Background: The inherent cytoprotective mechanism involved in repair of injured gastric mucosa is not clear.
Results: HO-1 is induced and translocated to mitochondria to favor repair of gastric mucosal injury induced by non-steroidal anti-inflammatory drug-mediated mitochondrial oxidative stress (MOS).
Conclusion: Mitochondrial localization of HO-1 is a novel cytoprotective mechanism against MOS-mediated gastric mucosal injury.
Significance: Induction of HO-1 in gastric mucosa is beneficial for gastroprotection.
Keywords: Heme Oxygenase, Mitochondrial Apoptosis, Nrf2, Oxidative Stress, Reactive Oxygen Species (ROS)
Abstract
The mechanism of action of heme oxygenase-1 (HO-1) in mitochondrial oxidative stress (MOS)-mediated apoptotic tissue injury was investigated. MOS-mediated gastric mucosal apoptosis and injury were introduced in rat by indomethacin, a non-steroidal anti-inflammatory drug. Here, we report that HO-1 was not only induced but also translocated to mitochondria during gastric mucosal injury to favor repair mechanisms. Furthermore, mitochondrial translocation of HO-1 resulted in the prevention of MOS and mitochondrial pathology as evident from the restoration of the complex I-driven mitochondrial respiratory control ratio and transmembrane potential. Mitochondrial translocation of HO-1 also resulted in time-dependent inhibition of apoptosis. We searched for the plausible mechanisms responsible for HO-1 induction and mitochondrial localization. Free heme, the substrate for HO-1, was increased inside mitochondria during gastric injury, and mitochondrial entry of HO-1 decreased intramitochondrial free heme content, suggesting that a purpose of mitochondrial translocation of HO-1 is to detoxify accumulated heme. Heme may activate nuclear translocation of NF-E2-related factor 2 to induce HO-1 through reactive oxygen species generation. Electrophoretic mobility shift assay and chromatin immunoprecipitation studies indicated nuclear translocation of NF-E2-related factor 2 and its binding to HO-1 promoter to induce HO-1 expression during gastric injury. Inhibition of HO-1 by zinc protoporphyrin aggravated the mucosal injury and delayed healing. Zinc protoporphyrin further reduced the respiratory control ratio and transmembrane potential and enhanced MOS and apoptosis. In contrast, induction of HO-1 by cobalt protoporphyrin reduced MOS, corrected mitochondrial dysfunctions, and prevented apoptosis and gastric injury. Thus, induction and mitochondrial localization of HO-1 are a novel cytoprotective mechanism against MOS-mediated apoptotic tissue injury.
Introduction
Mitochondrial dysfunction induces cell death in a myriad of pathological conditions (1). The major cause of mitochondrial dysfunction is the generation of reactive oxygen species (ROS)3 (1). Development of mitochondrial oxidative stress (MOS) due to increased production of ROS impairs respiratory chain function and depletes cellular ATP, leading to apoptosis (2). MOS disrupts cellular integrity and promotes cell death by down-regulating proliferating cell nuclear antigen, survivin, epidermal growth factor (EGF), basic fibroblast growth factor, Bcl2, and BclxL (3–5), ultimately leading to tissue injury, organ damage, and pathology. The mitochondrial apoptotic pathway contributes significantly to the pathogenesis of traumatic brain injury and sepsis (6), ischemic and hemorrhagic stroke, acute and degenerative cardiac myocyte death (7), alcoholic liver injury (8), gastric mucosal injury (2), and liver injury in malaria (9). However, subsequent to injury, a number of inherent cytoprotective factors are involved in repair mechanisms; one of these factors is heme oxygenase 1 (HO-1). HO-1, encoded by the Hmox1 gene, is an evolutionarily conserved enzyme (10). The Hmox1 gene exhibits a ubiquitous expression in most living organisms, which suggests that this enzyme appeared early in evolution. HO-1 is induced by a variety of stimuli such as free heme, oxidative stress, inflammation, heavy metals, and UV radiation (11–14) and is supposed to play an important role in the protection against tissue injury from oxidative stress (15, 16). HO-1 is overexpressed in neurons resisting oxidative stress-mediated cell death (17). Increased expression of HO-1 is evident in inflammatory diseases (18), cardiovascular diseases (19), non-cerebral forms of severe malaria (20), lung injury (21), and other pathological conditions. Previous studies designate a defensive role for HO-1 in heme- and non-heme-mediated models of acute renal injury using chemical inducers and inhibitors of HO-1 (22). HO-1 also protects against gastric mucosal tissue injury induced by non-steroidal anti-inflammatory drugs (NSAIDs) (23, 24). However, the mechanism of the cytoprotective role of HO-1 against mitochondrial oxidative stress has not been elucidated in sufficient detail. Thus, indomethacin-induced gastric injury and subsequent autohealing are an excellent in vivo model to follow the mechanism of the cytoprotective role of HO-1. Here, for the first time, we report the mitochondrial localization of HO-1 in vivo during NSAID-induced gastric mucosal injury as a novel cytoprotective mechanism. The mitochondrial translocation of HO-1 resulted in the prevention of NSAID-induced mitochondrial dysfunction and oxidative stress, gastric mucosal cell apoptosis, and gastric mucosal injury.
EXPERIMENTAL PROCEDURES
Indomethacin, thiobarbituric acid, 5,5′-dithiobis(nitrobenzoic acid), collagenase, hyaluronidase, the caspase-3 assay kit, NADPH, JC-1 (5,5′,6,6′-tetrachloro-1,1′,3,3′-tetraethylbenzimidazolylcarbocyanine iodide), glucose-6-phosphate dehydrogenase, glucose 6-phosphate, and hemin were obtained from Sigma. The caspase-9 assay kit was purchased from Biovision (Mountain View, CA). The Dead End Colorimetric TUNEL assay kit was purchased from Promega Corp. The RevertAid HMinus First Strand cDNA Synthesis kit, 2× PCR Master Mix, and nuclease-free water were purchased from Fermentas. HO-1 antibody was procured from Abcam. NF-E2-related factor 2 (Nrf2) antibody was obtained from Santa Cruz Biotechnology (Santa Cruz, CA). The custom-based primers and antioxidant response element (ARE) sequence were purchased from Sigma Genosys. The mitochondrial isolation kit was purchased from Biochain Institute (Hayward, CA). TRIzol and Alexa Fluor 647 anti-rabbit antibodies were purchased from Invitrogen. Cytochrome c oxidase (COX)-IV Alexa Fluor 488-conjugated primary antibody was obtained from Cell Signaling Technology. Zinc protoporphyrin and secondary anti-rabbit HRP-conjugated antibody were purchased from Calbiochem. The QuantiChromTM Heme Assay kit was purchased from Bioassay Systems (Hayward, CA). All other reagents were of analytical grade purity.
Animals and Indomethacin-induced Gastric Mucosal Injury
Sprague-Dawley rats (180–220 g) were used in all experiments. Animals were kept at 24 ± 2 °C with 12-h light and dark cycles. Before initiating the experimental procedure, the animals were fasted for 24 h with access only to water to avoid food-induced increased acid secretion and its indulging effect on gastric lesions. The animal ethics committee guidelines were stringently followed while carrying out all in vivo studies. Indomethacin-induced gastric mucosal injury or treatment with zinc protoporphyrin (ZnPP) and cobalt protoporphyrin (CoPP) were performed as described in the literature (24–26). Briefly, all the animals were divided into control, indomethacin, indomethacin plus ZnPP, only ZnPP, and indomethacin plus CoPP groups. Both the indomethacin and indomethacin plus ZnPP groups of rats were further subdivided into subgroups based on the time at which they were to be sacrificed. Gastric mucosal tissue injury was induced in the starved animals with oral administration of indomethacin at the dose of 48 mg kg−1 b.w. The indomethacin-treated rats were subdivided into several groups (six to eight rats in each group). The animals were sacrificed at different time points (0, 2, 4, 12, 24, 48, and 72 h, respectively) to obtain the stomachs, which were subsequently used for further studies. In only the ZnPP group and ZnPP-pretreated group, ZnPP (50 μg kg−1 b.w.) was injected intraperitoneally thrice at regular intervals of 8 h. The first treatment with ZnPP was given 30 min prior to indomethacin administration. Stomachs were obtained from animals 2, 4, 12, 24, 48, and 72 h after indomethacin treatment. The animals that were treated with indomethacin plus CoPP were sacrificed only at 4 h after indomethacin treatment. The indomethacin plus CoPP group was subdivided into four subgroups based on the four doses of CoPP (500 μg, 2.5 mg, 5 mg, and 10 mg kg−1 b.w.). The control animals received only vehicle (no indomethacin). The quantification of the mucosal injury or injury index was scored as follows: 0 = no pathology; 1 = one pinhead ulcer. The sum of the total scores divided by the number of animals gave the injury index (2, 3).
Histological Study
Stomachs from control, indomethacin-treated, and ZnPP plus indomethacin-treated rats at different time points were fixed in 10% formalin (in PBS) for 12 h at 25 °C and embedded in paraffin for preparing semithin sections, which were ultimately taken on poly-l-lysine-coated glass slides and subjected to eosin-hematoxylin double staining (3, 27). The slides were then observed under a microscope (Leica DM-2500) equipped with a high resolution digital camera.
PCR for HO-1 and Mitochondrial Ferritin (MtFt)
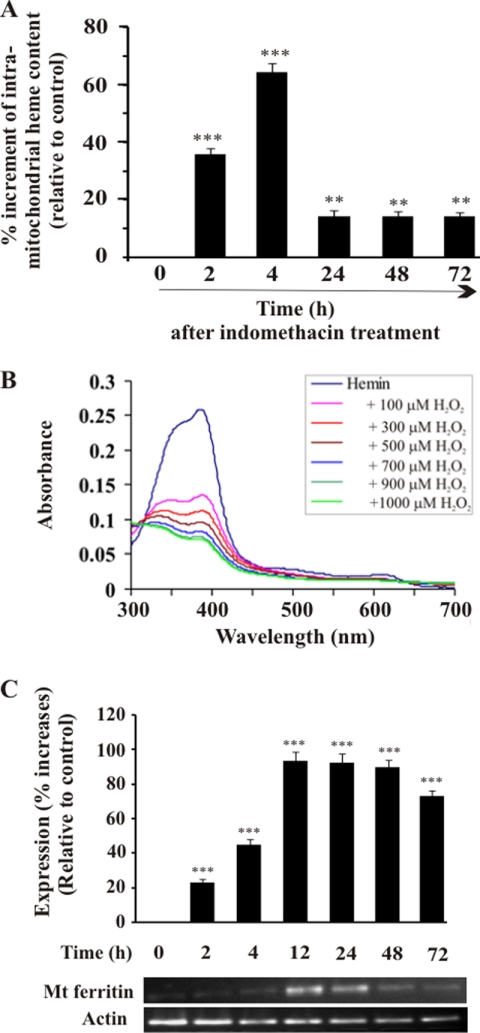
Equal amounts of tissues (75 mg) from both control and indomethacin-treated gastric mucosa were used for total RNA isolation using the TRIzol method of RNA isolation. RNA (2 μg) was used to prepare cDNA using the RevertAid HMinus First Strand cDNA Synthesis kit. Equal amounts of cDNA were used for PCR amplification using specific forward and reverse primers of HO-1, MtFt, and actin. Primers were made using the following sequences: for HO-1, 5′-TATGCCCCACTCTACTTCC-3′ (forward) and 5′-TCTTAGCCTCTTCTGTCACC-3′ (reverse); for MtFt, 5′-AATCAACATGGAGCTTTACG-3′ (forward) and 5′-CATGCAGGTAGTGAGTTTCC-3′ (reverse); and for β-actin, 5′-CTATGTTGCCCTAGACTTCG-3′ (forward) and 5′-TTGATCTTCATGGTGCTAGG-3′ (reverse). The PCR protocol consisted of 95 °C for 2 min for initial denaturation, 35 cycles of denaturation at 94 °C for 1 min, annealing for 45 s at respective annealing temperatures specific for each primer set, and then extension at 72 °C for 2 min followed by final extension at 72 °C for 10 min. The PCR-amplified products were resolved in a 2% agarose gel and documented in a Gel Doc system (Bio-Rad).
Isolation of Mitochondria
Mitochondria were isolated and purified by following the protocol reported earlier (28) with slight modifications. In brief, the scraped gastric mucosa from control and experimental rats (post-indomethacin, -ZnPP, and -CoPP treatments) obtained at 2-, 4-, 12-, 24-, 48-, and 72-h time intervals were minced, homogenized in isolation buffer, and subsequently centrifuged at 600 × g for 10 min to remove the nuclear pellet and cell debris. This was followed by centrifugation at 12,000 × g for 15 min to obtain the crude mitochondrial pellet. A 25–50% Percoll density gradient was prepared, and the crude mitochondrial pellet (resuspended in cold 15% Percoll solution) was layered above 25% Percoll. It was further centrifuged at 30,000 × g at 4 °C for 30 min to obtained pure mitochondria at the interface between the Percoll (25–50%) layers. The mitochondria were isolated from the interface, washed with isolation buffer, and centrifuged at 16,700 × g at 4 °C for 10 min. The supernatant was discarded, 10 mg ml−1 fatty acid-free BSA was added and mixed, and the sample was further centrifuged at 6900 × g at 4 °C for 10 min. The resultant pellet containing purified mitochondria was resuspended in storage buffer. Mitochondrial protein content was determined using the Lowry method (29). The identity of the purified mitochondria was confirmed by Western immunoblot analysis using a mitochondrion-specific marker, COX-IV.
Preparation of Nuclear Extracts
Nuclear extracts were isolated using nuclear extraction buffers I (10 mm HEPES, 1.5 mm MgCl2, 10 mm KCl, and 0.5% Triton X-100) and II (1 m NaCl, 0.2 m EDTA, 20% glycerol, and 0.5 mm DTT). The gastric mucosa collected at different time points after indomethacin treatment was first minced and homogenized with buffer I. The samples were centrifuged, and the homogenates were kept in separate tubes as cytosols. The pellets were resuspended in an equal volume of both buffers I and II, vortexed, kept on ice for 30 min, and then centrifuged to obtain the nuclear extracts in the supernatant.
Western Immunoblot
Samples were applied to 12% polyacrylamide-SDS gels and subjected to electrophoresis, and proteins were then immunoblotted. Mitochondrial protein (200 μg) was subjected to Western immunoblotting using HO-1 antibody. Although cytosolic and nuclear proteins (100 μg for both) were subjected to Western immunoblotting using Nrf2, COX-IV, β-actin, and histone 3 antibodies were used as internal controls for mitochondria, cytosol, and nucleus, respectively.
HO-1 Assay
HO-1 activity was assayed in both tissue homogenates and mitochondria obtained at different time points as described earlier (30, 31). Each sample was assayed at different time points to obtain linear kinetic data. For convenience in data representation, a common single time point (60 min) was selected for all the samples. Briefly, for tissue homogenates, gastric mucosal tissues obtained at different time points subsequent to indomethacin treatment were homogenized in 1 mm PBS (pH 7.4) and centrifuged at 12,000 × g for 30 min. The supernatants of samples obtained from different time points were used for the assay of HO-1. Mitochondria isolated as described above were also used for measuring the internal HO-1 activity. The assay system consisted of an equal volume of reaction mixture (2 mm MgCl2, 30 μm hemin, 30 mg of rat liver homogenate, 0.2 units of glucose-6-phosphate dehydrogenase, 2 mm glucose 6-phosphate, and 0.8 mm NADPH), and samples were incubated at 37 °C in the dark for 1 h. The formed bilirubin was measured by ΔA464–530 (ϵ453 nm = 40 mm−1 cm−1) (11) using a UV-1700 PharmaSpec UV-visible spectrophotometer (Shimadzu). HO-1 activity was expressed as nanomoles of bilirubin formed per milligram of protein per hour.
Confocal Microscopy
Gastric mucosal cells were isolated from experimental rats as described earlier (3). Mucosa from control and indomethacin-treated stomachs was scraped in Hanks' balanced salt solution (pH 7.4) containing 100 units/ml penicillin and 100 μg/ml streptomycin at different time points. Mucosa was further minced and suspended in Hanks' balanced salt solution (pH 7.4) containing 0.05% hyaluronidase and 0.1% collagenase. For 30 min, the suspension was incubated at 37 °C in 5% CO2 under shaking condition. The suspension was then filtered through a sterile nylon mesh. The filtrate was centrifuged at 600 × g for 5 min, and the cell pellet was washed with Hanks' balanced salt solution (pH 7.4) for subsequent studies. About 100 μl of cell suspension (approximately 105 cells) from the control and the two experimental groups was taken and spread on poly-l-lysine-coated slides kept at room temperature. Cells were then washed with Tris-buffered saline (TBS), fixed in 4% paraformaldehyde, and permeabilized with 0.25% Triton X-100. Cells were then washed with TBS and blocked with 1% bovine serum albumin in TBS-Tween 20. Immunostaining was performed using antibody against HO-1 developed in rabbit (1:100), COX-IV Alexa Fluor 488-conjugated primary antibody (1:400; Cell Signaling Technology), and goat anti-rabbit Alexa Fluor 647-conjugated secondary antibody (1:500; Invitrogen). Cells were imaged at 1024 × 1024 resolution with a Nikon A1R confocal imaging system. The colocalization was analyzed by the use of “color composite” and “colocalization” functions using Image Pro Plus software (32). The process involved the correction of background, generation of a scatter plot, and selection of the area of interest. Pearson's correlation was used to represent the level of colocalization, and values are expressed in percent for better understanding. Pearson's correlation takes into account the similarities between shapes and does not take into account image intensity. To overcome this drawback, an overlap coefficient was also calculated and compared with Pearson's correlation values. The data presented are a representation of one of four experiments showing similar values.
Measurement of Mitochondrial Oxidative Stress
MOS was measured as described earlier (9, 25, 26, 33) by following the formation of mitochondrial lipid peroxidation products and protein carbonyl. Mitochondria were isolated using a mitochondrial isolation kit as described above. An equal amount of mitochondrial protein was taken for all experiments. For quantifying protein carbonyl formation during MOS induced by indomethacin, mitochondrial protein was precipitated with 25% trichloroacetic acid. Furthermore, the protein samples from different time points were allowed to react with 0.5 ml of 10 mm 2,4-dinitrophenylhydrazine for 1 h. The samples were then washed thrice with ethanol:ethyl acetate (1:1) solvent, dissolved in 0.6 ml of a solution containing 6 m guanidine HCl in 20 mm potassium phosphate buffer (pH 2.3) with trifluoroacetic acid, and centrifuged, and the supernatants of each set were used for measurement of protein carbonyl at 362 nm. To measure mitochondrial lipid peroxidation, 2 ml of thiobarbituric acid-TCA mixture (0.375% (w/v) and 15% (w/v), respectively) in 0.25 n HCl was added to 1 ml of the mitochondrial fraction in 0.9% normal saline and boiled for 15 min. The solutions were then cooled and further centrifuged. The supernatants were taken, and the absorbance was read at 535 nm. Tetraethoxypropane was used as a standard (25, 33).
Mitochondrial Respiration
Mitochondrial oxygen consumption was measured using a Clark-type electrode in a Liquid-Phase Oxygen Measurement System (Oxygraph, Hansatech, Norfolk, UK) with a thermoregulated chamber set at 24 °C (34). Consumption of oxygen by complex I (Stage 3) was started by the incorporation of glutamate and malate (5 mm each) to 1 ml of respiratory medium containing 250 mm sucrose, 5 mm KH2PO4, 5 mm MgCl2, 0.1 mm EDTA, and 0.1% BSA in 20 mm HEPES (pH 7.2). After the addition of mitochondrial suspension, the basal respiration (State 2) was recorded. State 3 respiration was initiated with the addition of ADP (1 mm) to the respiration medium. State 4 respiration was measured in the absence of ADP. The respiratory control ratio (RCR) was calculated from the ratio of State 3 respiration (nmol of O2 consumed) and State 4 respiration (nmol of O2 consumed).
Mitochondrial Transmembrane Potential (Δψm)
Mitochondrial transmembrane potential was measured as described earlier (2, 9, 27). Isolated mitochondria in 100 μl of JC-1 assay buffer were incubated in the dark with JC-1 (300 nm) for 10 min at 25 °C. The fluorescence of each sample was measured in a Hitachi F-7000 fluorescence spectrophotometer (excitation, 490 nm; emission, 530 nm for JC-1 monomer and 590 nm for JC-1 aggregates).
Assay of Caspase-9 and Caspase-3
Caspase-9 and -3 activity was measured from the cytosolic fraction of gastric tissues as described earlier (2, 27, 35) using a commercially available kit according to the manufacturer's protocol (Biovision, Mountain View, CA and Sigma). For caspase-9 activity, the gastric mucosa was homogenized in cell lysis buffer (provided with the kit). The homogenate was centrifuged at 16,000 × g for 15 min. The collected supernatant in turn was mixed with 50 μl of 2× reaction buffer (provided with the respective kit). Then the substrate for caspase-9 (LEHD-p-nitroanilide; 200 μm final concentration), was added. The mixture was incubated at 37 °C for 1 h, and the absorbance was taken at 405 nm. In brief, for caspase-3, gastric mucosa was homogenized in caspase-3 lysis buffer followed by centrifugation at 16,000 × g for 15 min. 10 μl of supernatant was mixed with 85 μl of assay buffer in the presence of 5 μl of substrate (Ac-DEDV-p-nitroanilide; 200 μm final concentration) and then incubated at 37 °C, and the absorbance was taken at 405 nm.
Terminal Deoxynucleotidyltransferase dUTP Nick End Labeling (TUNEL) Assay
The TUNEL assay was performed following the instructions provided with the kit (Promega). For the detection of in situ apoptosis in the gastric mucosa, TUNEL staining was performed. Control and indomethacin-treated stomachs were obtained at different time points, fixed in 10% buffered formalin for 12 h at 25 °C, and embedded in paraffin for preparing semithin sections, which were ultimately taken on poly-l-lysine-coated glass slides and subjected to the TUNEL assay. A commercially available kit (Promega) was used for the TUNEL staining of the semithin tissue sections (5 μm).
Measurement of Intramitochondrial Heme
Intramitochondrial heme was quantified using the QuantiChrom Heme Assay kit (Bioassay Systems). Isolated mitochondria were used for heme quantitation. Equal amounts of the mitochondrial protein from control and experimental groups were mixed with 200 μl of reaction mixture (provided with the kit) and incubated for 5 min at room temperature, and optical density was measured at 400 nm in a microtiter plate reader (μQuant, BioTek Instruments). The concentration of heme was measured from a standard curve using the heme standard provided with the kit.
Soret Spectroscopy
The degradation of heme by H2O2 was followed by Soret spectroscopy. In brief, in a 1-ml quartz cuvette with a 1-cm path length, 2.5 μm hemin chloride was dissolved in 0.1 n NaOH and incubated with different concentration of H2O2 (0–1000 μm) in a final volume of 1 ml. The spectra were recorded in a Shimadzu UV-1700 UV-visible spectrophotometer in a scan range of 300–700 nm.
Electrophoretic Mobility Shift Assay (EMSA)
For the electrophoretic mobility shift assay, a consensus ARE sequence, 5′TTTATGCTGTGTCATGGTT-3′ (the core ARE sequence is underlined), was used (36). The sequence was 5′-end-labeled with [γ-32P]ATP using T4 polynucleotide kinase according to standard protocols. Unincorporated [γ-32P]ATP was removed by ethanol precipitation at −20 °C (overnight) followed by washing with 70% ethanol. Reactions were performed in a volume of 10 μl containing 32P-labeled oligonucleotide probes incubated with 10 μg of nuclear extract in binding buffer (10 mm HEPES buffer (pH 7.6), 50 mm NaCl, 1 mm EDTA, 5 mm MgCl2, 0.1 mm dithiothreitol, 1 mg/ml BSA, and 0.05% Triton X-100) for 30 min on ice. Complexes were separated by native gel electrophoresis (5%) running at 10 V/cm at 4 °C (0.5× Tris borate/EDTA and 1 mm EDTA). A supershift assay was performed using anti-Nrf2 antibody.
Chromatin Immunoprecipitation (ChIP) Assay
The ChIP assay was performed as described earlier (37) with slight modification. Tissue samples (100 mg for each sample) at different time points after indomethacin treatment were incubated with 1% formaldehyde in PBS for 15 min at room temperature to allow protein-DNA cross-linking. Cross-linking was checked by adding fresh glycine to a final concentration of 0.125 m. Samples were washed twice by centrifugation at 1000 rpm for 5 min with ice-cold PBS supplemented with a protease inhibitor mixture. Samples were then minced and homogenized in SDS lysis buffer (1% SDS, 10 mm EDTA, and 50 mm Tris (pH 8.1)) (500 μl) and kept on ice for 15–20 min. This was followed by centrifugation at 600 × g for 10 min at 4 °C to pellet the nuclei. The pellets were further subjected to lysis using SDS lysis buffer, kept on ice for 30 min, and sonicated at 40% maximum amplitude (10 s on, 60 s off) for 12 pulses to get fragments in the range of 200–1000 bp. The samples were then centrifuged for 10 min at 13,000 rpm at 4 °C.
An aliquot of the lysates used in the immunoprecipitation was processed along with the rest of the sample as an input DNA sample. Chromatin was precleared using rabbit IgG (1 μg/sample) bound to protein A-Sepharose for 2 h at 4 °C. Precleared chromatin was immunoprecipitated using rabbit anti-Nrf2 antibody and rabbit IgG as control. The reactions were incubated on a rotor at 4 °C overnight. This was followed by a wash with PBS and incubation with herring sperm DNA (5 μg/sample) for 2 h at 4 °C. DNA-cross-linked proteins were then immunoprecipitated by adding the precleared samples to the herring sperm DNA/protein A-Sepharose, and the mixture was kept at 4 °C overnight. Subsequently, the sample was washed serially once in (a) low salt immune complex wash buffer, (b) high salt immune complex wash buffer, and (c) LiCl immune complex wash buffer and finally twice in a buffer containing 10 mm Tris-HCl (pH 8.0) and 1 mm EDTA. To remove the precipitated complexes from the beads, the mixtures were incubated for 30 min in 250 μl of 1% SDS with 100 mm NaHCO3. The cross-links between protein and DNA were reversed by heating the samples at 65 °C overnight in 250 μl of Tris-EDTA with 2.5 μl of proteinase K (20 mg/ml). DNA was purified using a QIAquick PCR purification kit according to the manufacturer's protocol. DNA from ChIP and input samples was resuspended in 15 and 30 μl of elution buffer from the QIAquick kit, respectively. For PCR amplification of ARE of the HO-1 promoter region, 2 μl of the ChIP and 0.5 μl of input DNA were used as template using the forward primer 5′-CCACCATATCCGGACTTTGT-3′ (bp 4035–4015 relative to the ATG start codon) and the reverse primer 5′-GCTTCTGTGCCTTTCTAGAG-3′ (bp 3798–3778) (38).
Statistical Analysis
All data are presented as mean ± S.E. The level of significance was determined by unpaired Student's t test with one-way analysis of variance as applied. A p value ≤ 0.05 was considered as significant. The degree of colocalization in confocal images was determined by Pearson's correlation using Image Pro Plus software.
RESULTS
Expression Pattern of HO-1 during Gastric Mucosal Injury and Autohealing with Time
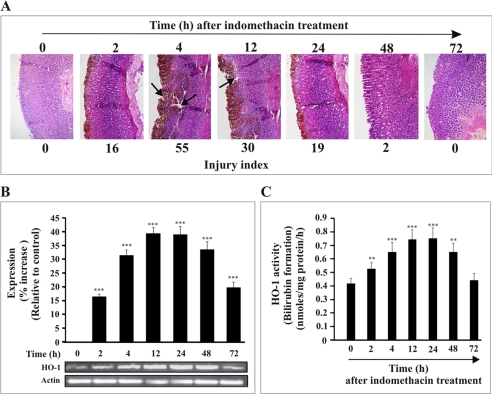
NSAID causes severe gastric mucosal injury in rats, and the injured mucosa possesses the ability to heal itself (autohealing) with time. The NSAID-induced gastric mucosal injury and subsequent “autohealing” have been used as a model to follow the expression of HO-1 and to evaluate its role in injury and healing. The expression pattern for HO-1 and the gastric mucosal injury indices were estimated at different time points subsequent to the administration of indomethacin. The injury indices are 0, 16 ± 2, 55 ± 5, 30 ± 5, 19 ± 2, 2 ± 1, and 0 for control, 2, 4, 12, 24, 48, and 72 h, respectively (Fig. 1A). The injury in gastric mucosa after indomethacin treatment was reflected by cell shedding, disintegration of mucosa, and intragastric bleeding (reddish brown color indicated by black arrow) in the superficial layer of the mucosa (Fig. 1A). The injury increased from 0 to 4 h after indomethacin treatment and then decreased as evident from the injury indices and histological studies, which document the restoration of mucosal integrity and re-epithelization in damaged mucosa with time (Fig. 1A). The recovery from the injury with time was evident from 46, 65, and 97% autohealing at 12-, 24-, and 48-h intervals, respectively (Fig. 1A). The healing or regeneration of cells in the injured site is related to the level of HO-1 mRNA expression. Despite a basal level of HO-1 mRNA expression, a dramatic increase in HO-1 expression was detected within about 2 h of indomethacin treatment with a peak between 12 and 24 h that was sustained during the healing stage (48–72 h) relative to control. It is notable that high expression of HO-1 persists even when the mucosal injury has been re-epithelized (72 h) (Fig. 1A) although at a reduced level compared with the 48-h time point (Fig. 1B). The increased expression of HO-1 was further monitored by measuring the activity of HO-1 in gastric mucosa at different time points. A marked increase in HO-1 activity was also found at the injured gastric mucosa at 4 h, and HO-1 activity remained at an elevated level from 12 to 24 h and decreased at 72 h (Fig. 1C). The enzymatic activity for “control” samples at different time points was not significantly changed from the activity found at 0 h. The “0”-h control represents rats sacrificed immediately after indomethacin treatment. Thus, the time course data clearly indicate that the expression of HO-1 is modulated in vivo in gastric mucosa during indomethacin-induced mucosal injury and subsequent autohealing with time.
FIGURE 1.
Relation of HO-1 expression with gastric mucosal injury at different times after administration of indomethacin. A, hematoxylin and eosin staining of gastric mucosal section. Histological evidence for induction of mucosal injury and subsequent healing with time after indomethacin treatment is shown. The arrow indicates severe mucosal damage at 4 and 12 h after indomethacin treatment. B, the expression of HO-1 mRNA upon induction of injury by indomethacin at different time points as followed by RT-PCR together with the densitometric analysis of RT-PCR data. Actin was used as an internal control. C, measurement of HO-1 activity. Gastric mucosal total homogenate was used to measure HO-1 activity in vivo by following the formation of bilirubin as described under “Experimental Procedures.” 0 h represents control. Data are presented as mean ± S.E. (***, p < 0.001 versus control; **, p < 0.05 versus control; n = 6).
Mitochondrial Localization of HO-1 during Gastric Mucosal Injury
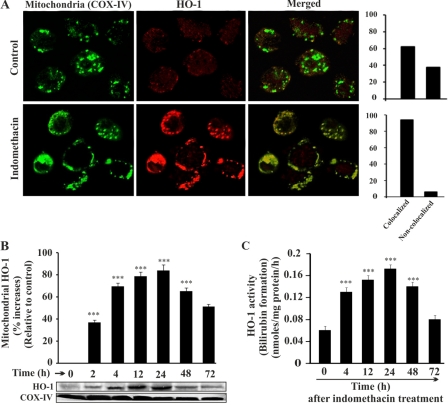
Our data from immunofluorescence studies reveal that indomethacin treatment not only up-regulated HO-1 expression but also triggered HO-1 translocation to mitochondria. A confocal microscopic study was performed using cells isolated from control rats and rats sacrificed at 4 h after indomethacin treatment (Fig. 2A). Green fluorescence represents mitochondria immunostained with Alexa Fluor 488-COX-IV antibody (Fig. 2A, first column). Red fluorescence represents the localization of HO-1 immunostained with HO-1 primary antibody and Alexa Fluor 647-conjugated secondary antibody (Fig. 2A, second column). In the control, a basal level of HO-1 in cytosol as well as in mitochondria is evident from the weak red fluorescence signal (Fig. 2A, second column), whereas in the mucosal cells from indomethacin-treated stomachs, red fluorescence indicates HO-1 overexpression and its increased mitochondrial translocation after indomethacin treatment (Fig. 2A, second column). The image analysis indicates that HO-1 (red) and COX-IV (green) colocalized (merged) in mitochondria. The level of colocalization was further measured as described under “Experimental Procedures” (Fig. 2A, side panels). In the indomethacin-treated group, the colocalization (merge) of HO-1 (red fluorescence) in mitochondria (green fluorescence; COX-IV) is excellent as evident from their colocalization score (Pearson's correlation or overlap coefficient, ∼0.94) when compared with the control group (Pearson's correlation or overlap coefficient, ∼0.62) (Fig. 2A, side panels). We further confirmed the mitochondrial translocation of HO-1 by Western immunoblot of isolated mitochondria from gastric mucosal tissue at different time points after the administration of indomethacin (Fig. 2B). Thus, all the experimental data support the translocation of HO-1 in mitochondria. The expression level of HO-1 was found to increase from 4 h, peaking between 12 and 24 h and then gradually decreasing with time (Fig. 2B). COX-IV was used as an internal control (Fig. 2B). Furthermore, to confirm whether the mitochondrial HO-1 retained its function while in mitochondria, the activity of HO-1 was measured with time (Fig. 2C). The result was consistent with the Western immunoblot analysis (Fig. 2B). The activity of HO-1 was prominent at 4 h during maximum injury. The activity reached its peak at 12–24 h and then declined subsequently (Fig. 2C).
FIGURE 2.
Mitochondrial localization of HO-1 in gastric mucosal cells during mucosal injury. A, colocalization of HO-1 with mitochondrion-specific COX-IV. The upper panel represents gastric mucosal cells from control rats; the lower panel represents gastric mucosal cells from indomethacin-treated rats. The first column shows cells immunostained with Alexa Fluor 488-COX-IV antibody giving green fluorescence specific for mitochondria, the second column represents HO-1 primary antibody- and Alexa Fluor 647 secondary antibody-stained cells giving red fluorescence specific for HO-1, and the third column shows the merged image of the first and second columns. The side panel illustrates the score values of colocalized and non-colocalized HO-1 in control and indomethacin-exposed cells. The score values indicate the level of colocalization expressed as Pearson's correlation. B, Western immunoblot for HO-1 localization in mitochondria at different time points. COX-IV was used as a loading control for the mitochondrial sample. C, HO-1 activity was measured in mitochondria obtained from rats sacrificed at different time points. 0 h represents control. Data are presented as mean ± S.E. (***, p < 0.001 versus control; n = 6).
Mitochondrial Localization of HO-1 Results in Prevention of MOS, Mitochondrial Pathology, and Apoptosis
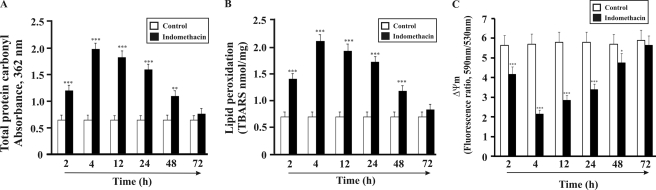
MOS plays a significant role in indomethacin-induced mitochondrial pathology and damage. The cytoprotective role of HO-1 against MOS and subsequent mitochondrial pathology was evaluated at different time points after indomethacin treatment. MOS was evaluated by measuring protein carbonyl and lipid peroxidation products in mitochondria at different time points after the administration of indomethacin (progression from injury to autohealing). The data indicate that MOS is maximal at 4 h as evident from significant protein carbonylation (71% relative to control) and lipid peroxidation (66% relative to control), respectively (Fig. 3, A and B). With time, both the formation of protein carbonyl and peroxidation of lipid decreased (Fig. 3, A and B). It is interesting to note that the localization of HO-1 in mitochondria was also prominent at 4 h after the administration of indomethacin (Fig. 2). This led us to consider the possibility of MOS triggering the translocation of HO-1 from the cytoplasm to mitochondria in response to gastric mucosal damage. HO-1 maintained a more or less increased level of expression up to 72 h. During this 4–72-h period, the protein carbonyl and lipid peroxidation status was also found to decline in accord with the temporal progress in autohealing. MOS is known to be associated with mitochondrial pathology. The generation of ROS, fall of Δψm, defect in electron transport chain, and prevention of ATP formation are the major events during mitochondrial pathology (39). It is reasonable to assume that if HO-1 in mitochondria prevents MOS then it will also restore mitochondrial function and prevent indomethacin-induced mitochondrial pathology. The maintenance of Δψm is vital for electron transport and ATP formation. The status of Δψm was checked using a lipophilic-cationic dye, JC-1. The data from these experiments indicate that indomethacin-induced gastric damage was associated with the maximum collapse of Δψm at 4 h (63% compared with control), which was recovered with time of healing when HO-1 was induced and remained up-regulated from 4 to 72 h (Fig. 3C). At higher membrane potential, JC-1 molecules generally exist as J-aggregates (emitting red fluorescence), whereas at lower potential, JC-1 molecules mainly subsist as J-monomers (green fluorescence). A drop in the ratio of red (J-aggregate; 590 nm) to green (J-monomer; 530 nm) JC-1 fluorescence is indicative of mitochondrial depolarization. We also evaluated complex I-mediated mitochondrial respiration, which exhibited time-dependent alteration (Table 1). RCR (State 3/State 4) reflects the functional status of mitochondria (Table 1). At 4 h, indomethacin significantly inhibited the electron transport chain as evident from a significant decrease of RCR (Table 1). However, the electron transport chain recovered its status with time as inferred from the restoration of RCR between 12 and 72 h (Table. 1). This was again consistent with the data found in the case of MOS recovery.
FIGURE 3.
Time course study of mitochondrial oxidative stress and mitochondrial pathology after indomethacin treatment. A, time course study of indomethacin-induced oxidation of mitochondrial proteins as measured by protein carbonyl formation. B, time course study of indomethacin-induced mitochondrial lipid peroxidation as measured by thiobarbituric acid-reactive substance (TBARS) formation. C, time course study of mitochondrial transmembrane potential as measured by JC-1 uptake (590/530 nm fluorescence ratio). Detailed descriptions are given under “Experimental Procedures.” Data are presented as mean ± S.E. (***, p < 0.001 versus control; **, p < 0.01 versus control; *, p < 0.05 versus control; n = 6).
TABLE 1.
Effect of indomethacin on mitochondrial oxygen consumption
Indomethacin was administered orally at 48 mg kg−1 b.w. to each group containing six to eight rats. Animals were sacrificed at different time points after indomethacin treatment to measure mitochondrial oxygen consumption as described under “Experimental Procedures.” Data are presented as mean ± S.E. (n = 6–8). 0 h represents control.
| Time after indomethacin treatment | State 3 | State 4 | RCR (State 3/State 4) |
|---|---|---|---|
| h | nmol O2/mg/ml | nmol O2/mg/ml | |
| 0 | 149 ± 7 | 25 ± 2 | 5.91 ± 0.6 |
| 2 | 110 ± 6a | 25 ± 1 | 4.38 ± 0.56a |
| 4 | 75 ± 5a | 23 ± 2b | 3.25 ± 0.43a |
| 12 | 78 ± 5a | 23 ± 3 | 3.29 ± 0.40a |
| 24 | 92 ± 6a | 24 ± 1 | 3.79 ± 0.39a |
| 48 | 118 ± 6a | 24 ± 2 | 4.74 ± 0.54a |
| 72 | 128 ± 6a | 25 ± 1 | 5.1 ± 0.58b |
a p < 0.001 versus control.
b p < 0.05.
Mitochondrial Localization of HO-1 Consequently Inhibits Gastric Mucosal Apoptosis
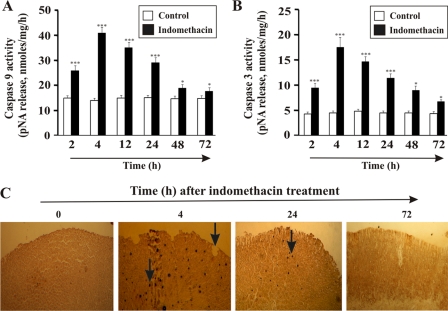
MOS plays a significant role in the induction of gastric mucosal cell apoptosis in NSAID injury. To find any link between HO-1 translocation and apoptosis, the activation of both caspase-9 (marker for the mitochondrial pathway of apoptosis) and caspase-3 (general marker for apoptosis) was measured in addition to performing a TUNEL assay to follow DNA fragmentation. The data indicate that when HO-1 is translocated to the mitochondria at 4 h the apoptosis is maximal as documented by caspase activation and DNA fragmentation (Fig. 4). However, although HO-1 expression peaked at 12–24 h and was maintained at an elevated status up to 72 h, the activity of caspases within this period decreased, suggesting an antiapoptotic role of HO-1 (Fig. 4). Caspase-9 activity was maximal at 4 h (63% augmentation relative to control) and then was reduced significantly (65 and 97% autohealing at 24 and 48 h with a 33 and 97% decrease in caspase-9 activity, respectively) with time (Fig. 4A). In the mitochondrial death pathway, activated caspase-9 ultimately leads to the activation of caspase-3. Our data confirm the maximum activation of caspase-3 at 4 h (75% increased activity compared with control) (Fig. 4B). The activity of caspase-3 also decreased with the passage of time of healing (65 and 97% autohealing at 24 and 48 h with a 31 and 54% decrease in caspase-3 activity, respectively) (Fig. 4B). DNA fragmentation is a hallmark of apoptosis. Thus, to further confirm the impact of HO-1 translocation on apoptosis in the gastric epithelial cells, a TUNEL assay was performed to track DNA fragmentation. TUNEL staining revealed that indomethacin considerably increased the apoptosis of gastric epithelial cells at 4 h (black staining; arrowhead) (Fig. 4C), and apoptosis was found to be under control at 72 h. At 24 h, however, mucosal apoptosis was also observed, which is consistent with the caspase activity (Fig. 4C).
FIGURE 4.
Time-dependent induction of apoptosis in gastric mucosal cells by indomethacin. A, time course study of indomethacin-induced activation of caspase-9. B, indomethacin-induced caspase-3 activity at different time points. C, TUNEL assay in vivo (deep brown staining suggests apoptotic DNA fragmentation, which is indicated by arrows) showing indomethacin-induced gastric mucosal cell apoptosis (which was maximum at 4 h but eventually recovered slowly) at different intervals. 0 h represents control. Data are presented as mean ± S.E. (***, p < 0.001 versus control; *, p < 0.05 versus control; n = 6). pNA, p-nitroanilide.
Plausible Mechanism for HO-1 Induction and Its Mitochondrial Localization: Heme Accumulation and Subsequent ROS Generation inside Mitochondria
Now the question is what instigated HO-1 to localize in mitochondria. Both heme (the substrate for HO-1) and heme-induced ROS can stimulate HO-1 expression (10, 40, 41). At 4 h, MOS was maximal, and the level of mitochondrial HO-1 was also concomitantly high. Because heme is the substrate of HO-1 and a potent pro-oxidant, we were motivated to quantify heme inside mitochondria at different time intervals. A significant increase of heme content was observed in mitochondria compared with control at 4 h (Fig. 5A). The heme content declined with the progression of healing starting from 24 to 72 h. The highest level of HO-1 inside mitochondria was found at 24 h. When the MOS and the mitochondrial heme content were maximal at 4 h, the HO-1 entry inside mitochondria was visible, attaining maximum localization at 24 h. Thus, we propose that HO-1, acting on its substrate heme, decreased the heme content at 24 h, and high levels of HO-1 were maintained (Fig. 5A) for persistent action. We hypothesize that increased intramitochondrial heme and subsequent ROS generation may be the driving force to mobilize HO-1 to mitochondria, and this translocation followed by the catabolism of heme may be an inherent cytoprotective mechanism against MOS. We have previously provided evidence that indomethacin treatment enhances the generation of H2O2 in mitochondria (2). Interaction of heme with H2O2 generates •OH and amplifies ROS (42). The degradation of heme by H2O2 at different concentrations was confirmed by Soret spectroscopy using pure heme (Fig. 5B). Moreover, degradation of heme by H2O2 may favor the release of iron from heme, or the iron still attached to the heme can be redox-active (43, 44). It is reasonable to assume that degradation of heme either by H2O2 or by the action of HO-1 can release free iron inside mitochondria. The question is how this redox-active free iron is detoxified in the mitochondria. It is known that MtFt, a nuclearly encoded protein, enters the mitochondria and scavenges mitochondrial free iron in a number of tissues under different pathological conditions (45–49). Therefore, we checked the expression of MtFt after indomethacin treatment. Initially up to 4 h, there was no significant increase in its expression (Fig. 5C). However, maximum expression was found at 12–24 h with a subsequent fall in expression at 48 and 72 h, respectively (Fig. 5C).
FIGURE 5.
Intramitochondrial heme accumulation and mitochondrial ferritin expression in gastric mucosal cells after administration of indomethacin. A, intramitochondrial heme content at different time points after indomethacin treatment. Indomethacin-treated rats were sacrificed at different time points, gastric mucosal mitochondria were isolated, and heme was quantitated as described under “Experimental Procedures.” 0 h represents control. B, H2O2 damages heme. The fall of Soret spectra at 403 nm with increasing concentrations of H2O2 indicates heme degradation. C, RT-PCR for mitochondrial (Mt) ferritin mRNA expression together with densitometric analysis of RT-PCR data. Actin was used as an internal control. 0 h represents control. Data are presented as mean ± S.E. (***, p < 0.001 versus control; **, p < 0.05 versus control; n = 6).
HO-1 Induction Is Mediated by ROS-dependent ARE/Nrf2 Signaling
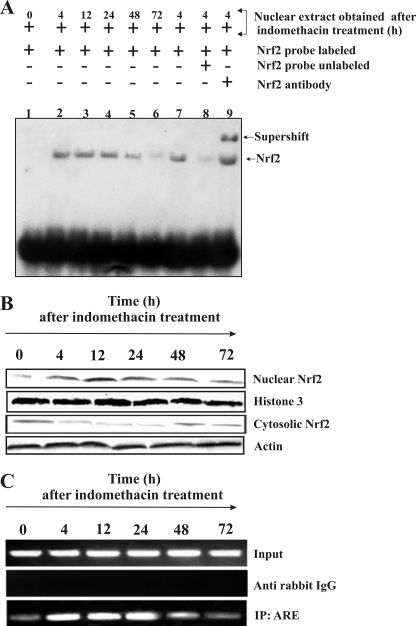
ROS mediates the translocation of redox-sensitive Nrf2 to the nucleus. Nrf2 is the known transcriptional activator of HO-1 because Nrf2 appears to be critical for the ARE-mediated gene expression. Therefore, we investigated whether indomethacin-induced oxidative stress stimulated the translocation of Nrf2 to the nucleus. The translocation of Nrf2 to the nucleus was analyzed by EMSA using an ARE probe corresponding to the rat HO-1 promoter. No complex was visible in labeled DNA (without nuclear extract) (Fig. 6A, lane 1). However, increased Nrf2 translocation was observed at 4, 12, and 24 h post-indomethacin treatment (Fig. 6A, lanes 2-4) in comparison with control (without indomethacin treatment) as evident from the band shift in EMSA (Fig. 6A, lane 1). The data indicate reduced Nrf2 translocation after 48 and 72 h of indomethacin treatment (Fig. 6A, lanes 5 and 6). The selective binding of Nrf2 was confirmed by competition (Fig. 6A, lane 8) and supershift assays (Fig. 6A, lane 9). For the competition assay, we used the nuclear extract obtained at 4 h after indomethacin treatment. In the competition assay, unlabeled Nrf2 probe competed with the labeled probe, producing a fainter band (Fig. 6A, lane 8) in comparison with sample without unlabeled probe (Fig. 6A, lane 7), whereas antibody specific to Nrf2 caused a further band shift (Fig. 6A, lane 9). If Nrf2 is translocated from the cytosol to the nucleus then it is reasonable to assume that the content of cytosolic Nrf2 will be decreased with the concomitant increase of nuclear Nrf2. To verify this, both the cytosolic fraction and nuclear extracts obtained from gastric mucosa of control and indomethacin-treated rats at different time points were subjected to Western immunoblot study. The data clearly suggest that the entry of Nrf2 to the nucleus at 4, 12, and 24 h correlates well with the decrease in the Nrf2 level in the cytosol (Fig. 6B).
FIGURE 6.
Nuclear translocation of Nrf2 during gastric mucosal injury by indomethacin. Nuclear fractions isolated from gastric mucosa from control and indomethacin-treated rats at different time points were used for EMSA and Western immunoblot analysis as described under “Experimental Procedures.” A, EMSA. Lane 1, labeled probe plus nuclear fraction from control; lanes 2-6, labeled probe plus nuclear fractions isolated at different time points after indomethacin treatment (4, 12, 24, 48, and 72 h, respectively); lane 7, labeled probe plus nuclear fraction (4 h after indomethacin treatment); lane 8, labeled probe plus nuclear fraction (4 h after indomethacin treatment) plus a 30-fold excess of cold competitor probe (ARE); lane 9, labeled probe plus nuclear fraction (4 h after indomethacin treatment) plus anti-Nrf2 antibody. Note that both lane 2 and lane 7 are similar. We reloaded the same 4-h sample that was used in lane 2 again in lane 7 for better comparative analysis. B, Western immunoblot analysis for Nrf2 (nuclear and cytosolic) translocation from the cytosol to the nucleus. The nuclear translocation of Nrf2 was increased at 4, 12, and 24 h following indomethacin treatment, whereas that in the cytosolic fraction was decreased at 4, 12, and 24 h. Histone 3 and β-actin were used as positive controls to check the purity of nuclear and cytosolic fractions, respectively. C, ChIP analysis to confirm nuclear translocation of Nrf2 and subsequent binding at the ARE sequence in the promoter region of HO-1 gene after indomethacin treatment. Immunoprecipitated DNA was analyzed by PCR amplification specific to the OH-1 promoter region. Input DNA material was used as positive control, whereas the material immunoprecipitated using rabbit IgG was used as a control. Nrf2 binding to the ARE is evident from the intensity of the PCR products obtained by amplifying the immunoprecipitated (IP) DNA using ARE-specific primers. The intensities of the PCR products were high at 4, 12, and 24 h, respectively, compared with 0- and 72-h samples, indicating the recruitment and binding of an increased amount of Nrf2 to the promoter region of HO-1 at 4, 12, and 24 h. 0 h represents control.
The translocation of Nrf2 inside the nucleus is not sufficient to conclude that the transcription of HO-1 is under the induction of Nrf2 binding to the ARE of HO-1 promoter. Therefore, a ChIP assay was performed to confirm the binding of Nrf2 to the ARE sequence of the HO-1 promoter (Fig. 6C). The data indicate that the PCR products for the ARE sequence at different time points showed an increase at 4, 12, and 24 h post-indomethacin treatment in comparison with the control (Fig. 6C). These findings suggest the recruitment and binding of Nrf2 to the promoter of HO-1 gene after indomethacin treatment.
Inhibition of HO-1 Aggravated Mucosal Injury and Delayed Repair Mechanism Due to Persistent MOS, Mitochondrial Pathology, and Apoptosis
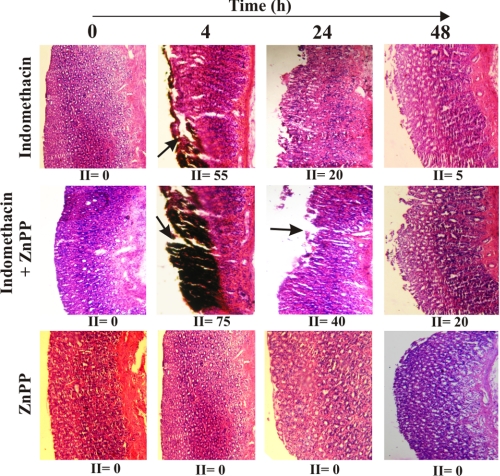
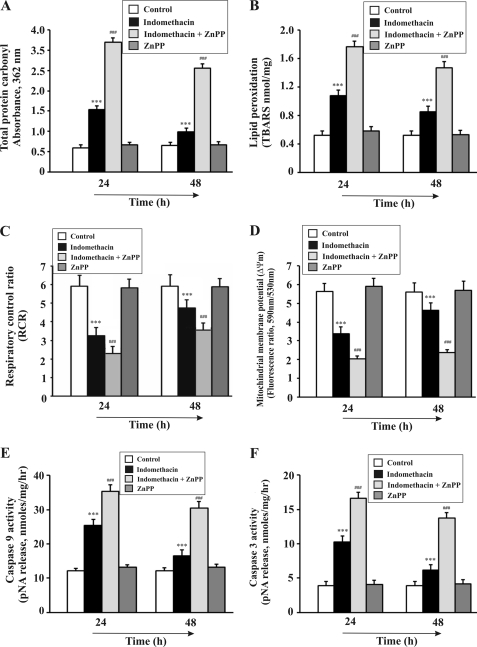
To confirm the cytoprotective role of HO-1 in MOS-mediated apoptotic injury by indomethacin, we examined the effect of ZnPP, a known HO-1 inhibitor, on indomethacin-induced gastric mucosal injury and its autohealing. Generally, the maximum injury was observed (injury index = 55) 4 h after indomethacin treatment (Fig. 7). But when pretreated with ZnPP, the gastric mucosal damage was higher (injury index = 75) than that in the indomethacin alone group (Fig. 7). The gastric injury was quite high even at 24 h (injury index = 40) in animals treated with indomethacin plus ZnPP as evident from the histological data (Fig. 7). The injury induced by indomethacin alone was autohealed almost completely at 48 h (injury index = 5), but animals treated with indomethacin plus ZnPP showed persistent injury (injury index = 20) as evaluated by histological studies (Fig. 7). No injury was observed in gastric mucosa treated with only ZnPP (injury index = 0) (Fig. 7). Now, to confirm whether HO-1 really prevents NSAID-induced MOS, dysfunction, and apoptosis in gastric mucosa, protein carbonylation, lipid peroxidation, RCR, Δψm, and activation of caspase-9 and caspase-3 were measured at two different times (24 and 48 h) in the presence and absence of ZnPP. Tissue samples (24 and 48 h) treated with both ZnPP and indomethacin showed higher mitochondrial oxidative stress compared with samples obtained only from indomethacin-treated rats as evident from both increased protein carbonylation and lipid peroxidation (Fig. 8, A and B). The persistent or higher rate of mitochondrial dysfunction was evident as measured by RCR and Δψm when HO-1 was inhibited by ZnPP compared with the indomethacin alone group (Fig. 8, C and D). Furthermore, a higher rate of apoptosis was evident from caspase-9 and caspase-3 assays in the presence of ZnPP but not in its absence (Fig. 8, E and F) (24 and 48 h). However, by 48 h, although the condition of MOS, dysfunction, and consequent apoptosis declined marginally, it was still severe in the presence of ZnPP. Thus, it is clear that ZnPP increases the toxic insult to the cell and delays healing by inhibiting HO-1, establishing the phenomenon of mitochondrial localization of HO-1 as an antioxidant strategy of the cell.
FIGURE 7.
ZnPP aggravates gastric mucosal injury and slows down healing process in vivo. Hematoxylin and eosin staining of gastric mucosal sections of control, indomethacin-, indomethacin + ZnPP-, and only ZnPP-treated groups at different time points is shown. 0 h represents control. II, injury index. Arrows indicate mucosal injury.
FIGURE 8.
ZnPP treatment increases mitochondrial oxidative stress, mitochondrial pathology, and apoptosis in gastric mucosa. A–F, measurement of protein carbonyl (A), lipid peroxidation (B), complex I-mediated RCR (C), mitochondrial membrane potential (D), caspase-9 activity (E), and caspase-3 activity (F) in control, indomethacin-treated, indomethacin + ZnPP-treated, and only ZnPP-treated groups of rats at 24 and 48 h after indomethacin treatment as described under “Experimental Procedures.” Data are presented as mean ± S.E. (***, p < 0.001 versus control; ###, p < 0.001 versus indomethacin; n = 6). TBARS, thiobarbituric acid-reactive substance; pNA, p-nitroanilide.
Induction of HO-1 by CoPP Ameliorates Gastric Mucosal Injury
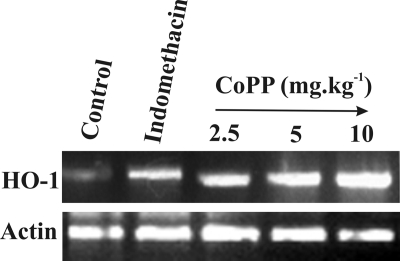
To further confirm the protective role of HO-1 against MOS and gastropathy induced by indomethacin, HO-1 was induced by CoPP administration at different doses 30 min prior to indomethacin treatment (Fig. 9 and Table 2). The data indicate dose-dependent induction of HO-1 by CoPP with a maximum induction at 5–10 mg kg−1 (Fig. 9). Moreover, the dose-dependent induction of HO-1 correlated well with a significant reduction of gastric mucosal injury, oxidative stress, apoptosis, and mitochondrial pathology (Table 2).
FIGURE 9.
CoPP treatment enhances HO-1 mRNA expression. CoPP treatment prior to indomethacin administration enhanced HO-1 mRNA expression. mRNA was prepared from gastric mucosal tissue of CoPP-pretreated and indomethacin-treated rats as described under “Experimental Procedures.” Increased HO-1 mRNA expression was found at 5 and 10 mg kg−1 doses of CoPP prior to indomethacin treatment.
TABLE 2.
Effect of CoPP on indomethacin-induced gastric mucosal injury, MOS, apoptosis, and mitochondrial function
CoPP was administered intraperitoneally in a single dose of 0.5, 2.5, 5, and 10 mg kg−1 b.w. to respective groups (containing six to eight rats in each group) 1 h prior to indomethacin treatment. Only vehicle was administered to the control group. Animals were sacrificed 4 h after indomethacin treatment to measure injury, lipid and protein oxidation, caspase-3, caspase-9, Δψm, and RCR as described under “Experimental Procedures.” pNA, p-nitroanilide; TBARS, thiobarbituric acid-reactive substance. All data are presented as mean ± S.E. (n = 6–8).
| Injury index | Lipid peroxidation (TBARS) | Total protein carbonyl formation (absorbance at 362 nm) | Caspase-3 activity (pNA release) | Caspase-9 activity (pNA release) | Δψm (590/530 nm fluorescence ratio) | RCR (State 3/State 4) | |
|---|---|---|---|---|---|---|---|
| nmol/mg | nmol/mg protein/h | nmol/mg protein/h | |||||
| Control (only vehicle) | 0 | 0.52 ± 0.02 | 0.62 ± 0.03 | 4 ± 0.2 | 15 ± 1 | 5.62 ± 0.53 | 5.91 ± 0.6 |
| Indomethacin | 50 ± 5a | 1.76 ± 0.05a | 2.06 ± 0.06a | 17 ± 2a | 41 ± 2a | 2.14 ± 0.21a | 3.25 ± 0.35a |
| Indomethacin + 0.5 mg kg−1 CoPP | 50 ± 5 | 1.72 ± 0.05 | 2.05 ± 0.05 | 17 ± 1.8 | 40 ± 2b | 2.18 ± 0.22 | 3.21 ± 0.33 |
| Indomethacin + 2.5 mg kg−1 CoPP | 35 ± 2c | 1.37 ± 0.08c | 1.97 ± 0.08c | 15 ± 0.9c | 37 ± 2c | 2.80 ± 0.26c | 3.95 ± 0.41c |
| Indomethacin + 5 mg kg−1 CoPP | 25 ± 2c | 1.3 ± 0.07c | 1.63 ± 0.07c | 13 ± 0.9c | 30 ± 2c | 3.10 ± 0.29c | 4.33 ± 0.43c |
| Indomethacin + 10 mg kg−1 CoPP | 15 ± 5c | 1.0 ± 0.06c | 1.20 ± 0.61c | 10 ± 0.7c | 26 ± 2c | 4.02 ± 0.38c | 4.87 ± 0.43c |
a p < 0.001 versus control.
b p < 0.01 versus indomethacin.
c p < 0.001 versus indomethacin.
DISCUSSION
The present study provides evidence for mitochondrial translocation of HO-1 in vivo in NSAID-induced gastric mucosal tissue injury and mitochondrial translocation of HO-1 in a time-dependent fashion, preventing MOS and mitochondrial dysfunction and promoting the repair of MOS-mediated apoptotic mucosal injury.
NSAIDs have been previously reported to interfere with mitochondrial energy production (50). NSAIDs are weak acids, are lipophilic in nature, and act as an uncoupling agent (51). Indomethacin with its acidic carboxyl group (pKa = 4.5) and lipid solubility has been found to damage both rat and human mitochondria (51). Moreover, indomethacin treatment has been reported to enhance mitochondrial ROS, which disrupt mitochondrial function (2). Thus, indomethacin is used as a natural choice in generating MOS and inducing apoptotic cell death in rat gastric mucosa. Furthermore, indomethacin is selected as the representative NSAID over others because it is the most frequently used NSAID in gastrointestinal toxicity studies in experimental animals (51). Although maximum injury was observed at 4 h after indomethacin treatment, the mucosa recovered from the damage very slowly (autohealing) with time without prior medication. This explains the existence of inherent cytoprotective machinery that was activated in response to the mucosal injury and mitochondrial oxidative insult induced by indomethacin.
The dose of indomethacin selected was 48 mg kg−1 as reported earlier (2, 26). This dose appears high when converted, according to the recommendation of the United States Food and Drug Administration (52), to the human equivalent, which is about 211 (3.5 mg kg−1) or 423 mg (7.05 mg kg−1). For therapeutic purposes, however, indomethacin is used at a dose of 25 mg two to three times daily or 150–200 mg daily to treat inflammatory disorders. This dose is sustained for a long period when treating chronic inflammatory disorders, resulting in a higher cumulative effective dose to cause gastric damage (53). Thus, a higher effective dose of indomethacin to induce mitopathology and gastric damage in rat is in good agreement with the therapeutic doses of indomethacin used in humans (2).
HO-1 has been reported to be up-regulated during gastric mucosal injury (23, 24, 30, 54–56). The role of HO-1 in tissue injury repair is well known in other models also (57). The mechanism of how HO-1 facilitates repair of NSAID-induced mucosal injury is not yet clear. A cellular model for autohealing of gastric mucosa injured by NSAID would be helpful in this regard, but no such model has been reported. Thus, an in vivo model was used to explore the role of HO-1. ROS has been reported to be the causative factor behind gastric mucosal damage of different origins (58, 59). Mitochondria are the major source and target of ROS generation. Excessive ROS generation leads to MOS. The role of MOS and the consequent apoptosis after NSAID-induced gastric mucosal injury is already well established, and MOS is considered to be the major player in the acid-independent (3) and COX-independent pathway of NSAID-mediated gastric injury (60, 61). But ROS is generated inside mitochondria, whereas HO-1 resides on the endoplasmic reticulum. How then does HO-1 favor repair by preventing ROS in mitochondria? Our time course study not only showed an up-regulation of HO-1 but also showed the mitochondrial localization of HO-1 in gastric epithelial cells during injury by NSAID. We also observed that HO-1 translocation resulted in the prevention of MOS and gastric injury induced by indomethacin.
Mitochondrial localization of HO-1 was verified by confocal microscopy using a specific mitochondrial marker, Western immunoblotting using HO-1 antibodies, and assaying the activity of HO-1 in mitochondria in the presence and absence of indomethacin. The score value for colocalized and non-colocalized fluorescence for indomethacin-treated sample was much higher than that of control. As evident from the colocalization score and other biochemical studies (Western immunoblot and activity assay), a basal level of HO-1 was found in the control sample. A basal level of HO-1 is always present in mitochondria in the control condition not only in gastric mucosal cells but also in a number of other cell types as reported earlier (31). There are several reports regarding the subcellular distribution of HO-1. HO-1 localization inside the nucleus not only protects cells against hydrogen peroxide-mediated injury but also is assumed to up-regulate genes involved in cytoprotection against oxidative stress (62). The mitochondrial localization of HO-1 has been associated with cytoprotection (31, 63–66). HO-1 lacks the N-terminal mitochondrial targeting presequence; therefore, as a transporter colocalized in the inner membrane, HO-1 probably requires internal hydrophobic targeting domains and specific import machinery like the Tim 23 transporter (65, 67, 68). However, rigorous experimental studies are required to explore the specific import machinery for mitochondrial entry of HO-1 in indomethacin-induced gastropathy. The important question is what is the impact of HO-1 entry on MOS. During maximum injury at 4 h after indomethacin treatment, HO-1 was localized inside mitochondria, but the highest level of HO-1 was observed between 12 and 24 h when the injury began to decline. This implies that the up-regulated status of HO-1 is associated with the amelioration of MOS, mitochondrial dysfunction, and apoptosis. However, HO-1 cannot scavenge ROS directly, prompting us to investigate the mechanism by which it controls MOS. It is assumed that HO-1 may prevent ROS generation by catabolizing the pro-oxidant heme accumulated in mitochondria. In mitochondria, maximum heme content was found during maximum injury at 4 h when maximum MOS was observed. On the contrary, although the HO-1 level was considerable inside mitochondria at 4 h, it was maximal at 24 h when heme content was reduced. Thus, the data indicate a possible role of HO-1 to act on its substrate heme to prevent MOS. The role of inducible HO-1 in the mitochondria to degrade accumulated heme has been suggested previously (14, 31).
The deleterious effect of heme is well known (42). Free heme is highly toxic (42) and can induce peroxidation in membranes and cause damage to cellular macromolecules such as proteins, DNA, and carbohydrates (43). By coupling oxidative stress to the stimulation of HO-1, cells would ensure that the labile heme generated does not act in a cytotoxic manner. Heme catabolism by HO-1 is an advantage that may be utilized by the cells. Once heme is catabolized by HO-1 it produces equimolar quantity of CO, biliverdin, and iron. Biliverdin is converted to bilirubin by the biliverdin reductase enzyme. Both CO and biliverdin IXa, two metabolites of heme formed by HO-1, were thought to be futile or noxious products, but recent facts are contrary to such assumptions. Both bilirubin and CO have been attributed to have antioxidant and cytoprotective roles (10, 24, 69–72). This notion is strongly supported by the observation that the cytotoxic effects of oxidative stress are worsened in cells that lack HO-1 (Hmox1−/−) and thus cannot increase the rate of heme catabolism in response to oxidative stress.
What are the possible sources of heme inside mitochondria during gastric injury by indomethacin? NSAID-induced gastropathy is characterized by subepithelial hemorrhages, erosions, and bleeding (73). When erythrocytes are lysed, extracellular hemoglobin is easily oxidized from ferrous to ferric hemoglobin (methemoglobin), which in turn readily releases heme. The hydrophobicity of free heme is responsible for intercalating into cell membranes, making the cell susceptible to oxidant-mediated insult as well as production of ROS (74). The transport of intracellular labile heme from the cytosol to the mitochondria is ensured by the mitochondrial heme transporters. Furthermore, the mitochondria are the major source of ROS, and they contain a number of heme-containing proteins (hemoproteins). The possibility of ROS attacking hemoproteins and releasing heme cannot be excluded (10). The reasons why cells respond to oxidative stress by increasing their ability to catabolize free heme are not clear. Oxidative stress can lead to heme release from some hemoproteins, an effect that produces cytotoxic free heme (10, 57). Mitochondria are potential sites for the generation of H2O2, which may act on heme to generate ROS. It has been reported that 0.1 mm/min H2O2 is produced continuously under physiological conditions. There will be an increment of this rate during adverse conditions such as hyperoxia and ischemia-reperfusion (75–77). Moreover, NSAID-induced gastric mucosal damage is an ischemia-reperfusion type of injury (78). Hemin, on the other hand, acts as a chemoattractant of neutrophils (79). The infiltration of neutrophils and generation of enhanced ROS in the injured site have already been attributed in NSAID-induced gastropathy. Thus, the possibility of heme playing a role in attracting neutrophils in the gastric mucosal injured site cannot be discarded. We found an increased myeloperoxidase activity (marker for neutrophil) in gastric mucosa at 4 h after indomethacin treatment compared with control (data not shown).
The catabolism of heme by HO-1 inside mitochondria produces iron, which can amplify the generation of highly toxic hydroxyl radical (•OH) by Fenton's reaction in the presence of H2O2 (80, 81). It has been reported that during oxidative stress the MtFt is up-regulated and can enter inside mitochondria to encounter mitochondrial iron (45, 82). MtFt prevents mitochondrial damage, reduces the production of ROS, and radically rescues cells from apoptosis (45, 82). MtFt expression is very tissue-specific, and MtFt has been reported to be overexpressed in different tissues of human and mouse (46–49). Thus, we searched for MtFt expression in gastric mucosa in the presence or absence of indomethacin. MtFt expression was detected in rat gastric mucosa. Moreover, to overcome iron overload, the role of mitochondrial iron exporters in mitochondrial iron homeostasis during indomethacin-induced gastric mucosal injury cannot be excluded (83).
We are interested in the transcriptional regulation of HO-1 at different time points. Different studies on transfection and deletion mutagenesis have revealed an element assigned as the ARE in the promoter regions of genes encoding phase II detoxification enzymes and antioxidant proteins like HO-1 (84). Nrf2 is a basic leucine zipper transcription factor that regulates a number of ARE-driven genes (85). Induction of HO-1 has been shown to occur via Nrf2 in a number of cell types (24, 55, 86, 87) including gastric mucosal epithelial cells (23), epithelial cells (88), monocytes (89), and mouse embryo fibroblasts (90). Under normal conditions, Nrf2 remains bound to Keap1 in the cytosol. Oxidative stress induces Keap1 ubiquitination-degradation. Once released from Keap1, the transcription factor Nrf2 moves into the nucleus. A number of kinases are also involved in the activation of Nrf2 in response to oxidative stress (91). HO-1-inducing stressors such as ROS mobilize Nrf2 inside the nucleus from the cytoplasm, and Nrf2 through a consensus cis-element can act as a transcriptional activator of HO-1 (24). We searched for Nrf2 translocation inside the nucleus in a time-dependent manner. Nrf2 together with small Maf proteins binds to ARE on the HO-1 promoter and initiates transcription (10, 91). Bach1 is the transcriptional repressor that resides inside the nucleus, and it competes with Nrf2 for binding to ARE, leading to repression of genes downstream of Nrf2 (92). However, Bach1 has heme-binding sites (10, 93). Heme can induce conformational alterations in Bach1. Heme is also attributed to play a significant role in microRNA processing. Bach1 has been reported to be regulated by microRNAs such as miR-122 and miR-196 (94, 95). Moreover, rapid alterations in heme availability can have a significant effect on the processing of crucial microRNA molecules including those that regulate HO-1 (11). As a result, Bach1 binding to ARE could be inhibited (93). The combined role of the two microRNAs miR-217 and miR-377 has been demonstrated to regulate HO-1 protein expression (11). Reactive oxygen species can also directly attack the sulfhydryl groups of Bach1 to check its binding to ARE (10, 96). Thus, the Bach1/Nrf2 transcriptional system most likely works together to control HO-1 transcription (10). Electrophoretic mobility shift assay and Western immunoblot studies confirmed the nuclear translocation of Nrf2. ChIP assay further confirmed the binding of Nrf2 to the HO-1 promoter and induction of HO-1 transcription. The cytoprotective function of HO-1 against MOS was reconfirmed using an inhibitor, ZnPP. Zinc protoporphyrin is a known inhibitor of HO-1 (97). Because the autohealing of the gastric mucosa after damage was prominent at 24 h and was almost complete at 48 h, these two time points were selected for testing the action of ZnPP on indomethacin-induced MOS, mitochondrial dysfunction, and apoptosis. It was clear from the time course data that HO-1 activity was significantly inhibited by ZnPP at these two time points (data not shown). Lingering injury was also evident in the ZnPP-treated samples when compared with only indomethacin. Inhibition of HO-1 with ZnPP enhanced MOS, mitochondrial pathology, and apoptosis (in vivo) at both 24 and 48 h at a significantly higher rate than that of indomethacin alone. The ZnPP-treated samples, however, showed a recovery at 48 h, but it was not as significant as that of the samples treated with only indomethacin. If inhibition of HO-1 aggravated gastropathy then induction of the same may have a reverse effect. To test the gain of function of HO-1, we administered CoPP, which is a popular inducer of HO-1 that has no toxicity (98). CoPP pretreatment has been reported to prevent apoptosis in human gastric mucosa (99). Treatment of CoPP enhanced HO-1 expression, reducing gastric mucosal injury induced by indomethacin. Thus, HO-1 up-regulation and time-dependent translocation to the mitochondria are a novel cytoprotective event in attenuating indomethacin-induced gastric mucosal MOS and subsequent apoptotic gastric mucosal injury.
Acknowledgments
We thank Prof. Rahul Banerjee (Saha Institute of Nuclear Physics, Kolkata, India) for critical reading of the manuscript. We also thank Dr. Anupam Banerjee for confocal microscopy.
This work was supported in part by grants from the Council of Scientific and Industrial Research, New Delhi.
- ROS
- reactive oxygen species
- HO-1
- heme oxygenase-1
- NSAID
- non-steroidal anti-inflammatory drug
- MOS
- mitochondrial oxidative stress
- RCR
- respiratory control ratio
- JC-1
- 5,5′,6,6′-tetrachloro-1,1′,3,3′-tetraethylbenzimidazolylcarbocyanine iodide
- Nrf2
- NF-E2-related factor 2
- ZnPP
- zinc protoporphyrin
- Δψm
- transmembrane potential
- ARE
- antioxidant response element
- COX
- cytochrome c oxidase
- CoPP
- cobalt protoporphyrin
- b.w.
- body weight
- MtFt
- mitochondrial ferritin.
REFERENCES
- 1. Ott M., Gogvadze V., Orrenius S., Zhivotovsky B. (2007) Apoptosis 12, 913–922 [DOI] [PubMed] [Google Scholar]
- 2. Maity P., Bindu S., Dey S., Goyal M., Alam A., Pal C., Mitra K., Bandyopadhyay U. (2009) J. Biol. Chem. 284, 3058–3068 [DOI] [PubMed] [Google Scholar]
- 3. Maity P., Bindu S., Choubey V., Alam A., Mitra K., Goyal M., Dey S., Guha M., Pal C., Bandyopadhyay U. (2008) J. Biol. Chem. 283, 14391–14401 [DOI] [PubMed] [Google Scholar]
- 4. González-Flecha B., Cutrin J. C., Boveris A. (1993) J. Clin. Investig. 91, 456–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dröge W. (2002) Physiol. Rev. 82, 47–95 [DOI] [PubMed] [Google Scholar]
- 6. Bayir H., Kagan V. E. (2008) Crit. Care 12, 206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hoye A. T., Davoren J. E., Wipf P., Fink M. P., Kagan V. E. (2008) Acc. Chem. Res. 41, 87–97 [DOI] [PubMed] [Google Scholar]
- 8. Bailey S. M., Cunningham C. C. (2002) Free Radic. Biol. Med. 32, 11–16 [DOI] [PubMed] [Google Scholar]
- 9. Guha M., Kumar S., Choubey V., Maity P., Bandyopadhyay U. (2006) FASEB J. 20, 1224–1226 [DOI] [PubMed] [Google Scholar]
- 10. Gozzelino R., Jeney V., Soares M. P. (2010) Annu. Rev. Pharmacol. Toxicol. 50, 323–354 [DOI] [PubMed] [Google Scholar]
- 11. Beckman J. D., Chen C., Nguyen J., Thayanithy V., Subramanian S., Steer C. J., Vercellotti G. M. (2011) J. Biol. Chem. 286, 3194–3202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Alam J., Shibahara S., Smith A. (1989) J. Biol. Chem. 264, 6371–6375 [PubMed] [Google Scholar]
- 13. Applegate L. A., Luscher P., Tyrrell R. M. (1991) Cancer Res. 51, 974–978 [PubMed] [Google Scholar]
- 14. Keyse S. M., Tyrrell R. M. (1989) Proc. Natl. Acad. Sci. U.S.A. 86, 99–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Inoue K., Takahashi T., Uehara K., Shimuzu H., Ido K., Morimatsu H., Omori E., Katayama H., Akagi R., Morita K. (2008) Shock 29, 252–261 [DOI] [PubMed] [Google Scholar]
- 16. Li X., Schwacha M. G., Chaudry I. H., Choudhry M. A. (2008) J. Immunol. 180, 6933–6940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chen K., Gunter K., Maines M. D. (2000) J. Neurochem. 75, 304–313 [DOI] [PubMed] [Google Scholar]
- 18. Pae H. O., Chung H. T. (2009) Immune Netw. 9, 12–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chan K. H., Ng M. K., Stocker R. (2011) Clin. Sci. 120, 493–504 [DOI] [PubMed] [Google Scholar]
- 20. Seixas E., Gozzelino R., Chora A., Ferreira A., Silva G., Larsen R., Rebelo S., Penido C., Smith N. R., Coutinho A., Soares M. P. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 15837–15842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Morse D., Lin L., Choi A. M., Ryter S. W. (2009) Free Radic. Biol. Med. 47, 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Agarwal A., Nick H. S. (2000) J. Am. Soc. Nephrol. 11, 965–973 [DOI] [PubMed] [Google Scholar]
- 23. Aburaya M., Tanaka K., Hoshino T., Tsutsumi S., Suzuki K., Makise M., Akagi R., Mizushima T. (2006) J. Biol. Chem. 281, 33422–33432 [DOI] [PubMed] [Google Scholar]
- 24. Ueda K., Ueyama T., Yoshida K., Kimura H., Ito T., Shimizu Y., Oka M., Tsuruo Y., Ichinose M. (2008) Am. J. Physiol. Gastrointest. Liver Physiol. 295, G460–G469 [DOI] [PubMed] [Google Scholar]
- 25. Bandyopadhyay U., Biswas K., Chatterjee R., Bandyopadhyay D., Chattopadhyay I., Ganguly C. K., Chakraborty T., Bhattacharya K., Banerjee R. K. (2002) Life Sci. 71, 2845–2865 [DOI] [PubMed] [Google Scholar]
- 26. Biswas K., Bandyopadhyay U., Chattopadhyay I., Varadaraj A., Ali E., Banerjee R. K. (2003) J. Biol. Chem. 278, 10993–11001 [DOI] [PubMed] [Google Scholar]
- 27. Pal C., Bindu S., Dey S., Alam A., Goyal M., Iqbal M. S., Maity P., Adhikari S. S., Bandyopadhyay U. (2010) Free Radic. Biol. Med. 49, 258–267 [DOI] [PubMed] [Google Scholar]
- 28. Sims N. R., Anderson M. F. (2008) Nat. Protoc. 3, 1228–1239 [DOI] [PubMed] [Google Scholar]
- 29. Lowry O. H., Rosebrough N. J., Farr A. L., Randall R. J. (1951) J. Biol. Chem. 193, 265–275 [PubMed] [Google Scholar]
- 30. Guo J. S., Cho C. H., Wang W. P., Shen X. Z., Cheng C. L., Koo M. W. (2003) World J. Gastroenterol. 9, 1767–1771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Slebos D. J., Ryter S. W., van der Toorn M., Liu F., Guo F., Baty C. J., Karlsson J. M., Watkins S. C., Kim H. P., Wang X., Lee J. S., Postma D. S., Kauffman H. F., Choi A. M. (2007) Am. J. Respir. Cell Mol. Biol. 36, 409–417 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 32. Manders E. M., Verbeek F. J., Aten J. A. (1993) J. Microsc. 169, 375–382 [DOI] [PubMed] [Google Scholar]
- 33. Guha M., Maity P., Choubey V., Mitra K., Reiter R. J., Bandyopadhyay U. (2007) J. Pineal Res. 43, 372–381 [DOI] [PubMed] [Google Scholar]
- 34. Rousou A. J., Ericsson M., Federman M., Levitsky S., McCully J. D. (2004) Am. J. Physiol. Heart Circ. Physiol. 287, H1967–H1976 [DOI] [PubMed] [Google Scholar]
- 35. Maity P., Bindu S., Dey S., Goyal M., Alam A., Pal C., Reiter R., Bandyopadhyay U. (2009) J. Pineal Res. 46, 314–323 [DOI] [PubMed] [Google Scholar]
- 36. Liu X. M., Peyton K. J., Ensenat D., Wang H., Schafer A. I., Alam J., Durante W. (2005) J. Biol. Chem. 280, 872–877 [DOI] [PubMed] [Google Scholar]
- 37. Freitas-Junior L. H., Hernandez-Rivas R., Ralph S. A., Montiel-Condado D., Ruvalcaba-Salazar O. K., Rojas-Meza A. P., Mâncio-Silva L., Leal-Silvestre R. J., Gontijo A. M., Shorte S., Scherf A. (2005) Cell 121, 25–36 [DOI] [PubMed] [Google Scholar]
- 38. Kim J. Y., Cho H. J., Sir J. J., Kim B. K., Hur J., Youn S. W., Yang H. M., Jun S. I., Park K. W., Hwang S. J., Kwon Y. W., Lee H. Y., Kang H. J., Oh B. H., Park Y. B., Kim H. S. (2009) Cardiovasc. Res. 82, 550–560 [DOI] [PubMed] [Google Scholar]
- 39. Duchen M. R. (2004) Diabetes 53, Suppl. 1, S96–S102 [DOI] [PubMed] [Google Scholar]
- 40. Lang D., Reuter S., Buzescu T., August C., Heidenreich S. (2005) Int. Immunol. 17, 155–165 [DOI] [PubMed] [Google Scholar]
- 41. Han Z., Varadharaj S., Giedt R. J., Zweier J. L., Szeto H. H., Alevriadou B. R. (2009) J. Pharmacol. Exp. Ther. 329, 94–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kumar S., Bandyopadhyay U. (2005) Toxicol. Lett. 157, 175–188 [DOI] [PubMed] [Google Scholar]
- 43. Nagababu E., Rifkind J. M. (2004) Antioxid. Redox Signal. 6, 967–978 [DOI] [PubMed] [Google Scholar]
- 44. Sugiyama K., Highet R. J., Woods A., Cotter R. J., Osawa Y. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 796–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Campanella A., Rovelli E., Santambrogio P., Cozzi A., Taroni F., Levi S. (2009) Hum. Mol. Genet. 18, 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Santambrogio P., Biasiotto G., Sanvito F., Olivieri S., Arosio P., Levi S. (2007) J. Histochem. Cytochem. 55, 1129–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Campanella A., Isaya G., O'Neill H. A., Santambrogio P., Cozzi A., Arosio P., Levi S. (2004) Hum. Mol. Genet. 13, 2279–2288 [DOI] [PubMed] [Google Scholar]
- 48. Missirlis F., Holmberg S., Georgieva T., Dunkov B. C., Rouault T. A., Law J. H. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 5893–5898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Corsi B., Cozzi A., Arosio P., Drysdale J., Santambrogio P., Campanella A., Biasiotto G., Albertini A., Levi S. (2002) J. Biol. Chem. 277, 22430–22437 [DOI] [PubMed] [Google Scholar]
- 50. Adams S. S., Cobb R. (1958) Nature 181, 773–774 [DOI] [PubMed] [Google Scholar]
- 51. Jacob M., Bjarnason I., Rafi S., Wrigglesworth J., Simpson R. J. (2001) Aliment. Pharmacol. Ther. 15, 1837–1842 [DOI] [PubMed] [Google Scholar]
- 52. Reagan-Shaw S., Nihal M., Ahmad N. (2008) FASEB J. 22, 659–661 [DOI] [PubMed] [Google Scholar]
- 53. Swift G. L., Arnold J., Williams G. T., Williams B. D., Rhodes J., Khan F. (1992) Digestion 53, 88–93 [DOI] [PubMed] [Google Scholar]
- 54. Gomes A. S., Gadelha G. G., Lima S. J., Garcia J. A., Medeiros J. V., Havt A., Lima A. A., Ribeiro R. A., Brito G. A., Cunha F. Q., Souza M. H. (2010) Eur. J. Pharmacol. 642, 140–145 [DOI] [PubMed] [Google Scholar]
- 55. Shibuya A., Onda K., Kawahara H., Uchiyama Y., Nakayama H., Omi T., Nagaoka M., Matsui H., Hirano T. (2010) Biochem. Biophys. Res. Commun. 398, 581–584 [DOI] [PubMed] [Google Scholar]
- 56. Becker J. C., Grosser N., Boknik P., Schröder H., Domschke W., Pohle T. (2003) Biochem. Biophys. Res. Commun. 312, 507–512 [DOI] [PubMed] [Google Scholar]
- 57. Takahashi T., Shimizu H., Akagi R., Morita K., Sassa S. (2006) Drug Dev. Res. 67, 130–153 [Google Scholar]
- 58. Chattopadhyay I., Bandyopadhyay U., Biswas K., Maity P., Banerjee R. K. (2006) Free Radic. Biol. Med. 40, 1397–1408 [DOI] [PubMed] [Google Scholar]
- 59. Maity P., Biswas K., Roy S., Banerjee R. K., Bandyopadhyay U. (2003) Mol. Cell. Biochem. 253, 329–338 [DOI] [PubMed] [Google Scholar]
- 60. Musumba C., Pritchard D. M., Pirmohamed M. (2009) Aliment. Pharmacol. Ther. 30, 517–531 [DOI] [PubMed] [Google Scholar]
- 61. Fornai M., Colucci R., Antonioli L., Awwad O., Ugolini C., Tuccori M., Fulceri F., Natale G., Basolo F., Blandizzi C. (2011) Pharmacol. Res. 63, 59–67 [DOI] [PubMed] [Google Scholar]
- 62. Lin Q., Weis S., Yang G., Weng Y. H., Helston R., Rish K., Smith A., Bordner J., Polte T., Gaunitz F., Dennery P. A. (2007) J. Biol. Chem. 282, 20621–20633 [DOI] [PubMed] [Google Scholar]
- 63. Sahni S. K., Saxena N., Tekwani B. L., Dutta G. P., Pandey V. C. (1991) Exp. Mol. Pathol. 55, 55–62 [DOI] [PubMed] [Google Scholar]
- 64. Srivastava P., Pandey V. C. (1996) Int. J. Biochem. Cell Biol. 28, 1071–1077 [DOI] [PubMed] [Google Scholar]
- 65. Converso D. P., Taillé C., Carreras M. C., Jaitovich A., Poderoso J. J., Boczkowski J. (2006) FASEB J. 20, 1236–1238 [DOI] [PubMed] [Google Scholar]
- 66. Turkseven S., Drummond G., Rezzani R., Rodella L., Quan S., Ikehara S., Abraham N. G. (2007) J. Cell. Biochem. 100, 815–823 [DOI] [PubMed] [Google Scholar]
- 67. Lithgow T. (2000) FEBS Lett. 476, 22–26 [DOI] [PubMed] [Google Scholar]
- 68. Neupert W., Brunner M. (2002) Nat. Rev. Mol. Cell Biol. 3, 555–565 [DOI] [PubMed] [Google Scholar]
- 69. Gopinathan V., Miller N. J., Milner A. D., Rice-Evans C. A. (1994) FEBS Lett. 349, 197–200 [DOI] [PubMed] [Google Scholar]
- 70. Sammut I. A., Foresti R., Clark J. E., Exon D. J., Vesely M. J., Sarathchandra P., Green C. J., Motterlini R. (1998) Br. J. Pharmacol. 125, 1437–1444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Ferreira A., Balla J., Jeney V., Balla G., Soares M. P. (2008) J. Mol. Med. 86, 1097–1111 [DOI] [PubMed] [Google Scholar]
- 72. Akamatsu Y., Haga M., Tyagi S., Yamashita K., Graça-Souza A. V., Ollinger R., Czismadia E., May G. A., Ifedigbo E., Otterbein L. E., Bach F. H., Soares M. P. (2004) FASEB J. 18, 771–772 [DOI] [PubMed] [Google Scholar]
- 73. Laine L. (1996) Gastrointest. Endosc. Clin. N. Am. 6, 489–504 [PubMed] [Google Scholar]
- 74. Belcher J. D., Beckman J. D., Balla G., Balla J., Vercellotti G. (2010) Antioxid. Redox Signal. 12, 233–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Kim N. H., Kang J. H. (2006) J. Biochem. Mol. Biol. 39, 452–456 [DOI] [PubMed] [Google Scholar]
- 76. Boveris A., Oshino N., Chance B. (1972) Biochem. J. 128, 617–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Britton R. S., Bacon B. R., Recknagel R. O. (1987) Chem. Phys. Lipids 45, 207–239 [DOI] [PubMed] [Google Scholar]
- 78. Maricic N., Ehrlich K., Gretzer B., Schuligoi R., Respondek M., Peskar B. M. (1999) Br. J. Pharmacol. 128, 1659–1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Porto B. N., Alves L. S., Fernández P. L., Dutra T. P., Figueiredo R. T., Graça-Souza A. V., Bozza M. T. (2007) J. Biol. Chem. 282, 24430–24436 [DOI] [PubMed] [Google Scholar]
- 80. Halliwell B., Gutteridge J. M. (1990) Methods Enzymol. 186, 1–85 [DOI] [PubMed] [Google Scholar]
- 81. Suttner D. M., Dennery P. A. (1999) FASEB J. 13, 1800–1809 [DOI] [PubMed] [Google Scholar]
- 82. Shi Z. H., Nie G., Duan X. L., Rouault T., Wu W. S., Ning B., Zhang N., Chang Y. Z., Zhao B. L. (2010) Antioxid. Redox Signal. 13, 783–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Richardson D. R., Lane D. J., Becker E. M., Huang M. L., Whitnall M., Rahmanto Y. S., Sheftel A. D., Ponka P. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 10775–10782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Niture S. K., Kaspar J. W., Shen J., Jaiswal A. K. (2010) Toxicol. Appl. Pharmacol. 244, 37–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Lee J. M., Johnson J. A. (2004) J. Biochem. Mol. Biol. 37, 139–143 [DOI] [PubMed] [Google Scholar]
- 86. Farombi E. O., Shrotriya S., Na H. K., Kim S. H., Surh Y. J. (2008) Food Chem. Toxicol. 46, 1279–1287 [DOI] [PubMed] [Google Scholar]
- 87. Takagi T., Naito Y., Okada H., Ishii T., Mizushima K., Akagiri S., Adachi S., Handa O., Kokura S., Ichikawa H., Itoh K., Yamamoto M., Matsui H., Yoshikawa T. (2009) J. Pharmacol. Exp. Ther. 331, 255–264 [DOI] [PubMed] [Google Scholar]
- 88. Balogun E., Hoque M., Gong P., Killeen E., Green C. J., Foresti R., Alam J., Motterlini R. (2003) Biochem. J. 371, 887–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Rushworth S. A., Ogborne R. M., Charalambos C. A., O'Connell M. A. (2006) Biochem. Biophys. Res. Commun. 341, 1007–1016 [DOI] [PubMed] [Google Scholar]
- 90. Andreadi C. K., Howells L. M., Atherfold P. A., Manson M. M. (2006) Mol. Pharmacol. 69, 1033–1040 [DOI] [PubMed] [Google Scholar]
- 91. Kang K. W., Lee S. J., Kim S. G. (2005) Antioxid. Redox Signal. 7, 1664–1673 [DOI] [PubMed] [Google Scholar]
- 92. Dhakshinamoorthy S., Jain A. K., Bloom D. A., Jaiswal A. K. (2005) J. Biol. Chem. 280, 16891–16900 [DOI] [PubMed] [Google Scholar]
- 93. Hira S., Tomita T., Matsui T., Igarashi K., Ikeda-Saito M. (2007) IUBMB Life 59, 542–551 [DOI] [PubMed] [Google Scholar]
- 94. Lanford R. E., Hildebrandt-Eriksen E. S., Petri A., Persson R., Lindow M., Munk M. E., Kauppinen S., Ørum H. (2010) Science 327, 198–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Hou W., Tian Q., Zheng J., Bonkovsky H. L. (2010) Hepatology 51, 1494–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Ishikawa M., Numazawa S., Yoshida T. (2005) Free Radic. Biol. Med. 38, 1344–1352 [DOI] [PubMed] [Google Scholar]
- 97. Maines M. D. (1981) Biochim. Biophys. Acta 673, 339–350 [DOI] [PubMed] [Google Scholar]
- 98. Benallaoua M., François M., Batteux F., Thelier N., Shyy J. Y., Fitting C., Tsagris L., Boczkowski J., Savouret J. F., Corvol M. T., Poiraudeau S., Rannou F. (2007) Arthritis Rheum. 56, 2585–2594 [DOI] [PubMed] [Google Scholar]
- 99. Lawson T. E., Redlak M. J., Yager D. R. (2011) Wound Repair Regen. 19, 241–249 [DOI] [PubMed] [Google Scholar]