Background: The signaling mechanisms involved in PGE2-induced MMP-9 expression in DCs are unknown.
Results: PGE2-induced MMP-9 expression is mediated through the EP2/EP4 → cAMP → PKA/PI3K → ERK signaling pathway.
Conclusion: PGE2-induced ERK activation is required for the binding of AP-1 to the MMP-9 promoter.
Significance: ERK inhibitors can be used to regulate DC migration.
Keywords: AP-1 Transcription Factor, Cyclic AMP (cAMP), Dendritic Cells, ERK, PI3K, MMP-9, PGE2, PKA, c-Jun
Abstract
Dendritic Cells (DCs) play an important role in the initiation of the immune response by migrating to regional lymph nodes and presenting antigen processed at the inflammatory site to antigen-specific naïve T cells. Prostaglandin E2 (PGE2) has been reported to play an essential role in DC migration. We reported previously that PGE2 induces matrix metalloproteinase 9 (MMP-9) expression in DCs and that PGE2-induced MMP-9 is required for DC migration in vivo and in vitro. In this study, we investigated the signaling mechanisms involved in PGE2-induced MMP-9 expression in DCs. We show that PGE2-induced MMP-9 expression is mediated primarily through the EP2/EP4 → cAMP → protein kinase A (PKA)/PI3K → ERK signaling pathway, leading to c-Fos expression, and through JNK-mediated activation of c-Jun in a PKA/PI3K/ERK-independent manner. EP2 and EP4 receptor agonists, as well as cAMP analogs, mimic the up-regulation of MMP-9 by PGE2. PKA, PI3K, and ERK inhibitors abolished PGE2- and cAMP-induced c-Fos and MMP-9 up-regulation, and ERK activation was required for the binding of activator protein 1 (AP-1) transcription factor to the MMP-9 promoter. Our results describe a new molecular mechanism for the effect of PGE2 on MMP-9 production in DCs that could lead to future therapeutic approaches using ERK inhibitors to regulate DC migration.
Introduction
Dendritic cells (DCs)2 exert their unique ability to present processed antigen to cognate naïve T cells upon migration to draining lymph nodes. Migration to secondary lymphoid organs requires the expression of the chemokine receptor CCR7 on DCs to respond to the chemokines CCL19 and CCL21 constitutively produced in lymph nodes. In addition to the CCR7 chemokine receptor, MMP-9 is also required for DC migration (1–3). Indeed, DC migration in vivo is impaired in MMP-9-deficient mice whose DCs express CCR7 chemokine receptors (4).
Matrix metalloproteinases (MMPs) represent a family of zinc-dependent proteinases that play a key role in facilitating cell migration by degrading the extracellular matrix (ECM) and basement membranes. Among MMPs, MMP-2 and MMP-9 are the major MMPs that cleave collagen IV, an essential component of basement membranes. MMP-9 is secreted as an inactive proenzyme by different cell types, including activated macrophages, monocytes, and DCs. Pro-MMP-9 is cleaved by MMPs such as MMP-2, MMP-3, and MMP-13 and can function both as a secreted and membrane-bound proteinase (5).
Prostaglandin E2 (PGE2), generated from arachidonic acid by cycloxygenases and prostaglandin E synthases, has been shown to induce migration in a variety of cell types, including DCs (6–8). The role of PGE2 in DC migration was established in mice deficient in PGE2 receptor EP4, which showed reduced DC migration to lymph nodes (9). In addition, the role of PGE2 in MMP-9 production has been shown in several other cell types. For instance, exogenous PGE2 increased MMP-9 expression in macrophages stimulated with phorbol myristate acetate (PMA) and in LPS-stimulated trophoblasts (10, 11). Moreover, endogenous PGE2 up-regulated MMP-9 expression in macrophages exposed to ECM and in tumor cells (12, 13). We showed previously for the first time that PGE2 also promoted MMP-9 up-regulation in DCs and that PGE2-induced MMP-9 production was required for DC migration (3). Although PGE2 was identified as a potent MMP-9 inducer in DCs, the molecular mechanisms of PGE2-induced MMP-9 expression are still unknown. Here we studied the signaling pathways involved in PGE2-induced MMP-9 expression. This study shows for the first time the involvement of EP2/EP4 receptor-mediated cAMP in the activation and binding of AP-1 to the MMP-9 promoter in DCs. We show that AP-1 activation involves c-Fos induction through PKA/PI3K signaling, leading to ERK activation, and c-Jun phosphorylation through JNK activation in a PKA/PI3K-independent manner.
MATERIALS AND METHODS
Mice
6- to 8-week-old B10.A mice were purchased from The Jackson Laboratory (Bar Harbor, Maine) and maintained in the Temple University School of Medicine animal facility under pathogen-free conditions. Mice were handled and housed in accordance with the guidelines of the Temple University Animal Care and Use Committee.
Reagents
PGE2 was purchased from Sigma. GM-CSF was purchased from Peprotech, Inc. (Rocky Hill, NJ). Butaprost, sulprostone, and CAY10580 were purchased from Cayman (Ann Arbor, MI). Dibutyryl-cAMP (dbcAMP), the EPAC-specific activator 8-CPT-2′-O-Me-cAMP, H89, LY294002, SB203580, JNK inhibitor II, and JSH-32 were purchased from Calbiochem. U0126 and antibodies to phospho-ERK, ERK, phospho-c-Jun, c-Jun, and c-Fos were obtained from Cell Signaling Technology, Inc. (Danvers, MA).
Generation and Purification of DC from Bone Marrow
DCs were generated in vitro from bone marrow cells as described previously (3).
FACS Analysis for Phospho-ERK, ERK, Phospho-c-Jun, c-Jun, and c-Fos
Cells treated as indicated were fixed, permeabilized, and incubated with anti-mouse phospho-ERK, anti-mouse ERK, anti-mouse phospho-c-Jun, anti-mouse c-Jun, or anti-mouse c-Fos for 40 min at room temperature followed by Alexa Fluor-conjugated goat anti-rabbit IgG (Invitrogen) for 30 min. Data were collected for 10,000 cells and analyzed by FACS.
Western Blot Analysis
3–6 × 106 DCs were serum-starved for 3 h before treatment with PGE2 at room temperature. Samples were lysed in radioimmunoprecipitation assay lysis buffer (50 mm Tris-HCl (pH 8), 150 mm NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS) plus a protease inhibitor mixture (5 mm phenylmethyl-sulfonyl fluoride, 1 mm sodium orthovanadate, 1 mm sodium fluoride, 0.2 μm okadaic acid (Sigma)). 20–30 μg of whole protein lysate were mixed with 6× sample buffer and boiled for 7 min, followed by loading on 10% SDS-PAGE gels. Separated protein were transferred onto polyvinylidene fluoride membranes (Bio-Rad) and probed with primary antibodies against phospho-p44/p42 MAP Kinase (Thr-202, Tyr-204), total p44/p42 MAP Kinase (L34F12), phospho-cJun (Ser-63) II, and total cJun (L70B11) (Cell Signaling Technology, Inc.) at 1:1000 dilution in 50:50 Odyssey blocking buffer:PBS (LiCor Biosciences). Goat anti-mouse IRDye 800CW and goat anti-rabbit IRDye 680CW antibodies (LiCor Biosciences) were used as secondary antibodies. Transferred proteins were visualized by using the Odyssey infrared imaging system (LiCor Biosciences).
Real-time RT-PCR
The expression of MMP-9 and c-Fos was detected by the SYBR Green-based real-time RT-PCR technique. RNA was isolated from purified CD11c+ DCs and treated with or without PGE2 for various time periods. cDNA was prepared, and the 20 μl (total volume) of the PCR mixture consisted of 4 μl of diluted cDNA, 10 μl of SYBR Green-containing PCR master mixture (2×) and 150 nm of each primer. The primers for real-time RT-PCR are as follows. MMP-9: sense, 5′-AAAACCTCC-AACCTCACGGA-3′ and antisense, 5′-GCGGT-ACAAGTATGCCTCTGC-3′. c-Fos: sense, 5′-AGCCCAGACCTGCAGTGGCT-3′ and antisense, 5′-GCGCTCTGCCTCCTGACACG-3′. Real-Time RT-PCR was performed using the Stratagene Mx3005P, and the cycling conditions used were 95 °C for 30 s, 55 °C for 1 min, 72 °C for 30 s, for 40 cycles, followed by a melting point determination or dissociation curves. The expression level of each gene is indicated by the cycle numbers needed for the cDNA to be amplified to reach a threshold. The amount of DNA is calculated from the cycle numbers by using standard curves, and the results are normalized to the housekeeping gene β-actin from the same sample.
Cytokine ELISA
Purified CD11c+ DCs (1 × 106 cells/ml) were seeded in 12-well plates and treated as described under “Results.” The amounts of pro-MMP-9 released in the medium were measured by sandwich ELISA with antibodies obtained from R&D Systems (Minneapolis, MN). The absorbance was determined using a Polarstar Optima plate reader at a wavelength of 450 nm.
Nuclear Extracts and EMSA
Purified CD11c+ DCs were cultured at a concentration of 1 × 106 cells/ml and stimulated with PGE2 10−6 m for 1 h. Nuclear extracts were prepared by using the NucBuster protein extraction kit (Novagen, Inc., Madison, WI) as recommended by the manufacturer. Double-stranded oligonucleotides binding to target transcriptional factors were end-labeled with [γ-32P]ATP using T4 polynucleotide kinase (Promega, Madison, WI). The radiolabeled AP-1 probe (10,000 cpm, 5′-CTTGATGAGTCAGC-CGGAA-3′) was incubated with 10 μg of nuclear extracts for 20 min at room temperature in 10 μl of total volume of binding solution containing 20 mm HEPES (pH 7.8), 0.1 mm EDTA, 1 mm dithiothreitol, 0.1 mm phenylmethylsulfonyl fluoride, 10% glycerol, and 1 μg of poly dI-dC.
Protein/DNA Array
Protein/DNA arrays were used to simultaneously screen a large number of transcription factors for DNA binding activity. Nuclear extracts were prepared, and the protein/DNA arrays were carried out with the TranSignalTM Protein/DNA Array I (Panomics, Redwood City, CA) according to the manufacturer's instructions.
ChIP Assay
Following various treatments, DCs were fixed with 1% formaldehyde (final concentration) for 30 min, followed by 125 mm glycine (final concentration) for 10 min. Cells were washed twice with ice-cold phosphate-buffered saline containing protease inhibitors, collected, resuspended in SDS lysis buffer containing protease inhibitors, incubated for 30 min on ice, and sonicated to shear DNA. After sonication, the lysates were centrifuged at 13,000 rpm for 15 min at 4 °C, and the supernatants were diluted in ChIP dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mm EDTA, 16.7 mm Tris-HCl (pH 8.1), 167 mm NaCl, plus protease inhibitors). The chromatin was immunoprecipitated with anti-c-Fos, anti-c-Jun, or anti-GST as a negative control (Santa Cruz Biotechnology, Inc., Santa Cruz, CA). After overnight incubation at 4 °C, protein A/G-agarose was added for 1 h, and the immune complexes were washed sequentially with low-salt wash buffer once (0.1% SDS, 1% Triton X-100, 2 mm EDTA, 20 mm Tris-HCl (pH 8.1), and 150 mm NaCl), high-salt wash buffer once (0.1% SDS, 1% Triton X-100, 2 mm EDTA, 20 mm Tris-HCl (pH 8.1), and 500 mm NaCl), LiCl wash buffer once (0.25 m LiCl, 1% octyl phenyl-polyethylene glycol, 1% deoxycholic acid, 1 mm EDTA, and 10 mm Tris-HCl (pH 8.1)), and Tris/EDTA buffer twice (10 mm Tris-HCl and 1 mm EDTA (pH 8.0)). Antibodies were eluted from the immune complexes with elution buffer (1% SDS, 10 mm EDTA, 50 mm Tris-HCl (pH 8.0)) and cross-linking was reversed by heating at 65 °C overnight. After Proteinase K digestion, input DNA and precipitated DNA were purified and real-time PCR-amplified with primers encompassing the MMP-9 promoter region containing AP-1 sites (sense, GACCCTGGGAA-CCGGGTCCA and antisense, CAGGGACCGGCCG-TGGAAAC).
Statistical Analysis
Results are given as mean ± S.E. Comparisons between multiple groups were done by analysis of variance. Statistical significance was determined as p values less than 0.05.
RESULTS
Involvement of EP2/EP4 and cAMP in PGE2-induced MMP-9 Production
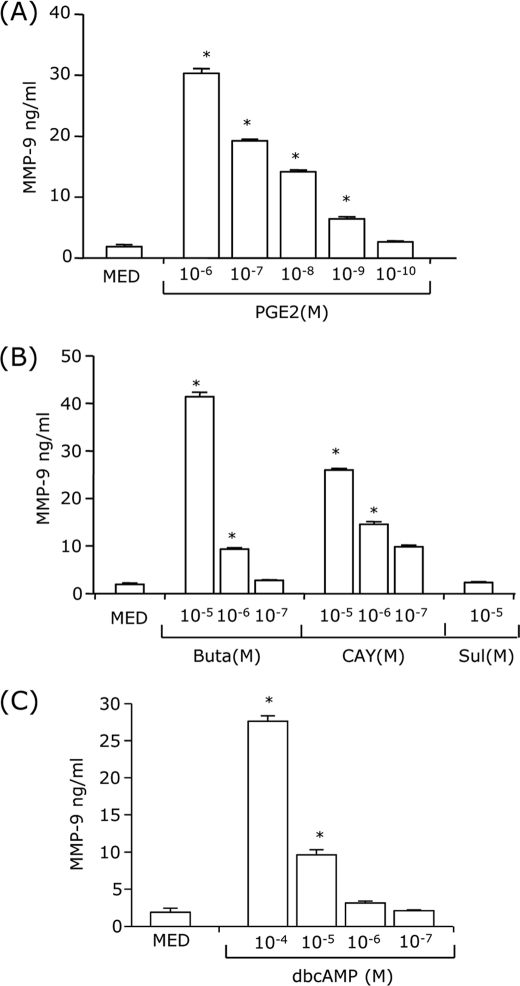
We have shown previously that PGE2 is a potent MMP-9 inducer in immature and TNF-α+IFN-α-matured bone marrow-derived DCs (3). In this study, we dissected the signaling pathways involved in PGE2-induced MMP-9. Immature CD11c+ bone marrow-derived DCs were treated with different concentrations of PGE2, and supernatants were tested for MMP-9 by ELISA. The results show that PGE2 induced MMP-9 production in a dose-dependent manner (Fig. 1A). To identify the specific EP receptors involved in MMP-9 up-regulation, DCs were treated with receptor agonists, i.e. butaprost (EP2), CAY10580 (EP4), and sulprostone (EP1/EP3). Butaprost and CAY10580 induced MMP-9 production in a dose-dependent manner, whereas sulprostone had no effect (Fig. 1B). This indicates that EP2 and EP4 mediate the effect of PGE2 on MMP-9 expression.
FIGURE 1.
PGE2 induction of MMP-9 is mediated through EP2/EP4 and cAMP. CD11c+ DCs (1 × 106 cells/ml) were treated with different concentrations of PGE2 (10−6-10−10 m) (A), butaprost (an EP2 agonist, 10−5-10−7 m), CAY10580 (an EP4 agonist, 10−5-10−7 m), and sulprostone (an EP1/EP3 agonist, 10−5 m) (B), or dbcAMP (10−4-10−7 m) (C) for 24 h followed by MMP-9 ELISA. MED, medium only. *, p < 0.01 compared with MED. Data are representative of three independent experiments.
A major effect of EP2/EP4 ligand binding is the stimulation of adenylate cyclase, resulting in an increase in intracellular cAMP. To assess whether MMP-9 expression is mediated through cAMP induction, we used dbcAMP, a stable cAMP analog. Similar to PGE2 and to the EP2/EP4 agonists, dbcAMP up-regulated MMP-9 production in a dose-dependent manner (Fig. 1C).
PGE2 Increases Nuclear AP-1
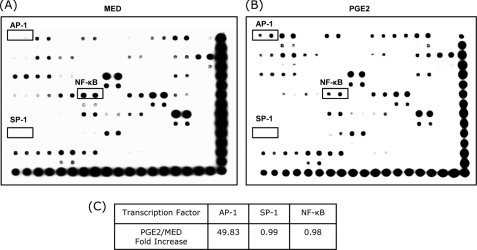
The MMP-9 promoter contains functional AP-1, Sp-1, and NF-κB binding sites (14–16). To investigate whether PGE2 activates these transcription factors, nuclear extracts from untreated and PGE2-treated DCs were subjected to protein/DNA arrays that detect and quantify transcription factors through interaction with specific nucleotides. PGE2 significantly increased nuclear AP-1 (Fig. 2). The level of nuclear NF-κB was not increased by PGE2 treatment. No significant SP-1 levels were observed in either untreated or PGE2-treated DCs (Fig. 2). These results suggest that PGE2-induced MMP-9 expression might be mediated primarily through AP-1 activation.
FIGURE 2.
Effects of PGE2 on the binding of nuclear transcription factors in DCs. CD11c+ DCs were treated with PGE2 10−6 m for 1 h. Nuclear extracts from untreated DCs (A) and PGE2-treated DCs (B) were prepared and subjected to protein/DNA arrays. C, the amounts of AP-1, SP-1, and NF-κB were normalized to controls on each membrane, and the normalized values for AP-1, SP-1, and NF-κB obtained for PGE2-treated DC were compared with the normalized values for the medium control. Results are representative of two independent experiments.
Both PKA and PI3K Pathways Are Involved in PGE2-induced MMP-9 Production
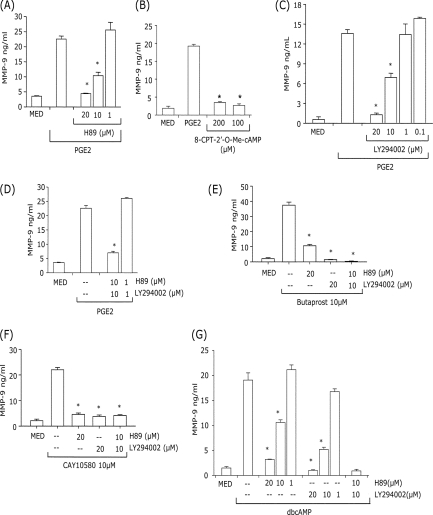
cAMP activates both PKA- and EPAC-dependent signal pathways (17). Treatment of DCs with the PKA inhibitor H89 resulted in a significant reduction of PGE2-, butaprost-, CAY10580- and dbcAMP-induced MMP-9 production (Fig. 3, A, E, and G). In contrast, treatment of DCs with the EPAC activator 8-CPT-2′-O-Me-cAMP did not lead to MMP-9 induction (Fig. 3B). These results support the role of PKA in the induction of MMP-9 by PGE2.
FIGURE 3.
Inhibition of PKA and/or PI3K prevents PGE2- or cAMP-induced MMP-9. CD11c+ DCs were pretreated with different concentrations of H89 (A), LY294002 (C), or H89+LY294002 (D) for 30 min, followed by 10−7 m PGE2 for 24 h. B, DCs were treated with 8-CPT-2′-O-Me-cAMP (200 and 100 μm) or 10−7 m PGE2 for 24 h. E and F, cells were pretreated with 20 μm H89, 20 μm LY294002, or 10 μm H89 and 10 μm LY294002 for 30 min followed by 10 μm butaprost (E) or 10 μm CAY10580 (F) for 24 h. G, cells were pretreated with different concentrations of H89, LY294002, or H89 and LY294002 for 30 min, followed by 10−4 m dbcAMP for 24 h. The supernatants were collected and subjected to MMP-9 ELISA. *, p < 0.01 compared with PGE2, butaprost, CAY10580, or dbcAMP without inhibitors. Data are representative of three independent experiments.
EP2 and especially EP4 can also activate PI3K (18, 19). To examine the role of PI3K in PGE2-induced MMP-9 production, DCs were pretreated with the PI3K inhibitor LY294002. Similar to H89, LY294002 significantly reduced MMP-9 induction by PGE2, butaprost, CAY10580 or dbcAMP (Fig. 3, C, E, F, and G). The involvement of both PKA and PI3K in the MMP-9 induction by PGE2 is supported by the fact that although lower concentrations (10 μm) of H89 and LY294002 were only partially inhibitory when used separately, the combined H89+LY294002 treatment resulted in complete inhibition (Fig. 3, A, C, and D). The same results were obtained for dbcAMP suggesting that, similar to PGE2, the effect of cAMP on MMP-9 expression is mediated through both PKA and PI3K (Fig. 3G). DC viability was not affected by the presence of the highest concentrations of inhibitors (> 90% propidium iodide-negative).
Inhibition of ERK but Not p38MAPK Signaling Abolished PGE2-induced MMP-9 Production
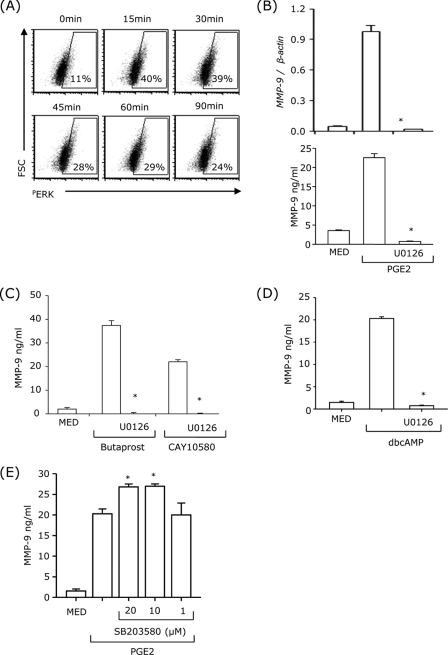
Treatment of DCs with PGE2 induced ERK phosphorylation with a maximum effect between 15 and 30 min (Fig. 4A). To examine the role of PGE2-induced ERK activation in MMP-9 mRNA expression and protein production, DCs were pretreated with the MEK1/2 inhibitor, U0126, for 30 min, followed by PGE2 treatment for 24 h. U0126 abolished PGE2-induced MMP-9 mRNA expression and protein production (Fig. 4B). Similar results were obtained for butaprost-, CAY10580-, and dbcAMP-induced MMP-9 production (Fig. 4, C and D). In contrast, inhibition of p38MAPK had no effect on PGE2-induced MMP-9 up-regulation (Fig. 4E). These results suggest that ERK, but not p38MAPK, is involved in PGE2-induced MMP-9 production.
FIGURE 4.
Inhibition of ERK, but not p38MAPK signaling, abolished PGE2-induced MMP-9 production. A, DCs were treated with 10−7 m PGE2. At different time points, DCs were fixed, permeabilized, and analyzed by intracellular staining with phospho-ERK antibody. B–D, DCs were pretreated with 10 μm U0126 for 30 min, followed by 10−7 m PGE2 (B), 10−5 m butaprost or 10−5 m CAY10580 (C), or 10−4 m dbcAMP (D) for 24 h. MMP-9 mRNA expression was analyzed by real-time RT-PCR (B, top panel), and MMP-9 secretion was measured by ELISA (B, bottom panel, C, and D). E, cells were pretreated with different concentrations of SB203580 for 30 min followed by 10−7 m PGE2 for 24 h. The supernatants were collected for MMP-9 ELISA. *, p < 0.01 compared with PGE2, butaprost, CAY10580, or dbcAMP without inhibitors. Data are representative of three independent experiments.
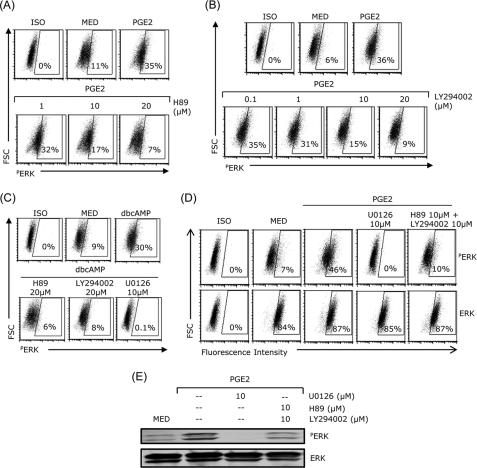
Inhibition of PKA or PI3K Blocked PGE2- or cAMP-induced ERK Activation
To determine whether ERK activation was mediated through PKA or PI3K signaling, DCs were treated with PGE2 or dbcAMP in the presence of H89 or LY294002 and then assessed for ERK phosphorylation by FACS. Both H89 and LY294002 dose-dependently inhibited PGE2-induced ERK phosphorylation (Fig. 5, A and B).
FIGURE 5.
Inhibition of PKA or PI3K pathway blocked PGE2- or cAMP-induced ERK activation. DCs were pretreated with H89, LY294002, or U0126 for 30 min, followed by 10−7 m PGE2 (A and B) or 10−4 m dbcAMP (C). After 20 min, DCs were fixed, permeabilized, and analyzed by intracellular staining with phospho-ERK antibody. D and E, DCs pretreated with U0126 or H89 plus LY294002 for 30 min followed by 10−7 m PGE2 for 20 min were subjected to flow cytometry (D) or Western blot analysis (E) for phospho-ERK and ERK expression. Data are representative of three independent experiments.
The two inhibitors also blocked dbcAMP-induced ERK activation (Fig. 5C). H89 and LY294002 and U0126 (ERK inhibitor used as control) blocked ERK phosphorylation without affecting the levels of total ERK in DCs treated with PGE2 (Fig. 5D). Similar results were observed in Western blot analysis (Fig. 5E). Taken together, these results indicate that PGE2-induced ERK phosphorylation is mediated through both PKA and PI3K activation.
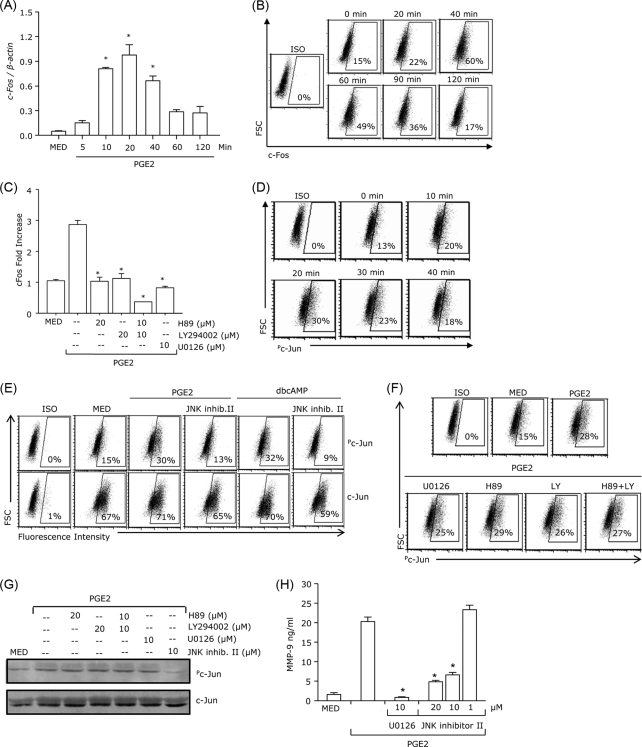
PGE2 Induces ERK-dependent c-Fos Expression and ERK-independent c-Jun Activation
ERK has been shown to induce the activation of the AP-1 family of transcription factors, including c-Fos and c-Jun (20, 21). In terms of c-Fos, activation of ERK has been linked to the up-regulation of c-Fos mRNA expression. To determine the effect of PGE2 on c-Fos expression, we treated DCs with PGE2 and measured c-Fos mRNA and protein expression at different time points. c-Fos mRNA expression was significantly induced 20 min after PGE2 treatment (Fig. 6A). In addition, c-Fos protein expression reached a peak at 40 min and decreased to basal levels 2 h after PGE2 treatment (Fig. 6B). To determine whether PGE2-induced ERK activation is required for c-Fos expression, DCs were pretreated with U0126 followed by PGE2 stimulation. In the presence of U0126, PGE2 did not induce c-Fos expression (Fig. 6C). Because the PKA/PI3K pathways are required for ERK activation (Fig. 5, A and B), we investigated their role in PGE2-induced c-Fos expression. H89 and LY294002 abolished PGE2-induced c-Fos expression, suggesting the involvement of both PKA and PI3K (Fig. 6C).
FIGURE 6.
PGE2 induces ERK-dependent c-Fos expression and ERK-independent c-Jun activation. DCs were treated with PGE2 10−7 m and analyzed for c-Fos mRNA expression by real-time RT-PCR (A) or intracellular c-Fos protein expression at different time points (B). C, DCs were pretreated with U0126, H89, LY294002, or H89 plus LY294002 for 30 min followed by PGE2 10−7 m for 40 min. Cells were fixed, permeabilized, and analyzed by intracellular staining with c-Fos antibody. D, intracellular phospho-c-Jun expression was measured at different time points after PGE2 10−7 m stimulation. E–G, DCs were pretreated with 10 μm JNK inhibitor II, 20 μm H89, 20 μm LY294002, or 10 μm H89 plus 10 μm LY294002 for 30 min, followed by 10−7 m PGE2 or 10−4 m dbcAMP for 20 min. Phosphorylation of c-Jun was analyzed by FACS (E and F) or Western blot analysis (G). H, DCs were pretreated with different concentrations of JNK inhibitor II or with 10 μm U0126 as control for 30 min followed by 10−7 m PGE2 for 24 h. Supernatants were subjected to MMP-9 ELISA. Data are representative of three independent experiments. *, p < 0.01 compared with medium only (MED) (A) or with PGE2 without inhibitors (C and H).
In DCs treated with PGE2, c-Jun phosphorylation was apparent at 10 min, reaching a maximum at 20 min (Fig. 6D). PGE2-induced c-Jun phosphorylation was prevented by the JNK inhibitor II (Fig. 6E) but not by U0126, H89, LY294002, or H89+LY294002 as determined by both flow cytometry and Western blot analysis (Fig. 6, F and G). This indicates that the effect of PGE2 on c-Jun is mediated through JNK but does not depend on PKA/PI3K signaling. Similar levels of phosphorylated c-Jun were observed in DCs treated with PGE2, EP2, and EP4 ligands, and the JNK inhibitor II prevented c-Jun phosphorylation by all three agents (supplemental Fig. 1). In addition, dbcAMP also induced c-Jun phosphorylation which was significantly reduced by the JNK inhibitor II (Fig. 6E). To further assess the role of c-Jun activation in PGE2-induced MMP-9 production, DCs were pretreated with JNK inhibitor II followed by PGE2. PGE2-induced MMP-9 production was inhibited in the presence of the JNK inhibitor (Fig. 6H). Altogether, these results suggest that PGE2 induces c-Fos expression through the PGE2 → PKA/PI3K → ERK axis, whereas PGE2-induced c-Jun phosphorylation is mediated by JNK, independent of PKA/PI3K signaling and ERK activation.
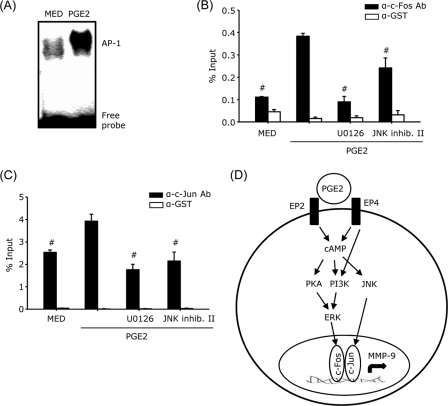
AP-1 Binding to the MMP-9 Promoter
The minimal MMP-9 promoter region contains two AP-1 binding sites (14, 22). To investigate whether treatment with PGE2 affects the DNA binding activity of AP-1, nuclear extracts from untreated and PGE2-treated DCs were subjected to EMSA. In agreement with the protein/DNA binding arrays (Fig. 2), PGE2 induced a significant increase in AP-1 binding (Fig. 7A). To assess the in vivo interaction of AP-1 with the MMP-9 promoter, we performed ChIP assays. Cell lysates from untreated or PGE2-treated DCs in the presence or absence of U0126 or JNK inhibitor II were immunoprecipitated with anti-c-Fos antibody, anti-c-Jun antibody, or anti-GST as a negative control, followed by real-time PCR amplification using primers encompassing the AP-1 sites in the murine MMP-9 promoter. The binding of both c-Fos and c-Jun to the MMP-9 promoter was increased significantly in DCs treated with PGE2, and the PGE2-induced binding was significantly reduced in the presence of U0126 or JNK inhibitor II (Fig. 7, B and C), suggesting that PGE2-induced ERK and JNK activation are required for the generation and binding of c-Fos/c-Jun heterodimers to the AP-1 site in the MMP-9 promoter.
FIGURE 7.
AP-1 binding to the MMP-9 promoter. A, CD11c+ DCs were treated with 10−6 m PGE2 for 1 h. Nuclear extracts were subjected to EMSA analysis with DNA probes containing the AP-1 consensus motifs. B and C, DCs were pretreated with U0126 10 μm or JNK inhibitor II 10 μm for 30 min followed by PGE2 10−7 m for 1 h. Cells were fixed, sonicated, and subjected to ChIP analysis using antibodies to c-Fos (B), c-Jun (C), or GST as a negative control. Precipitated DNA was isolated and evaluated by real-time PCR using the primers mentioned under “Materials and Methods.” #, p < 0.05 compared with PGE2. Data are representative of two independent experiments in A and C and three experiments in B. D, model for the signaling pathways that mediate PGE2 induction of MMP-9 in DCs. MED, medium only.
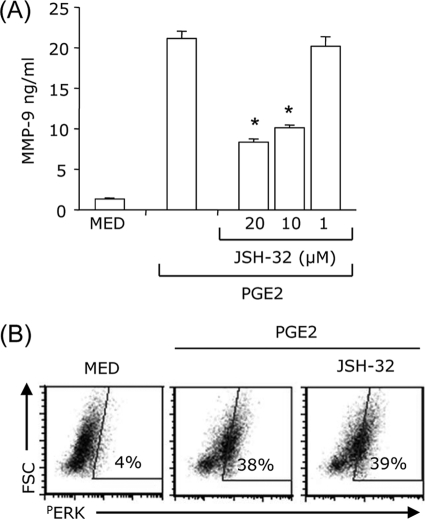
Blocking NF-κB Activation Partially Inhibited PGE2-induced MMP-9 Production and Had No Effect on PGE2-induced ERK Phosphorylation
We have reported previously that PGE2 does not have a substantial effect on NF-κB activation (23). In agreement with our previous observation, inhibition of NF-κB using JSH-32 reduced only partially PGE2-induced MMP-9 production (Fig. 8A). Also, JSH-32 did not affect PGE2-induced ERK activation (Fig. 8B). These results suggest that AP-1 is the major transcription factor promoting MMP-9 expressing by PGE2.
FIGURE 8.
NF-κB contributes to the induction of MMP-9 by PGE2 without affecting PGE2-induced ERK activation. A, DCs were pretreated with different concentrations of JSH-32 for 30 min and treated with 10−7 m PGE2 for 24 h. The supernatants were collected and subjected to the MMP-9 ELISA. B, DCs were pretreated with 10 μm JSH-32 for 30 min followed by 10−7 m PGE2 stimulation. After 20 min, DCs were fixed, permeabilized, and analyzed by intracellular staining with phospho-ERK antibody. *, p < 0.01 compared with PGE2. Data are representative of three independent experiments.
DISCUSSION
MMPs serve broad functions in immune defense, tissue repair and inflammation (24, 25). By promoting ECM turnover, the degradation of basement membranes, and increasing endothelial permeability, MMP-2 and MMP-9 play a major role in immune cell migration. DC transmigration into central and peripheral inflammatory sites and migration to draining lymph nodes for antigen presentation and activation of T cells was shown to require MMP-9 (1–3, 26, 27). Depending on the cell type, MMP-9 gene expression is induced following LPS or PMA stimulation, treatment with cytokines such as TNF-α and TGFβ, or with oxidized low-density lipoproteins (28–31). In addition, a direct correlation between induction of PGE2 and MMP-9 was reported in a number of cell types, including monocytes (32, 33). Exogenous and endogenous PGE2 was also shown to play a role in MMP-9 expression in macrophages stimulated with PMA, LPS, or exposed to ECM (10, 13).
We showed previously that exogenous PGE2 induced MMP-9 expression in immature DCs, up-regulated MMP-9 in DCs treated with TNF-α and IFN-α, and that PGE2-induced MMP-9 was essential for in vitro migration of DCs through Matrigel and for the in vivo migration of DCs in wild-type and MMP-9-deficient hosts (3). However, the molecular mechanisms by which PGE2 induces MMP-9 expression in DCs are not elucidated. Here we report that PGE2-induced MMP-9 expression in DCs is mediated primarily through the activation of AP-1 and involves c-Fos induction through PKA/PI3K signaling and subsequent ERK activation and through the PKA/PI3K-independent activation of JNK, leading to phosphorylation of c-Jun.
The biological activity of PGE2 is mediated through four types of G protein-coupled receptors, i.e. EP1 through EP4, which differ in terms of tissue distribution and signaling pathways (19, 34). In agreement with our previous observation that bone marrow-derived DCs express high levels of EP2 and EP4 (35), this study indicates that EP2 and EP4 mediate PGE2-induced MMP-9 expression in DCs. EP2 and EP4 were shown to associate with Gαs and activate adenylate cyclase, leading to increases in intracellular cAMP (19, 34). As expected, the effects of PGE2 were mimicked by EP2 and EP4 agonists and the stable cAMP analog dbcAMP.
cAMP signaling involves two major intracellular mediators, i.e. the classical PKA and the more recently discovered EPAC, which may act in an independent, synergistic, or opposing manner (17). We showed previously that the PGE2 effect on TNF-α production in DCs was mediated by PKA, whereas effects on CCL3/4 were mediated through EPAC (35, 36). This study indicates that PKA, not EPAC, mediates the induction of MMP-9 by PGE2.
In addition to adenylate cyclase, EP4 receptors were reported to also activate PI3K, presumably through coupling to Gαi (37). Although initial experiments using EP2 overexpressing cells did not show PI3K activation (37), recent reports indicate that EP2 can signal through PI3K in certain cell types (18, 38). This study suggests that both PKA and PI3K are involved in the induction of MMP-9 in DCs following treatment with PGE2, EP2, and EP4 agonists. A similar involvement of both kinases was reported for PGE2 signaling through EP2 in C6 rat brain cells in ischemic preconditioning (18). In our system, the activation of the two kinases appears to be downstream of cAMP because MMP-9 induction by dbcAMP was blocked by both H89 and LY294002. Similar effects of cAMP on PKA and PI3K activation were reported previously (39–42). At this point we cannot rule out the possibility that PI3K activation might occur through PKA, as reported previously in human carcinoma cells treated with PGE2 (40).
The involvement of ERK activation in MMP-9 induction has been reported in monocytes/macrophages, keratinocytes, and various tumor cell lines (13, 28, 43–45). Here we show that ERK phosphorylation in DCs was induced as early as 15 min after PGE2 treatment, and was maintained for at least 90 min. Treatment with U0126 abolished PGE2-induced MMP-9 mRNA and protein expression. Similar results were obtained when DCs were treated with EP2/EP4 agonists or dbcAMP in the presence of U0126. In contrast to studies that reported the involvement of p38MAPK in MMP-9 induction in macrophages (46), we did not observe MMP-9 reduction in DCs treated with PGE2 in the presence of SB203580. The slight but statistically significant increase in MMP-9 production in the presence of p38MPAK inhibitor might be actually be due to enhanced ERK activity.
The induction of MMP-9 by PGE2 requires activation of PKA, PI3K, and ERK because PGE2-induced MMP-9 production is prevented in the presence of inhibitors for these three proteins. To establish the hierarchy of the signaling pathway, we treated DCs with PGE2 or dbcAMP in the presence of PKA or PI3K inhibitors and measured ERK activation. In these conditions, PGE2- or dbcAMP-induced ERK phosphorylation was prevented, suggesting that PGE2/cAMP activates PKA/PI3K to stimulate ERK phosphorylation. Although we did not investigate the molecular mechanisms of PKA/PI3K-induced ERK activation, several studies reported that PKA and PI3K activate the Raf/Rap1 signaling pathway, resulting in MEK1/2 phosphorylation and ERK activation (47, 48).
Optimal expression of MMP-9 depends on a promoter region containing binding sites for AP-1, Sp-1, NF-κB, and Ets (14–16). In macrophages stimulated with PMA or TNF-α, AP-1 was shown to be essential but required NF-κB or Sp-1 for optimal MMP-9 expression (16). Our protein/DNA array for transcription factors indicated that PGE2 induced a significant increase in AP-1 without affecting the basal levels of NF-κB and without inducing detectable levels of Sp-1. As a result we focused on the effect of PGE2 on AP-1 activation in DCs as related to MMP-9 induction.
AP-1 complexes are c-Jun/c-Fos heterodimers that bind to the consensus DNA sequence 5′TGAG/CTCA-3′ as dimers. We found that PGE2 up-regulates c-Fos at both mRNA and protein levels and that ERK activation is required for the up-regulation of c-Fos expression. As expected, PGE2-induced c-Fos up-regulation was inhibited by PKA, PI3K, and ERK inhibitors. This indicates that the effect of PGE2 on c-Fos is mediated through PKA, PI3K, and ERK activation.
Activation of c-Jun is also required for functional AP-1 complexes. PGE2 as well as EP2 and EP4 ligands induced c-Jun phosphorylation through JNK activation. Although dependent on cAMP, c-Jun phosphorylation was not mediated through the PKA/PI3K → ERK pathway. A potential candidate is the homeodomain-interacting protein kinase 3 (HIPK3) shown to be involved in cAMP-induced stimulation of JNK in the adrenal gland (49).
By using ChIP assays we confirmed that the in vivo PGE2-induced AP-1 binding to the MMP-9 promoter in DCs was dependent on ERK and c-Jun activation. In addition to AP-1, NF-κB activation plays an important role in MMP-9 up-regulation. We confirmed the role of NF-κB by using the JSH-32 inhibitor. Although PGE2-induced ERK phosphorylation was not affected, JSH-32 partially inhibited MMP-9 production. We concluded that PGE2-induced expression of MMP-9 in DCs requires both AP-1 and NF-κB, with AP-1 as the essential transcription factor.
In conclusion, this study shows for the first time that PGE2 induces MMP-9 expression in DCs through signaling pathways initiated by EP2 and EP4 receptors, which in turn activate downstream sequences consisting on one hand of cAMP-mediated activation of PKA and PI3K, activation of ERK and induction of c-Fos expression and on the other hand of JNK-mediated, PKA/PI3K-independent phosphorylation of c-Jun (Fig. 7D). Our results indicate that although NF-κB is required for PGE2-induced MMP-9 expression in DCs, AP-1 represents the major transcription factor activated by PGE2.
Footnotes
- DC
- dendritic cell
- MMP
- matrix metalloproteinase
- ECM
- extracellular matrix
- PMA
- phorbol myristate acetate
- PKA
- protein kinase A
- EPAC
- exchange protein directly activated by cAMP
- dbcAMP
- dibutyryl-cAMP
- EP
- PGE2 receptor.
REFERENCES
- 1. Hollender P., Ittelett D., Villard F., Eymard J. C., Jeannesson P., Bernard J. (2002) Immunobiology 206, 441–458 [DOI] [PubMed] [Google Scholar]
- 2. Ratzinger G., Stoitzner P., Ebner S., Lutz M. B., Layton G. T., Rainer C., Senior R. M., Shipley J. M., Fritsch P., Schuler G., Romani N. (2002) J. Immunol. 168, 4361–4371 [DOI] [PubMed] [Google Scholar]
- 3. Yen J. H., Khayrullina T., Ganea D. (2008) Blood 111, 260–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ichiyasu H., McCormack J. M., McCarthy K. M., Dombkowski D., Preffer F. I., Schneeberger E. E. (2004) Am. J. Respir. Cell Mol. Biol. 30, 761–770 [DOI] [PubMed] [Google Scholar]
- 5. Van den Steen P. E., Dubois B., Nelissen I., Rudd P. M., Dwek R. A., Opdenakker G. (2002) Crit. Rev. Biochem. Mol. Biol. 37, 375–536 [DOI] [PubMed] [Google Scholar]
- 6. Luft T., Jefford M., Luetjens P., Toy T., Hochrein H., Masterman K. A., Maliszewski C., Shortman K., Cebon J., Maraskovsky E. (2002) Blood 100, 1362–1372 [DOI] [PubMed] [Google Scholar]
- 7. Scandella E., Men Y., Legler D. F., Gillessen S., Prikler L., Ludewig B., Groettrup M. (2004) Blood 103, 1595–1601 [DOI] [PubMed] [Google Scholar]
- 8. van Helden S. F., Krooshoop D. J., Broers K. C., Raymakers R. A., Figdor C. G., van Leeuwen F. N. (2006) J. Immunol. 177, 1567–1574 [DOI] [PubMed] [Google Scholar]
- 9. Kabashima K., Sakata D., Nagamachi M., Miyachi Y., Inaba K., Narumiya S. (2003) Nat. Med. 9, 744–749 [DOI] [PubMed] [Google Scholar]
- 10. Renò F., Cannas M. (2005) Prostaglandins Other Lipid Mediat. 75, 13–24 [DOI] [PubMed] [Google Scholar]
- 11. Li W., Unlugedik E., Bocking A. D., Challis J. R. (2007) Biol. Reprod. 76, 654–659 [DOI] [PubMed] [Google Scholar]
- 12. Itatsu K., Sasaki M., Yamaguchi J., Ohira S., Ishikawa A., Ikeda H., Sato Y., Harada K., Zen Y., Sato H., Ohta T., Nagino M., Nimura Y., Nakanuma Y. (2009) Am. J. Pathol. 174, 829–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pavlovic S., Du B., Sakamoto K., Khan K. M., Natarajan C., Breyer R. M., Dannenberg A. J., Falcone D. J. (2006) J. Biol. Chem. 281, 3321–3328 [DOI] [PubMed] [Google Scholar]
- 14. Farina A. R., Tacconelli A., Vacca A., Maroder M., Gulino A., Mackay A. R. (1999) Cell Growth & Differ. 10, 353–367 [PubMed] [Google Scholar]
- 15. Sato H., Kita M., Seiki M. (1993) J. Biol. Chem. 268, 23460–23468 [PubMed] [Google Scholar]
- 16. Sato H., Seiki M. (1993) Oncogene 8, 395–405 [PubMed] [Google Scholar]
- 17. Cheng X., Ji Z., Tsalkova T., Mei F. (2008) Acta Biochim. Biophys. Sin. 40, 651–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Park M. K., Kang Y. J., Ha Y. M., Jeong J. J., Kim H. J., Seo H. G., Lee J. H., Chang K. C. (2009) Biochem. Biophys. Res. Commun. 379, 1043–1047 [DOI] [PubMed] [Google Scholar]
- 19. Sugimoto Y., Narumiya S. (2007) J. Biol. Chem. 282, 11613–11617 [DOI] [PubMed] [Google Scholar]
- 20. Monje P., Hernández-Losa J., Lyons R. J., Castellone M. D., Gutkind J. S. (2005) J. Biol. Chem. 280, 35081–35084 [DOI] [PubMed] [Google Scholar]
- 21. Murphy L. O., Smith S., Chen R. H., Fingar D. C., Blenis J. (2002) Nat. Cell Biol. 4, 556–564 [DOI] [PubMed] [Google Scholar]
- 22. St-Pierre Y., Couillard J., Van Themsche C. (2004) Expert Opin. Ther. Targets 8, 473–489 [DOI] [PubMed] [Google Scholar]
- 23. Jing H., Yen J. H., Ganea D. (2004) J. Biol. Chem. 279, 55176–55186 [DOI] [PubMed] [Google Scholar]
- 24. Page-McCaw A., Ewald A. J., Werb Z. (2007) Nat. Rev. Mol. Cell Biol. 8, 221–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Manicone A. M., McGuire J. K. (2008) Semin. Cell Dev. Biol. 19, 34–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chabot V., Reverdiau P., Iochmann S., Rico A., Sénécal D., Goupille C., Sizaret P. Y., Sensebé L. (2006) J. Leukocyte Biol. 79, 767–778 [DOI] [PubMed] [Google Scholar]
- 27. Zozulya A. L., Reinke E., Baiu D. C., Karman J., Sandor M., Fabry Z. (2007) J. Immunol. 178, 520–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Holvoet S., Vincent C., Schmitt D., Serres M. (2003) Exp. Cell Res. 290, 108–119 [DOI] [PubMed] [Google Scholar]
- 29. Lee K. W., Kim J. H., Lee H. J., Surh Y. J. (2005) Antioxid. Redox Signal. 7, 1612–1620 [DOI] [PubMed] [Google Scholar]
- 30. Tobar N., Villar V., Santibanez J. F. (2010) Mol. Cell Biochem. 340, 195–202 [DOI] [PubMed] [Google Scholar]
- 31. Wang H. H., Hsieh H. L., Wu C. Y., Sun C. C., Yang C. M. (2009) Glia 57, 24–38 [DOI] [PubMed] [Google Scholar]
- 32. Corcoran M. L., Stetler-Stevenson W. G., Brown P. D., Wahl L. M. (1992) J. Biol. Chem. 267, 515–519 [PubMed] [Google Scholar]
- 33. Xue J., Hua Y. N., Xie M. L., Gu Z. L. (2010) Biomed. Pharmacother. 64, 118–123 [DOI] [PubMed] [Google Scholar]
- 34. Narumiya S., Sugimoto Y., Ushikubi F. (1999) Physiol. Rev. 79, 1193–1226 [DOI] [PubMed] [Google Scholar]
- 35. Jing H., Vassiliou E., Ganea D. (2003) J. Leukocyte Biol. 74, 868–879 [DOI] [PubMed] [Google Scholar]
- 36. Vassiliou E., Jing H., Ganea D. (2003) Cell Immunol. 223, 120–132 [DOI] [PubMed] [Google Scholar]
- 37. Fujino H., Xu W., Regan J. W. (2003) J. Biol. Chem. 278, 12151–12156 [DOI] [PubMed] [Google Scholar]
- 38. Sturm E. M., Schratl P., Schuligoi R., Konya V., Sturm G. J., Lippe I. T., Peskar B. A., Heinemann A. (2008) J. Immunol. 181, 7273–7283 [DOI] [PubMed] [Google Scholar]
- 39. Chernogubova E., Cannon B., Bengtsson T. (2004) Endocrinology 145, 269–280 [DOI] [PubMed] [Google Scholar]
- 40. Leone V., di Palma A., Ricchi P., Acquaviva F., Giannouli M., Di Prisco A. M., Iuliano F., Acquaviva A. M. (2007) Am. J. Physiol. Gastrointest. Liver Physiol. 293, G673–681 [DOI] [PubMed] [Google Scholar]
- 41. Sánchez S., Jiménez C., Carrera A. C., Diaz-Nido J., Avila J., Wandosell F. (2004) Neurochem. Int. 44, 231–242 [DOI] [PubMed] [Google Scholar]
- 42. Zhang B., Li S., Harbrecht B. G. (2011) Biochim. Biophys. Acta 1813, 73–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kim H. S., Kim M. H., Jeong M., Hwang Y. S., Lim S. H., Shin B. A., Ahn B. W., Jung Y. D. (2004) Anticancer Res. 24, 747–753 [PubMed] [Google Scholar]
- 44. Kim J. H., Choi C., Benveniste E. N., Kwon D. (2008) Biochem. Biophys. Res. Commun. 377, 195–199 [DOI] [PubMed] [Google Scholar]
- 45. Lin C. C., Tseng H. W., Hsieh H. L., Lee C. W., Wu C. Y., Cheng C. Y., Yang C. M. (2008) Toxicol. Appl. Pharmacol. 229, 386–398 [DOI] [PubMed] [Google Scholar]
- 46. Yue W., Huang Z. Q., Wang C. Q., Wang L. S., Shu M., Zhang Y. C., Ting C., Fan Y. Q. (2010) Clin. Exp. Pharmacol. Physiol. [Google Scholar]
- 47. Vuchak L. A., Tsygankova O. M., Prendergast G. V., Meinkoth J. L. (2009) Mol. Pharmacol. 76, 1123–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wang Z., Dillon T. J., Pokala V., Mishra S., Labudda K., Hunter B., Stork P. J. (2006) Mol. Cell. Biol. 26, 2130–2145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lan H. C., Li H. J., Lin G., Lai P. Y., Chung B. C. (2007) Mol. Cell. Biol. 27, 2027–2036 [DOI] [PMC free article] [PubMed] [Google Scholar]