Abstract
Several studies in humans or transgenic animals have reported that the 389 Arg or Gly polymorphic variation of the β1-adrenergic receptor (AR) is associated with differential responses to beta-blocker therapy and/or myocardial disease progression. Analysis of changes in gene expression is an important means of defining molecular differences associated with structural or functional phenotypic variations. To determine if structural and functional myocardial phenotypic differences between β1389 Arg vs. Gly transgenic overexpressors are associated with qualitative and/or quantitative differences in gene expression, a comprehensive analysis of mRNAs and miRNAs expressed in the hearts of 3 and 6–8 mo old β1-Arg389 and β1-Gly389 overexpressor transgenic mice was performed. Changes in mRNA and miRNA expression were analyzed by arrays and partially confirmed by RT-qPCR. Bioinformatic analysis demonstrated that several genes, including those involved in PKA and CaMK signaling pathways, are regulated in a temporal- or phenotype-specific manner. Furthermore, expression signature analyses indicated that miRNAs have the potential to target expression of a number of genes involved in multiple cardiomyopathy-related pathways, and changes in miRNA expression can precede the onset of disease. Differences in gene expression between β1-Arg389 and β1-Gly389 transgenic mice are largely quantitative rather than qualitative and are associated with the development of cardiomyopathy in a time-dependent manner. Chronic β1-AR overdrive results in increased expression of components of the CaMK pathway, with correspondingly decreased levels of components of the PKA pathway. Based on the temporal and genotype-specific pattern of miRNA expression, miRNAs are likely to be important predictors of disease states, especially when miRNA expression is paired with mRNA expression, and that miRNA/mRNA expression signatures have the potential to be useful in determining the underlying risk associated with cardiac disease progression.
Keywords: Ca2+/calmodulin-dependent protein kinase, signal transduction, genetic polymorphism
polymorphic variation in amino acid position 389 of the human β1-adrenergic receptor (β1-AR) results in a glycine (Gly, minor allele) for arginine (Arg, major allele) substitution, based on a C→G at nucleotide 1165 (15). Several studies in humans or in transgenic animals have demonstrated that this variation is associated with markedly different pharmacologic and biologic effects including clinical responses to certain beta-blockers (12), degrees of ischemia-reperfusion injury (1), exercise capacity (32), risk of hypertension (2), and heart failure associated left ventricular remodeling (29). Depending on the degree of overexpression of each variant in transgenic mice, there are marked differences in myocardial disease progression, with Arg389 mice progressing to cardiac dysfunction at a more rapid rate (14). By 6 mo of age, animals expressing the β1-Arg389 but not the β1-Gly389 variant develop signs of desensitization to β-agonist stimulation of contractility and adenylyl cyclase (17). The pharmacological phenotypic differences at 3 mo are likely the result of more efficient coupling of β1-Arg389 to Gαs and adenylyl cyclase stimulation (17), a higher affinity for catecholamine agonist including norepinephrine (15, 34) and/or a larger number of constitutively active receptors (12, 34). By 6 mo of age the higher degree of signal transduction in Arg vs. Gly animals has resulted in a greater degree of β-adrenergic desensitization (17). In addition, at 6 mo, β1-Arg389 but not Gly overexpressor mice exhibit induction of the “fetal” gene program (FGP) (17), a set of molecular markers associated with hypertrophic cardiomyopathies in murine models and humans. Finally, at 9 mo of age, Arg but not Gly β1-AR overexpressor mice develop a decompensated, dilated cardiomyopathy with increased systolic and diastolic volumes and markedly reduced echocardiographically measured fractional shortening (FS) (17).
These findings indicate that increased receptor functionality in β1-Arg389 overexpressor mice results in differential changes in gene expression that may cause differences in myocardial phenotype observed in older animals. To investigate these differences, Swift at al. (27) performed genome-wide mRNA expression profiling in 3 mo old transgenic mice overexpressing each of the β1389 AR variants. Overexpression of the β1-Arg389 variant resulted in a greater number of uniquely regulated genes that were specific to apoptosis, inflammation and extracellular matrix (27) categories. This observation suggests that β1-Arg and Gly389 ARs may access different downstream signaling pathways, i.e., that there may be qualitative differences in post receptor signaling between the two receptor variants that have pathophysiologic implications. Such differences would not be unprecedented, as this is the case for Gαs-coupled β1-and β2-ARs (36).
microRNAs (miRNAs or miR; small, 20- to 22-nucleotide RNAs) have been shown to be important regulators of gene expression (7). miRNAs can regulate gene expression by promoting RNA degradation or translation suppression, and can regulate development and disease processes (reviewed in Ref. 21). It is well documented that miRNAs are differentially regulated in and can be important modulators of cardiovascular disease (reviewed in Refs. 7, 21, 23). As shown in a recent study in which miRNA expression normalized in response to left ventricular assist device therapy, while expression of the vast majority of mRNAs did not mirror functional improvements (16) miRNAs maybe better molecular markers of the myocardial stress response than are mRNAs.
In the current study we performed a comprehensive analysis of mRNAs and miRNAs expressed in the hearts of 3 and 6–8 mo old β1-Arg389 and β1-Gly389 overexpressor transgenic mice, to determine if abnormal phenotype-associated differential gene dysregulation observed between the two models reflects qualitative (unique to the receptor variant) or quantitative (related to the degree or duration of signaling and common to both variants) differences. Based on the composite array results, miRNA-mRNA target prediction was performed using bioinformatic approaches.
The results indicate that differential gene expression in Arg vs. Gly animals observed at 3 mo are associated with the early development of cardiomyopathy in the Arg but not the Gly model. However, the differences in gene expression are minimized as disease progresses, such that by 6–8 mo changes in gene expression are similar. Furthermore, for both Arg and Gly animals, there is an increase in the expression of constituents of CaMK pathways with a corresponding downregulation in PKA pathways that is temporally and phenotypically related. Importantly, we also show that changes in miRNA and mRNA expression at 3 mo can precede changes in the pathologic phenotype observed at 6–8 mo, and that these early gene expression changes may therefore be useful as predictors of myocardial disease progression.
METHODS
Animal models.
Generation of β1-AR variant transgenic animals was as described previously (17). β1-Arg389 and β1-Gly389 transgenic mice were studied at two time points, 3 and 6–8 mo of age. For each genotype and time point, n = 4–8 animals were used in all RT-PCR and echocardiographic measurements. We used n = 2–4 randomly selected animals for each genotype and time point in miRNA and mRNA array experiments and compared with nontransgenic β1-Arg389 and β1-Gly389 littermate controls. A total of 13 β1-Arg389 and 10 β1-Gly389 animals were investigated, along with 16 littermate controls. Left ventricle was used in all molecular experiments.
RNA extraction and RT-PCR.
Total RNA was extracted using the Mirvana kit (ABI) according to manufacturer's condition. For mRNA detection 0.5 μg of RNA were reverse transcribed into cDNA using I-script (Bio-Rad) and RT-PCR was performed as previously described (25). The primers are described below, and expression was normalized to 18S rRNA (primer sequence described in Ref. 25).
miRNA reverse transcription was performed using the TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems) according to manufacturer's recommendations. We combined 5 ng of miRNA with dNTPs, MultiScribe reverse transcriptase, and the primer specific for the target miRNA. The resulting cDNA was diluted 15-fold and used in PCR reactions. PCR was performed according to manufacturer's recommendations (Applied Biosystems). Briefly, cDNA was combined with the TaqMan assay specific for the target miRNA, and PCR reaction was done using the ABI7300. α-Myosin heavy chain (MyHC) forward (F): 5′ cccaggatctctggattggtct; αMyHC reverse (R): 5′ggcaggaagaggagtagcaga; βMyHC F: 5′ gagcattctcctgctgtttcctt; βMyHC R: 5′ ctcctctgctgaggcttccttt; sarcoplasmic reticulum (SR) via a sarcoplasmic reticulum Ca2+ ATPase (Serca) F: 5′ gcattgcagtctggatcatcaaca; Serca R: 5′ gccaccatgaactgggtcatt; atrial natriuretic factor (ANF) F: 5′ gccggtagaagatgaggtcatg; ANF R: 5′ gcttcctcagtctgctcactca; B-type natriuretic peptide (BNP) F: 5′ cgctgggaggtcactcctat; BNP R: 5′ gctctggagactggctaggactt.
Echocardiography.
Cardiac function was assessed essentially as described previously (33). Mice were lightly sedated with isoflurane, and cardiac function was analyzed by a 2D-transthoracic echocardiography using a Visual Sonics Vevo 770 high-resolution ultrasound imager equipped with a 35-MHz transducer. Heart rates were maintained >500 beats/min. Parasternal long axis and multiple short axis B-mode videos and M-mode images (at the level of the midpapillary short axis) were routinely acquired. Determination of FS and ejection fraction (EF) values was performed off-line and in a blinded fashion.
miRNA arrays.
miRNA expression analysis was performed by Dharmacon (Lafayette, CO) using arrays based on the Sanger miRBase 10.1 database, capable of detecting 567 miRNAs. Statistical analysis was done using ANOVA.
mRNA arrays.
mRNA expression analysis was performed by the University of Colorado Denver array core facility using Affymetrix Mouse GeneChip 1.0 ST version 1, capable of detecting 28,000 probes. Statistical analysis was done using ANOVA. A P value of 0.05 was used as an initial cutoff followed by a second cutoff that excluded fold differences <1.25-fold.
β1-AR measurements.
β1-AR receptor density was determined by radioligand binding studies with 125[I]cyanopindolol ([125I]CYP), using right ventricular tissue obtained from transgenic mice and nontransgenic littermates, as previously described (5).
Network of diseases.
Only genes associated with diseases, as defined by Ingenuity, were included in generating networks. Disease gene networks were created and presented as figures in which each gene is represented by a geometrical symbol with the linkage between genes and a disease represented by a line connecting the two symbols. With this approach a line exists between two genes only if the same disease shares both genes. For each group of genes within a disease, a central node is created and labeled with the disease type and connected to the genes within that disease. Genes that are associated with multiple diseases are labeled as “complex.” For each complex gene a line to each of its associated disease is generated. Finally, each of the networks are color-coded based on disease and are visualized using Cytoscape an open-source, platform-independent environment for visualizing biological networks (6).
Statistical analyses.
For RT-PCR experiments and echocardiographic measurements, all analyses were performed using an ANOVA with a Bonferroni correction. An ANOVA with P < 0.05 in a two-tailed distribution was considered to be statistically significant. Statistical analyses of mRNA and miRNA arrays are described in the methodological section for each array. χ2-tests were used to compare the number of up- or downregulated mRNAs in Arg vs. Gly animals. Regression analysis was performed to determine the relationship between gene expression and myocardial dysfunction.
RESULTS
β1389-Arg animals show signs of myocardial dysfunction at 6–8 mo age.
Previous studies have shown that mice overexpressing the β1-Arg389 receptor begin to develop signs of myocardial contractile dysfunction at 6 mo of age and, by 9 mo, have typically developed a severe dilated cardiomyopathy. By comparison, Gly389 overexpresor animals typically do not develop at dilated cardiomyopathy until ≥9 mo (4, 8). In β1-389Arg overexpressor mice, changes in gene expression that are likely related to the development of myocardial dysfunction and dilated cardiomyopathy are present at 6–8 mo, but not detectable at 3 mo. Therefore, we chose 3 and 6–8 mo time points as being potentially informative in our attempt to uncover changes in mRNA and miRNA expression that are a predictive of functional changes.
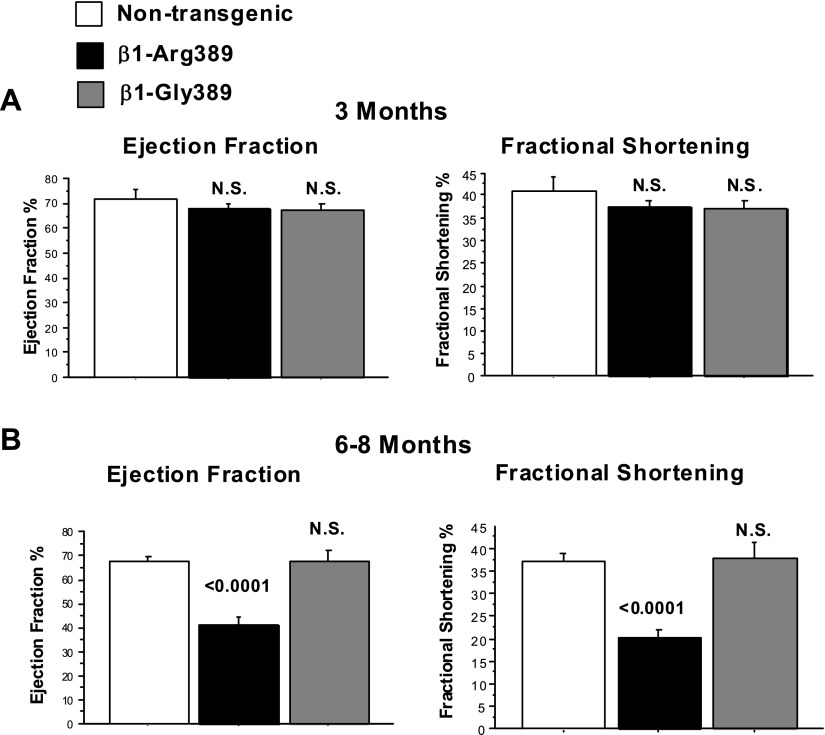
As shown in Fig. 1A, at 3 mo of age neither β1-389 polymorphic variant induces myocardial dysfunction and left ventricular structural remodeling as measured by FS or EF. However, by 6–8 mo β1-Arg389 animals have significantly decreased EF and FS, whereas β1-Gly389 animals show no signs of dysfunction or remodeling (Fig. 1B). To verify receptor expression equivalency, at 3 mo of age, right ventricular β1-AR receptor density averaged 600–700 fmol/mg in both Arg and Gly overexpressors, with both being increased ∼25-fold compared with nontransgenic littermates (data not shown).
Fig. 1.
Overexpression of β1-Arg389 results in myocardial dysfunction and remodeling in 6–8 mo animals. Echocardiograms were performed in animals over expressing β1-Arg389 or β1-Gly389 at 3 (A) and 6–8 mo (B) (n = 4). N.S., not significant.
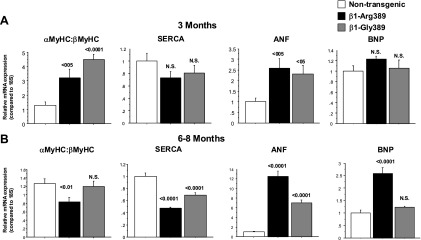
Changes in contractile protein gene expression from adult to the myocardial pathology-associated FGP are shown in Fig. 2. With the exception of ANF, neither β1-Arg389 nor β1-Gly389 3 mo old animals express pathology-associated changes in gene expression profile, such as a decrease in αMyHC/βMyHC ratio or SERCA expression, or an increase in BNP. In contrast, in 6–8 mo old β1-Arg389 animals significant changes in MyHC isoform, SERCA and BNP gene expression were observed along with the increase in ANF. In contrast, 6–8 mo old β1-Gly389 animals showed only a decrease in SERCA and an increase in ANF (Fig. 2) but no decline in the αMyHC/βMyHC ratio or increase in BNP. Interestingly, both 3 mo old Arg389 and Gly389 animals exhibited an increased αMyHC/βMyHC ratio, mostly due to upregulation of αMyHC (data not shown).
Fig. 2.
Expression of the hypertrophic gene program is activated in older β1-Arg389 transgenic animals. Total RNA was extracted from left ventricle of transgenic and control animals and analyzed by RT-PCR. Gene expression in transgenic animal groups was compared with control groups at 3 (A) and 6–8 mo (B); 6–8 mo old n = 6, 3 mo old n = 4.
Genome-wide expression profile of 3 and 6–8 mo old β1-Arg389 or β1-Gly389 animals reveals similarities between models, but more genes are regulated exclusively in Arg389 overexpressors.
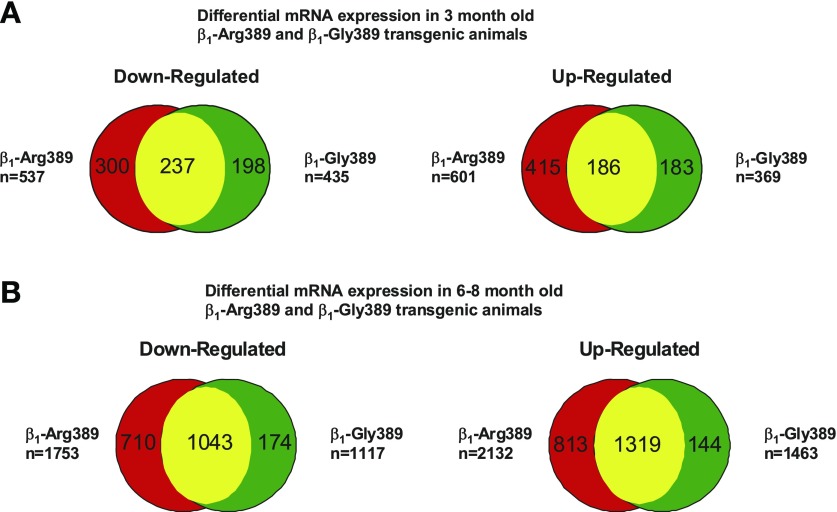
To compare changes in gene expression in younger, 3 mo old animals, mRNA array experiments were performed. Although mRNA array data have been previously reported for 3 mo age group mice (27), we thought it was critical to repeat analysis since genotype drifting, use of different array platforms, and other factors can result in model-dependent differences in gene expression. Furthermore, it was considered critical to perform the mRNA and miRNA interaction analysis from the same samples. Using a statistical cutoff of P < 0.05 as determined by ANOVA with a subsequent fold change cutoff of ≥1.25 compared with littermate controls, we considered ∼4,000 genes either up- or downregulated between the two β1-AR overexpressor models. As shown in Fig. 3A, in 3 mo old animals 601 genes were upregulated in β1-389-Arg, while 369 genes were upregulated in β1389-Gly [P < 0.0001, χ2, compared with wild type (WT)] mice, with 186 genes being commonly upregulated. Similarly, at 3 mo of age 537 genes were downregulated in β1-389-Arg, while 435 genes were downregulated in β1-389-Gly animals (P < 0.0001), with 237 genes being commonly downregulated in both genotypes.
Fig. 3.
Distribution of genes differentially regulated in 3 (A) and 6–8 (B) mo old β1-Arg389 and β1-Gly389 transgenic animals. Numbers in each partition represent the number of commonly or uniquely regulated genes in each animal model. The size of each partition is not to scale. The total number of regulated genes is defined on the left and right side of the diagrams.
In 6–8 mo old animals, 1,753 and 1,117 genes were downregulated in β1-Arg389 and β1-Gly389 hearts, respectively (P < 0.0001, χ2, compared with WT); 1,043 were downregulated in both models. The number of genes downregulated exclusively in Arg or Gly mice was 710 and 174, respectively. In β1-Arg389 overexpressors 2,132 genes were upregulated, while 1,463 genes were upregulated in β1-Gly389 animals (P < 0.0001) with 1,319 genes being upregulated in common (Fig. 3B). Genes upregulated exclusively in Arg vs. Gly animals were 813 and 144 (P < 0.0001), respectively. Paired comparisons showed that changes in gene expression are dependent on the genotype (P < 0.0001, 3 mo; P < 0.005 6–8 mo).
The percentage of mRNAs that change between β1-Arg389 and β1-Gly389 mice at 3 and 6 mo age is shown in Table 1. These analyses show that in β1-Arg389 mice, at either 3 or 6–8 mo, a substantial number of genes are uniquely up- or downregulated, a result concordant with the previous gene expression study performed in 3 mo β1-Arg389 vs. Gly overexpressor animals (27). In contrast, the majority of regulated genes in Gly389 mice are also regulated in Arg389 overexpressors. Interestingly, 70% of genes upregulated and 74.8% of genes downregulated in 3 mo old β1-Arg389 animals are also regulated in 3 and/or 6–8 mo old β1-Gly389 animals, suggesting that differences in gene expression are not unique to a specific polymorphism but are dependent on the state of disease progression. A comprehensive list of regulated genes in both polymorphisms and at all time points is given in Supplemental Table S1.1
Table 1.
Relative comparison of the percentage of up- and downregulated mRNAs in different genotypes and time points
| Upregulated | ARG 3 | ARG 6 | Upregulated | GLY 3 | GLY 6 |
| ARG 3 | 8.8% | 16.84% | GLY 3 | 12.62% | 9.6% |
| ARG 6 | 16.84% | 74.35% | GLY 6 | 9.6% | 77.78% |
| Upregulated | ARG 3 | GLY 3 | Upregulated | ARG 6 | GLY 6 |
| ARG 3 | 52.82% | 23.72% | ARG 6 | 35.76% | 57.95% |
| GLY 3 | 23.72% | 23.46% | GLY 6 | 57.95% | 6.27% |
| Downregulated | ARG 3 | ARG 6 | Downregulated | GLY 3 | GLY 6 |
| ARG 3 | 8.5% | 19.35% | GLY 3 | 16.3% | 13.7% |
| ARG 6 | 19.35% | 72.15% | GLY 6 | 13.7% | 70% |
| Downregulated | ARG 3 | GLY 3 | Downregulated | ARG 6 | GLY 6 |
| ARG 3 | 41.45% | 32.45% | ARG 6 | 36.74% | 54.35% |
| GLY 3 | 32.45% | 26.1% | GLY 6 | 54.35% | 8.91% |
The table shows percentage of genes uniquely or commonly regulated in different conditions.
Pathway analysis of mRNA changes.
To identify the representative biological/disease functions of genes differentially regulated in the two animal models of β1-AR overexpression, up- or downregulated genes were separately subjected to Ingenuity Pathway Analysis. In 6–8 mo old animals, genes for metabolic pathways were highly up- or downregulated in both transgenic models, whereas genes involved in cell growth, apoptosis, and immune response/disease pathways were substantially upregulated only in both models (Supplemental Table S2). In 3 mo old animals, genes related to metabolism, transcription regulation, and cardiovascular disease (including hypertrophy, myocardial infarction, and cardiac dilation) were proportionally more downregulated than in older Arg and Gly animals. The other pathway categories displayed substantial similarity in the proportion of regulated genes between 3 and 6–8 mo old β1-Arg389 overexpressor animals (Supplemental Tables S2 and S3). Biological function analysis of genes that are commonly regulated in all models revealed genes associated with different biological functions, with the greatest number being associated with the cardiovascular disease pathway (Supplemental Table S4).
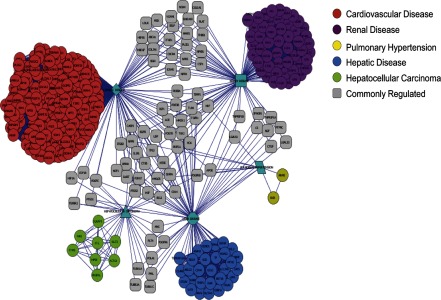
To determine if genes differentially regulated in the β1-Arg389 and β1-Gly389 overexpressor animals were also involved in other pathologic processes, a network of diseases was generated (Fig. 4) using the results of toxicology function from Ingenuity Pathway Analysis. This analysis is restricted to kidney, liver, and cardiovascular diseases. As shown in Fig. 4, Supplemental Fig. S1, and Supplemental Table S5, regulated genes are involved in various diseases, with the majority of regulated genes being involved in cardiac disease. There are several genes that are commonly regulated among the different diseases. Interestingly, pulmonary hypertension-related genes are mostly upregulated in 6 mo old β1-Arg389 animals, although 3 mo old β1-Gly389 animals show upregulation of three genes involved in pulmonary hypertension (see discussion). Downregulated genes strongly correlate with cardiac disease. It is also interesting to notice that although a network exists between cardiac and kidney diseases in most of the analyzed time points for both polymorphisms, only two downregulated genes in 6 mo β1-Gly389 animals are related to kidney disease and are not connected to the other diseases in the network.
Fig. 4.
Network of disease analysis of differentially regulated genes. Genes upregulated in 6 mo β1-Arg389 transgenic animals were subjected to Ingenuity Pathway Analysis. Based on the toxicology function results, a network of diseases was generated. Diseases are color coded according, and genes commonly regulated in multiples diseases are linked by a line.
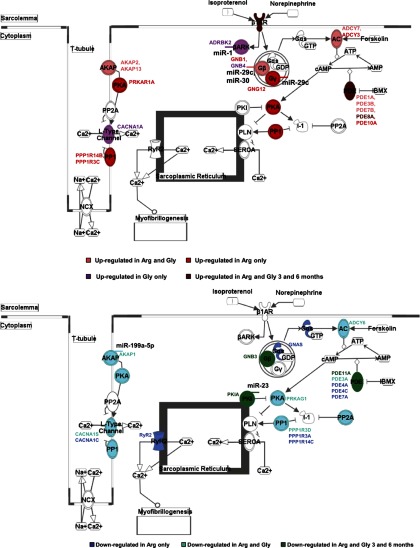
To understand the cellular context of the differentially expressed genes, pathway analysis was also performed. As shown in Fig. 5 and Supplemental Fig. S2, several genes in various pathways are upregulated in 6–8 mo Arg and Gly animals, while some genes are specifically regulated in each model. Also shown in the figures are genes that are differentially regulated in all models and time points. Specifically, Rock, IL6R, PLC4B, and PDE8A are upregulated in the cardiac hypertrophy and β-AR pathways, and Calpain 3, PDE11A, GNB3, α1-AR, and protein kinase A inhibitor (PKIA) are downregulated in these pathways. Although not shown, the glutamate receptor gene, Grinc2c, is upregulated, while calsequestrin 1 and myosin heavy chain 7B are downregulated in all models and time points as noted in the calcium pathway (data not shown).
Fig. 5.
Canonical pathway analysis of differentially regulated genes. Genes up- or downregulated in β1-Arg389 or β1-Gly389 transgenic animals were subjected to Ingenuity Pathway Analysis. The β-adrenergic receptor (AR) pathway is shown. Genes are color coded according to regulation in Arg or Gly only, in both animals at 6–8 mo old or in all ages and genotypes. microRNAs (miRNAs) that can potentially regulate specific genes are annotated in the figure.
Due to the small(er) number of genes uniquely upregulated in Arg and/or Gly 3 mo old animals, pathway analysis is not presented. However, these results revealed an increase in expression of genes involved in mitogen-activated protein kinase (MAPK) signaling and in the transcription factor, Hand2. Interestingly, CaMKIIδ expression was downregulated in these animals, whereas CaMKK genes were upregulated in 3 mo Arg and 6 mo Arg and Gly animals.
RT-PCR measurements of mRNA expression.
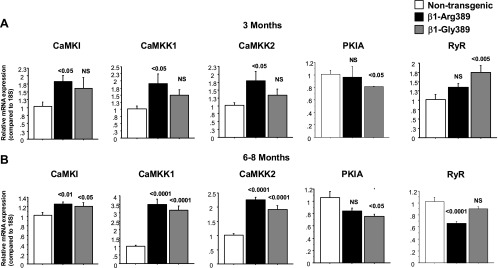
To determine the fidelity of array measurements in a larger sample number, expression of a subset of regulated genes was analyzed by RT-PCR. These genes were selected for their roles in pathways known to be activated by β1-AR signaling. As shown in Fig. 6, CaMKI, CaMKK1, and CaMKK2 were upregulated in 6–8 mo Arg and Gly overexpressor animals and in 3 mo Arg animals. In contrast, the PKIA gene was downregulated in 3 and 6–8 mo old Gly overexpressors. These data suggest an increase in components of the CaMK pathway in the Arg genotype and an increase in components of the PKA pathway in the Gly genotype. Expression of the ryanodine receptor (RyR2) gene was also analyzed. There was a significant increase in RyR2 mRNA in younger Gly animals and a trend toward an increase in younger Arg animals. Interestingly, in 6–8 mo old Arg animals there was a significant decrease in RyR2 expression. Furthermore, expression of the transcription factor NFATc4, which with the phosphatase calcineurin plays an important role in the hypertrophic process (35), is increased in 6–8 mo old Arg and Gly animals.
Fig. 6.
Differential expression of genes in Arg and Gly animals. Expression of a subset of mRNAs differentially regulated in the array analysis was analyzed by RT-PCR. A: 3 mo old animals. B: 6–8 mo old animals.
Changes in gene expression correlate with myocardial dysfunction.
To determine if there is an association between changes in gene expression and left ventricular functional and structural changes, regression analysis between left ventricular remodeling as determine by EF and changes in gene expression measured by RT-PCR was performed in 6 mo old WT control (n = 11), β1-Gly389 (n = 6) and β1-Arg389 (n = 9) mice. As shown in Table 2, the 8 of 12 genes measured showed a significant correlation between changes in gene expression and EF, and good correlations were demonstrated for left ventricular ejection fraction (LVEF) decrease and SERCA downregulation and CaMKK1 upregulation.
Table 2.
Regression analysis of changes in gene expression and myocardial dysfunction as determined by %EF
| αMyhC | SERCA | βMyhC | Sk-Act | ANF | BNP | CaMKI | CaMKK1 | CaMKK2 | PKIA | NFATc4 | RyR2 | NFATc4 | RyR2 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| R | −0.12 | −0.60 | 0.44 | 0.05 | 0.58 | 0.52 | 0.35 | 0.62 | 0.54 | −0.16 | 0.40 | −0.42 | 0.40 | −0.42 |
| P | 0.58 | 0.001 | 0.026 | 0.80 | 0.002 | 0.008 | 0.089 | 0.001 | 0.006 | 0.44 | 0.05 | 0.04 | 0.05 | 0.04 |
R and P values are shown. EF, ejection fraction.
miRNAs are regulated similarly in older β1-Arg389 and β1-Gly389 animals and differentially in younger animals.
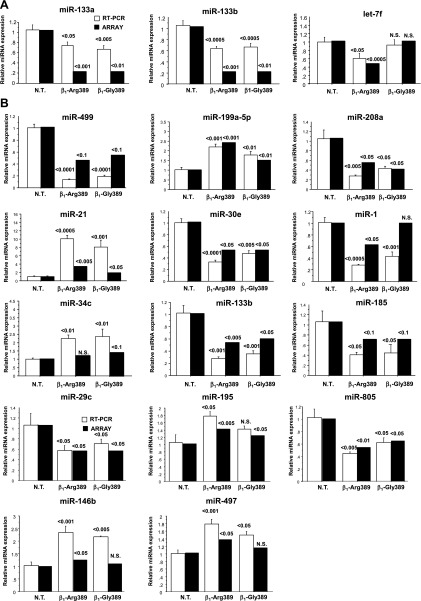
To analyze patterns of miRNA expression, array analysis was performed in Arg389 and Gly389 overexpressor mice at both time points. As shown in Table 3, at 3 mo, miR-133a and miR-133b were the only two miRNAs statistically changed (both downregulated) in β1-Gly389 animals, whereas in Arg389 overexpressors, a total of 8 miRNAs were differentially regulated. miR-133a and b were the only commonly regulated miRs, while miRs let-7f, let-7b, miR-805, miR-615–3p, miR-449a, and miR-711 were exclusively regulated in Arg overexpressors. In contrast, the great majority of changed miRNAs in older animals were commonly regulated.
Table 3.
miRNAs differentially regulated in 3 mo or 6-8 mo old β1-Arg389 and β1-Gly389 transgenic animals
| Upregulated miRNAs 3 mo old | ||
| β1389-Arg | commonly regulated | β1389-Gly |
| mmu-miR-615-3p | ||
| mmu-miR-449a | ||
| mmu-miR-711 | ||
| Downregulated miRNAs 3 mo old | ||
| β1389-Arg | commonly regulated | β1389-Gly |
| mmu-let-7f | mmu-miR-133a | |
| mmu-let-7b | mmu-miR-133b | |
| mmu-miR-805 | ||
| Upregulated miRNAs 6-8 mo old | ||
| β1389-Arg | commonly regulated | β1389-Gly |
| mmu-miR-497 | mmu-miR-195 | mmu-miR-690 |
| mmu-miR-214* | mmu-miR-199a-3p | |
| mmu-miR-199b* | mmu-miR-199a-5p | |
| mmu-miR-23a | mmu-miR-199b | |
| mmu-miR-23b | mmu-miR-21 | |
| mmu-miR-146b | mmu-miR-34c | |
| Downregulated miRNAs 6-8 mo old | ||
| β1389-Arg | commonly regulated | β1389-Gly |
| mmu-miR-378 | mmu-miR-133a | mmu-miR-293 |
| mmu-miR-133a* | mmu-miR-491 | |
| mmu-miR-133b | mmu-miR-669a | |
| mmu-miR-19a* | ||
| mmu-miR-208a | ||
| mmu-miR-29c | ||
| mmu-miR-30b | ||
| mmu-miR-30e | ||
| mmu-miR-378* | ||
| mmu-miR-805 | ||
| mmu-miR-1 | ||
The table describes all microRNAs (miRNAs) specifically and commonly regulated in each animal model. P value cutoff for described miRNAs (RT-PCR or arrays) was <0.05.
RT-PCR measurements of miRNA expression.
To validate the miRNA expression array results, expression of a subset of miRNAs was measured by RT-PCR. As shown in Fig. 7, directionality of expression was essentially the same between array and RT-PCR methodologies. However, of the miRNAs analyzed, the amplitude and significance of the measured differences were greatly increased with RT-PCR. Moreover, a few miRNAs that were differentially expressed only in β1-Arg389 or β1-Gly389 by arrays (miR-1 and miR-34, respectively) were concordantly regulated in both models when tested by RT-PCR. In general, however, the RT-PCR data support the validity of miRNA arrays as a methodology for measuring broad-based miRNA expression.
Fig. 7.
miRNA expression analysis was confirmed by RT-PCR. Expression of a subset of miRNAs differentially regulated in the array analysis was analyzed by RT-PCR. A: 3 mo old animals. B: 6–8 mo old animals. N.T., nontransgenic.
Differential expression of mRNAs potentially targeted by miRNAs differentially regulated in β1-Arg389 and β1-Gly389 transgenic mice.
To analyze potential mRNA targets for regulated miRNAs, mRNA and miRNA array data sets were simultaneously analyzed using the miRNA and mRNA integrated analysis (MMIA) software that integrates mRNA and miRNA expression data based on miRNA-predicted targets (19). mRNAs and miRNAs that were both differentially and antithetically regulated were input into the program. mRNAs predicted to be regulated by miRNAs according to the MMIA were subjected further to Ingenuity Pathway Analysis and clustered into common biological/disease themes for Arg and Gly models, described in Supplemental Table S6.
Because of the limited number of miRNAs differentially regulated in 3 mo old animals, only 44 up- and 14 downregulated genes were predicted to be potential miRNA targets (Supplemental Table S7). As shown in Fig. 8 and Supplemental Table S7, in 6–8 mo animals, far more (550 in β1-Arg389 and 366 in β1-Gly389) upregulated mRNAs were predicted miRNA targets. Similarly, 240 β1-Arg389 and 161 β1-Gly389 downregulated mRNAs were predicted miRNA targets. miRNA target prediction for various genes are annotated in the pathways in Fig. 4 and Supplemental Fig. S1. As shown in Supplemental Fig. S2, miRNAs known to be regulated in various models of cardiac hypertrophy (miR-1, miR-133, miR-29, miR-30) are predicted to target the expression of several genes in the cardiac hypertrophic pathway. The miR-30 and miR-29 families are predicted to target expression of CaMKK2, which suggests that downregulation of these miRNAs in the β1-AR overexpressor animals can be involved in the upregulation of CaMKK2 expression.
Fig. 8.
Distribution of potential miRNA-target mRNAs regulated in 6–8 mo old β1-Arg389 and β1-Gly389 transgenic animals. Numbers in each partition represent the number of commonly or uniquely regulated targets in each animal model. The size of each partition is not to scale. The total number of regulated target mRNAs is defined on the left and right side of the diagrams.
DISCUSSION
We analyzed changes in mRNA and miRNA expression in response to β1-AR overdrive as a function of disease progression that varied by genotype and time. The data indicate that 1) differences in gene expression in β1-Arg389 and β1-Gly389 transgenic mice are largely quantitative and associated in a time dependent way with the development of cardiomyopathy; 2) although relatively few, there appear to be a limited number of qualitative changes in mRNA and miRNA expression found exclusively in Arg389 overexpressors; 3) chronic β1-AR overdrive results in increased expression of components of the CaMK pathway with a corresponding decreased expression of components of the PKA pathway, in both Arg389 and Gly389 genotypes but with Gly animals developing these changes only at the later time point; 4) an miRNA signature, associated with putative target mRNAs, can potentially be used as a refined determinate of risk of cardiac disease.
Qualitative and quantitative differences in the expression profiles of 3 and 6–8 mo old mice.
Early development of cardiomyopathy is characteristic of β1-Arg389 overexpressor animals. In contrast, myocardial dysfunction only occurs in β1-Gly389 animals at time points >6–8 mo (4). Both Arg389 and Gly389 overexpressor animals demonstrated time-dependent dysregulation of mRNA and miRNA expression with Arg389 animals showing a greater number of differentially expressed mRNAs and miRNAs at all time points. Given the greater degree of pathophysiology for the Arg overexpressor mice, this is not unexpected.
For mRNA expression, at 3 mo the majority of regulated mRNAs were in β1-Arg389 animals, while at 6–8 mo most of the regulated mRNAs were in both genotypes. Moreover, >70% of the genes differentially regulated in 3 mo old β1-Arg389 animals are also regulated in 6–8 mo β1-Gly389 animals. This suggests that the β1-Arg389 animals develop a number of changes in gene expression prior to Gly animals, but as time-dependent excessive β1-AR signaling occurs in Gly hearts these differences diminish. In β1-Arg389 animals a likely consequence of this increase in adrenergic signaling is the development of myocardial systolic dysfunction and ventricular dilatation, collectively measured as the remodeling index LVEF. In this regard evidence of expression of the pathologic/FGP was present at 6–8 mo in Arg animals who had evidence of a dilated cardiomyopathy by LVEF, and partially present in Gly animals who had no evidence of a cardiomyopathy. The appearance of an FGP signature in Arg animals at 6–8 mo is identical to changes that have been described in humans with dilated cardiomyopathies, which then partially reverses with β-receptor blocker treatment that improves LVEF (14). However, β1-Gly389 overexpressing mice have been shown to develop myocardial dysfunction at 9 mo (4, 8), so the partial expression of the FGP at 6–8 mo in Gly animals likely would have presaged the development of a dilated cardiomyopathy. The miRNA studies also support this interpretation; at 6–8 mo changes in miRNA expression are very similar in the two animal models, suggesting that these changes precede the development of myocardial dysfunction. These observations argue that the majority of gene dysregulation in the two models is quantitatively different with few qualitative changes, with the hypofunctional β1-Gly389 isoform model requiring 3–5 mo additional time to achieve the degree of gene dysregulation observed in Arg animals at 3 mo.
Analysis of network of diseases revealed several common diseases regulated in all models. The pulmonary hypertension gene set was specific to 6–8 mo old β1-Arg animals. Although it was also noted in 3 mo β1-Gly389 animals, that correlation was due to upregulation of β2-AR. Of the three genes involved in pulmonary hypertension, two (ApoE and MIF) are also upregulated in 3 mo β1-Arg389 and 6 mo β1-Gly389 animals; however, upregulation of these two genes does not reach the necessary P value to be characterized as pulmonary hypertension on the disease list generated by Ingenuity Pathway. Upregulation of β2-AR, ApoE, and MIF do meet the P value necessary to establish pulmonary hypertension as a regulated disease in 3 mo old β1-Gly389. In contrast, 10 upregulated genes in 6 mo old β1-Arg389 are involved in pulmonary hypertension. It is not clear if this is a result of disease progression in the β1-Arg389 animals or a specific characteristic of this polymorphism. Of the downregulated genes, 6–8 mo β1-Gly389 animals do not show a network connection between kidney and other diseases. This suggests a characteristic of the β1-Gly389 polymorphism just before the onset of cardiomyopathy since 3 mo β1-Arg389 and β1-Gly389 animals (before any signs of myocardial pathology) show a connection with kidney disease that involves genes that are commonly downregulated in 6–8 mo β1-Arg389. Lack of regulation of these genes in older β1-Gly389 animals suggests a unique regulation of gene expression in response to this polymorphism that may not be directly related to myocardial disease progression.
Overexpression of β1-AR results in increased expression of genes that negatively regulate the PKA pathway and positively regulate the CaMK pathway.
β1-Arg389 and β1-Gly389 overexpressor animals offer unique model systems to study differences in myocardial gene expression in animals that develop early vs. late signs of cardiomyopathy via signaling through variants of the same receptor. Differential regulation of several pathways was observed when these comparisons were made. CaMKK1 and -2 were upregulated in Arg389 mice at 3 and 6 mo and in 6 mo old Gly389 animals, suggesting that CaMKK may play an early important role in myocardial disease development. Interestingly, miR-30b and -e and miR-29c were both downregulated in older Arg389 and Gly389 animals and are predicted to target CaMKK2 and could therefore have an important effect in disease development. We have recently shown that the CaMKII is responsible for the increase in cardiac myocyte size and expression of the pathologic FGP (26). Unpublished results suggest that pharmacological inhibition of CaMKK prevents β1-AR-mediated induction of the pathologic FGP (data not shown) in neonatal rat ventricular myocytes. These results underscore the importance of the CaMK pathway in the pathologic effects of β1-AR overstimulation.
Recent data (24) show that PKA protects against expression of the fetal gene isoforms by preventing phosphorylation of the histone deacetylase HDAC5 through activation of a phosphatase. Furthermore, an increase in αMyHC levels in response to cAMP treatment has been previously observed by others (10), similar to the increase in αMyHC/βMyHC ratio observed in 3 mo animals. These results indicate that in response to increased β1-AR signaling delivered prior to the onset of contractile dysfunction and ventricular chamber dilatation, upregulation of αMyHC may occur, likely mediated through the cAMP/PKA pathway. These results suggest that the cAMP/PKA pathway may be beneficial. In support of this hypothesis, array results showed increased expression of genes that negatively regulate PKA activity [regulatory subunit of PKA (6 mo Arg389 animals) and RSK (6 mo Gly389 animals) (9)]. Furthermore, downregulation of the PKA inhibitor PKIA was observed in Gly389 animals while no downregulation was observed in Arg389 animals. This could be a reflection of the initial onset of cardiomyopathy in Arg389 animals where an excessive stimulation resulting in desensitization of the receptor associated with an increase in pathologic pathways that can ultimately result in decreased PKA activity. Moreover, although a transgenic animal overexpressing adenylyl cyclase (AC) 5 displays increased contractility, it does not develop myocardial dysfunction (28), and our results show that expression of the cardiac specific AC6 is downregulated in 6 mo β1-Arg389 and β1-Gly389 animals. These observations are consistent with the hypothesis that downregulation of the downstream AC/cAMP/PKA signaling axis is biologically and physiologically detrimental, while PKA-independent signaling including CaMKK and CaMK is detrimentally upregulated. This would imply that, overall, PKA dependent signaling is beneficial, while PKA-independent β1-AR signaling predisposes to the development of cardiomyopathy.
Interestingly, RyR2 is downregulated exclusively in β1-Arg389 animals, while the L-type Ca2+ channel is upregulated exclusively in Gly389 animals. The decrease in RyR2 expression has also been observed in calsequestrin overexpressing transgenic mice (11), and the decrease in SR Ca2+ release and depressed [Ca2+]i transients have been observed in several models of heart failure (3). These changes affect excitation-contraction coupling and could potentially explain the decreased contractility observed in heart failure (3) that in turn triggers increased and sustained adrenergic activation, a feedback loop that contributes to disease progression (20). Decreased RyR2 receptor levels in the β1-Arg389 model can be a result of adrenergic overdrive in this system, and could play a major role in the genesis of decreased contractility and activation of CaMKII.
Expression of several miRNAs change in response to β1-AR overexpression.
The results of miRNA array experiments suggest that changes in miRNA expression precede changes in mRNA expression and ventricular remodeling. At the 3 mo time point, in β1-Gly389 animals changes in miRNA expression are limited to miR-133a and miR-133b, whereas an expanded set of 8 miRNAs changes expression in 3 mo β1-Arg389 animals. At the 6–8 mo time point, changes in miRNA expression are very similar between the two genotypes, suggesting again that these changes precede changes in mRNA expression and myocardial pathology since several differences in mRNA expression and in contractile parameters are still observed in the two genotypes. Dysregulation of miR-133a and miR-133b at all time points and in both overexpressor genotypes suggests that these miRNAs may be candidate diagnostic biomarkers for preclinical myocardial dysfunction. It also suggests that mRNA targets of miR-133a/b may be a focal point of cardiac disease progression.
Several of the miRNAs regulated in response to β1-AR overexpression (miR-133, miR-1, miR-199a-3p, miR-23, miR-21, miR-30, miR-29) are also differentially regulated in human myocardial failure and in other animal models of myocardial disease (7, 21). This suggests a common myocardial miRNA signature in heart failure that is common to both humans and mouse models. Proper regulation of expression of some of these miRNAs, such as miR-133 (13), is likely to have important effects on reversal of the dilated cardiomyopathy/heart failure phenotype. However, dysregulation of several miRs (miR-669a, miR-690, miR-491, miR-293, miR-214*, miR-133a*, among others) appear specific to this model and may be related to unique aspects of β1-AR overexpression.
MMIA of possible miRNA-targeted mRNAs was performed using the unique data sets from our combined mRNA and miRNA array analyses. As a predicate condition, only mRNAs and miRNAs whose expression changed both significantly and antithetically were subjected to further analysis. Based on the Targetscan 5.1 database, several regulated mRNAs were predicted to be targets for the regulated miRNAs. All miRNAs and possible targets are described in Supplemental Table S7. Some of these mRNAs are known to be involved in the hypertrophic/remodeling process. For example, miR-29 has been demonstrated to have an important role in the regulation of collagen expression and the development of fibrosis; specifically, downregulation of miR-29 results in upregulation of several collagen genes (31), which correlates with increased fibrosis observed in older β1-Arg389 animals (17). As described in Supplemental Table S3, 6–8 mo old β1-Arg389 animals show increased expression of several collagen genes, many of which are predicted targets for downregulated miRNAs (Supplemental Table S7). The miR-30 family is predicted to regulate expression of Smad1, which is upregulated in older β1-Arg389 and β1-Gly389 animals and along with other Smad family members, has been implicated in a wide range of cardiac disorders including fibrosis (37); miR-29c is predicted to regulate expression of NFATc4 (NFAT3), a transcription factor known to be involved in the hypertrophic response (18) and is upregulated in older β1-Arg389 and β1-Gly389 animals; increased expression of miR-195 results in the development of cardiac hypertrophy and dilatation (30) and is predicted to target components of the cardiac hypertrophy and apoptosis pathways (Supplemental Fig. S2); increased expression of miR-21 can also result in the development of fibrosis through regulation of phosphatase and tensin homolog that results in increased expression of matrix metalloproteinase 2 in fibroblasts (22), which is upregulated in older β1-Arg389 and β1-Gly389 animals (Supplemental Table S7). miR-21 has also been recently shown to regulate expression of Smad7, which negatively regulates lung fibrosis (13). The authors show that increased expression of miR-21 results in decreased Smad7 levels and increased fibrogenic activation of pulmonary fibroblasts (13).
Several other miRNAs are predicted to be target expression of regulated genes involved in β1-AR signaling (Fig. 5), apoptosis, and cardiac hypertrophy (Supplemental Fig. S2) pathways. These include miR-23 and miR199a-3p, which are predicted to target Ask1; miR-195 and miR-497, which are predicted to target cRaf; miR-23, miR-21, and miR-199a-3p, which are predicted to target members of the PKC family; miR-30 and miR-29c, which are predicted to target several G proteins; miR-133 and miR-208a, which are predicted to target TGF-βR1 and -2 and TGF-β1, -2, and -3; miR-133, miR-30, and miR-378, which are predicted to target IGF1 and IGFR1; miR-1 and miR-133, which are predicted to target PI3K; and miR-133 and miR-30, which are predicted to target several MEK kinases (for a full description of regulated miRNAs in each time point and genotype and predicted targets that are antithetically regulated in all time points and models please see Supplemental Table S7). Interestingly, the number of mRNAs predicted to be targeted by miRNAs is greater in β1-Arg389 animals than in β1-Gly389 animals, but very few qualitative differences were observed in miRNA expression between the two variants. As described in Table 3 and Fig. 7, of the miRNAs discussed above only miR-23a, miR-23b, and miR-378 are uniquely regulated in β1-Arg389 animals. It is possible that the miRNAs that change at the early time point contribute to the later mRNA expression changes. It is also possible that the quantitative differences in miRNA expression have an effect on mRNA levels. Lastly, because miRNAs can cause translation repression and it is likely that many predicted miRNA targets are regulated by this mechanism, these changes in gene expression may only be detected by protein measurements. Future studies of overexpression or downregulation of these miRNAs coupled with protein measurements will allow us to confirm these potential targets.
Previously, Swift et al. (27) have analyzed gene expression changes in 3 mo old β1-Arg389, β1-Gly389, and adenylyl cyclase 5 (AC5) transgenic animals. The authors identified a set of genes and pathways uniquely regulated at early stages of β1-Arg389 overexpression, prior to the onset of myocardial disease. Moreover, they identified genes uniquely regulated by the cAMP/PKA-dependent pathways through overexpression of AC5 as well as cAMP-independent pathways exclusively regulated in response to overexpression of the β1-AR. However, only early time points were characterized in that study. The main difference between the current study and the Swift et al. study (27) is that the previous study focused on gene expression changes at an early, “preclinical” time point, and between-genotype comparison of gene expression changes and their relationship to cAMP/PKA pathways. The current study analyzes changes in gene expression over time as disease progresses and compares these changes between genotypes to determine if the changes observed at an early stage are related to genotype (Arg or Gly) or to the subsequent development of a cardiomyopathy and LV dysfunction.
Since the onset of a dilated cardiomyopathy is different in β1-Arg389 and β1-Gly389 animals, it is important to determine if this is due to qualitative or quantitative changes in gene expression, or both. Based on a greater number of genes that are commonly regulated in Arg389 and Gly389 animals as they age and on the fact that most genes uniquely regulated in 3 mo β1-Arg389 animals are also regulated in older Gly animals, our results suggest that the observed differences in gene expression between genotypes are primarily quantitative. Moreover, of the programmed cell death genes uniquely regulated in 3 mo β1-Arg389 animals [Table 2, Swift et al. (27)], 50% are regulated in our 6–8 mo old β1-Gly389 animals (data not shown). As shown in Supplemental Table S1 and Supplemental Fig. S3, no upregulation of collagen genes was detected in 3 mo β1-Gly389 animals, whereas 16 collagen genes are upregulated in 6 mo β1-Gly389 animals. Of these, eight are upregulated in 3 and 6 mo old β1-Arg389 animals, and eight are upregulated in 6 mo old β1-Arg389 animals. These results strongly suggest that the development of fibrosis occurs in both genotypes but at an earlier time point in β1-Arg389 animals and corroborate the notion that differences in gene expression between the two polymorphisms are dependent on different rates of disease progression in the two models.
In summary, the results presented herein indicate that differential gene expression in Arg389 vs. Gly389 animals observed at 3 mo are associated with the early development of cardiomyopathy in the Arg model. However, these differences are minimized as disease progresses, such that at 6–8 mo changes in gene expression are similar between the two β1-AR models. The current study is limited by a lack of protein expression data and protein target validation of miRNAs. Future studies will address miRNA target proteins and their relevance for disease progression.
GRANTS
This work was supported by the Fondation Leducq and National Heart, Lung, and Blood Institute Grants 2R01 HL-48013 (M. R. Bristow), 1K01HL-088708-01 (C. C. Sucharov), and HL-051239 (J. D. Port).
DISCLOSURES
Michael R. Bristow: Employee, shareholder, and President/CEO, ARCA biopharma; Equity in miRagen, Inc. Carmen Sucharov: Equity in miRagen, Inc., J. David Port, Employee and shareholder, ARCA biopharma, Equity in miRagen. Stephen Liggett, consultant and equity in ARCA biopharma.
AUTHOR CONTRIBUTIONS
Author contributions: K. D., K. N., A. M., and P. N. performed experiments; K. D. and C. C. S. analyzed data; A. K.-F., M. R. B., and C. C. S. interpreted results of experiments; A. K.-F. and C. C. S. prepared figures; J. D. P., S. B. L., M. R. B., and C. C. S. edited and revised manuscript; J. D. P., M. R. B., and C. C. S. approved final version of manuscript; M. R. B. and C. C. S. conception and design of research; C. C. S. drafted manuscript.
Supplementary Material
Footnotes
The online version of this article contains supplemental material.
REFERENCES
- 1. Akhter SA, D'Souza KM, Petrashevskaya NN, Mialet-Perez J, Liggett SB. Myocardial beta1-adrenergic receptor polymorphisms affect functional recovery after ischemic injury. Am J Physiol Heart Circ Physiol 290: H1427–H1432, 2006. [DOI] [PubMed] [Google Scholar]
- 2. Bengtsson K, Melander O, Orho-Melander M, Lindblad U, Ranstam J, Rastam L, Groop L. Polymorphism in the beta(1)-adrenergic receptor gene and hypertension. Circulation 104: 187–190, 2001. [DOI] [PubMed] [Google Scholar]
- 3. Benitah JP, Kerfant BG, Vassort G, Richard S, Gomez AM. Altered communication between L-type calcium channels and ryanodine receptors in heart failure. Front Biosci 7: e263–e275, 2002. [DOI] [PubMed] [Google Scholar]
- 4. Bisognano JD, Weinberger HD, Bohlmeyer TJ, Pende A, Raynolds MV, Sastravaha A, Roden R, Asano K, Blaxall BC, Wu SC, Communal C, Singh K, Colucci W, Bristow MR, Port DJ. Myocardial-directed overexpression of the human beta(1)-adrenergic receptor in transgenic mice. J Mol Cell Cardiol 32: 817–830, 2000. [DOI] [PubMed] [Google Scholar]
- 5. Bristow MR, Minobe WA, Raynolds MV, Port JD, Rasmussen R, Ray PE, Feldman AM. Reduced beta 1 receptor messenger RNA abundance in the failing human heart. J Clin Invest 92: 2737–2745, 1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cline MS, Smoot M, Cerami E, Kuchinsky A, Landys N, Workman C, Christmas R, Avila-Campilo I, Creech M, Gross B, Hanspers K, Isserlin R, Kelley R, Killcoyne S, Lotia S, Maere S, Morris J, Ono K, Pavlovic V, Pico AR, Vailaya A, Wang PL, Adler A, Conklin BR, Hood L, Kuiper M, Sander C, Schmulevich I, Schwikowski B, Warner GJ, Ideker T, Bader GD. Integration of biological networks and gene expression data using Cytoscape. Nat Protoc 2: 2366–2382, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Condorelli G, Latronico MV, Dorn GW., 2nd microRNAs in heart disease: putative novel therapeutic targets? Eur Heart J 31: 649–658, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Engelhardt S, Hein L, Wiesmann F, Lohse MJ. Progressive hypertrophy and heart failure in beta1-adrenergic receptor transgenic mice. Proc Natl Acad Sci USA 96: 7059–7064, 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gao X, Chaturvedi D, Patel TB. p90 ribosomal S6 kinase 1 (RSK1) and the catalytic subunit of protein kinase A (PKA) compete for binding the pseudosubstrate region of PKAR1alpha: role in the regulation of PKA and RSK1 activities. J Biol Chem 285: 6970–6979, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gupta MP, Gupta M, Dizon E, Zak R. Sympathetic control of cardiac myosin heavy chain gene expression. Mol Cell Biochem 157: 117–124, 1996. [DOI] [PubMed] [Google Scholar]
- 11. Jones LR, Suzuki YJ, Wang W, Kobayashi YM, Ramesh V, Franzini-Armstrong C, Cleemann L, Morad M. Regulation of Ca2+ signaling in transgenic mouse cardiac myocytes overexpressing calsequestrin. J Clin Invest 101: 1385–1393, 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liggett SB, Mialet-Perez J, Thaneemit-Chen S, Weber SA, Greene SM, Hodne D, Nelson B, Morrison J, Domanski MJ, Wagoner LE, Abraham WT, Anderson JL, Carlquist JF, Krause-Steinrauf HJ, Lazzeroni LC, Port JD, Lavori PW, Bristow MR. A polymorphism within a conserved beta(1)-adrenergic receptor motif alters cardiac function and beta-blocker response in human heart failure. Proc Natl Acad Sci USA 103: 11288–11293, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liu G, Friggeri A, Yang Y, Milosevic J, Ding Q, Thannickal VJ, Kaminski N, Abraham E. miR-21 mediates fibrogenic activation of pulmonary fibroblasts and lung fibrosis. J Exp Med 207: 1589–1597, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lowes BD, Gilbert EM, Abraham WT, Minobe WA, Larrabee P, Ferguson D, Wolfel EE, Lindenfeld J, Tsvetkova T, Robertson AD, Quaife RA, Bristow MR. Myocardial gene expression in dilated cardiomyopathy treated with beta-blocking agents. N Engl J Med 346: 1357–1365, 2002. [DOI] [PubMed] [Google Scholar]
- 15. Mason DA, Moore JD, Green SA, Liggett SB. A gain-of-function polymorphism in a G-protein coupling domain of the human beta1-adrenergic receptor. J Biol Chem 274: 12670–12674, 1999. [DOI] [PubMed] [Google Scholar]
- 16. Matkovich SJ, Van Booven DJ, Youker KA, Torre-Amione G, Diwan A, Eschenbacher WH, Dorn LE, Watson MA, Margulies KB, Dorn GW., 2nd Reciprocal regulation of myocardial microRNAs and messenger RNA in human cardiomyopathy and reversal of the microRNA signature by biomechanical support. Circulation 119: 1263–1271, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mialet Perez J, Rathz DA, Petrashevskaya NN, Hahn HS, Wagoner LE, Schwartz A, Dorn GW, Liggett SB. Beta 1-adrenergic receptor polymorphisms confer differential function and predisposition to heart failure. Nat Med 9: 1300–1305, 2003. [DOI] [PubMed] [Google Scholar]
- 18. Molkentin JD, Lu JR, Antos CL, Markham B, Richardson J, Robbins J, Grant SR, Olson EN. A calcineurin-dependent transcriptional pathway for cardiac hypertrophy. Cell 93: 215–228, 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nam S, Li M, Choi K, Balch C, Kim S, Nephew KP. MicroRNA and mRNA integrated analysis (MMIA): a web tool for examining biological functions of microRNA expression. Nucleic Acids Res 37: W356–W362, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Port JD, Bristow MR. Altered beta-adrenergic receptor gene regulation and signaling in chronic heart failure. J Mol Cell Cardiol 33: 887–905, 2001. [DOI] [PubMed] [Google Scholar]
- 21. Port JD, Sucharov C. Role of MicroRNAs in cardiovascular disease: therapeutic challenges and potentials. J Cardiovasc Pharmacol 56: 444–453, 2010. [DOI] [PubMed] [Google Scholar]
- 22. Roy S, Khanna S, Hussain SR, Biswas S, Azad A, Rink C, Gnyawali S, Shilo S, Nuovo GJ, Sen CK. MicroRNA expression in response to murine myocardial infarction: miR-21 regulates fibroblast metalloprotease-2 via phosphatase and tensin homologue. Cardiovasc Res 82: 21–29, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Small EM, Frost RJ, Olson EN. MicroRNAs add a new dimension to cardiovascular disease. Circulation 121: 1022–1032, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sucharov CC, Dockstader K, Nunley K, McKinsey TA, Bristow M. β-Adrenergic receptor stimulation and activation of protein kinase A protect against α1-adrenergic-mediated phosphorylation of protein kinase D and histone deacetylase 5. J Card Fail 17: 592–600, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sucharov CC, Dockstader K, McKinsey TA. YY1 protects cardiac myocytes from pathologic hypertrophy by interacting with HDAC5. Mol Biol Cell 19: 4141–4153, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sucharov CC, Mariner PD, Nunley KR, Long C, Leinwand L, Bristow MR. A beta1-adrenergic receptor CaM kinase II-dependent pathway mediates cardiac myocyte fetal gene induction. Am J Physiol Heart Circ Physiol 291: H1299–H1308, 2006. [DOI] [PubMed] [Google Scholar]
- 27. Swift SM, Gaume BR, Small KM, Aronow BJ, Liggett SB. Differential coupling of Arg- and Gly389 polymorphic forms of the beta1-adrenergic receptor leads to pathogenic cardiac gene regulatory programs. Physiol Genomics 35: 123–131, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tepe NM, Lorenz JN, Yatani A, Dash R, Kranias EG, Dorn GW, 2nd, Liggett SB. Altering the receptor-effector ratio by transgenic overexpression of type V adenylyl cyclase: enhanced basal catalytic activity and function without increased cardiomyocyte beta-adrenergic signalling. Biochemistry 38: 16706–16713, 1999. [DOI] [PubMed] [Google Scholar]
- 29. Terra SG, Hamilton KK, Pauly DF, Lee CR, Patterson JH, Adams KF, Schofield RS, Belgado BS, Hill JA, Aranda JM, Yarandi HN, Johnson JA. Beta1-adrenergic receptor polymorphisms and left ventricular remodeling changes in response to beta-blocker therapy. Pharmacogenet Genomics 15: 227–234, 2005. [DOI] [PubMed] [Google Scholar]
- 30. van Rooij E, Sutherland LB, Liu N, Williams AH, McAnally J, Gerard RD, Richardson JA, Olson EN. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc Natl Acad Sci USA 103: 18255–18260, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. van Rooij E, Sutherland LB, Thatcher JE, DiMaio JM, Naseem RH, Marshall WS, Hill JA, Olson EN. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc Natl Acad Sci USA 105: 13027–13032, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wagoner LE, Craft LL, Zengel P, McGuire N, Rathz DA, Dorn GW, 2nd, Liggett SB. Polymorphisms of the beta1-adrenergic receptor predict exercise capacity in heart failure. Am Heart J 144: 840–846, 2002. [DOI] [PubMed] [Google Scholar]
- 33. Walker LA, Walker JS, Ambler SK, Buttrick PM. Stage-specific changes in myofilament protein phosphorylation following myocardial infarction in mice. J Mol Cell Cardiol 48: 1180–1186, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Walsh RFR, Kelly M, Nelson P, Morrison J, Port JD, Bristow MR. Human Myocardial β1 389 Arg/Arg adrenergic receptors exhibit a propensity for constitutively active, high affinity agonist binding and are selectively inactivated by bucindolol. J Cardiac Fail 14: S8, 2008. [Google Scholar]
- 35. Wang WC, Juan AH, Panebra A, Liggett SB. MicroRNA let-7 establishes expression of beta2-adrenergic receptors and dynamically down-regulates agonist-promoted down-regulation. Proc Natl Acad Sci USA 108: 6246–6251, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Xiao RP. Beta-adrenergic signaling in the heart: dual coupling of the beta2-adrenergic receptor to G(s) and G(i) proteins. Sci STKE 2001: re15, 2001. [DOI] [PubMed] [Google Scholar]
- 37. Yuan SM, Jing H. Cardiac pathologies in relation to Smad-dependent pathways. Interact Cardiovasc Thorac Surg 11: 455–460, 2010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.