Abstract
The morphological concept of Penicillium sclerotiorum (subgenus Aspergilloides) includes strains with monoverticillate, vesiculate conidiophores, and vivid orange to red colony colours, with colourful sclerotia sometimes produced. Multigene phylogenetic analyses with the nuclear ribosomal internal transcribed spacer (ITS) region, cytochrome c oxidase subunit 1 (cox1), β-tubulin (benA), translation elongation factor 1-α (tef1-α), and calmodulin (cmd), reveal that the P. sclerotiorum morphospecies is a complex of seven phylogenetically distinct species, three of which were recently described, namely P. guanacastense, P. mallochii, and P. viticola. Three previously unidentified species are described here as P. cainii, P. jacksonii, and P. johnkrugii. The phylogenetic species are morphologically similar, but differ in combinations of colony characters, sclerotium production, conidiophore stipe roughening and branching, and conidial shape. Ecological characters and differences in geographical distribution further characterise some of the species, but increased sampling is necessary to confirm these differences. The fungal DNA barcode, the ITS, and the animal DNA barcode, cox1, have lower species resolving ability in our phylogenetic analyses, but still allow identification of all the species. Tef1-α and cmd were superior in providing fully resolved, statistically well-supported phylogenetic trees for this species complex, whereas benA resolved all species but had some issues with paraphyly. Penicillium adametzioides and P. multicolor, considered synonyms of P. sclerotiorum by some previous authors, do not belong to the P. sclerotiorum complex.
Taxonomic novelties:
New species: Penicillium cainii K.G. Rivera, Malloch & Seifert, P. jacksonii K.G. Rivera, Houbraken & Seifert, P. johnkrugii K.G. Rivera, Houbraken & Seifert.
Keywords: DNA barcoding, multigene phylogeny, sclerotia, soil-borne hyphomycetes
INTRODUCTION
Penicillium sclerotiorum was first isolated from air in Java, Indonesia, by K.B. Boedijn, and then described by van Beyma (1937). The species has monoverticillate, vesiculate conidiophores and vivid orange to red colony colours, and some strains produce orange sclerotia that give the species its name. Cultures identified as P. sclerotiorum have been isolated from many countries in Africa, Asia, and North America, suggesting a cosmopolitan distribution, but it has been reported infrequently (Pitt 1980, Ramírez 1982). Strains commonly originate from soil, and occasionally textiles, but are also isolated from house dust (Vesper et al. 2005), diseased grape fruit and stems (De Lucca et al. 2008), and as a potential endophyte of Coffea arabica berries (Vega et al. 2006). Air sampling revealed higher concentrations of P. sclerotiorum outdoors than in indoor environments in India (Sawane & Saoji 2004).
Penicillium sclerotiorum is now classified in Penicillium subgenus Aspergilloides, section Sclerotiora by Houbraken & Samson (2011). As noted by Peterson (2000) and discussed at length by Houbraken & Samson (2011), the monoverticillate conidiophore, although a useful phenotypic character for identification, is phylogenetically uninformative and the concept of subgenus Aspergilloides promoted by Pitt (1980) has been substantially revised. Species with such conidiophores are phylogenetically intermingled with species with divaricate or symmetrically biverticillate conidiophores. Pitt (1980) classified P. sclerotiorum in his broadly circumscribed subgenus Aspergilloides series Glabra, which Peterson's phylogenetic studies distributed among his Clades 2, 3, and 5 (Peterson 2000). Peterson's ‘Clade 3’ of Penicillium included the predominantly monoverticillate species P. adametzii, P. adametzioides, P. bilaiae, and the biverticillate P. herquei, all now classified in section Sclerotiora by Houbraken & Samson (2011). Three biverticillate species were included by Houbraken & Samson (2011) in section Sclerotiora, namely P. herquei, classified by Pitt (1980) in Penicillium subgenus Furcatum section Furcatum, and the subsequently described P. malachiteum and P. nodositatum. Sclerotia are sporadically produced by species across many sections of Penicillium, with the orange sclerotia of P. thomii (section Aspergilloides) perhaps the most conspicuous and commonly encountered example.
Our recent studies of Penicillium strains isolated from the guts of tropical leaf-eating caterpillars in Costa Rica (Rivera et al. 2011) led to the description of two phylogenetically distinct species, P. mallochii and P. guanacastense, which although they do not produce sclerotia, otherwise conform to the morphological concepts of P. sclerotiorum of Pitt (1980) and Stolk & Samson (1983). This led to the realisation that the morphological concept of P. sclerotiorum includes a complex of phylogenetically distinct species, and to the taxonomic revision presented in this paper. Among the modern revisions of this taxon by Raper & Thom (1949), Pitt (1980), Ramírez (1982), and Stolk & Samson (1983), only the latter proposed synonyms for P. sclerotiorum; we reconsidered the status of P. adametzioides and P. multicolor as possible names for some of the phylogenetic lineages we observed. This paper provides a polyphasic taxonomic revision of 29 strains from this complex, based on morphological and microscopic characters, and phylogenetic analyses of five genes: β-tubulin (benA), cytochrome c oxidase subunit 1 (cox1), the internal transcriber spacer (ITS) region, translation elongation factor 1-α (tef1-α), and calmodulin (cmd). The ITS and cox1 genes are of particular relevance as DNA barcodes, with ITS now functioning as the sanctioned DNA barcode for fungi (Schoch et al., in prep.), and cox1 as the DNA barcode for animals (Hebert et al. 2003).
MATERIALS AND METHODS
Fungal isolates and herbarium specimens
We focused on cultures identified as P. sclerotiorum, and the related species in Clade 3 from the phylogenetic study of Peterson (2000). They were obtained from the CBS Fungal Biodiversity Centre (CBS) and USDA-ARS, National Center for Agricultural Utilization Research (NRRL) culture collections, and the personal research collections of K.A. Seifert and C. André Lévesque (Ottawa, ON, Canada), D. Malloch (formerly University of Toronto, ON, Canada), and J. Houbraken and R.A. Samson (CBS). Representative strains are deposited in the Canadian Collection of Fungal Cultures (DAOM) and CBS. Table 1 includes the metadata for the 29 strains used in this study. We note that cultures of this complex do not preserve well in sterile water at 4 °C, often dying within one year. Some P. sclerotiorum strains (DAOM 239931, 239932) and all P. johnkrugii strains were isolated from ethanol treated soils from Australia and Malaysia (Houbraken, pers. comm.).
Table 1.
Accession numbers of cultures, isolation details and GenBank accession numbers for the five genes used for phylogenetic analysis of the Penicillium sclerotiorum complex.
| Species | Accession number | Location | Host or substrate | GenBank Accession numbers |
||||
|---|---|---|---|---|---|---|---|---|
| ITS | benA | tef1-α | cmd | cox1 | ||||
| P. adametzii | CBS 209.28 (T) | Poland, Poznan | Soil under conifers | JN714929 | JN625957 | JN626136 | – | JN626049 |
| KAS 3463 | Malaysia, Kedah | Forest soil | JN714930 | JN625958 | JN626137 | – | JN626050 | |
| KAS 3464 | Malaysia, Kedah | Forest soil | JN714931 | JN625959 | JN626138 | – | JN626051 | |
| KAS 3465 | Malaysia, Kedah | Forest soil | JN714932 | JN625960 | JN626139 | – | JN626052 | |
| KAS 3466 | Malaysia, Kedah | Forest soil | JN714933 | JN625961 | JN626140 | – | JN626053 | |
| P. adametziodes | DAOM 239916 | Canada, Ontario, Vineland | Riesling grapes | JN686434 | JN799643 | – | JN686387 | JN686410 |
| CBS 313.59 (T) | Japan | Soil | JN686433 | JN799642 | – | JN686388 | JN686411 | |
| P. bilaiae | NRRL 3391 (T) | Ukraine, Kiev | Soil | JN714937 | JN625966 | JN626145 | JN626009 | JN626058 |
| ATCC 20851 | Canada, Alberta, Lethbridge | Soil | JN714934 | JN625964 | JN626141 | JN626005 | JN626054 | |
| ATCC 22348 | Ukraine, Kiev | Soil | JN714935 | JN625962 | JN626143 | JN626007 | JN626056 | |
| DAOM 197974 | Canada, Alberta | Soil | JN714936 | JN625965 | JN626144 | JN626008 | JN626057 | |
| P. cainii | DAOM 239914 (T) | Canada, Ontario, Niagara, Niagara Falls, Fireman's Park | Nuts of Juglans nigra | JN686435 | JN686366 | JN686456 | JN686989 | JN686412 |
| DAOM 239915 | Canada, Ontario, Niagara, Niagara Falls, Fireman's Park | Nuts of Carya ovata | JN686436 | JN686367 | JN686457 | JN686390 | JN686413 | |
| P. guanacastense | DAOM 239912(T) | Costa Rica, Santa Rosa, Área de Conservación Guanacaste | Gut of the caterpillar Eutelia sp. reared on leaves of Spondias mombin | JN626098 | JN625967 | JN626146 | JN626010 | JN626059 |
| DAOM 239913 | Costa Rica, Santa Rosa, Área de Conservación Guanacaste | Gut of the caterpillar Eutelia sp. reared on leaves of Spondias mombin | JN626099 | JN625968 | JN626147 | JN626011 | JN626060 | |
| P. herquei | CBS 336.48 (T) | France | Leaf of Agauria pirifolia | JN626101 | JN625970 | JN626149 | JN626013 | JN626062 |
| CBS 136.22 | France | JN626100 | JN625969 | JN626148 | JN626012 | JN626061 | ||
| CBS 347.51 | Japan, Nehira | Wakamoto corn and rice cake | JN626102 | JN625971 | JN626150 | JN626014 | JN626063 | |
| CBS 110644 | USA, Wisconsin | Forest soil | JN626103 | JN625972 | JN626151 | JN626015 | JN626064 | |
| P. hirayamae | CBS 229.60 (T) | Thailand | Milled rice | JN626095 | JN625955 | JN626135 | JN626003 | JN626046 |
| CBS 238.65 | South Africa | Corn meal | JN626096 | JN625956 | JN626134 | JN626004 | JN626047 | |
| P. jacksonii | DAOM 239937 (T) | Australia, Queensland, Barrine Lake | Forest soil | JN686437 | JN686368 | JN686458 | JN686391 | JN686414 |
| DAOM 239938 | Australia, Queensland, Barrine Lake | Forest soil | JN686438 | JN686369 | JN686459 | JN686392 | JN686415 | |
| P. johnkrugii | DAOM 239939 | Malaysia, Kedah, Langkawi, Gunung Raya Rainforest | Rainforest soil | JN686443 | JN686374 | JN792190 | JN686397 | JN686420 |
| DAOM 239940 | Malaysia, Kedah, Langkawi, Gunung Raya Rainforest | Forest soil | JN686444 | JN686375 | JN792191 | JN686398 | JN686421 | |
| DAOM 239941 | Malaysia, Kedah, Langkawi, Gunung Raya Rainforest | Forest soil | JN686445 | JN686376 | JN792192 | JN686399 | JN686422 | |
| DAOM 239942 | Malaysia, Kedah, Langkawi, Gunung Raya Rainforest | Forest soil | JN686446 | JN686377 | JN792193 | JN686400 | JN686423 | |
| DAOM 239943 (T) | Malaysia, Kedah, Langkawi, Gunung Raya Rainforest | Forest soil | JN686447 | JN686378 | JN792194 | JN686401 | JN686424 | |
| DAOM 239944 | Malaysia, Kedah, Langkawi, Gunung Raya Rainforest | Forest soil | JN686448 | JN686779 | JN792195 | JN686402 | JN686425 | |
| DAOM 239945 | Malaysia, Kedah, Langkawi, Gunung Raya Rainforest | Forest soil | JN686449 | JN686780 | JN792196 | JN686403 | JN686426 | |
| DAOM 239946 | Malaysia, Kedah, Langkawi, Gunung Raya Rainforest | Forest soil | JN686450 | JN686781 | JN792197 | JN686404 | JN686427 | |
| KAS 3479 | Malaysia, Kedah, Langkawi, Gunung Raya Rainforest | Forest soil | JN686451 | JN686782 | JN792198 | JN686405 | JN686428 | |
| P. levitum | NRRL 705 (T) | USA, New York | Modeling clay | JN626097 | JN714938 | JN714928 | JN714939 | JN626048 |
| P. mallochii | DAOM 239917 (T) | Costa Rica, Santa Rosa, Área de Conservación Guanacaste | Caterpillar on Spondias mombin | JN626104 | JN625973 | JN626152 | JN626016 | JN626065 |
| DAOM 239919 | Costa Rica, Santa Rosa, Área de Conservación Guanacaste | Midgut of the caterpillar Citheronia lobesis feeding on Spondias mombin | JN626106 | JN625975 | JN626154 | JN626018 | JN626067 | |
| DAOM 239922 | Costa Rica, Santa Rosa, Área de Conservación Guanacaste | Hindgut of the caterpillar Rothschildia lebeau reared on leaves of Spondias mombin | JN626109 | JN625978 | JN626157 | JN626020 | JN626070 | |
| DAOM 239925 | Costa Rica, Santa Rosa, Área de Conservación Guanacaste | Guts of the caterpillar Citheronia lobesis reared on leaves of Cochlospermum vitifolium | JN626112 | JN625980 | JN626159 | JN626023 | JN626072 | |
| DAOM 239926 | Costa Rica, Santa Rosa, Área de Conservación Guanacaste | Frass of the caterpillar Rothschildia lebeau reared on leaves of Spondias mombin | JN626111 | JN625981 | JN626160 | JN626024 | JN626073 | |
| DAOM 239927 | Costa Rica, Santa Rosa, Área de Conservación Guanacaste | Gut of the caterpillar Rothschildia lebeau reared on leaves of Spondias mombin | JN626113 | JN625982 | JN626161 | JN626025 | JN626074 | |
| P. multicolor | CBS 501.73 (T) | USSR | Soil | JN799647 | JN799645 | JN799648 | JN799646 | JN799644 |
| P. sclerotiorum | NRRL 2074 (T) | Indonesia, Java, Buitenzorg | Air | JN626132 | JN626001 | JN626180 | JN626044 | JN626093 |
| NRRL 32583 | USA, Hawai'i, Kuauai | Coffee seeding crown | JN626133 | JN626002 | JN626181 | JN626045 | JN626094 | |
| DAOM 239930 | Thailand, Hua Hin | Forest soil | JN626129 | JN625998 | JN626177 | JN626041 | JN626090 | |
| DAOM 239931 | Australia, Queensland, Barron Falls | Forest soil | JN626130 | JN625999 | JN626178 | JN626042 | JN626091 | |
| DAOM 239932 | Australia, Queensland, Barron Falls | Forest soil | JN626131 | JN626000 | JN626179 | JN626043 | JN626092 | |
| CBS 128.65 | Zaire, Leopoldville, Nsang–Ngidinga River | Forest litter | JN686452 | JN686783 | JN686464 | JN686406 | JN686429 | |
| CBS 258.55 | Turkey, Istanbul | Culture contaminant | JN686453 | JN686784 | JN686465 | JN686407 | JN686430 | |
| CBS 118889 | Korea | Soil | JN686454 | JN686785 | JN686466 | JN686408 | JN686431 | |
| P. viticola | DAOM 239933 | Australia, Queensland, Barron Falls | Forest soil | JN686439 | JN686370 | JN686460 | JN686393 | JN686416 |
| DAOM 239934 | Australia, Queensland, Atherton | Forest soil | JN686440 | JN686371 | JN686461 | JN686394 | JN686417 | |
| DAOM 239935 | Australia, Queensland, Atherton | Rainforest soil | JN686441 | JN686372 | JN686462 | JN686395 | JN686418 | |
| DAOM 239936 | Australia, Queensland, Atherton | Rainforest soil | JN686442 | JN686373 | JN686463 | JN686396 | JN686419 | |
| Penicillium sp. | CBS 248.65 | South Africa | Corn meal | JN686455 | JN686786 | JN686467 | JN686409 | JN686432 |
Abbreviations: CBS - CBS-KNAW Biodiversity Centre culture collection, Utrecht, the Netherlands. DAOM - culture collection and herbarium of the National Mycological Collections, Agriculture & Agri-Food Canada, Ottawa. NRRL - culture collection of USDA-ARS, National Center for Agricultural Utilization Research, Peoria, IL, USA. ATCC - American Type Culture Collection, Manassas, VA, USA. KAS - personal culture collection of Keith A. Seifert.
Morphological analysis
All strains were inoculated at three points onto Blakeslee's Malt Extract Agar (MEA, for microscopic analysis and colony characters), Czapek Yeast Agar (CYA, for colony characters; Pitt 1973), Czapek Agar (CZ, for ability to grow and sporulate in the absence of ammonia; Raper & Thom 1949), Yeast Extract Sucrose Agar (YES to stimulate colony pigmentation by enhancing secondary metabolite production; Filtenborg et al. 1990), Oatmeal Agar (OA, to stimulate sclerotial or ascomatal development) and Creatine Sucrose Agar (CREA, to test for acid production; Frisvad 1993). All measurements and observations were performed in duplicate from cultures inoculated at different times. Plates were incubated in the dark at 25 °C for 7 days.
Microscopic observations employed a BX 50 light microscope (Olympus Canada, Richmond Hill, ON), using tissue removed from 7 d old colonies grown on MEA, and mounted in 85 % lactic acid. Microphotographs were taken and characters measured with an Evolution MP Camera using Image-Pro Plus v. 6 (both from Media Cybernetics, MD, USA). For each strain, automated measurements of 25 conidia were made from phase contrast images using the count/size algorithm of Image-Pro, and manual measurements were made for ten phialides, stipes, branches, vesicles, sclerotia and sclerotial cells. Variations in microscopic dimensions are presented as mean ± standard error. Alphanumeric colony colour codes are based on Kornerup & Wanscher (1978). Colony photographs were taken using a copy stand and a Coolpix P5000 camera (Nikon Canada, Mississauga, ON) under incandescent light.
DNA extractions, PCR and DNA sequencing
Genomic DNA was extracted from strains grown on MEA or CYA using the UltraClean™ Microbial DNA Isolation Kit (MoBio Laboratories, Montreal, Canada) following the manufacturer's protocol. BenA was amplified and sequenced with primers Bt2a and Bt2b (Glass & Donaldson 1995); ITS with primers ITS1 and ITS4 (White et al. 1990); cox1 with primers PF and AR (Seifert et al. 2007); tef1-α with primers EF1c and EF6 (Peterson et al. 2004); and cmd with primers CMD5 and CMD6 (Hong et al. 2006).
PCR reactions were performed in 10 μl reaction mixtures containing 1 μl genomic DNA, 1X PCR Buffer, 0.1 mm of dNTPs, 0.08 μM of each primer, and 0.5X Taq polymerase. Amplifications were performed in a TGradient (Biometra, Montreal, Canada) or a Genius (Techne, Duxford, Cambridge, UK) thermocycler. The PCR parameters for ITS, cox1, benA were denaturation at 95 °C for 1 min, followed by primer annealing at 56 °C for 45 s, and primer extension at 72 °C for 90 s for 35 cycles, plus a final 10 min elongation step at 72 °C. The profile for tef1-α and cmd was denaturation at 94 °C for 1 min, annealing at 62 °C for 30 s, primer extension at 72 °C for 90 s for 42 cycles, then a final elongation step at 72 °C for 10 min. PCR products were visualised by gel electrophoresis in a 1 % agarose gel containing ethidium bromide (0.05 μg/mL).
Sequencing reactions were performed directly on PCR amplicons using forward and reverse primers. Reactions with a total volume of 10 μl contained 1 μl amplicon, 0.5 μl of ready-made BigDye and terminator mix v. 7 and 0.125X BigDye buffer (Applied Biosystems, Foster City, CA, USA), and 0.161 μM of primer. Reactions were performed in the thermocyclers noted above, programmed for denaturation at 95 °C for 1 min, followed by primer annealing at 56 °C for 30 s and primer extension at 72 °C for 1 min for 30 cycles, plus a final 10 min elongation step at 72 °C. Sequence reaction mixtures were precipitated using ethanol/EDTA/sodium acetate precipitation. Samples were analysed on an ABI 3130XL Genetic Analyzer (Applied Biosystems, Foster City, CA, USA).
Two strains of P. herquei (CBS 336.48T, 110644) produced two amplicons (ca. 360 bp and 470 bp long) for benA. PCR products with multiple bands were cloned using Promega's pGEM®-T Vector Kit and JM109 High Efficiency Competent Cells (Madison, WI, USA) following the manufacturer's protocol. Fifteen transformed colonies were selected for PCR and sequencing.
Consensus sequences were assembled using Sequencher v. 4.8 (Genes Codes Corporation, Ann Arbor, MI, USA) and SeqMan in the LASERGENE package v. 8 (DNASTAR Inc., Madison, WI, USA). Alignments were constructed using the online version of MAFFT v. 6 (Katoh et al. 2009), and adjusted to optimise homology using BioEdit 7.0.9 (Hall 1999).
Maximum parsimony (MP) analyses were performed using heuristic searches in PAUP v. 4 (Swofford 2002) with the tree bisection-reconnection (TBR) branch swapping algorithm. Uninformative characters were removed for all analyses, gaps were treated as missing data, and maxtrees were set to 5000. Consensus trees were calculated, and the robustness of the gene trees was tested using a full heuristic search, saving 10 trees per replicate (1000 replicates).
Maximum likelihood (ML) analyses were performed using Phylogenies for Maximum Likelihood (PhyML) v. 2.4.4 (Guindon et al. 2005). Tree searches for each alignment were run under the nucleotide substitution models obtained from ModelTest 3.7, namely GTR+G (benA, cox1, ITS, tef1-a), and GTR+I+G (cmd), using the Alkaike Information Criteria (AIC) (Posada & Crandall 1998). The starting tree for branch swapping was obtained using the modified neighbour joining algorithm BIONJ (Gascuel 1997), as implemented in PhyML. The robustness for each tree was tested by performing 1000 bootstrap replicates.
Bayesian inference (BI) analyses were performed using MrBayes 3.1.2 (Huelsenbeck & Ronquist 2001), using the same models noted for ML above. All Bayesian analyses were performed using random starting trees, and were run for four chains with one million generations for all genes, sampling every 100 generations, generating 10,001 trees, with the first 2,500 discarded as ‘burn-in’ for each chain.
Individual gene trees and a combined data set with all five genes were analysed by MP, ML and BI individually. A partition homogeneity test (PHT) was performed in PAUP v. 4 (Swofford 2002) using all genes to determine whether combining the genes was advisable. Missing cmd sequences for P. adametzii strains and missing tef1-α sequences for DAOM 239916 were replaced by N's to indicate missing data. Parsimony parameters were set to TBR, 1000 replicates, 10 trees saved per replicate, and maxtrees were set to 5000.
All new sequences used for our analyses are deposited in GenBank under accession numbers JN625955–JN799648 (Table 1). Alignments are in TreeBase under study S12031.
Table 2.
Comparative summary of colony and microscopic characters of species in the P. sclerotiorum complex.
| Species | Colony characters on CYA |
Morphological characters |
Substrate/host | Geographical distribution | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Conidium colours | Diam (mm) | Sclerotia | Conidiophores |
Conidia |
||||||
| Roughening | Length > 135 μm | Branching | Roughening | Shape | ||||||
| P. sclerotiorum | Greenish grey/orange | 18–40 | Present | Smooth to finely rough | All | Unbranched | Finely rough | Subglobose to ellipsoidal | Soil, litter, coffee beans, air | Pantropical and subtropical |
| P. cainii | Deep blue to deep green | 23–29 | Absent | Rough | No | Unbranched or ∼10 % branched | Finely rough | Globose | Nuts of Juglandaceae | Canada |
| P. guanacastense | Greenish grey | 25–33 | Absent | Finely rough | No | ∼10 % branched | Finely rough | Globose | Caterpillars feeding on leaves | Costa Rica |
| P. jacksonii | Deep green | 30–33 | Absent | Smooth to rough | No | Unbranched or < 65 % branched | Finely rough | Globose | Soil | Australia |
| P. johnkrugii | White | 30–38 | Present | Smooth to finely rough | Most | Unbranched | Finely rough | Globose to subglobose | Soil | Malaysia |
| P. mallochii | Turquoise grey/greenish grey | 29–39 | Absent | Smooth to finely rough | Most | ∼10 % branched | Finely rough | Globose to subglobose | Caterpillars feeding on leaves | Costa Rica |
| P. viticola | Greenish grey | 26–36 | Absent | Rough | No | Unbranched or ∼10 % branched | Smooth | Globose | Grape vines, soil | Australia, Japan |
RESULTS
Analysis of morphological characters
Morphological species descriptions for P. cainii, P. jacksonii, P. johnkrugii, P. viticola, and the revised species description for P. sclerotiorum are provided in the Taxonomy section. Together with the descriptions of P. guanacastense and P. mallochii in Rivera et al. (2011), this constitutes a monographic revision of the P. sclerotiorum species complex. Most species can be identified by subtle phenotypic characters with the aid of Table 2 and the dichotomous key at the end of the paper. Several species seem to be ecologically distinct, but the apparent geographical disjunctions are preliminary and more sampling is needed to confirm these patterns.
Table 3.
Statistical support for species clades for individual gene trees using Maximum Parsimony (MP), Maximum Likelihood (ML) and Bayesian Inference (BI).
| P. cainii | P. guanacastense | P. jacksonii | P. johnkrugii | P. mallochii |
P. sclerotiorum |
P. viticola | |||
|---|---|---|---|---|---|---|---|---|---|
| A | B | ||||||||
| ITS | MP bootstrap (%) | < 70 | 92.00 | < 70 | < 70* | < 70* | 85.00 | < 70 | < 70 |
| ML probability | 0.82 | 0.99 | 0.93 | 0.718* | 0.772* | 0.19 | 0.76 | 0.49 | |
| BI posterior probability | 0.63 | 1.00 | 0.99 | 0.00 | 0.00 | 1.00 | 0.89 | 0.00 | |
| cox1 | MP bootstrap (%) | 98.00 | 96.00 | 92.00 | 0.00 | < 70 | < 70 | < 70 | 72.00 |
| ML probability | 1.00 | 1.00 | 1.00 | 0.237* | 0.502* | 0.00 | 0.00 | 0.79 | |
| BI posterior probability | 1.00 | 1.00 | 0.94 | 0.68* | 0.99* | 1.00 | 0.99 | 0.94 | |
| tef1-α | MP bootstrap (%) | 100.00 | 100.00 | 100.00 | <70 | 99.00 | 100.00 | 84.00 | 100.00 |
| ML probability | 1.00 | 1.00 | 1.00 | 0.66 | 0.83 | 0.99 | 0.85 | 0.98 | |
| BI posterior probability | 1.00 | 1.00 | 1.00 | 0.55 | 0.95 | 1.00 | 0.86 | 1.00 | |
| benA | MP bootstrap (%) | 100.00 | 100.00 | 100.00 | < 70* | < 70* | 99.00 | 100.00 | 100.00 |
| ML probability | 1.00 | 1.00 | 1.00 | 1.00* | 0.975* | 1.00 | 1.00 | 1.00 | |
| BI posterior probability | 1.00 | 1.00 | 1.00 | 1.00* | 1.00* | 1.00 | 1.00 | 1.00 | |
| cmd | MP bootstrap (%) | 100.00 | 100.00 | 100.00 | <70 | 100.00 | 77.00 | 87.00 | 82.00 |
| ML probability | 1.00 | 1.00 | 1.00 | 0.68 | 1.00 | 0.94 | 1.00 | 0.85 | |
| BI posterior probability | 1.00 | 1.00 | 1.00 | 0.92 | 1.00 | 1.00 | 1.00 | 0.99 | |
indicates analyses where P. johnkrugii and P. mallochii were paraphyletic.
In general, the species of this complex grow 20–40 mm diam in 7 d on CYA, and slightly slower, about 15–35 mm, on MEA. The conidial colours are green, often with grey or turquoise tinges; some species can be distinguished by subtle colour differences. Reverse colours are typically orange or red on CYA, and variations from this can be useful for species identification. Conidiophores are usually monoverticillate, but in several species up to about 10 % of conidiophores may have a single branch, and in some strains of P. jacksonii more than half of the conidiophores may be branched. Stipe roughening is usually slight, except for the conspicuous roughening in P. cainii and P. viticola. Stipe length varies considerably among the species; although ranges overlap, there seems to be a division between species with most conidiophores shorter or longer than 135 μm. We evaluated the width of the vesicles of monoverticillate conidiophores; they vary between 3–6 μm wide, and although there are minor statistical differences, we do not think this is a useful character for species recognition. Similarly, the number of phialides per vesicle, a highly variable character that is difficult to count confidently, is not useful as a species character. The conidia of most species in the complex are globose or subglobose and about 2–3 μm diam (L/W ratio 1:1 to about 1.2:1). Conidia of P. sclerotiorum sensu stricto are the most ellipsoidal in the complex. The conidia of most species are slightly roughened, although this is rather inconspicuous, and does not seem to be a useful character for species recognition, at least as observed with the light microscope.
Sclerotium production is inconsistent within some species, particularly P. sclerotiorum, where only four of eight strains produced them. The ex-type strain is very unpredictable, sometimes producing colonies completely covered with sclerotia, and in other transfers producing none at all. Sclerotia are normally visible after 7 d in fresh strains, but production is sometimes delayed to 10–14 d in some transfers of older strains. All strains of P. johnkrugii produced abundant sclerotia on all media, visible within 7 d. The colourful sclerotia often dramatically affect colony appearance, particularly, as occurs in P. sclerotiorum, when sclerotial colonies have much reduced, or absent, conidiation. Penicillium cainii, P. guanacastense, P. jacksonii, P. mallochii, and P. viticola strains produced no sclerotia on the media tested. All strains of all species were left for eight months in the dark at 25 °C on OA; ascospores were not produced in any of the sclerotial strains.
On MEA, several species, notably P. guanacastense and P. mallochii, have a tendency towards crustose colonies, with planar sheets of conidia dislodging en masse onto the Petri dish lid or slipping sideways across the agar. The phenomenon is less dramatic than in the terverticillate species P. crustosum and similar to that seen in monoverticillate species related to P. glabrum.
We included CREA to determine whether it had any diagnostic value for this group, but most strains of all species produced abundant acid, turning the entire plate yellow within 1 wk, and they did not produce any base to restore the original purple medium. All strains sporulated poorly and grew weakly or moderately. Only the slightly different but overlapping growth rates, and the presence of sclerotia, are recorded in the species descriptions. We do not consider this medium helpful for diagnosis in this group.
Phylogeny
Multigene phylogenetic analyses (ITS, cox1, benA, cmd, and tef1-α) revealed that strains previously identified as P. sclerotiorum comprise a complex of phylogenetically distinct species. The complex includes P. sclerotiorum, the newly described species in this paper, P. cainii, P. jacksonii, and P. johnkrugii, and the recently described P. guanacastense, P. mallochii (Rivera et al. 2011), and P. viticola (Nomura et al. 2011). Representative ML trees for each gene are presented in Fig. 1; the log likelihood values for these trees are benA -3725.72122, cmd -4537.96193, cox1 -1919.56534, ITS -1846.917238, and tef1-α -4663.651570. The MP and BI trees are not shown, but differences in topology are discussed and support values for each analysis are mapped onto the ML trees and included in Table 3. Sequences were obtained for all genes and strains, except no cmd or tef1-α PCR products were obtained for the outgroup species P. adametzii and P. adametzioides. We experimented with denaturation temperatures as low as 40°C for 1 min, and for cmd with the primer combinations CF4 and CF5, CF4 and CMD6, and CMD5 and CF5 (Peterson et al. 2004, Hong et al. 2006), without success. Two strains of the outgroup species P. herquei, CBS 336.48T and CBS 110644, yielded multiple bands after benA amplification; the most similar copy to that of the other species was selected for alignment.
Fig. 1.
Maximum likelihood (ML) trees generated for cox1, ITS, benA, tef1-α and cmd with PhyML using the GRT+G model (except cmd, GTR+I+G model), showing the clades representing the seven species of the P. sclerotiorum complex. Bold black branches have ML support > 0.90, MP bootstrap support > 90 %, and BI > 0.95; bold grey branches have ML support > 0.700, MP bootstrap support > 70 %, and BI > 0.95 (see Table 3 for details).
We examined two of the species proposed as synonyms of P. sclerotiorum by Stolk & Samson (1983) and found that they did not belong to the revised complex. The ex-type strain of P. adametzioides, CBS 313.59, is sterile, and individual gene trees (Fig. 1) and the combined gene trees (Fig. 2) reveals that this is a distinct species, not part of the P. sclerotiorum complex, but still part of section Sclerotiora as defined by Houbraken & Samson (2011). Preliminary phylogenetic analyses with all five genes excluded the ex-type strain of P. multicolor, CBS 501.73, from the P. sclerotiorum complex, and BLAST results with benA, ITS, tef1-α, and cmd sequences had 100 % sequence similarity to P. fellutanum, classified by Houbraken & Samson (2011) in subgenus Aspergilloides section Charlesii. Morphologically, this strain grew slower, 13–15 mm after 7 d on all media, and lacked the vivid red to orange colony colours and vesiculate conidiophores usually observed in the P. sclerotiorum complex. We observed only monoverticillate conidiophores in the culture, not the metulate conidiophores emphasised in the description of P. fellutanum by Pitt (1980), but our strain of the ex-type was degenerated and sporulated poorly. Penicillium multicolor is not accepted as a synonym for P. sclerotiorum here, and it is probably a synonym of P. fellutanum.
Fig. 2.
Bayesian inference (BI) tree for the combined cox1, ITS, BenA, and tef1-α data set generated using MrBayes using the GTR model (except cmd, GTR+I+G model), showing the relationships among the seven species of the P. sclerotiorum complex. Bold black branches have ML support > 0.90, MP bootstrap support > 90 %, and BI > 0.95 (see Table 3 for details).
TAXONOMY
Penicillium sclerotiorum J.F.H. Beyma, Zentralbl. Bakteriol., 2 Abt. 96: 416. 1937. MycoBank MB277708. Figs 3A, 4.
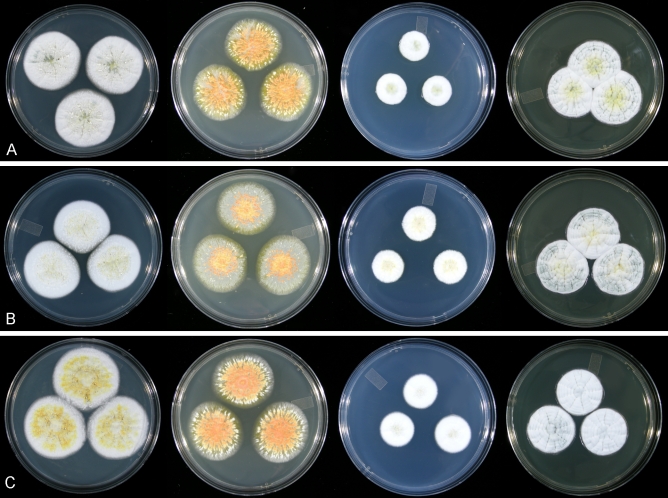
Fig. 3.
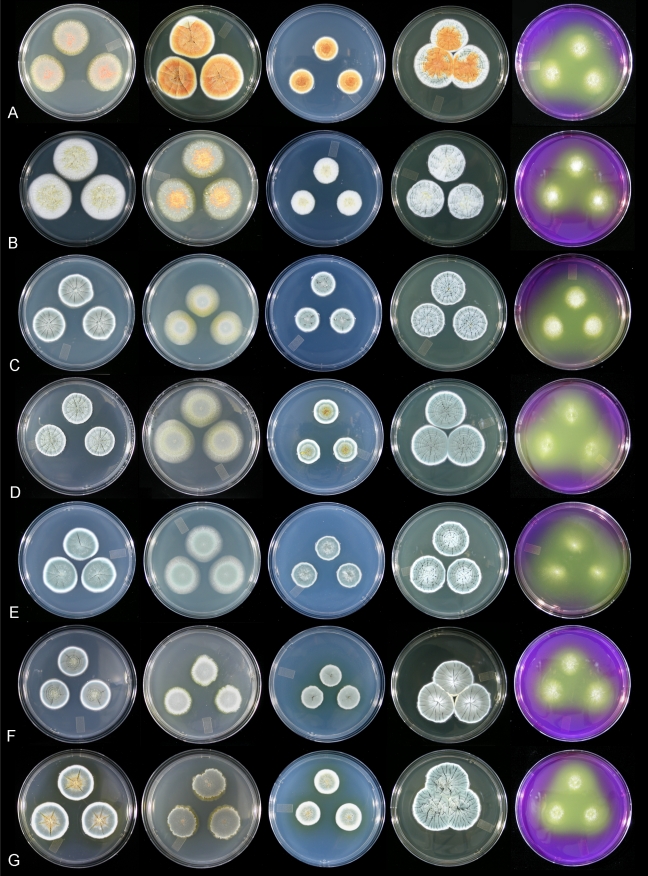
Colonies of the seven species of the P. sclerotiorum complex grown for 7 d at 25 °C on five media, right to left CYA, MEA, CZ, YES and CREA. A. P. sclerotiorum. B. P. johnkrugii. C. P. viticola. D. P. cainii. E. P. jacksonii. F. P. mallochii. G. P. guanacastense. All ex-type strains except A DAOM 239931, C DAOM 239935.
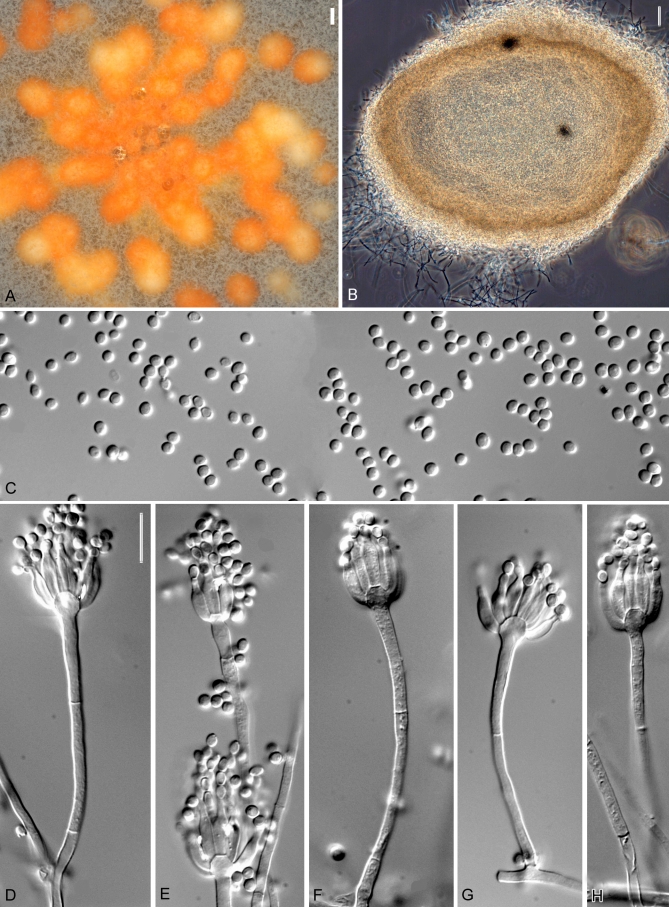
Fig. 4.
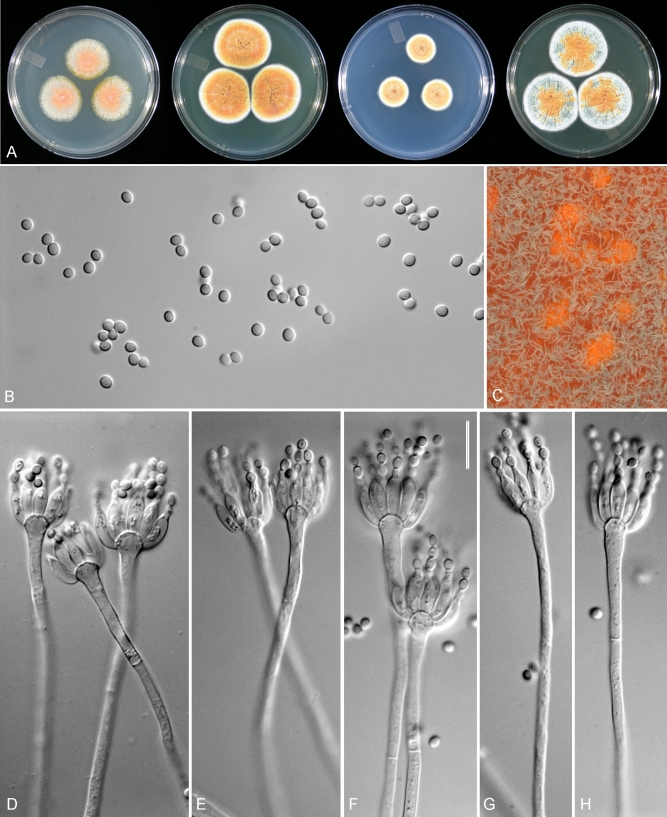
Penicillium sclerotiorum, DAOM 239931. A. Colonies grown for 7 d on at 25 °C CYA, MEA, CZ, YES. B. Conidia. C. Close-up of colony on MEA, showing conidial columns and orange hyphae around sclerotia. D–H. Conidiophores. Scale bar in F = 10 μm for all micrographs.
Colonies on CYA after 7 d at 25 °C: 18–40 mm diam, low and velutinous, ca. 1 mm deep, with 8–11 sulcae and 2–3 wrinkles in some strains, sporulation dense in absence of sclerotia, very sparse or absent when orange sclerotia produced, conidia Greenish Grey (25–27E2), aerial mycelium sparse or absent, exudate moderately produced and light yellow in some strains, margin entire, reverse Orange to Reddish Yellow (3A7–4A6), Brown (7E4–8) or pale, some strains with darker colours turning orange towards the centre, vivid orange soluble pigments present in some strains. Colonies on CZ resembling those on CYA, 15–30 mm diam, with fewer sulcae, the sclerotia tending to be darker or reddish orange when present, exudate droplets light yellow to red when present, reverse less pigmented in some strains and then Yellowish White (1–4A2–3).
Colonies on MEA after 7 d at 25 °C: 15–32 mm diam, planar, strictly velutinous when sporulating, orange sclerotia produced by some strains, in some strains with yellow sclerotia towards the centre, conidia of medium abundance, Grey (25–28D–F1) or Greyish Green (25–28C2), aerial mycelium sparse or absent, exudates not produced, margin entire, reverse Orange (6B7–8), Reddish Orange (7A–B7–8), Reddish Yellow (4A6) or Pale (1A2), soluble pigments not produced.
Colonies on YES after 7 d at 25 °C: (20–) 33–38 (–44) mm diam, dense and with 16–20 sulcae and 6–8 wrinkles present in some strains, light orange and orange sclerotia present in some strains, conidia Turquoise Grey (24B–F2), Grey (27B1), Greenish Grey (25–27C–D2), with raised concentric rings of aerial mycelium, exudates not produced, margin entire, reverse Reddish Yellow (4A6–7), Orange (7C8) turning Pastel Yellow (3A4) near the margins or Pale Yellow (4A3), soluble pigments not produced.
Colonies on CREA after 7 d at 25 °C: (13–) 20–28 mm diam, sclerotia present in some strains.
Conidiophores monoverticillate on MEA, borne directly from agar surface, stipes smooth to finely roughened, septate, (88–) 190–400 μm × 2–3 μm, unbranched, vesicles 4–7 μm wide (mean for different strains 4.6–5.9 ± 0.6). Phialides ampulliform to cylindrical, 8–11 × 2.5–3 μm, with short to long necks, periclinal thickenings not obvious. Conidia produced in columns, ellipsoidal, finely roughened, 2–3 μm diam (means for different strains = 2.5–3.0 ± 0.1 × 2–2.5 ± 0.1 μm, n = 25), mean L/W ratio 1.3:1. Sclerotia yellow, orange or reddish orange, subglobose to ellipsoidal, 180–320 μm diam; sclerotial cells 7–11 × 5–8 μm.
Habitat: Air, forest soil, berries of Coffea arabica.
Distribution: Asia (Indonesia), Australia, Pacifica (Hawai'i), but see notes below.
Typification: Indonesia, Java, Buitenzorg, isol. ex air, K.B. Boedijn, holotype IMI 40569 (not seen) ex-type NRRL 2074*, with equivalent culture collection numbers CBS 287.36, ATCC 10494. DNA barcodes: ITS JN626132, cox1 JN626093.
Other cultures examined (Clade A): NRRL 32583, DAOM 239930, 239931*, 239932*. (Clade B): CBS 258.55, 126.65, 118889 (see Table 1).
* indicates sclerotium producing strains.
Notes: Penicillium sclerotiorum is the only species in the complex with clearly ellipsoidal conidia. About half the strains we examined produced orange sclerotia, and as noted above in the analysis of morphological characters, the ex-type strain produces abundant sclerotia in some transfers, and few or none in others. When sclerotia are observed, strains need only be compared with P. johnkrugii, which differs in colony colours on CYA and CZ, and by its subglobose conidia. Because of the revision of the complex here, it is difficult to evaluate the geographical distribution of P. sclerotiorum as reported in the literature. However, even from our limited sampling it is clear that this species is relatively widely distributed in tropical and subtropical areas, and that it is usually associated with soil. Similarly, as reviewed in more detail in the Discussion, the metabolites attributed to this morphological species need to be reevaluated in light of the revised species concept.
A phylogenetic division within P. sclerotiorum was suggested by all gene trees, the first comprising the ex-type group (marked A in Figs 1, 2), and group B comprising strains that did not produce conidia or sclerotia. We excluded the latter from our morphological description of P. sclerotiorum above, which thus describes Clade A. Description of Clade B as a new species may be warranted if sporulating strains can be isolated.
Penicillium cainii K.G. Rivera, Malloch & Seifert, sp. nov. MycoBank MB563159. Figs 3D, 5.
Fig. 5.
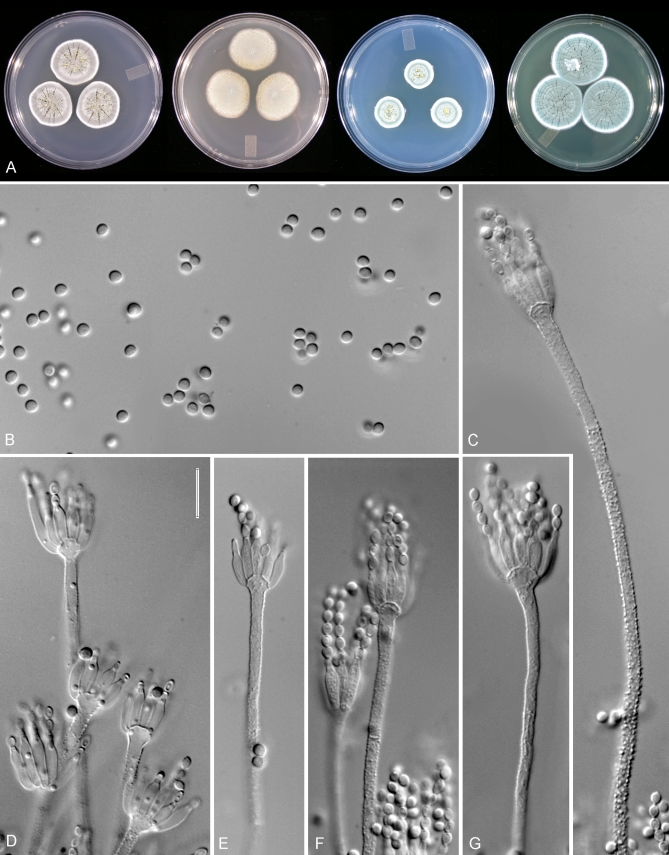
Penicillium cainii. A. Colonies of DAOM 239915 grown for 7 d on at 25 °C CYA, MEA, CZ, YES. B–G. Microphotographs of ex-type strain. B. Conidia. C–G. Conidiophores. Scale bar in D = 10 μm for all micrographs.
Etymology: Named for Roy F. Cain, a faculty member of the University of Toronto, a world authority on coprophilous Ascomycota, who was the Ph.D. supervisor of David Malloch and John Krug.
Coloniae in agaro CYA post 7 dies ad 25 °C 23–29 mm diam, conidia veneta vel viridia, exudatum flavum, in reverso flaviscentes vel flavi-brunneum. Coloniae in agaro MEA post 7 dies ad 25 °C 28–35 mm diam. Conidiophora monoverticillata vel raro metula singula, stipites 69–79 μm × 2.5–3 μm, parietibus asperulatis, ad apicem in vesiculam 4–6 μm latam inflati; metulae adsunt 31–34 μm longae. Cellulae conidiogenae phialidicae, ampulliformes, 7.5–10 × 2–3 μm. Conidia globosa vel subglobosa, levia vel plusminusve asperulata, 2.0–2.5 μm diam.
Colonies on CYA after 7 d at 25 °C: 23–29 mm diam, dense and velutinous, ca. 1 mm deep, with 14–18 sulcae and 1–2 radial wrinkles in some strains, sclerotia absent, conidia medium in abundance, Deep Blue to Deep Green (23–27E2), 1–2 mm of white mycelia at the margin, clear yellow exudates produced moderately by some strains, margin entire, reverse Golden Yellow (5B7) and Brownish Yellow (5C8–9), Yellowish White (2–3A2) towards the edges, soluble pigments not produced, margin entire. Colonies on CZ after 7 d at 25 °C: 18–21 mm diam, similar to colonies on CYA but lacking sulcae and radial wrinkles, conidia Turquoise Grey (24B–F2), with a concentric ring of paler shades of this colour towards the centre, margin entire, reverse Brownish Yellow (5–6C8), turning Yellowish Grey (3–4B2) towards the edges.
Colonies on MEA after 7 d at 25 °C: 28–35 mm diam, planar, dense and velutinous, sclerotia not produced, conidia Greenish Grey (28E2) and Dull Green (27C–D2–4), aerial mycelium not present, exudate not produced, margin entire, reverse Orange to Dark Orange (5A–B8), Yellowish White (2–3A2) towards the edges, and soluble pigments not produced.
Colonies on YES after 7 d at 25 °C: 31–34 mm diam, dense and velutinous, with 21–23 sulcae and 8–10 radial wrinkles, sclerotia not produced, conidia Greenish Grey (27D–F2), 2 mm of white mycelia at the margin, clear dark orange exudates droplets produced in negligible amounts in some strains, margin entire, reverse Brownish Orange to Reddish Orange (7B–D8), Greyish Brown (2B3) towards the edges, wrinkled towards the centre, soluble pigments not produced.
Colonies on CREA after 7 d at 25 °C: (13–) 25–31 mm diam.
Conidiophores mostly monoverticillate on MEA, borne from agar surface, stipes rough-walled, septate, 70–80 μm × 2.5–3 μm, vesicle 3.5–5.0 μm wide, unbranched or in some strains with ca. 10 % of conidiophores with a single branch 31–34 μm long. Phialides ampulliform, 7.5–10 × 2–3 μm, with a distinguishable neck and inconspicuous periclinal thickening. Conidia globose, finely roughened, 2.0–2.5 μm diam (mean for different strains = 2.3–2.4 x 2.1–2.2 ± 0.035 μm), mean L/W ratio 1.07:1.
Habitat: Nuts of Juglans nigra and Carya ovata.
Distribution: North America (Canada: Ontario).
Typification: canada, Ontario, Niagara, Niagara Falls, Fireman's Park, N43° 08' 49” W79° 07' 04, isol. ex nuts of black walnut, Juglans nigra, D. Malloch W-10, May 1996, holotype DAOM 239914 (dried culture). The ex-type strain has the same accession number and is maintained in the Canadian Collection of Fungal Cultures. DNA barcodes: ITS JN686435, cox1 JN686412.
Other culture examined: Same location and date as type, isol. ex nuts of shagbark hickory, Carya ovata, D. Malloch W-15, DAOM 239915.
Notes: Penicillium cainii produces short conidiophores with conspicuously roughened stipes, and these microscopic characters combined with the substrate of nuts of various species of the family Juglandaceae make the species easily recognisable. Neither strain produced sclerotia. Colonies resemble those of P. sclerotiorum, P. mallochii, P. guanacastense, and P. viticola on CYA, MEA, and YES, but colonies on CZ differ by the production of a concentric ring of a light shade of turquoise grey and clear yellow exudate droplets in the centre of the colony. The vesicles terminating the monoverticillate conidiophores tend to be more clavate than swollen, but this is a difficult character to interpet.
Penicillium jacksonii K.G. Rivera, Houbraken & Seifert, sp. nov. MycoBank MB563160. Figs 3E, 6.
Fig. 6.
Penicillium jacksonii. A. Colonies of DAOM 239938 grown for 7 d on at 25 °C CYA, MEA, CZ, YES. B–H. Microphotographs of ex-type strain. B. Conidia. C–H. Conidiophores. Scale bar in F = 10 μm for all micrographs.
Etymology: Named for H.S. Jackson, faculty member at the University of Toronto, an authority on rusts, but an avid collector of all fungi, and the Ph.D. advisor of R.F. Cain.
Coloniae in agaro CYA post 7 dies ad 25 °C 30–33 mm diam, conidia viridia, exudatum flavum, in reverso flaviscentes. Coloniae in agaro MEA post 7 dies ad 25 °C 31–37 mm diam. Conidiophora monoverticillata vel modice metula singula, stipites 83–134 × 2–3 μm, parietibus plusminusve asperulatis, apicem in vesiculam 3–6 μm latam inflati; metulae adsunt 28–48 μm longae. Cellulae conidiogenae phialidicae, ampulliformes, 6–5 (–13) × 2–3 μm. Conidia globosa vel subglobosa, levia vel plusminusve asperulata, 2.5–3 μm diam.
Colonies on CYA after 7 d at 25 °C: 30–33 mm diam, dense and velutinous, ca. 1 mm deep, with 10–11 sulcae but no radial wrinkles, conidia produced abundantly, Deep Green (25–26E2), with concentric rings of paler shades of these colours, aerial mycelium present towards the centre, clear yellow exudate produced sparsely by some strains, with a margin 1–3 mm of white mycelia, margin entire, reverse Yellow (3A6–7), Vivid Yellow (2–3A8), or near the edges Yellowish White (2A2), soluble pigments not produced. Colonies on CZ after 7 d at 25 °C: 19–23 (–30) mm diam, similar to colonies on CYA but lacking sulcae, conidia Greenish Grey (26–27E2), aerial mycelium present throughout the colony, one strain floccose near the centre, with 1–2 mm of white mycelia near the margin, reverse Brownish Yellow (5C8–9), Deep Yellow (4A8), Yellowish White towards the edges (1A2), soluble pigments not produced
Colonies on MEA after 7 d at 25 °C: 31–37 mm diam, planar and velutinous, moderately dense, sporulation dense, conidia Dull Green (26–27E3), with concentric ring near the edges of paler shades of these colours, margin entire, reverse pale.
Colonies on YES after 7 d at 25 °C: 30–32 mm diam, dense, velutinous, with 8–10 sulcae in the centre and 19–23 sulcae at the margin, and 6–9 radial wrinkles, sporulation good, conidia Greenish Grey (25E2), with concentric rings of paler shades of this colour present in some strains, aerial mycelia absent or white patches present occasionally, margin entire, reverse Yellowish White (3A2) or Reddish Yellow (4A6), soluble pigments not produced.
Colonies on CREA after 7 d at 25 °C: 26–33 mm diam.
Conidiophores monoverticillate and/or once-branched on MEA, borne from agar surface, stipes smooth to rough, septate, 80–135 × 2–3 μm, vesicle 3–6 μm wide (mean for different strains 4.4–4.7 ± 0.4 μm), in some strains all unbranched, in others with ca. 65 % of conidiophores with a single branch 28–48 μm long. Phialides ampulliform, 6–5(–13) × 2–3 μm wide, with distinguishable collarette and inconspicuous periclinal thickening. Conidia globose, walls finely roughened, 2.5–3 μm diam (mean for different strains 2.8 × 2.5–2.6 ± 0.035 μm), mean L/W ratio 1.1:1.
Habitat: Forest soil.
Distribution: Queensland, Australia.
Typification: Australia, Queensland, Barrine Lake, S17° 15′ 1″ E145° 38′ 7″ E, isol. from soil pretreated with ethanol, Sept. 2006, leg. J. Houbraken, L. Janson, holotype DAOM 239937 (dried culture). The ex-type strain has the same accession number and is maintained in the Canadian Collection of Fungal Cultures. DNA barcodes: ITS JN686437, cox1 JN686414.
Other culture examined: Same data as type, DAOM 239938.
Notes: Penicillium jacksonii produces short conidiophores, but otherwise has few microscopic characters that clearly distinguish it from related species. It is unusual in the P. sclerotiorum complex for the production of a high proportion of conidiophores with a single branch, but this is an inconsistent character and some transfers are strictly monoverticillate. Colonies of P. jacksonii colonies are similar to those of P. mallochii, P. guanacastense, P. viticola, and P. cainii on CYA, MEA, and on YES, but the conidia are perhaps the darkest green of the species. The most closely related species, P. cainii (Fig. 2), has conspicuously roughened conidiophores.
Penicillium johnkrugii K.G. Rivera, Houbraken & Seifert, sp. nov. MycoBank MB563161. Figs 3B, 7, 8.
Fig. 7.
Penicillium johnkrugii, ex-type strain A. Sclerotia on MEA. B. Micrograph of sclerotium. C. Conidia. D–H. Conidiophores. Scale bars A = 200 μm, B = 20 μm, in D = 10 μm for micrographs (C–H).
Fig. 8.
Penicillium johnkrugii, three strains grown for 7 d on at 25 °C CYA, MEA, CZ, YES. A. DAOM 239942. B. DAOM 239943 (T). C. DAOM 239944.
Etymology: Named for John Krug, faculty member at the University of Toronto and a research associate at the Royal Ontario Museum (ROM). Like his PhD supervisor, RF Cain, he was a specialist on coprophilous fungi, but also an avid lichen collector. He introduced KGR to the world of fungal taxonomy.
Coloniae in agaro CYA post 7 dies ad 25 °C 30–38 mm diam, alba, conidia sparsa, sclerotia abundans, grisea vel aurantia, ca. 135–550 × 130–430 μm, exudatum plusminusve flavum, in reverso aurantiae vel flaviscentes. Coloniae in agaro MEA post 7 dies ad 25 °C 26–36 mm diam. Conidiophora monoverticillata, stipites 88–229 μm × 2–2.5 μm, parietibus plusminusve asperulatis, apicem in vesiculam 4–6 μm latam inflati. Cellulae conidiogenae phialidicae, ampulliformes, 7–11 × 2–3(–5) μm. Conidia globosa vel subglobosa, levia vel plusminusve asperulata, 2–3 μm diam.
Colonies on CYA after 7 d at 25 °C: 30–38 mm diam, planar, surface mycelia White (1–3A1), ca. 1 mm deep, with 5–9 sulcae and 2–3(–9) radial wrinkles, sclerotia white, conidia not produced, light yellow exudate droplets produced in low to moderate amounts, margin entire, reverse Reddish Yellow to Orange (4–6AB7–8) and Yellowish Grey (3–4B2) towards the edges, stellate with radial wrinkles, soluble pigments absent. Colonies on CZ after 7 d at 25 °C: 15–23 mm diam, occasionally 10–19 mm diam, velutinous and dense, 4–6 sulcae and 1–2 radial wrinkles present in some strains, sclerotia white, with sparse to negligible sporulation, conidia Greenish Grey (25–27D2), light yellow exudates droplets sparsely produced, reverse Yellow to Deep Yellow (3A6–8) and Orange (5–6B8), some strains Yellowish White (3A3) or Pastel Yellow (3A4) towards the centre, soluble pigments absent.
Colonies on MEA after 7 d at 25 °C: 26–36 mm diam, planar, low and velutinous, sclerotia orange, yellow towards the centre, and white towards the margin, conidia Greenish Grey (24–27DE 3–5), conidia and sclerotia sectoring in some strains, aerial mycelium not present, exudates not produced, margin entire, reverse Orange (5–6B8), Vivid Yellow (3A8), Yellowish Grey (3–4B2) towards the centre, soluble pigment not produced.
Colonies on YES after 7 d at 25 °C: 28–38 mm diam, velutinous, with 11–17 sulcae and 6–10 radial wrinkles, sclerotia white, sporulation poor, conidia Greenish Grey (25D2), yellow exudate droplets produced sparsely by some strains, white mycelia at the marginal 1 mm, margin entire, reverse Yellow (3A3–4) and Orange (5–6B8), some strains Pastel Yellow (3A3–4) towards the edges, stellate with radial wrinkles or wrinkled, soluble pigments not produced.
Colonies on CREA after 7 d at 25 °C: 14–22 mm diam, sclerotia present.
Conidiophores strictly monoverticillate on MEA, borne from agar surface, stipes smooth to finely roughened, septate, 85–230 × 2–2.5 μm, vesiculate, 4–6 μm wide (means for different strains 4.6–5.4 ± 0.3). Phialides ampulliform to cylindrical, 7–11 × 2–3(–5) μm, with short to distinguishable necks, periclinal thickenings not obvious. Conidia globose to subglobose, finely roughened, 2–3 μm diam (means for different strains 2.7–2.9 ± 0.01 × 2.3–2.5 ± 0.01 μm, n = 25), mean L/W ratio 1.1:1. Sclerotia produced on all media, subglobose to ellipsoidal, at first white, becoming orange or yellow, 136–552 × 131–433 μm, sclerotial cells 5–8 × 3–6 μm.
Habitat: Forest soil.
Distribution: Langkawi, Malaysia.
Typification: Malaysia, Kedah, Langkawi, N 6° 19' 24” E 99° 51' 45”, isol. ex soil after ethanol treatment, Nov. 2007, leg. R.A. Samson, isol. J. Houbraken, holotype DAOM 239943 (dried culture). The ex-type strain has the same accession number and is maintained in the Canadian Collection of Fungal Cultures. DNA barcodes: ITS JN686447, cox1 JN686424.
Other cultures examined: Same data as type and presumably from the same soil sample, DAOM 239939, DAOM 239940, DAOM 239941, DAOM 239942, DAOM 239944, DAOM 239945, DAOM 239946.
Notes: Penicillium johnkrugii is distinct within the P. sclerotiorum complex for its production of white colonies and abundant grayish sclerotia on CYA and CZ; on other media, the sclerotia tend to be yellow or orange. The species is morphologically similar to P. sclerotiorum. Both have vivid yellow to orange reverse colony colours although P. johnkrugii differs by subglobose conidia and more conspicuously vesiculate conidiophores.
Penicillium johnkrugii and P. mallochii were paraphyletic in some analyses of benA, cox1, and ITS. Monophyletic recognition of the species occurred in all tef1-α analyses, and in the MP and ML trees for ITS, and ML and BI trees for cmd. Despite these phylogenetic issues, all genes provided diagnostic sequences for P. johnkrugii, and the species is morphologically distinct.
The strain DAOM 239944 did not cause any colour changes when grown on CREA.
Penicillium viticola Nonaka & Masuma, Mycoscience 52: 339. 2011. MycoBank MB516048. Figs 3C, 9.
Fig. 9.
Penicillium viticola. A. Colonies of DAOM 239933 grown for 7 d on at 25 °C CYA, MEA, CZ, YES. B–G. Microphotographs of DAOM 239935. B. Conidia. C–G. Conidiophores. Scale bar in C 10 μm for all micrographs.
Colonies on CYA after 7 d at 25 °C: 26–30 (–36) mm diam, dense and velutinous, ca. 1 mm deep, with 6–13(–17) sulcae and 2–3 radial wrinkles, sclerotia absent, conidia Greenish Grey (25–27E2), with Bluish Grey (22D–F2) concentric rings, pale yellow exudate droplets produced sparsely by some strains, with white aerial mycelium in the central 2–3 mm and marginal 2–3 mm, margin entire, reverse Light Yellow (5C8), Yellowish Grey (3–4B2), Orange Grey (6B7–8), sometime Yellowish Grey (3–4B2) or Yellowish White (1A2–3) near the margin, stellate with some radial wrinkles, soluble pigments not produced. Colonies on CZ after 7 d at 25 °C: 17–25 mm diam, similar to those on CYA, but some strains floccose at the centre, with 5–6 sulcae and 2–3 wrinkles in some strains, mycelia at margin 1–2 mm, reverse Reddish Yellow (4A7) and Brownish Yellow (5C8) in some strains.
Colonies on MEA after 7 d at 25 °C: 23–35 (–40) mm diam, planar and moderately dense, velutinous, sclerotia not observed, conidia Greenish Grey (25–27E2), aerial mycelium not observed, exudates not produced, margin entire, reverse Orange Yellow to Orange (4–6B7–8), Brownish Orange (7C7–8), Pastel Yellow or Reddish Yellow (4A6–7) near the edges, soluble pigment not produced.
Colonies on YES after 7 d at 25 °C: 29–33 mm diam, less commonly 34–36 mm diam, dense and velutinous, densely sulcate, especially near the edges (with 15–21 sulcae), with 7–9 radial wrinkles, sclerotia not produced, conidia Greenish Grey (25–26D–E2) and Turquoise Grey (24D2), concentric rings of different shades of turquoise grey present in some strains, white mycelia 2–3 mm at margins, clear vivid yellow exudate droplets produced sparsely by some strains, margin entire, reverse Reddish Yellow (7B8), Orange (6B8), Greyish Yellow (2–4B3), some strains Yellowish Grey (2B–C3) or Dull Yellow (3B3–4) towards the edges, wrinkled towards the centre with visible sulcae and radial wrinkles towards the margin, soluble pigment not observed.
Colonies on CREA after 7 d at 25 °C: 23–29 mm diam.
Conidiophores predominantly monoverticillate on MEA, borne from agar surface, stipes rough, septate, 30–135 × 2–3 μm wide, moderately vesiculate, vesicles 4–6 μm wide (means for different strains 4.6–5.0 ± 0.3 μm), mostly unbranched, sometimes with about 10 % of conidiophores with a single branch 22–55 × 2–3 μm wide. Phialides ampulliform, 7–10 × 2–3 μm, with a distinguishable collarette. Conidia borne in columns, globose, smooth, 2–3 μm diam (means for different strains 2.6–2.7 × 2.3–2.6 ± 0.01 μm), mean L/W ratio 1.1:1.
Typification: Japan, Yamanashi, isol. ex grape (Vitus sp.), 11 Jul 2006, K. Nonaka, holotype TNS-F-38702, ex-type culture JCM 17636 = FKI-4410 (not seen). DNA barcode: ITS AB606414 (Nonaka et al. 2011).
Other cultures examined: DAOM 239933, DAOM 239934, DAOM 239935, DAOM 239936, see Table 1 for details.
Habitat: Rainforest soil, and vines of Vitus sp.
Distribution: Asia (Japan), Australia (Queensland).
Notes: Penicillium viticola produces short, rough-walled conidiophores, slightly less roughened than those of the closely related species P. cainii, but rougher than all other species of the complex. The colony characters of P. viticola colony morphology resemble those of P. mallochii, P. guanacastense, P. cainii, and P. sclerotiorum on CYA and MEA. On CZ, it resembles P. mallochii and P. guanacastense. Microscopically it lacks the more vesiculate conidiophore apices observed in P. mallochii, P. guanacastense, and P. sclerotiorum.
Penicillium viticola was described by Nonaka et al. (2011) when our own revision was completed, and we were unable to examine the type strain. Their description matches ours in most respects, with only minor differences in growth rates, colony colours and exudate production that may be attributed to subtle variations in media. The published calmodulin tree in Nonaka et al. (2011) inadvertently included a mislabeled sequence for P. viticola that indicated a relationship with P. angulare; the sequence deposited by the authors (AB540173) is correct (R. Masuma, pers. comm.). With the benA sequence for the type strain (AB540174), this species is firmly established as a member of the P. sclerotiorum complex, and conspecific with our Australian strains, as it is treated here.
Iwatsuki et al. (2010) reported the production of tropolone compounds by the ex-type strain of P. viticola (in the publication named only as Penicillium sp.), including the anti-malarial compound puberulic acid, stipitatic acid and their novel analogues, viticolins A–C.
DISCUSSION
The Penicillium sclerotiorum complex now includes seven phylogenetically distinct but morphologically similar species. In this paper, three species in the P. sclerotiorum complex were described, added to the two species recently described from Costa Rica (P. guanacastense, P. mallochii; Rivera et al. 2011) and one recently described from Japan (P. viticola, Nonaka et al. 2011). Members of the complex can be recognised by the general suite of characters used by previous authors to delineate the species, namely moderately fast growing colonies on CYA and MEA, with a tendency to produce orange or reddish colony reverses, monoverticillate conidiophores with some conidiophores having a single branch, vesiculate conidiophore apices, and globose to ellipsoidal conidia in greyish green, dull green, or turquoise green colours. Careful examination of colony and microscopic characters revealed subtle morphological differences among species. The species differ by some conidial colours, variation in the roughening of the stipe, stipe lengths, and inconspicuously in the extent of vesiculation of the conidiophore apex and in conidial shape. Sclerotia are produced abundantly and consistently by P. johnkrugii, in most but not all fresh strains of P. sclerotiorum, and have not been seen in the other species. Sclerotium production is a difficult taxonomic character, because they may be constant or inconstant within a species or even a strain. Physiological experiments with Aspergillus caelatus show that a pH of 6–10 and temperatures of 28–30 °C are optimal for sclerotia formation, production peaks at a C:N ratio of 8.6, and the sugar used in CYA (sucrose) suppresses production by 12 % (McAlpin 2004). Similar results have been seen in other Aspergillus species (Rudolph 1962), Sclerotinia rolfsii (Wheeler & Sharan 1965), and Verticillium species (Wyllie & DeVay 1970).
Five of the species so far have restricted geographical distributions, but sampling is still very meagre. Penicillium johnkrugii was isolated from soil from Langkawi, Malaysia, P. jacksonii from soil from Queensland, Australia, P. cainii from nuts collected in Niagara Falls, Ontario, while P. mallochii and P. guanacastense were isolated from the guts of two different caterpillar families reared in the Área de Conservación Guanacaste, Costa Rica. Penicillium sclerotiorum has the broadest known distribution (Table 1), while P. viticola has a disjunct distribution (soil from Australia, grapes from Japan) that hints at a broader distribution and a so-far undefined ecology.
Extrolite profiling is a common practice for characterising Penicillium species and contributed significantly to developing polyphasic species concepts in Penicillium subgenus Penicillium (Frisvad & Samson 2004). We were unable to study extrolites in this study, but there are indications that the P. sclerotiorum complex may be metabolically diverse, providing further characters for delimiting these morphologically similar species. Secondary metabolites with antibacterial and antifungal activities were reported for some strains identified as P. sclerotiorum sensu lato, including the antimicrobial compounds sclerotin (Curtin & Reilly 1940), isochromphilone VI and pencolide (De Lucca et al. 2008). Pairet et al. (1995) reported two azapholines from P. sclerotiorum as antagonists of endothelin-A and endothelin-B receptors, vasoconstriction peptides implicated in hypertension, heart and renal failure, ischemia, and cerebral vasospasms. We could not trace strains from these studies and therefore it is unclear what phylogenetic species produce these compounds. As noted above, only the production of puberulic acid, stipitatic acid and viticolins A–C can be attributed with any certainly to the phylogenetically defined P. viticola, but only the ex-type has been examined for these metabolites (Iwatsuki et al. 2010).
Phylogenetic analyses using maximum parsimony, maximum likelihood, and Bayesian inference algorithms, and DNA sequence data benA, ITS, cox1, tef1-α, and cmd, gave consistent results. The less variable genes did not provide robust statistical support for all species of the complex, but the results did not conflict with the other gene trees. The recognised species generally conform to the Genealogical Concordance Phylogenetic Species Recognition (Taylor et al. 2000) concept, although some of the species are only represented by two strains. There were problems with paraphyly of P. johnkrugii and P. mallochii in some benA, cox1, and ITS analyses, but all five genes yielded species specific sequences for all species, and each species formed a cohesive (if not strictly monophyletic) group that reaffirmed the species concepts proposed here. We have listed accession numbers of ITS and cox1 barcodes in the paragraphs on Typification above, although all tested genes would work as barcodes in this complex.
The partition homogeneity test (PHT) indicated that the cmd was incongruent with the other genes, and this gene was not included in combined phylogenetic analyses. Incongruency in phylogenetic signal between genes suggests different evolutionary histories for these genes (Scott et al. 2006). Visual comparisons of the cmd results suggest that the variable position of P. guanacastense within the ingroup, and a generally discordant arrangement of outgroups, probably led to the failure of the PHT. From the perspective of recognising phylogenetic species, the species groupings remained constant for all genes. From the perspective of accurately determining sister group relationships among species, the rejection of cmd is unfortunate.
The designation of P. adametzioides and P. multicolor as synonyms of P. sclerotiorum, as proposed by Stolk & Samson (1983), was not accepted here. Penicillium adametzioides is a distinct phylogenetic species, also noted by Peterson (2000) and accepted by Houbraken & Samson (2011). According to our molecular data, P. multicolor is a synonym of P. fellutanum, but the degenerated morphology of the ex-type strain we examined leaves this conclusion tentative.
KEY TO SPECIES
1. Conidiophore stipes distinctly roughened.................................................................................................................................................. 2
1. Conidiophore stipes at most finely roughened.......................................................................................................................................... 3
2. On CYA conidial colours with a blue tinge, colonies usually less than 30 mm diam; on nuts.......................................................... P. cainii
2. On CYA, conidia colours lacking blue tinge, colonies 26–36 mm diam; on grape vines or in soil................................................ P. viticola
3. Sclerotia produced.................................................................................................................................................................................... 4
3. Sclerotia not produced.............................................................................................................................................................................. 5
4. Colonies on CYA white, sclerotia white or grey, turning orange on MEA; conidia globose or subglobose............................... P. johnkrugii
4. Colonies on CYA with obvious green areas, sclerotia orange; conidia subglobose to ellipsoidal....................................... P. sclerotiorum
5. Associated with caterpillars feeding on leaves in the neotropics; colonies on MEA crustose................................................................... 6
5. Soilborne; colonies on MEA not crustose.................................................................................................................................................. 7
6. Associated with Saturnid caterpillars; conidiophores 50–380 μm long..................................................................................... P. mallochii
6. Associated with Noctuid caterpillars; conidiophores 85–100 μm long............................................................................ P. guanacastense
Note: Colony photographs of P. guanacastense and P. mallochii on CYA, MEA, CZ, YES and CREA are included in Fig. 3; the species are described in Rivera et al. (2011).
7. Colony reverse colours on CYA and MEA pale; some strains with > 50 % of conidiophores with a single branch, other strains all monoverticillate; conidia globose................................................................................................................................................ P. jacksonii
7. Colony reverse colours on CYA and MEA yellow, orange or in vivid red colours; conidiophores strictly monoverticillate; conidia subglobose to ellipsoidal...................................................................................................................................... P. sclerotiorum (non-sclerotial strains)
Acknowledgments
We are grateful for critical reviews of the thesis version of this manuscript by Drs Frances Pick, Guy Drouin and Myron Smith, to Rafik Assabgui for processing sequences, to Gerry Louis-Seize and Tom Gräfenhan for their advice on PCR and sequencing, and the curators of the CBS, DAOM, and NRRL for providing cultures. Jos Houbraken, Rob Samson and David Malloch kindly provided many isolates for this study. This study was funded by the Natural Science and Engineering and Research Council of Canada and Genome Canada through the Ontario Genomics Institute, and other sponsors listed at www.BOLNET.ca.
REFERENCES
- Beyma JFH van. (1937). Penicillium sclerotiorum nov. spec. Centrablatt für Bakteriologie, Parastenkunde und Infektionskrankheiten Abt. II, 96: 481–491 [Google Scholar]
- Curtin TP, Reilly J. (1940). Sclerotiorine, C20H20O5Cl, a chlorine-containing metabolic product of Penicillium sclerotiorum van Beyma. Biochemical Journal 34: 1418–1421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lucca AJ, Klich M, Boue S, Cleveland TE, Sien T, Walsh TJ. (2008). Fungicidal activity of plant saponin CAY-1 for fungi isolated from diseased Vitus fruit and stems. American Journal of Enology and Viticuluture 59: 67–72 [Google Scholar]
- Filtenborg O, Frisvad JC, Trane U. (1990). The significance of yeast extract composition of metabolite production in Penicillium. In: Modern concepts in Penicillium and Aspergillus classification. (Samson RA, Pitt JI, eds). Plenum Press, New York: 433–441 [Google Scholar]
- Frisvad JC. (1993). Modifications on media based on creatine for use in Penicillium and Aspergillus taxonomy. Letters in Applied Microbiology 16: 154–157 [Google Scholar]
- Frisvad JC, Samson RA. (2004). Polyphasic taxonomy of Penicillium subgenus Penicillium. A guide to identification of the food and air-borne terverticillate Penicillia and their mycotoxins. Studies in Mycology 49: 1–174 [Google Scholar]
- Gascuel O. (1997). BIONJ: An improved version of the NJ algorithm based on a simple model of sequence data. Molecular Biology and Evolution 14: 685–695 [DOI] [PubMed] [Google Scholar]
- Glass NL, Donaldson GC. (1995) Development of primer sets deigned for use with the PCR to amplify conserved genes from filamentous ascomycetes. Applied and Environmental Microbiology 61: 1323–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Lethiec F, Duroux P, Gascuel O. (2005). PhyML online: A web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Research 33 (Web Server issue): W557–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall TA. (1999). BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series 41: 95–98 [Google Scholar]
- Hebert PDN, Cywinska A, Ball SL, de Waard JR. (2003). Biological identifications through DNA barcodes. Proceedings of the Royal Society of London Series B-Biological Sciences 270: 313–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong SB, Cho HS, Shin HD, Frisvad JC, Samson RA. (2006). Novel Neosartorya species isolated from soil in Korea. International Journal of Systematics and Evolutionary Microbiology 2: 477–486 [DOI] [PubMed] [Google Scholar]
- Houbraken J, Samson RA. (2011). Phylogeny of Penicillium and the segregation of Trichocomaceae into three families. Studies in Mycology 70: 1–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. (2001). MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics 17: 754–755 [DOI] [PubMed] [Google Scholar]
- Iwatsuki M, Takada S, Ishiyama A, Namatame M, Tukashima-Nishihara A, Nonaka K, Masuma R, Mori M, Shiomi K, Otoguro K, Omura S. (2010). In vitro and in vivo antimalarial activities of puberulic acid and its new analogs, viticolins A-C, produced by Penicillium sp. FKI-4410. Journal of Antibiotics (Tokyo) 64: 183–188 [DOI] [PubMed] [Google Scholar]
- Katoh K, Asimenos G, Toh H. (2009). Multiple alignment of DNA sequences with MAFFT. Methods in Molecular Biology 537: 39–64 [DOI] [PubMed] [Google Scholar]
- Kornerup A, Wanscher JH. (1978). Methuen handbook of color, 3rd ed. Denmark, Sankt Jørgen Tryk; [Google Scholar]
- McAlpin CE. (2004). Synnema and sclerotium production in Aspergillus caelatus and the influence of substrate composition on their development in selected strains. Mycologia 5: 937–947 [PubMed] [Google Scholar]
- Nonaka K, Masuma R, Iwatsuki M, Shiomi K, Otoguro K. (2011). Penicillium viticola, a new species isolated from a grape in Japan. Mycoscience 52: 338–343 [Google Scholar]
- Pairet L, Wrigley SK, Chetland I, Reynolds EE, Hayes MA, Holloway J, Ainsworth AM, Katzer W, Cheng XM, Hupe DJ, Charleton PDAM. (1995). Azapilones with endothelin receptor binding activity produced by Penicillium sclerotiorum: Taxonomy, isolation, structure elucidation and biological activity. The Journal of Antibiotics 48: 913–929 [DOI] [PubMed] [Google Scholar]
- Peterson SW. (2000). Phylogenetic analyses of Penicillium species based on ITS and LSU -rDNA nucleotide sequences. In: Integration of modern taxonomic methods for Penicillium and Aspergillus. (Samson RA, Pitt JI, eds). Hardwood Academic Publishers, Amsterdam: 163–178 [Google Scholar]
- Peterson SW, Bayer EM, Wicklow DT. (2004). Penicillium thiersii, Penicillium angulare and Penicillium decaturense, new species isolated from wood-decay fungi in North America and their phylogenetic placement from multilocus DNA sequence analysis. Mycologia 96: 1280–1293 [PubMed] [Google Scholar]
- Pitt JI. (1973). An appraisal of identification methods for Penicillium species: Novel taxonomic criteria based on temperature and water relations. Mycologia 65: 1135–1157 [PubMed] [Google Scholar]
- Pitt JI. (1980). The Genus Penicillium and its Teleomorphic States Eupenicillium and Talaromyces. Academic Press, London, UK: [Google Scholar]
- Posada D, Crandall KA. (1998). Modeltest: testing the model of DNA substitution. Bioinformatics 14: 817–818 [DOI] [PubMed] [Google Scholar]
- Ramírez C. (1982). Manual and atlas of the Penicillia. Elsevier Biomedical Press, Amsterdam: [Google Scholar]
- Raper KB, Thom C. (1949). A Manual of the Penicillia. Williams & Wilkins Co., Baltimore: [Google Scholar]
- Rivera KG, Díaz J, Chavarría-Díaz F, Garcia M, Urb M, Thorn RG, Louis-Seize G, Janzen DH, Seifert KA. (2011). Penicillium mallochii and P. guanacastense, two new species isolated from Costa Rican caterpillars. Mycotaxon (in press) [Google Scholar]
- Ronquist F, Huelsenbeck JP. (2003). MrBayes v. 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574 [DOI] [PubMed] [Google Scholar]
- Rudolph ED. (1962). The effect of some physiological and environmental factors on sclerotial Aspergilli. American Journal of Botany 49: 71–78 [Google Scholar]
- Samson RA, Seifert KA, Kuijpers AFA, Houbraken JAMP, Frisvad JC. (2004). Phylogenetic analysis of Penicillium subgenus Penicillium using partial beta-tubulin sequences. Studies in Mycology 48: 175–200 [Google Scholar]
- Sawane AM, Saoji AA. (2004). A report on Penicillium in the intramural and extramural air of residential areas of Nagpur city (India). Aerobiologia 20: 229–236 [Google Scholar]
- Scott JB, Chkraborty S. (2006). Multilocus sequence analyses of Fusarium pseudograminearum reveals a single phylogenetic species. Mycological Research 110: 1413–1425 [DOI] [PubMed] [Google Scholar]
- Seifert KA, Samson RA, deWaard J, Houbraken J, Levesque CA, Moncalvo JM, Louis-Seize G, Hebert PDN. (2007). Prospects for fungus identification using CO1 DNA barcodes. Proceedings of the National Academy of Science 104: 3901–3906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolk AC, Samson RA. (1983). The Ascomycete genus Eupenicillium and related Penicillium anamorphs. Studies in Mycology 23: 1–149 [Google Scholar]
- Swofford DL. (2003). Phylogenetic Analysis Using Parsimony (*and Other Methods). v. 4, Sinauer Associates, Sunderland, Massachusetts: [Google Scholar]
- Taylor JW, Jacobson D, Kroken S, Kasuga T, Geiser DM, Hibbitt DS, Fisher MC. (2000). Phylogenetic species recognition and species concepts in fungi. Fungal Genetics and Biology 31: 21–32 [DOI] [PubMed] [Google Scholar]
- Vega FE, Posada F, Peterson SW, Gianfagna TJ, Chavez F. (2006). Penicillium species endophytic in coffee plants and ochratoxin A production. Mycologia 98: 31–42 [DOI] [PubMed] [Google Scholar]
- Vesper SJ, Wymer LJ, Meklin T, Varma M, Stott R, Richardson M, Haugland RA. (2005). Comparison of populations of mould species in homes in the UK and USA using mould–specific quantitative PCR. Letters in Applied Microbiology 41: 367–373 [DOI] [PubMed] [Google Scholar]
- Wheeler BEJ, Sharon N. (1965). The production of sclerotia by Sclerotium rolfsii. Transactions of the British Mycological Society 48: 291–301 [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor JW. (1990). Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: PCR protocols: A Guide to Methods and Applications. (Innis MA, Gelfand DH, Sninsky JJ, White TJ, eds). Academic Press, London, UK: 315–322 [Google Scholar]
- Wyllie TD, DeVay JE. (1970). Growth characteristics of several isolates of Verticillium albo-atrum and Verticillium nigrescens from cotton. Phytopathology 60: 907–910 [Google Scholar]