Abstract
Macroautophagy (hereafter autophagy) is a lysosomal catabolic pathway that controls cellular homeostasis and survival. It has recently emerged as an attractive target for the treatment of a variety of degenerative diseases and cancer. The targeting of autophagy has, however, been hampered by the lack of specific small molecule inhibitors. Thus, we screened two small molecule kinase inhibitor libraries for inhibitors of rapamycin-induced autophagic flux. The three most potent inhibitors identified conferred profound inhibition of autophagic flux by inhibiting the formation of autophagosomes. Notably, the autophagy inhibitory effects of all three compounds were independent of their established kinase targets, i.e. ataxia telangiectasia mutated for KU55933, protein kinase C for Gö6976, and Janus kinase 3 for Jak3 inhibitor VI. Instead, we identified phosphatidylinositol 3-kinase (PtdIns3K) as a direct target of KU55933 and Gö6976. Importantly, and in contrast to the currently available inhibitors of autophagosome formation (e.g. 3-methyladenine), none of the three compounds inhibited the cell survival promoting class I phosphoinositide 3-kinase-Akt signaling at the concentrations required for effective autophagy inhibition. Accordingly, they proved to be valuable tools for investigations of autophagy-associated cell death and survival. Employing KU55399, we demonstrated that autophagy protects amino acid-starved cells against both apoptosis and necroptosis. Taken together, our data introduce new possibilities for the experimental study of autophagy and can form a basis for the development of clinically relevant autophagy inhibitors.
Introduction
Autophagy is an intracellular degradative process by which cells recycle macromolecules and organelles (1–4). In this process, cellular material is sequestered in double membrane vesicles termed autophagosomes that fuse with lysosomes to form autolysosomes, in which the cargo is exposed to acidic hydrolases. Autophagy is essential for energy homeostasis and removal of damaged organelles and protein complexes during various kinds of stresses, such as starvation, growth factor deprivation, hypoxia, and DNA damage. It is also involved in physiological processes like development, immunity, and aging as well as in various diseases including neurodegenerative disorders and cancer. Whereas autophagy clearly has a beneficial effect in preventing many degenerative disorders, its role in cancer is more complex. It can function as a tumor suppressor mechanism, and yet it can also promote tumor growth by protecting cancer cells against the hostile tumor environment and antineoplastic drugs (5, 6).
The mammalian target of rapamycin complex 1 (mTORC1)3 serine/threonine kinase integrates information on cell metabolic, growth, and stress status to regulate biosynthetic pathways and autophagy (7, 8). It activates biosynthetic pathways and inhibits autophagy in response to various growth factors via MAPK/ERK and class I phosphoinositide 3-kinase (PI3K)/Akt-dependent pathways. On the other hand, when the energy levels are low or cells are exposed to a wide range of other stresses, AMP-activated protein kinase (AMPK) represses mTORC1 activity thereby inducing autophagy and inhibiting protein synthesis (9). mTORC1 controls autophagy partly by inhibiting unc51-like kinases (ULK1 and ULK2), whose activation is essential for the nucleation of the isolation-membrane that eventually forms the autophagosome (10). This early step is dependent on the generation of phosphatidylinositol 3-phosphate (PtdIns(3)P) synthesized by the autophagy-specific phosphatidylinositol 3-kinase (PtdIns3K) complex, which consists of the catalytic subunit Vps34 and its regulators Vps15, Beclin1, and Atg14L (11). The ubiquitin-like molecules Atg12 and microtubule-associated protein 1 light chain 3 (LC3 or Atg8) together with their corresponding conjugation systems are essential for the expansion of the isolation membrane. LC3 is present on the membranes of the completed autophagosome and gets degraded in the autolysosome along with the membranes. The degradation of LC3 can thus serve as a marker for the autophagic flux (12, 13).
Because of its involvement in many pathological processes, autophagy is an utmost attractive drug target. Rapamycin, lithium, and chloroquine are the first examples of old drugs that are entering the clinics for new indications as regulators of autophagy (14, 15). Rapamycin and lithium are mTORC1 dependent and independent inducers of autophagy, respectively. As relatively safe drugs, they may prove useful in the treatment of various degradative disorders. The anti-malaria drug chloroquine inhibits autolysosomal degradation by disrupting the lysosomal pH gradient and it is presently the preferred drug for autophagy inhibition in clinical trials for cancer treatment. In experimental studies, the potent vacuolar H+-ATPase inhibitors concanamycin A and bafilomycin A are commonly used to block the autolysosomal degradation, whereas 3-methyladenine (3-MA), LY-294002 and wortmannin that inhibit PtdIns3K and class I PI3Ks, are the standard drugs for the inhibition of autophagosome formation (12). Chloroquine and vacuolar H+-ATPase inhibitors block the lysosomal function and are therefore very unspecific autophagy inhibitors with major negative impact on cell growth and survival. On the other hand, the above-mentioned PtdIns3K/PI3K inhibitors show little or no selectivity toward PtdIns3K over class I PI3Ks greatly complicating their use in studies related to cell growth and survival (16, 17). Taken together, there is an acute need for more specific autophagy inhibitors both in the autophagy research community and the clinic.
To identify novel autophagy inhibitors, we screened two small molecule kinase inhibitor libraries containing a total of 159 compounds for inhibitors of autophagic flux by a Renilla luciferase (RLuc)-based assay for LC3 turnover (13). We further validated and characterized the three most potent autophagy inhibitors identified in the screen; KU55933 that was originally introduced as a specific inhibitor of ataxia telangiectasia-mutated (ATM) (18), a broad spectrum protein kinase C inhibitor Gö6976 and Janus 3 kinase (Jak3) inhibitor VI. All inhibitors effectively inhibited rapamycin-induced LC3 translocation at low micromolar concentrations indicating that they function downstream of mTORC1 and upstream of LC3 translocation. Notably, none of the compounds inhibited autophagy through their known target kinases. Employing PtdIns3K activity assays in living cells and in vitro, we identified KU55933 and Gö6976 as direct and effective inhibitors of PtdIns3K. Contrary to the known inhibitors of autophagosome formation discussed above, the effective concentrations of the three inhibitors identified here did not affect the activity of class I PI3K or cell viability. Exploiting these properties we demonstrated that they are highly suitable for studies related to the role of autophagy on cell death and survival.
EXPERIMENTAL PROCEDURES
Cell Culture, Transfections, and Retroviral Infections
MCF7 human breast carcinoma cells, MCF7 cells expressing Renilla luciferase (RLuc) carrying a C124A mutation fused to wild type (MCF7-RLuc-LC3wt) and G120A-mutated (MCF-RLuc-LC3G120A) LC3 (13) and MCF7 cell expressing enhanced green fluorescent protein (eGFP) fused to LC3 (MCF7-eGFP-LC3; (19)) were grown in RPMI 1640 (Invitrogen, 61870) supplemented with 6% fetal calf serum, penicillin, and streptomycin. MCF7, HeLa human cervical adenocarcinoma and U-2-OS human osteosarcoma cells expressing the ecotropic receptor (EcoR) were grown in DMEM (Invitrogen, 31966) supplemented with 10% fetal calf serum, non-essential amino acids (Invitrogen, 11140), penicillin, and streptomycin.
The human cell lines expressing EcoR were produced by retroviral infection of amphotropically packed virus produced from the plasmid pWZL-Neo-EcoR (Gift from Kristian Helin, BRIC, Copenhagen, Denmark). Retroviral infection of EcoR-positive cells with pBabe-vectors containing RLuc-LC3wt and RLuc-LC3G120A encoding inserts, were performed essentially as described (20). Similarly, ATM RNAi was accomplished by retroviral expression of a short hairpin specific for ATM in U-2-OS-EcoR cells (21). MCF7-eGFP-2xFYVE-D4 single cell clone was created by limiting dilution of MCF7 cells transfected with a plasmid encoding for eGFP fused to two PtdIns(3)P-binding FYVE domains (kindly provided by Harald Stenmark, Norwegian Radium Hospital, Oslo, Norway) and selected in 400 μg/ml geneticin (Invitrogen, G-418).
Reagents and Treatments
The following reagents were used: Small molecule kinase inhibitor library-I and II (Calbiochem, Cat. 539744 and 539745), KU55933 (Tocris Biosciences, Cat. 3544), Gö6976 (Tocris Biosciences, Cat. 2253), CP466722 (Axon Medchem, axon 1495), LY294002 (Sigma, L9908), Jak3 Inhibitor VI (Calbiochem, 420126), Rapamycin (Sigma R0395), 3-MA (Sigma, M9281), zVAD-fmk (Bachem, Switzerland), concanamycin A (Sigma) Necrostatin-1 (Sigma N9037). Ionizing radiation (IR) was delivered with an x-ray generator (Pantak, Berkshire, UK; HF160; 150 kV, 15 mA and dose rate of 2.18 Gy/min). Starvation was performed in Hanks balanced salt solution (HBSS) (Invitrogen, 14025) following two washes in HBSS. When inhibitors were used they were included in the respective washing buffers.
Autophagy Assays
RLuc reporter assays for autophagic flux were performed either in cell lysates or live cells essentially as described previously (13). Briefly, RLuc-LC3wt and RLuc-LC3G120A-expressing cells were plated 24 h before treatment at a density of 2.4 × 104 cells/cm2 in the even and uneven numbered columns in 96 wells plates respectively. The cells lysed in 40 μl of passive lysis buffer (Promega; E1941) at the end of the treatment were subjected to a single freeze/thaw cycle, and 6 μl of each lysate was transferred to corresponding wells in a white half-volume 96-well dish (Costar 3693). The luminescence reaction was started by addition of 80 μl of freshly made assay buffer (100 mm Tris/HCl pH 7.4, 300 mm sodium ascorbate, 25 μm Coelenterazine (Synchem s053), and the luminescence was measured in an EnSpire™ 2300 multilabel reader (Perkin Elmer). The live cell assay was performed by similar plating of 3.0 × 104 cells/cm2 cells in white 96-well dishes (No. 136101, Nunc). EndurenTM was used at a final concentration of 50 nm. The luminescence was measured in a Varioskan Flash plate-reader (Thermo Electron Corporation).
The accumulation of autophagosomes (eGFP-positive puncta) was analyzed in MCF7-eGFP-LC3 cells fixed in 3.7% formaldehyde applying inverted Zeiss Axiovert microscope with 20× magnification. The average number of eGFP-positive puncta/cell was obtained by counting a minimum of 39 randomly chosen cells/sample.
Screening of Small Molecule Libraries for Autophagy Inhibitors
Screens of small molecule kinase inhibitor libraries were performed using the RLucLC3-assays described above. The reporter cells were incubated with 100 nm rapamycin in combination with library compounds (2 μm) in duplicate. Reporter activities in samples treated with library I and II were measured in live cells after 12 h of treatment and in lysates after 6 h of treatment, respectively.
Plasmid Construction
The plasmid encoding VPS34wt with an NH2-terminally located Strep-tag (pStrep-VPS34wt) was created by PCR amplification of the VPS34 open reading frame from an MCF7 cDNA library using primers with EcoR1 and Xho1 restriction sites (underlined) included as 5′-extensions. Upstream primer: ACGTGAATTCGATGGGGGAAGCAGAGAAGTTTCAC; downstream primer: TGCATCTCGAGTCATTTTCTCCAGTACTGGGCAAAC. The PCR product was cut with EcoR1 and Xho1 and inserted into the corresponding sites in the pEXPR-IBA105 vector (Iba BioTAGnology). Amino acid substitutions in the kinase dead pStrep-VPS34D743N mutant and pStrep-VPS34L750M mutant were made by site directed mutagenesis using CTATATACTTGGAGTTGGAAACAGGCACCTGGATAAC and CACCTGGATAACCTTATGCTAACAAAAACAGGCAAAC primers together with corresponding antiparallel primers, respectively.
PBabeHygro-RLuc-LC3wt and pBabeHygroRLuc-LC3G120A were created by transfer of the corresponding inserts in pRLucLC3wt and pRLucLC3G120A (13) into pBabeHygro (22) using PCR and the following primers with BamHI and EcoR1 restriction sites in their 5′-ends (underlined): Rluc; TAGCGCTGGATCCCGCCACCATGACTTCGAAAGTTTATG and LC3; TAGACGTGAATTCTCACAAGCATGGCTCTCTTC.
EGFP-2xFYVE Relocalization Assay
PtdIns(3)P in cells were analyzed with the GFP-2XFYVE relocalization assay (23). MCF7-eGFP-2xFYVE cells were grown on glass coverslips and fixed in PBS containing 3.7% formaldehyde. eGFP was visualized using LSM 510 Meta confocal microscope. The average number of eGFP-2xFYVE puncta/cell was obtained by counting a minimum 50 cells/sample from printed images.
In Vitro VPS34 Activity Assay
Purification of Strep-tagged Vps34 and the lipid kinase assay were performed essentially as described in the One-STrEP kit manual (Iba BioTAGnology) and in Ref. 24, respectively. Briefly, MCF7-cells were transiently transfected with pStrep-VPS34wt, pStrep-VPS34D743N, pStrep-VPS34L750M, or pEXPR-IBA105 using Fugene HD (Roche). 48 h later, cells were collected in 1.5-ml Eppendorf tubes by scraping in PBS. Cell pellets were frozen in liquid nitrogen and thawed on ice for 15 min before addition of 300 μl of lysis buffer (50 mm Tris pH 7.4, 7.5% glycerol, 150 mm NaCl, 1 mm EDTA, protease inhibitors (Complete, Mini; Roche))/100 μl pellet. Cells were subjected to two freeze/thaw cycles and passed 40 times through a 26-gauge needle (20 times up and down). The obtained lysates were centrifuged at 20,000 × g for 20 min at 4 °C, and the obtained supernatant was allowed to bind to Strep-Tactin beads in Eppendorf tubes for 60 min at 4 °C with gentle end over end mixing. The beads were washed three times in lysis buffer and finally once in 2.5× substrate buffer (75 mm Tris (pH 7.5), 125 mm NaCl, 12.5 mm MnCl2, 250 μg/ml phosphoinositol (Sigma, 79403)). The beads were resuspended in 2.5× substrate buffer and incubated in half-volume 96-well dish on ice for 15 min. Kinase reactions were started by the addition of ATP (Sigma, A6559) to a final concentration of 10 μm either with or without simultaneous addition of kinase inhibitors. Kinase reactions were allowed to proceed for 30 min at 20 °C. Next, reactions were spotted onto a nitrocellulose membrane, blocked for 1 h in PBS containing 1% low fat dry milk, and incubated sequentially with 0.5 μg/ml PtdIns(3)P Grip (Echelon, G-0302) dissolved in PBS containing 0.1% Tween-20 and 3% BSA (Sigma, A7030) for 2 h, Anti-GST antibody (Sigma, G1160) for 2 h and horseradish peroxidase-coupled anti-mouse secondary antibody (Dako). All incubations were done at 20 °C with gentle agitation and separated by washing steps in PBS containing 0.1% Tween 20. The membrane was developed with ECL plus (Amersham BiosciencesTM) in an ImageQuantTM Las 4000 mini and quantified using ImageJ software.
Immunoblotting
Cells were extracted in SDS lysis buffer. Extracts were separated by SDS-PAGE and blotted onto nitrocellulose membranes. The primary antibodies used were anti-ATM (Cell Signaling, no. 2873), anti-p62 (Enzo life Sciences, BML-PW9860), anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH; Serotec, Cat no. 4699-9555), Anti-LC3 (Nanotools, 5F10) anti-β-Actin (Sigma, A2228), Anti-TP53 (DO-1), anti-Phospho-TP53 (Ser-15) (Cell Signaling, 9284), anti-Beclin1 (BD, 612112), anti Vps34 (cell Signaling, no 3358). Appropriate peroxidase-conjugated mouse (DAKO A/S, P0260) and rabbit (Vector Laboratories, PI-1000) secondary antibodies and ECL Western blotting Detection Reagents (GE Healthcare, RPN2132) were used for the detection. Immunoblot signals were quantified using ImageJ software.
RNAi
Transfection of siRNA was performed with Oligofectamine (invitrogen) according to manufacturer's instructions using 27 nm of siRNA. The siRNAs were from Qiagen. The siRNA against Ulk1 was a mix of Hs-Ulk1–5 and Hs-Ulk1–6 (13.5 nm each). The “AllStars neg. Control siRNA” was used as negative control.
Cell Viability and Cell Death Assays
Cell death and viability were monitored by lactate dehydrogenase (LDH) release (Roche) and 3-(4,5-dimethylthiazole-2-yl)-2,5-diphenyltetrazolium bromide (MTT) reduction assays, respectively, essentially as described previously (25).
Statistical Analysis
One-tailed paired t test (Figs. 1A, 3B (top), and 4B), one-tailed unpaired t test (Figs. 1B and 4, C and D), two-tailed paired t test (Figs. 2, B and C and 4A), two-tailed unpaired t test (Figs. 1C and 3B (bottom)) were used to asses the statistical significance of the data.
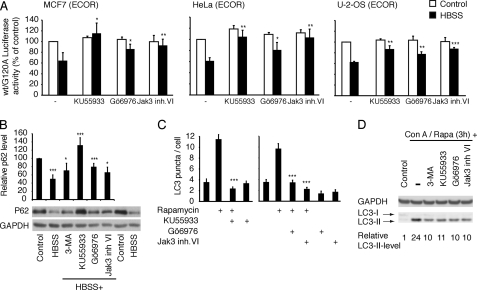
FIGURE 1.
Gö6976, Jak3 inhibitor VI and KU55933 inhibit autophagosome formation. A, MCF7, HeLa, and U-2-OS cells infected with RLuc-LC3wt or RLuc-LC3G120A expressing retroviruses were left untreated (Control) or starved for 4 h (HBSS) in the presence or absence (−) of 8 μm Gö6976, 2 μm KU55933 or 5 μm Jak3 inhibitor VI. The data represent average ± S.D. of at least three independent experiments. B, quantification and a representative immunoblot with antibodies against p62 and GAPDH (loading control) in total extracts from MCF7 cells cultured in normal growth medium (control), starved for 7 h in the absence (HBSS) or presence of 10 mm 3-MA, 2 μm KU55933, 8 μm Gö6976, or 5 μm Jak3 inhibitor VI. The data represent the p62-signal normalized to GAPDH. The untreated control was defined as 100%. The data represent average ± S.D. of four independent experiments. C, average number of autophagosomes/cell ± S.E. in MCF7-eGFP-LC3 cells left untreated or treated as indicated with 200 nm rapamycin, 2 μm KU55933, 10 μm Gö6976, or 5 μm Jak3 inhibitor VI for 6 h. D, immunoblotting with antibodies against LC3 and GAPDH in total extracts from MCF7 cells cultured in normal growth medium (control) or treated for 3h with rapamycin (100 nm)+ concanamycin A (2 nm) in the absence or presence of 10 mm 3-MA, 2 μm KU55933, 8 μm Gö6976, or 5 μm Jak3 inhibitor VI. Quantification of the LC3-II signal normalized to GAPDH signal is shown below. *, p value <0.05; **, p value <0.01; and ***, p value <0.001 as compared with control (A) or control (B, HBSS) or HBSS (B, HBSS+inhibitors) or rapamycin (C) in the absence of the inhibitors.
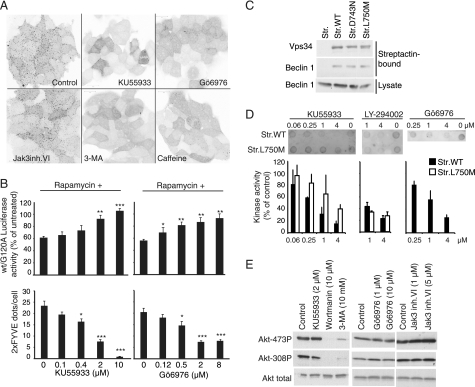
FIGURE 3.
KU55933 and Gö6976 inhibit Vps34. A, confocal microscopy images of GFP-2XFYVE expressing MCF7 cells either untreated (Control) or treated for 2 h with 2 μm KU55933, 8 μm Gö6976, 5 μm Jak3 inhibitor VI, 10 mm 3-MA, or 10 mm caffeine. The images were converted to grayscale and inverted using Photoshop. B, comparison of the effects of KU55933 (left) and Gö6976 (right) on rapamycin-induced autophagy and on eGFP-2XFYVE-relocalization. (B, top panels) MCF7 cells expressing RLucLC3wt or RLuc-LC3G120A were left untreated or treated with 100 nm rapamycin alone or in combination with indicated concentrations of KU55933 or Gö6976 for 6 h. The data represent average ± S.D. of at least three independent experiments. (B, bottom panels) MCF7- eGFP-2XFYVE cells were treated with indicated concentrations of KU55933 or Gö6976 for 2 h and the number of eGFP-positive dots/cell were calculated in confocal images. *, p value <0.05; **, p value <0.01; and ***, p value <0.001 as compared with cells treated with rapamycin alone (B, top) or control (B, bottom). C, immunoblotting with antibodies against Vps34 and Beclin 1 of streptactin-pulldowns of MCF7 cells transiently transfected with pEXPR-IBA105 (Str.), pStrep-VPS34wt (Str.WT), pStrep-VPS34D743N (Str. D743N), or pStrep-VPS34L750M (Str. L750M). The bottom row shows immunoblotting of the corresponding lysates with antibody against Beclin 1. D, dot blotting of PtdIns(3)P produced in vitro by strep-tagged Vps34WT (Str.WT) or strep-tagged Vps34L750M (Str.L750M). Kinase reactions were performed in the absence of inhibitors (Control) or with either KU55933, LY-294002, or Gö6976 at the indicated concentrations. The bar graph shows the Quantification of PtdIns(3)P dot-blots from Vps34 in vitro kinase reactions analyzing either Strep-tagged Vps34wt or Strep-tagged Vps34L750M. Kinase reactions were performed in the absence of inhibitors (Control) or with KU55933 (n = 3), LY-294002 (Vps34wt) (n = 3) LY-294002 (Vps34L750M) (n = 2), or Gö6976 (n = 2) as indicated. E, immunoblotting with antibodies against Akt and phospho-Akt in total extracts from MCF7 cells incubated with the indicated concentrations of inhibitors for 6 h.
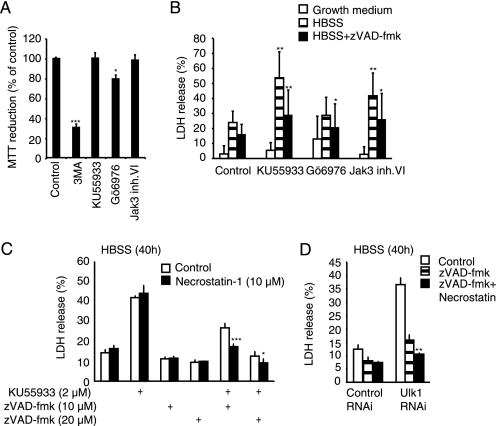
FIGURE 4.
Induction of apoptosis and necroptosis via small-molecule inhibition of autophagy. A, MCF7 cells left untreated (Control) or treated with 10 mm 3-MA, 2 μm KU55933, 8 μm Gö6976, or 5 μm Jak3 inhibitor VI for 40 h were analyzed for cell viability by the MTT reduction assay. Values represent average ± S.D. of three independent experiments. B, MCF7 cells in either normal growth medium, HBSS or HBSS+ zVAD-fmk (20 μm) were treated with the indicated inhibitors for 40 h as in A, and cell death was analyzed by the LDH release assay. Values represent average ± S.D. of at least three independent experiments. C, cells were starved in HBSS for 40 h in the presence of KU55933, zVAD-fmk, and/or Necrostatin-1 as indicated and cell death was measured by the LDH release assay. The values represent average ± S.D. of a representative experiment performed in quadruplicate. D. Cells pretreated with control siRNA or Ulk1 siRNA for 48 h, were starved in HBSS for 40 h in presence of zVAD-fmk and/or Necrostatin-1 as indicated and cell death was analyzed by the LDH-release assay. The values represent average ± S.D. of a representative experiment performed in triplicate. *, p value <0.05; **, p value <0.01; and ***, p value <0.001 as compared with Control (A), Treatment with HBSS (B, striped columns), Treatment with HBSS + kinase inhibitor (B, white columns), corresponding control (C), or treatment with Ulk1-siRNA + zVAD-fmk (D).
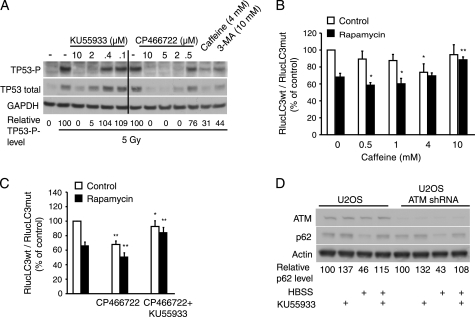
FIGURE 2.
ATM is not the target of KU55933-mediated autophagy inhibition. A, immunoblotting with antibodies against phospho-Ser15-TP53 (TP53-P), total TP53 and GAPDH in total extracts from MCF7 cells left untreated (No IR) or irradiated (5 Gy) in the absence (−) or presence of the indicated concentrations of the inhibitors. Cells were harvested 30 min after irradiation. The level of TP53-P normalized to GAPDH is shown below. B and C, MCF7 cells expressing RLucLC3wt or RLucLC3G120A were left untreated (Control) or treated with 100 nm rapamycin alone or in combination with indicated concentrations of caffeine (B) or with 5 μm CP466722 alone or with 2 μm KU55933 (C) for 6 h. The data represent average ± S.D. of at least three independent experiments. *, p value <0.05; **, p value <0.01; and ***, p value <0.001 as compared with cells treated similarly in absence of caffeine (B) CP466722 (C, middle) or KU55933 (C, right). D, immunoblotting with antibodies against ATM, p62 or β-actin in total extracts from normal or ATM-depleted U-2-OS cells. Cells were left untreated, treated with 2 μm KU55933, or starved in HBSS without or with KU55933 for 7 h. The level of p62 normalized to β-actin is shown below.
RESULTS
Identification of Small Molecule Inhibitors of Autophagy
We recently described a luciferase reporter-based assay for autophagic flux (13). The assay measures autophagy-dependent changes in LC3 turnover. This is accomplished by monitoring changes in the ratio of luciferase activities in two parallel cell lines expressing luciferase fused to wild type and mutant LC3, respectively. Using this assay, we screened two Calbiochem small molecule kinase inhibitor libraries for inhibitors of rapamycin-induced autophagy in MCF7 human breast carcinoma cells (supplemental Figs. S1 and S2).
Two compounds from library I (Gö6976 and Jak3 inhibitor VI) and one from library II (KU55933) completely inhibited rapamycin-induced autophagic flux (supplemental Figs. S1 and S2), and therefore they were selected for further characterization. After obtaining new batches of the inhibitors, we performed dose-response studies for their ability to inhibit rapamycin-induced autophagic flux and thereafter used Gö6976 at 8 or 10 μm, Jak3 inhibitor VI at 5 μm and KU55933 at 2 μm. At these concentrations, all three compounds effectively inhibited amino acid starvation-induced autophagic flux in MCF7 cells as well as in HeLa human cervix carcinoma and U-2-OS human osteosarcoma cells as analyzed by the luciferase reporter-based assay for autophagic flux (Fig. 1A) and immunoblot analysis of p62/sequestesome (p62; Fig. 1B), an autophagic cargo protein whose loss serves as an indicator of autophagic flux (12). Strikingly, KU55933 completely prevented the massive p62 loss upon amino acid starvation, and also Gö6976 and Jak3 inhibitor VI were effective in this assay similar to the widely used autophagy inhibitor 3-MA. To determine whether the compounds inhibited the formation of autophagosomes or the further maturation of autophagosomes to autolysosomes, we tested their effect on rapamycin-induced eGFP-LC3 puncta formation. All three inhibitors inhibited rapamycin-induced eGFP-LC3 puncta formation completely (Fig. 1C) indicating that they inhibited autophagic flux downstream of mTORC1 but upstream of LC3 translocation and autophagosome formation. To further evaluate the efficacy of the compounds we tested their ability to inhibit endogenous LC3-II formation upon rapamycin treatment. To avoid LC3-II-degradation in the autolysosome, we co-treated the cells with concanamycin A that inhibits lysosomal acidification and function. We observed that the three compounds inhibited the LC3-II formation as effectively as 3-MA (Fig. 1D).
ATM Is Dispensable for Autophagy Signaling Downstream of mTORC1
KU55933 is an ATP competitive ATM kinase inhibitor previously described to be highly selective for ATM even within the PtdIns3/PI3 kinase superfamily (18, 26). The robust autophagy inhibitory effect of KU55933 prompted us to investigate the role of ATM in autophagy signaling. To that end, we tested the autophagy interfering capacity of two other compounds known to inhibit ATM kinase, i.e. caffeine (27) and CP466722, a recently introduced highly specific and potent ATM inhibitor with a structure distinct from that of KU55933 (28). As analyzed by their ability to inhibit ionizing radiation (IR)-induced phosphorylation (Ser-15) and subsequent stabilization of a well-described ATM target, tumor protein 53 (TP53) in MCF7 cells, 4 mm caffeine and 5 μm CP466722 were as effective in inhibiting ATM activity as 2 μm KU55933 (Fig. 2A). Notably, also 3-MA, which is widely used at 10–20 mm as an inhibitor of PtdIns3K and autophagy, effectively inhibited TP53 phosphorylation at 10 mm concentration (Fig. 2A). Contradicting the role of ATM as a positive regulator of autophagy, caffeine was recently shown to induce autophagy in mammalian cells (29). In line with this, caffeine induced autophagic flux in MCF7 cells at 4 mm and enhanced rapamycin-induced autophagic flux at 0.5 and 1 mm (Fig. 2B). However, when applied at 10 mm, caffeine had no effect on constitutive autophagy and inhibited rapamycin-induced autophagic flux (Fig. 2B). The induction of autophagy and the failure to reduce rapamycin-induced autophagy by 4 mm caffeine, which was sufficient to inhibit the IR-induced Ser-15 phosphorylation of TP53 and subsequent stabilization of TP53 argues, however, against an essential role for ATM in autophagy signaling (Fig. 2A). Supporting this hypothesis, 5 μm CP466722 induced autophagic flux and augmented rapamycin-induced autophagy in MCF7 cells (Fig. 2C). Notably, KU55933 effectively inhibited autophagy induced by CP466722 alone or in combination with rapamycin further indicating that the essential target of KU55933 for autophagy inhibition is distinct from ATM (Fig. 2C). To challenge this hypothesis by a genetic approach, we created an U-2-OS cell line expressing a short hairpin RNA targeting ATM and compared the effect of amino acid starvation (7 h) on autophagic flux (p62 levels) in parental and ATM-depleted cells. Starvation was as effective in reducing p62 levels and KU55933 completely inhibited this reduction in both cell lines (Fig. 2D). Thus, we conclude that KU55933 exerts its autophagy inhibiting effect via another target than ATM.
KU55933 and Gö6976 Inhibit PtdIns3K
To investigate the effect of our newly identified autophagy inhibitors on the activity of PtdIns3K, which controls the formation of autophagosomes, we created MCF7 cells that stably express a fusion protein consisting of eGFP and two PtdIns(3)P-binding FYVE domains (eGFP-2xFYVE) (23). As a result of the high constitutive activity of the endosomal PtdIns3K complex, PtdIns(3)P is enriched in early endosomes resulting in a punctate cytoplasmic localization of eGFP-2xFYVE (Fig. 3A). Remarkably, a 2-h incubation of MCF7 cells with 2 μm KU55933 or 8 μm Gö6976 inhibited the punctate distribution of GFP-2xFYVE almost as effectively as 10 mm 3-MA, whereas a 2-h treatment with 5 μm Jak3 inhibitor VI left the punctate pattern practically intact (Fig. 3, A and B). In concordance with its autophagy inhibiting effect shown in Fig. 2B, 10 mm caffeine also significantly reduced the number of eGFP-2xFYVE puncta (Fig. 3A). To challenge the relationship between the effect of KU55933 and Gö6976 on autophagy and GFP-2xFYVE puncta, we compared the concentration dependence of these effects. In favor of a causal relationship of the two responses, the concentrations of KU55933 and Gö6976 required to inhibit LC3 turnover and GFP-2xFYVE puncta fell within the same range (Fig. 3B). These data suggest that KU55933 and Gö6976 inhibit either PtdIns(3)K itself or signaling pathways required for its activation.
To test whether KU55933 and Gö6976 are direct inhibitors of PtdIns(3)K, we expressed Vps34, the catalytic subunit of the PtdIns(3)K complex, as a strep-tagged fusion protein in MCF7 cells. In addition we expressed a kinase-dead catalytic loop mutant strep-Vps34-D743N and strep-Vps34-L750M mutant with a methionine substitution of a leucine residue in the ATP-binding pocket. The leucine residue is conserved between PtdIns(3)K and ATM, whereas class I PI3Ks have a methionine at this position, which is predicted to be important for the drug binding characteristics of the pocket (30). All three fusion proteins bound beclin 1 when isolated on streptactin beads from MCF7 cell lysates suggesting that they fold correctly and form proper complexes (Fig. 3C). Streptactin-purified kinase was then incubated with PtdIns followed by the addition of ATP alone or in combination with the inhibitors. The reactions were analyzed by dot-blotting using a PtdIns(3)P-specific probe. As expected, the PtdIns(3)P synthesis depended on an intact Vps34 catalytic loop since no activity was detected using StrepVps34-D743N (data not shown). When the Strep-Vps34wt kinase reaction was performed in the presence of KU55933 or Gö6976, we observed a concentration dependent inhibition of the activity (Fig. 3D). Notably, StrepVps34-L750M, was clearly less sensitive for the inhibition by KU55933 at all tested concentrations (Fig. 3D) confirming the suggested importance of the pocket-residue for drug binding and indicating that KU55933 inhibits Vps34 directly by ATP competition. To compare the effect of KU55933 and Gö6976 with a known inhibitor of class I PI3Ks and PtdIns(3)K, we included LY294002 in the study. LY294002 is structurally related to KU55933 (18), but is required at much higher concentrations (10–100 μm) for efficient inhibition of autophagy as assessed by long-lived protein degradation assay in hepatocytes (31) or by RLucLC3 assay in MCF7 cells (supplemental Fig. S3). It was therefore notable that LY294002 inhibited strep-Vps34 kinase activity in vitro almost as efficiently as KU55933 (Fig. 3D and supplemental Fig. S3). Consistent with its ability to inhibit class I PI3Ks, LY294002 also inhibited the L750M mutant of Vps34 as effectively as the wild-type Vps34 (Fig. 3D). In light of the quantitative similarity in direct inhibition of Vps34, the reason for the discrepancy in autophagy inhibitory capacity between KU55933 and LY294002 is unclear but might reflect differences in cellular uptake, other factors affecting the free intracellular pool or inhibition of class I PI3Ks.
The data presented above reveal a key molecular difference that confers the specificity of KU55933 for PtdIns(3)K and ATM over for class I PI3Ks. Accordingly, and contrary to the unspecific PtdIns3K inhibitors (3-MA and wortmannin), the autophagy inhibitory concentrations of KU55933 did not inhibit the activity of class I PI3K as analyzed by the phosphorylation status of its important downstream effector Akt (Fig. 3E). Akin to KU55933, autophagy inhibitory concentrations of Gö6976 and Jak3 inhibitor VI also failed to inhibit the class I PI3K activity (Fig. 3E). Thus, all the identified compounds inhibit autophagy without affecting the class I PI3K signaling pathway.
Autophagy Inhibition Induces Apoptosis and Necroptosis in Starved MCF7 Cells
It is essential that pharmacological inhibitors used to investigate the role of autophagy in cell death and survival have a minimal impact on cell survival in the control situation. The lack of an experimental window for autophagy inhibition without concomitant inhibition of class I PI3K-Akt-mediated survival pathways is thus a major obstacle in using 3-MA, LY294002, and wortmannin for such studies. Thus, we tested the usefulness of KU55933, Gö6976, and Jak3 inhibitor VI in studying starvation-induced autophagy, a model associated with autophagy-mediated protection from apoptosis (32). First, we incubated MCF7 cells with the inhibitors for 40 h and tested their viability by the MTT assay that gives an indication of the number of viable cells. 3-MA that was included in the study for comparison caused a 70% reduction in viable cells thereby illustrating the problem with this compound (Fig. 4A). In contrast, KU55933 and Jak3 inhibitor VI caused no reduction in cell number, while the cells treated with Gö6976 displayed ∼20% reduction in viable cells (Fig. 4A). The lack of toxicity of KU55933 and Jak3 inhibitor VI as well as the mild toxicity of Gö6976 were confirmed by microscopy (data not shown) and the LDH release assay (Fig. 4B), which measures cell death. Next, we tested the effect of KU55933, Gö6976, and Jak3 inhibitor VI on the survival of MCF7 cells exposed to amino acid starvation. As expected for autophagy inhibitors, both KU55933 and Jak3 inhibitor VI significantly increased cell death induced by 40 h amino acid starvation (Fig. 4B). This death was partially inhibited by the pan-caspase inhibitor zVAD-fmk supporting the notion that autophagy protects cells from starvation-induced caspase-dependent apoptosis. To investigate the zVAD-fmk insensitive residual cell death, we treated MCF7 cells with KU55933 and zVAD-fmk with or without Necrostatin-1, a specific inhibitor of programmed necrosis termed necroptosis (33). Strikingly, Necrostatin-1 significantly inhibited the death of starved cells when autophagy and apoptosis were inhibited by KU55933 and zVAD-fmk, respectively (Fig. 4C). The quantitative effect of necrostatin-1 on cell death was higher when zVAD-fmk was used at 10 μm than at 20 μm. This suggests that zVAD-fmk has several targets, with different effects on death signaling or execution (see discussion). To learn if necroptosis induced by KU55933 is a consequence of autophagy inhibition, we replaced drug treatment with RNAi targeting the essential autophagy kinase Ulk1. As expected, Ulk1 RNAi sensitized the cells to amino acid starvation and furthermore, when combined with zVAD-fmk the cell death became sensitive to necrostatin-1 (Fig. 4D). Thus, autophagy protects starved cells from both apoptosis and necroptosis.
DISCUSSION
We screened two small-molecule kinase-inhibitor libraries for inhibitors of rapamycin induced autophagic flux. Three potent inhibitors, KU55933, Gö6976 and Jak3 inhibitor VI, were selected for further characterization. KU55933 and Gö6976 were identified as direct inhibitors of PtdIns(3)K, whereas the critical autophagy-related cellular target of Jak3 inhibitor VI remains to be identified. Importantly, and in contrast to 3-MA, LY294002, and wortmannin, the three identified compounds could be readily used to inhibit autophagy without inhibiting class I PI3K signaling.
Interestingly the three inhibitors all block autophagosome formation. The reason for this apparent bias toward initiation of the degradation pathway is uncertain, but it is possible that the events downstream of autophagosome formation, e.g. autophagosome-lysosome fusion and autolysosomal degradation, are less prone for inhibition simply because kinase signaling plays a less prominent role for these processes or because of great redundancy in signaling pathways. Emphasizing that maturation is potentially difficult to target via kinase inhibition, we recently published a screen of a siRNA library targeting the human kinome (726 known and putative kinases) for siRNAs that induce an accumulation of LC3-positive autophagosomes in MCF7 cells, and WNK lysine-deficient protein kinase 2 (WNK2) was the sole kinase identified as a positive regulator of maturation in this screen (34).
Our finding that KU55933 inhibits autophagy at concentrations comparable to that required for the inhibition of IR-induced TP53 phosphorylation was surprising considering the reported high specificity of the drug toward ATM kinase (18, 26). We, however, excluded ATM kinase as a target for autophagy inhibition and instead identified the key autophagy-regulator Vps34 as a direct target of KU55933. ATM kinase belongs to the PtdIns3/PI3 kinase superfamily and cross-inhibition within this family is a common problem when using kinase inhibitors (16). This aspect is particularly challenging within the autophagy field since the family in addition to ATM and PtdIns(3)K includes the notorious autophagy inhibitors mTor and class I PI3Ks. The lack of specificity may also explain the paradoxical effects of caffeine observed herein where low and high concentrations promote and inhibit autophagy respectively, probably reflecting different thresholds for inhibition of the Akt/mTor/p70S6 pathway and PtdIns3K (29).
Gö6976 was originally described as a selective inhibitor of calcium-dependent members of the protein kinase C family (35). Since then, equally efficient inhibition of a large number of unrelated kinases was reported (36). To our knowledge this is, however, the first report describing Vps34 as a direct target of Gö6976. We notice that the concentration gradient of Gö6976 for autophagy inhibition is rather shallow displaying weak but significant effect even at 0.125 μm (Fig. 3C). At this concentration the effect on GFP-2xFYVE puncta formation is insignificant. It is therefore possible that the effect on autophagy by Gö6976 is a consequence of inhibition of other kinases in addition to Vps34. It is however unlikely that protein kinase C is involved, because we previously identified other inhibitors of this kinase-family as potent inducers of autophagy (13).
As the name implies, Jak3 inhibitor VI was developed as a specific inhibitor of Jak3 (37), and we are not aware of any published off-targets for this inhibitor. However, Jak3 is not likely to be the critical autophagy-inducing targets of Jak3 inhibitor VI in our assay system. This assumption is based on our data showing that 3 non-overlapping siRNA sequences targeting Jak3 fail to inhibit rapamycin-induced autophagy (data not shown). Furthermore, CP690550 a Jak inhibitor with a slight preference for Jak3 within the Jak-family (38), has no effect on autophagy at concentrations that inhibit Oncostatin induced Stat3-phosphorylation (supplemental Fig. S4, A and B), a phosphorylation event primarily caused by Jak1 (data not shown). In contrast, Jak3 inhibitor VI does not inhibit Oncostatin-induced Stat3 phosphorylation at a concentration sufficient to inhibit autophagy (supplemental Fig. S4, A and B). Even though Jak inhibitor VI is not inhibiting Vps34, we cannot exclude that it targets autophagy specific PtdIns(3)P production. The eGFP-2XFYVE probe used in this study is not suitable to differentiate between endosomal and autophagosomal PtdIns(3)P. Variations in autophagy specific PtdIns(3)P production have, instead, been observed with a probe containing the FYVE domain-containing protein 1 (DFCP1) that has been reported to localize to the early autophagic membranes (so called omegasomes) at the ER and Golgi apparatus (39). Unfortunately, our attempts to detect autophagy-dependent relocalization of DFCP1 or translocation of the autophagy specific PtdIns(3)K complex subunit Atg14L (40) have not been successful. We have therefore not been able to test if Jak3 inhibitor VI functions at those steps of the autophagic pathway. Even with no defined autophagy-related target identified yet, Jak3 inhibitor VI may emerge as a useful compound for inhibiting autophagy both in vitro and in vivo. To this end it is interesting to note that Jak3 inhibitor VI has recently been identified as an inhibitor of left-right assymmetri formation in zebrafish (41), a developmental process in which autophagy has been suggested to play a role (42).
During metabolic stress, autophagy inhibition leads to apoptotic cell death (32). However, if apoptosis-defective cells are exposed to metabolic stress, the outcome of autophagy inhibition is necrosis (43). Our data demonstrating that necrostatin-1 effectively inhibits the residual cell death in amino acid-starved MCF7 cells treated with KU55933 or Ulk1 siRNA and zVAD-fmk further defines this necrotic death as programmed necrosis or necroptosis. These data implicate that the necrostatin-1 target receptor-interacting protein 1 (RIP1) or a related kinase mediates this non-apoptotic cell death (44). When used at high concentration (20 μm), zVAD-fmk reduces also the necrostatin-1 sensitive cell death. However, concentrations of zVAD-fmk beween 10 μm and 20 μm reveal a window for inhibition by necrostatin-1. This is in contrast to the 100 μm zVAD-fmk used by Degterev et al. in their necroptosis defining paper (33). It is possible that the cell death inhibitory effect of zVAD-fmk when used at increasing concentrations observed here reflects the progressive inhibition of caspases and other cysteine proteases such as calpains and cathepsins that are also targets of zVAD-fmk (45). In agreement with a role for cathepsins in the cell death mechanism, lysosomal membrane permeabilization was recently described as a common denominator in several necrotic death modes including necroptosis (46).
To our knowledge, no specific autophagy or PtdIns3K inhibitors have been reported. The data presented above identify, however, KU55933 as a superior alternative for PtdIns3K and autophagy inhibition and Jak3 inhibitor VI as an attractive autophagy inhibitor suitable also for in vivo studies. Because of their low toxicity and lack of class I PI3K inhibition they can serve as useful reagents especially for the investigation of the role of autophagy in cell death and survival.
Supplementary Material
Acknowledgments
We thank Elisabeth Corcelle for technical assistance, Pekka Kallunki for kinase inhibitor libraries, Harald Stenmark for the EGFP-2xFYVE construct, and Kristian Helin for the pWZL-Neo-EcoR construct.
This work was supported by grants from the Danish National Research Foundation, the Danish Cancer Society, the European Commission FP7 (APO-SYS), the Lundbeck Foundation, the Meyer Foundation, and the Novo Nordisk Foundation (to M. J.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
- mTORC
- mammalian target of rapamycin complex
- AMPK
- AMP-activated kinase
- ATM
- ataxia telangiectasia mutated
- EcoR
- ecotropic receptor
- eGFP
- enhanced green fluorescent protein
- LC3
- microtubule-associated protein 1 light chain 3
- LDH
- lactate dehydrogenase
- MTT
- 3-(4,5-dimethylthiazole-2-yl)-2,5-diphenyltetrazolium bromide
- PI
- phosphoinositide
- PI3K
- phosphoinositide 3-kinase
- PtdIns
- phosphatidylinositol
- PtdIns3K
- phosphatidylinositol 3-kinase
- PtdIns(3)P
- phosphatidylinositol 3-phosphate
- p62
- p62/sequestesome
- RLuc
- Renilla luciferase
- 3-MA
- 3-methyladenine.
REFERENCES
- 1. Mizushima N., Levine B. (2010) Nat. Cell Biol. 12, 823–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kroemer G., Mariño G., Levine B. (2010) Mol. Cell 40, 280–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Weidberg H., Shvets E., Elazar Z. (2011) Annu. Rev. Biochem. 80, 125–156 [DOI] [PubMed] [Google Scholar]
- 4. Yang Z., Klionsky D. J. (2010) Nat. Cell Biol. 12, 814–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Høyer-Hansen M., Jäättelä M. (2008) Autophagy 4, 574–580 [DOI] [PubMed] [Google Scholar]
- 6. Mathew R., White E. (2011) Curr. Opin. Genet. Dev. 21, 113–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Corcelle E. A., Puustinen P., Jäättelä M. (2009) FEBS J 276, 6084–6096 [DOI] [PubMed] [Google Scholar]
- 8. Zoncu R., Efeyan A., Sabatini D. M. (2011) Nat. Rev. Mol. Cell Biol. 12, 21–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Høyer-Hansen M., Jäättelä M. (2007) Autophagy 3, 381–383 [DOI] [PubMed] [Google Scholar]
- 10. Chan E. Y. (2009) Sci. Signal 2, pe51. [DOI] [PubMed] [Google Scholar]
- 11. Simonsen A., Tooze S. A. (2009) J. Cell Biol. 186, 773–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Klionsky D. J., Abeliovich H., Agostinis P., Agrawal D. K., Aliev G., Askew D. S., Baba M., Baehrecke E. H., Bahr B. A., Ballabio A., Bamber B. A., Bassham D. C., Bergamini E., Bi X., Biard-Piechaczyk M., Blum J. S., Bredesen D. E., Brodsky J. L., Brumell J. H., Brunk U. T., Bursch W., Camougrand N., Cebollero E., Cecconi F., Chen Y., Chin L. S., Choi A., Chu C. T., Chung J., Clarke P. G., Clark R. S., Clarke S. G., Clave C., Cleveland J. L., Codogno P., Colombo M. I., Coto-Montes A., Cregg J. M., Cuervo A. M., Debnath J., Demarchi F., Dennis P. B., Dennis P. A., Deretic V., Devenish R. J., Di Sano F., Dice J. F., Difiglia M., Dinesh-Kumar S., Distelhorst C. W., Djavaheri-Mergny M., Dorsey F. C., Droge W., Dron M., Dunn W. A., Jr., Duszenko M., Eissa N. T., Elazar Z., Esclatine A., Eskelinen E. L., Fesus L., Finley K. D., Fuentes J. M., Fueyo J., Fujisaki K., Galliot B., Gao F. B., Gewirtz D. A., Gibson S. B., Gohla A., Goldberg A. L., Gonzalez R., Gonzalez-Estevez C., Gorski S., Gottlieb R. A., Häussinger D., He Y. W., Heidenreich K., Hill J. A., Hoyer-Hansen M., Hu X., Huang W. P., Iwasaki A., Jäättelä M., Jackson W. T., Jiang X., Jin S., Johansen T., Jung J. U., Kadowaki M., Kang C., Kelekar A., Kessel D. H., Kiel J. A., Kim H. P., Kimchi A., Kinsella T. J., Kiselyov K., Kitamoto K., Knecht E., Komatsu M., Kominami E., Kondo S., Kovacs A. L., Kroemer G., Kuan C. Y., Kumar R., Kundu M., Landry J., Laporte M., Le W., Lei H. Y., Lenardo M. J., Levine B., Lieberman A., Lim K. L., Lin F. C., Liou W., Liu L. F., Lopez-Berestein G., Lopez-Otin C., Lu B., Macleod K. F., Malorni W., Martinet W., Matsuoka K., Mautner J., Meijer A. J., Melendez A., Michels P., Miotto G., Mistiaen W. P., Mizushima N., Mograbi B., Monastyrska I., Moore M. N., Moreira P. I., Moriyasu Y., Motyl T., Munz C., Murphy L. O., Naqvi N. I., Neufeld T. P., Nishino I., Nixon R. A., Noda T., Nurnberg B., Ogawa M., Oleinick N. L., Olsen L. J., Ozpolat B., Paglin S., Palmer G. E., Papassideri I., Parkes M., Perlmutter D. H., Perry G., Piacentini M., Pinkas-Kramarski R., Prescott M., Proikas-Cezanne T., Raben N., Rami A., Reggiori F., Rohrer B., Rubinsztein D. C., Ryan K. M., Sadoshima J., Sakagami H., Sakai Y., Sandri M., Sasakawa C., Sass M., Schneider C., Seglen P. O., Seleverstov O., Settleman J., Shacka J. J., Shapiro I. M., Sibirny A., Silva-Zacarin E. C., Simon H. U., Simone C., Simonsen A., Smith M. A., Spanel-Borowski K., Srinivas V., Steeves M., Stenmark H., Stromhaug P. E., Subauste C. S., Sugimoto S., Sulzer D., Suzuki T., Swanson M. S., Tabas I., Takeshita F., Talbot N. J., Talloczy Z., Tanaka K., Tanida I., Taylor G. S., Taylor J. P., Terman A., Tettamanti G., Thompson C. B., Thumm M., Tolkovsky A. M., Tooze S. A., Truant R., Tumanovska L. V., Uchiyama Y., Ueno T., Uzcategui N. L., van der Klei I., Vaquero E. C., Vellai T., Vogel M. W., Wang H. G., Webster P., Wiley J. W., Xi Z., Xiao G., Yahalom J., Yang J. M., Yap G., Yin X. M., Yoshimori T., Yu L., Yue Z., Yuzaki M., Zabirnyk O., Zheng X., Zhu X., Deter R. L. (2008) Autophagy 4, 151–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Farkas T., Høyer-Hansen M., Jäättelä M. (2009) Autophagy 5, 1018–1025 [DOI] [PubMed] [Google Scholar]
- 14. Rubinsztein D. C., Gestwicki J. E., Murphy L. O., Klionsky D. J. (2007) Nat. Rev. Drug Discov. 6, 304–312 [DOI] [PubMed] [Google Scholar]
- 15. Amaravadi R. K., Lippincott-Schwartz J., Yin X. M., Weiss W. A., Takebe N., Timmer W., DiPaola R. S., Lotze M. T., White E. (2011) Clin. Cancer Res. 17, 654–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Knight Z. A., Shokat K. M. (2007) Biochem. Soc. Trans. 35, 245–249 [DOI] [PubMed] [Google Scholar]
- 17. Wu Y. T., Tan H. L., Shui G., Bauvy C., Huang Q., Wenk M. R., Ong C. N., Codogno P., Shen H. M. (2010) J. Biol. Chem. 285, 10850–10861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hickson I., Zhao Y., Richardson C. J., Green S. J., Martin N. M., Orr A. I., Reaper P. M., Jackson S. P., Curtin N. J., Smith G. C. (2004) Cancer Res. 64, 9152–9159 [DOI] [PubMed] [Google Scholar]
- 19. Høyer-Hansen M., Bastholm L., Szyniarowski P., Campanella M., Szabadkai G., Farkas T., Bianchi K., Fehrenbacher N., Elling F., Rizzuto R., Mathiasen I. S., Jäättelä M. (2007) Mol. Cell 25, 193–205 [DOI] [PubMed] [Google Scholar]
- 20. Dietrich N., Thastrup J., Holmberg C., Gyrd-Hansen M., Fehrenbacher N., Lademann U., Lerdrup M., Herdegen T., Jäättelä M., Kallunki T. (2004) Cell Death Differ 11, 301–313 [DOI] [PubMed] [Google Scholar]
- 21. Di Micco R., Fumagalli M., Cicalese A., Piccinin S., Gasparini P., Luise C., Schurra C., Garre M., Nuciforo P. G., Bensimon A., Maestro R., Pelicci P. G., d'Adda di Fagagna F. (2006) Nature 444, 638–642 [DOI] [PubMed] [Google Scholar]
- 22. Morgenstern J. P., Land H. (1990) Nucleic Acids Res. 18, 3587–3596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gillooly D. J., Morrow I. C., Lindsay M., Gould R., Bryant N. J., Gaullier J. M., Parton R. G., Stenmark H. (2000) EMBO J. 19, 4577–4588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Knight Z. A., Feldman M. E., Balla A., Balla T., Shokat K. M. (2007) Nat. Protoc. 2, 2459–2466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Foghsgaard L., Wissing D., Mauch D., Lademann U., Bastholm L., Boes M., Elling F., Leist M., Jäättelä M. (2001) J. Cell Biol. 153, 999–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Knight Z. A., Gonzalez B., Feldman M. E., Zunder E. R., Goldenberg D. D., Williams O., Loewith R., Stokoe D., Balla A., Toth B., Balla T., Weiss W. A., Williams R. L., Shokat K. M. (2006) Cell 125, 733–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sarkaria J. N., Busby E. C., Tibbetts R. S., Roos P., Taya Y., Karnitz L. M., Abraham R. T. (1999) Cancer Res. 59, 4375–4382 [PubMed] [Google Scholar]
- 28. Rainey M. D., Charlton M. E., Stanton R. V., Kastan M. B. (2008) Cancer Res. 68, 7466–7474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Saiki S., Sasazawa Y., Imamichi Y., Kawajiri S., Fujimaki T., Tanida I., Kobayashi H., Sato F., Sato S., Ishikawa K., Imoto M., Hattori N. (2011) Autophagy 7, 176–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Miller S., Tavshanjian B., Oleksy A., Perisic O., Houseman B. T., Shokat K. M., Williams R. L. (2010) Science 327, 1638–1642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Blommaart E. F., Krause U., Schellens J. P., Vreeling-Sindelárová H., Meijer A. J. (1997) Eur. J. Biochem. 243, 240–246 [DOI] [PubMed] [Google Scholar]
- 32. Boya P., González-Polo R. A., Casares N., Perfettini J. L., Dessen P., Larochette N., Métivier D., Meley D., Souquere S., Yoshimori T., Pierron G., Codogno P., Kroemer G. (2005) Mol. Cell. Biol. 25, 1025–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Degterev A., Huang Z., Boyce M., Li Y., Jagtap P., Mizushima N., Cuny G. D., Mitchison T. J., Moskowitz M. A., Yuan J. (2005) Nat. Chem. Biol. 1, 112–119 [DOI] [PubMed] [Google Scholar]
- 34. Szyniarowski P., Corcelle-Termeau E., Farkas T., Hoyer-Hansen M., Nylandsted J., Kallunki T., Jaattela M. (2011) Autophagy 7, 892–903 [DOI] [PubMed] [Google Scholar]
- 35. Martiny-Baron G., Kazanietz M. G., Mischak H., Blumberg P. M., Kochs G., Hug H., Marmé D., Schachtele C. (1993) J. Biol. Chem. 268, 9194–9197 [PubMed] [Google Scholar]
- 36. Bain J., Plater L., Elliott M., Shpiro N., Hastie C. J., McLauchlan H., Klevernic I., Arthur J. S., Alessi D. R., Cohen P. (2007) Biochem. J. 408, 297–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Adams C., Aldous D. J., Amendola S., Bamborough P., Bright C., Crowe S., Eastwood P., Fenton G., Foster M., Harrison T. K., King S., Lai J., Lawrence C., Letallec J. P., McCarthy C., Moorcroft N., Page K., Rao S., Redford J., Sadiq S., Smith K., Souness J. E., Thurairatnam S., Vine M., Wyman B. (2003) Bioorg. Med. Chem. Lett. 13, 3105–3110 [DOI] [PubMed] [Google Scholar]
- 38. Chrencik J. E., Patny A., Leung I. K., Korniski B., Emmons T. L., Hall T., Weinberg R. A., Gormley J. A., Williams J. M., Day J. E., Hirsch J. L., Kiefer J. R., Leone J. W., Fischer H. D., Sommers C. D., Huang H. C., Jacobsen E. J., Tenbrink R. E., Tomasselli A. G., Benson T. E. (2010) J. Mol. Biol. 400, 413–433 [DOI] [PubMed] [Google Scholar]
- 39. Axe E. L., Walker S. A., Manifava M., Chandra P., Roderick H. L., Habermann A., Griffiths G., Ktistakis N. T. (2008) J. Cell Biol. 182, 685–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Matsunaga K., Morita E., Saitoh T., Akira S., Ktistakis N. T., Izumi T., Noda T., Yoshimori T. (2010) J. Cell Biol. 190, 511–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cao Y., Semanchik N., Lee S. H., Somlo S., Barbano P. E., Coifman R., Sun Z. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 21819–21824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cecconi F., Levine B. (2008) Dev. Cell 15, 344–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Degenhardt K., Mathew R., Beaudoin B., Bray K., Anderson D., Chen G., Mukherjee C., Shi Y., Gélinas C., Fan Y., Nelson D. A., Jin S., White E. (2006) Cancer Cell 10, 51–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Degterev A., Hitomi J., Germscheid M., Ch'en I. L., Korkina O., Teng X., Abbott D., Cuny G. D., Yuan C., Wagner G., Hedrick S. M., Gerber S. A., Lugovskoy A., Yuan J. (2008) Nat. Chem. Biol. 4, 313–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kroemer G., Jäättelä M. (2005) Nat. Rev. Cancer 5, 886–897 [DOI] [PubMed] [Google Scholar]
- 46. Berghe T. V., Vanlangenakker N., Parthoens E., Deckers W., Devos M., Festjens N., Guerin C. J., Brunk U. T., Declercq W., Vandenabeele P. (2010) Cell Death Differ 17, 922–930 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.