Background: ELMO complexes with DOCK180 and contributes to Rac signaling.
Results: Arl4A binds ELMO and is a membrane localization signal that triggers DOCK180-Rac-dependent actin cytoskeleton remodeling.
Conclusion: ELMO, via its versatile Ras-binding domain, binds its effector Arl4A, and this novel interaction facilitates Rac signaling.
Significance: This is the first demonstration of a Ras-binding domain that binds Arf or Rho family GTPases.
Keywords: Cytoskeleton, Guanine Nucleotide Exchange Factor (GEF), Protein-Protein Interactions, Rac, Small GTPases, Arl4A GTPase, Cytoskeletal Reorganization, DOCK180, ELMO, Ras-binding Domain
Abstract
The prototypical DOCK protein, DOCK180, is an evolutionarily conserved Rac regulator and is indispensable during processes such as cell migration and myoblast fusion. The biological activity of DOCK180 is tightly linked to its binding partner ELMO. We previously reported that autoinhibited ELMO proteins regulate signaling from this pathway. One mechanism to activate the ELMO-DOCK180 complex appears to be the recruitment of this complex to the membrane via the Ras-binding domain (RBD) of ELMO. In the present study, we aimed to identify novel ELMO-interacting proteins to further define the molecular events capable of controlling ELMO recruitment to the membrane. To do so, we performed two independent interaction screens: one specifically interrogated an active GTPase library while the other probed a brain cDNA library. Both methods converged on Arl4A, an Arf-related GTPase, as a specific ELMO interactor. Biochemically, Arl4A is constitutively GTP-loaded, and our binding assays confirm that both wild-type and constitutively active forms of the GTPase associate with ELMO. Mechanistically, we report that Arl4A binds the ELMO RBD and acts as a membrane localization signal for ELMO. In addition, we report that membrane targeting of ELMO via Arl4A promotes cytoskeletal reorganization including membrane ruffling and stress fiber disassembly via an ELMO-DOCK1800-Rac signaling pathway. We conclude that ELMO is capable of interacting with GTPases from Rho and Arf families, leading to the conclusion that ELMO contains a versatile RBD. Furthermore, via binding of an Arf family GTPase, the ELMO-DOCK180 is uniquely positioned at the membrane to activate Rac signaling and remodel the actin cytoskeleton.
Introduction
An elaborate cast of players is directed to coordinate Rho GTPase signaling during numerous basic biological processes such as cell migration, polarity, and adhesion. The evolutionarily conserved family of DOCK proteins mediates guanine nucleotide exchange on a subset of these Rho GTPases to control active remodeling of the actin cytoskeleton (1). Among the 11 mammalian proteins, DOCK1 (also known as DOCK180) through DOCK5 and DOCK9 through DOCK11 are characterized as specific activators of Rac and Cdc42 GTPases, respectively (1, 2). Deviants of the distinctive Dbl Homology-Pleckstrin Homology region (DH-PH) family of Rho GEFs,6 the DOCK proteins rely on the DOCK homology region-2 for guanine nucleotide exchange activity and a lipid-binding DOCK homology region-1 for membrane targeting (3–5).
Amid the DOCK proteins, the CDM members (Caenorhabditis elegans Ced-5, Drosophila Myoblast City, and mammalian DOCK1/2/5) have been reported to regulate a number of Rac-dependent biological events including cell migration, cell polarization, myoblast fusion, and engulfment of apoptotic cells (6–11). The interaction of DOCK180 with various proteins is critical in regulating Rac signaling. ELMO family members are established binding partners of DOCK180, and genetic analyses in worms and flies suggest that ELMO is crucial for the biological functions of DOCK180 (1, 2). Likewise, cell biology studies in mammalian cells suggest that disrupting the ELMO-DOCK180 interaction blocks signaling from this complex (12, 13).
ELMO proteins exist in a repressed state. Our recent work identified an autoinhibitory switch in ELMO occurring through three previously uncharacterized protein modules: a Ras-binding domain (RBD), ELMO inhibitory domain, and ELMO autoregulatory domain (14). De-regulation of ELMO autoinhibition promotes DOCK180- and Rac-dependent cell elongation and migration, highlighting the importance of tight conformational control of ELMO (14).
Because of its ability to interact with membrane-localized and signaling proteins, the N terminus of ELMO is a strong candidate for proper targeting of the GEF, DOCK180 (15–18). Indeed, a functional RBD in ELMO is required for membrane targeting upon integrin engagement (14). The RBD of ELMO proteins recognize GTP-loaded RhoG, and this interaction recruits ELMO-DOCK180 to the membrane to induce Rac-dependent cytoskeletal changes (17, 19–22). However, a later study demonstrated that RhoG is not required for integrin-mediated Rac signaling and motility (23), implying that additional proteins may bind the ELMO RBD to target the protein to the membrane. It is evident that understanding the molecular events that regulate ELMO-DOCK180 recruitment to the membrane is an important area of investigation to fully comprehend how these proteins are controlled.
This study aimed to identify novel ELMO-interacting proteins to define the molecular events capable of controlling ELMO recruitment to the membrane. Using two complementary approaches, we identified an Arf-related GTPase, Arl4A, as a novel ELMO binding partner and membrane recruitment signal. Moreover, ELMO localization via Arl4A promoted cytoskeletal reorganization via an ELMO-DOCK180-Rac signaling pathway. Our data reveal that the ELMO N terminus has the ability to interact with GTPases from Rho and Arf families leading to the conclusion that ELMO contains a versatile RBD. To our knowledge, this is the first study to identify a RBD with dual specificity for Rho and Arf family GTPases.
EXPERIMENTAL PROCEDURES
Antibodies, Cell Culture, and Transfections
The following antibodies were obtained commercially: anti-DOCK180 (C-19, H-4, and H-70) and anti-Myc (9E10) were from Santa Cruz Biotechnologies, anti-Rac was from Millipore, and anti-FLAG M2 and anti-FLAG-M2-HRP were from Sigma. The Arl4A antibody was described previously (24). HEK293T and HeLa cells were cultured in DMEM supplemented with 10% fetal bovine serum, penicillin, and streptomycin (Invitrogen). The cells were transfected by calcium phosphate or Lipofectamine 2000 (Invitrogen) using standard procedures. Biochemical and cell biological studies were performed 24–48 h after transfection.
Plasmid Constructs
pCNX2 FLAG-DOCK180 was a gift from M. Matsuda (Kyoto University, Kyoto, Japan). pcDNA3.1 Myc-ELMO1 was previously described (3). Plasmids coding for Myc-ELMO1 proteins (residues 1–113 and 212–727) were generated by PCR and cloned into the BamHI/XhoI sites of pcDNA3.1Myc. The yeast constructs for ELMO1 (WT and residues 1–113, 1–212, 1–315, 1–495, 315–727, 212–727, 113–727, Δ114–524, Δ213–524, and Δ310–492) were generated via PCR and cloned into the BamHI/XhoI sites of pEG202 (LexA tagged vector (gift from Dr. J. Archambault, Institut de Recherches Cliniques de Montréal, Montreal, Canada)). Myc-ELMO11–212 (L43A) has been described previously (14). FLAG-Arl4AWT was generated via PCR and cloned into the EcoRI/XhoI sites of the pcDNA-FLAG vector. FLAG-Arl4AQ79L and FLAG-Arl4AT34N were generated via site-directed mutagenesis. Arl4A-3XFLAG was generated via PCR and cloned into the KpnI/BamHI sites of the p3XFLAG-CMV-14 vector. Arl4AQ79L-3XFLAG and Arl4AT34N-3XFLAG were generated via site-directed mutagenesis. Nontagged ARL4A (WT, T34N, and Q79L) constructs have been described previously (24). The yeast constructs of Arl4 (Arl4AWT, Arl4AT34N, and Arl4AQ79L; Arl4CWT, Arl4CT27N, and Arl4CQ72L; and Arl4DWT, Arl4DT35N, and Arl4DQ80L) were generated via PCR and cloned into the EcoRI/XhoI sites of pJG4–5alt (B42-tagged vector (gift from Dr. J. Archambault)). pJG4–5alt-Arl4AL43A was generated via site-directed mutagenesis.
Construction of an Activated GTPase Library
Sequences coding for all members of Rho, Ras, and Arf families (84 GTPases) were retrieved via PubMed. cDNAs coding for these Rho, Ras, and Arf subfamily members, in their constitutively active forms, were generated by gene synthesis and cloned in pUC57 (GenScript). AttB sites were added at both ends of the gene coding for the GTPases to enable use in the Gateway system. Each clone was recombined with pDONR221 to generate a collection of 84 GTPases in an ENTRY vector. Site-specific recombination was used to generate the yeast two-hybrid compatible plasmids pJG4–5alt-GTPase (84 of Ras superfamily members). The pJG4–5alt-ccdB plasmid used for the LR recombination was constructed as follows: a DNA fragment coding for the ccdB protein and flanked with attR sites was generated by PCR and cloned into the EcoRI/XhoI sites of pJG4–5alt.
Immunoprecipitation and GST Fusion Protein Pulldowns
Immunoprecipitation and pulldown experiment protocols have been described previously (12). Briefly, the cells were lysed for 10 min in a buffer consisting of 50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1% Nonidet P-40, and 1× Complete protease inhibitor (Roche). For immunoprecipitation, clarified cell lysates were incubated with the appropriate antibody, and immune complexes were allowed to form for 1 h at 4 °C. Protein A-Sepharose was added for 30 min to isolate the immune complex. For cross-linking prior to immunoprecipitation, the cells were treated with DSP (2 mm) (Pierce) for 30 min according to the manufacturer's instructions. For GST fusion protein pulldowns, the GST fusion proteins were expressed in bacteria and purified on glutathione-Sepharose 4B according to the manufacturer's instructions (Amersham Biosciences). Equal amounts of the various GST fusion proteins bound to glutathione-Sepharose 4B were next incubated with cell extracts (500 μg of protein/condition). In both types of assays, the beads were washed three times with lysis buffer, and the bound proteins were analyzed by SDS-PAGE and immunoblotting.
Yeast Two-hybrid Interaction Assay
Two separate screens were performed to identify ELMO-binding partners. First, a yeast two-hybrid screen was performed by Hybrigenics Services (Paris, France) using full-length ELMO1 as a bait to probe an embryonic mouse brain cDNA library. More specifically, the coding sequence for full-length mouse ELMO1 (GenBankTM accession number gi: 17933765) was PCR-amplified and cloned into pB27 as a C-terminal fusion to LexA (N-LexA-ELMO1-C). The construct was checked by sequencing the entire insert and used as a bait to screen a random-primed mouse embryo brain cDNA library constructed into pP6. pB27 and pP6 derive from the original pBTM116 (25) and pGADGH (26) plasmids, respectively. 83 million clones (8-fold the complexity of the library) were screened using a mating approach with Y187 (matα) and L40ΔGal4 (mata) yeast strains as previously described (27). 252 His+ colonies were selected on a medium lacking tryptophan, leucine, and histidine. The prey fragments of the positive clones were amplified by PCR and sequenced at their 5′ and 3′ junctions. The resulting sequences were used to identify the corresponding interacting proteins in the GenBankTM data base (NCBI) using a fully automated procedure. A confidence score (predicted biological score) was attributed to each interaction as previously described (28).
Second, a yeast two-hybrid screen was developed to specifically interrogate ELMO/GTPases interactions using the collection of activated Ras GTPases generated in our lab (for more details, see “Construction of an Activated GTPase Library” under “Experimental Procedures”). Yeast two-hybrid experiments were performed as previously mentioned (14).
Cell Spreading and Colocalization Assay
For the cell spreading assay on fibronectin, HeLa cells transfected with the indicated plasmids were subject to cell morphology as previously described (4). Briefly, the cells were transfected with the indicated plasmids and serum-starved (0.5% FBS) overnight. The cells were gently detached (0.01% trypsin and 5 mm EDTA in Hanks' balanced solution) and washed in DMEM supplemented with 0.5% BSA, and 40,000 cells were then allowed to spread for the indicated time (50 min or 2 h) before fixing with 4% paraformaldehyde. The cells were permeabilized with 0.2% Triton X-100 in PBS and blocked in PBS-1% BSA prior to staining with DAPI and phalloidin. The remainder of the cells was lysed to verify the expression levels of the exogenous proteins by Western blotting. For colocalization assays, experiments were performed as previously described (14). Statistical differences between groups of data were analyzed using an analysis of variance test and Bonferroni's multiple comparison procedures (minimum of n = 3).
Stress Fiber Disassembly Assay
Stress fiber disassembly assays were performed as previously described (24). Statistical differences between groups of data were analyzed using Student's t test.
PAK Pulldowns
PAK-PBD pulldown assays were performed as previously described (12).
RESULTS
An Arf-related GTPases, Arl4A, Binds the ELMO1 RBD
We previously reported that the formation of an ELMO-DOCK180 complex is essential for Rac GTP-induced cytoskeletal changes but not for Rac GTP-loading per se, the latter being solely dependent on the intrinsic GEF activity of DOCK180 (12). Our recent data also highlighted that the RBD of ELMO is essential for targeting ELMO to the membrane upon integrin activation (14). These results suggested that additional GTPase(s) might bind the RBD. To further explore the molecular events regulating the localization of ELMO at the membrane, we undertook two independent but complementary approaches to identify novel partners of ELMO1. First, using ELMO1 as bait in a yeast two-hybrid system, we scanned an embryonic mouse brain cDNA library. Second, we investigated specifically whether ELMO can interact with additional GTP-loaded GTPases. To do so, we constructed a library of the superfamily of Ras GTPases in their active conformation (see “Experimental Procedures” and supplemental Fig. S1 and Table S1). We individually tested the ability of all Rho, Ras, and Arf GTPases (84 members) to interact with ELMO in a yeast two-hybrid assay. The combination of these experiments confirmed active RhoG, DOCK proteins (DOCK1-DOCK5), and BAI family receptors (BAI1 and BAI3) as known ELMO binding partners. Interestingly, our two approaches also converged on Arl4A, a small GTPase of the Arf family, as a putative novel ELMO interactor. The Arf family member, Arf6, is heavily implicated in cytoskeletal reorganization via numerous signaling events (8, 24, 29–31). One such pathway places Arf6 upstream of DOCK180 and ELMO and ensuing Rac activation and signaling (8). However, the exact mechanism by which Arf6 controls ELMO-DOCK180-mediated Rac signaling is unclear. Arl4A belongs to the large family of Arf-related proteins. The Arl4 family consists of three closely related members: A, C, and D (32). Unlike the other members of this family, Arl4s are preferentially GTP-loaded because of their weak affinity for nucleotides and therefore exhibit high spontaneous nucleotide exchange rates (33). Recent reports suggest that Arl4s are involved in cytoskeletal rearrangement through their ability to bind the Arf6 guanine exchange factor, ARNO, and localize it to the cell periphery for Arf6 activation (24, 30).
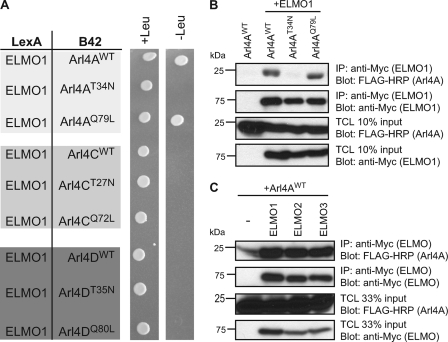
We used the yeast two-hybrid system to verify which isoforms of Arl4s interact with ELMO proteins. Using ELMO1 as bait revealed specificity for the Arl4A protein because no interaction was noted for Arl4C or Arl4D (Fig. 1A). Moreover, ELMO1 binding to Arl4A was nucleotide state-dependent, with ELMO1 selectively interacting with the WT and constitutively active (Q79L) forms but not with dominant negative (T34N) Arl4A (Fig. 1A). To validate these results in a mammalian cell context, we coexpressed Myc-ELMO1 with the WT, Q79L, and T34N forms of Arl4A in 293T cells. Similar to what was observed in yeast, specific interaction between ELMO1 and both Arl4AWT and Arl4AQ79L was observed (Fig. 1B). Finally, we could generalize the binding of Arl4A to all isoforms of ELMO because we found a specific interaction in coimmunoprecipitation between Arl4AWT and Myc-ELMO1, Myc-ELMO2, and Myc-ELMO3 (Fig. 1C).
FIGURE 1.
Arl4A is a novel ELMO-interacting partner. A, Arl4AWT and Arl4AQ79L interact with ELMO1 in a yeast two-hybrid system. Yeast strain EGY48 cotransformed with LexA BD fusion construct of ELMO1 and the B42 fusion constructs of the indicated Arl4s were grown on selective (−histidine, −tryptophan, −leucine) and nonselective (−histidine, −tryptophan) medium for a nutrient selective growth assay. B, Arl4A and ELMO1 interact in vivo in cells. HEK293T cells transfected with the indicated plasmids were subjected to a cross-linker, lysed, and immunoprecipitated (IP) with an anti-Myc antibody (ELMO1). Immunoblot analysis using anti-Myc and FLAG-HRP antibodies established the coprecipitation of ELMO and Arl4A proteins. C, Arl4A interacts with all forms of ELMO. HEK293T cells transfected with the indicated plasmids were cross-linked, lysed, and immunoprecipitated with an antibody against the Myc epitope (ELMO1–3). The coprecipitation of the various ELMO proteins and Arl4A was analyzed via immunoblotting with anti-Myc (ELMO1–3) and anti-FLAG-HRP (Arl4A) antibodies, respectively. TCL, Total Cell Lysate.
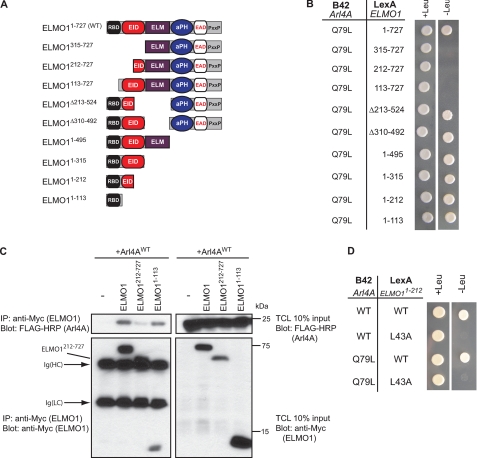
To map the active ELMO1 binding interface for Arl4A, we used a panel of ELMO1 truncation mutants (Fig. 2A) in the yeast two-hybrid system. Our data suggest that the first 113 amino acids of ELMO1 is the smallest fragment with the ability to complex with Arl4A (Fig. 2B). Similarly, a deletion of the first 113 amino acids was sufficient to abrogate binding of ELMO1 to Arl4A (Fig. 2B). Moreover, we also found that ELMO11–113 was sufficient to bind Arl4A in 293T cells (Fig. 2C). A mutant of ELMO1 lacking the first 113 amino acids was difficult to express, but we could demonstrate that the first 212 amino acids of ELMO1 are required for interacting with Arl4A because ELMO1212–727 is unable to coprecipitate Arl4A (Fig. 2C).
FIGURE 2.
Arl4A binds the ELMO1 RBD through a key evolutionarily conserved RBD residue. A, schematic representation of ELMO1 deletion mutants used in yeast two-hybrid experiments. B, the ELMO1 N terminus is required for Arl4A binding. Yeast strain EGY48 cotransformed with LexA BD fusion construct of ELMO1WT and deletion mutants, and the B42 fusion constructs of the indicated Arl4As were grown on selective (−histidine, −tryptophan, −leucine) and nonselective (−histidine, −tryptophan) medium for a nutrient selective growth assay. C, the Arl4A-ELMO1 interaction in cellulo requires the ELMO1 RBD. HEK293T cells transfected with the indicated plasmids were cross-linked, lysed, and immunoprecipitated (IP) with an antibody against the Myc epitope (ELMO1). The coprecipitation of the various ELMO1 proteins and Arl4A was analyzed via immunoblotting with anti-Myc (ELMO1) and anti-FLAG-HRP (Arl4A) antibodies, respectively. D, mutation of a key conserved residue in the ELMO RBD (L43A) abolishes Arl4A binding. Yeast strain EGY48 cotransformed with LexA BD fusion construct of ELMO1WT and ELMO1L43A and the B42 fusion construct of Arl4AWT were grown on selective (−histidine, −tryptophan, −leucine) and nonselective (−histidine, −tryptophan) medium for a nutrient selective growth assay.
Our discovery that the Arl4A-binding site encompasses the ELMO RBD led to the following possibility: similar to active RhoG, Arl4A may also have specific affinity for this protein module. Our prior findings disclosed ELMO1 residue leucine 43 as the cornerstone for forging the ELMO1 RBD/RhoG-GTP contact (14). In a yeast two-hybrid assay, we found that both Arl4AWT and Arl4AQ79L were unable to interact with ELMO11–212 (L43A) in comparison with its wild-type counterpart (Fig. 2D). Our data identifies, for the first time, an RBD with the ability to interact with both Rho and Arf family GTPases and supports the notion that the ELMO RBD may have the potential to attract GTPases of different families.
Arl4A Targets ELMO to the Membrane
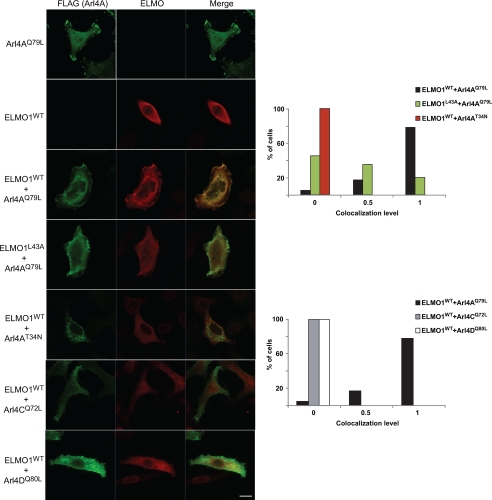
Our results are consistent with the model that Arl4A interacts with the RBD of ELMO proteins to favor membrane recruitment. RhoG was previously reported to activate the ELMO-DOCK180 pathway by localizing ELMO to the membrane in a manner dependent on its nucleotide state (22). To test whether Arl4A also promotes membrane recruitment of ELMO1, we analyzed the cellular distribution of ELMO1 in cells expressing the various forms of Arl4A (WT, Q79L and T34N). Although Myc-ELMO1 alone is cytoplasmic, coexpression with WT and active Arl4A led to its membrane recruitment (Fig. 3). Arl4AT34N displayed a punctate staining in cells and, when expressed with ELMO1, did not redistribute ELMO to the cell periphery (Fig. 3, see figure legend for more details on the scoring system). Moreover, the Arl4A binding-defective mutant of ELMO (ELMO1L43A) was unable to localize to the cell periphery and instead demonstrated a diffuse cytosolic expression when coexpressed with Arl4AQ79L (Fig. 3). The specificity of ELMO binding to Arl4A was further demonstrated by colocalization experiments revealing that ELMO1 does not colocalize at the membrane with either Arl4CQ72L or Arl4DQ80L, with these cells showing less membrane ruffles (Fig. 3). We additionally observed that although Arl4D has been noted to induce membrane protrusions in COS-7 cells (24), in our hands, Arl4D in HeLa cells does not extensively promote lamellipodia-like structures, suggesting that the effect of Arl4 proteins on membrane remodeling may be context-dependent. This suggests that ELMO proteins are bona fide effectors of active Arl4A and act by localizing ELMO-DOCK180 to the membrane for potential Rac activation.
FIGURE 3.
ELMO1 colocalizes with Arl4A at membrane protrusions. Transfected HeLa cells were fixed, permeabilized, and labeled with anti-FLAG M2 (Arl4A) and anti-ELMO antibody and analyzed by confocal microscopy. Coexpression of Arl4AQ79L-FLAG with Myc-ELMO1WT promotes membrane ruffling and localization of ELMO1 to membrane protrusions, whereas Myc-ELMO1L43A does not colocalize with Arl4AQ79L at membrane ruffles. The images shown are representative of multiple cells of three independent experiments. Quantification of colocalization of variants of both Arl4 with ELMO1 at peripheral membrane ruffles. Experiment was performed in triplicate, and at least 20–30 cells were analyzed for each condition. The data in the graph represent one of three experiments, where a score of 0 (no colocalization) to 1 (complete colocalization) grades each cell. Cells showing minimal colocalization in few membrane ruffles were scored as 0.5 (indicating some partial colocalization). Scale bar, 20 μm.
Arl4A Induces Actin Cytoskeleton Remodeling in an Arf6-independent Manner
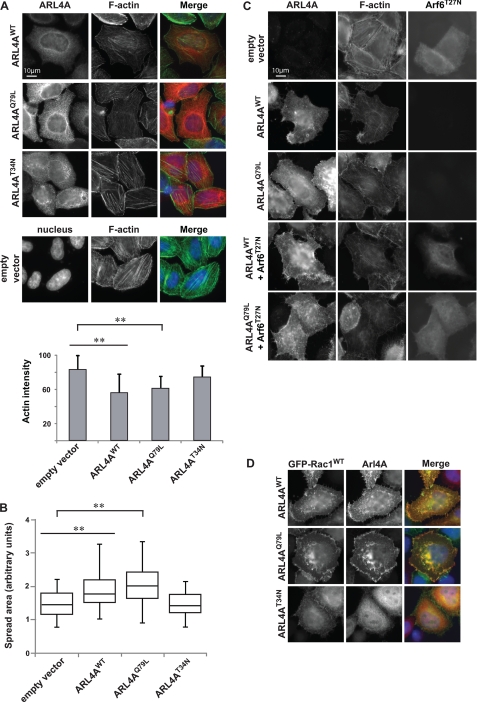
Previous studies demonstrated that activated Arl4A, C, and D could recruit ARNO GEFs to the membrane (24, 30). Arl4D was studied in more detail and was reported to induce actin stress fiber disassembly through an ARNO-Arf6 dependent pathway (24). Independent work also reported that the ARNO-Arf6 pathway might facilitate membrane recruitment of ELMO-DOCK180 and promote Rac-dependent migration (8). Similar to what was reported for expression of Arl4DQ80L in HeLa cells, we found that both Arl4AWT and Arl4AQ79L promotes actin stress fiber disassembly, whereas this was not observed for Arl4AT34N (Fig. 4A). Additionally, Arl4AWT and Arl4AQ79L, but not Arl4AT34N, induced cell spreading compared with control cells (Fig. 4B). In contrast to what was observed with Arl4D (24), coexpression of Arf6T27N did not fully block Arl4A-induced stress fiber disassembly (Fig. 4C). Furthermore, compared with exogenous expression of Arl4AT34N, Arl4AWT expression in Arf6-depleted HeLa cells demonstrated actin stress fiber disassembly (supplemental Fig. S2). These results suggest that Arl4A may have an alternative pathway to regulate remodeling of the actin cytoskeleton.
FIGURE 4.
Arl4A induces cytoskeletal changes in an Arf6-independent manner. A, overexpression of Arl4AWT and Arl4AQ79L alters actin structure in HeLa cells, but Alr4AT34N does not. Wild-type and active Arl4A were overexpressed in HeLa cells and stained for Arl4A (red) and phalloidin (green) using anti-Arl4A and Fluor 488 phalloidin, respectively. The bar chart indicates quantification results for each condition. The area of each cell was delineated, and the average fluorescence intensity of Fluor 488 phalloidin was measured in pixels using ImageJ (National Institutes of Health). More than 40 cells were assessed in each experiment, and the data are the means ± S.E. of at least triplicate experiments. Student's t test was performed to compare each condition (**, p < 0.01). Scale bar, 10 μm. B, expression of Arl4AWT and ARL4AQ79L promotes cell spreading on fibronectin. For quantification, the area of each cell was delineated, and more than 40 cells were estimated for each condition using ImageJ. The box plot shows the distribution of cell size for each condition. Student's t test was performed to compare each condition (**, p < 0.01). C, coexpression of dominant negative Arf6 (Arf6T27N) with Arl4AWT or Arl4AQ79L does not fully hinder actin cytoskeletal reorganization. HeLa cells were cotransfected with Arf6T27N and either Arl4AWT or Arl4AQ79L. Transfected cells were fixed and stained with anti-Arl4A and anti-Arf6 antibodies. Scale bar, 10 μm. D, ARL4AWT and Arl4AQ79L colocalize with GFP-Rac1WT at membrane ruffles. Arl4AT34N fails to relocalize GFP-Rac1WT and to promote membrane ruffles. Transfected cells were fixed and stained with anti-Arl4A.
We next investigated whether Arl4A can regulate localization and activity of Rac in HeLa cells. We found that exogenous Arl4AQ79L colocalizes with GFP-Rac1 in HeLa cells (Fig. 4D). Moreover, we noted a more cytosolic distribution of GFP-Rac1 with a marked decrease in membrane protrusiveness in cells coexpressing Rac1 and dominant negative Arl4A (Arl4AT34N) (Fig. 4D). In addition, by using the pSUPER RNAi system, down-regulation of Arl4A levels with a specific Arl4A shRNA decreased the level of active Rac when compared with HeLa cells transfected with a control vector (supplemental Fig. S3). Cells treated with shRNA against Arf6 did not lead to decreased active Rac levels. Interestingly, Hu et al. (34) demonstrated that active Arf6 promotes Rac1 activation through a complex with IQGAP1 in metastatic glioma cells. Depletion of Arf6 suppressed Rac1 activation when cells were stimulated with hepatocyte growth factor or FBS, resulting in cell migration defects (34). However, this study also indicated that the mechanism of Arf6-dependent Rac1 activation is cell type- and stimulus-dependent (34). In support of this, another study reported that in stimulated HEK293 cells stably expressing angiotensin type I receptor, depletion of Arf6 increased basal Rac1 activation (35). Our results suggest that, at least in this context, Rac activation is Arf6-independent (supplemental Fig. S3). Together, these results suggest that active Arl4A can impact the actin cytoskeleton, Rac localization, and Rac GTP loading in an Arf6-independent manner.
Arl4A-mediated Actin Cytoskeleton Reorganization Occurs through ELMO-DOCK180 and Rac
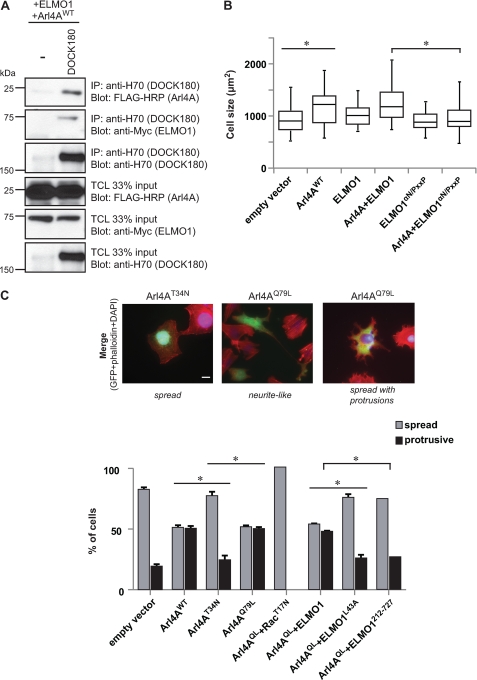
To investigate whether Arl4A modulates the actin cytoskeleton via an ELMO-DOCK180-Rac signaling pathway, we first tested whether Arl4A/ELMO can enter into a trimolecular complex with DOCK180. We found that Arl4A, alongside ELMO1, can be specifically coprecipitated with DOCK180 (Fig. 5A). Next, we examined whether the ELMO1-DOCK180-Rac signaling pathway mediates ARL4A signaling. In an integrin-independent cell spreading assay, we found that overexpression of ARL4A results in an increase in cell spreading as compared with control cells (Fig. 5B). Although coexpression of ARL4A with ELMO1WT did not affect cell size, cells coexpressing ARL4A and ELMO1αN/PxxP (a DOCK180 binding-defective mutant (12)) display a distinct reduction in cell spreading comparable with mock-treated cells (Fig. 5B). Moreover, to test the role of Rac and ELMO1 in Arl4A-mediated cytoskeleton rearrangements, we developed an assay implicating HeLa cell spreading on fibronectin. Although Arl4AT34N did not affect HeLa cell morphology, two major phenotypes were identified in Arl4AWT and Arl4AQ79L spreading cells. We found that 44–50% of the cells expressing the active GTPases (WT or Q79L) displayed: (i) formation of neurite-like extension or (ii) membrane ruffles. In cells showing a phenotype, the neurite-like phenotype is less frequent and is observed in 10–15% of the cells. Therefore, for quantification purposes, these two phenotypes were pooled and termed “protrusive” (Fig. 5C). In contrast, dominant negative Arl4A-expressing cells looked identical to control cells with the majority of cells displaying a spread, nonprotrusive phenotype (Fig. 5C). We next tested whether Arl4A is mediating cytoskeletal changes via ELMO and Rac. Coexpression of either a dominant negative Rac1 or ELMO1 lacking Arl4A binding activity (ELMO1212–727 or ELMO1L43A) with Arl4AWT prevented cytoskeletal reorganization in HeLa cells (Fig. 5C). Together, these results suggest that the ELMO-DOCK180-Rac pathway mediates Arl4A-induced remodeling of the actin cytoskeleton.
FIGURE 5.
Arl4A induces cellular protrusions through an ELMO-DOCK180-Rac signaling module. A, ELMO is the common denominator for DOCK180-ELMO1-Arl4A trimeric complex formation. HEK293T cells transfected with the indicated plasmids were subject to a cross-linker, lysed, and immunoprecipitated (IP) with H-70 (DOCK180). The coprecipitation of DOCK180, ELMO1 WT and mutants, and Arl4A was analyzed via immunoblotting with anti-H-70 (DOCK180), anti-Myc (ELMO1), and anti-FLAG-HRP (Arl4A) antibodies, respectively. B, Arl4A signaling induces cell spreading through an ELMO-DOCK180 pathway. HeLa cells transfected with the indicated plasmids were fixed and stained with anti-Arl4A and anti-Myc antibodies (ELMO1). The box plot shows the distribution of cell size for each condition. The size of more than 20 transfected cells for each condition was measured by ImageJ (*, p < 0.05). C, quantification of the effect on cell morphology in response to overexpression of Arl4A and other proteins. HeLa cells were transfected with the indicated plasmids, and cell morphology was analyzed by fluorescence microscopy. Several independent fields were photographed at a magnification of 40×, and cells were scored for two phenotypes: spread (clearly spread and flat cells) and spread with protrusions (subdivided into (i) protrusive and (ii) neurite-like elongated cells). For each condition, >35 cells were measured. Analysis of variance tests and Bonferroni's multiple comparison was performed to compare each condition (*, p < 0.05; error bars represent S.E., n = 3). Scale bar, 20 μm.
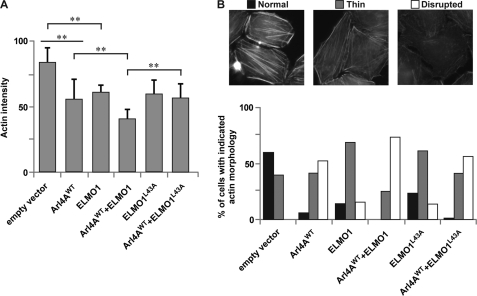
Additionally, we used a stress fiber disassembly assay to examine the contribution of ELMO1 during Arl4A-induced cytoskeletal rearrangement. Expression of either Arl4AWT or ELMO alone promoted actin fiber disassembly. However, coexpression of ELMO1 and Arl4AWT led to a maximal decrease in stress fiber formation (Fig. 6A). Importantly, an ELMO1 mutant defective in Arl4A binding, ELMO1L43A, was unable to synergize with Arl4A in actin stress fibers disassembly (Fig. 6A). For further quantification, we classified the actin morphology into three groups: normal (long thick actin fibers), thin (thinner but ordered actin fibers), and disrupted (disordered thin actin fibers) (Fig. 6B). Compared with untreated cells, HeLa cells expressing ARL4A resulted in thin-to-disrupted morphologies. Coexpression of ELMO1WT but not ELMO1L43A increased the percentage of cells exhibiting disrupted actin morphology (Fig. 6B). These results highlight the importance of Arl4A binding via the ELMO RBD for ELMO-induced restructuring of the actin cytoskeleton, possibly via the DOCK180-Rac pathway.
FIGURE 6.
Hindrance of the Arl4A-ELMO1 complex reduces stress fiber disassembly. A, coexpression of Myc-ELMO1WT and Arl4AWT reduced F-actin intensity, indicating stress fiber disassembly, whereas coexpression of Myc-ELMO1L43A and Arl4AWT did not induce substantial stress fiber disassembly. Quantification of average fluorescence intensity of F-actin in cells expressing the indicated plasmids. Student's t test was performed to compare each condition (**, p < 0.01). B, hindrance of the Arl4A-ELMO complex reduces stress fiber disassembly. There are three different filamentous actin morphologies: (i) normal (long thick actin fibers), (ii) thin (thinner but ordered actin fibers), and (iii) disrupted (disordered thin actin fibers). Quantification of cells with different actin morphologies is shown. HeLa cells transfected with the indicated plasmids were fixed, and filamentous actin was visualized with FITC-phalloidin. ARL4A-expressing cells show thin-to-disrupted actin morphologies, with coexpression of ELMO1WT but not ELMO1L43A increasing the percentage of cells exhibiting disrupted actin morphology. For each condition, more than 60 cells were analyzed.
DISCUSSION
The role of ELMO during DOCK180-induced Rac signaling and cellular restructuring is not completely understood. Our recent work demonstrated that ELMO, via intramolecular interactions between its newly identified ELMO inhibitory domain and ELMO autoregulatory domain regions, exists as an autoinhibited molecule at basal level. Cell stimulation leads to RBD engagement, and signaling for (i) membrane localization of ELMO-DOCK180-mediated Rac signaling and/or (ii) autoinhibition relief (14). In the present study, we identify the RBD of ELMO as a versatile GTPase-binding region capable of interacting with different Ras GTPase family members. Although it was well known that ELMO has the ability to bind active RhoG through its extreme N terminus, our recent work revealed that the minimal RhoG binding site was an evolutionarily conserved RBD (14). The work from the present study identifies Arl4A, a member of the Arf subfamily of Ras GTPases, as a novel ELMO RBD binding partner, and the evolutionarily conserved feature of the GTPase-ELMO interaction is demonstrated by critical point mutation of a conserved residue in the ELMO RBD. This discovery opens up a gateway of possibilities where various Ras superfamily GTPases may converge to regulate ELMO membrane localization and/or relief of ELMO autoinhibition. Surprisingly, our data from a systematic screen of the Rho, Ras, and Arf superfamily of GTPases only uncovered Arl4A, apart from the already established active RhoG, as a novel ELMO interactor (supplemental Fig. S1).
Membrane Targeting of the DOCK180-ELMO Complex
Targeting of the ELMO-DOCK180 complex to the membrane may be fine tuned by various inputs. Although initial studies pointed to a role for the pleckstrin homology domain of ELMO as being instrumental for the targeting of the ELMO-DOCK180 complex to the plasma membrane, we uncovered that the ELMO pleckstrin homology domain displays no such activity (12). Membrane targeting of this complex has also been attributed to the lipid binding properties of the DOCK homology region-1 of DOCK180 (4). Also, the localization of DOCK2 to the neutrophil pseudopod requires sequential binding of two signaling lipids: first a global recruitment to the membrane via the DOCK homology region-1 domain engaging phosphatidylinositol-3,4,5-phosphates, followed by polybasic region binding to phosphatidic acid (36). Further studies are required to expose whether localization of DOCK180 can be regulated in a similar manner. Additionally, adaptor proteins such as CrkII and Nck associate with DOCK180 and may contribute to the localization of the GEF-ELMO complex.
The functional importance of the N terminus of ELMO1 for ELMO-DOCK180 subcellular localization has been previously noted (14, 17, 18). Now, with the discovery of Arl4A as a novel binding partner of the ELMO RBD, our study demonstrates for the first time a RBD that can bind both a Rho and Arf family GTPase. The Arl4 proteins (Arl4A, Arl4C, and Arl4D) have recently emerged as important cytoskeletal regulators. In terms of structure, these three proteins are similar to other Arf family members yet are unique by virtue of a short basic extension at the C terminus (37). Interestingly, the N-terminal amphipathic helices of the Arl4 proteins are shorter and less hydrophobic than those of other Arf family members. Deletion of the basic extension in cells results in displacement of Arl4A from the plasma membrane, advocating that the C-terminal basic extension may function as a support system for the N-terminal amphipathic helices and aid in the localization of Arl4 proteins to membranes (38).
Our findings demonstrate that ELMO1 alone is cytoplasmic, and coexpression specifically with active Arl4A led to its membrane recruitment to sites of membrane ruffling, whereas expression with either Arl4C or Arl4D did not induce a protrusive phenotype. We therefore propose ELMO proteins as bona fide effectors of Arl4A that can target the ELMO-DOCK180 module to the membrane for localized Rac activation and signaling.
Arl4A Induces Cytoskeletal Remodeling through ELMO-DOCK180 and Rac
Our findings support a role for Arl4A in actin cytoskeleton rearrangement through a pathway that stimulates DOCK180-ELMO-induced Rac signaling. Studies have already demonstrated that Arl4A and its close relatives Arl4C and Arl4D promote actin restructuring through recruitment of ARNO, an Arf6 GEF, to the plasma membrane (24, 30). Interestingly, Arf6 is positioned upstream of Rac activation in various biological processes. One model advocates that Arf6 activation will recruit the DOCK180-ELMO complex to the leading edge of a cell to promote lamellipodia formation (8, 29, 31). An interesting question that arises is whether there is a direct interaction between Arf6 and ELMO that guides its membrane localization. Our investigation found that there is no evidence for such an interaction, either through yeast two-hybrid or pull-down assays (supplemental Fig. S1 and data not shown). Additionally, our studies demonstrate that Arl4A-induced cytoskeletal remodeling occurs via an Arf6-independent pathway. Intriguingly, this may signify that Arl4A can act as a central signaling node for two divergent GTPase pathways.
We investigated whether Arl4A can act in a similar manner to RhoG and promote migration and phagocytosis when coexpressed with ELMO. In marked contrast to the RhoG-ELMO interaction, Arl4A was incapable of synergizing with ELMO-DOCK180 in both cell migration and engulfment assays (data not shown). The exact role of the cytoskeleton modification triggered by Arl4A-ELMO remains to be explored. Interestingly, these data also suggest the possibility that Arl4A and RhoG are not located in the same membrane microdomain because their interaction with ELMO led to different biological output(s).
In conclusion, we identify a novel RBD in ELMO displaying, for the first time, selectivity for both Arf and Rho GTPases. In contrast to the Dia formins, the effector binding to the ELMO RBD is not proven to be a release mechanism for the autoinhibited ELMO molecule. Rather, similar to the FHOD1 protein, this signal seems to target ELMO to distinct areas of the plasma membrane. It will be interesting to investigate whether additional members of the Ras superfamily, such as Rab proteins, can bind the ELMO RBD and whether they also act as membrane localization signals and/or relieve ELMO autoinhibition. It is possible that the ELMO RBD is strictly required for subcellular localization, whereas binding of additional partners at distinct sites in the protein (i.e. ELMO inhibitory domain or ELMO autoregulatory domain) acts to release the closed conformation of ELMO. The unleashing of ELMO can result in the exposure of otherwise masked regions of ELMO, such as the ELM domain. The work from Bowzard et al. (39) demonstrate that the ELMOD family of proteins display GAP activity on selected Arf family members, and they attribute this enzymatic function to the ELM domain. To date, the ELM region of ELMO has been poorly investigated and has no ascribed function. Clearly, further studies are required to identify components that will open up the ELMO molecule and how its hidden regions contribute toward actin cytoskeleton remodeling.
Supplementary Material
Acknowledgments
We acknowledge the help of Dr. Jacques Archambault (and members of his lab) for sharing yeast assays expertise. We thank Dr. Andrew Craig for commenting on this work and Barbara Ruggiero at Hybrigenics for helpful advice and for setting up the yeast two-hybrid screen on ELMO1.
This work was funded by a Canadian Institutes of Health Research operating grant (to J.-F. C.). This work was also supported by grants from the National Science Council, Taiwan, (NSC-100 2325-B-022-028), and National Taiwan University Hospital (99P21-1, to F.-J. S. L.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1–S3.
- GEF
- guanine nucleotide exchange factor
- RBD
- Ras-binding domain
- ELMO
- engulfment and cell motility.
REFERENCES
- 1. Côté J. F., Vuori K. (2007) Trends Cell Biol. 17, 383–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Meller N., Merlot S., Guda C. (2005) J. Cell Sci. 118, 4937–4946 [DOI] [PubMed] [Google Scholar]
- 3. Côté J. F., Vuori K. (2002) J. Cell Sci. 115, 4901–4913 [DOI] [PubMed] [Google Scholar]
- 4. Côté J. F., Motoyama A. B., Bush J. A., Vuori K. (2005) Nat. Cell Biol. 7, 797–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yang J., Zhang Z., Roe S. M., Marshall C. J., Barford D. (2009) Science 325, 1398–1402 [DOI] [PubMed] [Google Scholar]
- 6. Laurin M., Fradet N., Blangy A., Hall A., Vuori K., Côté J. F. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 15446–15451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhou Z., Caron E., Hartwieg E., Hall A., Horvitz H. R. (2001) Dev. Cell 1, 477–489 [DOI] [PubMed] [Google Scholar]
- 8. Santy L. C., Ravichandran K. S., Casanova J. E. (2005) Curr. Biol. 15, 1749–1754 [DOI] [PubMed] [Google Scholar]
- 9. Pajcini K. V., Pomerantz J. H., Alkan O., Doyonnas R., Blau H. M. (2008) J. Cell Biol. 180, 1005–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sanematsu F., Hirashima M., Laurin M., Takii R., Nishikimi A., Kitajima K., Ding G., Noda M., Murata Y., Tanaka Y., Masuko S., Suda T., Meno C., Côté J. F., Nagasawa T., Fukui Y. (2010) Circ. Res. 107, 1102–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Vives V., Laurin M., Cres G., Larrousse P., Morichaud Z., Noel D., Côté J. F., Blangy A. (2011) J. Bone Miner. Res. 26, 1099–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Komander D., Patel M., Laurin M., Fradet N., Pelletier A., Barford D., Côté J. F. (2008) Mol. Biol. Cell 19, 4837–4851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Grimsley C. M., Kinchen J. M., Tosello-Trampont A. C., Brugnera E., Haney L. B., Lu M., Chen Q., Klingele D., Hengartner M. O., Ravichandran K. S. (2004) J. Biol. Chem. 279, 6087–6097 [DOI] [PubMed] [Google Scholar]
- 14. Patel M., Margaron Y., Fradet N., Yang Q., Wilkes B., Bouvier M., Hofmann K., Côté J. F. (2010) Curr. Biol. 20, 2021–2027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Grimsley C. M., Lu M., Haney L. B., Kinchen J. M., Ravichandran K. S. (2006) J. Biol. Chem. 281, 5928–5937 [DOI] [PubMed] [Google Scholar]
- 16. Handa Y., Suzuki M., Ohya K., Iwai H., Ishijima N., Koleske A. J., Fukui Y., Sasakawa C. (2007) Nat. Cell Biol. 9, 121–128 [DOI] [PubMed] [Google Scholar]
- 17. Katoh H., Negishi M. (2003) Nature 424, 461–464 [DOI] [PubMed] [Google Scholar]
- 18. Park D., Tosello-Trampont A. C., Elliott M. R., Lu M., Haney L. B., Ma Z., Klibanov A. L., Mandell J. W., Ravichandran K. S. (2007) Nature 450, 430–434 [DOI] [PubMed] [Google Scholar]
- 19. deBakker C. D., Haney L. B., Kinchen J. M., Grimsley C., Lu M., Klingele D., Hsu P. K., Chou B. K., Cheng L. C., Blangy A., Sondek J., Hengartner M. O., Wu Y. C., Ravichandran K. S. (2004) Curr. Biol. 14, 2208–2216 [DOI] [PubMed] [Google Scholar]
- 20. Hiramoto K., Negishi M., Katoh H. (2006) Exp. Cell Res. 312, 4205–4216 [DOI] [PubMed] [Google Scholar]
- 21. Katoh H., Yasui H., Yamaguchi Y., Aoki J., Fujita H., Mori K., Negishi M. (2000) Mol. Cell Biol. 20, 7378–7387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Katoh H., Hiramoto K., Negishi M. (2006) J. Cell Sci. 119, 56–65 [DOI] [PubMed] [Google Scholar]
- 23. Meller J., Vidali L., Schwartz M. A. (2008) J. Cell Sci. 121, 1981–1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li C. C., Chiang T. C., Wu T. S., Pacheco-Rodriguez G., Moss J., Lee F. J. (2007) Mol. Biol. Cell 18, 4420–4437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Vojtek A. B., Hollenberg S. M. (1995) Methods Enzymol. 255, 331–342 [DOI] [PubMed] [Google Scholar]
- 26. Bartel P. L., Chien C.-T., Stemglanz R., Fields S. (1993) in Cellular Interactions in Development: A Practical Approach (Hartley D. A. ed.) pp. 153–179, Oxford University Press, Oxford [Google Scholar]
- 27. Fromont-Racine M., Rain J. C., Legrain P. (1997) Nat. Genet. 16, 277–282 [DOI] [PubMed] [Google Scholar]
- 28. Formstecher E., Aresta S., Collura V., Hamburger A., Meil A., Trehin A., Reverdy C., Betin V., Maire S., Brun C., Jacq B., Arpin M., Bellaiche Y., Bellusci S., Benaroch P., Bornens M., Chanet R., Chavrier P., Delattre O., Doye V., Fehon R., Faye G., Galli T., Girault J. A., Goud B., de Gunzburg J., Johannes L., Junier M. P., Mirouse V., Mukherjee A., Papadopoulo D., Perez F., Plessis A., Rossé C., Saule S., Stoppa-Lyonnet D., Vincent A., White M., Legrain P., Wojcik J., Camonis J., Daviet L. (2005) Genome Res. 15, 376–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. White D. T., McShea K. M., Attar M. A., Santy L. C. (2010) Mol. Biol. Cell 21, 562–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hofmann I., Thompson A., Sanderson C. M., Munro S. (2007) Curr. Biol. 17, 711–716 [DOI] [PubMed] [Google Scholar]
- 31. Santy L. C., Casanova J. E. (2001) J. Cell Biol. 154, 599–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kahn R. A., Cherfils J., Elias M., Lovering R. C., Munro S., Schurmann A. (2006) J. Cell Biol. 172, 645–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jacobs S., Schilf C., Fliegert F., Koling S., Weber Y., Schürmann A., Joost H. G. (1999) FEBS Lett. 456, 384–388 [DOI] [PubMed] [Google Scholar]
- 34. Hu B., Shi B., Jarzynka M. J., Yiin J. J., D'Souza-Schorey C., Cheng S. Y. (2009) Cancer Res. 69, 794–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cotton M., Boulay P. L., Houndolo T., Vitale N., Pitcher J. A., Claing A. (2007) Mol. Biol. Cell 18, 501–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nishikimi A., Fukuhara H., Su W., Hongu T., Takasuga S., Mihara H., Cao Q., Sanematsu F., Kanai M., Hasegawa H., Tanaka Y., Shibasaki M., Kanaho Y., Sasaki T., Frohman M. A., Fukui Y. (2009) Science 324, 384–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gillingham A. K., Munro S. (2007) Annu. Rev. Cell Dev. Biol. 23, 579–611 [DOI] [PubMed] [Google Scholar]
- 38. Heo W. D., Inoue T., Park W. S., Kim M. L., Park B. O., Wandless T. J., Meyer T. (2006) Science 314, 1458–1461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bowzard J. B., Cheng D., Peng J., Kahn R. A. (2007) J. Biol. Chem. 282, 17568–17580 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.