Background: Cdt1 is a DNA replication factor that loads MCM helicase onto chromatin. SNF2H is a chromatin remodeler.
Results: Recruitment of SNF2H to replication origins is Cdt1-dependent and promotes MCM loading.
Conclusion: SNF2H may contribute to maintenance of genome integrity through promotion of MCM loading.
Significance: Clarifying the roles of SNF2H in the process of MCM loading is crucial to understanding DNA replication.
Keywords: Cell Cycle, Chromatin Immunoprecipitation (ChIP), Chromatin Remodeling, DNA Replication, Mammal, Cdt1, MCM, SNF2H, Pre-replication Complex
Abstract
From late mitosis to the G1 phase of the cell cycle, ORC, CDC6, and Cdt1 form the machinery necessary to load MCM2–7 complexes onto DNA. Here, we show that SNF2H, a member of the ATP-dependent chromatin-remodeling complex, is recruited onto DNA replication origins in human cells in a Cdt1-dependent manner and positively regulates MCM loading. SNF2H physically interacted with Cdt1. ChIP assays indicated that SNF2H associates with replication origins specifically during the G1 phase. Binding of SNF2H at origins was decreased by Cdt1 silencing and, conversely, enhanced by Cdt1 overexpression. Furthermore, SNF2H silencing prevented MCM loading at origins and moderately inhibited S phase progression. Although neither SNF2H overexpression nor SNF2H silencing appeared to impact rereplication induced by Cdt1 overexpression, Cdt1-induced checkpoint activation was inhibited by SNF2H silencing. Collectively, these data suggest that SNF2H may promote MCM loading at DNA replication origins via interaction with Cdt1 in human cells. Because efficient loading of excess MCM complexes is thought to be required for cells to tolerate replication stress, Cdt1- and SNF2H-mediated promotion of MCM loading may be biologically relevant for the regulation of DNA replication.
Introduction
In eukaryotes, DNA replication is strictly controlled so that the genome is replicated once per single cell cycle. From late mitosis to the G1 phase, the sequential assembly of multiple proteins, including ORC1–6 (origin recognition complex 1–6), CDC6, Cdt1 (Cdc10-dependent transcript 1), and MCM2–7 (minichromosome maintenance 2–7), results in the formation of a pre-replication complex (pre-RC)3 that is “licensed” for replication. In the latter period of the cell cycle, whereas MCM helicase is activated in a Cdk (cyclin-dependent kinase)-dependent manner, the activity of the MCM loaders is carefully regulated by multiple mechanisms so as to prohibit inappropriate reassembly of pre-RC and subsequent rereplication (1–3). Cdt1 strongly stimulates the licensing reaction in human cells (4, 5), and its activity is very tightly restricted by multiple mechanisms (3, 6–12). The overexpression of Cdt1, ORC1, or CDC6 alone induces no detectable rereplication. However, the overexpression of Cdt1 + ORC1 or Cdt1 + CDC6 yields a moderate level of rereplication, whereas joint overexpression of Cdt1 + ORC1 + CDC6 yields strong rereplication (5). In addition, the overexpression of Cdt1 induces ATM (ataxia telangiectasia mutated)/Chk2 checkpoint activation. This activation is apparently independent of rereplication induction, although its precise mechanism is unknown (13). Therefore, deregulation of Cdt1 is harmful to genome stability (13, 14). However, it is not fully understood how Cdt1 strongly stimulates the MCM loading reaction.
The initiation of DNA replication is strongly influenced by chromatin structure, as is expected for any molecular events involving DNA in eukaryotic cells. For example, histone acetylation is linked to the control of initiation of DNA replication. Early firing origins are typically localized in genomic regions that are transcribed and contain hyperacetylated chromatin, whereas late firing origins lie in silenced heterochromatic domains (15–19). In addition, histone acetylation is involved in origin activation during early development in Xenopus (20) and at the chorion gene loci in Drosophila follicle cells (21, 22).
In pre-RC formation, the efficient loading of multiple MCM complexes is required for the toleration of replication stresses and activation of checkpoint pathways (23–25). In general, chromatin-remodeling proteins, histone chaperones, and histone acetylation enzymes are thought to act synergistically to stimulate transcription on chromatin templates (26). The situation may be the same for efficient MCM loading. In this regard, HBO1 (a MYST family histone acetyltransferase that binds to ORC), originally identified through its physical interactions with human ORC1 (27), was recently found to associate with replication origins through interaction with Cdt1 and to enhance licensing and DNA replication through its acetylation activity (28–30).
Chromatin-remodeling complexes that utilize energy derived from ATP hydrolysis alter chromatin structure by disrupting and/or mobilizing nucleosomes. This large group of complexes can be subdivided into four subfamilies that include the SWI/SNF-type complex, the ISWI (imitation switch)-type complex, the INO80-type complex, and the CHD-type complex. Each complex contains a major catalytic component that possesses DNA-dependent ATPase activity, such as Brg1 (in the SWI/SNF-type complex) or SNF2H (sucrose nonfermenting 2 homolog; in the ISWI-type complex) (31–35). The selection of catalytic ATPase subunits, combined with other complex components, defines the role of these complexes in various nuclear events, including transcription, DNA replication, and DNA repair.
The ISWI-type nucleosome-remodeling factor SNF2H and Williams syndrome transcription factor (WSTF) were identified previously as novel human Cdt1-binding proteins (12). However, the biological significance of the interaction with Cdt1 remains unclear. SNF2H uses ATP hydrolysis to regulate chromatin structure and modulate nucleosome spacing (31–35). It is a constituent of several multiprotein remodeling complexes. These include WICH (WSTF/ISWI chromatin-remodeling complex), ACF, CHRAC, RSF, and NoRC (31–35). The presence of these distinct complexes suggests that SNF2H performs multiple functions in chromatin regulation. Moreover, several previous reports implicate SNF2H in stimulating the initiation of DNA replication. For example, CHRAC allows binding of T-antigen and efficient initiation in an in vitro replication system that employs SV40 DNA reconstituted into chromatin (36). Furthermore, SNF2H is apparently recruited to the Epstein-Barr virus origin of plasmid replication (oriP), and depletion of SNF2H with siRNAs reduces MCM3 loading at oriP (37). However, it remains unclear whether this is also the case for cellular replication origins, and, if so, how SNF2H is recruited.
In this study, the hypothesis was explored that SNF2H proteins might play a role in the stimulation of MCM loading onto cellular replication origins and that the Cdt1-SNF2H interaction is important in this context. On the basis of the data obtained, we propose that SNF2H promotes MCM loading at cellular replication origins through interaction with Cdt1.
EXPERIMENTAL PROCEDURES
Cell Culture and Synchronization
HEK293T, T98G, and HeLa cells were grown in Dulbecco's modified Eagle's medium with 8% fetal calf serum. For cell cycle synchronization, T98G cells were rendered quiescent by serum starvation for 48 h and then released into the cell cycle by serum stimulation. Synchronization was verified by analysis of DNA contents with a flow cytometer.
Plasmids
Mammalian expression vectors pCLMSCVhyg-T7-Cdt1, pcDNA3.1–3HA-Cdt1, and pcDNA3.1-zeo-FLAG-ORC1 and bacterial expression vector pGEX-6P1-Cdt1 were described previously (5, 12). The pEGFP-C1 expression vector was purchased from Clontech. pCMV-FLAG-HBO1 was described previously (38). The pCMV6-XL4-SNF2H expression vector, which expresses human SNF2H, was purchased from OriGene Technologies (Rockville, MD). To construct the T7 promoter-driven SNF2H expression vector, pCMV6-XL4-SNF2H was digested with NotI, and the SNF2H fragment was subcloned into pCMV6-XL5. The SNF2H protein was then synthesized by in vitro transcription-translation with rabbit reticulocyte lysate (TnT T7 quick coupled transcription/translation system, Promega, Madison, WI) according to the manufacturer's instructions.
Immunoprecipitation
Nuclear extracts were prepared from HEK293T cells with modified cytoskeleton buffer containing 500 mm NaCl, 0.1% Triton X-100, 0.1 mm ATP, 1 mm DTT, and multiple protease inhibitors as described previously (12, 39). Aliquots of the extracts were then immunoprecipitated using anti-Cdt1 or anti-SNF2H antibody bound to protein G-Sepharose beads (Amersham Biosciences). The beads were washed four times with NET gel buffer (12) containing 200 mm NaCl. The immunoprecipitates were eluted with 1× SDS sample buffer (62.5 mm Tris-HCl (pH 6.8), 2% SDS, 5% β-mercaptoethanol, 10% glycerol, and 0.01% bromphenol blue) and subjected to immunoblotting.
GST-Cdt1 Pulldown Assay
For Fig. 1E, GST-Cdt1 (0.6 μg) was mixed with SNF2H produced by in vitro translation in 50 μl of reaction mixture, diluted with binding buffer A (25 mm Tris-HCl (pH 7.4), 150 mm NaCl, 0.01% Nonidet P-40, 1 mm DTT, 0.5 mm PMSF, and 10% glycerol) containing 10 mm glycerophosphate and 5 mm NaF, and incubated at 4 °C for 3 h. GST-Cdt1 and associated SNF2H proteins were collected on glutathione beads and washed with buffer A. The bound proteins were eluted and analyzed by immunoblotting or Coomassie Brilliant Blue staining.
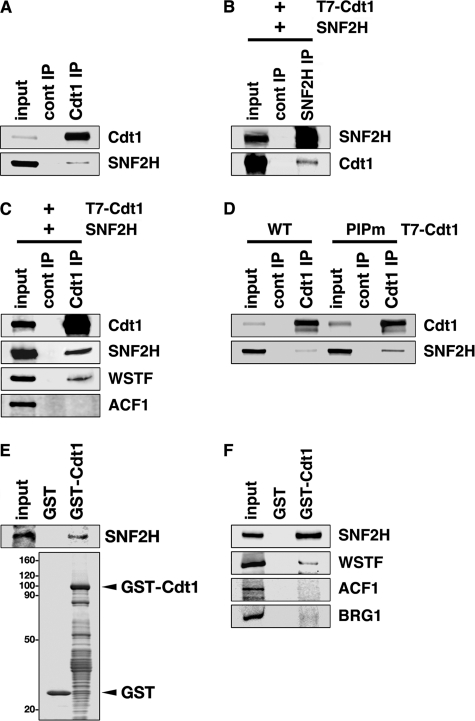
FIGURE 1.
Cdt1 interacts with SNF2H. A, nuclear extracts were prepared from HEK293T cells and immunoprecipitated with anti-Cdt1 antibody (Cdt1 IP) or control rabbit IgG (cont IP). Immunoprecipitates were subjected to immunoblotting with the indicated antibodies. Three percent of the input was also loaded. B, HEK293T cells were transfected with T7-Cdt1 and SNF2H expression vectors, and nuclear extracts were prepared. After immunoprecipitation with anti-SNF2H antibody, the precipitates were blotted with the indicated antibodies. One percent of the input for SNF2H or 0.3% of the input for Cdt1 was loaded. C, HEK293T cells were transfected with T7-Cdt1 and SNF2H expression vectors, and nuclear extracts were prepared. After immunoprecipitation with anti-Cdt1 antibody, the precipitates were immunoblotted with the indicated antibodies. One percent of the input was loaded. D, HEK293T cells were transfected with the indicated expression vectors, and nuclear extracts were prepared. After immunoprecipitation with anti-Cdt1 antibody or control rabbit IgG, the precipitates were immunoblotted with the indicated antibodies. Three percent of the input was loaded. E, GST-Cdt1 or GST was incubated with SNF2H protein synthesized by in vitro transcription-translation with rabbit reticulocyte lysate. GST-Cdt1 and the associated SNF2H protein were then collected on glutathione beads and subjected either to immunoblotting with anti-SNF2H antibody (upper panel) or to Coomassie Brilliant Blue staining (lower panel). Twenty percent of the input sample was analyzed. F, GST-Cdt1 or GST was incubated with HeLa nuclear extracts, and bound proteins were analyzed by immunoblotting with the indicated antibodies. Fifteen percent of the input was loaded.
For Fig. 1F, GST-Cdt1 was bound to glutathione beads, and the beads were mixed with diluted nuclear extracts prepared from HeLa cells. After washing three times with buffer A, the bound proteins were eluted and analyzed by immunoblotting.
Transfection
For Fig. 1 (B–D), expression plasmids (total ∼6 μg) were transiently transfected into 3 × 106 HEK293T cells in 100-mm culture dishes using TransIT-293 (Mirus, Madison, WI) according to the manufacturer's instructions. Forty-eight hours after transfection, cells were harvested and subjected to immunoprecipitation analysis.
For Fig. 3 (D–F), expression plasmids (total ∼12 μg) and 1.8 μg of pMSCVpuro (Clontech) were transiently transfected into 6 × 106 HEK293T cells in 150-mm culture dishes using TransIT-293. Twenty-four hours after transfection, cells were selected with 2 μg/ml puromycin for 2 days. Cells were then harvested and subjected to ChIP assay.
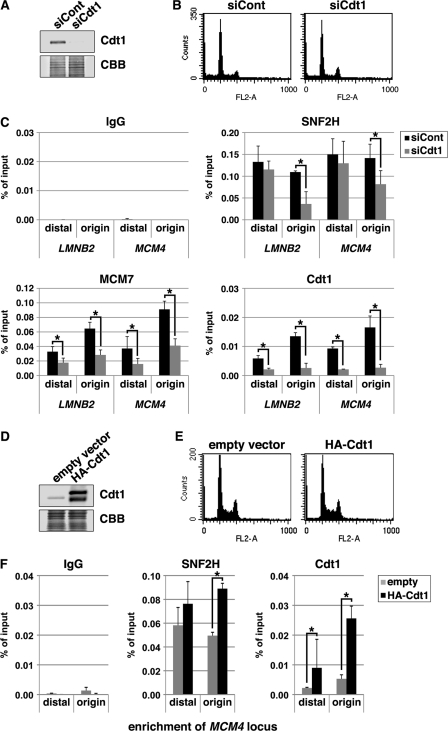
FIGURE 3.
SNF2H binding to origins is inhibited by Cdt1 silencing and enhanced by Cdt1 overexpression. A–C, HeLa cells transfected with control (DS scrambled-Neg; siCont) or Cdt1 (siCdt1) siRNAs for 48 h were subjected to immunoblotting (A), FACS analysis (B), or ChIP assay (C). D--F, HEK293T cells transfected with the indicated expression vectors were analyzed by immunoblotting (D), FACS analysis (E), or ChIP assay (F). Coomassie Brilliant Blue (CBB) staining served as the loading control. For ChIP data, the means ± S.D. are shown (n = 3). *, p < 0.05.
For Fig. 5, expression plasmids (total ∼1.6 μg) and siRNA (40 pmol) were transiently transfected into 2 × 105 HEK293T cells in 12-well culture plates using Lipofectamine 2000 (Invitrogen). Forty-eight hours after transfection, cells were lysed in 1× SDS sample buffer containing multiple protease and phosphatase inhibitors and then subjected to immunoblotting.
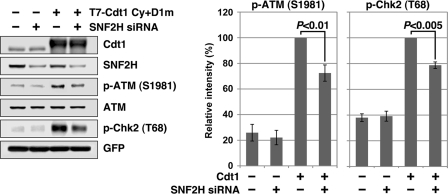
FIGURE 5.
Cdt1 overexpression-induced ATM activation is decreased by SNF2H silencing. HEK293T cells were transfected with the T7-Cdt1 Cy+D1m expression vector and SNF2H siRNAs using Lipofectamine 2000 along with a plasmid expressing GFP to allow monitoring of the transfection efficiency. GL2 siRNAs were used as the control. Whole cell lysates were prepared 48 h after transfection and immunoblotted with the indicated antibodies. The signal intensity of each band was quantified, with the intensity of the band corresponding to cells transfected with T7-Cdt1 and treated with control siRNAs set at 100%. The means ± S.D. are shown (n = 3).
siRNA Experiments
HeLa and T98G cells were transfected with siRNA duplexes using HiPerFect transfection reagent (Qiagen) according to the manufacturer's instructions. siRNA oligonucleotides were synthesized (IDT, Coralville, IA, or Dharmacon, Lafayette, Co) with the following sequences (sense strands): Cdt1, 5′-GCUGUUGUACUAUCAUGAGCCCUGGdAdC-3′; SNF2H, 5′-GAGGAGGAUGAAGAGCUAUdTdT-3′; CDC6-1, 5′-AGACUAUAACUCUACAGAUUGUGdAdA-3′; CDC6-3, 5′-GGAGGACACUGGUUAAAGAAUUUdAdT-3′; ORC1, 5′-CUGCACUACCAAACCUAUAdTdT-3′; HBO1, 5′-GGGAUAAGCAGAUAGAAGAAAGGAT-3′; control DS scrambled-Neg, 5′-CUUCCUCUCUUUCUCUCCCUUGUdGdA-3′; and control GL2, 5′-CGUACGCGGAAUACUUCGAdTdT-3′.
FACS
Cells were treated with a CycletestTM Plus DNA reagent kit (BD Biosciences) for propidium iodide staining and then analyzed with a BD FACSCalibur.
ChIP
ChIP assays were carried out essentially as described previously (40). Briefly, 6 × 106 cells were fixed with 1% formaldehyde and lysed in SDS lysis buffer (1% SDS, 10 mm EDTA, and 50 mm Tris-HCl (pH 8.0)). Chromatin was sonicated to yield an average fragment size of 0.5 kb. Each sample was incubated overnight at 4 °C with 5 μg of the indicated antibodies or control IgGs and then collected using Dynabeads (Invitrogen). After formaldehyde reversal, phenol/chloroform extraction, and ethanol precipitation, the DNA was dissolved in Tris/EDTA buffer.
Quantitative Real-time PCR Analysis
SYBR® Premix Ex TaqTM II (Takara, Kyoto, Japan) was used according to the manufacturer's instructions. All PCRs were carried out using an iCycler iQ real-time PCR detection system (Bio-Rad). The annealing temperature was optimized for each primer (supplemental Table S1). For each reaction, the cycling parameters were set as follows: 1 min at 95 °C; five cycles at 95 °C for 30 s, annealing temperature plus 4 °C for 30 s, and 72 °C for 30 s; five cycles at 95 °C for 30 s, annealing temperature plus 2 °C for 30 s, and 72 °C for 30 s; and 50 cycles at 95 °C for 30 s, annealing temperature for 30 s, and 72 °C for 30 s.
Chromatin Binding Assay
The chromatin binding assay was performed as described previously (5, 39).
Immunoblotting and Antibodies
Immunoblotting and quantification of band signals were performed as described previously (12). Preparation of rabbit polyclonal antibodies against human Cdt1, ORC1, CDC6, MCM7, and MCM3 was described previously (10, 13, 41). Anti-human Brg1 antibody was obtained by immunizing rabbits with a peptide corresponding to the 50-amino acid C-terminal region of Brg1.
Other antibodies were purchased from various companies: GFP (catalog no. 46-0092, Invitrogen), Thr68-phosphorylated Chk2 (catalog no. 2661) and WSTF (catalog no. 2152) (Cell Signaling Technology, Danvers, MA), ATM (clone 2C1, GeneTex, San Antonio, TX), Ser1981-phosphorylated ATM (catalog no. 200-301-400, Rockland Biosciences, Gilbertsville, PA), SNF2H (clone 1B9/D12, Upstate Biotechnology, Lake Placid, NY; or clone H-300, catalog no. sc-13054, Santa Cruz Biotechnology, Santa Cruz, CA), ACF1 (catalog no. A301–319A, Bethyl Laboratories, Montgomery, TX), cyclin E (catalog no. sc-247) and HBO1 (catalog no. sc-13284) (Santa Cruz Biotechnology), and Ser780-phosphorylated Rb (catalog no. 555S, MBL International Corp.).
Statistical Analysis
Statistical analysis was performed using Student's two-tailed t test to validate the data. p values <0.05 were considered statistically significant.
RESULTS
SNF2H Interacts with Cdt1 in Vivo and in Vitro
The chromatin-remodeling proteins SNF2H and WSTF were identified previously as novel Cdt1-binding proteins by a combination of affinity chromatography and tandem mass spectrometry analyses (12). In addition, SNF2H and WSTF were shown to bind to GST-Cdt1 by pulldown assays with HeLa cell nuclear extracts, and ectopically expressed T7-Cdt1 was shown to co-immunoprecipitate with endogenous SNF2H (12). Here, we further confirmed the association between endogenous SNF2H and Cdt1. As shown in Fig. 1A, SNF2H co-immunoprecipitated with Cdt1. Reciprocally, ectopically expressed T7-Cdt1 co-immunoprecipitated with SNF2H (Fig. 1B).
The WICH complex binds to proliferating cell nuclear antigen (PCNA) through a PCNA-WSTF interaction (42). In addition, Cdt1 binds to PCNA (11). Therefore, the observed interaction between Cdt1 and SNF2H may be mediated by PCNA-WSTF binding. Indeed, WSTF was also detected in the T7-Cdt1 immunoprecipitates (Fig. 1C). However, WSTF was not enriched to the same extent as SNF2H (Fig. 1, C and F). Moreover, the Cdt1 PIPm mutant, which is unable to interact with PCNA (11), also co-immunoprecipitated with SNF2H (Fig. 1D). These results suggest that Cdt1 likely binds to SNF2H without mediation by other factors. To evaluate this hypothesis, purified GST-Cdt1 fusion proteins were incubated with SNF2H synthesized by in vitro transcription-translation with rabbit reticulocyte lysate. A direct interaction between the two proteins in vitro was demonstrated (Fig. 1E).
SNF2H is partnered with divergent proteins in various chromatin-remodeling complexes such as WSTF in the WICH complex (31–35). In addition to previous and current findings of WSTF co-purification with GST-Cdt1 (Fig. 1F) (12), this study showed that WSTF co-immunoprecipitated with T7-Cdt1 (Fig. 1C). WSTF is also a subunit of the WINAC complex, a subclass of the Brg1-dependent chromatin-remodeling complexes (43). Therefore, the possibility that Cdt1 interacts with Brg1 was also investigated. As shown in Fig. 1F, Brg1 did not interact with GST-Cdt1. ACF1 is another SNF2H partner, forming ACF or CHRAC complexes with SNF2H. However, ACF1 was not detected in either T7-Cdt1 immunoprecipitation or GST-Cdt1 pulldown assays (Fig. 1, C and F). Finally, SNF2H and SNF2L are the two ISWI proteins found in human cells (44). However, considering that SNF2L protein is reportedly undetectable because of its low expression levels in HeLa cells (45), SNF2L binding to Cdt1 was not explored. Taken together, these data demonstrate that SNF2H specifically interacts with Cdt1 and that the WSTF-containing WICH complex may be the predominant SNF2H-related complex involved. However, the possibility cannot be ruled out that additional SNF2H-containing complexes may also interact with Cdt1.
SNF2H Binds to Replication Origins in Human Cells
Cdt1 is a key component of pre-RC that has been shown to be indispensable during the initiation steps of DNA replication. The interaction between Cdt1 and SNF2H observed in this work may thus indicate a functional link between chromatin remodeling and pre-RC formation. To assess this possibility directly, the presence of SNF2H at regions of known replication origins was first examined by ChIP assay. Chromatin prepared from asynchronously growing cells was precipitated with the indicated antibodies, and the presence of the well characterized lamin B2 and MCM4 origin sequences (46–50) was assessed by subsequent quantitative real-time PCR analysis using specific sets of primers (Fig. 2A). In two different human cell lines, HeLa and T98G, Cdt1-, MCM7-, or MCM3-specific antibodies efficiently immunoprecipitated DNA sequences that included origin regions of both loci (Fig. 2, B and C). Although some enrichment for the regions distal to the origins was also observed, it was less efficient compared with the enrichment of the origin regions. In human cells, replication origins are not determined as strictly as those in budding yeast (51). Thus, “origin regions” may in fact represent “preferential origins.” In other words, pre-RCs may be formed at the origin-distal regions with lower frequency than the origins. Overall, these data are in good agreement with previous reports (49, 50, 52, 53).
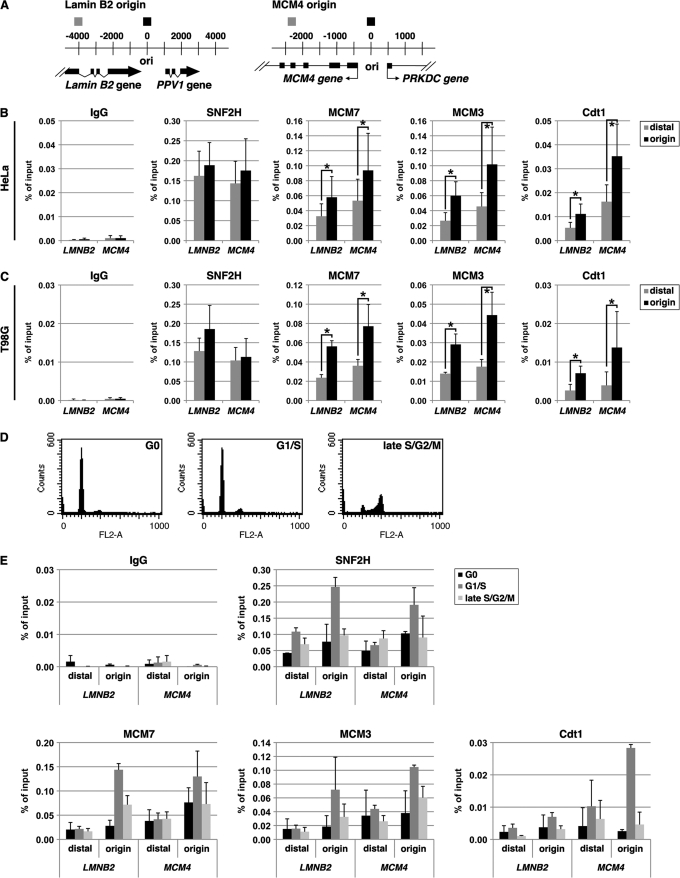
FIGURE 2.
SNF2H interacts with cellular replication origins during the G1 phase. ChIP assay was performed as described under “Experimental Procedures.” Quantitative real-time PCR was performed with DNA templates extracted from chromatin precipitated with control IgGs or antibody against SNF2H, MCM7, MCM3, or Cdt1. A, schematic diagram of the human MCM4 and lamin B2 origin loci. Positions of primer pairs to detect the origins and distal sequences are shown above (black and gray boxes, respectively). Coordinates flanking the origin are given in base pairs. B, results with asynchronously growing HeLa cells are shown as the percent of input DNA. The means ± S.D. are shown (n = 8). C, results with asynchronously growing T98G cells. The means ± S.D. are shown (n = 6). *, p < 0.05. D and E, T98G cells synchronized at the G0 (serum starvation for 48 h), G1/S (10 h after serum addition), and late S/G2/M (24 h in the presence of 50 ng/ml nocodazole after serum addition) phases were examined by FACS analysis (D) or ChIP assay (E). For ChIP data, the means ± S.D. are shown (n = 3).
On the other hand, the anti-SNF2H antibodies efficiently enriched both the origin and distal sequences of lamin B2 and MCM4 loci (Fig. 2, B and C). The binding of SNF2H to the origin-distal regions may be due to its multifunctionality, i.e. SNF2H binds to chromatin not only for the purpose of regulating replication but also to mediate transcription and other functions. Nevertheless, it seems clear that SNF2H does indeed bind to replication origins. Furthermore, SNF2H silencing with siRNAs decreased the origin enrichment in ChIP assay (see Fig. 4), confirming the validity of the assay. Therefore, the origin binding of SNF2H was the focus of further analysis.
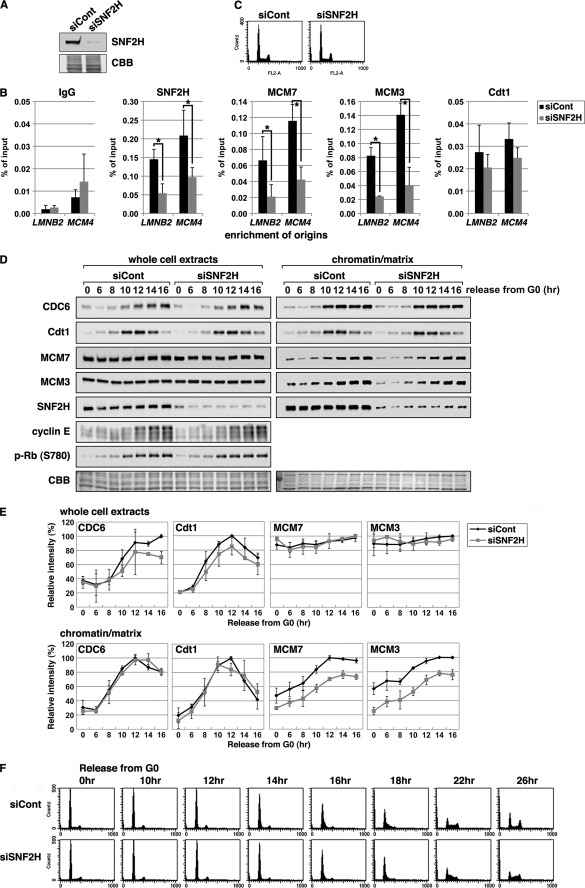
FIGURE 4.
Silencing of SNF2H suppresses MCM, but not Cdt1, loading at origins. A–C, HeLa cells transfected with control (GL2; siCont) or SNF2H (siSNF2H) siRNAs for 48 h were subjected to immunoblotting (A), ChIP assay (B), or FACS analysis (C). Coomassie Brilliant Blue (CBB) staining served as the loading control. For ChIP data, the means ± S.D. are shown (n = 3). *, p < 0.05. D–F, T98G cells were transfected with the siRNAs for 24 h, rendered quiescent by serum starvation for 48 h, and then stimulated to reenter the cell cycle by serum addition. Cells were harvested at the indicated times and subjected to chromatin binding assay (D and E) or FACS analysis (F). D, whole cell extracts and chromatin/matrix-binding fractions were immunoblotted with the indicated antibodies. E, the signal intensities of the pre-RC proteins were quantified, normalized to the signals of Coomassie Brilliant Blue staining, and are shown with the maximal values set at 100%. The means ± S.D. are shown (n = 2).
SNF2H Associates with Replication Origins Specifically during the G1 Phase
Chromatin association of licensing factors reaches its maximum in the G1 phase when DNA licensing is complete. Levels are reduced upon progression into the S phase or cell cycle exit. To monitor the cell cycle dependence of SNF2H localization to replication origins, ChIP assays were performed using T98G cells synchronized in the G1/S phase, and the data were compared with those from cells synchronized in the G0 or late S/G2/M phase (Fig. 2D). The specific enrichment of MCM7, MCM3, or Cdt1 at the lamin B2 and MCM4 origins was maximal in the G1/S phase (Fig. 2E), as reported previously (49, 50, 52, 53). Interestingly, SNF2H levels at replication origins were low in chromatin derived from cells synchronized in the G0 or late S/G2/M phase but were significantly elevated in chromatin derived from cells synchronized in the G1/S phase (Fig. 2E). The association of SNF2H at the origin-distal regions was not significantly altered during the cell cycle progression (Fig. 2E). The pattern of SNF2H binding was similar to that of licensing factors, a finding that is similar to previous results obtained with oriP of Epstein-Barr virus (37). These results suggest that SNF2H is associated with cellular replication origins in the G1 phase in a cell cycle-dependent manner.
Origin Binding of SNF2H Is Dependent on Cdt1
Given the potential role of a Cdt1-SNF2H interaction in SNF2H-origin binding, the dependence of SNF2H loading at the origins on Cdt1 was examined. Chromatin was prepared from HeLa cells treated with control siRNAs or siRNAs targeting Cdt1 and subjected to ChIP assay. Down-regulation of Cdt1 with the Cdt1 siRNA was confirmed by immunoblotting (Fig. 3A). The cell cycle distribution pattern was not remarkably affected by Cdt1 silencing at this level (Fig. 3B). The specific enrichment of the origin sequences with the anti-Cdt1 antibody was also reduced by siRNA treatment (Fig. 3C and supplemental Fig. S1), demonstrating the validity of ChIP assay.
Cdt1 is essential for the loading of hexameric MCM2–7 complexes onto chromatin (1–3). As expected, MCM7 binding at replication origins was decreased by Cdt1 silencing (Fig. 3C). Importantly, SNF2H binging at origins was also significantly decreased by Cdt1 silencing (Fig. 3C and supplemental Fig. S1). The levels of Cdt1 and MCM7 at the origin-distal regions were also lowered by Cdt1 silencing (Fig. 3C). However, SNF2H binding at the origin-distal regions was not remarkably changed by Cdt1 silencing (Fig. 3C). Similar results were obtained with other siRNAs targeting Cdt1 (data not shown).
The hypothesis that Cdt1 overexpression affects origin binding of SNF2H was also investigated. For this purpose, Cdt1 was overexpressed in HEK293T cells (Fig. 3D). The cell cycle distribution pattern was not remarkably affected by Cdt1 overexpression (Fig. 3E), as we reported previously (5). SNF2H binding at the MCM4 origin was enhanced upon Cdt1 overexpression (Fig. 3F). However, SNF2H binding at the distal region was not significantly affected (Fig. 3F).
Taken together, these results suggest that the origin binding of SNF2H is, at least partly, dependent on Cdt1. On the other hand, recruitment of SNF2H to the origin-distal regions may be mediated mostly by other factors.
Silencing SNF2H Suppresses MCM but Not Cdt1 Loading at Origins
The possibility that origin recruitment of SNF2H might affect MCM loading was next investigated. For this purpose, chromatin was prepared from HeLa cells treated with siRNA targeting SNF2H and then subjected to ChIP assay with the indicated antibodies. The efficient depletion of SNF2H was confirmed by both immunoblotting and ChIP assays (Fig. 4, A and B). Remarkable change in cell cycle distribution pattern was not observed in SNF2H-silenced asynchronously growing HeLa cells (Fig. 4C). In line with the previous observation on oriP of Epstein-Barr virus (37), the binding of MCM7 and MCM3 to the cellular origins was suppressed by SNF2H silencing (Fig. 4B). On the other hand, Cdt1 binding at the origins was not affected by SNF2H silencing. These results suggest that SNF2H association with cellular replication origins occurs after the loading of Cdt1 but prior to the loading of the MCM complex and that both Cdt1 and SNF2H are required for the efficient loading of MCM complexes.
The effect of SNF2H silencing on MCM loading was also investigated using a chromatin binding assay. Control and SNF2H-silenced T98G cells were synchronized in the G0 phase by serum deprivation and then reentered into the cell cycle by serum stimulation. Cells were harvested sequentially at the indicated times and subjected to chromatin loading assay to estimate the binding of pre-RC components to intact chromatin (Fig. 4, D and E). In control siRNA-treated cells, Cdt1 and CDC6 were absent after serum starvation. The levels of both proteins increased ∼10 h after serum addition (Fig. 4, D and E, siCont whole cell extracts), coincident with the loading of MCM7 and MCM3 onto chromatin (Fig. 4, D and E, chromatin/nuclear matrix-binding fractions). This result is in agreement with previous findings (12, 54).
Silencing of SNF2H modestly but significantly reduced the recruitment of MCM7 and MCM3 onto chromatin, whereas CDC6 and Cdt1 loading were not affected (Fig. 4, D and E, siSNF2H chromatin/nuclear matrix-binding fractions). Although SNF2H is also involved in transcription regulation (31–35), its silencing did not affect the expression levels of CDC6, Cdt1, MCM7, MCM3, or cyclin E proteins under these experimental conditions (Fig. 4, D and E, whole cell extracts). In addition, Rb phosphorylation at Ser780 by cyclin D-Cdk4/Cdk6 was also investigated. Silencing of SNF2H had no notable effect on the kinetics of the Rb phosphorylation (Fig. 4D). These data give further support for the role of SNF2H in MCM loading.
In SNF2H-silenced cells, a slight delay of the S phase progression was observed (Fig. 4F), which could result from the partial reduction of MCM loading by SNF2H silencing. However, it has been reported that cells with depleted MCM replicate at normal rates (25). SNF2H is also reportedly recruited onto replication foci via a physical interaction between WSTF and PCNA, where it stimulates replication fork progression (42, 55). In the absence of further experimental evidence, it is difficult to distinguish between these two possibilities.
SNF2H Is Involved in Activation of the ATM/Chk2 Checkpoint Pathway Induced by Cdt1 Overexpression
Given that SNF2H promotes MCM loading via the interaction with Cdt1, manipulations in its level of expression could in turn influence Cdt1 overexpression-induced rereplication. As reported previously (5), Cdt1 overexpression alone did not induce detectable rereplication in HEK293T cells. However, simultaneous deregulation of Cdt1 plus ORC1 synergistically induced rereplication (supplemental Fig. S2) (5). Under the same experimental conditions, co-overexpression of Cdt1 and SNF2H did not induce detectable rereplication (supplemental Fig. S2). Although earlier work demonstrated that the co-overexpression of Cdt1 and HBO1 induced overt rereplication (29), no significant rereplication was observed with Cdt1 and HBO1 overexpression in this study (supplemental Fig. S2).
Whether the silencing of SNF2H or HBO1 diminishes rereplication induced by overexpression of the degradation-resistant Cdt1 mutant Cy+D1m (5, 12) was next examined. Cdt1 Cy+D1m-induced rereplication was not affected by the silencing of SNF2H or HBO1 (supplemental Fig. S3, A and B), but it was significantly reduced by ORC1 or CDC6 silencing (supplemental Fig. S3C). These data suggest that SNF2H (and probably HBO1) may not be necessary for the minimal licensing reaction required for DNA replication. However, SNF2H may be essential for the maximal licensing reaction required for genome stability (discussed below).
We previously reported that the overexpression of Cdt1 induces ATM/Chk2 checkpoint activation (13), although the mechanism remains unclear. It has been proposed that ATM is activated not only by DNA strand breaks but also by inappropriate changes in chromatin structure (56). Therefore, when Cdt1 is overexpressed, the protein may recruit excess SNF2H. Excess SNF2H might in turn cause chromatin hyper-remodeling and subsequent ATM activation. To test this possibility, whether the overexpression of SNF2H activates the ATM/Chk2 checkpoint was first examined. However, no significant checkpoint activation was induced by SNF2H overexpression (supplemental Fig. S4). Also, coexpression of Cdt1 and SNF2H did not enhance the Cdt1-induced checkpoint activation (supplemental Fig. S4). This could be because endogenous SNF2H protein levels are high enough to support Cdt1. Therefore, whether the SNF2H silencing inhibits Cdt1-induced checkpoint activation was next investigated. HEK293T cells were transfected with the T7-Cdt1 Cy+D1m expression vector along with control or SNF2H siRNAs, and cells were harvested and analyzed by immunoblotting (Fig. 5). When T7-Cdt1 Cy+D1m was overexpressed, ATM and Chk2 were phosphorylated. SNF2H silencing partially but significantly reduced Cdt1-induced ATM/Chk2 activation (Fig. 5). This result indicates the involvement of SNF2H in Cdt1 overexpression-induced ATM activation and an additional functional link between Cdt1 and SNF2H.
DISCUSSION
The licensing reaction for DNA replication occurs on chromatin. As such, diverse chromatin-modifying molecules may associate with pre-RC components to facilitate the reaction. Here, we have demonstrated that Cdt1 physically interacts with the chromatin-remodeling protein SNF2H (Fig. 1). SNF2H is recruited onto cellular replication origins in a cell cycle-dependent manner, with peak in the G1 phase (Fig. 2). The recruitment of SNF2H is impaired by Cdt1 silencing and stimulated by Cdt1 overexpression (Fig. 3). Furthermore, SNF2H is required for efficient MCM loading (Fig. 4). Thus, it is conceivable that SNF2H is recruited to DNA replication origins through interaction with Cdt1 and promotes MCM loading by regulating nucleosomal structure. This finding is consistent with previous reports that SNF2H is recruited onto oriP of Epstein-Barr virus in the late G1 phase and promotes MCM loading at oriP (37). In this case, however, the mechanism by which SNF2H is recruited onto oriP remains to be clarified.
On the other hand, depletion of SNF2H by the siRNA has only a marginal effect on the S phase progression in synchronized T98G cells, and neither its depletion nor its coexpression affects Cdt1 overexpression-induced rereplication. Thus, SNF2H may function to promote excess MCM loading via an interaction with Cdt1, but SNF2H may not be essential for the minimal loading of MCM helicase required for DNA replication. In accordance with this finding, SNF2H-WSTF complexes appear not to be required for DNA replication in Xenopus egg extracts (57). However, several previous studies have demonstrated that excess MCM loading is crucial for maintaining the genome integrity. For example, although cells with depleted MCM replicate at normal rates, they are hypersensitive to replicative stress and defective in Rad17-dependent ATR-mediated checkpoint activation (23–25). Moreover, a mutation of MCM4 termed Chaos3 (chromosome aberrations occurring spontaneously 3) is a viable allele and causes adenocarcinoma (58–60). Similarly, although mice with reduced expression of MCM2 develop normally, their life span is greatly reduced because of lymphomas (61). In other studies, a reduction in MCM levels caused DNA damage with activation of ATR and ATM (62). These findings suggest that excess MCM loading is critical for the toleration of replication stress and activation of the checkpoint. Therefore, the SNF2H-mediated promotion of MCM loading reported herein may be biologically relevant for the regulation of DNA replication. On the other hand, SNF2H is a multifunctional protein involved in transcription (31–35), replication (36, 37, 42, 43, 55), and repair/recombination (63, 64). Therefore, the production of separation-of-function mutants of Cdt1 and/or SNF2H that specifically lose their interaction would be required for further analysis of the SNF2H-mediated promotion of MCM loading.
Several works have indicated that HBO1, a histone H4 acetylase, is recruited on replication origins via interaction with Cdt1 and stimulates MCM loading by inducing H4 acetylation during the G1 phase (28–30). In addition, the coexpression of HBO1 enhances Cdt1-induced rereplication (29). However, in our experimental system, coexpression of SNF2H or HBO1 did not enhance Cdt1-induced rereplication (supplemental Fig. S2). The apparent difference in the impact of overexpression of HBO1 on Cdt1-induced rereplication might be due to the difference in its expression level. In addition, silencing of SNF2H or HBO1 did not impact Cdt1-induced rereplication (supplemental Fig. S3). Therefore, like SNF2H, HBO1 may be dispensable as far as the minimal MCM loading required for DNA replication is concerned but essential for excess MCM loading. Indeed, both DNA replication and cell cycle progression proceed normally in HBO1 knock-out mice (65), and proliferation occurs in flies with reduced levels of HBO1 (66). Nevertheless, these results do not rule out the involvement of HBO1 in the promotion of MCM loading required for tolerance to replication stress.
It has been reported that SNF2H-WSTF complexes preferentially associate with acetylated histones in chromatin (67). Taking all of the data into consideration, one possible model is that 1) HBO1 is recruited onto replication origins through interaction with Cdt1 and acetylates histone H4, and 2) SNF2H is then recruited to the origin sites through interaction with both Cdt1 and acetylated H4, where it increases the accessibility or fluidity of chromatin. Finally, 3) MCM is efficiently recruited during the G1 phase. Further experimental analysis will be required to confirm this model.
In addition to histone acetylation and ATP-dependent chromatin remodeling, histone chaperones are thought to contribute to transcriptional control. In this regard, ubiquitylation of the histone chaperone FACT by Rtt101, the cullin subunit of E3 ubiquitin ligase, promotes MCM loading onto origins in Saccharomyces cerevisiae (68). On the other hand, FACT is reported to facilitate DNA replication elongation through interaction with MCM complexes in human cells (69). Furthermore, histone methylation is also involved in chromatin transactions. The histone methyltransferase PR-Set7/Set8 is the sole enzyme to catalyze monomethylation of histone H4 at Lys20 (H4K20me1). Recently, PR-Set7 was shown to be regulated by PCNA- and CRL4Cdt2-dependent ubiquitylation and destruction during the S phase. This regulation of PR-Set7 was suggested to contribute to the disappearance of H4K20me1 at origins and the inhibition of licensing (70–74). However, it remains unclear how PR-Set7 is recruited to the replication origins and regulates the licensing reaction.
Although remodeling of chromatin structure is essential for several critical nuclear functions, aberrant activity in chromatin remodeling emerges as a contributor to checkpoint activation. Thus, as proposed previously, ATM is activated not only by DNA strand breaks but also by inappropriate changes in chromatin structure (56). In this study, Cdt1-induced ATM/Chk2 checkpoint activation (13) was shown to depend on SNF2H. Therefore, excess Cdt1 may recruit excess SNF2H and cause alterations in chromatin architecture, thereby activating ATM. This possibility is open to further investigation.
Supplementary Material
Acknowledgment
We appreciate the technical support from the Research Support Center, Graduate School of Medical Sciences, Kyushu University.
This work was supported in part by grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to M. F. and M. I.) and a grant from the Science Research Promotion Fund from the Promotion and Mutual Aid Corporation for Private Schools of Japan (to M. I.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4 and Table S1.
- pre-RC
- pre-replication complex
- WSTF
- Williams syndrome transcription factor
- PCNA
- proliferating cell nuclear antigen.
REFERENCES
- 1. Bell S. P., Dutta A. (2002) Annu. Rev. Biochem. 71, 333–374 [DOI] [PubMed] [Google Scholar]
- 2. Diffley J. F. (2004) Curr. Biol. 14, R778–R786 [DOI] [PubMed] [Google Scholar]
- 3. Fujita M. (2006) Cell Div. 1, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Vaziri C., Saxena S., Jeon Y., Lee C., Murata K., Machida Y., Wagle N., Hwang D. S., Dutta A. (2003) Mol. Cell 11, 997–1008 [DOI] [PubMed] [Google Scholar]
- 5. Sugimoto N., Yoshida K., Tatsumi Y., Yugawa T., Narisawa-Saito M., Waga S., Kiyono T., Fujita M. (2009) J. Cell Sci. 122, 1184–1191 [DOI] [PubMed] [Google Scholar]
- 6. McGarry T. J., Kirschner M. W. (1998) Cell 93, 1043–1053 [DOI] [PubMed] [Google Scholar]
- 7. Wohlschlegel J. A., Dwyer B. T., Dhar S. K., Cvetic C., Walter J. C., Dutta A. (2000) Science 290, 2309–2312 [DOI] [PubMed] [Google Scholar]
- 8. Tada S., Li A., Maiorano D., Méchali M., Blow J. J. (2001) Nat. Cell Biol. 3, 107–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lee C., Hong B., Choi J. M., Kim Y., Watanabe S., Ishimi Y., Enomoto T., Tada S., Kim Y., Cho Y. (2004) Nature 430, 913–917 [DOI] [PubMed] [Google Scholar]
- 10. Sugimoto N., Tatsumi Y., Tsurumi T., Matsukage A., Kiyono T., Nishitani H., Fujita M. (2004) J. Biol. Chem. 279, 19691–19697 [DOI] [PubMed] [Google Scholar]
- 11. Nishitani H., Sugimoto N., Roukos V., Nakanishi Y., Saijo M., Obuse C., Tsurimoto T., Nakayama K. I., Nakayama K., Fujita M., Lygerou Z., Nishimoto T. (2006) EMBO J. 25, 1126–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sugimoto N., Kitabayashi I., Osano S., Tatsumi Y., Yugawa T., Narisawa-Saito M., Matsukage A., Kiyono T., Fujita M. (2008) Mol. Biol. Cell 19, 1007–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tatsumi Y., Sugimoto N., Yugawa T., Narisawa-Saito M., Kiyono T., Fujita M. (2006) J. Cell Sci. 119, 3128–3140 [DOI] [PubMed] [Google Scholar]
- 14. Arentson E., Faloon P., Seo J., Moon E., Studts J. M., Fremont D. H., Choi K. (2002) Oncogene 21, 1150–1158 [DOI] [PubMed] [Google Scholar]
- 15. Kemp M. G., Ghosh M., Liu G., Leffak M. (2005) Nucleic Acids Res. 33, 325–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhou J., Chau C., Deng Z., Stedman W., Lieberman P. M. (2005) Cell Cycle 4, 889–892 [DOI] [PubMed] [Google Scholar]
- 17. Karnani N., Taylor C., Malhotra A., Dutta A. (2007) Genome Res. 17, 865–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lucas I., Palakodeti A., Jiang Y., Young D. J., Jiang N., Fernald A. A., Le Beau M. M. (2007) EMBO Rep. 8, 770–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Goren A., Tabib A., Hecht M., Cedar H. (2008) Genes Dev. 22, 1319–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Danis E., Brodolin K., Menut S., Maiorano D., Girard-Reydet C., Méchali M. (2004) Nat. Cell Biol. 6, 721–730 [DOI] [PubMed] [Google Scholar]
- 21. Aggarwal B. D., Calvi B. R. (2004) Nature 430, 372–376 [DOI] [PubMed] [Google Scholar]
- 22. Hartl T., Boswell C., Orr-Weaver T. L., Bosco G. (2007) Chromosoma 116, 197–214 [DOI] [PubMed] [Google Scholar]
- 23. Tsao C. C., Geisen C., Abraham R. T. (2004) EMBO J. 23, 4660–4669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Woodward A. M., Göhler T., Luciani M. G., Oehlmann M., Ge X., Gartner A., Jackson D. A., Blow J. J. (2006) J. Cell Biol. 173, 673–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ge X. Q., Jackson D. A., Blow J. J. (2007) Genes Dev. 21, 3331–3341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Aalfs J. D., Kingston R. E. (2000) Trends Biochem. Sci. 25, 548–555 [DOI] [PubMed] [Google Scholar]
- 27. Iizuka M., Stillman B. (1999) J. Biol. Chem. 274, 23027–23034 [DOI] [PubMed] [Google Scholar]
- 28. Iizuka M., Matsui T., Takisawa H., Smith M. M. (2006) Mol. Cell. Biol. 26, 1098–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Miotto B., Struhl K. (2008) Genes Dev. 22, 2633–2638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Miotto B., Struhl K. (2010) Mol. Cell 37, 57–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Varga-Weisz P. (2001) Oncogene 20, 3076–3085 [DOI] [PubMed] [Google Scholar]
- 32. Tsukiyama T. (2002) Nat. Rev. Mol. Cell Biol. 3, 422–429 [DOI] [PubMed] [Google Scholar]
- 33. Corona D. F., Tamkun J. W. (2004) Biochim. Biophys. Acta 1677, 113–119 [DOI] [PubMed] [Google Scholar]
- 34. Dirscherl S. S., Krebs J. E. (2004) Biochem. Cell Biol. 82, 482–489 [DOI] [PubMed] [Google Scholar]
- 35. Eberharter A., Becker P. B. (2004) J. Cell Sci. 117, 3707–3711 [DOI] [PubMed] [Google Scholar]
- 36. Alexiadis V., Varga-Weisz P. D., Bonte E., Becker P. B., Gruss C. (1998) EMBO J. 17, 3428–3438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhou J., Chau C. M., Deng Z., Shiekhattar R., Spindler M. P., Schepers A., Lieberman P. M. (2005) EMBO J. 24, 1406–1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Iizuka M., Sarmento O. F., Sekiya T., Scrable H., Allis C. D., Smith M. M. (2008) Mol. Cell. Biol. 28, 140–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fujita M., Kiyono T., Hayashi Y., Ishibashi M. (1997) J. Biol. Chem. 272, 10928–10935 [DOI] [PubMed] [Google Scholar]
- 40. Yugawa T., Handa K., Narisawa-Saito M., Ohno S., Fujita M., Kiyono T. (2007) Mol. Cell. Biol. 27, 3732–3742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Fujita M., Yamada C., Tsurumi T., Hanaoka F., Matsuzawa K., Inagaki M. (1998) J. Biol. Chem. 273, 17095–17101 [DOI] [PubMed] [Google Scholar]
- 42. Poot R. A., Bozhenok L., van den Berg D. L., Steffensen S., Ferreira F., Grimaldi M., Gilbert N., Ferreira J., Varga-Weisz P. D. (2004) Nat. Cell Biol. 6, 1236–1244 [DOI] [PubMed] [Google Scholar]
- 43. Kitagawa H., Fujiki R., Yoshimura K., Mezaki Y., Uematsu Y., Matsui D., Ogawa S., Unno K., Okubo M., Tokita A., Nakagawa T., Ito T., Ishimi Y., Nagasawa H., Matsumoto T., Yanagisawa J., Kato S. (2003) Cell 113, 905–917 [DOI] [PubMed] [Google Scholar]
- 44. Lazzaro M. A., Picketts D. J. (2001) J. Neurochem. 77, 1145–1156 [DOI] [PubMed] [Google Scholar]
- 45. Bozhenok L., Wade P. A., Varga-Weisz P. (2002) EMBO J. 21, 2231–2241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Giacca M., Zentilin L., Norio P., Diviacco S., Dimitrova D., Contreas G., Biamonti G., Perini G., Weighardt F., Riva S. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 7119–7123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Abdurashidova G., Deganuto M., Klima R., Riva S., Biamonti G., Giacca M., Falaschi A. (2000) Science 287, 2023–2026 [DOI] [PubMed] [Google Scholar]
- 48. Ladenburger E. M., Keller C., Knippers R. (2002) Mol. Cell. Biol. 22, 1036–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Schaarschmidt D., Ladenburger E. M., Keller C., Knippers R. (2002) Nucleic Acids Res. 30, 4176–4185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Gerhardt J., Jafar S., Spindler M. P., Ott E., Schepers A. (2006) Mol. Cell. Biol. 26, 7731–7746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Masai H., Matsumoto S., You Z., Yoshizawa-Sugata N., Oda M. (2010) Annu. Rev. Biochem. 79, 89–130 [DOI] [PubMed] [Google Scholar]
- 52. Ghosh M., Kemp M., Liu G., Ritzi M., Schepers A., Leffak M. (2006) Mol. Cell. Biol. 26, 5270–5283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Rampakakis E., Zannis-Hadjopoulos M. (2009) Nucleic Acids Res. 37, 5714–5724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Mailand N., Diffley J. F. (2005) Cell 122, 915–926 [DOI] [PubMed] [Google Scholar]
- 55. Collins N., Poot R. A., Kukimoto I., García-Jiménez C., Dellaire G., Varga-Weisz P. D. (2002) Nat. Genet. 32, 627–632 [DOI] [PubMed] [Google Scholar]
- 56. Bakkenist C. J., Kastan M. B. (2003) Nature 421, 499–506 [DOI] [PubMed] [Google Scholar]
- 57. MacCallum D. E., Losada A., Kobayashi R., Hirano T. (2002) Mol. Biol. Cell 13, 25–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Shima N., Alcaraz A., Liachko I., Buske T. R., Andrews C. A., Munroe R. J., Hartford S. A., Tye B. K., Schimenti J. C. (2007) Nat. Genet. 39, 93–98 [DOI] [PubMed] [Google Scholar]
- 59. Shima N., Buske T. R., Schimenti J. C. (2007) Cell Cycle 6, 1135–1140 [DOI] [PubMed] [Google Scholar]
- 60. Chuang C. H., Wallace M. D., Abratte C., Southard T., Schimenti J. C. (2010) PLoS Genet. 6, e1001110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Pruitt S. C., Bailey K. J., Freeland A. (2007) Stem Cells 25, 3121–3132 [DOI] [PubMed] [Google Scholar]
- 62. Orr S. J., Gaymes T., Ladon D., Chronis C., Czepulkowski B., Wang R., Mufti G. J., Marcotte E. M., Thomas N. S. (2010) Oncogene 29, 3803–3814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Lan L., Ui A., Nakajima S., Hatakeyama K., Hoshi M., Watanabe R., Janicki S. M., Ogiwara H., Kohno T., Kanno S., Yasui A. (2010) Mol. Cell 40, 976–987 [DOI] [PubMed] [Google Scholar]
- 64. Nakamura K., Kato A., Kobayashi J., Yanagihara H., Sakamoto S., Oliveira D. V., Shimada M., Tauchi H., Suzuki H., Tashiro S., Zou L., Komatsu K. (2011) Mol. Cell 41, 515–528 [DOI] [PubMed] [Google Scholar]
- 65. Kueh A. J., Dixon M. P., Voss A. K., Thomas T. (2011) Mol. Cell. Biol. 31, 845–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Grienenberger A., Miotto B., Sagnier T., Cavalli G., Schramke V., Geli V., Mariol M. C., Berenger H., Graba Y., Pradel J. (2002) Curr. Biol. 12, 762–766 [DOI] [PubMed] [Google Scholar]
- 67. Hakimi M. A., Bochar D. A., Schmiesing J. A., Dong Y., Barak O. G., Speicher D. W., Yokomori K., Shiekhattar R. (2002) Nature 418, 994–998 [DOI] [PubMed] [Google Scholar]
- 68. Han J., Li Q., McCullough L., Kettelkamp C., Formosa T., Zhang Z. (2010) Genes Dev. 24, 1485–1490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Tan B. C., Chien C. T., Hirose S., Lee S. C. (2006) EMBO J. 25, 3975–3985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Abbas T., Shibata E., Park J., Jha S., Karnani N., Dutta A. (2010) Mol. Cell 40, 9–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Centore R. C., Havens C. G., Manning A. L., Li J. M., Flynn R. L., Tse A., Jin J., Dyson N. J., Walter J. C., Zou L. (2010) Mol. Cell 40, 22–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Lee J., Zhou P. (2010) Mol. Cell 40, 345–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Oda H., Hübner M. R., Beck D. B., Vermeulen M., Hurwitz J., Spector D. L., Reinberg D. (2010) Mol. Cell 40, 364–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Tardat M., Brustel J., Kirsh O., Lefevbre C., Callanan M., Sardet C., Julien E. (2010) Nat. Cell Biol. 12, 1086–1093 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.