Background: The oncoprotein NUP98-HOXA9 deregulates transcription and induces cell proliferation leading to acute myeloid leukemia (AML).
Results: The transcription factor amino-terminal enhancer of split (AES) interacts with NUP98-HOXA9 and augments its ability to deregulate transcription and proliferation.
Conclusion: The data indicate a role for AES in the induction of AML by NUP98-HOXA9.
Significance: Understanding the interactions between oncoproteins and transcription factors is important for elucidating the mechanisms of leukemogenesis.
Keywords: Hematopoiesis, Leukemia, Myeloid Cell, Nuclear Oncogene, Transcription Factors, AES, CD34, HOXA9, NUP98, NUP98-HOXA9
Abstract
NUP98-HOXA9 is the prototype of NUP98 fusion oncoproteins that cause acute myeloid leukemia. It consists of an N-terminal FG-rich portion of the nucleoporin NUP98 fused to the homeodomain region of the homeobox protein HOXA9, and acts as an aberrant transcription factor. To identify interacting partners of NUP98-HOXA9, we used a cytoplasmic yeast two-hybrid assay to avoid the nonspecific trans-activation that would occur with the traditional yeast two-hybrid assay due to the transactivating properties of NUP98-HOXA9. We identified amino-terminal enhancer of split (AES), a transcriptional regulator of the transducin-like enhancer/Groucho family as a novel interaction partner of NUP98-HOXA9. The interaction was confirmed by in vitro pulldown and co-immunoprecipitation assays and was shown to require the FG repeat region of NUP98-HOXA9. Immunofluorescence analysis showed that AES localizes primarily to the interior of the nucleus. AES also showed a strong interaction with wild-type NUP98. AES augmented the transcriptional activity of NUP98-HOXA9. In the presence of NUP98-HOXA9, AES caused an increase in long-term proliferation of primary human CD34+ cells with a marked increase in the numbers of primitive cells. These effects of AES were not observed in the absence of NUP98-HOXA9. AES knockdown diminished the transcriptional and proliferative effects of NUP98-HOXA9. AES caused a shift away from the erythroid lineage in cells expressing NUP98-HOXA9. These data establish AES as an interacting partner of NUP98-HOXA9 and show that it cooperates with NUP98-HOXA9 in transcriptional regulation and cell transformation.
Introduction
NUP98 belongs to a group of nucleoporins characterized by multiple FG repeats that function in nucleocytoplasmic transport by interacting with import and export carriers (1). The FG repeats of NUP98 are concentrated in its N-terminal half (1–3). At least 27 chromosomal rearrangements affecting the NUP98 gene have been reported in hematopoietic malignancies, particularly acute myeloid leukemia (AML)2 (4–17). Interestingly, in all NUP98 gene rearrangements, the N terminus containing the FG repeats is retained in the oncogenic fusion protein. The best characterized NUP98 fusion is NUP98-HOXA9. NUP98-HOXA9 and several other NUP98 fusions have the ability to induce cell proliferation and block differentiation in both human and mouse hematopoietic precursors (18–23). Limited information is available regarding the protein interactions of NUP98-HOXA9 and their role in leukemic transformation. NUP98-HOXA9 interacts with CREB-binding protein/p300, HDAC, and CRM1, resulting in transcriptional activation, transcriptional repression, and inhibition of nuclear export, respectively (22, 24–29).
In this study we sought to identify NUP98-HOXA9-interacting proteins and determine their effects on the function of NUP98-HOXA9. Traditional yeast two-hybrid assays that rely on intranuclear transcriptional activation are likely to deliver a high number of false positives due to the transactivating properties of the FG repeat region of NUP98-HOXA9 (24, 30). Therefore, the Cytotrap yeast two-hybrid method, which relies on cytoplasmic interactions, was used instead. The transcriptional regulator amino-terminal enhancer of split (AES) was identified as a novel interaction partner of NUP98-HOXA9 by this method. This interaction was found to enhance the ability of NUP98-HOXA9 to transform primary human hematopoietic cells.
EXPERIMENTAL PROCEDURES
K562 cDNA Library and Plasmid Construction
Total RNA was isolated from 32 × 106 K562 cells using the RNAaqueous kit (Ambion) according to the manufacturer's instructions. Total RNA (350 μg) was obtained and poly(A) RNA was purified using the Poly(A) Purist kit (Ambion) according to the manufacturer's instructions. A total of 5 μg of poly(A) RNA was recovered and a cDNA library was synthesized and cloned into pMyr XR vector using the Cytotrap XR library construction kit (Stratagene). The control plasmids for the two-hybrid assay were provided with the kit. The bait plasmid pSOS-HA-NUP98-HOXA9 was constructed by subcloning HA-tagged NUP98-HOXA9 in-frame from pcDNA3-HA-NUP98-HOXA9 (25). pGEX6P1-HA-NUP98-HOXA9 and pGEX6P1-HA-HOXA9 were constructed by subcloning from pcDNA3-HA-NUP98-HOXA9 and pcDNA3-HA-HOXA9 (25). pGEX6P1-AES, pcDNA3-FLAG-AES, and MSCV-IRES-YFP-FLAG-AES were constructed by subcloning PCR-amplified AES cDNA from pMyr-AES. The construction of NUP98-HOXA9 deletion mutants (28), pGL4.11-KBTBD10 (22), and MSCV-IRES-GFP-HA-NUP98-HOXA9 (25) are described elsewhere. The construction of full-length and deletion mutants of NUP62 and NUP153 are described elsewhere (31). All PCR products were verified by DNA sequencing.
Cytotrap Yeast Two-hybrid Analysis
Cytotrap yeast two-hybrid analysis was performed according to the manufacturer's instructions. Briefly, the K562 cDNA library in pMyr vector and pSOS-HA-NUP98-HOXA9 were co-transformed into cdc25H yeast strain, plated into selective medium containing glucose as the carbon source, and incubated at room temperature until the colonies appeared. Colonies were replicated into selective medium containing galactose as the carbon source and incubated at 37 °C until the colonies appeared. Temperature-resistant colonies were isolated, patched into fresh selective plates, grown, and replica plated again to confirm the positive growth and eliminate false positives. The patches were grown in liquid medium to isolate pMyr plasmid DNA containing the positive clones. The isolated plasmids were amplified in Escherichia coli and purified DNA was sequenced using primers on both sides of the cDNA insert regions. The cDNA inserts were identified using the National Center for Biotechnology Information BLAST application.
Recombinant Proteins
The recombinant proteins glutathione S-transferase (GST), as well as GST-tagged HA-NUP98-HOXA9, HOXA9, and AES were produced from the pGEX-6P1 vector in E. coli BL21(DE3) bacteria and purified using glutathione-Sepharose 4B beads (GE Healthcare). Recombinant AES was released by digestion with Pre-Scission protease (GE Healthcare).
Protein Binding Assays
NUP98, NUP98-HOXA9 and its variants, NUP62 full-length and deletions, and NUP153 full-length and deletions were produced using the TnT T7 quick coupled transcription/translation system (Promega) in the presence of Tran35S-label (MP Biomedicals). Binding assays were carried out essentially as described previously (31). For a binding reaction, 10 μl of beads from the immobilized recombinant protein (GST control or GST-AES) were incubated for 1 h at 4 °C with 34-μl mixtures consisting of in vitro translated protein, transport buffer-Tween 20 (TB-T, which consists of 20 mm HEPES-KOH, pH 7.4, 110 mm potassium acetate, and 2 mm MgCl2 with 0.1% Tween 20), and 0.5 mg/ml of Pefabloc (Roche Applied Science). The unbound fraction was removed and the beads were washed three times in cold TB-T. One-fourth of the unbound and all of the bound fractions were visualized by SDS-PAGE and autoradiography.
For the AES binding assays, purified recombinant AES was incubated with immobilized GST, GST-NUP98-HOXA9, or GST-HOXA9 proteins in 34-μl mixtures in TB-T and processed as described above. The samples were subjected to SDS-PAGE and AES was detected with anti-AES antibody.
AES Antibody Preparation
Recombinant AES protein was submitted to Harlan Laboratories where rabbit immunizations and bleedings were done according to their standard procedures. Bleeds with the highest concentration of antibody as detected by immunoblotting were selected for antibody purification. At each step, serum and purified antibody fractions were tested by immunoblotting of a K562 lysate (supplemental Fig. S2). Antiserum (supplemental Fig. S2, lane 1) was first precleared using 1 ml of Affi-Gel 15 beads (Bio-Rad) loaded with lysate containing 30 mg of protein from E. coli BL21 bacteria expressing empty pGEX6P1 vector for 4 h at 4 °C (supplemental Fig. S2, lane 2). The pre-cleared serum was added to a column containing 1 ml of Affi-Gel 15 beads loaded with 1 mg of AES protein and incubated overnight with rotation at 4 °C. The flow-through fraction was depleted of AES antibody as shown under supplemental Fig. S2, lane 3. The column was washed 6 times with 10 ml of cold DPBS at 4 °C. Bound antibody was eluted once with 4 ml of 1 m NaCl in DPBS, once with 4 ml of 0.1 m glycine, pH 2.5, and once with 4 ml of 0.1 m glycine, pH 2.5, 1 m NaCl, 1% CHAPS. For each elution, 0.5-ml fractions were collected, and glycine-containing fractions were neutralized with 25 μl of 1 m Tris base. Antibody concentration in the various fractions was determined by Amido Black staining and immunoblotting of purified recombinant AES or K562 lysate (supplemental Fig. S2, lanes 4–12). More than 90% of the antibody was detected in fractions 6 and 7 of the 0.1 m glycine, pH 2.5, elution (supplemental Fig. S2, lanes 9 and 10); these fractions were stored at −80 °C.
To test the specificity of the anti-AES antibody, K562 cells were nucleofected with empty vector or vector containing HA-tagged AES. Lysates were prepared as described below and AES was immunoprecipitated with the anti-AES antibody. The immunoprecipitated complexes were subjected to SDS-PAGE and probed for HA-tagged AES with anti-HA antibody (12CA5) (supplemental Fig. S3A). To further test the anti-AES antibody K562 cells were nucleofected with empty pRFP-C-RS vector, vector containing nonspecific shRNA, or vectors containing shRNA against AES (Origene) (see supplemental Table S3 for sequences). Lysates were subjected to SDS-PAGE and AES was detected with the anti-AES antibody. (supplemental Fig. S3B). To test the specificity of the antibody in immunofluorescence, cytospin smears were prepared from the transfected cells and AES was detected by immunofluorescence with the anti-AES antibody (supplemental Fig. S3C). To quantitate the specific knockdown of AES, the average fluorescence intensity of 35 cells for each condition was measured using Metamorph version 6.3r2 software (supplemental Fig. S3D).
Immunofluorescence Microscopy
For localization of AES, 25,000 K562 cells were washed twice with Hanks' balanced salt solution and centrifuged onto slides by cytospin centrifugation, fixed with 4% paraformaldehyde in DPBS for 20 min, and permeabilized with 0.1% Triton X-100 for 20 min at room temperature. For blocking and washing, 2% normal goat serum in DPBS with 1% BSA, 0.1% Tween 20 was used. Primary antibodies were rabbit anti-AES along with mouse mAb 414 (Abcam). The fluorescent secondary antibodies were FITC-conjugated anti-rabbit IgG from goat (Santa Cruz Biotechnology) and rhodamine-conjugated anti-mouse IgG from goat (Millipore Corp.). The images were captured using an Eclipse 80i fluorescent microscope (Nikon) and processed using Metamorph version 6.3r2 software (Molecular Devices).
Co-immunoprecipitation
For NUP98-HOXA9 immunoprecipitation with AES, 107 K562 cells were nucleofected with pcDNA3-HA-NUP98-HOXA9 using a Nucleofector device (Lonza). The co-immunoprecipitations of NUP62, NUP153, and NUP98 with AES were done in untransfected K562 cells. Cells were harvested 16 h postnucleofection, washed with cold DPBS, and lysed for 30 min on ice with 0.5 ml of lysis buffer (10 mm Tris-HCl, pH 7.5, 0.4 m NaCl, 1% Nonidet P-40, 0.4% Triton X-100, 0.2% sodium deoxycholate, 1 mm EDTA, protease inhibitors (Roche Applied Science), 1 mm PMSF). Dilution buffer (10 mm Tris-HCl, pH 7.5, 1 mm EDTA, protease inhibitors, 1 mm PMSF) (0.5 ml) was added, followed by centrifugation at 17,000 × g for 30 min. The supernatants were transferred to new tubes and the appropriate antibodies were added. For AES immunoprecipitation either rabbit IgG or rabbit anti-AES antibody was added. After overnight incubation at 4 °C, 30 μl of Protein G beads were added to lysates with control rabbit IgG or anti-AES antibodies and incubated for another 1 h. Beads were washed with 700 μl of wash buffer (10 mm Tris-HCl, pH 7.5, 0.2 m NaCl, 0.5% Nonidet P-40, 0.2% Triton X-100, 0.1% sodium deoxycholate, 1 mm EDTA, 1× protease inhibitors, 1 mm PMSF) five times, 3 min each time followed by centrifugation at 1,800 × g for 3 min at 4 °C. Beads were washed with cold DPBS and bound proteins were eluted by boiling with 30 μl of 2× SDS buffer (0.1 m Tris-HCl, pH 6.8, 3.5% SDS, 10% glycerol, 2 mm DTT, 0.004% bromphenol blue) for 10 min. Proteins were subjected to SDS-PAGE (7.5% gel) followed by immunoblotting. NUP98-HOXA9 was detected with an anti-HA antibody, NUP98 with anti-NUP98 antibody (Cell Signaling Technology), and NUP62 or NUP153 with mouse MAb414 antibody. The input amounts were determined by immunoblotting the cell lysates with the appropriate antibodies. To confirm coimmunoprecipitation of AES with HA-NUP98-HOXA9, 107 K562 cells were nucleofected with pcDNA3 or pcDNA3-HA-NUP98-HOXA9. Cell lysates were incubated with anti-HA antibody and AES in the immunoprecipitated complex was detected with the anti-AES antibody.
Luciferase Assay
K562 cells were transfected by electroporation using a Bio-Rad GenePulser with 5 μg of pGL4.11 vector or pGL4.11 driven by the KBTBD10 promoter in combination with 10 μg of either empty pcDNA3, or pcDNA3 expressing HA-NUP98-HOXA9, without or with pcDNA3 vector expressing FLAG-AES. To control for efficiency of transfection, 0.5 μg of pRL-TK (Promega), which expresses Renilla luciferase was included. Five million cells were incubated with the DNA at room temperature for 10 min before electroporation and 10 min after electroporation, and cultured in 10 ml of Iscove's modified Dulbecco's medium (IMDM) with 10% FBS, 2 mm l-glutamine, and 100 units/ml of penicillin/streptomycin. Luciferase activity was measured 48 h after electroporation using the Dual Luciferase Reporter Assay System (Promega) and the results were normalized to Renilla luciferase. For the AES knockdown experiment, K562 cells were nucleofected with 5 μg of pGL4.11 vector driven by the KBTBD10 promoter in combination with 10 μg of either empty pcDNA3, or pcDNA3 expressing HA-NUP98-HOXA9. In addition, either empty pRFP-C-RS vector, vector containing nonspecific shRNA, or vectors containing shRNA against AES (Origene) (see supplemental Table S3 for sequences) were included. Expression of HA-NUP98-HOXA9 and knockdown of AES was verified by immunoblotting with anti-HA or anti-AES antibody, respectively.
Retroviral Production
The GP293 packaging cell line was transiently transfected with 4.4 μg of control MSCV-IRES-GFP retroviral vector, control MSCV-IRES-YFP vector, or vector expressing FLAG-tagged AES along with 1.1 μg of the vesicular stomatitis virus glycoprotein plasmid (Clontech) using Lipofectamine and Plus reagents (Invitrogen). After 48 h, the culture supernatant containing vesicular stomatitis virus glycoprotein-pseudotyped retrovirus was collected and used for transduction of PG13 packaging cells (American Type Culture Collection) by spinoculation in the presence of 8 μg/ml of Polybrene (hexadimethrine bromide; Sigma) to produce GaLV-pseudotyped retrovirus. PG13 cells producing GaLV-pseudotyped MSCV-IRES-GFP/HA-NUP98-HOXA9 retrovirus were previously described (21). The PG13 culture supernatants containing GaLV-pseudotyped retrovirus were used for transduction of K562 cells and human primary CD34+ cells by co-culturing for 48 h as described previously (21). For knockdown of AES in CD34+ cells, GIPZ lentiviral shRNAmir particles were purchased from Open Biosystems.
Retroviral Transduction and Analysis of Primary Human CD34+ Cells
Frozen human CD34+ cells from mobilized peripheral blood of healthy volunteers (purchased from the Fred Hutchinson Cancer Research Center) were prestimulated and transduced with retrovirus as described (21). After 48 h, GFP/YFP double positive cells were isolated using a MoFlo high-speed sorter (Dako) and expression of the transfected gene was confirmed by immunoblotting with anti-HA tag antibody for NUP98-HOXA9 and anti-AES antibody for AES. For long-term liquid culture, sorted cells were seeded at 105 cells/ml of IMDM containing 20% FBS, 100 ng/ml of Fms-related tyrosine kinase 3 (Flt 3)-ligand, 20 ng/ml of granulocyte/macrophage colony-stimulating factor (GM-CSF), 100 ng/ml of stem cell factor, 100 ng/ml of thrombopoietin, 50 ng/ml of IL-3, 100 ng/ml of IL-6 (all cytokines were from Peprotech, Rocky Hill, NJ), 2 mm l-glutamine, and 100 units/ml of penicillin/streptomycin. Cells were periodically counted and resuspended in fresh media at 105 cells/ml. Cytospin preparations of cells harvested from the liquid cultures at weeks 4, 5, and 6 were stained with Giemsa and 500 cell differential counts were performed using an Olympus BX51 microscope (Olympus America). Cells with blast and promyelocyte morphology were counted as primitive; those with myelocyte/metamyelocyte morphology as intermediate myeloid; those with band, segmented neutrophil, monocyte, and macrophage morphology as mature myeloid. Photomicrographs were taken with an Olympus DP71 camera with a ×20 and ×60 oil objective. For colony-forming cell (CFC) assays, sorted cells were resuspended in IMDM containing 2% FBS and mixed with Methocult GF+ (H4435, StemCell Technologies) that consists of 1% methylcellulose, 30% FBS, 1% bovine serum albumin, 10−4 m 2-mercaptoethanol, 2 mm l-glutamine, 50 ng/ml of stem cell factor, 20 ng/ml of GM-CSF, 20 ng/ml of IL-3, 20 ng/ml of IL-6, 20 ng/ml of granulocyte colony-stimulating factor (G-CSF), and 3 units/ml of erythropoietin in IMDM. One thousand cells were plated in each 35-mm dish and cultured for 14 days. Colonies were counted at ×40 magnification and classified into 3 categories: erythroid, myeloid, and mixed/branched.
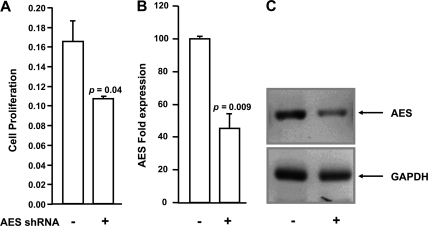
For AES knockdown using lentivirus, prestimulated CD34+ cells were transduced with MSCV-IRES-YFP retrovirus expressing HA-NUP98-HOXA9. After 48 h, YFP positive cells were isolated and grown in culture medium for 2 weeks. Cells were then transduced by spinoculation with GIPZ Lentiviral shRNAmir particles that express nonspecific shRNA or AES shRNA and a GFP marker (see supplemental Table S3 for shRNA sequences). After 48 h, GFP/YFP positive cells were isolated by sorting. One week later, knockdown of AES was confirmed by quantitative PCR and semi-quantitative RT-PCR (Fig. 10, B and C) using primers specific for AES (expression was normalized to GAPDH). One week later an MTS cell proliferation assay was done using CellTiter 96® AQueous NonRadioactive Cell Proliferation Assay (Promega G5421) according to the manufacturer's instructions. Twenty-five thousand cells were seeded into a 96-well plate in triplicate and grown for 72 h in 100 μl of culture medium. Twenty μl of MTS/phenazine methosulfate solution was added and the cells were incubated at 37 °C for 2 h. The absorbance at 490 nm was measured using an ELISA plate reader.
FIGURE 10.
Knockdown of AES inhibits proliferation of cells expressing NUP98-HOXA9. NUP98-HOXA9-expressing human primary CD34+ cells were transfected with vectors containing nonspecific shRNA (−) or AES-specific shRNA (+) and grown in liquid culture. A, sorted cells were seeded into a 96-well plate in triplicate and cell proliferation was measured 72 h later using CellTiter 96® AQueous Nonradioactive Cell Proliferation Assay. The y axis (cell proliferation) represents the absorbance at 490 nm. B, knockdown of AES was quantified by quantitative PCR using primers specific for AES and normalized to GAPDH. C, knockdown of AES is shown by RT-PCR. PCR products were run on 1.2% agarose gels and images were acquired in Chemidoc XRS (Bio-Rad).
Flow Cytometry
Flow cytometry was performed on a FACScan flow cytometer (BD Biosciences) upgraded to 5 colors and two lasers, and analyzed using FlowJO version 8.6.3 software (Tree Star). The antibodies used for these studies were anti-CD235a (allophycocyanin-conjugated clone GA-R2) (BD Biosciences) and anti-CD45 (phycoerythrin-Cy7-conjugated clone J.33) (Beckman Coulter).
RESULTS
Cytotrap Two-hybrid Analysis Shows That NUP98-HOXA9 Interacts with AES
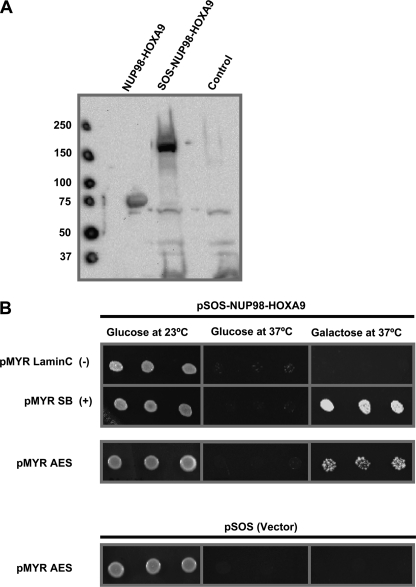
To identify possible interaction partners of the oncoprotein NUP98-HOXA9, we used a Cytotrap yeast two-hybrid assay in which the interaction between target and the bait proteins occurs in the cytoplasm. The traditional two-hybrid assay relies on intranuclear interactions that result in transactivation and is therefore likely to show many false positives when the bait contains a transactivation domain as is the case with NUP98-HOXA9 (24). In the Cytotrap yeast two-hybrid assay, the bait protein was fused to hSOS and the target library was subcloned downstream of a myristylation signal that targets the proteins to the plasma membrane. A physical interaction between the bait protein and one of the targets recruits the hSOS protein to the plasma membrane. This activates the Ras signaling pathway allowing the cdc25H strain to grow and form colonies at 37 °C in galactose containing medium. NUP98-HOXA9 was fused to hSOS as a bait protein and the target library was obtained from human K562 cells. Expression of hSOS-NUP98-HOXA9 in the cdc25H yeast strain was confirmed by immunoblotting (Fig. 1A). Controls for the assay were performed according to the manufacturer's instructions (supplemental Fig. S1). The bait protein and the target library were co-transformed into the cdc25H heat-sensitive yeast strain. Several hundred colonies appeared and were replica-plated onto galactose-containing medium to induce expression of the target library. Colonies were isolated and selection in stringent temperature and media was repeated for isolation of true positive clones. Inserts were sequenced and several of the positive clones encoded full-length AES.
FIGURE 1.
Cytoplasmic yeast two-hybrid analysis identifies AES as an interacting partner for NUP98-HOXA9. A, immunoblot showing expression of SOS-NUP98-HOXA9 in yeast cells. cdc25H yeast cells were transformed with either pSOS vector (Control) or pSOS-NUP98-HOXA9. Lysates were subjected to SDS-PAGE and immunoblotting with anti-HOXA9 antibody. Lysates from K562 cells expressing NUP98-HOXA9 were used as a positive control. B, K562 cDNA library in pMyr vector was co-transformed with pSOS-NUP98-HOXA9 into cdc25H yeast strain. Isolated colonies were patched on galactose-containing medium and transferred to 37 °C. Patches growing at 37 °C were re-spotted on plates containing selective medium supplemented with either glucose (repressing) or galactose (de-repressing) as the sole carbon source and incubated either at 23 (permissive) or 37 °C (restrictive) for 3–5 days. Plasmids were isolated from yeast cells growing at 37 °C in galactose containing medium and retransformed along with pSOS-NUP98-HOXA9 to confirm the interactions. Several positive cDNA clones were sequenced and identified as AES. Cells containing pSOS-NUP98-HOXA9 with pMYR-Lamin C or empty pSOS vector with pMyr-AES were spotted as negative controls. Cells containing pSOS-NUP98-HOXA9 with pMYR-SB were spotted as a positive control. Growth at 37 °C in galactose containing medium indicates a positive interaction.
To confirm the interaction between NUP98-HOXA9 and AES, the plasmid encoding AES along with either pSOS vector or vector expressing hSOS-NUP98-HOXA9 was reintroduced into the cdc25H yeast strain. Colonies were spotted into selective medium and subjected to the restrictive temperature. Cells were transfected with the pSOS-NUP98-HOXA9 bait construct along with a plasmid expressing myristylated Lamin C as a negative control, and with a plasmid expressing myristylated SOS-binding protein (SB) as a positive control. As shown in Fig. 1B, growth was observed in all transformants at 23 °C. Growth was not observed at 37 °C in glucose containing medium because the cDNA library was cloned under a galactose-inducible promoter. When transferred to 37 °C, there was no growth in cells co-expressing pMyr-Lamin C and pSOS-NUP98-HOXA9 indicating that NUP98-HOXA9 does not activate Ras by targeting itself to the plasma membrane. Growth was present in cells co-expressing myristylated SB and pSOS-NUP98-HOXA9 because the SB binds to the hSOS portion and targets hSOS-NUP98-HOXA9 to the plasma membrane. A similar growth pattern was observed in cells co-expressing hSOS-NUP98-HOXA9 with AES suggesting a specific interaction between NUP98-HOXA9 and this protein. As expected, there was no growth in cells that co-expressed the empty hSOS vector with AES indicating that the positive interaction observed with hSOS-NUP98-HOXA9 cannot be explained by binding AES to the hSOS portion of the bait (Fig. 1B).
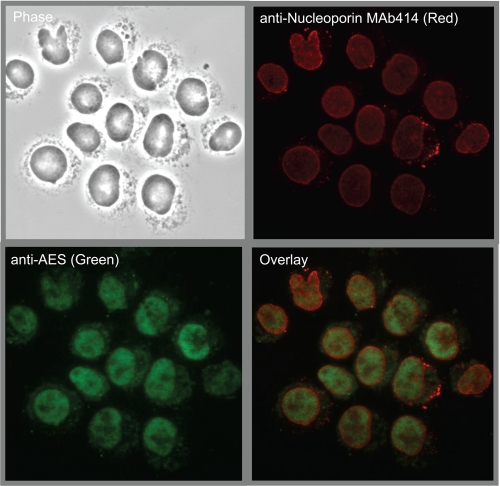
AES Localizes to the Nucleus
AES Is a Transcriptional Regulator of the TLE family, and is also known as TLE5 (32). To determine the localization of endogenous AES, a rabbit polyclonal antibody was raised against recombinant AES and affinity purified (supplemental Figs. S2 and S3). K562 cells were immunostained with the anti-AES antibody together with mAb414 that marks the nuclear periphery by staining nucleoporins. Consistent with its reported function in transcriptional regulation (33–35), AES localized primarily to the interior of the nucleus (Fig. 2).
FIGURE 2.
AES localizes to the interior of the nucleus. Cytospin smears of K562 cells were immunostained with the anti-AES (green) antibody together with mAb414 (red) that marks the nuclear periphery by staining nucleoporins.
AES Interacts with the FG Repeat Region of NUP98-HOXA9
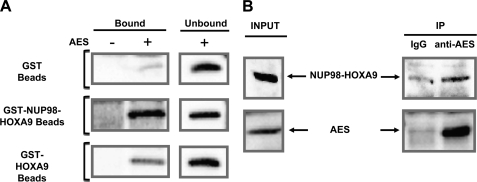
To establish a direct interaction between AES and NUP98-HOXA9 an in vitro binding assay was carried out using purified recombinant proteins. GST, GST-NUP98-HOXA9, or GST-HOXA9 were immobilized on beads and incubated with purified recombinant AES protein. As shown in Fig. 3A, AES interacted with GST-NUP98-HOXA9 and to a lesser degree with GST-HOXA9, whereas no significant interaction was observed with GST. The entire immunoblot is shown under supplemental Fig. S4A and a corresponding Coomassie Blue-stained gel is shown under supplemental Fig. 4B.
FIGURE 3.
AES interacts with NUP98-HOXA9 in vitro and in vivo. A, GST, GST-NUP98-HOXA9, or GST-HOXA9 were immobilized on beads and incubated with purified recombinant AES protein. The samples were subjected to SDS-PAGE and AES was detected by immunoblotting with anti-AES antibody. B, co-immunoprecipitation of NUP98-HOXA9 with AES in K562 cells transfected with HA-NUP98-HOXA9. Whole cell lysates were subjected to immunoprecipitation with either rabbit IgG or anti-AES antibody. NUP98-HOXA9 in the cell lysates (Input) and immunoprecipitated complexes (IP) was detected by immunoblotting with anti-HA antibody. AES was detected by immunoblotting with anti-AES antibody.
To demonstrate that these interactions occur in vivo, co-immunoprecipitation experiments were carried out in K562 cells expressing HA-NUP98-HOXA9. Whole cell lysates were subjected to immunoprecipitation with either rabbit IgG (negative control) or anti-AES antibody, followed by anti-HA immunoblotting. As shown in Fig. 3B, NUP98-HOXA9 co-immunoprecipitated with endogenous AES (the full gel is shown under supplemental Fig. S4C). To further confirm the in vivo interaction, a reverse co-immunoprecipitation was carried out. Whole cell lysates of K562 cells expressing HA-NUP98-HOXA9 were subjected to immunoprecipitation with anti-HA antibody followed by anti-AES immunoblotting. As shown under supplemental Fig. S4D, AES co-immunoprecipitated with NUP98-HOXA9.
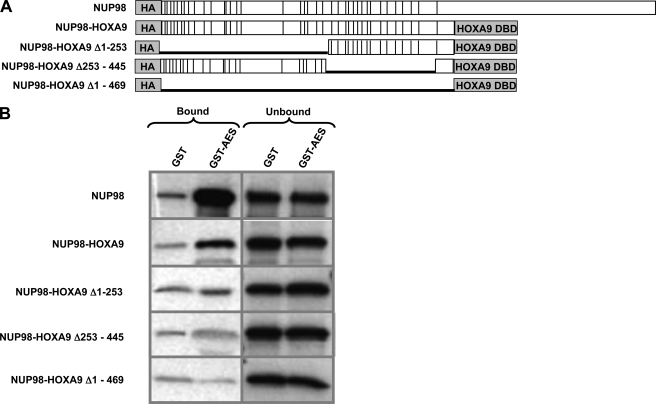
To identify the domain in NUP98-HOXA9 responsible for interaction with AES, GST-AES was immobilized on beads and incubated with in vitro translated full-length NUP98, NUP98-HOXA9, or deletion mutants of the NUP98 portion of NUP98-HOXA9. The NUP98-HOXA9 mutants were deleted for the N terminus (Δ1–253), C terminus (Δ253–445), or the entire NUP98 portion (Δ1–469) (Fig. 4A). AES bound to both NUP98 and NUP98-HOXA9. Deletions encompassing part or all of the NUP98 portion of NUP98-HOXA9 resulted in loss of AES binding, indicating that the interaction between AES and NUP98-HOXA9 is mediated by the NUP98 portion of the latter (Fig. 4B). As illustrated in Fig. 4A, this NUP98 portion contains all of the FG repeats of NUP98, which raised the question of whether AES would bind to other nucleoporins that contain FG repeats.
FIGURE 4.
AES interacts with the FG repeat region of NUP98-HOXA9. A, full-length NUP98, NUP98-HOXA9, and NUP98-HOXA9 deletion mutants. Vertical lines represent FG repeats. DBD, DNA-binding domain. B, GST-AES or GST were immobilized on beads and incubated with in vitro translated full-length NUP98, NUP98-HOXA9, or deletion mutants of NUP98-HOXA9. Bound and unbound fractions were resolved by SDS-PAGE and subjected to autoradiography.
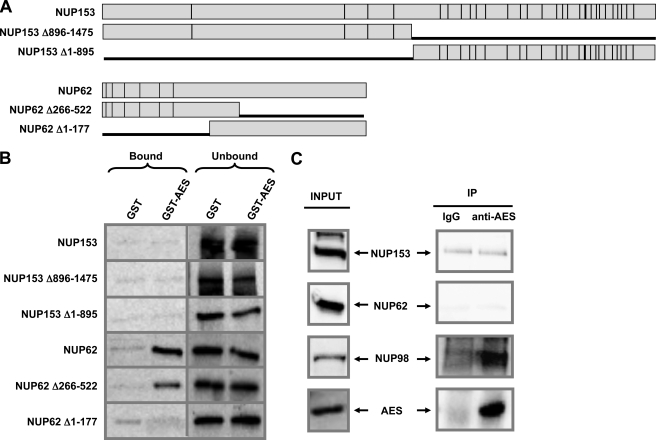
To determine whether AES interacts with other FG repeat nucleoporins, GST or GST-AES immobilized on beads were incubated with in vitro translated NUP153, NUP62, or their deletion mutants as illustrated in Fig. 5A. As shown in Fig. 5B, AES interacted with NUP62, and removal of the FG-repeat containing N terminus of NUP62 totally abolished the interaction with AES. Thus in the context of both NUP98-HOXA9 and NUP62, the presence of FG repeats appears to be necessary for AES binding. On the other hand, NUP153, despite having FG repeats, did not bind to AES indicating that the presence of FG repeats is not in itself sufficient to confer AES binding.
FIGURE 5.
AES interacts in vivo with NUP98 but not with other FG repeat nucleoporins. A, full-length NUP153, NUP153 deletion mutants, NUP62, and NUP62 deletion mutants. Vertical lines represent FG repeats. B, GST or GST-AES immobilized on beads were incubated with in vitro translated NUP153, NUP62, or their deletion mutants. Bound and unbound fractions were resolved by SDS-PAGE and subjected to autoradiography. C, co-immunoprecipitation of NUP153, NUP62, and NUP98 with AES in K562 cells. Whole cell lysates were subjected to immunoprecipitation with either rabbit IgG or anti-AES antibody. Both input and immunoprecipitates (IP) were probed for NUP153 or NUP62 using the mAb414 antibody or NUP98 using anti-NUP98 antibody. AES was detected with anti-AES antibody.
Consistent with the in vitro binding data, NUP98 co-immunoprecipitated with AES from K562 cell lysates and NUP153 did not (Fig. 5C). On the other hand, NUP62 did not co-immunoprecipitate with AES despite their in vitro interaction. Given the intranuclear localization of AES, this may reflect the different intracellular localizations of NUP98 and NUP62: whereas both are present at the nuclear pore complex, NUP98 has been shown to localize to the inside of the nucleus as well (36–38), which would allow it to interact with AES. Another difference between NUP98 and other FG nucleoporins is that many of the FG repeats of NUP98 are GLFG repeats (1); it is not clear whether this contributes to the ability of NUP98 to bind to AES in vivo. Overall, the data provide solid evidence that AES interacts with NUP98. Whether it also interacts with other nucleoporins under physiologic conditions remains to be determined.
AES Enhances Transcriptional Activation by NUP98-HOXA9
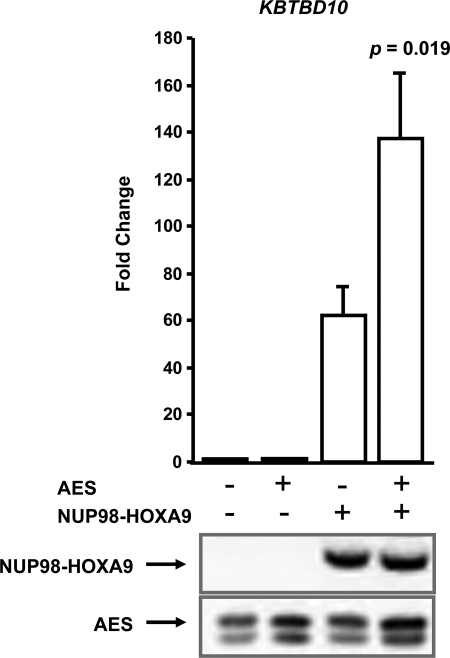
NUP98-HOXA9 acts as an aberrant transcription factor (24, 25). A well characterized target gene is KBTBD10 whose promoter is transcriptionally up-regulated by NUP98-HOXA9 (22). To determine the role of AES in the regulation of transcription by NUP98-HOXA9, a luciferase reporter construct driven by the KBTBD10 promoter was introduced into K562 cells along with a construct expressing NUP98-HOXA9, with or without AES. Overexpression of AES resulted in a significant increase in transcriptional activation of the reporter construct by NUP98-HOXA9 (Fig. 6). Immunoblotting of the cell lysates with anti-HA and anti-AES antibody confirmed the expression of NUP98-HOXA9 and elevated the levels of AES compared with endogenous AES (Fig. 6, bottom panel).
FIGURE 6.
AES cooperates with NUP98-HOXA9 in transcriptional activation. K562 cells were transfected with a luciferase construct driven by the KBTBD10 promoter in addition to either empty vector or vector expressing NUP98-HOXA9, without or with AES. Firefly luciferase activity was measured 48 h after transfection and normalized to a Renilla luciferase internal control. The numbers represent fold-change over control (average of three independent experiments); error bars represent S.D. The p value indicated was obtained by a two-tailed t test. Bottom panel, expression of HA-NUP98-HOXA9 and AES was verified by immunoblotting with anti-HA and anti-AES antibody, respectively.
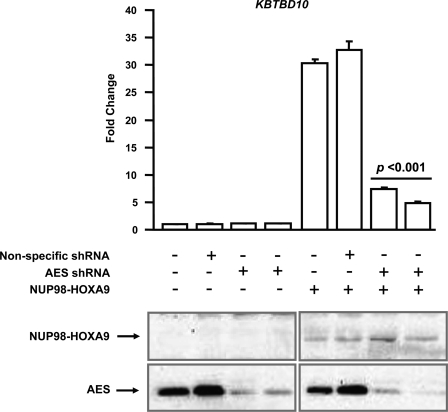
To further demonstrate the role of AES in downstream gene regulation by NUP98-HOXA9, the effect of AES knockdown on NUP98-HOXA9-driven transcription from the KBTBD10 promoter was assayed. The KBTBD10-luciferase construct was introduced into K562 cells along with empty vector or vector expressing NUP98-HOXA9, without or with vectors expressing two different shRNAs specific to AES. Knockdown of AES resulted in a significant decrease in transcriptional activation of the reporter construct by NUP98-HOXA9 (Fig. 7). Immunoblotting of the cell lysates with anti-HA and anti-AES antibodies confirmed expression of NUP98-HOXA9 and knockdown of AES, respectively (Fig. 7, bottom panel).
FIGURE 7.
shRNA-mediated knockdown of AES inhibits downstream gene regulation by NUP98-HOXA9. K562 cells were transfected with a firefly luciferase construct driven by the KBTBD10 promoter with either empty vector or vector expressing NUP98-HOXA9. In addition, the transfections included empty vector, vector expressing nonspecific shRNA, or 2 vectors expressing different AES shRNAs. Firefly luciferase activity was measured 48 h after transfection and normalized to a Renilla luciferase internal control. The numbers represent fold-change over control (average of three independent experiments); error bars represent S.D. The p value indicated was obtained by a two-tailed t test. Expression of HA-NUP98-HOXA9 and knockdown of AES were verified by immunoblotting with anti-HA or anti-AES antibody, respectively.
AES Enhances the Proliferation of CD34+ Cells in the Presence of NUP98-HOXA9
The finding that AES augments the transcriptional activity of NUP98-HOXA9 raised the possibility that AES may cooperate with NUP98-HOXA9 in cell transformation. We have previously shown that NUP98-HOXA9 induces long-term proliferation of primary human hematopoietic CD34+ cells and that this proliferation is dependent on the transcriptional activity of NUP98-HOXA9 (21, 22). Because AES increased the transcriptional activity of NUP98-HOXA9 (Fig. 6), we asked whether AES might also enhance the ability of NUP98-HOXA9 to induce the proliferation of human primary CD34+ cells. An MSCV-IRES-YFP retroviral vector that expresses AES and an MSCV-IRES-GFP vector expressing NUP98-HOXA9 were used to address this question. Human CD34+ primary cells were retrovirally transduced to express either NUP98-HOXA9 or empty MSCV-IRES-GFP in combination with either AES or empty MSCV-IRES-YFP. Doubly transduced cells were isolated by sorting for both GFP and YFP positivity and expression of NUP98-HOXA9 and AES was confirmed by immunoblotting (Fig. 8). The sorted cells were continually grown in liquid culture in the presence of cytokines with periodic cell counting as previously described (21–23, 39) (see “Experimental Procedures”). NUP98-HOXA9 was detectable by anti-HA immunoblotting up to the 3rd week after transduction (supplemental Fig. S5). However, the effects of NUP98-HOXA9 on proliferation and self-renewal of primary human CD34+ cells have been documented well beyond that point by us and others (21, 22, 39, 40). It may be that at later time points NUP98-HOXA9 is expressed at levels below the limit of detection by the anti-HA tag antibody. On the other hand, transduced AES was still clearly detectable by immunoblotting up to the 5th week after transduction (supplemental Fig. S5).
FIGURE 8.
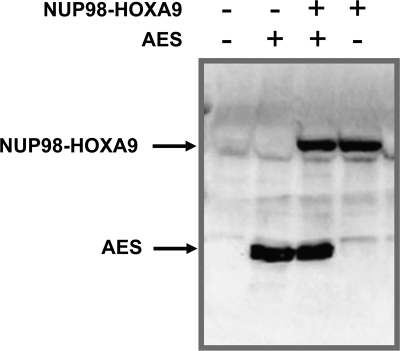
Expression of AES and NUP98-HOXA9 in primary human CD34+ cells. Primary human CD34+ cells were retrovirally transduced with either control MSCV-IRES-GFP vector or vector expressing NUP98-HOXA9 with or without MSCV-IRES-YFP vector or vector expressing human AES. Cells positive for both GFP and YFP were sorted. NUP98-HOXA9 was detected with an anti-HA antibody and AES was detected with an anti-AES antibody.
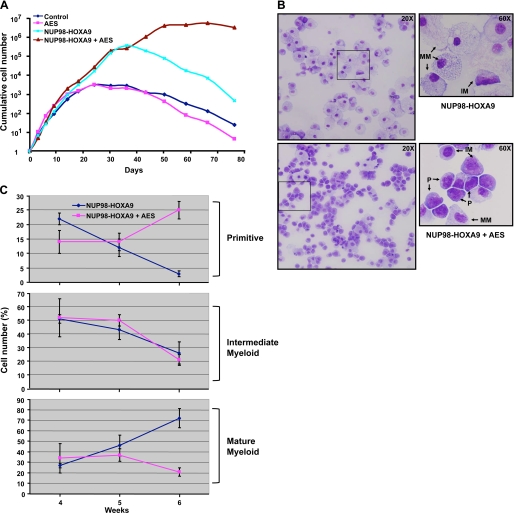
As previously reported (21), NUP98-HOXA9 increased the long-term proliferation of primary human CD34+ cells by several orders of magnitude (Fig. 9A). AES alone did not cause an increase in long-term proliferation. Interestingly, cells expressing both NUP98-HOXA9 and AES exhibited a steep increase in proliferation and continued to grow for more than 70 days in liquid culture, whereas cells expressing NUP98-HOXA9 alone began a steep decline after an average of ∼40 days. These data show that AES induces the proliferation of primary human CD34+ cells in the presence, but not absence, of NUP98-HOXA9.
FIGURE 9.
AES enhances the proliferation of CD34+ cells in the presence of NUP98-HOXA9. A, primary human CD34+ cells were retrovirally transduced to express AES, NUP98-HOXA9, both, or neither as described in the legend to Fig. 8. Cells positive for both GFP and YFP were sorted and continually cultured in the presence of cytokines with periodic cell counting and feeding. The cumulative fold-increase in cell numbers compared with day 0 is plotted on a logarithmic scale. B, cells growing in liquid culture were subjected to morphological evaluation by Giemsa staining at weeks 4–6. Representative fields are shown from cells expressing NUP98-HOXA9 alone or NUP98-HOXA9 with AES at week 6 of culture. P, primitive cells; IM, intermediate myeloid cells; MM, mature myeloid cells. Photomicrographs were taken with ×20 and 60 oil objectives. C, a 500-cell differential count was carried out on the Giemsa-stained slides and average numbers from three independent experiments for weeks 4–6 were plotted. For quantification of the different cell types see supplemental Table S1.
The cells growing in liquid culture were subjected to morphological evaluation by Giemsa staining at weeks 4–6, when cells expressing NUP98-HOXA9 alone begin to decline, whereas those expressing both NUP98-HOXA9 and AES continue to proliferate. In Fig. 9B representative fields are shown from cells expressing NUP98-HOXA9 alone or with AES at week 6 of culture. Primitive, intermediate, and mature myeloid cells were distinguished, with the NUP98-HOXA9 sample showing a preponderance of mature cells, whereas the NUP98-HOXA9 + AES sample contained many primitive cells. A 500-cell differential count was carried out on the Giemsa-stained slides and the average numbers from three independent experiments for weeks 4–6 are shown under supplemental Table S1 and a time course comparison between the NUP98-HOXA9 and NUP98-HOXA9 + AES samples is shown in Fig. 9C. The data clearly show a decrease in primitive cells and an increase in mature cells over time in the NUP98-HOXA9 samples, whereas the opposite trend is seen in samples expressing both NUP98-HOXA9 and AES (Fig. 9C). These data show that in the presence of NUP98-HOXA9 AES induces proliferation accompanied by an increase in the numbers of primitive cells.
To further demonstrate the role of AES in NUP98-HOXA9-mediated aberrant cell proliferation, CD34+ cells expressing NUP98-HOXA9 were subjected to shRNA-mediated AES knockdown. The cell proliferation assay for CD34+ cells was done using an MTS assay. CD34+ cells expressing NUP98-HOXA9 along with either nonspecific shRNA or AES shRNA were grown for 72 h prior to the assay. As shown in Fig. 10A, depletion of AES resulted in a significant inhibition of cell proliferation mediated by NUP98-HOXA9. The shRNA-mediated knockdown of AES was confirmed by quantitative and semi-quantitative PCR (Fig. 10, B and C). PCR was used to quantify AES expression because its level in normal primary human CD34+ cells is below the threshold of detection by immunoblotting. In this context it was of interest to note that recent data show an increase in the expression of AES in some patients with AML and cooperation between AES and another oncogene, AML1-ETO, in the induction of self-renewal of hematopoietic progenitors (41).
AES Counteracts the Erythroid Hyperplasia Caused by NUP98-HOXA9
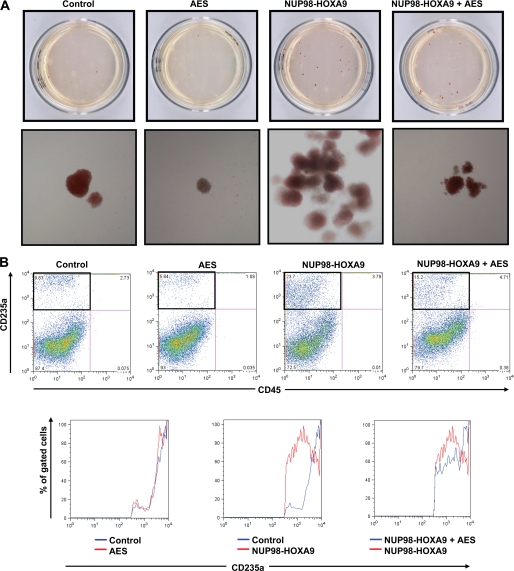
We have previously shown that NUP98-HOXA9 causes erythroid hyperplasia and shift to immaturity (21, 22). To determine the effect of AES overexpression on the differentiation of primary human CD34+ cells in the presence or absence of NUP98-HOXA9, CFC assays were carried out. Sorted cells were plated in semisolid methylcellulose-based media for 14 days and the resulting colonies were examined and counted (Fig. 11A, and supplemental Fig. S6 and Table S2). As previously reported (21), many prominent large erythroid colonies appeared on the CFC plates with cells expressing NUP98-HOXA9, when observed without magnification. The control plates expressing empty vectors did not show such large colonies. When observed under low magnification (×40), the cells on the control plates showed small, tight, uniformly red erythroid colonies, whereas the red colonies in the NUP98-HOXA9-transduced plates were large in size. Many of those large colonies showed irregular contours with a mixed red/colorless and branched morphology. Interestingly, overexpression of AES in the presence of NUP98-HOXA9 resulted in fewer red colonies on the CFC plates. These colonies showed a more uniformly red color indicating more maturity. This suggests that overexpression of AES counteracts the erythroid hyperplasia and shift to immaturity caused by NUP98-HOXA9.
FIGURE 11.
AES counteracts the erythroid hyperplasia caused by NUP98-HOXA9. A, primary human CD34+ cells were retrovirally transduced to express AES, NUP98-HOXA9, both, or neither as described in the legend to Fig. 8. Cells positive for both GFP and YFP were sorted and 1,000 cells were seeded into each of two duplicate plates for CFC assay. Representative plates without magnification (top row) and low power photomicrographs of representative erythroid colonies (bottom row) are shown. For quantification of the number of colonies see supplemental Table S2. B, flow cytometry for erythroid differentiation. Cells from the CFC plates were harvested and stained with antibodies to CD45 and CD235a. The CD235a+ gate was plotted on a histogram (lower panels) to show the level of expression of CD235a relative to control cells.
To further confirm and quantitate this finding, erythroid differentiation was evaluated by subjecting cells harvested from the CFC plates to flow cytometry. As previously reported (21), NUP98-HOXA9 caused an increase in CD235a+ CD45− erythroid cells compared with empty vector control and overexpression of AES counteracted this effect (Fig. 11B). As previously shown, the levels of CD235a expression on erythroid cells are reduced by NUP98-HOXA9, consistent with an erythroid shift to immaturity (22). Overexpression of AES with NUP98-HOXA9 increased the expression levels of CD235a suggesting a shift toward a more mature stage of differentiation (Fig. 11B). Overall, the data show that AES overexpression counteracts the effects of NUP98-HOXA9 on erythroid differentiation of primary human CD34+ cells by decreasing the percentage of erythroid cells and increasing their maturity.
DISCUSSION
AES is a 197-amino acid protein that belongs to the TLE/Groucho family of co-repressors (32). The human TLE proteins and the homologous Drosophila Groucho proteins repress several transcription regulatory complexes (33, 42–49). The TLE repressor complex is believed to function by either modifying histone acetylation through interactions with histones and histone deacetylases or contacting components of the basal transcription machinery (35, 49–51). The larger members of this family contain several common domains (52). AES (also known as TLE5 or Grg5) is the shortest member of the family and contains only the glutamine-rich region (Q domain) and a glycine/proline-rich region (GP domain) (52–54). The Q-domain of AES can bind to other members of the TLE complex and inhibit their function either by preventing them from forming functional multimers that interact with the transcription machinery or by titrating them away from the repressor complex (33, 46, 48, 55–57).
AES is also able to modulate transcription directly. For example, AES physically interacts with the human androgen receptor and NFκB-p65 and acts as a co-repressor of their target genes (34, 35). In addition, mouse AES (Grg5) interacts with RUNX2 and enhances its transcriptional activation function (58). The ability of AES to augment transcriptional activation by NUP98-HOXA9 (Fig. 6) may be due to the direct interaction between the two proteins (Figs. 3 and 4).
AES showed a stronger interaction with NUP98 than with NUP98-HOXA9 (Fig. 4B). This suggests that the C-terminal portion of NUP98, which is missing in NUP98-HOXA9, plays a role in the interaction with AES. NUP98 exists both at the nuclear pore complex and within the nucleus (2, 36, 37, 60–65). The sequences responsible for targeting NUP98 to the nuclear pore complex reside in its C-terminal portion (61) that may be inaccessible in the nuclear pore complex-bound NUP98. Thus AES may interact only with the intranuclear pool of NUP98; this is consistent with the observation that AES is completely intranuclear and does not localize to the nuclear pore complex (Fig. 2).
Overexpression of AES in primary human CD34+ cells in the presence of NUP98-HOXA9 induced proliferation and caused a significant increase in the numbers of primitive cells (Fig. 9 and supplemental Table S1). In contrast, AES had no significant effect on cell proliferation or the numbers of primitive cells in the absence of NUP98-HOXA9 (Fig. 9A and supplemental Table S1). The ability of AES to cooperate with NUP98-HOXA9 in the induction of cell proliferation may be mediated by counteracting the effects of other TLE proteins. TLE1 and TLE4 act as tumor suppressors that are down-regulated by hypermethylation and/or deletion in a subset of human AML (66, 67). Growth of myeloid cell lines is inhibited by overexpression of TLE proteins, and enhanced by their knockdown (66, 67). It is of particular interest to note that TLE1 and TLE4 interact with the AML-associated oncoprotein AML1-ETO and that knockdown of TLE1 and TLE4 cooperates with AML1-ETO by promoting cell growth and survival (67). In addition, knockdown of the TLE homolog, gro3, in zebrafish induces abnormal hematopoiesis in the presence of AML1-ETO (67). These data suggest that AES cooperates with NUP98-HOXA9 in the induction of leukemogenesis by inhibiting the functions of other TLE proteins such as TLE1 and TLE4. Another example of the role of TLE proteins in hematopoiesis involves the hematopoietically expressed homeobox (HHEX) protein (also known as PRH or Hex). HHEX functions as a transcriptional repressor in early hematopoiesis by recruiting TLE1 (56, 68). The interaction with TLE1 is mediated by the proline-rich N-terminal domain of HHEX (56). This domain is lost as a result of the AML-associated NUP98-HHEX fusion (12). These data suggest that loss of the ability of HHEX to recruit TLE1 plays a role in leukemogenesis. In this context, it is of interest to note that AES counteracts transcriptional repression by HHEX, presumably by titrating away TLE1 (56). Thus it is possible that the ability of AES to cooperate with NUP98-HOXA9 is due in part to titrating TLE1 away from HHEX transcriptional repression complexes.
The opposing effects of AES and NUP98-HOXA9 on NFκB-mediated transcription may play a role in their opposing effects on erythroid differentiation (Fig. 11 and supplemental Fig. S6). We have previously shown that NUP98-HOXA9 up-regulates NFκB-mediated transcription (28). NFκB plays a role in erythroid differentiation: it is overexpressed in early erythroid progenitors but is down-regulated during erythroid maturation (59, 69). Consistent with its up-regulation of NFκB, NUP98-HOXA9 induces erythroid hyperplasia with a shift to erythroid immaturity (21) (Fig. 11B). These effects are counteracted by AES, which is known to inhibit NFκB-mediated transcription (34).
To summarize, we have identified AES as a novel interacting partner for NUP98-HOXA9 and show that it can cooperate with NUP98-HOXA9 in transcriptional regulation and cell transformation. These data provide a rationale for future studies to explore the role of AES and other TLE proteins in aberrant transcriptional regulation by NUP98 fusions and determine whether abnormalities in the TLE/Groucho system contribute to human AML caused by NUP98 gene rearrangements. In addition, recent data showing cooperation between AES and the AML1-ETO oncogene in inducing self-renewal of hematopoietic progenitor cells suggest that AES may have a more general role in the pathogenesis of AML (41).
Supplementary Material
Acknowledgments
We thank Dr. Akiko Takeda for useful comments on the manuscript. We thank the Alvin J. Siteman Cancer Center at Washington University School of Medicine and Barnes-Jewish Hospital, St. Louis, MO, for the use of the High Speed Cell Sorter Core, which provided flow cytometry analysis and sorting services.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 HL082549 and K02 HL084179 (to N. R. Y.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S6 and Tables S1–S3.
- AML
- acute myeloid leukemia
- CFC
- colony-forming cell
- AES
- amino-terminal enhancer of split
- TLE
- transducin-like enhancer of split
- HHEX
- hematopoietically expressed homeobox
- IMDM
- Iscove's modified Dulbecco's medium
- SB
- SOS-binding protein
- DPBS
- Dulbecco's phosphate-buffered saline
- MTS
- 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-M2-(4-sulfophenyl)-2H-tetrazolium.
REFERENCES
- 1. Terry L. J., Wente S. R. (2009) Eukaryot. Cell 8, 1814–1827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Radu A., Moore M. S., Blobel G. (1995) Cell 81, 215–222 [DOI] [PubMed] [Google Scholar]
- 3. Powers M. A., Forbes D. J., Dahlberg J. E., Lund E. (1997) J. Cell Biol. 136, 241–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Romana S. P., Radford-Weiss I., Ben Abdelali R., Schluth C., Petit A., Dastugue N., Talmant P., Bilhou-Nabera C., Mugneret F., Lafage-Pochitaloff M., Mozziconacci M. J., Andrieu J., Lai J. L., Terre C., Rack K., Cornillet-Lefebvre P., Luquet I., Nadal N., Nguyen-Khac F., Perot C., Van den Akker J., Fert-Ferrer S., Cabrol C., Charrin C., Tigaud I., Poirel H., Vekemans M., Bernard O. A., Berger R. (2006) Leukemia 20, 696–706 [DOI] [PubMed] [Google Scholar]
- 5. Tosi S., Ballabio E., Teigler-Schlegel A., Boultwood J., Bruch J., Harbott J. (2005) Genes Chromosomes Cancer 44, 225–232 [DOI] [PubMed] [Google Scholar]
- 6. Nebral K., Schmidt H. H., Haas O. A., Strehl S. (2005) Clin. Cancer Res. 11, 6489–6494 [DOI] [PubMed] [Google Scholar]
- 7. van Zutven L. J., Onen E., Velthuizen S. C., van Drunen E., von Bergh A. R., van den Heuvel-Eibrink M. M., Veronese A., Mecucci C., Negrini M., de Greef G. E., Beverloo H. B. (2006) Genes Chromosomes Cancer 45, 437–446 [DOI] [PubMed] [Google Scholar]
- 8. Panagopoulos I., Kerndrup G., Carlsen N., Strömbeck B., Isaksson M., Johansson B. (2007) Br. J. Haematol. 136, 294–296 [DOI] [PubMed] [Google Scholar]
- 9. Reader J. C., Meekins J. S., Gojo I., Ning Y. (2007) Leukemia 21, 842–844 [DOI] [PubMed] [Google Scholar]
- 10. Pan Q., Zhu Y. J., Gu B. W., Cai X., Bai X. T., Yun H. Y., Zhu J., Chen B., Weng L., Chen Z., Xue Y. Q., Chen S. J. (2008) Oncogene 27, 3414–3423 [DOI] [PubMed] [Google Scholar]
- 11. Ishikawa M., Yagasaki F., Okamura D., Maeda T., Sugahara Y., Jinnai I., Bessho M. (2007) Int. J. Hematol. 86, 238–245 [DOI] [PubMed] [Google Scholar]
- 12. Jankovic D., Gorello P., Liu T., Ehret S., La Starza R., Desjobert C., Baty F., Brutsche M., Jayaraman P. S., Santoro A., Mecucci C., Schwaller J. (2008) Blood 111, 5672–5682 [DOI] [PubMed] [Google Scholar]
- 13. Yamamoto M., Kakihana K., Kurosu T., Murakami N., Miura O. (2005) Cancer Genet. Cytogenet. 157, 104–108 [DOI] [PubMed] [Google Scholar]
- 14. Gorello P., Brandimarte L., La Starza R., Pierini V., Bury L., Rosati R., Martelli M. F., Vandenberghe P., Wlodarska I., Mecucci C. (2008) Haematologica 93, 1398–1401 [DOI] [PubMed] [Google Scholar]
- 15. Kaltenbach S., Soler G., Barin C., Gervais C., Bernard O. A., Penard-Lacronique V., Romana S. P. (2010) Blood 116, 2332–2335 [DOI] [PubMed] [Google Scholar]
- 16. Petit A., Ragu C., Della-Valle V., Mozziconacci M. J., Lafage-Pochitaloff M., Soler G., Schluth C., Radford I., Ottolenghi C., Bernard O. A., Penard-Lacronique V., Romana S. P. (2010) Leukemia 24, 654–658 [DOI] [PubMed] [Google Scholar]
- 17. Slape C., Lin Y. W., Hartung H., Zhang Z., Wolff L., Aplan P. D. (2008) J. Natl. Cancer Inst. Monogr. 2008, 64–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang G. G., Cai L., Pasillas M. P., Kamps M. P. (2007) Nat. Cell Biol. 9, 804–812 [DOI] [PubMed] [Google Scholar]
- 19. Kroon E., Thorsteinsdottir U., Mayotte N., Nakamura T., Sauvageau G. (2001) EMBO J. 20, 350–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Calvo K. R., Sykes D. B., Pasillas M. P., Kamps M. P. (2002) Oncogene 21, 4247–4256 [DOI] [PubMed] [Google Scholar]
- 21. Takeda A., Goolsby C., Yaseen N. R. (2006) Cancer Res. 66, 6628–6637 [DOI] [PubMed] [Google Scholar]
- 22. Yassin E. R., Sarma N. J., Abdul-Nabi A. M., Dombrowski J., Han Y., Takeda A., Yaseen N. R. (2009) PLoS One 4, e6719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yassin E. R., Abdul-Nabi A. M., Takeda A., Yaseen N. R. (2010) Leukemia 24, 1001–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kasper L. H., Brindle P. K., Schnabel C. A., Pritchard C. E., Cleary M. L., van Deursen J. M. (1999) Mol. Cell. Biol. 19, 764–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ghannam G., Takeda A., Camarata T., Moore M. A., Viale A., Yaseen N. R. (2004) J. Biol. Chem. 279, 866–875 [DOI] [PubMed] [Google Scholar]
- 26. Bai X. T., Gu B. W., Yin T., Niu C., Xi X. D., Zhang J., Chen Z., Chen S. J. (2006) Cancer Res. 66, 4584–4590 [DOI] [PubMed] [Google Scholar]
- 27. Bei L., Lu Y., Eklund E. A. (2005) J. Biol. Chem. 280, 12359–12370 [DOI] [PubMed] [Google Scholar]
- 28. Takeda A., Sarma N. J., Abdul-Nabi A. M., Yaseen N. R. (2010) J. Biol. Chem. 285, 16248–16257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Oka M., Asally M., Yasuda Y., Ogawa Y., Tachibana T., Yoneda Y. (2010) Mol. Biol. Cell 21, 1885–1896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Vidalain P. O., Boxem M., Ge H., Li S., Vidal M. (2004) Methods 32, 363–370 [DOI] [PubMed] [Google Scholar]
- 31. Zhong H., Takeda A., Nazari R., Shio H., Blobel G., Yaseen N. R. (2005) J. Biol. Chem. 280, 10675–10682 [DOI] [PubMed] [Google Scholar]
- 32. Miyasaka H., Choudhury B. K., Hou E. W., Li S. S. (1993) Eur. J. Biochem. 216, 343–352 [DOI] [PubMed] [Google Scholar]
- 33. Ren B., Chee K. J., Kim T. H., Maniatis T. (1999) Genes Dev. 13, 125–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tetsuka T., Uranishi H., Imai H., Ono T., Sonta S., Takahashi N., Asamitsu K., Okamoto T. (2000) J. Biol. Chem. 275, 4383–4390 [DOI] [PubMed] [Google Scholar]
- 35. Yu X., Li P., Roeder R. G., Wang Z. (2001) Mol. Cell. Biol. 21, 4614–4625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fontoura B. M., Dales S., Blobel G., Zhong H. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 3208–3213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Powers M. A., Macaulay C., Masiarz F. R., Forbes D. J. (1995) J. Cell Biol. 128, 721–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schwartz T. U. (2005) Curr. Opin. Struct. Biol. 15, 221–226 [DOI] [PubMed] [Google Scholar]
- 39. Abdul-Nabi A. M., Yassin E. R., Varghese N., Deshmukh H., Yaseen N. R. (2010) PLoS One 5, e12464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chung K. Y., Morrone G., Schuringa J. J., Plasilova M., Shieh J. H., Zhang Y., Zhou P., Moore M. A. (2006) Cancer Res. 66, 11781–11791 [DOI] [PubMed] [Google Scholar]
- 41. Steffen B., Knop M., Bergholz U., Vakhrusheva O., Rode M., Köhler G., Henrichs M. P., Bulk E., Hehn S., Stehling M., Dugas M., Bäumer N., Tschanter P., Brandts C., Koschmieder S., Berdel W. E., Serve H., Stocking C., Müller-Tidow C. (2011) Blood 117, 4328–4337 [DOI] [PubMed] [Google Scholar]
- 42. Cavallo R. A., Cox R. T., Moline M. M., Roose J., Polevoy G. A., Clevers H., Peifer M., Bejsovec A. (1998) Nature 395, 604–608 [DOI] [PubMed] [Google Scholar]
- 43. Dubnicoff T., Valentine S. A., Chen G., Shi T., Lengyel J. A., Paroush Z., Courey A. J. (1997) Genes Dev. 11, 2952–2957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jiménez G., Paroush Z., Ish-Horowicz D. (1997) Genes Dev. 11, 3072–3082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Paroush Z., Finley R. L., Jr., Kidd T., Wainwright S. M., Ingham P. W., Brent R., Ish-Horowicz D. (1994) Cell 79, 805–815 [DOI] [PubMed] [Google Scholar]
- 46. Roose J., Molenaar M., Peterson J., Hurenkamp J., Brantjes H., Moerer P., van de Wetering M., Destrée O., Clevers H. (1998) Nature 395, 608–612 [DOI] [PubMed] [Google Scholar]
- 47. Tolkunova E. N., Fujioka M., Kobayashi M., Deka D., Jaynes J. B. (1998) Mol. Cell. Biol. 18, 2804–2814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gasperowicz M., Otto F. (2005) J. Cell Biochem. 95, 670–687 [DOI] [PubMed] [Google Scholar]
- 49. Courey A. J., Jia S. (2001) Genes Dev. 15, 2786–2796 [DOI] [PubMed] [Google Scholar]
- 50. Chen G., Fernandez J., Mische S., Courey A. J. (1999) Genes Dev. 13, 2218–2230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhang H., Emmons S. W. (2002) Genetics 160, 799–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Stifani S., Blaumueller C. M., Redhead N. J., Hill R. E., Artavanis-Tsakonas S. (1992) Nat. Genet. 2, 119–127 [DOI] [PubMed] [Google Scholar]
- 53. Fisher A. L., Caudy M. (1998) Genes Dev. 12, 1931–1940 [DOI] [PubMed] [Google Scholar]
- 54. Mallo M., Franco del Amo F., Gridley T. (1993) Mech. Dev. 42, 67–76 [DOI] [PubMed] [Google Scholar]
- 55. Pinto M., Lobe C. G. (1996) J. Biol. Chem. 271, 33026–33031 [DOI] [PubMed] [Google Scholar]
- 56. Swingler T. E., Bess K. L., Yao J., Stifani S., Jayaraman P. S. (2004) J. Biol. Chem. 279, 34938–34947 [DOI] [PubMed] [Google Scholar]
- 57. Wang J. C., Waltner-Law M., Yamada K., Osawa H., Stifani S., Granner D. K. (2000) J. Biol. Chem. 275, 18418–18423 [DOI] [PubMed] [Google Scholar]
- 58. Wang W., Wang Y. G., Reginato A. M., Glotzer D. J., Fukai N., Plotkina S., Karsenty G., Olsen B. R. (2004) Dev. Biol. 270, 364–381 [DOI] [PubMed] [Google Scholar]
- 59. Zhang M. Y., Sun S. C., Bell L., Miller B. A. (1998) Blood 91, 4136–4144 [PubMed] [Google Scholar]
- 60. Griffis E. R., Xu S., Powers M. A. (2003) Mol. Biol. Cell 14, 600–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Hodel A. E., Hodel M. R., Griffis E. R., Hennig K. A., Ratner G. A., Xu S., Powers M. A. (2002) Mol. Cell 10, 347–358 [DOI] [PubMed] [Google Scholar]
- 62. Griffis E. R., Altan N., Lippincott-Schwartz J., Powers M. A. (2002) Mol. Biol. Cell 13, 1282–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Radu A., Blobel G., Moore M. S. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 1769–1773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Griffis E. R., Craige B., Dimaano C., Ullman K. S., Powers M. A. (2004) Mol. Biol. Cell 15, 1991–2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Nakielny S., Shaikh S., Burke B., Dreyfuss G. (1999) EMBO J. 18, 1982–1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Fraga M. F., Berdasco M., Ballestar E., Ropero S., Lopez-Nieva P., Lopez-Serra L., Martín-Subero J. I., Calasanz M. J., Lopez de Silanes I., Setien F., Casado S., Fernandez A. F., Siebert R., Stifani S., Esteller M. (2008) Cancer Res. 68, 4116–4122 [DOI] [PubMed] [Google Scholar]
- 67. Dayyani F., Wang J., Yeh J. R., Ahn E. Y., Tobey E., Zhang D. E., Bernstein I. D., Peterson R. T., Sweetser D. A. (2008) Blood 111, 4338–4347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Guiral M., Bess K., Goodwin G., Jayaraman P. S. (2001) J. Biol. Chem. 276, 2961–2970 [DOI] [PubMed] [Google Scholar]
- 69. Liu J. J., Hou S. C., Shen C. K. (2003) J. Biol. Chem. 278, 19534–19540 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.