Background: Respiratory supercomplexes are known to exist, but their function remains to be revealed.
Results: Plant supercomplexes are affected by hypoxia and a concomitant drop in pH.
Conclusion: Respiratory supercomplexes are dynamic structures that are affected by the intracellular environment.
Significance: Supercomplexes could have a regulatory function in guiding electrons through alternative respiratory pathways.
Keywords: Hypoxia, Mitochondria, Plant, Respiration, Respiratory Chain, Supercomplexes
Abstract
Respiratory supercomplexes are large protein structures formed by various enzyme complexes of the mitochondrial electron transport chain. Using native gel electrophoresis and activity staining, differential regulation of complex activity within the supercomplexes was investigated. During prolonged hypoxia, complex I activity within supercomplexes diminished, whereas the activity of the individual complex I-monomer increased. Concomitantly, an increased activity was observed during hypoxia for complex IV in the smaller supercomplexes that do not contain complex I. These changes in complex activity within supercomplexes reverted again during recovery from the hypoxic treatment. Acidification of the mitochondrial matrix induced similar changes in complex activity within the supercomplexes. It is suggested that the increased activity of the small supercomplex III2+IV can be explained by the dissociation of complex I from the large supercomplexes. This is discussed to be part of a mechanism regulating the involvement of the alternative NADH dehydrogenases, known to be activated by low pH, and complex I, which is inhibited by low pH. It is concluded that the activity of complexes within supercomplexes can be regulated depending on the oxygen status and the pH of the mitochondrial matrix.
Introduction
Mitochondrial supercomplexes are associations of different respiratory protein complexes (1, 2). In plants, the proton-translocating complexes I2 (NADH-dehydrogenase), III (cytochrome c reductase), and IV (cytochrome c oxidase) have been documented to be organized in supercomplexes with various configurations and stoichiometries. Supercomplexes can be composed of different combinations of only two complexes, like the supercomplex I+III2, I2+III2, III2+IV, and III2+IV2, or various combinations of the three complexes I, III, and IV in so-called respirasomes. Moreover, it was suggested that these supercomplexes may be organized in even larger linear structures, so-called megacomplexes or respiratory strings (3). However, it should be noted that apart from the conglomeration of respiratory complexes into supercomplexes, a considerable population of the complexes I and IV present in the inner mitochondrial membrane occur as individual complexes (4).
Since their first discovery in animals (5), fungi (6), and plants (4), the function of supercomplexes for respiratory electron transport has been discussed, but no conclusive explanation is formulated. Several roles of mitochondrial supercomplexes have been discussed in the literature. First, supercomplexes would increase the stability of individual complexes (7, 8). Second, supercomplexes would affect the membrane structure and increase the dense packing of proteins within the membranes (9). Third, electrons could be efficiently channeled between the reactive sites of the complexes within supercomplexes (10). This electron channeling model is supported by the three-dimensional structure analysis of plant (11), yeast (12), and animal (1) supercomplexes, which revealed that the binding sites for the electron carriers ubiquinone and cytochrome c within the complexes I and III or III and IV are in relative proximity to each other. Finally, in plants the organization of respiratory complexes into supercomplexes could affect the direction of the electron flow from and toward the various alternative components of the mitochondrial electron transport chain, such as the type II dehydrogenases (alternative NAD(P)H dehydrogenase (ND2)) or the alternative oxidase (AOX) (2). This could play a role in the fine-tuning of energy metabolism and ATP yield when environmental conditions change (13).
The latter putative functions of respiratory supercomplexes imply a dynamic recombination of complexes within the supercomplexes rather than that supercomplexes exist as static structures. As a consequence, it would mean that the distribution and activity of specific complexes among the various supercomplexes can vary upon specific environmental conditions (14). The aim of this study was to analyze the variation in activity of specific complexes within the various supercomplexes of the mitochondrial electron transport chain when the oxygen availability changes. We used blue native PAGE and determined changes in the activity of complex I and complex IV within the respective supercomplexes and related these findings to the activity of oxygen consumption and changes of the mitochondrial membrane potential. From our studies it is concluded that supercomplex formation is a dynamic process and that supercomplex composition can be readily and reversibly modified.
EXPERIMENTAL PROCEDURES
Plant Cultivation
Tubers from potato (Solanum tuberosum cv. desiree) were collected from plants cultivated in the fields of the Max Planck Institute of Molecular Plant Physiology in Potsdam-Golm, Germany. After harvesting, tubers were stored in the dark at 4 °C. Tobacco wild type plants (Nicotiana sylvestris) and the CMSII mutant line (kindly provided by R. de Paepe, Paris, France) were grown at 20 °C in the greenhouse with an irradiation of at least 300 μmol/m2 and a light period of 16 h per day.
The hypoxic treatment to potato tubers was performed by flooding tubers for 24–36 h (as indicated in the legends) in oxygen-deprived water (oxygen concentration varied between 2.5 and 10 μm). Recovery from this treatment was done in normal air for another 36 h.
Tissue pH Measurements
Changes of the pH of potato tuber tissue as induced by flooding-induced hypoxia were determined on the cell sap that was set free on the cutting surface immediately after cutting a potato tuber ∼1 cm below the surface using a micro-pH electrode (NMPH2 Beetrode Micro pH electrode; WPI). The pH electrode and a reference electrode (DRIREF-2, WPI) were connected to a pH meter (S20-SevenEasy, Mettler-Toledo Intl. Inc.) via a ZBEECAL Beetrode offset (WPI).
Isolation of Mitochondria
Potato mitochondria were isolated at 4 °C by homogenizing 0.5–1 cm3 potato tuber pieces in homogenization buffer (100 ml of buffer freshly supplemented with cysteine per 100 g of tissue, 25 mm MOPS (KOH), pH 7.8, 400 mm mannitol, 1 mm EGTA, 10 mm Tricine, 0.1% w/v BSA, 1% w/v Polyvinylpyrrolidone 40k (Sigma-Aldrich, St. Louis, MO), 8 mm cysteine) in a blender (8010EG, Waring Commercial) in the cold. The homogenate was filtered through four layers of cheesecloth and one layer of Miracloth (Calbiochem) and centrifuged 5 min at 1000 × g. The pellet was discarded, and the supernatant was centrifuged for 10 min at 9000 × g. The pellet was resuspended in 3–5 ml of wash medium (10 mm MOPS (KOH), pH 7.2, 400 mm mannitol, 1 mm EGTA, and 0.1% w/v BSA), loaded on top of a Percoll (GE Healthcare) gradient (40, 28, and 20% (v/v) Percoll in wash medium), and centrifuged 45 min at 40,000 × g. The enriched mitochondrial phase, located in the interphase between 40 and 28% Percoll, was resuspended in wash medium and centrifuged 10 min at 10,000 × g. This washing step was repeated once. Extraction of mitochondria from N. sylvestris leaves was essentially performed according to the method described previously (15).
Blue Native PAGE
Blue native polyacrylamide gel electrophoresis (BN-PAGE) was essentially performed as described in Schägger (10). In brief, a continuous 1.5-mm-thick polyacrylamide gradient gel (12–4.5% acrylamide) was cast at 4 °C from the bottom upwards into a PROTEAN II xi/XL vertical electrophoresis cell system (Bio-Rad) using a gradient mixer (MX40, Itf Labortechnik GmbH & Co KG, Wasserburg, Germany) and a peristaltic pump with a speed of 20 rpm (Pumpdrive 5101 SP Quick, Heidolph Instruments GmbH & Co. KG, Schwabach, Germany). The acrylamide concentration of the stacking gel was 4%.
Mitochondrial proteins were solubilized as described in Eubel et al. (16). Isolated mitochondria were solubilized using 2.8 mg of digitonin in 50 μl of 30 mm HEPES, pH 7.4, supplemented with 150 mm potassium acetate per 400 μg of protein. Per lane, 400 μg of solubilized protein was loaded on the gradient acrylamide gel. After loading the samples, 300 μl of 5% (w/v) Coomassie Blue in 750 mm aminocaproic acid was pipetted on top of the gel to fill empty wells. The gel ran with Coomassie-free cathode buffer for 13 h at 4 °C with a current of 9 mA per gel and a maximal voltage of 500 V using an Electrophoresis Power Supply EPS 3500 XL (Amersham Biosciences).
Activity Staining of Respiratory Complexes
Activity staining methods for complex I and complex IV of the mtETC were done as described in Zerbetto et al. (17) with modifications. The concentrations of NADH and nitrotetrazolium blue in the complex I staining were 10.5 μm and 50 μg/ml, respectively. The concentrations of 3,3′-diaminobenzidine tetrahydrochloride and cytochrome c for the complex IV staining were 1 mg/ml and 20 μg/ml, respectively. No catalase was added to the staining solution. The gels were scanned when an activity signal appeared after 10–90 min with an Epson Perfection V10 scanner (Seiko Epson Corp., Nagano, Japan). The pictures were saved in TIFF format, and densitographic analysis of the lanes was performed using the BioDocAnalyze software from Biometra (Göttingen, Germany). The peaks of the bands from the densitographic curves were distinguished manually using the Adobe Photoshop software (Adobe Systems, San José, CA), and the area of each peak was calculated with ImageJ (18). The peak area was quantified in relative units. The peak area of each band was compared between treatments using either Student's t test or an analysis of variance as indicated in the legends. For the calculation of correlation between bands the Pearson Correlation-Free Statistics Software (19) was used.
Respiration Measurements
Mitochondrial oxygen uptake rates were determined by loading a fresh mitochondrial suspension equivalent of 500–1000 μg of mitochondrial protein in 2 ml of incubation media (20) (pH 7.5 if not stated otherwise). The rate of respiration was determined by measuring the change of the oxygen concentration in the incubation medium at 25 °C using a Fibox-3 optical oxygen sensor (Presens, Regensburg, Germany). When substrates or inhibitors were added, the following final concentrations were used: 4 mm NADH, 10 mm succinate, 750 μm ADP, 750 μm ATP, 20 μm myxothiazol in DMSO, 100 μm rotenone in DMSO, 500 μm 2,4-dinitrophenol (DNP) in ethanol, 500 μm KCN.
Mitochondrial Membrane Potential Measurements
Isolated mitochondria (600 μg of protein) were suspended in 2 ml of incubation media (see “Respiration Measurements” above) supplemented with 10 mm tetraphenyl phosphonium (TPP+). A TPP+ selective electrode (WPI) connected to a voltmeter pH 209 (Hanna Instruments Inc.) was used to monitor changes in TPP+ concentrations. Data acquisition and recording were done with Lab-Trax-4/16 (WPI) operated with the Data-Trax software from the manufacturer. Calculation of the membrane potential from mitochondrial TPP+ accumulation was done as described in Refs. 21–24.
Supercomplex Identification Using LC-MSMS
The method used is essentially as described in Olsen et al. (25). Briefly, protein bands were cut from a BN-PAGE, and protein was reduced, carbamidomethylated, and extracted after trypsin digestion. The resulting peptides were analyzed by LC-MSMS as described in Kierszniowska et al. (26) using nanoflow HPLC (Proxeon Biosystems, Denmark) and an Orbitrap hybrid mass spectrometer (LTQ Orbitrap, Thermo Electron) as the mass analyzer. The mass spectra were analyzed using the MASCOT algorithm, and the polypeptides found were compared with the sequences in S. tuberosum EST data base from the J. Craig Venter Institute (Solanum_tuberosum_release_2.fasta).
The following search parameters were applied: trypsin as the cleaving enzyme, peptide mass tolerance 10 ppm, MS/MS tolerance 0.8 Da, 1 missed cleavage allowed. Carbamidomethylation of cysteine was set as a fixed modification, and methionine oxidation was chosen as a variable modification. In general, peptides were accepted without manual interpretation as described in Kierszniowska et al. (26), when the score was higher than 31 (Mascot p < 0.01 significance threshold). Peptides with a score between 24 and 31 were manually inspected, requiring a series of three consecutive y or b ions to be accepted. Mass accuracy and Δ scores were taken into account when single peptides were accepted.
RESULTS
Identification of Supercomplexes
The occurrence of respiratory supercomplexes was investigated using BN-PAGE. After separation of the supercomplexes in the native gel, the presence of complexes I and IV within the supercomplexes was determined using in gel enzyme activity staining (17, 27) (supplemental Fig. S1). This method not only allows qualitative identification of the various supercomplexes but also allows semiquantitative determination of complex activity within each supercomplex. The proportionality of protein amount of the complex or supercomplex bands and the activity stain of complexes I and IV was shown by Sabar et al. (27) and confirmed for our experimental settings (supplemental Figs. S2 and S3). Therefore, it was determined that the activity staining methods can be used to perform semiquantitative assessments of the activity of individual respiratory enzymes within each supercomplex of the mitochondrial electron transport chain.
For potato mitochondrial protein, the identity of the bands was further confirmed by LC/MSMS, as potato was the main research object in this study. These results confirmed the identity of the complexes and supercomplexes as described in Eubel et al. (4). The identified peptides from subunits of the expected complexes contained in each supercomplex and the abbreviations used are listed in supplemental Table S1. The complexes are abbreviated with Latin numbers (I–V), and the sub-indices indicate how many copies of complex are present within the supercomplex. The nomenclature used for the BN gels of N. sylvestris mitochondria was based on extensive previous studies available from the literature (11, 16, 28, 29).
Supercomplexes Are Affected in a Complex I Knock-out
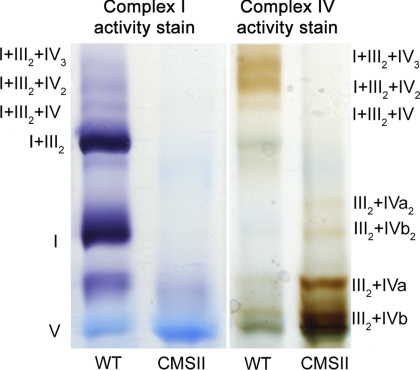
The activity of complex I and IV within supercomplexes was investigated for plants with changed expression of different proteins of the mETC. The tobacco CMSII mutant is impaired in its activity of complex I as this complex cannot associate properly due to the deletion of the subunit 7 gene (30, 31). Activity staining for complex IV isolated from the CMSII mutant revealed that its activity increased considerably in supercomplex III2+IV as compared with wild type (Fig. 1, supplemental Fig. S4). This result shows that in tobacco complexes III and IV remained associated and functional in a supercomplex structure, even when no complex I was present anymore. This demonstrates the stability of these specific supercomplexes (III2+IV) and emphasizes its functional relevance.
FIGURE 1.
Supercomplex analysis of plants with altered levels of complex I. shown is activity staining of complex I and IV within supercomplexes isolated from N. sylvestris wild type (WT) and the CMSII mutant deficient in complex I. Additional determinations on biologically independent material are provided in supplemental Fig. S4.
The Effect of the Oxygen Concentration on Supercomplex Stability
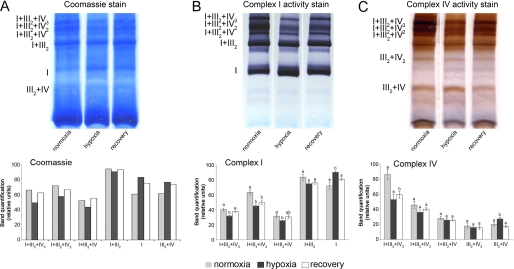
Hypoxia reduces respiratory activity (13). To test the effect of hypoxia on the respiratory supercomplexes, potato tubers were flooded for 36 h. The oxygen concentration of the water was reduced before the flooding treatment and kept low (between 2.5 and 10 μm) during the entire course of the treatment. Part of the tubers was subsequently transferred to normal air for 36 h to recover from the hypoxic treatment. During hypoxia, the distribution of activity of complex I or IV between the various supercomplexes revealed a shift toward a higher activity of the complex I monomer as well as an increased activity of complex IV within the supercomplexes III2+IV2 and III2+IV as compared with the control tissue sampled before the hypoxia treatment (Fig. 2, supplemental Figs. S5 and S6). In contrast, the activity, as determined for the enzymes of supercomplexes I+III2+IV1–3, remained unaltered, although it should be reminded here that slight changes of the activity of supercomplexes I+III2+IV1–3 when highly abundant are difficult to quantify. During recovery from the hypoxia treatment, the changes in complex activity within the supercomplexes as observed during hypoxia were reverted toward the situation as observed before the hypoxia treatment commenced (Fig. 2, supplemental Fig. S5).
FIGURE 2.
Effect of hypoxia on respiratory supercomplexes. A, shown is blue native PAGE of supercomplexes from mitochondria that were isolated from potato tubers after 36 h of flooding-induced hypoxia ([O2] ≤ 10 μm) or from tubers that regenerated during 36 h in normal air from the hypoxic treatment. As the control, samples were prepared from tubers that were kept in normal air during the entire course of the experiment. The first three lanes (A) show Coomassie-stained proteins from mitochondria isolated from tubers after a normoxic, hypoxic (flooded), or recovery treatment, respectively. The second (B) and third sets (C) of three lanes show activity staining of complex I and complex IV, respectively. B, shown is quantification of color intensity of the bands stained for complex I activity. C, shown is quantification of color intensity of the bands stained for complex IV activity. Bars represent mean values ± S.E. of at least three technical replicates. Significant differences were determined using single-way analysis of variance (p ≤ 0.05), and mean values that are significantly different from each other were marked with different letters above the bars. The results from additional biological repetitions are provided in supplemental Figs. S5 and S6.
Supercomplexes Destabilize When the pH Decreases
It is well documented that hypoxic conditions lead to the accumulation of organic acids like lactate or succinate (32, 33) and acidification of the cytosol (34). Indeed, confirmation of the pH of tubers from flooded and well aerated tubers, as determined using a micro pH electrode, revealed a pH of 5.73 ± 0.155 (mean ± S.D., n = 4) after flooding, whereas the pH values before flooding (pH 6.47 ± 0.116) and after 36 h of recovery from the hypoxic treatment (6.06 ± 0.094) were significantly higher (supplemental Fig. S7). This confirms an increase of the tissue acidity caused by hypoxia even though the determination does not directly reveal pH changes of specific (sub)cellular compartments.
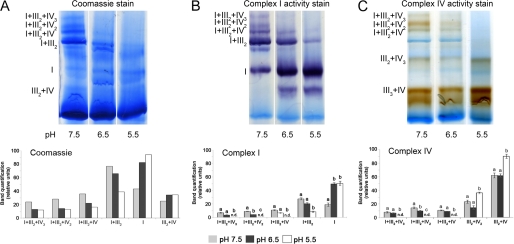
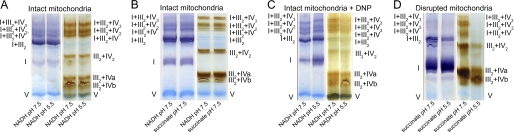
The effect of pH on the activity of complexes within the supercomplexes was further investigated by incubating isolated mitochondria for 10 min at various pH levels between 5.5 and 7.5 using 10 mm succinate as the respiratory substrate before supercomplex separation from the mitochondrial membranes using the standard protocol at pH 7.5. Coomassie staining of the BN gels showed that the large supercomplexes containing complexes I+III2+IV1–3 dissociated at pH 6.5 and below, whereas singular complex I accumulated concomitantly. This effect was also observed in the in-gel activity stainings of the complexes I and IV (Fig. 3, supplemental Fig. S8).
FIGURE 3.
Effect of pH on supercomplex composition in mitochondria oxidizing succinate. A, proteins isolated from mitochondria that were pretreated at various pH in the presence of 10 mm succinate were separated on a BN-PAGE gel. The same protein extracts were used for staining with Coomassie, complex I, and complex IV activity. B, quantification of color intensity of the bands stained for complex I activity is shown. C, quantification of color intensity of the bands stained for complex IV activity is shown. Bars represent mean values ± S.E. of at least three technical replicates. Significantly different mean values as determined by a 2-way analysis of variance followed by the Holm-Sidak post hoc test are marked by different letters (n = 3, p ≤ 0.05). The results from additional biological repetitions are provided in supplemental Fig. S8.
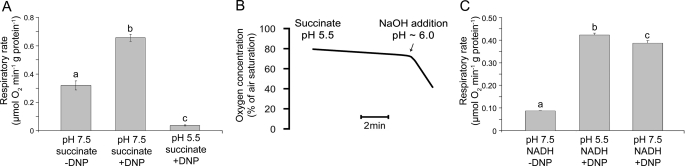
Decreasing the pH of the buffer inhibited respiratory activity of the isolated mitochondria when succinate was used as respiratory substrate (Fig. 4A). However, this inhibition was reversible, as the rate of oxygen consumption by the mitochondria increased again at pH 6 when the incubation buffer was titrated with NaOH from pH 5.5 to 7.5 (Fig. 4b). In contrast, neither the rate of state 3 respiration (Fig. 4C) nor supercomplex stability (Fig. 5A, supplemental Fig. S9) diminished with decreasing pH when NADH was used as respiratory substrate. Also, no differences in the activity pattern of the various supercomplexes (Fig. 5b, supplemental Fig. S10) could be observed when either succinate or NADH was tested at pH 7.5. Hence, the supercomplex composition did not depend on the substrate activation of either complex I or II, but supercomplex disassembly was induced by the combination of succinate availability and low pH.
FIGURE 4.
The effect of various combinations of respiratory substrates and different pH on respiratory activity. A, shown is respiratory activity of isolated potato tuber mitochondria supplied with 750 μm ADP and either 4 mm NADH (pH 7.5), 10 mm succinate (pH 7.5), or 10 mm succinate (pH 5.5). B, oxygen consumption of isolated potato tuber mitochondria was determined at pH 5.5 in the presence of 10 mm succinate and 500 μm DNP. Subsequently, NaOH was added to increase the pH of the incubation buffer. At pH 6, the rate of respiration is indicated by the slope of the graph increased rapidly. C, respiratory activity is shown of isolated potato tuber mitochondria supplied with 4 mm NADH at pH 7.5 or 5.5 in the presence or absence of 250 μm DNP to activate state 3-respiration. Significantly different mean values as determined by a 2-way analysis of variance followed by the Holm-Sidak post hoc test are marked by different letters (n = 3, p ≤ 0.05).
FIGURE 5.
The effect of various combinations of respiratory substrates and different pH values on supercomplex composition. A, blue native gels stained for the activity of complex I (left) and complex IV (right) within supercomplexes isolated from mitochondria were preincubated at either pH 5.5 or 7.5 for 10 min in a buffer containing 4 mm NADH and 750 μm ADP. B, shown is the effect of NADH or succinate at pH 7.5 on supercomplex composition. Activity staining of complex I (left) and complex IV (right) on supercomplexes isolated from mitochondria pretreated with either 750 μm ADP and 4 mm NADH or 10 mm succinate is shown. C, shown is the effect of DNP-induced membrane depolarization on the stability of supercomplexes at pH 5.5 and 7.5. Mitochondria were preincubated for 10 min in a buffer (pH 5.5 or 7.5) with 500 μm DNP, 4 mm NADH, and 750 μm ADP. D, determination of supercomplex stability in disrupted mitochondrial membranes is shown. Isolated potato tuber mitochondria were treated with a freeze-thaw cycle, and supercomplexes were isolated from the disrupted membranes. Complex activity within blue native gels was determined after preincubation of the mitochondria for 10 min in a buffer containing 10 mm succinate and 750 μm ADP at a pH of 5.5 or 7.5. Bar charts showing quantification data of the activity staining for the experiments shown in these figures as well as the results of biological replicate determinations are provided in the supplemental Figs. S9–S11.
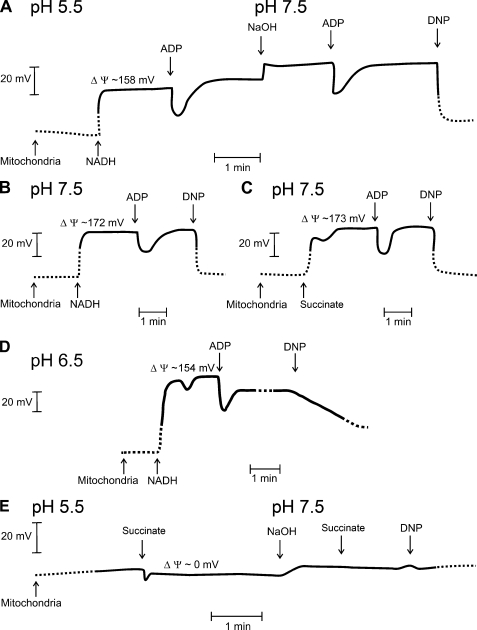
Incubation of isolated mitochondria in buffers with different pH could potentially affect the membrane potential of the mitochondrial inner membrane too. Therefore, the membrane potential was analyzed using the membrane-permeable cation TPP+ (Fig. 6). When NADH was used as respiratory substrate, the pH of the incubation solution had only little effect on the membrane potential of isolated mitochondria (around 160–170 mV) (Fig. 6A). Also, mitochondria treated with succinate at pH 7.5 had a similar membrane potential as when NADH was used (Fig. 6, B and C). As expected, the protonophore DNP induced a complete membrane depolarization under all conditions tested. Moreover, no electrochemical polarization of the membrane was observed in mitochondria that were treated with succinate at pH 5.5 (Fig. 6E). Even neutralizing the pH of the buffer by titrating NaOH to the solution did not lead to reestablishing of the membrane potential. Apparently, this treatment leads to endured uncoupling of the membrane in isolated mitochondria. However, respiratory activity was shown to recover by this treatment (Fig. 4b). When the pH of buffer was only reduced to pH 6.5, the membrane remained polarized (Fig. 6d), whereas the supercomplexes already dissociated (Fig. 3). This indicates that the dissociation of supercomplexes is caused by the pH shift rather than by the depolarization of the membrane potential.
FIGURE 6.
Determination of the mitochondrial membrane potential. A, shown is determination of the membrane potential of potato tuber mitochondria via measuring changes in TPP+ accumulation. The initial pH of the incubation solution was 5.5. Subsequently, 50 μl of 160 mm NADH was added to the incubation solution (volume 2 ml) followed by 15 μl of 15 mm ADP, 3 μl of 10 m NaOH to increase the pH of the buffer to pH 7.3, 15 μl of 15 mm ADP, or DNP (final concentration of 500 μm). B, shown is determination of the membrane potential of isolated mitochondria in medium with pH 7.5 using 4 mm NADH as respiratory substrate. Subsequently, 15 μl of 15 mm ADP or DNP were added. C, shown is determination of the membrane potential of isolated mitochondria incubated at pH 7.5 using 10 mm succinate as the respiratory substrate. Subsequently, 15 μl of 15 mm ADP or DNP were added. D, shown is determination of the membrane potential of isolated mitochondria incubated at pH 6.5 using 4 mm NADH as respiratory substrate. Subsequently, 15 μl of 15 mm ADP or DNP were added. E, shown is determination of the membrane potential of mitochondria incubated in buffer (pH 5.5) with 10 mm succinate as respiratory substrate. Subsequently, the pH of the solution was adjusted to 7.5 using 4 μl of 10 m NaOH, and succinate (final concentration: 20 mm) and DNP were added. ΔΨ, membrane potential.
To further differentiate between the effect of pH and the membrane potential on the dissociation of supercomplexes, the activity of complex I and IV within the various supercomplexes was analyzed from mitochondria in which no membrane potential could be generated. For this, membrane polarization was prevented via either a pretreatment with the protonophore DNP (Fig. 5C) or via disruption of the mitochondrial membranes by a repeated freeze-thaw treatment (Fig. 5D). Both experiments showed that the pH-induced shift in complex activity within supercomplexes occurred independently of the electrical polarization of the membrane. However, it should be noted here that freezing the mitochondrial membranes already reduced the staining of the largest supercomplexes as compared with the supercomplex pattern observed in freshly isolated mitochondria. Nevertheless, it can be concluded that the stability of the supercomplexes depends on the pH rather than on the membrane potential of the mitochondrial inner membrane.
Respiration Rate Does Not Affect Supercomplex Stability
To investigate the effect of the respiratory activity on the supercomplex composition, we determined the respiration rate as well as the activity of complex I and IV in the various supercomplexes in dormant and sprouting potato tubers. The net mitochondrial protein yield per tuber fresh weight was two to three times higher from sprouting potato tubers as compared with dormant tissue, and concomitantly the respiratory activity as referred to equal amounts of fresh weight of tuber tissue was about two times higher as well (supplemental Fig. 12A). However, respiratory activity normalized to equal amounts of isolated mitochondrial protein did not show a significant difference between the two developmental stages using NADH as respiratory substrate. Any change in complex activity related to a comparable change in the protein abundance as determined by Coomassie staining (supplemental Fig. 12, B–H), but no significant change in the activity of specific supercomplexes was observed. Rather, a general increase in the bands of all detected supercomplexes could be seen. Apparently, the net respiratory activity is not reflected in specific changes in the supercomplex composition.
The relation between respiratory activity and supercomplex formation was further characterized using various inhibitors of specific components of the mETC. Isolated mitochondria were incubated for 10 min in the presence or absence of substrate (NADH and ADP), 20 μg ml−1 alamethicin (permeabilizes membranes to NADH, thus feeding substrate to complex I), 100 μm rotenone (inhibitor of complex I), 20 μm myxothiazol (inhibitor of complex III), 500 μm potassium cyanide (inhibitor of complex IV), or 750 μm ATP before protein extraction and BN-PAGE. None of the treatments led to a change in the composition of the supercomplexes (supplemental Fig. S13) even though respiratory activity was reduced (supplemental Fig. S14). These observations indicate that the stability of the supercomplexes was not affected by inhibition of the electron flow through the supercomplex by any of the inhibitory treatments.
DISCUSSION
The respiratory complexes I, III, and IV of the mETC associate into supercomplexes with various individual stoichiometric compositions (9), but a functional role of these supercomplexes for respiration is still being discussed. Studies investigating the supercomplex composition or stability using specific respiratory inhibitors (supplemental Fig. S13 and S14) did not provide evidence for any differential regulation of supercomplexes. However, further results presented here reveal a role of the supercomplexes during hypoxia in the differential regulation of electron supply via alternative components of the respiratory pathway.
For studying supercomplex stability in plants, we investigated the occurrence of the various supercomplexes in the tobacco CMSII mutant, in which complex I is completely absent. Several studies revealed before that CMSII has no detectable complex I activity (35, 36) and that no supercomplexes exist anymore that contain complex I (29). Here, we show that supercomplexes III2+IV2 and III2+IV remained present and that complex IV activity in these supercomplexes even increased in the CMSII line (Fig. 1). Similarly, increased abundance of supercomplexes III2+IV2 and III2+IV was observed when complex I dissociated from the respirasomes during hypoxic conditions (Fig. 2, supplemental Figs. S5 and S6) or when succinate was supplied at low pH (Fig. 3, supplemental Fig. S8). It should be noted that isolated mitochondria that were supplied with succinate at pH 5.5 lost their membrane potential (Fig. 6D). However, membrane depolarization could not explain the dissociation of the supercomplexes as the addition of DNP or disrupting the membrane with a freeze-thaw cycle did not change the supercomplex activity under neutral pH conditions (Fig. 5, C and D).
Apparently, complex I can dissociate from a supercomplex, whereas the complexes III and IV are kept together in a smaller supercomplex. In humans it was shown that supercomplexes are essential for the stability of complex I. Cell lines from patients that are deficient in complex III do not form supercomplexes, and as a secondary effect, also complex I disintegrated (37). Also, proper assembly of complex IV appeared to be essential for the stability of complex I in both human and mice mitochondria (38, 39). In contrast to animals, complex I stability was not affected in the Cyc1–1 mutant of the fungus Podospora anserina, which is lacking complex III and, therefore, depends on alternative oxidase to keep the electron flow in the mitochondrial electron transport chain upright (39). Probably, the differences in respirasome stability between mammals, plants, and fungi relate to the presence of alternative components in the mETC, like alternative oxidase or the type-2 dehydrogenases in the latter organisms.
It is most striking that the change of the supercomplex composition as induced by hypoxia (Fig. 2, supplementary Figs. S5 and S6) resembles the changes induced by decreasing the pH in the presence of succinate as respiratory substrate (Fig. 3, supplementary Fig. S8). Therefore, we suggest that in both cases complex I dissociates from the large supercomplexes, whereas supercomplex III2+IV accumulates. However, also alternative post-translational modifications or allosteric regulation of complex I activity cannot be ruled out at this stage to explain the changes in complex activity within the supercomplexes that we observed during hypoxia. During reoxygenation the supercomplexes reassemble (Fig. 2), indicating that the dissociation of supercomplexes indeed relates to the oxygen availability. It is well described that the respiratory activity of plant tissue changes with the oxygen availability of the cells; at decreasing oxygen availability the respiratory activity declines (13, 40, 41). Hypoxia is also known to induce a decrease of the cellular pH in plants as well as in other organisms (42). Consistent with that, the pH at the cutting surface of potato tubers was determined to drop significantly in flooded tubers as compared with normoxic tubers, and these pH values reverted again during recovery in air after the flooding treatment (supplemental Fig. S7). Of course, these pH determinations cannot be attributed directly to any cellular compartment but, rather, combine changes in both apoplast and intracellular compartments. Nevertheless, these data confirm that our flooding treatment of the tubers resulted in the typical cellular acidification as induced by hypoxia in plants (40). The pH of the mitochondrial matrix is generally assumed to be slightly more basic than the cytosol but is documented to follow fluctuations of the cytosolic pH (42–46). Experiments using pH-sensitive fluorescent probes that were specifically targeted toward the mitochondria of HeLa cells revealed a marked decrease in the pH of the matrix after providing the cells with organic acids like lactate, pyruvate, or acetate (44, 45). As the cytosolic acidification of a hypoxic cell is caused by a combination of low ATP (43) and accumulation of organic acids like lactate and succinate (32), it must be assumed that the pH in the mitochondrial matrix of the hypoxic potato tubers that were used in our experiments decreased as well. This suggests that the dissociation of complex I from complexes III and IV that is observed during hypoxia and after decreasing the pH results from the changing pH in both cases.
For mammalian mitochondria, a role of respiratory supercomplexes was suggested in determining respiratory efficiency (47). After induced ischemic heart failure in dogs, dissociation of the large supercomplexes containing complex I, III, and IV and a concomitant decrease of respiratory activity of intact heart mitochondria was observed even though the capacity of the individual respiratory complexes was not affected by the treatment. In plants, the consequence of the separation between complex I and the combination of complexes III and IV for the flow of electrons through the mETC would be a shift toward supercomplex III2+IV cooperating with the alternative NADH dehydrogenases rather than with complex I. Consistently, the alternative NADH dehydrogenases are activated by lowered pH, whereas complex I is inhibited by a low pH (48–50). Moreover, complex I was also shown to be reversibly deactivated during hypoxia of mammalian mitochondria in a reactive nitrogen species-dependent manner (51). In plants, the dissociation of complex I from the large supercomplexes as observed here during hypoxia is suggested to favor the electron transmission from the alternative type 2 NAD(P)H dehydrogenases and succinate dehydrogenase to ubiquinone. This could help in oxidizing the pool of cytosolic NADH during hypoxia, which is required to prevent inhibition of glycolysis due NAD+ limitation (39).
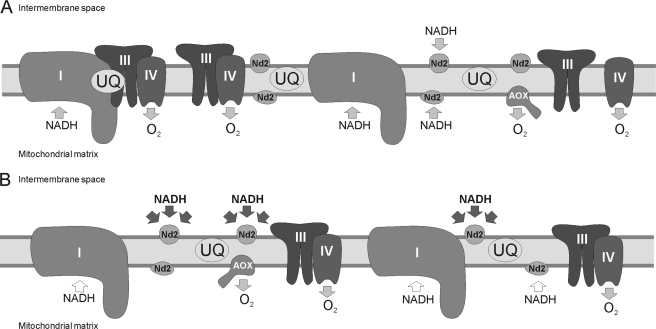
Summarizing, our results suggest that respiratory supercomplexes could be involved in directing the flow of electrons between different respiratory electron transport pathways of the plant mETC (Fig. 7). Dissociation of complex I from complexes III and IV occurs under hypoxic conditions, during which succinate accumulates as substrate of complex II, and alternative NAD(P)H dehydrogenases are favored above complex I as electron donors to ubiquinone due to cellular acidification. This response of the supercomplex composition could probably act as a regulatory mechanism involved in acclimation of respiration to stress conditions in which the cellular pH changes, such as hypoxia and anoxia.
FIGURE 7.
Model of the role of supercomplex formation for the channeling of electrons through the oxidative phosphorylation pathway. A, under standard conditions respirasomes as well as smaller supercomplexes and single complexes are observed simultaneously in the inner mitochondrial membrane. B, dissociation of complex I from the respirasomes supports electron transfer from cytosolic NAD(P)H into the mitochondrial electron transport chain via the external alternative dehydrogenases. Supercomplex dissociation can be induced by acidification of the cell and accumulation of organic acids such as it occurs during hypoxia. These same conditions were previously described to inhibit the enzyme activity of complex I and stimulate activity of the alternative dehydrogenases (45–47). The shift from complex I to the alternative dehydrogenases would support oxidation of cytosolic NADH to keep the glycolytic flux upright when oxidative phosphorylation is reduced upon hypoxia. UQ, ubiquinone; Nd2, alternative NAD(P)H dehydrogenase; AOX, alternative oxidase.
Supplementary Material
Acknowledgments
Seeds of the N. sylvestris wild type and CMSII mutant were kindly provided by Rosine de Paepe (Université de Paris-Sud, France). We thank Alisdair Fernie for discussions and Hans-Peter Braun and Lutz Eichacker for introducing us to the details of BN-PAGE.
This work was supported by the Max Planck Society, the German Research Foundation (SFB429), and the Swedish Research Council.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1–S14.
- I
- III, IV, respiratory complexes I, III, and IV respectively
- BN
- blue native
- DNP
- 2,4-dinitrophenol
- TPP+
- tetraphenyl phosphonium
- mtETC
- mitochondrial electron transport chain.
REFERENCES
- 1. Schäfer E., Dencher N. A., Vonck J., Parcej D. N. (2007) Biochemistry 46, 12579–12585 [DOI] [PubMed] [Google Scholar]
- 2. Eubel H., Heinemeyer J., Sunderhaus S., Braun H. P. (2004) Plant Physiol. Biochem. 42, 937–942 [DOI] [PubMed] [Google Scholar]
- 3. Bultema J. B., Braun H. P., Boekema E. J., Kouril R. (2009) Biochim. Biophys. Acta 1787, 60–67 [DOI] [PubMed] [Google Scholar]
- 4. Eubel H., Heinemeyer J., Braun H. P. (2004) Plant Physiol. 134, 1450–1459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Schägger H., Pfeiffer K. (2000) EMBO J. 19, 1777–1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Krause F., Scheckhuber C. Q., Werner A., Rexroth S., Reifschneider N. H., Dencher N. A., Osiewacz H. D. (2004) J. Biol. Chem. 279, 26453–26461 [DOI] [PubMed] [Google Scholar]
- 7. Acín-Pérez R., Fernández-Silva P., Peleato M. L., Pérez-Martos A., Enriquez J. A. (2008) Mol. Cell 32, 529–539 [DOI] [PubMed] [Google Scholar]
- 8. Diaz F., Fukui H., Garcia S., Moraes C. T. (2006) Mol. Cell. Biol. 26, 4872–4881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Boekema E. J., Braun H. P. (2007) J. Biol. Chem. 282, 1–4 [DOI] [PubMed] [Google Scholar]
- 10. Schägger H. (2001) IUBMB Life 52, 119–128 [DOI] [PubMed] [Google Scholar]
- 11. Dudkina N. V., Eubel H., Keegstra W., Boekema E. J., Braun H. P. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 3225–3229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Heinemeyer J., Braun H. P., Boekema E. J., Kouril R. (2007) J. Biol. Chem. 282, 12240–12248 [DOI] [PubMed] [Google Scholar]
- 13. Gupta K. J., Zabalza A., van Dongen J. T. (2009) Physiol. Plant. 137, 383–391 [DOI] [PubMed] [Google Scholar]
- 14. Chien L. F., Wu Y. C., Chen H. P. (2011) Plant Physiol. Biochem. 49, 449–457 [DOI] [PubMed] [Google Scholar]
- 15. Michalecka A. M., Agius S. C., Møller I. M., Rasmusson A. G. (2004) Plant J. 37, 415–425 [DOI] [PubMed] [Google Scholar]
- 16. Eubel H., Jänsch L., Braun H. P. (2003) Plant Physiol. 133, 274–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zerbetto E., Vergani L., Dabbeni-Sala F. (1997) Electrophoresis 18, 2059–2064 [DOI] [PubMed] [Google Scholar]
- 18. Collins T. J. (2007) Biotechniques 43, 25–30 [DOI] [PubMed] [Google Scholar]
- 19. Wessa P. (2008) Pearson Correlation (v1.0.3) in Free Statistics Software (v1.1.23-r6), Office for Research Development and Education, http://www.wessa.net/rwasp_correlation.wasp [Google Scholar]
- 20. Millar A. H., Sweetlove L. J., Giegé P., Leaver C. J. (2001) Plant Physiol. 127, 1711–1727 [PMC free article] [PubMed] [Google Scholar]
- 21. Douce R. (1985) Mitochondria in Higher Plants; Structure, Function, and Biogenesis, Academic Press, New York [Google Scholar]
- 22. Jensen B. D., Gunter K. K., Gunter T. E. (1986) Arch. Biochem. Biophys. 248, 305–323 [DOI] [PubMed] [Google Scholar]
- 23. Kamo N., Muratsugu M., Hongoh R., Kobatake Y. (1979) J. Membr. Biol. 49, 105–121 [DOI] [PubMed] [Google Scholar]
- 24. Mandolino G., Desantis A., Melandri B. A. (1983) Biochim. Biophys. Acta 723, 428–439 [Google Scholar]
- 25. Olsen J. V., Ong S. E., Mann M. (2004) Mol. Cell. Proteomics 3, 608–614 [DOI] [PubMed] [Google Scholar]
- 26. Kierszniowska S., Walther D., Schulze W. X. (2009) Proteomics 9, 1916–1924 [DOI] [PubMed] [Google Scholar]
- 27. Sabar M., Balk J., Leaver C. J. (2005) Plant J. 44, 893–901 [DOI] [PubMed] [Google Scholar]
- 28. Dudkina N. V., Heinemeyer J., Sunderhaus S., Boekema E. J., Braun H. P. (2006) Trends Plant Sci. 11, 232–240 [DOI] [PubMed] [Google Scholar]
- 29. Pineau B., Mathieu C., Gérard-Hirne C., De Paepe R., Chétrit P. (2005) J. Biol. Chem. 280, 25994–26001 [DOI] [PubMed] [Google Scholar]
- 30. De Paepe R., Chétrit P., Vitart V., Ambard-Bretteville F., Prat D., Vedel F. (1990) Mol. Gen. Genet. 222, 206–210 [DOI] [PubMed] [Google Scholar]
- 31. Vidal G., Ribas-Carbo M., Garmier M., Dubertret G., Rasmusson A. G., Mathieu C., Foyer C. H., De Paepe R. (2007) Plant Cell 19, 640–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. van Dongen J. T., Fröhlich A., Ramírez-Aguilar S. J., Schauer N., Fernie A. R., Erban A., Kopka J., Clark J., Langer A., Geigenberger P. (2009) Ann. Bot. 103, 269–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Narsai R., Rocha M., Geigenberger P., Whelan J., van Dongen J. T. (2011) New Phytol. 190, 472–487 [DOI] [PubMed] [Google Scholar]
- 34. Couldwell D. L., Dunford R., Kruger N. J., Lloyd D. C., Ratcliffe R. G., Smith A. M. (2009) Ann. Bot. 103, 249–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gutierres S., Sabar M., Lelandais C., Chetrit P., Diolez P., Degand H., Boutry M., Vedel F., de Kouchkovsky Y., De Paepe R. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 3436–3441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sabar M., De Paepe R., de Kouchkovsky Y. (2000) Plant Physiol. 124, 1239–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Schägger H., de Coo R., Bauer M. F., Hofmann S., Godinot C., Brandt U. (2004) J. Biol. Chem. 279, 36349–36353 [DOI] [PubMed] [Google Scholar]
- 38. Li Y., D'Aurelio M., Deng J. H., Park J. S., Manfredi G., Hu P., Lu J., Bai Y. (2007) J. Biol. Chem. 282, 17557–17562 [DOI] [PubMed] [Google Scholar]
- 39. Maas M. F., Krause F., Dencher N. A., Sainsard-Chanet A. (2009) J. Mol. Biol. 387, 259–269 [DOI] [PubMed] [Google Scholar]
- 40. Geigenberger P. (2003) Curr. Opin. Plant Biol. 6, 247–256 [DOI] [PubMed] [Google Scholar]
- 41. van Dongen J. T., Gupta K. J., Ramírez-Aguilar S. J., Araújo W. L., Nunes-Nesi A., Fernie A. R. (2011) J. Plant Physiol. 168, 1434–1443 [DOI] [PubMed] [Google Scholar]
- 42. Felle H. H. (2001) Plant Biol. 3, 577–591 [Google Scholar]
- 43. Felle H. H. (2005) Ann. Bot. 96, 519–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Llopis J., McCaffery J. M., Miyawaki A., Farquhar M. G., Tsien R. Y. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 6803–6808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Abad M. F., Di Benedetto G., Magalhães P. J., Filippin L., Pozzan T. (2004) J. Biol. Chem. 279, 11521–11529 [DOI] [PubMed] [Google Scholar]
- 46. Orij R., Postmus J., Ter Beek A., Brul S., Smits G. J. (2009) Microbiology 155, 268–278 [DOI] [PubMed] [Google Scholar]
- 47. Rosca M. G., Vazquez E. J., Kerner J., Parland W., Chandler M. P., Stanley W., Sabbah H. N., Hoppel C. L. (2008) Cardiovasc. Res. 80, 30–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Geisler D. A., Broselid C., Hederstedt L., Rasmusson A. G. (2007) J. Biol. Chem. 282, 28455–28464 [DOI] [PubMed] [Google Scholar]
- 49. Rasmusson A. G., Geisler D. A., Møller I. M. (2008) Mitochondrion 8, 47–60 [DOI] [PubMed] [Google Scholar]
- 50. Rasmusson A. G., Møller I. M. (1991) Physiol. Plant. 83, 357–365 [Google Scholar]
- 51. Galkin A., Abramov A. Y., Frakich N., Duchen M. R., Moncada S. (2009) J. Biol. Chem. 284, 36055–36061 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.