator complex mediates transcriptional activation and AP-1 inhibition by nuclear receptors. Cell. 1996;85:403–414. doi: 10.1016/s0092-8674(00)81118-6. [DOI] [PubMed] [Google Scholar]
- 9.Zhang JJ, Vinkemeier U, Gu W, Chakravarti D, Horvath CM, Darnell JE., Jr Two contact regions between Stat1 and CBP/p300 in interferon γ signaling. Proc Natl Acad Sci USA. 1996;93:15092–15096. doi: 10.1073/pnas.93.26.15092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klampfer L, Huang J, Swaby LA, Augenlicht L. Requirement of histone deacetylase activity for signaling by STAT1. J Biol Chem. 2004;279:30358–30368. doi: 10.1074/jbc.M401359200. [DOI] [PubMed] [Google Scholar]
- 11.Agalioti T, Chen G, Thanos D. Deciphering the transcriptional histone acetylation code for a human gene. Cell. 2002;111:381–392. doi: 10.1016/s0092-8674(02)01077-2. [DOI] [PubMed] [Google Scholar]
- 12.Nusinzon I, Horvath CM. Interferon-stimulated transcription and innate antiviral immunity require deacetylase activity and histone deacetylase 1. Proc Natl Acad Sci USA. 2003;100:14742–14747. doi: 10.1073/pnas.2433987100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sakamoto S, Potla R, Larner AC. Histone deacetylase activity is required to recruit RNA polymerase II to the promoters of selected interferon-stimulated early response genes. J Biol Chem. 2004;279:40362–40367. doi: 10.1074/jbc.M406400200. [DOI] [PubMed] [Google Scholar]
- 14.Chang HM, Paulson M, Holko M, Rice CM, Williams BR, Marié I, Levy DE. Induction of interferon-stimulated gene expression and antiviral responses require protein deacetylase activity. Proc Natl Acad Sci USA. 2004;101:9578–9583. doi: 10.1073/pnas.0400567101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leoni F, Fossati G, Lewis EC, Lee JK, Porro G, Pagani P, Modena D, Moras ML, Pozzi P, Reznikov LL, et al. The histone deacetylase inhibitor ITF2357 reduces production of pro-inflammatory cytokines in vitro and systemic inflammation in vivo. Mol Med. 2005;11:1–15. doi: 10.2119/2006-00005.Dinarello. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bode KA, Schroder K, Hume DA, Ravasi T, Heeg K, Sweet MJ, Dalpke AH. Histone deacetylase inhibitors decrease Toll-like receptor-mediated activation of proinflammatory gene expression by impairing transcription factor recruitment. Immunology. 2007;122:596–606. doi: 10.1111/j.1365-2567.2007.02678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Joseph J, Mudduluru G, Antony S, Vashistha S, Ajitkumar P, Somasundaram K. Expression profiling of sodium butyrate (NaB)-treated cells: identification of regulation of genes related to cytokine signaling and cancer metastasis by NaB. Oncogene. 2004;23:6304–6315. doi: 10.1038/sj.onc.1207852. [DOI] [PubMed] [Google Scholar]
- 18.Mesplède T, Navarro S, Génin P, Morin P, Island ML, Bonnefoy E, Civas A. Positive and negative control of virus-induced interferon-A gene expression. Autoimmunity. 2003;36:447–455. doi: 10.1080/08916930310001602119. [DOI] [PubMed] [Google Scholar]
- 19.Barnes BJ, Moore PA, Pitha PM. Virus-specific activation of a novel interferon regulatory factor, IRF-5, results in the induction of distinct interferon alpha genes. J Biol Chem. 2001;276:23382–23390. doi: 10.1074/jbc.M101216200. [DOI] [PubMed] [Google Scholar]
- 20.Barnes BJ, Kellum MJ, Field AE, Pitha PM. Multiple regulatory domains of IRF-5 control activation, cellular localization, and induction of chemokines that mediate recruitment of T lymphocytes. Mol Cell Biol. 2002;22:5721–5740. doi: 10.1128/MCB.22.16.5721-5740.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barnes BJ, Richards J, Mancl M, Hanash S, Beretta L, Pitha PM. Global and distinct targets of IRF-5 and IRF-7 during innate response to viral infection. J Biol Chem. 2004;279:45194–45207. doi: 10.1074/jbc.M400726200. [DOI] [PubMed] [Google Scholar]
- 22.Schoenemeyer A, Barnes BJ, Mancl ME, Latz E, Goutagny N, Pitha PM, Fitzgerald KA, Golenbock DT. The interferon regulatory factor, IRF5, is a central mediator of toll-like receptor 7 signaling. J Biol Chem. 2005;280:17005–17012. doi: 10.1074/jbc.M412584200. [DOI] [PubMed] [Google Scholar]
- 23.Takaoka A, Yanai H, Kondo S, Duncan G, Negishi H, Mizutani T, Kano S, Honda K, Ohba Y, Mak TW, Taniguchi T. Integral role of IRF-5 in the gene induction programme activated by Toll-like receptors. Nature. 2005;434:243–249. doi: 10.1038/nature03308. [DOI] [PubMed] [Google Scholar]
- 24.Yanai H, Chen HM, Inuzuka T, Kondo S, Mak TW, Takaoka A, Honda K, Taniguchi T. Role of IFN regulatory factor 5 transcription factor in antiviral immunity and tumor suppression. Proc Natl Acad Sci USA. 2007;104:3402–3407. doi: 10.1073/pnas.0611559104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mancl ME, Hu G, Sangster-Guity N, Olshalsky SL, Hoops K, Fitzgerald-Bocarsly P, Pitha PM, Pinder K, Barnes BJ. Two discrete promoters regulate the alternatively spliced human interferon regulatory factor-5 isoforms. Multiple isoforms with distinct cell type-specific expression, localization, regulation, and function. J Biol Chem. 2005;280:21078–21090. doi: 10.1074/jbc.M500543200. [DOI] [PubMed] [Google Scholar]
- 26.Dent CL, Macbride SJ, Sharp NA, Gewert DR. Relative transcriptional inducibility of the human interferon-alpha subtypes conferred by the virus-responsive enhancer sequence. J Interferon Cytokine Res. 1996;16:99–107. doi: 10.1089/jir.1996.16.99. [DOI] [PubMed] [Google Scholar]
- 27.Yeow WS, Au WC, Juang YT, Fields CD, Dent CL, Gewert DR, Pitha PM. Reconstitution of virus-mediated expression of interferon alpha genes in human fibroblast cells by ectopic interferon regulatory factor-7. J Biol Chem. 2000;275:6313–6320. doi: 10.1074/jbc.275.9.6313. [DOI] [PubMed] [Google Scholar]
- 28.Cheung SC, Chattopadhyay SK, Hartley JW, Morse HC, III, Pitha PM. Aberrant expression of cytokine genes in peritoneal macrophages from mice infected with LP-BM5 MuLV, a murine model of AIDS. J Immunol. 1991;146:121–127. [PubMed] [Google Scholar]
- 29.Hu G, Barnes BJ. IRF-5 is a mediator of the death receptor-induced apoptotic signaling pathway. J Biol Chem. 2009;284:2767–2777. doi: 10.1074/jbc.M804744200. [DOI] [PubMed] [Google Scholar]
- 30.Barnes BJ, Field AE, Pitha-Rowe PM. Virus-induced heterodimer formation between IRF-5 and IRF-7 modulates assembly of the IFNA enhanceosome in vivo and transcriptional activity of IFNA genes. J Biol Chem. 2003;278:16630–16641. doi: 10.1074/jbc.M212609200. [DOI] [PubMed] [Google Scholar]
- 31.Jepsen K, Rosenfeld MG. Biological roles and mechanistic actions of co-repressor complexes. J Cell Sci. 2002;115:689–698. doi: 10.1242/jcs.115.4.689. [DOI] [PubMed] [Google Scholar]
- 32.Li J, Lin Q, Wang W, Wade P, Wong J. Specific targeting and constitutive association of histone deacetylase complexes during transcriptional repression. Genes Dev. 2002;16:687–692. doi: 10.1101/gad.962502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weaver BK, Kumar KP, Reich NC. Interferon regulatory factor 3 and CREB-binding protein/p300 are subunits of double-stranded RNA-activated transcription factor DRAF1. Mol Cell Biol. 1998;18:1359–1368. doi: 10.1128/mcb.18.3.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yoneyama M, Suhara W, Fukuhara Y, Fukuda M, Nishida E, Fujita T. Direct triggering of the type I interferon system by virus infection: activation of a transcription factor complex containing IRF-3 and CBP/p300. EMBO J. 1998;17:1087–1095. doi: 10.1093/emboj/17.4.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Savitsky D, Tamura T, Yanai H, Taniguchi T. Regulation of immunity and oncogenesis by the IRF transcription factor family. Cancer Immunol Immunother. 2010;59:489–510. doi: 10.1007/s00262-009-0804-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clément JF, Bibeau-Poirier A, Gravel SP, Grandvaux N, Bonneil E, Thibault P, Meloche S, Servant MJ. Phosphorylation of IRF-3 on Ser 339 generates a hyperactive form of IRF-3 through regulation of dimerization and CBP association. J Virol. 2008;82:3984–3996. doi: 10.1128/JVI.02526-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tang X, Gao JS, Guan YJ, McLane KE, Yuan ZL, Ramratnam B, Chin YE. Acetylation-dependent signal transduction for type I interferon receptor. Cell. 2007;131:93–105. doi: 10.1016/j.cell.2007.07.034. [DOI] [PubMed] [Google Scholar]
- 38.Cheng TF, Brzostek S, Ando O, Van Scoy S, Kumar KP, Reich NC. Differential activation of IFN regulatory factor (IRF)-3 and IRF-5 transcription factors during viral infection. J Immunol. 2006;176:7462–7470. doi: 10.4049/jimmunol.176.12.7462. [DOI] [PubMed] [Google Scholar]
- 39.Caillaud A, Prakash A, Smith E, Masumi A, Hovanessian AG, Levy DE, Marié I. Acetylation of interferon regulatory factor-7 by p300/CREB-binding protein (CBP)-associated factor (PCAF) impairs its DNA binding. J Biol Chem. 2002;277:49417–49421. doi: 10.1074/jbc.M207484200. [DOI] [PubMed] [Google Scholar]
- 40.Chen W, Royer WE. Structural insights into interferon regulatory factor activation. Cell Signal. 2010;22:883–887. doi: 10.1016/j.cellsig.2009.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qin BY, Liu C, Srinath H, Lam SS, Correia JJ, Derynck R, Lin K, Lin K. Crystal structure of IRF-3 in complex with CBP. Structure. 2005;13:1269–1277. doi: 10.1016/j.str.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 42.Masumi A, Yamakawa Y, Fukazawa H, Ozato K, Komuro K. Interferon regulatory factor-2 regulates cell growth through its acetylation. J Biol Chem. 2003;278:25401–25407. doi: 10.1074/jbc.M213037200. [DOI] [PubMed] [Google Scholar]
- 43.Wathelet MG, Lin CH, Parekh BS, Ronco LV, Howley PM, Maniatis T. Virus infection induces the assembly of coordinately activated transcription factors on the IFN-beta enhancer in vivo. Mol Cell. 1998;1:507–518. doi: 10.1016/s1097-2765(00)80051-9. [DOI] [PubMed] [Google Scholar]
- 44.Au WC, Pitha PM. Recruitment of multiple interferon regulatory factors and histone acetyltransferase to the transcriptionally active interferon a promoters. J Biol Chem. 2001;276:41629–41637. doi: 10.1074/jbc.M105121200. [DOI] [PubMed] [Google Scholar]
- 45.Génin P, Lin R, Hiscott J, Civas A. Differential regulation of human interferon A gene expression by interferon regulatory factors 3 and 7. Mol Cell Biol. 2009;29:3435–3450. doi: 10.1128/MCB.01805-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Baek SH, Ohgi KA, Rose DW, Koo EH, Glass CK, Rosenfeld MG. Exchange of N-CoR corepressor and Tip60 coactivator complexes links gene expression by NF-kappaB and beta-amyloid precursor protein. Cell. 2002;110:55–67. doi: 10.1016/s0092-8674(02)00809-7. [DOI] [PubMed] [Google Scholar]
- 47.Hong SH, Privalsky ML. The SMRT corepressor is regulated by a MEK-1 kinase pathway: inhibition of corepressor function is associated with SMRT phosphorylation and nuclear export. Mol Cell Biol. 2000;20:6612–6625. doi: 10.1128/mcb.20.17.6612-6625.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hoberg JE, Yeung F, Mayo MW. SMRT derepression by the IkappaB kinase alpha: a prerequisite to NF-kappaB transcription and survival. Mol Cell. 2004;16:245–255. doi: 10.1016/j.molcel.2004.10.010. [DOI] [PubMed] [Google Scholar]
- 49.Kozyrev SV, Alarcon-Riquelme ME. The genetics and biology of Irf5-mediated signaling in lupus. Autoimmunity. 2007;40:591–601. doi: 10.1080/08916930701510905. [DOI] [PubMed] [Google Scholar]
- 50.Moser KL, Kelly JA, Lessard CJ, Harley JB. Recent insights into the genetic basis of systemic lupus erythematosus. Genes Immun. 2009;10:373–379. doi: 10.1038/gene.2009.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Feng D, Stone RC, Eloranta ML, Sangster-Guity N, Nordmark G, Sigurdsson S, Wang C, Alm G, Syvänen AC, Rönnblom L, Barnes BJ. Genetic variants and disease-associated factors contribute to enhanced interferon regulatory factor 5 expression in blood cells of patients with systemic lupus erythematosus. Arthritis Rheum. 2010;62:562–573. doi: 10.1002/art.27223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Niewold TB, Kelly JA, Flesch MH, Espinoza LR, Harley JB, Crow MK. Association of the IRF5 risk haplotype with high serum interferon-α activity in systemic lupus erythematosus patients. Arthritis Rheum. 2008;58:2481–2487. doi: 10.1002/art.23613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kariuki SN, Kirou KA, MacDermott EJ, Barillas-Arias L, Crow MK, Niewold TB. Cutting edge: autoimmune disease risk variant of STAT4 confers increased sensitivity to IFN-alpha in lupus patients in vivo. J Immunol. 2009;182:34–38. doi: 10.4049/jimmunol.182.1.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rönnblom L, Eloranta ML, Alm GV. The type I interferon system in systemic lupus erythematosus. Arthritis Rheum. 2006;54:408–420. doi: 10.1002/art.21571. [DOI] [PubMed] [Google Scholar]
- 55.Lee PY, Kumagai Y, Li Y, Takeuchi O, Yoshida H, Weinstein J, Kellner ES, Nacionales D, Barker T, Kelly-Scumpia K, et al. TLR7-dependent and FcgammaR-independent production of type I interferon in experimental mouse lupus. J Exp Med. 2008;205:2995–3006. doi: 10.1084/jem.20080462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim WU, Sreih A, Bucala R. Toll-like receptors in systemic lupus erythematosus; prospects for therapeutic intervention. Autoimmun Rev. 2009;8:204–208. doi: 10.1016/j.autrev.2008.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lenert P. Nucleic acid sensing receptors in systemic lupus erythematosus: development of novel DNA- and/or RNA-like analogues for treating lupus. Clin Exp Immunol. 2010;161:208–222. doi: 10.1111/j.1365-2249.2010.04176.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Leoni F, Zaliani A, Bertolini G, Porro G, Pagani P, Pozzi P, Donà G, Fossati G, Sozzani S, Azam T, et al. The antitumor histone deacetylase inhibitor suberoylanilide hydroxamic acid exhibits anti-inflammatory properties via suppression of cytokines. Proc Natl Acad Sci USA. 2002;99:2995–3000. doi: 10.1073/pnas.052702999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mishra N, Reilly CM, Brown DR, Ruiz P, Gilkeson GS. Histone deacetylase inhibitors modulate renal disease in the MRL-lpr/lpr mouse. J Clin Invest. 2003;111:539–552. doi: 10.1172/JCI16153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Reddy P, Teshima T, Kukuruga M, Ordemann R, Liu C, Lowler K, Ferrara JL. Interleukin-18 regulates acute graft-versus-host disease by enhancing Fas-mediated donor T cell apoptosis. J Exp Med. 2001;194:1433–1440. doi: 10.1084/jem.194.10.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chung YL, Lee MY, Wang AJ, Yao LF. A therapeutic strategy uses histone deacetylase inhibitors to modulate the expression of genes involved in the pathogenesis of rheumatoid arthritis. Mol Ther. 2003;8:707–717. doi: 10.1016/s1525-0016(03)00235-1. [DOI] [PubMed] [Google Scholar]
- 62.Nishida K, Komiyama T, Miyazawa S, Shen ZN, Furumatsu T, Doi H, Yoshida A, Yamana J, Yamamura M, Ninomiya Y, et al. Histone deacetylase inhibitor suppression of autoantibody-mediated arthritis in mice via regulation of p16INK4a and p21(WAF1/Cip1) expression. Arthritis Rheum. 2004;50:3365–3376. doi: 10.1002/art.20709. [DOI] [PubMed] [Google Scholar]
- 63.Reilly CM, Mishra N, Miller JM, Joshi D, Ruiz P, Richon VM, Marks PA, Gilkeson GS. Modulation of renal disease in MRL/lpr mice by suberoylanilide hydroxamic acid. J Immunol. 2004;173:4171–4178. doi: 10.4049/jimmunol.173.6.4171. [DOI] [PubMed] [Google Scholar]
- 64.Present DH, Rutgeerts P, Targan S, Hanauer SB, Mayer L, van Hogezand RA, Podolsky DK, Sands BE, Braakman T, DeWoody KL, et al. Infliximab for the treatment of fistulas in patients with Crohn’s disease. N Engl J Med. 1999;340:1398–1405. doi: 10.1056/NEJM199905063401804. [DOI] [PubMed] [Google Scholar]
- 65.Leonardi CL, Powers JL, Matheson RT, Goffe BS, Zitnik R, Wang A, Gottlieb AB Etanercept Psoriasis Study Group. . Etanercept as mono-therapy in patients with psoriasis. N Engl J Med. 2003;349:2014–2022. doi: 10.1056/NEJMoa030409. [DOI] [PubMed] [Google Scholar]
- 66.Lee JY, Kim NA, Sanford A, Sullivan KE. Histone acetylation and chromatin conformation are regulated separately at the TNF-α promoter in monocytes and macrophages. J Leukoc Biol. 2003;73:862–871. doi: 10.1189/jlb.1202618. [DOI] [PubMed] [Google Scholar]
- 67.Chen W, Lam SS, Srinath H, Jiang Z, Correia JJ, Schiffer CA, Fitzgerald KA, Lin K, Royer WE., Jr Insights into interferon regulatory factor activation from the crystal structure of dimeric IRF5. Nat Struct Mol Biol. 2008;15:1213–1220. doi: 10.1038/nsmb.1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Balkhi MY, Fitzgerald KA, Pitha PM. Functional regulation of MyD88 activated interferon regulatory factor 5 by K63-linked poly-ubiquitination. Mol Cell Biol. 2008;28:7296–7308. doi: 10.1128/MCB.00662-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vogelauer M, Wu J, Suka N, Grunstein M. Global histone acetylation and deacetylation in yeast. Nature. 2000;408:495–498. doi: 10.1038/35044127. [DOI] [PubMed] [Google Scholar]
- 70.Yamagoe S, Kanno T, Kanno Y, Sasaki S, Siegel RM, Lenardo MJ, Humphrey G, Wang Y, Nakatani Y, Howard BH, Ozato K. Interaction of histone acetylases and deacetylases in vivo. Mol Cell Biol. 2003;23:1025–1033. doi: 10.1128/MCB.23.3.1025-1033.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cai R, Kwon P, Yan-Neale Y, Sambuccetti L, Fischer D, Cohen D. Mammalian histone deacetylase 1 protein is posttranslationally modified by phosphorylation. Biochem Biophys Res Commun. 2001;283:445–453. doi: 10.1006/bbrc.2001.4786. [DOI] [PubMed] [Google Scholar]
- 72.Pflum MK, Tong JK, Lane WS, Schreiber SL. Histone deacetylase 1 phosphorylation promotes enzymatic activity and complex formation. J Biol Chem. 2001;276:47733–47741. doi: 10.1074/jbc.M105590200. [DOI] [PubMed] [Google Scholar]
- 73.David G, Neptune MA, DePinho RA. SUMO-1 modification of histone deacetylase 1 (HDAC1) modulates its biological activities. J Biol Chem. 2002;277:23658–23663. doi: 10.1074/jbc.M203690200. [DOI] [PubMed] [Google Scholar]
- 74.Boggio R, Chiocca S. Gam1 and the SUMO pathway. Cell Cycle. 2005;4:533–535. doi: 10.4161/cc.4.4.1605. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.