Abstract
Objective
There is evidence that both environmental and genetic factors contribute to pelvic organ prolapse (POP). We conducted a genome-wide association study to investigate whether common genetic variants modify the risk of POP.
Methods
We recruited women who had been evaluated and treated for POP at the University of Utah from 1996–2008 and their affected female relatives. Cases were genotyped on the Illumina 550K platform. We genetically matched 2,976 Caucasian iControls available from Illumina as controls. Association tests were adjusted for related subjects using two different software programs: Efficient Mixed-Model Association eXpedited (EMMAX) and Genie. Confirmation of findings was performed in a cohort of Dutch women (n=76) with recurrent POP and family history of POP.
Results
The Utah study sample included 115 cases treated for POP, in most cases with surgery (n=78) or repeat surgery (n=35). Results from association analyses using EMMAX software identified five single nucleotide polymorphisms (SNPs) significantly associated with POP (p<1×10−7). Independent association analysis with Genie software identified three of the same SNPs and one additional SNP. The six SNPs were located at 4q21 (rs1455311), 8q24 (rs1036819), 9q22 (rs430794), 15q11 (rs8027714), 20p13 (rs1810636), and 21q22 (rs2236479). Nominally significant findings (p<0.05) or findings trending towards significance (p<0.1) were observed for five of the six SNPs in the Dutch cohort.
Conclusion
Six SNPs have been identified that are significantly associated with POP in high-risk familial cases and provide evidence for a genetic contribution to POP.
Introduction
Pelvic floor disorders including stress incontinence, urge incontinence, and pelvic organ prolapse (POP) are common conditions that have been shown to affect approximately 27 to 50% of women aged 40 years and greater.1 Some 3–6% of women have pelvic organ prolapse that descends beyond the vaginal opening on routine pelvic examination,2 and an estimated 11% of women will undergo surgery for POP sometime in their lifespan.3 The etiology of POP is thought to be multifactorial with contributions from both genetic and environmental risk factors. Environmental factors that contribute to POP include vaginal delivery, chronic increases in intra-abdominal pressure, obesity, advanced age, and estrogen deficiency.4, 5
Evidence for a genetic contribution to POP has been found in family-based studies, candidate gene association studies, expression studies, and linkage studies.6 Family studies have shown an increased risk of POP when mothers and sisters are affected with the disease,7, 8 and after menopause, familial risk may be more important than contributions from childbirth.8 A segregation analysis of early onset POP suggested a dominant mode of inheritance with incomplete penetrance that can involve either maternal or paternal transmission.9 Candidate gene studies have focused on collagen and elastin biosynthesis,10,11,12 extracellular matrix metabolism,13 and hormone receptors14–16 with encouraging results. Expression studies have found a number of genes that are over and under expressed in women with POP compared to women without POP.17–21 Linkage studies have implicated a region near the LAMC1 gene in a single Filipino family,22 and our group has found significant genome-wide linkage evidence on chromosome 9q21 in 70 women with POP from 32 families.23
To further understand POP genetics, we performed a genome-wide association study (GWAS) for POP to identify common genetic variants that contribute to POP. Association is tested by comparing allele frequency differences between cases and controls for thousands of single nucleotide polymorphisms (SNPs). The purpose of this study was to identify SNPs associated with POP in a cohort of women with a family history of pelvic floor disorders who have been treated for moderate-to-severe POP.
Material and Methods
Subjects
Subjects included in this study had been evaluated and treated for POP, usually with surgery. Probands underwent surgery for POP at the University of Utah between 1996 and 2008. To be included in our original Utah Pelvic Floor Disorder Genetic Resource, a proband was required to have a sister who also had moderate to severe POP, received treatment for POP, and was willing to participate in the study. The Utah Pelvic Floor Disorder Genetic Resource has since been expanded to include more distant affected relatives. Documentation of surgical management was required for women who had surgery outside our system. Subjects were white and of Northern European descent, similar to Utah demographics. We included three subject groups for this analysis: i.) women who were treated for POP, had a close female family member who was similarly affected, and were originally part of our linkage analysis (n=70, 32 families);23 ii.) women treated for POP who were part of pedigrees of interest identified in the linkage analysis and recruited after the linkage analysis (n=33, sampled from 11 of the linkage families); and iii.) subjects who were the only POP case sampled from their family, although other members of their family had a pelvic floor disorder (n=23). Exclusion criteria were genetic diseases with a known increased risk of POP (such as Ehlers Danlos, Marfan and Steinert’s disease) or neurological conditions (e.g., multiple sclerosis or stroke) that might affect tissue weakness or incontinence. This study was approved by the University of Utah IRB, and informed consent was obtained from all study participants.
Controls
We selected controls from 3,293 iControls from Illumina iControlDB who had available Illumina 550K array genotype data and who self-reported as Caucasian. These controls are submitted as anonymous individuals from various different groups; information on presence of pelvic floor disorders is not available on these subjects. Because phenotype status is unknown, we would expect results using these controls to be conservative but not biased. We checked for evidence of duplicate samples and closely related subjects (i.e., parent-offspring relationships) using the pair-wise estimated proportion of alleles shared identity-by-descent in the genetic association analysis software tool, PLINK;24 20 subjects who fit this criterion were removed from the iControl list.
Validation Cohort
We identified a cohort of 76 independent Caucasian females from a Dutch genetic resource with recurrent POP and who had a mother, a sister or a daughter who also had POP. Blood samples were collected from consecutive Dutch women with POP who presented at the Department of Obstetrics and Gynecology of the Radboud University Nijmegen Medical Centre between January 2007 and August 2008. Only POP cases reporting a positive family history of POP were included in the present study. Exclusion criteria were genetic diseases with a known increased risk of POP (such as Ehlers Danlos, Marfan and Steinert’s disease). Control data for the Dutch POP samples were not available, hence association results were compared to Utah POP cases, Utah-matched iControls, and HapMap CEU independent individuals who are Utah residents with Northern and Western European ancestry from the Centre d’Etude du Polymorphisme Humain (CEPH) collection and selected as one of the populations genotyped for the International HapMap project. The CEU cohort consists of a set of 30 trios (mother-father-offspring); only the 60 independent parents were included in this analysis. All individuals from the Dutch cohort gave informed consent and the study was approved by the Medical Ethics Review Committee Arnhem/Nijmegen.
Genotyping and Quality Control
DNA was extracted from all Utah study subjects and genome-wide genotyping was performed at deCODE Genetics (Iceland). Samples were genotyped on the Illumina HumanHap550 (~550,000 SNPs) or 610Q platforms (~610,000 SNPs). The Illumina 610Q contains the great majority of the HumanHap 550 set of SNP markers plus additional copy number variants. We identified a set of SNP markers common to both the Illumina 610Q platform and the HumanHap 550 platform as our genotype set. All samples had a minimum call rate of 98%. We started with 126 strictly defined POP cases; 10 cases were removed because they failed to reach the minimum call rate.
For quality control purposes, SNPs were required to have a call rate of 95%, minor allele frequency of 0.01, and a p-value for Hardy-Weinberg equilibrium < 0.001. Call rate for a SNP refers to the percent of subjects for which genotypes can be assigned. The final number of SNP markers analyzed that met all of the quality control thresholds was 499,948.
Confirmation of significant SNPs in a second cohort
SNPs that met the significance criterion in the genome-wide association analyses were then studied in the confirmation set of recurrent familial POP cases from Holland. SNP genotyping was performed using the fluorogenic 5′ nuclease TaqMan Assay (Applied Biosystems) and PCR amplification was performed according to Applied Biosystems protocol. The 7900HT Sequence Detection System (Applied Biosystems) was used to measure each fluorescent dye-labeled probe specific for each allele studied, and results were analyzed with the Sequence Detection Software (Applied Biosystems). One of the SNPs (rs8027714) was custom ordered from Applied Biosystems because it was not available in their resource of pre-designed TaqMan assays; quality control measures for custom SNPs are not as rigorous as the Applied Biosystem’s pre-designed assays.
Population Stratification
Spurious association results can be attained because of population stratification or systematic differences in ancestry between cases and controls if not taken into account. Details of our method to control for population stratification are supplied in the Appendix. To control for population stratification we removed 1 POP case and 297 iControls. Our genomic inflation factor (λGC), or a measure of population stratification, was 1.05, as calculated using the software package EMMAX(Efficient Mixed Model Association eXpedited).25 Levels of population stratification are generally considered acceptable for λGC ≤ 1.05.26 Hence, our working sample size was 115 cases and 2,976 controls.
Statistical Analysis
Standard association methods typically assume independence of cases and of controls. Our inclusion of related subjects necessitates the use of methodology that adjusts for relatedness. Two different analytical methods were used. The software program EMMAX uses linear mixed model regression and a variance component matrix approach to account for the relatedness, even cryptic relatedness, between pairs of subjects.25 For the second method, we performed an initial naïve genome-wide screen ignoring family relationships using the program PLINK.24 SNPs meeting an initial p-value < 5×10−7 in PLINK were then re-analyzed using Genie, a Monte-Carlo simulation platform that performs association analyses and accounts for complete pedigree structure by computing an empirical p-value.27 These two methods both perform association tests among related individuals, but the method used is not the same. EMMAX accounts for relatedness through analysis of pairs of individuals, whereas Genie accounts for relatedness by genotype simulation through the known pedigree structure, which can take considerable computing time. Only markers meeting an initial naïve threshold were analyzed using Genie. As standard methodology for analyzing related data does not exist, we have opted to report results using these two different methodologies. Any SNP meeting a genome-wide significance threshold of 1×10−7 for either method was considered significant after accounting for multiple testing by Bonferroni correction (i.e., 0.05/499,948 tests). To search for systematic bias (e.g., unrecognized population stratification), we plotted a quantile-quantile plot. Genome-wide significance results were graphed using a Manhattan plot. The following R codes were used to generate the plots.28, 29 For the validation arm of the study using the Dutch POP cases, we considered a threshold of p<0.05 for significance and p<0.1 to be a trend towards significance.
Results
Characteristics of the Utah POP subjects are presented in Table 1. We analyzed 115 Utah POP cases and 2,976 iControls. Most cases had been treated for POP with surgery (98.2%); however, many also had been treated for stress urinary incontinence (62.6%) or overactive bladder (32.2%). Approximately 30% of the Utah cases had recurrent POP and required repeat surgery.
Table 1.
Phenotype Characteristics of Familial Pelvic Organ Prolapse Study Participants
| Characteristic | Utah Cohort | Dutch Cohort |
|---|---|---|
| Number of POP subjects | 115 | 76 |
| Treated with pessary | 2 (1.7%) | Not available |
| Treated with one surgery | 78 (67.8%) | 0 |
| Required repeat surgery | 35 (30.4%) | 76 (100%) |
| Mean ± SD age at diagnosis (range) | 48.8 ± 14.2 yrs (range 21–75, n=85) | 49.3 ± 12.4 yrs (range 24–86, n=67) |
| Mean ± SD parity | 4.3 ± 2.4 (range 0–13, n=104) | 2.4 ± 0.7 (range 1–4, n=76) |
| Mean BMI at time of phenotyping | 26.6 ± 5.1 kg/m2 (range 16–47, n=105) | 25.2 ± 3.5 kg/m2 (range 16.3–36.5, n=73) |
| Other pelvic floor disorders diagnosed in POP subjects | ||
| Treated for stress urinary incontinence | 72 (62.6%) | Not available |
| Treated for overactive bladder | 37 (32.2%) | Not available |
| Presence of hernia | 13 (11.3%) | Not available |
POP, pelvic organ prolapse; SD, standard deviation, BMI, body mass index.
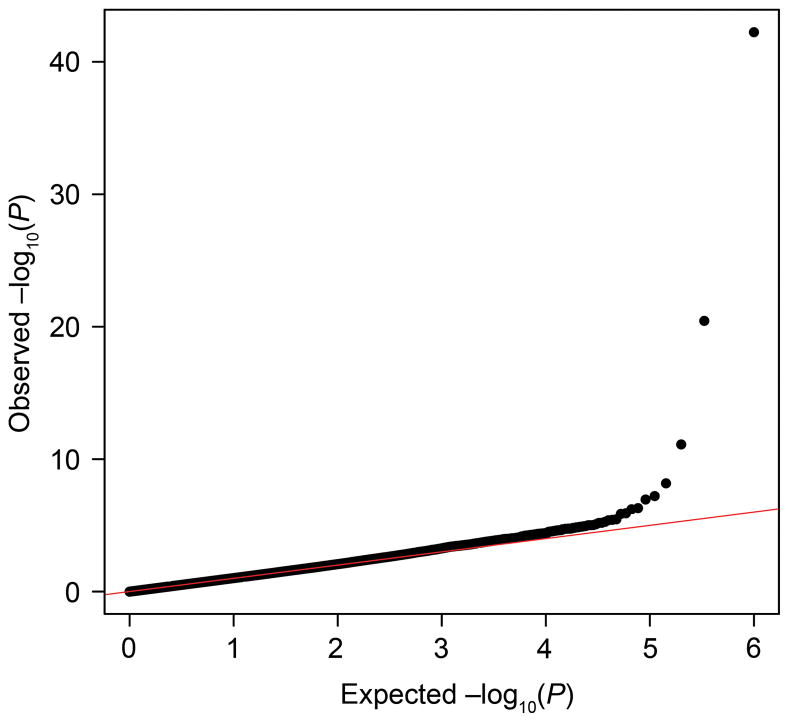
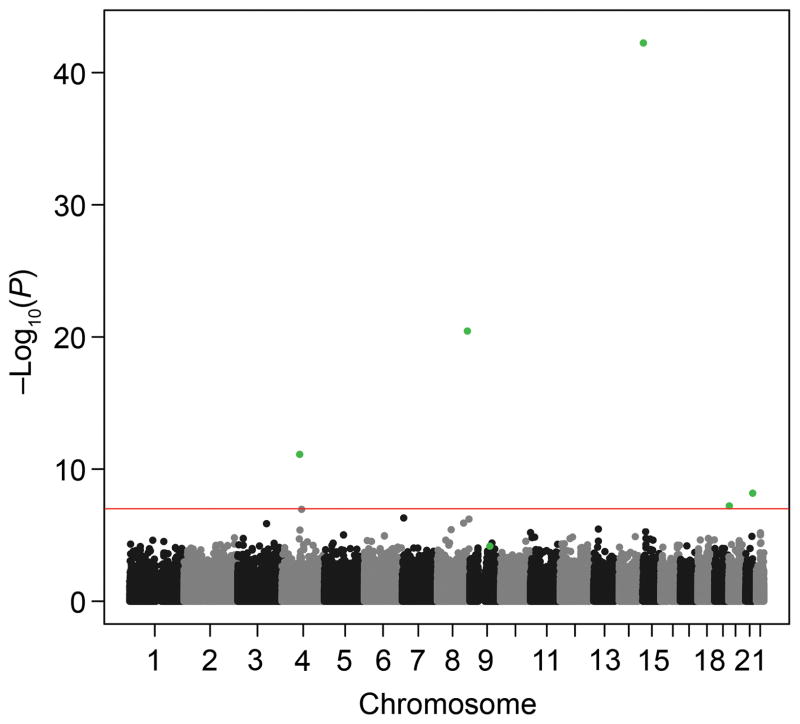
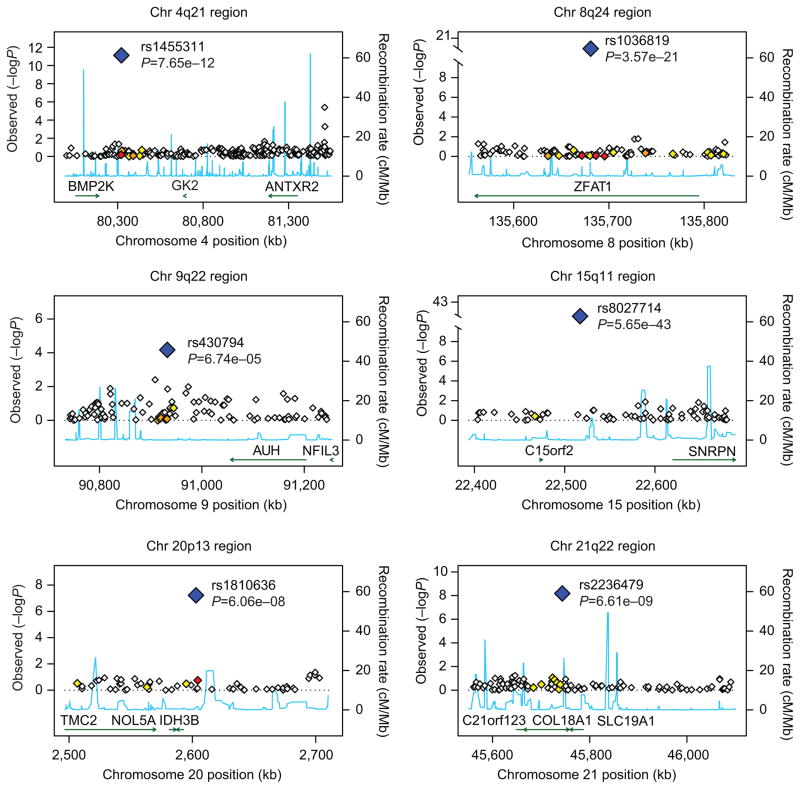
The quantile-quantile plot in Figure 1 showed little evidence of inflation of the test statistic after correction for relatedness using EMMAX. Results of the genome-wide association analyses using EMMAX are shown in Figure 2. Five SNPs met genome-wide significance at p-value < 1×10−7 (see list of SNPs in Table 2) using EMMAX. For the second genome-wide association analysis, we identified 133 unique markers from the naïve association analysis performed using PLINK with a p-value < 5×10−7. After analysis using Genie to account for relatedness, four markers remained significant at a p-value < 1×10−7. Three of the markers from the Genie analysis were also found to be significant using EMMAX (rs1455311, rs1036819, and rs8027714). Genie identified one SNP that was not found to be significant using EMMAX (rs430794). These six SNPs were considered of interest for validation. Results for these six SNPs are shown in Table 2 and Figure 3.
Figure 1.
The quantile–quantile plot for this analysis shows little evidence for inflation of the test statistic because of hidden population substructure, cryptic relatedness, or differential genotype calling between cases and controls. The genomic inflation factor (λGC) for this study was 1.05. The plot was made using association results from Efficient Mixed Model Association eXpedited software.
Figure 2.
Genome-wide Manhattan association results for pelvic organ prolapse. P-values are represented as −log (P-value). Results are taken from Efficient Mixed Model Association eXpedited software. The line represents the genome-wide significance threshold of 1×10−7. The green dots represent the six significant markers in the study.
Table 2.
Six Single Nucleotide Polymorphisms s Found to Be Significant Using EMMAX or Genie for Utah Pelvic Organ Prolapse Cases
| Marker | Location | Gene | P | OR† | Minor Allele Frequency | |||
|---|---|---|---|---|---|---|---|---|
| EMMAX | Genie* | Cases | iControls | CEU Participants (n=60) | ||||
| rs1455311 | 4q21.21 | Intergenic | 7.65×10−12 | < 1×10−9 | 2.58 | 0.34 | 0.17 | 0.21 |
| rs1036819 | 8q24.22 | ZFAT | 3.57×10−21 | < 1×10−9 | 4.03 | 0.31 | 0.1 | 0.14 |
| rs430794 | 9q22.2 | Intergenic | 6.74×10−05 | 8.0×10−8 | 0.35 | 0.13 | 0.3 | 0.33 |
| rs8027714 | 15q11.2 | Intergenic | 5.65×10−43 | < 1×10−9 | 9.04 | 0.26 | 0.04 | 0.04 |
| rs1810636 | 20p13 | Intergenic | 6.06×10−08 | 3.3×10−7 | 2.32 | 0.57 | 0.36 | 0.36 |
| rs2236479 | 21q22.3 | COL18A1 | 6.61×10−09 | 2.8×10−7 | 2.23 | 0.59 | 0.39 | 0.31 |
EMMAX, Efficient Mixed-Model Association eXpedited; OR, odds radio; CEU, HapMap CEPH Utah- independent individuals
Level of significance detected in Genie is determined by the number of simulations performed. For this analysis we ran 1 billion simulations, hence the minimum p-value that could be detected is 1×10−9.
Odds ratio reported for the allele test.
Figure 3.
Plots of the six chromosomal regions found to be of interest. Gene plots are based on NCBI35/hg17. Estimated underlying recombination rates for a region are based on HapMap CEU individuals and are shown in blue. Markers in high linkage disequilibrium with one of the six single nucleotide polymorphisms (SNPs) of interest are determined using r-squared values obtained from HapMap CEU and are coded by the color of the diamond shape. White indicates no linkage disequilibrium (r2<0.2), yellow indicates weak linkage disequilibrium (r2 between 0.2 and 0.5), orange indicates moderate linkage disequilibrium (r2 between 0.5 and 0.8), and red indicates strong linkage disequilibrium (r2 0.8 or greater).
Validation results of the six associated SNPs in the Dutch POP case set are shown in Table 3. For results to be of interest we would expect similar results between the Utah POP cases and the Dutch POP cases (i.e., no significant association difference), and we would expect significant differences between the Dutch POP cases and the control populations (i.e., the Utah matched iControl individuals and the HapMap CEU subjects). We observed that five of the six SNPs showed nominally significant findings (p<0.05) or trending towards significance (p<0.1) for one of the three comparison groups. For example, Dutch POP cases were significantly different from Utah matched iControls for SNP rs2236479 at 21q22.3 (p=0.029). One SNP was not significantly different than the Utah POP cases, rs430794 (p=0.892), which is in the direction expected for validation using the Utah POP cases as the comparison group. We note, however, that all of these results were observed for only one comparison group; there were no results that were consistent across all three comparison groups.
Table 3.
Analysis of the Six Single Nucleotide Polymorphisms of Interest in Recurrent Pelvic Organ Prolapse Cases From Holland
| Marker | Location | Compared to Utah POP Cases* | Compared to HapMap CEU (n=60)† | Compared to Utah-Matched iControls† | Minor Allele Frequency of Holland Cases | Minor Allele Frequency of Utah POP Cases | |
|---|---|---|---|---|---|---|---|
| Best P-value | Best P-value | Best P-value | OR | ||||
| rs1455311 | 4q21.21 | 0.015 | 0.09§ | 0.198 | 0.72 | 0.14 | 0.34 |
| rs1036819 | 8q24.22 | 1.2×10−3 | 0.086§ | 0.126 | 3.15 | 0.081 | 0.31 |
| rs430794 | 9q22.2 | 0.892‡ | 0.278 | 0.328 | 0.59 | 0.291 | 0.13 |
| rs8027714 | 15q11.2 | 6.4×10−3 | 0.588 | 0.085‡ | 13.57 | 0.061 | 0.26 |
| rs1810636 | 20p13 | 0.012 | 0.416 | 0.588 | 1.2 | 0.36 | 0.57 |
| rs2236479 | 21q22.3 | 1.4×10−3 | 0.616 | 0.029§ | 0.59 | 0.32 | 0.59 |
POP, pelvic organ prolapse; OR, odds ratio.
Genie was used for this analysis. We used 10,000 simulations for the analysis. Compared to the Utah POP cases, and the best P-value was considered the largest P-value.
Compared to the two control populations, the best P-value was considered the smallest P-value.
Recessive model
Dominant model
Discussion
We have identified six SNPs that are significantly associated with POP in a dataset of 115 women with a strong family history of pelvic floor disorders and diagnosed with strictly defined POP.
Genome-wide association studies test the hypothesis that common variants in the population (i.e., variants with typically 1– 5% frequency) increase the susceptibility to a common disease such as POP. An association study using large pedigrees that segregate a small set of variants increases the likelihood that some of these more rare variants become detectable. While our case sample size in this study is small compared to the thousands of individuals typically used in a GWAS, the use of women from high-risk POP families increases the likelihood that these cases have a genetic component to their disease, and hence increases the likelihood that rare disease-contributing variants could be detected in an association analysis.
We previously performed a linkage analysis using some of the same affected individuals who were included in this GWAS, and reported significant linkage of POP to chromosome region 9q21.23 One of 6 SNPs identified in the association analysis at 9q22 (rs430794) is just outside this previous significant linkage region. Linkage analysis is most ideal for detecting highly-penetrant rare loci in high-risk families whereas association analysis is most useful for detecting common loci in a case-control cohort. We assume that a complex disease such as POP involves multiple loci, some common and some rare. Linkage analysis and an association analysis have different strengths and can provide complementary information for study of a complex disease such POP.
Two of the six SNPs were located within genes; rs1036819 is located in the ZFAT gene [MIM: 610931] and rs2236479 is located in the COL18A1 gene [MIM: 120328]. The ZFAT gene has been found to play a transcriptional regulator role for immune regulation and apoptosis.30 ZFAT might be involved in development of mesodermal cells,31 and hence may affect development of muscle and connective tissue of the pelvic floor. The collagen XVIII (COL18A1) gene is another interesting candidate gene for POP. Endostatin, the C-terminal fragment of collagen XVIII, and its precursor collagen XVIII may play a role in the structural organization of basement membranes.32, 33 In mouse models of wound healing, mice overexpressing endostatin showed delayed healing, delayed formation of the epidermal basement membrane, and a more disorganized epidermal and capillary basement membrane structure.34 In the pelvic floor, endostatin and collagen XVIII may play a role in basement membrane organization and response to both minor and major insults.
The other four SNPs identified are intergenic, but one of them is also close to a gene of interest for POP. The SNP rs1455311 at 4q21 is approximately 0.85 Mb away from the anthrax toxin receptor 2 (ANTXR2) gene [MIM: 608041] which binds to collagen IV and laminin, suggesting that it may be involved in extracellular matrix adhesion.35
Although we were unable to conclusively validate the Utah findings, we obtained some nominally significant results in the validation set from Holland. There are a number of factors that might explain the lack of strong validation findings including lack of available control data matched to the Dutch POP cases, a small sample size for the Dutch cohort, and phenotype differences between the Utah and the Dutch POP cases (e.g., Utah cases were more likely to have a mixed pelvic floor disorder phenotype). Another explanation for the lack of strong validation findings is that some or all of the Utah results may be false positives; the Utah sample size is small relative to most other GWAS. Despite these limitations, we note that the Dutch Genetic Resource is a close match to the Utah subjects used in this study: the majority of Utahns are of Northern European descent similar to the Caucasian residents of Holland, and the Dutch resource included familial POP cases as did the Utah cohort. There are very few investigators in the world who are collecting blood from POP cases for genetic studies and even fewer who are collecting blood from families with an excess of POP cases. Future replication studies will need to include more familial cases and use appropriate controls.
While these results still require additional replication, it is likely that one day genetic screening tests will be available to assess genetic risk for POP. Understanding more about the genetic etiology of POP could improve prevention and treatment of this condition, such as studying at-risk groups for preventative strategies (e.g., managing constipation or changing delivery modes). Patients might benefit from changing management algorithms, such as whether surgery is considered early or late in an individual woman’s clinical course.
In conclusion, this work provides evidence for a genetic contribution to POP. We have identified six SNPs that are significantly associated with POP in women with a strong family history of pelvic floor disorders. While we were unable to conclusively replicate our results, we have identified at least three strong candidate genes for POP that warrant follow-up. This association study furthers our understanding of the genetic underpinnings of POP.
Acknowledgments
Funded by grants from the Eunice Kennedy Shriver Institute of Child Health and Human Development R01 HD0041163 (Peggy A. Norton) and RO1 HD061821 (Lisa Cannon-Albright and Peggy A. Norton).
The authors thank Shirley Ranke, RN, for help with recruitment, and Kim Nguyen and Jeroen R. Dijkstra for their assistance in the laboratory.
Appendix
Population Stratification
To account for population stratification and to select a set of genetically matched iControls for our case population, we selected a random POP case with genotype data from each pedigree to represent our case population. Using these independent subjects, we then performed multi-dimensional scaling (MDS) in PLINK25 to produce two-dimensional coordinates for each individual. We calculated the mean of each of the two dimensions for cases only and then computed a Euclidean distance from this case centroid measurement for each individual (i.e., both cases and controls). The genomic inflation factor (λGC) indicates the level of population stratification in the dataset. A value of λGC approximately equal to 1 indicates no stratification and values of λGC > 1 indicate population stratification or other confounders. One POP case and 297 iControls were considered outliers based on their distance from the case centroid measurement and removed from the analysis. Our initial unadjusted-for-relatedness λGC was 1.15 using one POP case per pedigree and all iControls. However, after removal of the outliers our λGC was reduced to 1.05, as calculated using the software package EMMAX, which does account for relatedness of subjects.26 Levels of population stratification are generally considered acceptable for λGC ≤ 1.05.27
Footnotes
Financial Disclosure: The authors did not report any potential conflicts of interest.
Presented at the American Urogynecology Society 2010 annual meeting, Long Beach, California, Sept 29-Oct 2, 2010.
References
- 1.Nygaard I, Barber MD, Burgio KL, Kenton K, Meikle S, Schaffer J, et al. Prevalence of symptomatic pelvic floor disorders in US women. JAMA. 2008;300(11):1311–6. doi: 10.1001/jama.300.11.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Swift S, Woodman P, O’Boyle A, Kahn M, Valley M, Bland D, et al. Pelvic Organ Support Study (POSST): the distribution, clinical definition, and epidemiologic condition of pelvic organ support defects. Am J Obstet Gynecol. 2005;192(3):795–806. doi: 10.1016/j.ajog.2004.10.602. [DOI] [PubMed] [Google Scholar]
- 3.Olsen AL, Smith VJ, Bergstrom JO, Colling JC, Clark AL. Epidemiology of surgically managed pelvic organ prolapse and urinary incontinence. Obstet Gynecol. 1997;89(4):501–6. doi: 10.1016/S0029-7844(97)00058-6. [DOI] [PubMed] [Google Scholar]
- 4.Brown JS, Waetjen LE, Subak LL, Thom DH, Van den Eeden S, Vittinghoff E. Pelvic organ prolapse surgery in the United States, 1997. Am J Obstet Gynecol. 2002;186(4):712–6. doi: 10.1067/mob.2002.121897. [DOI] [PubMed] [Google Scholar]
- 5.Rortveit G, Brown JS, Thom DH, Van Den Eeden SK, Creasman JM, Subak LL. Symptomatic pelvic organ prolapse: prevalence and risk factors in a population-based, racially diverse cohort. Obstet Gynecol. 2007;109(6):1396–403. doi: 10.1097/01.AOG.0000263469.68106.90. [DOI] [PubMed] [Google Scholar]
- 6.Norton P, Milsom I. Genetics and the lower urinary tract. Neurourol Urodyn. 2010;29(4):609–11. doi: 10.1002/nau.20908. [DOI] [PubMed] [Google Scholar]
- 7.Chiaffarino F, Chatenoud L, Dindelli M, Meschia M, Buonaguidi A, Amicarelli F, et al. Reproductive factors, family history, occupation and risk of urogenital prolapse. Eur J Obstet Gynecol Reprod Biol. 1999;82(1):63–7. doi: 10.1016/s0301-2115(98)00175-4. [DOI] [PubMed] [Google Scholar]
- 8.Buchsbaum GM, Duecy EE, Kerr LA, Huang LS, Perevich M, Guzick DS. Pelvic organ prolapse in nulliparous women and their parous sisters. Obstet Gynecol. 2006;108(6):1388–93. doi: 10.1097/01.AOG.0000245784.31082.ed. [DOI] [PubMed] [Google Scholar]
- 9.Jack GS, Nikolova G, Vilain E, Raz S, Rodriguez LV. Familial transmission of genitovaginal prolapse. Int Urogynecol J Pelvic Floor Dysfunct. 2006;17(5):498–501. doi: 10.1007/s00192-005-0054-x. [DOI] [PubMed] [Google Scholar]
- 10.Chen HY, Chung YW, Lin WY, Wang JC, Tsai FJ, Tsai CH. Collagen type 3 alpha 1 polymorphism and risk of pelvic organ prolapse. Int J Gynaecol Obstet. 2008;103(1):55–8. doi: 10.1016/j.ijgo.2008.05.031. [DOI] [PubMed] [Google Scholar]
- 11.Kluivers KB, Dijkstra JR, Hendriks JC, Lince SL, Vierhout ME, van Kempen LC. COL3A1 2209G>A is a predictor of pelvic organ prolapse. Int Urogynecol J Pelvic Floor Dysfunct. 2009;20(9):1113–8. doi: 10.1007/s00192-009-0913-y. [DOI] [PubMed] [Google Scholar]
- 12.Ferrell G, Lu M, Stoddard P, Sammel MD, Romero R, Strauss JF, 3rd, et al. A single nucleotide polymorphism in the promoter of the LOXL1 gene and its relationship to pelvic organ prolapse and preterm premature rupture of membranes. Reprod Sci. 2009;16(5):438–46. doi: 10.1177/1933719108330567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen HY, Lin WY, Chen YH, Chen WC, Tsai FJ, Tsai CH. Matrix metalloproteinase-9 polymorphism and risk of pelvic organ prolapse in Taiwanese women. Eur J Obstet Gynecol Reprod Biol. 2010;149(2):222–4. doi: 10.1016/j.ejogrb.2009.12.014. [DOI] [PubMed] [Google Scholar]
- 14.Chen HY, Chung YW, Lin WY, Chen WC, Tsai FJ, Tsai CH. Estrogen receptor alpha polymorphism is associated with pelvic organ prolapse risk. Int Urogynecol J Pelvic Floor Dysfunct. 2008;19(8):1159–63. doi: 10.1007/s00192-008-0603-1. [DOI] [PubMed] [Google Scholar]
- 15.Chen HY, Chung YW, Lin WY, Chen WC, Tsai FJ, Tsai CH. Progesterone receptor polymorphism is associated with pelvic organ prolapse risk. Acta Obstet Gynecol Scand. 2009;88(7):835–8. doi: 10.1080/00016340902822073. [DOI] [PubMed] [Google Scholar]
- 16.Chen HY, Wan L, Chung YW, Chen WC, Tsai FJ, Tsai CH. Estrogen receptor beta gene haplotype is associated with pelvic organ prolapse. Eur J Obstet Gynecol Reprod Biol. 2008;138(1):105–9. doi: 10.1016/j.ejogrb.2007.12.013. [DOI] [PubMed] [Google Scholar]
- 17.Connell KA, Guess MK, Tate A, Andikyan V, Bercik R, Taylor HS. Diminished vaginal HOXA13 expression in women with pelvic organ prolapse. Menopause. 2009;16(3):529–33. doi: 10.1097/gme.0b013e31818fb0c2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bukovsky A, Copas P, Caudle MR, Cekanova M, Dassanayake T, Asbury B, et al. Abnormal expression of p27kip1 protein in levator ani muscle of aging women with pelvic floor disorders - a relationship to the cellular differentiation and degeneration. BMC Clin Pathol. 2001;1(1):4. doi: 10.1186/1472-6890-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hundley AF, Yuan L, Visco AG. Skeletal muscle heavy-chain polypeptide 3 and myosin binding protein H in the pubococcygeus muscle in patients with and without pelvic organ prolapse. Am J Obstet Gynecol. 2006;194(5):1404–10. doi: 10.1016/j.ajog.2006.01.049. [DOI] [PubMed] [Google Scholar]
- 20.Hundley AF, Yuan L, Visco AG. Gene expression in the rectus abdominus muscle of patients with and without pelvic organ prolapse. Am J Obstet Gynecol. 2008;198(2):220, e1–7. doi: 10.1016/j.ajog.2007.08.047. [DOI] [PubMed] [Google Scholar]
- 21.Visco AG, Yuan L. Differential gene expression in pubococcygeus muscle from patients with pelvic organ prolapse. Am J Obstet Gynecol. 2003;189(1):102–12. doi: 10.1067/mob.2003.372. [DOI] [PubMed] [Google Scholar]
- 22.Nikolova G, Lee H, Berkovitz S, Nelson S, Sinsheimer J, Vilain E, et al. Sequence variant in the laminin gamma1 (LAMC1) gene associated with familial pelvic organ prolapse. Hum Genet. 2007;120(6):847–56. doi: 10.1007/s00439-006-0267-1. [DOI] [PubMed] [Google Scholar]
- 23.Allen-Brady K, Norton PA, Farnham JM, Teerlink C, Cannon-Albright LA. Significant linkage evidence for a predisposition gene for pelvic floor disorders on chromosome 9q21. Am J Hum Genet. 2009;84(5):678–82. doi: 10.1016/j.ajhg.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kang HM, Sul JH, Service SK, Zaitlen NA, Kong SY, Freimer NB, et al. Variance component model to account for sample structure in genome-wide association studies. Nat Genet. 2010;42(4):348–54. doi: 10.1038/ng.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Price AL, Zaitlen NA, Reich D, Patterson N. New approaches to population stratification in genome-wide association studies. Nat Rev Genet. 2010;11(7):459–63. doi: 10.1038/nrg2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Allen-Brady K, Wong J, Camp NJ. PedGenie: an analysis approach for genetic association testing in extended pedigrees and genealogies of arbitrary size. BMC Bioinformatics. 2006;7:209. doi: 10.1186/1471-2105-7-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Plotting genome-wide association results. 2011 May 17; http://www.broadinstitute.org/diabetes/scandinavs/figures.html.
- 29.Turner S. Annotated Manhattan plots and QQ plots for GWAS using R. Revisited. 2011 May 23; http://gettinggeneticsdone.blogspot.com/2011/04/annotated-manhattan-plots-and-qq-plots.html.
- 30.Koyanagi M, Nakabayashi K, Fujimoto T, Gu N, Baba I, Takashima Y, et al. ZFAT expression in B and T lymphocytes and identification of ZFAT-regulated genes. Genomics. 2008;91(5):451–7. doi: 10.1016/j.ygeno.2008.01.009. [DOI] [PubMed] [Google Scholar]
- 31.Tsunoda T, Takashima Y, Tanaka Y, Fujimoto T, Doi K, Hirose Y, et al. Immune-related zinc finger gene ZFAT is an essential transcriptional regulator for hematopoietic differentiation in blood islands. Proc Natl Acad Sci U S A. 2010;107(32):14199–204. doi: 10.1073/pnas.1002494107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Utriainen A, Sormunen R, Kettunen M, Carvalhaes LS, Sajanti E, Eklund L, et al. Structurally altered basement membranes and hydrocephalus in a type XVIII collagen deficient mouse line. Hum Mol Genet. 2004;13(18):2089–99. doi: 10.1093/hmg/ddh213. [DOI] [PubMed] [Google Scholar]
- 33.Elamaa H, Sormunen R, Rehn M, Soininen R, Pihlajaniemi T. Endostatin overexpression specifically in the lens and skin leads to cataract and ultrastructural alterations in basement membranes. Am J Pathol. 2005;166(1):221–9. doi: 10.1016/S0002-9440(10)62246-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seppinen L, Sormunen R, Soini Y, Elamaa H, Heljasvaara R, Pihlajaniemi T. Lack of collagen XVIII accelerates cutaneous wound healing, while overexpression of its endostatin domain leads to delayed healing. Matrix Biol. 2008;27(6):535–46. doi: 10.1016/j.matbio.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 35.Bell SE, Mavila A, Salazar R, Bayless KJ, Kanagala S, Maxwell SA, et al. Differential gene expression during capillary morphogenesis in 3D collagen matrices: regulated expression of genes involved in basement membrane matrix assembly, cell cycle progression, cellular differentiation and G-protein signaling. J Cell Sci. 2001;114(Pt 15):2755–73. doi: 10.1242/jcs.114.15.2755. [DOI] [PubMed] [Google Scholar]