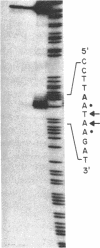
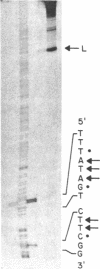
Abstract
We have determined the nucleotide sequence of a region of the vaccinia virus genome encoding RNA polymerase subunits of 22 and 147 kDa and have mapped the 5' and 3' ends of the two mRNAs. The predicted amino acid sequence of the vaccinia 147-kDa subunit shows extensive homology with the largest subunit of Escherichia coli RNA polymerase, yeast RNA polymerases II and III, and Drosophila RNA polymerase II. The regions of homology between the five RNA polymerases are subdivided into five separate domains that span most of the length of each. A sixth domain shared by the vaccinia and the eukaryotic polymerases is absent from the E. coli sequence. In all specified regions, the vaccinia large subunit has greater homology with eukaryotic RNA polymerases II and III than with the E. coli polymerase. Vaccinia virus and eukaryotic RNA polymerases may therefore have evolved from a common ancestral gene after the latter diverged from prokaryotes.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allison L. A., Moyle M., Shales M., Ingles C. J. Extensive homology among the largest subunits of eukaryotic and prokaryotic RNA polymerases. Cell. 1985 Sep;42(2):599–610. doi: 10.1016/0092-8674(85)90117-5. [DOI] [PubMed] [Google Scholar]
- Bajszár G., Wittek R., Weir J. P., Moss B. Vaccinia virus thymidine kinase and neighboring genes: mRNAs and polypeptides of wild-type virus and putative nonsense mutants. J Virol. 1983 Jan;45(1):62–72. doi: 10.1128/jvi.45.1.62-72.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baroudy B. M., Moss B. Purification and characterization of a DNA-dependent RNA polymerase from vaccinia virions. J Biol Chem. 1980 May 10;255(9):4372–4380. [PubMed] [Google Scholar]
- Biggs J., Searles L. L., Greenleaf A. L. Structure of the eukaryotic transcription apparatus: features of the gene for the largest subunit of Drosophila RNA polymerase II. Cell. 1985 Sep;42(2):611–621. doi: 10.1016/0092-8674(85)90118-7. [DOI] [PubMed] [Google Scholar]
- Coulter D. E., Greenleaf A. L. A mutation in the largest subunit of RNA polymerase II alters RNA chain elongation in vitro. J Biol Chem. 1985 Oct 25;260(24):13190–13198. [PubMed] [Google Scholar]
- Foglesong P. D. In vitro transcription of a cloned vaccinia virus gene by a soluble extract prepared from vaccinia virus-infected HeLa cells. J Virol. 1985 Mar;53(3):822–826. doi: 10.1128/jvi.53.3.822-826.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda R., Ishihama A. Subunits of RNA polymerase in function and structure; Maturation in vitro of core enzyme from Escherichia coli. J Mol Biol. 1974 Aug 15;87(3):523–540. doi: 10.1016/0022-2836(74)90102-8. [DOI] [PubMed] [Google Scholar]
- Golini F., Kates J. R. A soluble transcription system derived from purified vaccinia virions. J Virol. 1985 Jan;53(1):205–213. doi: 10.1128/jvi.53.1.205-213.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross G., Fields D. A., Bautz E. K. Characterization of a ts beta' mutant RNA polymerase of Escherichia coli. Mol Gen Genet. 1976 Sep 23;147(3):337–341. doi: 10.1007/BF00582886. [DOI] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984 Jun;28(3):351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- Isle H. B., Venkatesan S., Moss B. Cell-free translation of early and late mRNAs selected by hybridization to cloned DNA fragments derived from the left 14 million to 72 million daltons of the vaccinia virus genome. Virology. 1981 Jul 15;112(1):306–317. doi: 10.1016/0042-6822(81)90636-x. [DOI] [PubMed] [Google Scholar]
- Lipman D. J., Pearson W. R. Rapid and sensitive protein similarity searches. Science. 1985 Mar 22;227(4693):1435–1441. doi: 10.1126/science.2983426. [DOI] [PubMed] [Google Scholar]
- Mahr A., Roberts B. E. Arrangement of late RNAs transcribed from a 7.1-kilobase EcoRI vaccinia virus DNA fragment. J Virol. 1984 Feb;49(2):510–520. doi: 10.1128/jvi.49.2.510-520.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahr A., Roberts B. E. Organization of six early transcripts synthesized from a vaccinia virus EcoRI DNA fragment. J Virol. 1984 Feb;49(2):497–509. doi: 10.1128/jvi.49.2.497-509.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J., Vieira J. A new pair of M13 vectors for selecting either DNA strand of double-digest restriction fragments. Gene. 1982 Oct;19(3):269–276. doi: 10.1016/0378-1119(82)90016-6. [DOI] [PubMed] [Google Scholar]
- Morrison D. K., Carter J. K., Moyer R. W. Isolation and characterization of monoclonal antibodies directed against two subunits of rabbit poxvirus-associated, DNA-directed RNA polymerase. J Virol. 1985 Sep;55(3):670–680. doi: 10.1128/jvi.55.3.670-680.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevins J. R., Joklik W. K. Isolation and properties of the vaccinia virus DNA-dependent RNA polymerase. J Biol Chem. 1977 Oct 10;252(19):6930–6938. [PubMed] [Google Scholar]
- Ovchinnikov YuA, Monastyrskaya G. S., Gubanov V. V., Guryev S. O., Salomatina I. S., Shuvaeva T. M., Lipkin V. M., Sverdlov E. D. The primary structure of E. coli RNA polymerase, Nucleotide sequence of the rpoC gene and amino acid sequence of the beta'-subunit. Nucleic Acids Res. 1982 Jul 10;10(13):4035–4044. doi: 10.1093/nar/10.13.4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panny S. R., Heil A., Mazus B., Palm P., Zillig W., Mindlin S. Z., Ilyina T. S., Khesin R. B. A temperature sensitive mutation of the beta'-subunit of DNA-dependent RNA polymerase from E. coli T 16. FEBS Lett. 1974 Nov 15;48(2):241–245. doi: 10.1016/0014-5793(74)80477-1. [DOI] [PubMed] [Google Scholar]
- Plucienniczak A., Schroeder E., Zettlmeissl G., Streeck R. E. Nucleotide sequence of a cluster of early and late genes in a conserved segment of the vaccinia virus genome. Nucleic Acids Res. 1985 Feb 11;13(3):985–998. doi: 10.1093/nar/13.3.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puckett C., Moss B. Selective transcription of vaccinia virus genes in template dependent soluble extracts of infected cells. Cell. 1983 Dec;35(2 Pt 1):441–448. doi: 10.1016/0092-8674(83)90177-0. [DOI] [PubMed] [Google Scholar]
- Rohrmann G., Moss B. Transcription of vaccinia virus early genes by a template-dependent soluble extract of purified virions. J Virol. 1985 Nov;56(2):349–355. doi: 10.1128/jvi.56.2.349-355.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer E., Shuman S., Hurwitz J. Purification and properties of vaccinia virus DNA-dependent RNA polymerase. J Biol Chem. 1980 Jun 10;255(11):5388–5395. [PubMed] [Google Scholar]
- Stone T. W., Potter K. N. A DNA analysis program designed for computer novices working in an industrial-research environment. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):367–378. doi: 10.1093/nar/12.1part1.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier F. W., Dunn J. J. Organization and expression of bacteriophage T7 DNA. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 2):999–1007. doi: 10.1101/sqb.1983.047.01.114. [DOI] [PubMed] [Google Scholar]
- Venkatesan S., Baroudy B. M., Moss B. Distinctive nucleotide sequences adjacent to multiple initiation and termination sites of an early vaccinia virus gene. Cell. 1981 Sep;25(3):805–813. doi: 10.1016/0092-8674(81)90188-4. [DOI] [PubMed] [Google Scholar]
- Venkatesan S., Gershowitz A., Moss B. Complete nucleotide sequences of two adjacent early vaccinia virus genes located within the inverted terminal repetition. J Virol. 1982 Nov;44(2):637–646. doi: 10.1128/jvi.44.2.637-646.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir J. P., Moss B. Nucleotide sequence of the vaccinia virus thymidine kinase gene and the nature of spontaneous frameshift mutations. J Virol. 1983 May;46(2):530–537. doi: 10.1128/jvi.46.2.530-537.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilbur W. J., Lipman D. J. Rapid similarity searches of nucleic acid and protein data banks. Proc Natl Acad Sci U S A. 1983 Feb;80(3):726–730. doi: 10.1073/pnas.80.3.726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarbrough L. R., Hurwitz J. The isolation of subunits of deoxyribonucleic acid-dependent ribonucleic acid polymerase of Escherichia coli. J Biol Chem. 1974 Sep 10;249(17):5400–5404. [PubMed] [Google Scholar]