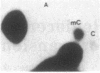
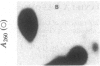
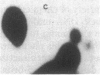
Abstract
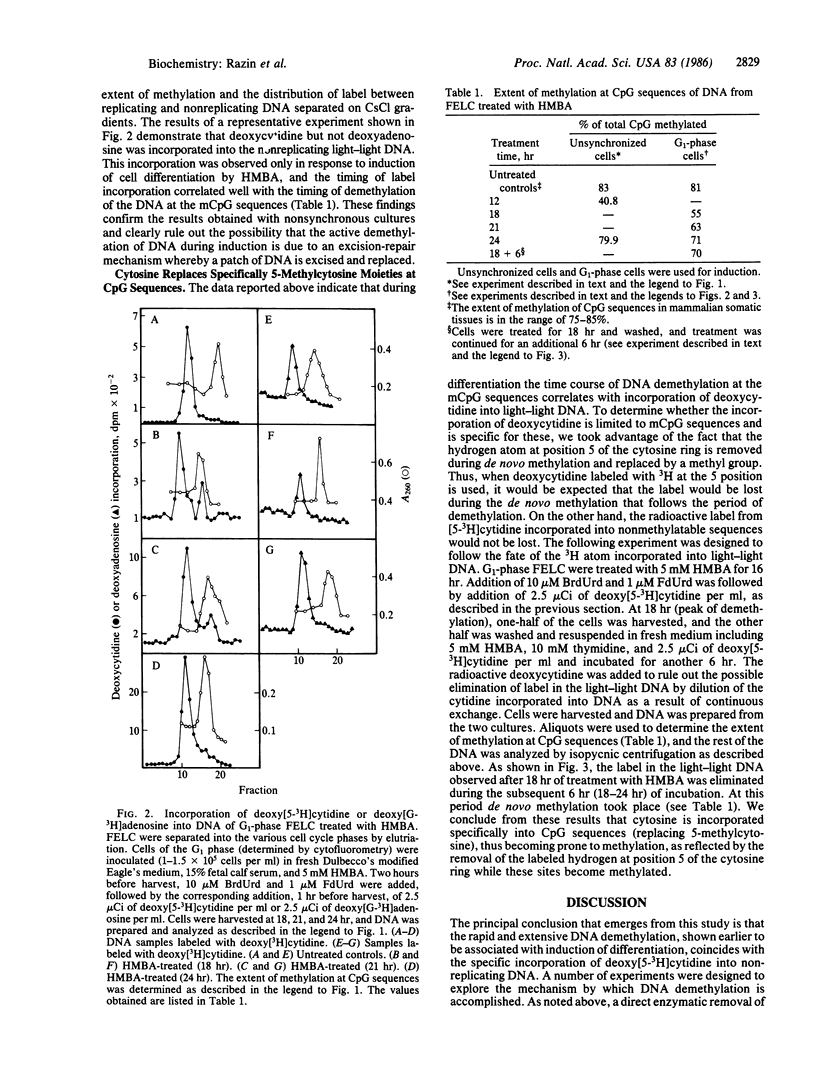
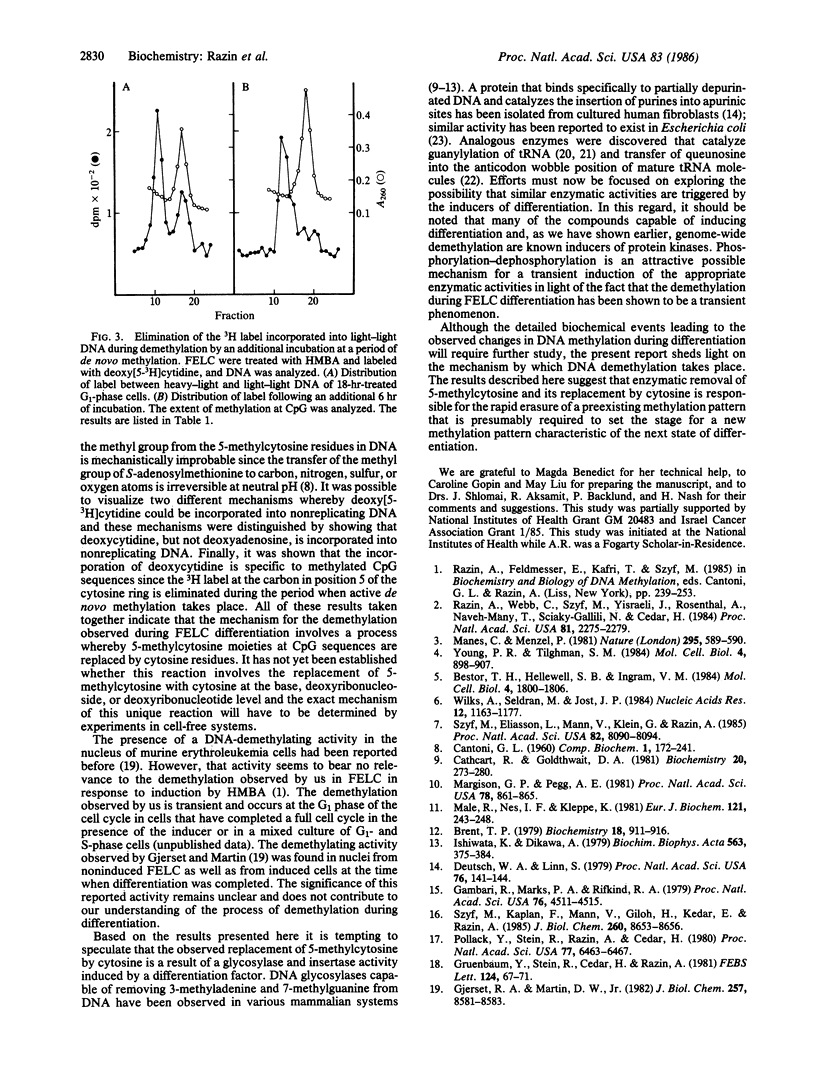
In an earlier study it was discovered that when Friend erythroleukemia cells (FELC) were exposed to a variety of chemical agents capable of inducing differentiation, their DNA underwent genome-wide transient demethylation. In an attempt to elucidate the biochemical mechanism responsible for this phenomenon we have induced FELC with 5 mM hexamethylenebisacetamide and labeled the DNA in vivo with a density label, 5-bromodeoxyuridine, and a radioactive label, deoxy[5-3H]cytidine. Newly replicated DNA (heavy-light) was separated from parental DNA (light-light) by isopycnic centrifugation. Incorporation of deoxy[5-3H]cytidine into light-light duplex DNA has been observed only in induced cells concomitantly with the demethylation of the DNA, whereas, in parallel experiments, deoxy[G-3H]adenosine was not incorporated into light-light DNA. It was also found that the labeling of light-light DNA with deoxy[5-3H]cytidine is transient since the 3H label was removed from the DNA during the period of de novo DNA methylation that follows the demethylation. These results, taken together, strongly suggest that the demethylation of the DNA during differentiation is achieved by an enzymatic mechanism whereby 5-methylcytosine is replaced by cytosine.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bestor T. H., Hellewell S. B., Ingram V. M. Differentiation of two mouse cell lines is associated with hypomethylation of their genomes. Mol Cell Biol. 1984 Sep;4(9):1800–1806. doi: 10.1128/mcb.4.9.1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brent T. P. Partial purification and characterization of a human 3-methyladenine-DNA glycosylase. Biochemistry. 1979 Mar 6;18(5):911–916. doi: 10.1021/bi00572a028. [DOI] [PubMed] [Google Scholar]
- Cathcart R., Goldthwait D. A. Enzymatic excision of 3-methyladenine and 7-methylguanine by a rat liver nuclear fraction. Biochemistry. 1981 Jan 20;20(2):273–280. doi: 10.1021/bi00505a007. [DOI] [PubMed] [Google Scholar]
- Deutsch W. A., Linn S. DNA binding activity from cultured human fibrolasts that is specific for partially depurinated DNA and that inserts purines into apurinic sites. Proc Natl Acad Sci U S A. 1979 Jan;76(1):141–144. doi: 10.1073/pnas.76.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas W. R., Singh R. D. Guanylation of transfer ribonucleic acid by a cell-free lysate of rabbit reticulocytes. J Biol Chem. 1973 Nov 25;248(22):7780–7785. [PubMed] [Google Scholar]
- Gambari R., Marks P. A., Rifkind R. A. Murine erythroleukemia cell differentiation: relationship of globin gene expression and of prolongation of G1 to inducer effects during G1/early S. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4511–4515. doi: 10.1073/pnas.76.9.4511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gjerset R. A., Martin D. W., Jr Presence of a DNA demethylating activity in the nucleus of murine erythroleukemic cells. J Biol Chem. 1982 Aug 10;257(15):8581–8583. [PubMed] [Google Scholar]
- Gruenbaum Y., Stein R., Cedar H., Razin A. Methylation of CpG sequences in eukaryotic DNA. FEBS Lett. 1981 Feb 9;124(1):67–71. doi: 10.1016/0014-5793(81)80055-5. [DOI] [PubMed] [Google Scholar]
- Ishiwata K., Oikawa A. Actions of human DNA glycosylases on uracil-containing DNA, methylated DNA and their reconstituted chromatins. Biochim Biophys Acta. 1979 Jul 26;563(2):375–384. doi: 10.1016/0005-2787(79)90056-x. [DOI] [PubMed] [Google Scholar]
- Katze J. R., Farkas W. R. A factor in serum and amniotic fluid is a substrate for the tRNA-modifying enzyme tRNA-guanine transferase. Proc Natl Acad Sci U S A. 1979 Jul;76(7):3271–3275. doi: 10.1073/pnas.76.7.3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livneh Z., Elad D., Sperling J. Enzymatic insertion of purine bases into depurinated DNA in vitro. Proc Natl Acad Sci U S A. 1979 Mar;76(3):1089–1093. doi: 10.1073/pnas.76.3.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Male R., Nes I. F., Kleppe K. Purification and properties of 3-methyladenine-DNA glycosylase from L-cells. Eur J Biochem. 1981 Dec;121(1):243–248. doi: 10.1111/j.1432-1033.1981.tb06455.x. [DOI] [PubMed] [Google Scholar]
- Manes C., Menzel P. Demethylation of CpG sites in DNA of early rabbit trophoblast. Nature. 1981 Oct 15;293(5833):589–590. doi: 10.1038/293589a0. [DOI] [PubMed] [Google Scholar]
- Margison G. P., Pegg A. E. Enzymatic release of 7-methylguanine from methylated DNA by rodent liver extracts. Proc Natl Acad Sci U S A. 1981 Feb;78(2):861–865. doi: 10.1073/pnas.78.2.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada N., Harada F., Nishimura S. Specific replacement of Q base in the anticodon of tRNA by guanine catalyzed by a cell-free extract of rabbit reticulocytes. Nucleic Acids Res. 1976 Oct;3(10):2593–2603. doi: 10.1093/nar/3.10.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollack Y., Stein R., Razin A., Cedar H. Methylation of foreign DNA sequences in eukaryotic cells. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6463–6467. doi: 10.1073/pnas.77.11.6463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razin A., Feldmesser E., Kafri T., Szyf M. Cell specific DNA methylation patterns; formation and a nucleosome locking model for their function. Prog Clin Biol Res. 1985;198:239–253. [PubMed] [Google Scholar]
- Razin A., Webb C., Szyf M., Yisraeli J., Rosenthal A., Naveh-Many T., Sciaky-Gallili N., Cedar H. Variations in DNA methylation during mouse cell differentiation in vivo and in vitro. Proc Natl Acad Sci U S A. 1984 Apr;81(8):2275–2279. doi: 10.1073/pnas.81.8.2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szyf M., Eliasson L., Mann V., Klein G., Razin A. Cellular and viral DNA hypomethylation associated with induction of Epstein-Barr virus lytic cycle. Proc Natl Acad Sci U S A. 1985 Dec;82(23):8090–8094. doi: 10.1073/pnas.82.23.8090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szyf M., Kaplan F., Mann V., Giloh H., Kedar E., Razin A. Cell cycle-dependent regulation of eukaryotic DNA methylase level. J Biol Chem. 1985 Jul 25;260(15):8653–8656. [PubMed] [Google Scholar]
- Wilks A., Seldran M., Jost J. P. An estrogen-dependent demethylation at the 5' end of the chicken vitellogenin gene is independent of DNA synthesis. Nucleic Acids Res. 1984 Jan 25;12(2):1163–1177. doi: 10.1093/nar/12.2.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young P. R., Tilghman S. M. Induction of alpha-fetoprotein synthesis in differentiating F9 teratocarcinoma cells is accompanied by a genome-wide loss of DNA methylation. Mol Cell Biol. 1984 May;4(5):898–907. doi: 10.1128/mcb.4.5.898. [DOI] [PMC free article] [PubMed] [Google Scholar]