Abstract
Allergic reactions to peanuts and tree nuts are major causes of anaphylaxis in the United States. We compare different properties of natural and recombinant versions of Ara h 1, a major peanut allergen, through structural, immunologic, and bioinformatics analyses. Small angle x-ray scattering studies show that natural Ara h 1 forms higher molecular weight aggregates in solution. In contrast, the full-length recombinant protein is partially unfolded and exists as a monomer. The crystal structure of the Ara h 1 core (residues 170–586) shows that the central part of the allergen has a bicupin fold, which is in agreement with our bioinformatics analysis. In its crystalline state, the core region of Ara h 1 forms trimeric assemblies, while in solution the protein exists as higher molecular weight assemblies. This finding reveals that the residues forming the core region of the protein are sufficient for formation of Ara h 1 trimers and higher order oligomers. Natural and recombinant variants of proteins tested in in vitro gastric and duodenal digestion assays show that the natural protein is the most stable form, followed by the recombinant Ara h 1 core fragment and the full-length recombinant protein. Additionally, IgE binding studies reveal that the natural and recombinant allergens have different patterns of interaction with IgE antibodies. The molecular basis of cross-reactivity between vicilin allergens is also elucidated.
Keywords: Allergen, Allergy, Protein Aggregation, Structural Biology, X-ray Crystallography, Ara h 1, IgE Binding, Food Allergen, In Vitro Digestion
Introduction
Soybean, peanut, kidney bean, pea, lentil, and tree nuts are sources of a significant number of food allergens (1–3). While many types of food allergies are outgrown, the chances of outgrowing peanut and tree nut allergies are 20 and 10%, respectively (4, 5). In the United States, where peanuts are used as ingredients in many foods, the incidence of peanut allergies is still increasing, and ∼1% of the population are affected (6). Not only does peanut allergy persist in 80% of the afflicted individuals, but for many of them contact with even minute amounts of the allergens results in severe reactions including anaphylaxis (7, 8).
Globulins (7 S and 11 S) represent the majority of the total protein in many seeds that are consumed by humans (9). Vicilins (7 S globulins) and legumins (11 S globulins) share similar folds and belong to the cupin superfamily of proteins (10–12). Vicilins and legumins are classified as bicupins because of the presence of two domains with the characteristic cupin β-barrel fold. Cupins form one of the most functionally diverse protein superfamilies (12) and are unusually thermostable (13–15). Their thermostability, in connection with their resistance to digestion in the human gastrointestinal tract (16–18), suggests that both vicilins and legumins should be treated as potential allergenic proteins. In the case of peanut allergens, both Ara h 1 (a vicilin) and Ara h 3 (a legumin) are considered to be major allergens eliciting immune responses in the majority of peanut allergic individuals (19–21). Ara h 1 is recognized by serum IgE from more than 90% of peanut-allergic patients (22). Ara h 1 forms homotrimers that may oligomerize further forming larger assemblies (23). The formation of the higher molecular weight complexes may be driven by interaction of the protein with small molecular compounds (24, 25). Presented here are the results of structural, immunological and bioinformatics analyses of Ara h 1 that were performed to compare the properties of the natural allergens and their full-length and truncated recombinant versions.
EXPERIMENTAL PROCEDURES
Cloning, Expression, and Purification
Natural Ara h 1 (nAra h 1) was purified as described previously (16). The DNA expression construct for recombinant full-length Ara h 1 (rAra h 1) was synthesized by EZbiolab (Carmel, IN), and the protein was purified using the same protocol as for nAra h 1. The insert of rAra h 1 in the pET9a vector was used as a template for PCR amplification of a shorter version of Ara h 1 (amino acids 170–586), referred to as recombinant, short Ara h 1 (rsAra h 1). This fragment of Ara h 1 was chosen, as it was shown to be stable and could be crystallized (26). The pET9a vector was used for rsAra h 1 production and purification of rsAra h 1 was performed according to the protocol reported by Cabanos et al. (26).
SDS-PAGE and IgE Western Blot Analysis
Purified proteins (300 ng/protein) were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on a 4–20% Novex Tris-HCl precast gel (Invitrogen) or spotted on a PVDF membrane and allowed to dry. For IgE Western blots and Spot Blots, membranes were blocked in 2% Blotto (2% dry milk dissolved into phosphate-buffered saline (PBS) containing 0.5% TWEEN (PBST)) for 30 min and incubated overnight with 1:10 dilutions in PBST of sera from individuals with a convincing history of peanut allergy or documented positive peanut ImmunoCAP (Phadia, Uppsala, Sweden) and skin prick test results. All sera were obtained in accordance with the regulations of Tulane IRB. After the incubation with patients' sera, the membranes were washed three times with PBST and incubated with anti-human IgE conjugated to horseradish peroxidase (HRP)-labeled secondary antibody (Sigma-Genosys) at 1:10,000, diluted in 2% Blotto for 30 min. The membrane was then washed three times with PBST and 2 times with PBS and incubated with ECL-Plus Western substrate (Amersham Biosciences). The signal was visualized using a CCD camera system (Fuji Photo Film Co., Ltd., Duluth, GA). SeeBlue Plus2 molecular weight standard (Invitrogen) was used according to the manufacturer's instructions.
In Vitro Gastric and Duodenal Digestions
In vitro gastric and duodenal digestions were performed as described by Moreno et al. (27). In phase 1 (in vitro gastric digestion), nAra h 1, rAra h 1, and rsAra h 1 (0.5 mg/ml) were subjected to pepsin (Porcine pepsin, enzymatic activity of 4230 units/mg protein, Sigma-Aldrich; product No. P6887) digestion using an enzyme/substrate ratio of 1:20 (w/w). The digestion was performed in simulated gastric fluid (0.15 m NaCl adjusted with 1 m HCl to pH 2) at 37 °C, and aliquots were taken at 0, 15, and 30 s and at 1, 2, 4, 16, 30, and 60 min. The digestion was stopped by raising the pH to 6.5 by addition of 1 m NaOH.
In phase 2 (in vitro duodenal digestion) digestion was performed using the 60 min gastric digesta as starting material. The following reagents were added: 0.125 m bile salt mixture, 9.2 mm CaCl2, 25 mm Bis-Tris-HCl, pH 6.5. Subsequently, solutions of trypsin and chymotrypsin were added at ratios of protein/trypsin/chymotrypsin = 1:400:100 (w/w/w). The digestion was performed at 37 °C, and aliquots were taken at 0 and 30 s and at 1, 2, 4, and 16 min. The digestion was stopped by adding a solution of a trypsin-chymotrypsin inhibitor (Sigma-Aldrich). The samples were analyzed using manually prepared 16% Tris-glycine polyacrylamide gels.
Crystallization
The natural and both recombinant versions of Ara h 1 were tested for crystallization. Tracking and analysis of the crystallization experiments were performed with Xtaldb (28, 29). Despite testing ∼1500 conditions, no crystals of natural Ara h 1 were obtained. At this point, crystallization attempts concentrated on the shorter recombinant protein (rsAra h 1). The protein was obtained and crystallized using conditions described previously by Cabanos et al. (26). Prior to crystallization, the protein dissolved in buffer containing 10 mm Tris-HCl, 500 mm NaCl, pH 7.5 was passed through a Superdex 200 column attached to an AKTA FPLC system (GE Healthcare). After gel filtration, fractions containing rsAra h 1 were pooled and concentrated to 7 mg/ml. Crystals were grown using the vapor diffusion method in hanging drops. The single crystal used to collect data for the initial structure was obtained from a drop created after mixing 1 μl of protein solution and 1 μl of well solution (15% v/v PEG 400, 100 mm NaCl, 100 mm sodium citrate, pH 5.6). Crystals were grown overnight at 20–25 °C. Prior to data collection, the crystal was transferred to a solution containing a 2:1 mixture of well solution and ethylene glycol and immediately cooled in a cold nitrogen stream. Further optimization of the crystal growth conditions (increase of NaCl concentration to 150 mm in well solution), application of PEG 200 instead of ethylene glycol for cryo-protection and flash-cooling in liquid nitrogen resulted in crystals and data of better quality.
Data Collection, Structure Determination, and Refinement
Data collection for the initially obtained crystals was performed using a Rigaku Corp. MicroMax 007 system equipped with a Saturn 92 CCD detector. HKL-3000 (30, 31) was used to control the diffractometer and process data. Analysis of diffraction images revealed that all tested crystals diffracted in a highly anisotropic way. Diffraction maxima up to 2.3 Å resolution were observed at some crystal orientations, while at other orientations only reflections corresponding to resolution below 3 Å were present. After data processing, a resolution limit of 2.7 Å was chosen, and the resulting data set was used for structure solution and refinement. Analysis of the intensities revealed that the crystal used for data collection was twinned. The structure was solved by molecular replacement using HKL-3000 in combination with MOLREP (32) with the structure of 7 S globulin from Adzuki bean (Protein Data Bank (PDB)3 code: 2ea7) as a starting model. Refinement was done using the programs HKL-3000, REFMAC (33), COOT (34), and CCP4 (35). REFMAC was used to treat twinned data (twin operator: -h-k, k, -l) and the twin fraction refined to 0.24. NCS restraints were used during refinement. TLS refinement (with whole protein chains treated as distinct TLS groups) was performed in the last stages of the refinement. Validation of the structures was performed using MOLPROBITY (36, 37) and ADIT (38).
Whereas optimization of crystal growth and protocol for cryo-protection resulted in a data set of higher quality, the Diffraction Anisotropy Server still indicated it was significantly affected by anisotropy (39). Data from optimized crystals were collected at the 19-ID beamline of the Structural Biology Center (40) at the Advanced Photon Source. A previously determined model was used for structure solution, and the same methodology used in the case of the lower resolution structure was applied for refinement and structure validation. In this case, the twin fraction was lower and refined to 0.05. The coordinates, structure factors, and intensities were deposited in PDB (41). Statistics describing the data and refinement are summarized in Table 1.
TABLE 1.
Data collection and refinement statistics
Ramachandran plot was calculated using MOLPROBITY. Numbers in parentheses refer to the highest resolution shell. In both cases, calculations of the Rfree values were performed using a subset containing 5% of the total number of reflections, chosen at random.
| PDB code | 3s7e | 3s7i |
|---|---|---|
| Data collection | ||
| Wavelength (Å) | 1.5418 | 0.9792 |
| Unit cell (Å) | a = b = 93.4, c = 237.1 | a = b = 92.9, c = 231.6 |
| Space group | R3 | R3 |
| Solvent content (%) | 41 | 39 |
| Resolution range (Å) | 50.0–2.7 | 50.0–2.35 |
| Highest resolution shell (Å) | 2.70–2.75 | 2.39–2.35 |
| Unique reflections | 19169(680) | 31048(1563) |
| Redundancy | 3.1(3.1) | 5.6(5.3) |
| Completeness (%) | 92.1(64.8) | 100.0(100.0) |
| Rmerge (%) | 5.3(15.9) | 8.8(62.8) |
| Average I/σ(I) | 18.5(6.6) | 30.6(3.4) |
| Refinement | ||
| R (%) | 23.6 | 20.5 |
| Rfree (%) | 26.0 | 24.4 |
| Mean B value (Å2) | 29.0 | 54.1 |
| B from Wilson plot (Å2) | 34.1 | 49.3 |
| RMS deviation bond lengths (Å) | 0.011 | 0.019 |
| RMS deviation bond angles (°) | 1.3 | 1.7 |
| Number of amino acid residues | 732 | 733 |
| Number of water molecules | 28 | 133 |
| Number of ions/ligands | 2 | 2 |
| Ramachandran plot | ||
| Most favored regions (%) | 95.1 | 98.3 |
| Additional allowed regions (%) | 4.9 | 1.7 |
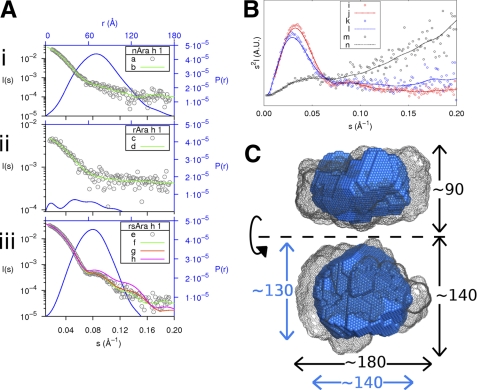
Small Angle X-ray Scattering
SAXS data were collected at Rigaku S-MAX3000 (λ = 1.54Å) setup at 4 °C. Two dimensional scattering data were integrated using SAXS GUI with Ag Behenate to calibrate momentum transfer. Three versions of Ara h 1, as well as the corresponding buffer solutions (20 mm HEPES, pH 7.5, 150 mm NaCl for nAra h 1 and 10 mm HEPES, pH 7.5, 500 mm NaCl for rAra h 1 and rsAra h 1), were analyzed. For nAra h 1, one sample with a concentration of 8.8 mg/ml was analyzed for 2 h and one sample of 2.2 mg/ml was analyzed for 4 h. For rsAra h 1, three samples with concentrations of 5 mg/ml, 2.5 mg/ml, and 1.25 mg/ml were analyzed with run times of 2, 2.6, and 2.7 h respectively. Three concentrations (6 mg/ml, 3 mg/ml, and 1.5 mg/ml) of rAra h 1 were analyzed for 7 h, 2 h, and 2 h, respectively. Normalization of data to beam intensity and buffer subtraction were conducted using PRIMUS (42). Sample folding was assessed using Kratky analysis by plotting the square of the scattering angle times the intensity versus the scattering angle (s2I(s) versus s) (43). Sample aggregation was assessed with autoRg (44). The radius of gyration (Rg) and the forward intensity (I(0)) were estimated using the Guinier interval with sRg < 1.3. The molecular mass was calculated using autoPOROD (44) and SAXS MOW (45). The scattering curves for nAra h 1 and rAra h 1 were extrapolated to zero concentration, while the average of both curves was taken for rsAra h 1 using PRIMUS. The indirect transform program GNOM (46) was used to calculate the distance distribution function (P(r)) and the largest dimension (Dmax) for each version. Ab initio bead modeling was performed with DAMMIF (47) for each version of Ara h 1, which were then averaged using DAMAVER (48). Additional bead modeling was performed on rsAra h 1 using smaller bead sizes. These models were then refined using SASREF (49). Scattering curves and χ values were computed for these rigid body models using CRYSOL (50).
Sequence Database Search and Sequence Clustering
The Ara h 1 sequence was used as a query in a PSI-BLAST (51, 52) search of the non-redundant (nr) data base. The search was run until convergence, with the expectation (e) value threshold for the retrieval of related sequences set to 10−3. An analysis of the available allergen databases was conducted in order to find related allergens, and sequences of vicilins and legumins classified as allergens were added to create the final dataset. CLANS (CLuster ANalysis of Sequences), a Java utility that applies a version of the Fruchterman-Reingold graph layout algorithm (53), was used to identify subgroups of closely related sequences and visualize pairwise similarities between and within the identified groups of sequences. CLANS uses the p values of highly scoring segment pairs (HSPs), obtained from an N × N BLAST search, to compute attractive and repulsive forces between each sequence pair in a user-defined dataset. A 2- or 3-dimensional representation of sequence families is achieved by randomly seeding the sequences in the arbitrary distance space, then moving them within this environment according to the force vectors resulting from all pairwise interactions, and repeating the process until convergence. Clustering of the Ara h 1 homologous sequences was performed based on their pairwise BLAST similarity scores, using CLANS (53), with a p value threshold of 10−3.
Other Computational Methods
PISA (54) was used to calculate information on the oligomeric assemblies. The SSM algorithm (55) implemented in COOT was used to superpose macromolecular models. All figures presenting macromolecular structures were prepared with PYMOL (56).
RESULTS
Clustering Analysis of the Closest Ara h 1 Homologs
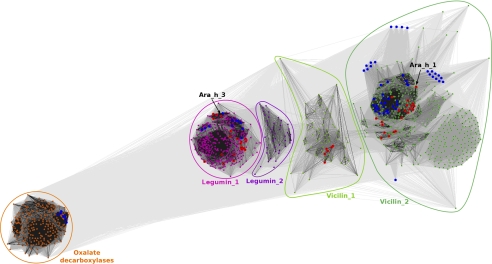
The clustering classification was carried out to identify groups of similar sequences and to determine the distribution of allergens and proteins with known structures among the identified proteins. This classification produced a separation of the sequences into clusters corresponding to the originally defined groups (Fig. 1). Bacterial oxalate decarboxylases, including Bacillus subtilis oxalate decarboxylase structures (PDB codes: 2uy9, 2v09, 2uy8, 2uya, 2uyb, 1j58, 1l3j, 1uw8), created the most distant clusters among Ara h 1 homologs. Other identified protein sequences (all seed storage globulins) were divided between several clusters. The most compact cluster (indicated as Legumin_1 in Fig. 1) is comprised of sequences of legumin-like (11S) proteins, including structures of the peanut allergen Ara h 3 (PDB code: 3c3v), soybean proglycinin and glycinin (Gly m 6) (PDB codes: 1fxz, 1ucx, 1ud1, 2d5f, 1od5), procruciferin (PDB code: 3kgl), pea prolegumin (PDB code: 3ksc) and pumpkin seed globulin (PDB code: 2evx). This cluster also enclosed known legumin-like allergens (Ana o 2, Ara h 4, Ber e 2, Car i 4, Cor a 9, Jug r 4, Pis v 2, Pis v 5, Pru du 6, Ses i 6, Ses i 7, Sin a 2). Adjacent the Legumin_1 cluster, there is a small dispersed cluster (Legumin_2) of proteins annotated mainly as legumin-like hypothetical proteins, with no known structural representatives or any classified allergens.
FIGURE 1.
Two-dimensional projection of the CLANS clustering results. Proteins are indicated by dots. Lines indicate sequence similarity detectable with BLAST and are colored by a spectrum of gray shades according to the BLAST p value (black: p value < 10−225, light gray: p value < 10−5). Sequences corresponding to structures in the PDB are indicated by blue dots, sequences of known allergens are indicated by red dots. Peanut allergens Ara h 1 and Ara h 3 are marked with arrows. Because the sequences were downloaded as they are in the nr database, sequences of separate, nonidentical chains in PDB deposits are represented separately. For example, Jack Bean canavalin structures were obtained by crystallization of the proteolytic product of the protein, and both domains are represented by separate sequences, which is why they moved away from the central cluster containing the full-length protein.
The two other large, dispersed clusters are composed of vicilin-like (7 S) proteins. The cluster indicated as Vicilin_1 not only encloses mostly proteins annotated as unknown or hypothetical, but also the soybean allergen Gly m Bd 28K. The largest cluster (Vicilin_2) encloses vicilin-like proteins, including numerous proteins with known structures: canavalin (PDB codes: 1dgr, 1dgw, 1cau, 1caw, 1cax, 1cav, 2cau, 2cav), phaseolin (PDB codes: 2phl, 1phs), soybean β-conglycinin (PDB codes: 1uik, 1ipk, 1ipj, 1uij), mungbean 8S alpha globulin (PDB code: 2cv6) and Azuki Bean 7S globulin 1 and 3 (PDB codes: 2ea7 and 2eaa, respectively). Several vicilin-like allergens can also be found in the most dense subcluster, including Ara h 1, Ana o 1, Cor a 11, Gly m 5, Jug n 2, Jug r 2, Len c 1, Lup an 1, Pis v 3, Pis s 1, Pis s 2, Ses i 3. Other subgroups in this cluster, with no structural or known allergen representatives, include vicilin-like globulins mainly from Zea mays, Oryza sativa, and different Theobroma species.
Structural Analysis of the Ara h 1 Core
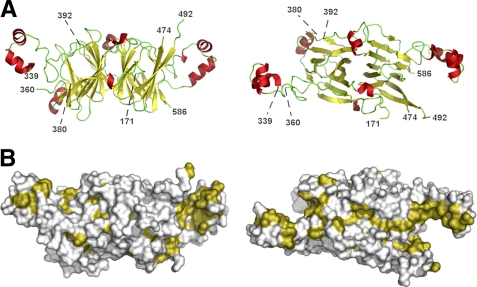
The Ara h 1 core (residues 170–586) crystallized in rhombohedral crystal form and in the space group R3 with two chains in the asymmetric unit, with each chain being a part of a different trimer. The trimer 3-fold axis coincides with the crystallographic 3-fold axis. While the determined crystal structures do not differ significantly in conformation of the main chains of the protein molecules, they differ slightly in the unit cell parameters (Table 1). Their Cα atoms superpose with root mean square deviation (rmsd) values of 0.5 Å. However, one of the molecules in the asymmetric unit is ordered better than the other one. In the structure reported here (PDB code: 3s7e), both chains contain residues 171–586, with three loop regions (residues 340–359, 381–391 and 475–491 for chain A and residues 339–360, 383–391 and 471–492 for chain B) that are missing, likely due to their disorder. Ara h 1 has an overall fold that is characteristic for bicupins (Fig. 2A). Superposition of N- and C-terminal domains results in a 1.9 Å rmsd value for 153 aligned residues despite sequence identity of only 15% between the superposed protein fragments. Hence, each Ara h 1 molecule may be described as having two modules that are related by a pseudo-dyad axis perpendicular to the 3-fold axes of the trimer. Taking this into account, the Ara h 1 oligomer may be described as having pseudo-D3 point group symmetry. Cupin domains are flanked by regions containing α-helices. These regions share similarities with helix-turn-helix (HTH) motifs that often interact with DNA. Analysis of the Ara h 1 structure performed with PROFUNC (57) identified a fragment composed of residues 555–576 as a possible HTH DNA-binding motif on the basis of its similarity to the DNA binding motif from an initiator binding protein (PDB code: 1pp7). Moreover, it was shown that the corresponding region in the 7 S globulin-1 from Adzuki bean (structure with PDB code: 2ae7) binds a calcium ion though carbonyl oxygen atoms from the main chain. Comparison of these regions in both Ara h 1 and 7S globulin-1 shows that the conformations of the main chain are very similar, indicating that Ara h 1 could bind calcium ions as well.
FIGURE 2.
Structure of rsAra h 1. A, ribbon diagram of the Ara h 1 core fragment presented in two different orientations. Numbers indicate the ends of fragments that could be traced. α-Helices are shown in red, β-strands in yellow, and loop regions are shown in green. B, molecular surface of Ara h 1 with hydrophobic residues are shown in olive. Orientation of the molecules is the same as for the ribbon diagrams. Figures from the right side of the panel show molecules as seen from the center of trimer.
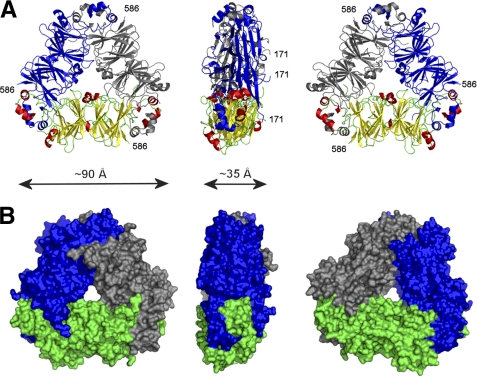
Helical regions flanking the cupin domains are important for trimer formation. They interact through their hydrophobic parts (Fig. 2B) with helical regions from the other molecules forming trimers, and they also interact with residues forming cupin domains (Fig. 3). Monomer-monomer interfaces in the trimer are very large as many residues participate in their formation. In its crystalline state, Ara h 1 molecules form trimeric assemblies and the buried area after trimer formation is ∼15,000 Å2. This corresponds to an interface area of almost 2500 Å2 per chain, which is similar to the interface area values observed for other 7 S and 11 S globulins reported in the PDB. In addition, the trimer is stabilized by multiple hydrogen bonds.
FIGURE 3.
Trimer formed by rsAra h 1. Ara h 1 trimer shown in three different orientations corresponding to 0°, 90°, and 180° rotation along the horizontal axis. Part A of the panel shows the trimer in ribbon representation with one protein molecule in the same orientation and color schema as in Fig. 2A, and two other molecules are shown in blue and gray. B, molecular surface of Ara h 1 trimer.
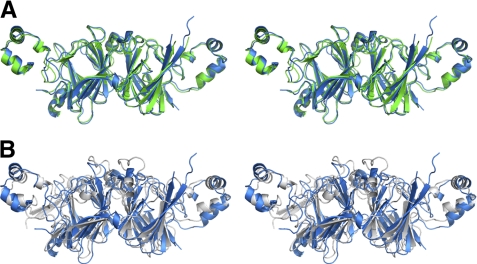
Soybean beta-conglycinin, Gly m 5, is the closest known homolog of Ara h 1 whose structure is reported in the PDB. The proteins have 51% sequence identity and their overall fold is very similar (Fig. 4A). The sequence identity for the corresponding regions of the proteins' core is even higher (57%) and their structures superpose with rmsd value of ∼1 Å. Similarly, comparison of Ara h 1 and Ara h 3 structures reveals that, despite significantly lower sequence identity (21%), the overall fold of the core region is quite similar, and the structures superpose with 2.5 Å rmsd value (Fig. 4B). Similar rmsd values are obtained after superposing Ara h 1 and oxalate decarboxylase (OxdC) from Bacillus subtilis (58).
FIGURE 4.
Stereoviews showing alignment of Ara h 1 (blue) with (A) β-conclycinin (green) and (B) Ara h 3 (gray). For β-conclycinin and Ara h 3 structures with PDB codes 1uij and 3c3v, respectively, were used.
Oxalate decarboxylase is a manganese-dependent enzyme that decomposes oxalate into formate and carbon dioxide. The metal ion is coordinated by three histidine residues and one glutamic acid residue. Ara h 1 is unable to bind metal ions the way oxalate decarboxylase does, as the corresponding regions of these proteins differ significantly in amino acid composition. Only one of these metal binding residues is conserved in Ara h 1. However, Ara h 1 His229 from the first cupin domain, which corresponds to His95 of Bacillus subtilis oxalate decarboxylate, does not point toward the center of the domain but in the opposite direction. In contrast, the side chain of His447 (equivalent of OxdC His273) faces the inside of the second cupin domain. Moreover, in the structure of rsAra h 1, His-447 together with Arg-540 are involved in binding of a chloride ion.
Oligomeric State of Ara h 1 in Solution
The structures of nAra h 1, rAra h 1, and rsAra h 1 in solution were investigated using small angle x-ray scattering. Estimates of the molecular mass given by autoPOROD and SAXS MOW indicate that both nAra h 1 (Molecular Mass (MM) = 610 kDa, 560 kDa) and rsAra h 1 (MM = 465 kDa, 475 kDa) form large oligomers in solution. However, according to both methods, it is plausible that rAra h 1 is monomeric in solution (MM = 76 kDa, 61 kDa). As shown in Fig. 5B, the “closed” nature of both the nAra h 1 (blue, k/l) and rsAra h 1 (red i/j) curves indicate that they are folded (43). The “open” nature of the rAra h 1 (black, m/n) curve suggests that in solution, this version is partially unfolded.
FIGURE 5.
Summary of SAXS data. A, experimental scattering data of each version of Ara h 1 (in gray, black axes) along with the fits of the most probable bead models (in green) and the particle distance distribution functions (in blue, blue axes). Aiii, in addition, shows the theoretical scattering curves of the four trimer (g) and the three trimer (h) SASREF models. B, Kratky plot with experimental points for each version. Where i, k, and m represent rsAra h 1, nAra h 1, and rAra h 1, respectively, along with the corresponding trend lines (j,l,n) determined with the smooth bezier function in GNUplot. C, representation of the most probable bead models of rsAra h 1 (in blue) and nAra h 1 (in gray mesh). The dimensions in Angstroms of each are given in blue and black, respectively. The bottom figure was obtained by rotating the top figure 90 degrees around the specified axis.
In addition to changes in molecular masses, the radius of gyrations and the largest diameter of each version also differed, with the radius of gyration of nAra h 1 being the largest at 56.6 ± 0.3 Å (Dmax 177Å), followed by rsAra h 1 at 49.8 ± 0.4 Å (Dmax 133Å) and rAra h 1 at 34.0 ± 0.7 Å (Dmax 108Å). The radius of gyration of both the rsAra h 1 trimer and monomer were calculated using CRYSOL and found to be 32.2 Å and 24.5 Å, respectively.
The average normalized spatial discrepancy (NSD) of 10 runs of DAMMIF for each version of Ara h 1 are 0.78 ± 0.03, 1.22 ± 0.08, and 0.77 ± 0.02 for nAra h 1, rAra h 1 and rsAra h 1, respectively. The fit of the most probable run for each version is plotted along with the scattering curves in Fig. 5A for nAra h 1 (Fig. 5, Ai), rAra h 1 (Fig. 5, Aii), and rsAra h 1 (Fig. 5, Aiii). The most probable bead models for nAra h 1 and rsAra h 1 were aligned using SUPCOMB (59) and are depicted in Fig. 5C. After rsAra h 1 models were filtered with DAMFILT, triangular features were observed. The scattering curves were computed using CRYSOL for both the four (χ = 1.22) and three (χ = 2.71) subunit models (please see supplemental Fig. S1) and are depicted in Fig. 5, Aiii, curves g and h, respectively.
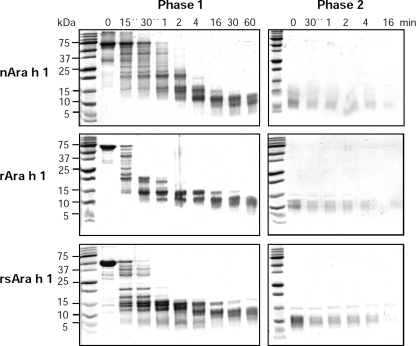
Digestion of Ara h 1 with Gastric Enzymes
The digestion studies indicate that nAra h 1 is the most stable form of the protein, followed by rsAra h 1 and rAra h 1 (Fig. 6), as shown during the in vitro gastric digestion test. However, from the 4th minute of the test the differences become significantly smaller, and all forms of the allergen are hydrolyzed in the duodenal digestion.
FIGURE 6.
Gastric (phase 1) and duodenal (phase 2) digestion of nAra h 1, rAra h 1 and rsAra h 1. Three micrograms of each sample taken at the indicated times were analyzed by 16% SDS-PAGE under reducing conditions and detected by Coomassie staining.
Immunologic Properties of Ara h 1
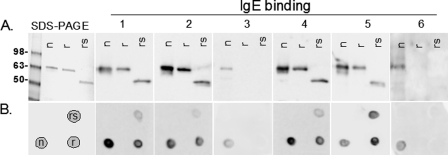
Purified, nAra h 1, rAra h 1, and rsAra h 1 were subjected to SDS-PAGE and IgE Western blot analysis (Fig. 7A) with sera from 6 peanut allergic individuals (indicated as 1–6). For all patients the linear native Ara h 1 bound higher levels of IgE than both recombinant versions (r and rs) (Fig. 7A). In all cases where the rAra h 1 was recognized, rsAra h 1 showed equal or reduced IgE binding, due to having a reduced number of IgE binding sites. In two cases (3 and 6), the patients only recognized nAra h 1. Spot blot analysis (Fig. 7B) of folded n, r, and rsAra h 1 showed similar results, with the exception of patient number 5, whose IgE bound with higher levels of the recombinant Ara h 1 proteins than the native Ara h 1.
FIGURE 7.
IgE binding to folded and unfolded, native and recombinant Ara h 1. A, SDS-PAGE and Western blot analysis of patient sera (1–6) IgE binding to linear native (n), full length recombinant (r), and recombinant shortened version of Ara h 1 (rs). B, spot blot of patient sera-IgE binding to folded n, r, and rsAra h 1 according to the template shown in the bottom left box.
DISCUSSION
The structural analysis revealed that the overall fold of the Ara h 1 core is very similar to the reported structures of other vicilins and legumins. These proteins share a bicupin fold, which most likely arose from the duplication and fusion of a single cupin domain (12). Despite the fact that many proteins having cupin folds are involved in enzymatic reactions (11, 12), there are no reports of vicilins having catalytic activity. While Ara h 1 cannot bind metal ions in the same way as OxdC, it is possible that Ara h 1 binds some small molecular ligands (24, 25). Moreover, as the binding cavities of both cupin domains are different, it is also possible that Ara h 1 binds more than one ligand.
In its crystalline state only trimeric rsAra h 1 assemblies could be identified. The structures reported here were determined in space group R3 with two protein chains in the asymmetric unit, whereas Cabanos et al. (26) reported that crystals obtained under very similar conditions belonged to the monoclinic space group C2 and contained two trimers of Ara h 1 in the asymmetric unit. However, SAXS data strongly support that this variant of the protein forms higher order oligomers, with the oligomeric states most likely being between nine and twelve. Similar composition of the oligomers is observed for the natural allergen, with assemblies that could be described as trimer of trimers or tetramer of trimers. This result is consistent with observation made by van Boxtel et al. (23) for native Ara h 1 purified from peanuts. Such results permit speculation that the core fragment of the protein is sufficient for trimer formation and formation of higher order oligomeric assemblies. It was also reported that the formation of the higher order oligomeric assemblies is promoted by the presence of some small molecules, whereas higher ionic strength promoted their dissociation into trimers (23). The SAXS results also show that rAra h 1 used in our experiments is most likely partially folded and, in the condition used for measurements, exists predominantly as a monomer. This degree of folding is consistent with the results of digestion studies that indicate that nAra h 1 is the most stable, followed by rsAra h 1 and rAra h 1.
Despite a significant difference in the length of the protein chain, rAra h 1 and rsAra h 1 display very similar patterns of interactions with IgE from sera of peanut allergic patients. Although rAra h 1does not exhibit fully native structure/conformation and is partially unfolded, it is recognized to a greater extent as it contains three additional IgE epitopes (19) that are missing in rsAra h 1, which was designed mainly for structure determination purposes. It seems that the presence of three additional epitopes is more important for IgE binding than the folding of rAra h 1. IgE from patient #5 binds to rAra h 1 at a higher level than to nAra h 1, which implies that fewer conformational epitopes are preferentially recognized by the IgE of some patients. The recombinant variants of the allergens are not recognized by two out of six analyzed sera (Fig. 7). This lack of recognition may be of consequence for the newly emerging component-resolved diagnostic methods that rely on the use of recombinant allergens, and may be explained by the lack of a carbohydrate component of the allergen (60) or by the presence of modified amino acids that have been shown to be present in food proteins, including peanut allergens (61). This difference in the interaction with IgE indicates that the recombinant allergen may not always be an adequate replacement of the natural protein in allergy diagnostics.
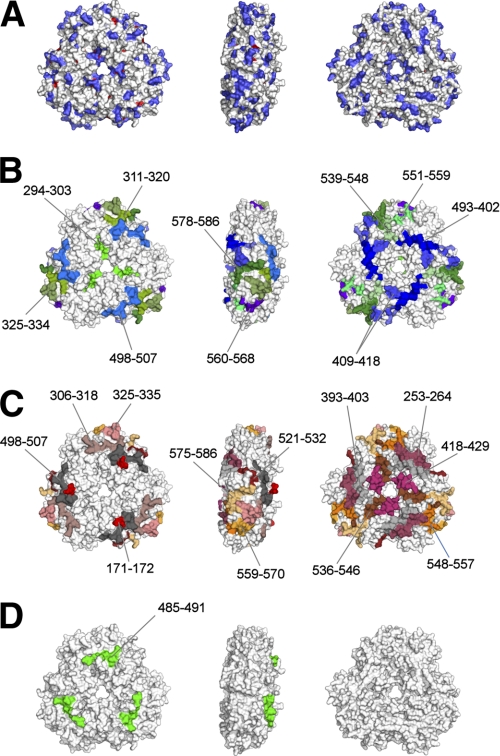
Peanuts and tree nuts are responsible for a significant number of anaphylactic reactions in the U.S (62, 63). It is estimated that 34% of patients with peanut allergies also react with tree nuts (9). It was demonstrated that vicilin allergens of peanut and tree nuts share IgE-binding epitopes (3), in agreement with bioinformatics analyses presented here (Fig. 1), which show that the cross-reacting proteins cluster together. Moreover, molecular modeling studies and clinical reports show that there is a structural basis for the cross-reactivity between Ara h 1, Len c 1 (lentil) and Pis s 1 (pea) (64, 65). It was also shown that Ara h 1 is cross-reactive with an allergen from lupine (66). Structural analysis of the rsAra h 1, sequence conservation analysis of the vicilin group allergens (Fig. 3C), and the results of IgE-binding studies (Fig. 8) reveal several regions that are both conserved in terms of the sequence and interaction with IgE antibodies. These regions are located close to the center and are also found on the “edge” of the Ara h 1 trimer. For example, residues 294–303 identified by Burks et al. (19) are partially conserved in vicilin allergens analyzed in this report and are located close to the center of the Ara h 1 trimer (Fig. 8B, left part). IgE-binding regions comprising residues 311–320, 325–334, 409–418, 498–507, 561–568, 578–587 are also partially conserved between vicilin allergens and are located on the “edge” of the trimer (Fig. 8B, central part). Some of these fragments (i.e. 409–418, 498–507 and 525–534) were identified as immunodominant epitopes (21). Two epitopes identified by Burks et al. (19) (residues 525–534) and Cong et al. (20) (residues 448–458), are almost completely buried and are not visible in Fig. 8. In addition, another epitope which was identified by both Burks et al. (residues 344–353) and Cong et al. (residues 342–353) is also not shown in Fig. 8, as the loop fragment on which it is located could not be modeled in the crystal structure of rsAra h 1. In summary, it is possible that the Ara h 1 fragments listed here, both of which bind IgE and are conserved between vicilin allergens, are at least partially responsible for the cross-reactivity between peanuts and tree nuts.
FIGURE 8.
Composite of the antibody binding studies and sequence conservation mapped on rsAra h 1 structure. A, sequence conservation between different allergens belonging to the vicilin group (Ana o 1, Ara h 1, Cor a 11, Gly m 5, Jug n 2, Jug r 2, Len c 1, Lup an 1, Pis s 1, and Ses i 3) are mapped onto the molecular surface of the rsAra h 1 trimer. Invariant residues are shown in red, and conserved residues are shown in blue. B–D, molecular surface of rsAra h 1 trimer is depicted with peptides that were shown to interact with human antibodies. B, epitopes identified by Burks et al. (19): each epitope is shown using a different color. Immunodominant epitopes are shown in shades of purple and blue, and other epitopes in shades of green. C, epitopes identified by Cong et al. (20): immunodominant epitopes are colored using different shades of gray, and other epitopes in shades of red, orange, and yellow. D, amino acids from an IgM binding epitope as reported by Shinmoto et al. (67) are shown in green. Only residues present in the rsAra h 1 structure are labeled.
Supplementary Material
Acknowledgments
We thank Steve Almo and Alexander Wlodawer for valuable comments on the manuscript and Michal Sabat for help with SAXS experiments. Some of the methodology used in this publication was developed for the New York Structural Genomics Consortium supported by Grant GM094662. The structural results shown in this report are derived from work performed at Argonne National Laboratory, at the Structural Biology Center of the Advanced Photon Source. Argonne is operated by University of Chicago Argonne, LLC, for the U.S. Dept. of Energy, Office of Biological and Environmental Research under Contract DE-AC02-06CH11357.
This work was supported by Grant GM53163, the Austrian Research Promotion Agency (FFG) BRIDGE Grant 820127, and the US Dept. of Agriculture-Agricultural Research Service.
The atomic coordinates and structure factors (codes 3s7e and 3s7i) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- PDB
- Protein Data Bank
- SAXS
- small angle x-ray scattering
- HSP
- highly scoring segment pair
- HTH
- helix-turn-helix
- CLANS
- CLuster ANalysis of Sequences
- Rg
- radius of gyration.
REFERENCES
- 1. Mills E. N., Jenkins J., Marigheto N., Belton P. S., Gunning A. P., Morris V. J. (2002) Biochem. Soc. Trans. 30, 925–929 [DOI] [PubMed] [Google Scholar]
- 2. Willison L. N., Tawde P., Robotham J. M., Penney R. M., 4th, Teuber S. S., Sathe S. K., Roux K. H. (2008) Clin. Exp. Allergy 38, 1229–1238 [DOI] [PubMed] [Google Scholar]
- 3. Barre A., Sordet C., Culerrier R., Rancé F., Didier A., Rougé P. (2008) Mol. Immunol. 45, 1231–1240 [DOI] [PubMed] [Google Scholar]
- 4. Skolnick H. S., Conover-Walker M. K., Koerner C. B., Sampson H. A., Burks W., Wood R. A. (2001) J. Allergy Clin. Immunol. 107, 367–374 [DOI] [PubMed] [Google Scholar]
- 5. Fleischer D. M. (2007) Curr. Allergy Asthma Rep 7, 175–181 [DOI] [PubMed] [Google Scholar]
- 6. Sicherer S. H., Sampson H. A. (2007) J. Allergy Clin. Immunol. 120, 491–503 [DOI] [PubMed] [Google Scholar]
- 7. de Leon M. P., Rolland J. M., O'Hehir R. E. (2007) Expert Rev. Mol. Med. 9, 1–18 [DOI] [PubMed] [Google Scholar]
- 8. Khodoun M., Strait R., Orekov T., Hogan S., Karasuyama H., Herbert D. R., Köhl J., Finkelman F. D. (2009) J. Allergy Clin. Immunol. 123, 342–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Teuber S. S., Beyer K. (2004) Curr. Opin. Allergy Clin. Immunol. 4, 201–203 [DOI] [PubMed] [Google Scholar]
- 10. Agarwal G., Rajavel M., Gopal B., Srinivasan N. (2009) PLoS One 4, e5736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dunwell J. M., Culham A., Carter C. E., Sosa-Aguirre C. R., Goodenough P. W. (2001) Trends Biochem. Sci. 26, 740–746 [DOI] [PubMed] [Google Scholar]
- 12. Dunwell J. M., Purvis A., Khuri S. (2004) Phytochemistry 65, 7–17 [DOI] [PubMed] [Google Scholar]
- 13. Koppelman S. J., Bruijnzeel-Koomen C. A., Hessing M., de Jongh H. H. (1999) J. Biol. Chem. 274, 4770–4777 [DOI] [PubMed] [Google Scholar]
- 14. Schmitt D. A., Nesbit J. B., Hurlburt B. K., Cheng H., Maleki S. J. (2010) J Agric Food Chem. 58, 1138–1143 [DOI] [PubMed] [Google Scholar]
- 15. Mondoulet L., Paty E., Drumare M. F., Ah-Leung S., Scheinmann P., Willemot R. M., Wal J. M., Bernard H. (2005) J. Agric Food Chem. 53, 4547–4553 [DOI] [PubMed] [Google Scholar]
- 16. Maleki S. J., Kopper R. A., Shin D. S., Park C. W., Compadre C. M., Sampson H., Burks A. W., Bannon G. A. (2000) J. Immunol. 164, 5844–5849 [DOI] [PubMed] [Google Scholar]
- 17. van Boxtel E. L., Koppelman S. J., van den Broek L. A., Gruppen H. (2008) J. Agric. Food Chem. 56, 2223–2230 [DOI] [PubMed] [Google Scholar]
- 18. Koppelman S. J., Hefle S. L., Taylor S. L., de Jong G. A. (2010) Mol. Nutr Food Res. 54, 1711–1721 [DOI] [PubMed] [Google Scholar]
- 19. Burks A. W., Shin D., Cockrell G., Stanley J. S., Helm R. M., Bannon G. A. (1997) Eur. J. Biochem. 245, 334–339 [DOI] [PubMed] [Google Scholar]
- 20. Cong Y. J., Lou F., Xue W. T., Li L. F., Chen M. H. (2008) Food Agric. Immunol. 19, 175–185 [Google Scholar]
- 21. Shin D. S., Compadre C. M., Maleki S. J., Kopper R. A., Sampson H., Huang S. K., Burks A. W., Bannon G. A. (1998) J. Biol. Chem. 273, 13753–13759 [DOI] [PubMed] [Google Scholar]
- 22. Burks A. W., Cockrell G., Stanley J. S., Helm R. M., Bannon G. A. (1995) J. Clin. Invest. 96, 1715–1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. van Boxtel E. L., van Beers M. M., Koppelman S. J., van den Broek L. A., Gruppen H. (2006) J. Agric Food Chem. 54, 7180–7186 [DOI] [PubMed] [Google Scholar]
- 24. Chung S. Y., Champagne E. T. (2007) J. Agric Food Chem. 55, 9054–9058 [DOI] [PubMed] [Google Scholar]
- 25. van Boxtel E. L., van den Broek L. A., Koppelman S. J., Vincken J. P., Gruppen H. (2007) J. Agric Food Chem. 55, 8772–8778 [DOI] [PubMed] [Google Scholar]
- 26. Cabanos C., Urabe H., Masuda T., Tandang-Silvas M. R., Utsumi S., Mikami B., Maruyama N. (2010) Acta Crystallogr Sect F Struct. Biol. Cryst Commun. 66, 1071–1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Moreno F. J., Mellon F. A., Wickham M. S., Bottrill A. R., Mills E. N. (2005) Febs J. 272, 341–352 [DOI] [PubMed] [Google Scholar]
- 28. Cymborowski M., Klimecka M., Chruszcz M., Zimmerman M. D., Shumilin I. A., Borek D., Lazarski K., Joachimiak A., Otwinowski Z., Anderson W., Minor W. (2010) J. Struct. Funct. Genomics 11, 211–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zimmerman M. D., Chruszcz M., Koclega K. D., Otwinowski Z., Minor W. (2005) Acta Crystallogr. Sect. A 61, c178-c179 [Google Scholar]
- 30. Minor W., Cymborowski M., Otwinowski Z., Chruszcz M. (2006) Acta Crystallogr. D Biol. Crystallogr. 62, 859–866 [DOI] [PubMed] [Google Scholar]
- 31. Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 32. Vagin A., Teplyakov A. (1997) J. Appl. Crystallogr. 30, 1022–1025 [Google Scholar]
- 33. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 34. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Acta Crystallogr. D66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Collaborative Computational Project, Number 4 (1994) Acta Crystallogr. D50, 760–763 [Google Scholar]
- 36. Lovell S. C., Davis I. W., Arendall W. B., 3rd, de Bakker P. I., Word J. M., Prisant M. G., Richardson J. S., Richardson D. C. (2003) Proteins 50, 437–450 [DOI] [PubMed] [Google Scholar]
- 37. Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B., 3rd, Snoeyink J., Richardson J. S., Richardson D. C. (2007) Nucleic Acids Res. 35, W375–W383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yang H., Guranovic V., Dutta S., Feng Z., Berman H. M., Westbrook J. D. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 1833–1839 [DOI] [PubMed] [Google Scholar]
- 39. Strong M., Sawaya M. R., Wang S., Phillips M., Cascio D., Eisenberg D. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 8060–8065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rosenbaum G., Alkire R. W., Evans G., Rotella F. J., Lazarski K., Zhang R. G., Ginell S. L., Duke N., Naday I., Lazarz J., Molitsky M. J., Keefe L., Gonczy J., Rock L., Sanishvili R., Walsh M. A., Westbrook E., Joachimiak A. (2006) J. Synchrotron Radiat. 13, 30–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Berman H. M., Battistuz T., Bhat T. N., Bluhm W. F., Bourne P. E., Burkhardt K., Feng Z., Gilliland G. L., Iype L., Jain S., Fagan P., Marvin J., Padilla D., Ravichandran V., Schneider B., Thanki N., Weissig H., Westbrook J. D., Zardecki C. (2002) Acta Crystallogr. D58, 899–907 [DOI] [PubMed] [Google Scholar]
- 42. Konarev P. V., Volkov V. V., Sokolova A. V., Koch M. H. J., Svergun D. I. (2003) J. Appl. Crystallogr. 36, 1277–1282 [Google Scholar]
- 43. Putnam C. D., Hammel M., Hura G. L., Tainer J. A. (2007) Q. Rev. Biophys. 40, 191–285 [DOI] [PubMed] [Google Scholar]
- 44. Petoukhov M. V., Konarev P. V., Kikhney A. G., Svergun D. I. (2007) J. Appl. Crystallogr. 40, S223-S228 [Google Scholar]
- 45. Fischer H., Neto M. D., Napolitano H. B., Polikarpov I., Craievich A. F. (2010) J. Appl. Crystallogr. 43, 101–109 [Google Scholar]
- 46. Svergun D. I. (1992) J. Appl. Crystallogr. 25, 495–503 [Google Scholar]
- 47. Franke D., Svergun D. I. (2009) J. Appl. Crystallogr. 42, 342–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Volkov V. V., Svergun D. I. (2003) J. Appl. Crystallogr. 36, 860–864 [Google Scholar]
- 49. Petoukhov M. V., Svergun D. I. (2005) Biophys. J. 89, 1237–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Svergun D. I., Barberato C., Koch M. H. J. (1995) J. Appl. Crystallogr. 28, 768–773 [Google Scholar]
- 51. Altschul S. F., Koonin E. V. (1998) Trends Biochem. Sci. 23, 444–447 [DOI] [PubMed] [Google Scholar]
- 52. Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. (1997) Nucleic Acids Res. 25, 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Frickey T., Lupas A. (2004) Bioinformatics 20, 3702–3704 [DOI] [PubMed] [Google Scholar]
- 54. Krissinel E., Henrick K. (2007) J. Mol. Biol. 372, 774–797 [DOI] [PubMed] [Google Scholar]
- 55. Krissinel E., Henrick K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2256–2268 [DOI] [PubMed] [Google Scholar]
- 56. DeLano W. L. (2002) The PyMOL Molecular Graphics System [Google Scholar]
- 57. Laskowski R. A., Watson J. D., Thornton J. M. (2005) Nucleic Acids Res. 33, W89–W93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Anand R., Dorrestein P. C., Kinsland C., Begley T. P., Ealick S. E. (2002) Biochemistry 41, 7659–7669 [DOI] [PubMed] [Google Scholar]
- 59. Kozin M. B., Svergun D. I. (2001) J. Appl. Crystallogr. 34, 33–41 [Google Scholar]
- 60. Kolarich D., Altmann F. (2000) Anal. Biochem. 285, 64–75 [DOI] [PubMed] [Google Scholar]
- 61. Li J., Shefcheck K., Callahan J., Fenselau C. (2010) Protein Sci. 19, 174–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Neugut A. I., Ghatak A. T., Miller R. L. (2001) Arch Intern Med. 161, 15–21 [DOI] [PubMed] [Google Scholar]
- 63. Keet C. A., Wood R. A. (2007) Immunol. Allergy Clin. North Am. 27, 193–212, vi [DOI] [PubMed] [Google Scholar]
- 64. Barre A., Borges J. P., Rouge P. (2005) Biochimie 87, 499–506 [DOI] [PubMed] [Google Scholar]
- 65. Wensing M., Knulst A. C., Piersma S., O'Kane F., Knol E. F., Koppelman S. J. (2003) J. Allergy Clin. Immunol. 111, 420–424 [DOI] [PubMed] [Google Scholar]
- 66. Dooper M. M., Plassen C., Holden L., Lindvik H., Faeste C. K. (2009) J. Investig Allergol Clin. Immunol. 19, 283–291 [PubMed] [Google Scholar]
- 67. Shinmoto H., Takeda M., Matsuo Y., Naganawa Y., Tomita S., Takano-Ishikawa Y. (2010) Hum. Antibodies 19, 101–105 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.