Background: Lysosomal storage disorders are caused by ER-associated degradation (ERAD) of mutated unstable lysosomal enzymes.
Results: ERAD inhibition enhances folding and activity of unstable lysosomal protein by prolonging ER retention.
Conclusion: ERAD is the rate-limiting step in the folding of mutated lysosomal proteins.
Significance: ERAD inhibition ameliorates the progression of multiple lysosomal storage disorders caused by protein misfolding and degradation.
Keywords: Apoptosis, ER-associated Degradation, Lysosomal Storage Disease, Molecular Chaperone, Protein Folding, Unfolded Protein Response, Glucocerebrosidase, Protein Misfolding Diseases, Proteostasis
Abstract
Lysosomal storage disorders are often caused by mutations that destabilize native folding and impair trafficking of secretory proteins. We demonstrate that endoplasmic reticulum (ER)-associated degradation (ERAD) prevents native folding of mutated lysosomal enzymes in patient-derived fibroblasts from two clinically distinct lysosomal storage disorders, namely Gaucher and Tay-Sachs disease. Prolonging ER retention via ERAD inhibition enhanced folding, trafficking, and activity of these unstable enzyme variants. Furthermore, combining ERAD inhibition with enhancement of the cellular folding capacity via proteostasis modulation resulted in synergistic rescue of mutated enzymes. ERAD inhibition was achieved by cell treatment with small molecules that interfere with recognition (kifunensine) or retrotranslocation (eeyarestatin I) of misfolded substrates. These different mechanisms of ERAD inhibition were shown to enhance ER retention of mutated proteins but were associated with dramatically different levels of ER stress, unfolded protein response activation, and unfolded protein response-induced apoptosis.
Introduction
Lysosomal storage disorders (LSD)2 are a group of inherited diseases characterized by deficiencies of specific hydrolytic functions and aberrant storage of metabolites in the lysosomes (1). Gaucher disease (GD), the most common LSD, is caused by loss of lysosomal glucocerebrosidase (GC) activity and consequent accumulation of glucosylceramide (2). A number of characterized mutations in the GC-encoding gene (GBA) (3) consist of single amino acid substitutions that do not directly impair protein activity but destabilize its native structure, leading to misfolding and ER-associated degradation (ERAD (4)). Patient-derived cells harboring different GC variants present mutation-specific phenotypes, which depend on the extent of enzyme ERAD, trafficking, and residual activity (5). The enzyme residual activity, in turn, has been linked to the severity of GD manifestations (6). For instance, L444P GC, one of the most common alleles, is associated with complete loss of activity and, in homozygous patients, severe neuronopathic symptoms (7). The N370S substitution, another highly prevalent GC mutation, is associated with residual enzymatic activity (∼10% of WT) and less severe, non-neuronopathic forms of the disease (4).
Most unstable GC variants retain catalytic activity if forced to fold into their native three-dimensional structure. This was demonstrated by enhancing the cellular folding capacity of patient-derived cells through the use of small molecules that influence general cellular folding pathways that maintain protein homeostasis, such as chaperones and the proteasome-ubiquitin system (8, 9) or Ca2+ homeostasis (10, 11). Mechanistic studies conducted to investigate changes in the cellular folding network that enhance mutated GC activity led to the observation that augmenting the pool of mutated GC in the ER is critical to promote rescue of its folding and trafficking (10, 11). Specifically, we demonstrated that an increase in mutated GC activity correlates with (i) up-regulation of the main ER chaperone BiP, which is known to enhance ER retention and prevent ERAD of misfolding intermediates (12), and (ii) up-regulation of GBA, most likely caused by UPR-induced changes in lipid metabolism (10, 11). Both strategies, BiP and GC up-regulation, cause an increase in the pool of GC folding intermediates that escapes ERAD and is amenable to folding rescue.
Extensive ERAD of mutated, unstable proteins is a common theme in the cellular pathogenesis of LSD. For instance, mutations in β-hexosaminidase A (HexA) cause storage of GM2 (N-AcGalβ1,4(NeuAcα2,3)Galβ1,4Glc-ceramide) gangliosides and development of Tay-Sachs disease (13). One of the most prevalent HexA mutations, the G269S substitution, destabilizes native folding of the HexA α subunit, which is rapidly processed by ERAD (14). Similar to what was described for GC variants, if G269S HexA is forced to fold into its native structure, it escapes ERAD and retains catalytic activity (8, 15).
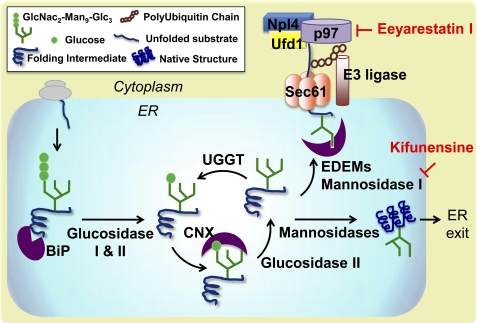
We hypothesized that native folding of mutated enzyme variants is limited by the rapid disposal of unstable folding intermediates via ERAD. Hence, we sought to investigate ERAD inhibition in LSD patient-derived cells and establish the role of this pathway in the development of loss-of-function phenotypes. The ERAD pathway is part of a complex quality control network that ensures correct folding and processing of active proteins and eliminates non-native, off-pathway products (16, 17). A simplified schematic is reported in Fig. 1. As newly synthesized proteins are translocated into the ER, they immediately interact with BiP, which facilitates their folding while preventing aggregation (18). Substrates are marked with oligosaccharide precursors (GlcNAc2-Man9-Glc3) and subsequently trimmed by ER glucosidases to allow recognition by the lectin chaperones (CNX and CRT) (17). Upon removal of the outermost glucose residue (GlcNAc2-Man9), natively folded proteins exit the ER and proceed through the secretory pathway, whereas misfolded intermediates are reglucosylated by UDP-glucose:glycoprotein glucosyltransferase. This cycle repeats itself until substrates either reach native folding or are recognized as irreversibly misfolded by ER degradation-enhancing α-mannosidase-like lectins (19–21). Removal of three to four mannose residues by ER mannosidases, and particularly mannosidase I, marks misfolded substrates for degradation (21, 22), which proceeds through polyubiquitination and retrotranslocation via the p97 complex (23, 24) (Fig. 1).
FIGURE 1.
ERAD pathways and mechanisms of ERAD inhibition. As newly synthesized polypeptides are translocated into the ER, they are immediately recognized by BiP, which promotes substrate folding and solubility. They are then marked with an oligosaccharide precursor (GlcNAc2-Man9-Glc3), which is sequentially trimmed to allow substrate interaction with the lectin chaperones CNX and CRT. Specifically, interaction with the lectin chaperones occurs upon cleavage of the two terminal glucoses by glucosidase I and II and is terminated by removal of the last outermost glucose residue (GlcNAc2-Man9) by glucosidase II. At this point, natively folded proteins exit the ER, whereas partially folded intermediates are reglucosylated by UDP-glucose:glycoprotein glucosyltransferase (UGGT) and re-enter the lectin folding cycle. In order to prevent excessive accumulation of folding intermediates, unstable misfolding-prone substrates are processed by ER mannosidase I, which cleaves three to four mannose residues from the oligosaccharidic group and promotes substrate binding with the ER degradation-enhancing α-mannosidase-like lectins (EDEM). ERAD substrates are then retrotranslocated to the cytoplasm via the Sec61 retrotranslocon and polyubiquitinated. Substrate retrotranslocation is mediated by the p97 complex, which includes ubiquitin fusion degradation 1 (Ufd1) and nuclear protein localization 4 (Npl4). p97 ATPase provides the driving force for substrate extraction and shuffling to the proteasome. As shown in the schematic, kifunensine and eeyarestatin I, small molecules that function as ERAD inhibitors, block different steps of the ERAD pathway. Kifunensine inhibits ER mannosidase I, and eeyarestatin I inhibits p97 ATPase activity.
Although rescue of mutated GC in patient-derived cells treated with proteasome inhibitors has been previously reported (8, 9), the role of the ERAD pathway in the processing of mutated GC variants remains elusive. We identified two small molecules ERAD inhibitors: kifunensine (Kif), which inhibits ER mannosidase I (21, 25) and thus interferes with early substrate recognition, and eeyarestatin I (EerI), which inhibits p97 ATPase activity (23, 24), thereby limiting retrotranslocation of misfolded substrates (Fig. 1). We report herein that treatment with ERAD inhibitors partially restores folding and activity of mutated GC variants in GD patient-derived fibroblasts. Rescue of mutated HexA was also observed in Tay-Sachs disease cells upon treatment with EerI, demonstrating the generality of this approach. Furthermore, we investigated the transcriptional changes that occur in GD fibroblasts in response to these two different mechanisms of ERAD inhibition, with particular attention to the expression of ER chaperones, the GC-encoding gene, and UPR activation. Results from this study show that ERAD regulates the processing of unstable secretory proteins and that ERAD inhibition is a viable strategy to rescue native folding and activity of mutated lysosomal enzymes associated with the development of LSD. EerI-mediated inhibition of substrate retrotranslocation, although possibly more efficient in rescuing mutated GC folding, was observed to cause UPR induction and cytotoxicity. Kif-mediated inhibition of early substrate recognition, however, which is likely to prolong ER retention and substrate folding without causing accumulation of irremediably misfolded proteins, caused minimal activation of the UPR and did not result in induction of apoptosis.
EXPERIMENTAL PROCEDURES
Enzyme Activity Assays
The intact cell GC activity assay and HexA activity assay were performed as described previously (8) and in the supplemental material.
Quantitative RT-PCR
RT-PCR was conducted as described previously (11) and in the supplemental material using the primers listed in supplemental Table S1.
Western Blot Analysis and Immunofluorescence Microscopy
Details are provided in the supplemental material.
Subcellular Fractionation
Subcellular fractionations were conducted as described previously (26). Briefly, cells were incubated with small molecules for 48 h, collected, and centrifuged at 1,000 rpm for 5 min. Cell pellets were resuspended in sucrose buffer (0.25 m sucrose, 10 mm Hepes, and 1 mm EDTA, pH 7.4) on ice. Cells were homogenized with a Potter-Elvehjem Teflon-glass homogenizer and centrifuged at 1,000 × g for 5 min. The supernatant was layered on top of a 40% Percoll solution in sucrose buffer and centrifuged at 25,000 × g for 1 h. Cell homogenates were collected into eight fractions from the top of the centrifuge tube and used for enzymatic assays.
Toxicity Assay
Toxicity assays were conducted as described previously (11) and in the supplemental material.
Statistical Analysis
All data are presented as mean ± S.D., and statistical significance was calculated using a two-tailed Student's t test.
RESULTS
ERAD Inhibition Enhances Mutated GC Folding, Trafficking, and Activity in GD Patient-derived Fibroblasts
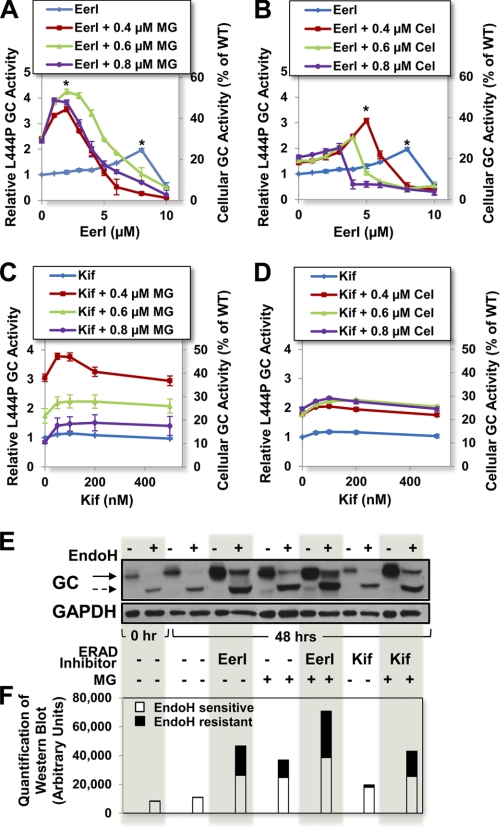
In order to investigate the role of the ERAD pathway on the folding of mutated GC variants, L444P GC fibroblasts were cultured in the presence of ERAD inhibitors (EerI and Kif) for 5 days, and GC activities were evaluated every 24 h with the intact cell GC activity assay (8). Culturing conditions resulting in maximal rescue of L444P GC activity are reported in Fig. 2 (blue lines). L444P GC activity was observed to increase up to 2.0-fold in cells treated with EerI (8 μm final medium concentration, p < 0.001) for 48 h compared with untreated cells, which corresponds to about 25% of the WT cellular activity (Fig. 2A), and is expected to ameliorate GD symptoms (2).
FIGURE 2.
Cell treatment with ERAD inhibitors enhances L444P GC activity in GD patient-derived fibroblasts. Relative L444P GC activities were evaluated in cells treated with a range of concentrations of ERAD inhibitors (EerI or Kif) and constant doses of proteostasis regulators (MG-132 or celastrol: 0.4, 0.6, or 0.8 μm). Shown are relative GC activities of L444P cells treated with EerI and MG-132 for 48 h (A), EerI and celastrol for 48 h (B), Kif and MG-132 for 120 h (C), and Kif and celastrol for 120 h (D). Relative GC activities were evaluated by normalizing GC activities measured in treated cells to the activity of untreated cells (left y axis) (p < 0.01 if not specified; *, p < 0.001). The corresponding fraction of WT GC activity is also reported (right y axis). E, Western blot analyses of Endo H-treated and -untreated total protein content from L444P GC fibroblasts cultured with EerI (6 μm), Kif (50 nm), and MG-132 (0.6 μm) for 48 h and detected using GC-specific antibody. The solid and dashed arrows indicate Endo H-resistant and Endo H-sensitive bands, respectively. F, quantification of GC bands detected by Western blot in Endo H-treated samples. Quantification of lower Mr, Endo H-sensitive bands corresponding to ER-retained GC are reported in the white portion of the bars, and quantification of higher Mr, Endo H-resistant bands corresponding to lysosomal GC are reported in the black top portions. Band analyses and quantifications were conducted using National Institutes of Health ImageJ analysis software. Experiments were repeated three times, and data points are reported as mean ± S.D. (error bars). MG, MG-132; Cel, celastrol.
We hypothesized that EerI-mediated ERAD inhibition prolongs ER retention of mutated GC, thereby enhancing the pool of GC folding intermediates amenable to folding rescue. Hence, we asked whether combining ERAD inhibition with enhancement of the cellular folding capacity could further increase the pool of natively folded GC that traffics to the lysosomes. To investigate this question, EerI-treated cells were cultured in the presence of MG-132 and celastrol, small molecules previously reported to function as proteostasis regulators and rescue GC folding through a mechanism distinct from ERAD modulation (8, 10). Experiments were designed to explore the addition of a constant concentration of proteostasis regulator (0.4, 0.6, or 0.8 μm) to a range of EerI concentrations. Co-administration of EerI (2 μm) and MG-132 (0.6 μm) for 48 h resulted in a dramatic 4.2-fold increase in L444P GC activity (p < 0.001; Fig. 2A) compared with untreated cells, which corresponds to 52.5% of WT GC activity and is significantly higher than the activity of cells treated only with EerI (2.0-fold) or MG-132 (2.4-fold) under the same conditions. Co-treatment with EerI and celastrol was also observed to enhance L444P GC activity rescue (Fig. 2B). Specifically, GD cells treated with EerI (5 μm) and celastrol (0.4 μm) for 48 h displayed a 3.1-fold increase in L444P GC activity (p < 0.001), which is higher than what was observed in cells treated only with EerI (1.3-fold) or celastrol (1.4-fold) under the same conditions.
A similar set of experiments was conducted using Kif, the other ERAD inhibitor selected. Cell treatment with Kif (100 nm) modestly increased L444P GC activity (1.2-fold, 15% of WT activity; p < 0.01), whereas co-administration of Kif (50 nm) and MG-132 (0.4 μm) led to a dramatic 3.8-fold increase in L444P GC activity (47.5% of WT activity; p < 0.01) after 120 h (Fig. 2C). Co-treatment with Kif and celastrol was also explored (Fig. 2D). Optimal culturing conditions (100 nm Kif and 0.8 μm celastrol) resulted in a 2.3-fold increase in L444P GC activity (28.8% of WT activity, p < 0.01), again higher than what was measured upon treatment with either molecule alone. Taken together, these results suggest that ERAD limits the folding and trafficking of L444P GC and that ERAD inhibition is a viable strategy to promote native folding and trafficking of this mutated, degradation-prone enzyme variant. In order to confirm that the increase in GC activity measured in cells treated with EerI and Kif is caused by partial restoration of L444P GC folding and lysosomal trafficking, we investigated the L444P GC glycosylation state and its intracellular localization and trafficking.
The L444P GC glycosylation state was investigated by endoglycosidase H (Endo H) treatment as described previously (11), using culturing conditions that resulted in maximal GC activity rescue (6 μm EerI, 50 nm Kif, 0.6 μm MG-132 for 48 h). The total protein content was subjected to Endo H treatment, which hydrolyzes immature high mannose N-linked glycoproteins. GC detection by Western blot typically reveals a low Mr band corresponding to partially glycosylated, ER-retained GC (Endo H-sensitive) and a high Mr band corresponding to fully glycosylated, lysosomal GC (Endo H-resistant) (27). A representative Western blot (Fig. 2E) and quantification of Endo H-resistant and Endo H-sensitive GC bands (Fig. 2F) are reported. In untreated cells, nearly all L444P GC was detected as Endo H-sensitive, as expected (11). A band corresponding to Endo H-resistant L444P GC was detected in cells treated with EerI, and its intensity was 1.6-fold higher than that detected in cells treated with MG-132 (results obtained using MG-132 were reported previously (8) and are included here for comparison). Kif treatment caused a mild increase in the GC Endo H-resistant pool, corresponding to about 13% of that of MG-132-treated cells. Co-treatment with MG-132 and EerI (2 μm) or Kif resulted in a 2.6- and 1.4-fold increase, respectively, in the Endo H-resistant L444P GC, compared with treatment with MG-132 only, which is in agreement with results obtained from enzymatic assays (Fig. 2, A and C).
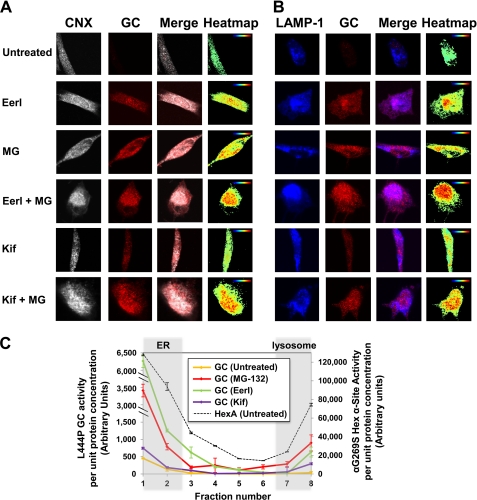
L444P GC intracellular localization was evaluated using immunofluorescence microscopy and subcellular fractionations of L444P GC patient-derived fibroblasts treated with small molecules at concentrations corresponding to maximum activity rescue (6 μm EerI, 50 nm Kif, and 0.6 μm MG-132) for 48 h. Immunofluorescence microscopy was conducted using antibodies specific for GC, for an ER marker (CNX), and for a lysosomal marker (LAMP-1) to evaluate GC localization in the ER and in the lysosome, respectively. Co-localizations of GC and CNX (Fig. 3A) and of GC and LAMP-1 (Fig. 3B) are reported, respectively, in pink and purple (merged colors) and analyzed with ImageJ software to provide a co-localization heat map. L444P GC was barely detectable in untreated cells due to extensive ERAD, as reported previously (28). In agreement with the results obtained from GC enzymatic assays (Fig. 2A), a significantly larger pool of GC was detected in the ER and in the lysosome upon EerI treatment. Furthermore, GC accumulation was observed to increase with increasing concentration of EerI and to be further enhanced by co-treatment with EerI and MG-132 (Fig. 3, A and B). Kif treatment also enhanced GC localization in the ER and in the lysosomes compared with untreated cells, albeit to a lower extent than EerI treatment (Fig. 3, A and B). Co-treatment with Kif and MG-132 increased GC accumulation in the ER and in the lysosomes, again supporting the results obtained from GC enzymatic assays (Fig. 2C).
FIGURE 3.
ERAD inhibitors promote L444P GC folding and trafficking in GD patient-derived fibroblasts. Shown is immunofluorescence microscopy of GC and CNX (an ER marker) (A) and GC and LAMP-1 (a lysosomal marker) (B) in L444P GC fibroblasts. Cells were treated with EerI (2 and 6 μm), Kif (50 nm), and MG-132 (0.6 μm) for 48 h. Colocalization of CNX (gray, column 1) and GC (red, column 2) is shown in pink (column 3). Colocalization of LAMP-1 (blue, column 1) and GC (red, column 2) is shown in purple (column 3). Heat maps of co-localization images were obtained with National Institutes of Health ImageJ analysis software (column 4). Hot colors represent positive correlation (co-localization), whereas cold colors represent negative correlation (exclusion). C, GC activity of subcellular homogenate fractions. Untreated cells and cells cultured with EerI (6 μm), Kif (50 nm), and MG-132 (0.6 μm) for 48 h were fractionated. Subcellular fractions were collected, numbered from low to high density (from 1 to 8), and subjected to GC activity assays. HexA activity was also measured in each fraction obtained from untreated cells to identify fractions containing the ER (fractions 1 and 2) and lysosomes (fractions 7 and 8). The total protein concentration of each fraction was determined by Nanodrop. GC enzyme activity (left y axis) and HexA enzyme activity (right y axis) of each fraction were normalized to the corresponding protein concentrations. Experiments were repeated three times, and data points are reported as mean ± S.D. MG, MG-132.
Subcellular fractions of cell homogenates were collected upon Percoll density gradient centrifugation, and GC enzyme activity assay for each fraction was performed to evaluate L444P GC intracellular localization. Because β-hexosaminidase A (HexA) trafficking and activity are not altered in GD fibroblasts compared with WT fibroblasts, HexA activity was first evaluated in each fraction to distinguish fractions containing ER and lysosomes. HexA activity was detected in both low density (fractions 1 and 2) and high density (fractions 7 and 8) fractions in untreated L444P GC cells (Fig. 3C, dashed line, right y axis), which comprise the ER and the lysosomes, respectively, as reported previously (26). In untreated cells, L444P GC activity (Fig. 3C, left y axis) was barely detectable in low density fractions (ER) and undetectable in high density fractions (lysosomes). Treatment with MG-132 resulted in a significant increase in L444P GC activity in both low and high density fractions, confirming that MG-132 promotes rescue of mutant GC folding and trafficking (Fig. 3C). A significant increase in GC activity was also detected in EerI-treated cells, particularly in low density ER fractions, confirming that EerI functions by inhibiting ERAD and prolonging ER retention. Kif treatment also enhanced GC activity both in the low and high density fractions compared with untreated cells, although to a lower extent than EerI treatment, in agreement with the results obtained from GC enzymatic assays (Fig. 2, A and C).
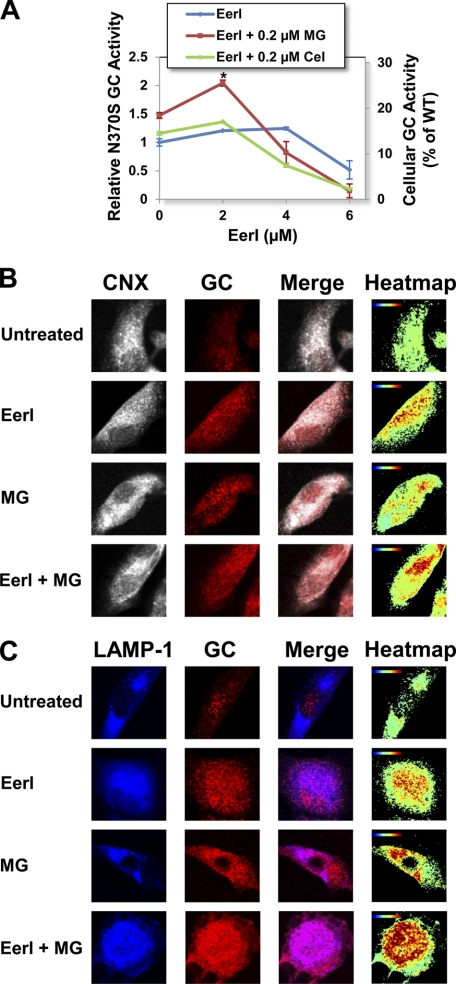
In order to investigate whether ERAD inhibition is a mutation-dependent strategy for the rescue of unstable GC variants, N370S GC fibroblasts were cultured in the presence of EerI and a proteostasis regulator, and GC activities were evaluated every 24 h for up to 3 days (Fig. 4A and supplemental Fig. S1). EerI treatment (4 μm) for 72 h resulted in a 1.25-fold increase in GC activity (15.6% WT activity; p < 0.01). Similar to what was described for L444P GC cells, a lower concentration of EerI (2 μm) combined with MG-132 (0.2 μm) further enhanced N370S GC rescue, resulting in a 2.1-fold increase in activity compared with untreated cells (26.3% of WT activity, p < 0.001), which is higher than what observed in the presence of either molecule alone (EerI, 1.2-fold; MG-132, 1.5-fold). When the same experiment was conducted using celastrol as a proteostasis modulator, co-treatment with EerI (2 μm) and celastrol (0.2 μm) resulted in a 1.4-fold increase in activity (17.5% of WT activity, p < 0.01), which again is significantly higher than what was observed using either molecule alone (EerI, 1.2-fold; celastrol, 1.2-fold).
FIGURE 4.
EerI facilitates N370S GC folding, lysosomal trafficking, and activity in GD patient-derived fibroblasts. A, relative N370S GC activities were measured in cells treated with proteostasis regulators (MG-132 (0.2 μm) and celastrol (0.2 μm)) and a range of EerI concentrations for 72 h. Relative GC activities were evaluated as described in the legend to Fig. 2 (p < 0.01 if not specified; *, p < 0.001). Experiments were repeated three times, and data points are reported as mean ± S.D. (error bars). MG, MG-132; Cel, celastrol. Shown are immunofluorescence microscopy images of GC and CNX (an ER marker) (B) and GC and LAMP-1 (a lysosomal marker) (C) in cells treated with EerI (2 μm) and MG-132 (0.2 μm) for 48 h. Colocalization images were analyzed as described in the legend to Fig. 3.
To confirm that the observed increase in GC activity is due to rescue of the enzyme folding and trafficking, N370S GC cellular localization was evaluated by immunofluorescence microscopy in cells treated with EerI (2 μm) and MG-132 (0.2 μm). EerI treatment resulted in an increase in N370S GC accumulation in the ER and in the lysosomes compared with untreated cells. Co-administration of EerI and MG-132 further enhanced N370S GC concentration both in the ER and in the lysosomes (Fig. 4, B and C), confirming the results obtained from enzymatic assays.
In summary, these results demonstrate that ERAD prevents native folding of mutated, unstable GC and provide compelling evidence that ERAD inhibition is a viable strategy to rescue lysosomal activity of degradation-prone GC variants containing destabilizing, non-inactivating mutations. Interestingly, the activity rescue measured in N370S GC fibroblasts was consistently less pronounced than that observed in L444P GC fibroblasts. We suggest that this difference is due to the different destabilizing effect of the N370S and L444P substitutions. L444P GC is normally completely targeted to ERAD, whereas the N370S GC variant partially escapes degradation and can be detected throughout the secretory pathway (29). Hence, ERAD is likely to have a more direct and rate-limiting role in L444P GC processing, and, not surprisingly, ERAD inhibition results in more efficient rescue of L444P GC than N370S GC folding.
ERAD Inhibition Enhances HexA Activity in Tay-Sachs Patient-derived Fibroblasts
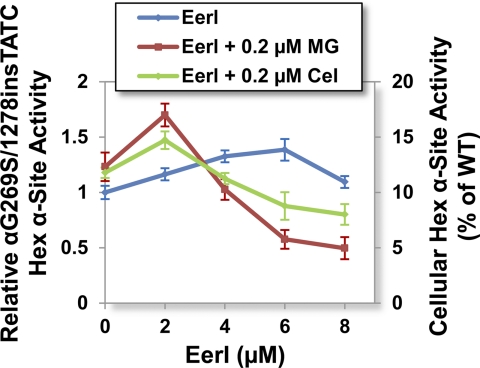
A number of loss-of-function LSD are caused by destabilizing mutations and degradation of secretory proteins. As reported above, we demonstrated that ERAD inhibition enhances folding of mutated GC variants. We then asked whether ERAD inhibition is a general strategy to rescue activity of mutated proteins containing misfolding, non-inactivating mutations associated with the development of LSD. Thus, we investigated the folding of HexA, deficiency of which causes Tay-Sachs disease. Specifically, we focused on one of the most prevalent mutations, the G269S substitution in the HexA α subunit, which destabilizes the protein native structure causing loss of activity to ∼10% of WT (15). Patient-derived fibroblasts harboring G269S HexA were cultured in the presence of an ERAD inhibitor and a proteostasis modulator, and HexA activity was measured as previously described (8). Administration of EerI (6 μm) for 96 h led to a 1.4-fold increase in G269S HexA α activity (14% of WT HexA activity; p < 0.01; Fig. 5). The addition of MG-132 (0.2 μm) to cells treated with EerI (2 μm) for 96 h caused a further increase in HexA activity (1.7-fold, ∼17% of WT; p < 0.01; Fig. 5).
FIGURE 5.
EerI enhances G269S HexA activities in Tay-Sachs patient-derived fibroblasts. Cells were cultured with proteostasis regulators (MG-132 (0.2 μm) and celastrol (0.2 μm)) and a range of EerI concentrations for 96 h. Relative αG269S/1278insTATC HexA activities (p < 0.01) were evaluated by normalizing HexA activity of treated cells to that of untreated cells (left y axis). The corresponding fraction of WT HexA activity is also reported (right y axis). Experiments were repeated three times, and data points are reported as mean ± S.D. (error bars). MG, MG-132; Cel, celastrol.
These findings suggest that ERAD inhibition facilitates folding of destabilized enzyme variants prone to degradation. In summary, results from studies in Gaucher and Tay-Sachs disease patient-derived cells indicate that the activity rescue observed upon treatment with ERAD inhibitors is inversely proportional to the loss of lysosomal activity normally associated with each enzyme variant.
ERAD Inhibition via EerI Treatment Causes Up-regulation of BiP Expression
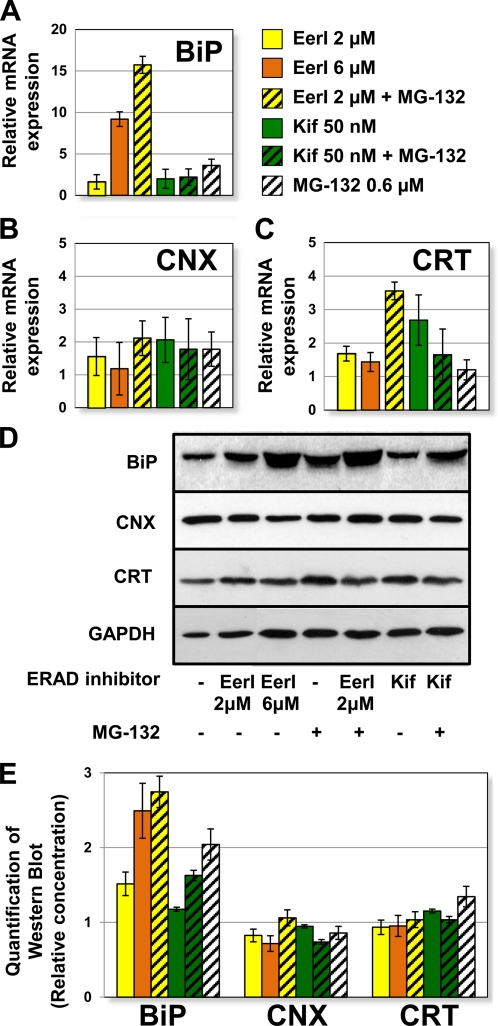
We speculated that small molecule-mediated ERAD inhibition, by inducing accumulation of misfolded proteins in the ER, could lead to up-regulation of ER chaperones. We previously reported that the ER luminal chaperone BiP plays a critical role in the rescue of L444P GC folding (10, 11). We asked whether the increase in mutated GC activity observed upon ERAD inhibition could be attributed to up-regulation of BiP or other ER chaperones induced in response to the sudden load of misfolded proteins in the ER. Quantitative RT-PCR experiments were conducted to evaluate the expression levels of representative ER chaperones (BiP, CNX, and CRT) in L444P GC fibroblasts treated with EerI (2 and 6 μm), Kif (50 nm), MG-132 (0.6 μm), or a combination thereof (Fig. 6, A–C).
FIGURE 6.
Up-regulation of BiP expression in L444P GC fibroblasts treated with ERAD inhibitors. Relative mRNA expression levels of BiP (p < 0.01) (A), CNX (p < 0.05) (B), and CRT (p < 0.05) (C) in L444P GC fibroblasts treated with EerI (2 and 6 μm), Kif (50 nm), and MG-132 (0.6 μm) for 24 h were obtained by quantitative RT-PCR, corrected by the expression of the housekeeping gene GAPDH, and normalized to those of untreated cells. The data are reported as mean ± S.D. (error bars) D, Western blot analyses of BiP, CNX, CRT, and GAPDH (used as loading control) in cells treated with EerI (2 and 6 μm), Kif (50 nm), and MG-132 (0.6 μm) for 48 h. E, quantification of Western blot bands. ER chaperone band intensities were quantified with National Institutes of Health ImageJ analysis software, corrected by GAPDH band intensities, and divided by the values obtained in untreated samples.
BiP expression (Fig. 6A) was mildly up-regulated by EerI treatment at 2 μm (1.6-fold, p < 0.01) but highly up-regulated at 6 μm (9.2-fold) and upon co-treatment with EerI (2 μm) and MG-132 (15.7-fold). Hence, the highest up-regulation of BiP expression was observed at conditions causing maximal rescue of GC activity, namely upon cell treatment with a high concentration of EerI (6 μm) or co-treatment with MG-132 and a low concentration of EerI (2 μm), suggesting a correlation between BiP transcriptional regulation and GC activity rescue. The dramatic increase in BiP expression observed could be part of UPR induction, which was previously shown to facilitate GC folding rescue (10, 11) and is analyzed below in more detail.
Interestingly, cell treatment with Kif under conditions observed to maximize L444P GC activity rescue only resulted in a moderate increase of BiP expression (2.0-fold, p < 0.01), even when used in combination with MG-132 (2.2-fold, p < 0.01). Hence, under the conditions tested, Kif-mediated ERAD inhibition does not lead to ER stress and chaperone up-regulation. We suggest that the lower increase in BiP expression observed upon treatment with Kif compared with EerI is not an indication of the lower rescue of L444P GC activity but rather of the two molecules' different mechanisms of action. In support of this hypothesis is evidence that even the addition of MG-132, which dramatically increases Kif-mediated L444P GC rescue (Fig. 2C), does not cause up-regulation of BiP expression.
CNX was mildly up-regulated by treatment with EerI or Kif, alone or in combination with MG-132 (Fig. 6B). CRT expression was not substantially altered by EerI treatment, although it was up-regulated (3.5-fold) by co-treatment with MG-132 and EerI (Fig. 6C).
Western blot analyses (Fig. 6D) were conducted to confirm ER chaperone expression, and bands were quantified with ImageJ software (Fig. 6E). BiP protein accumulation was enhanced by treatment with EerI in a concentration-dependent fashion (2 μm EerI caused a 1.5-fold increase, and 6 μm EerI resulted in 2.5-fold increase). Co-treatment with EerI and MG-132 further enhanced BiP accumulation (2.7-fold) compared with untreated cells. Kif treatment, however, caused a very modest increase in BiP accumulation when used alone (1.2-fold) or in combination with MG-132 (1.6-fold). CNX and CRT protein levels did not seem to be considerably altered upon small molecule treatment. Overall, results from Western blot analyses are consistent with RT-PCR experiments, with the exception of CRT expression, for which transcriptional changes are not reflected at the translational level, suggesting that CRT up-regulation by cell treatment with MG-132 and EerI does not translate into enhanced accumulation of CRT protein.
Induction of UPR Depends on the Mechanism of ERAD Inhibition
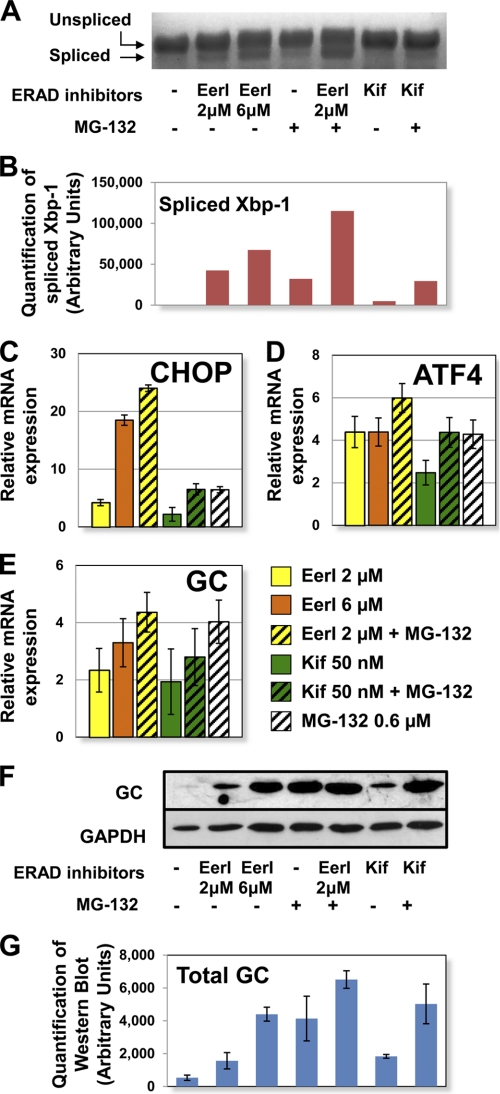
Accumulation of misfolded proteins triggers ER stress, which in turn leads to UPR induction. UPR manifests as a series of attempts to restore a physiologic balance between folded and misfolded proteins in the ER (30, 31). Specifically, UPR induction is regulated by three proximal membrane signal transducers, namely inositol-requiring kinase 1 (IRE1), activating transcription factor 6 (ATF6), and double-stranded RNA-activated ER kinase (PERK). Their activation results in transcriptional modulation of a series of downstream genes involved in enhancement of chaperone capacity, reduction of protein synthesis, and, eventually, induction of apoptosis (30, 31). In order to evaluate UPR induction in cells treated with ERAD inhibitors, we measured the expression of three representative target proteins: X-box-binding protein-1 (Xbp-1) which is activated by IRE1; activating transcription factor 4 (ATF4), which is part of the PERK signaling cascade; and C/EBP homologous protein (CHOP), which is up-regulated in response to ATF6 activation (30). Quantitative RT-PCR was conducted to evaluate the expression of Xbp-1, ATF4, and CHOP in cells treated with EerI (2 and 6 μm), Kif (50 nm), MG-132 (0.6 μm), or a combination thereof.
Activation of the IRE1 signaling cascade involves splicing of Xbp-1 mRNA. Spliced Xbp-1 mediates induction of UPR genes, whereas the unspliced Xbp-1 precursor functions as a repressor (31). To evaluate activation of the IRE1 arm of the UPR, spliced and unspliced Xbp-1 mRNA were quantified by RT-PCR followed by gel electrophoresis (Fig. 7, A and B). MG-132 was previously shown to enhance Xbp-1 splicing (8) and is reported here for comparison. Treatment with EerI resulted in Xbp-1 splicing in a concentration-dependent fashion (2 μm EerI, 1.3-fold increase in splicing; 6 μm EerI, 2.1-fold increase). Co-administration of EerI (2 μm) and MG-132 further increased Xbp-1 splicing (3.6-fold compared with MG-132 treatment). Kif treatment did not seem to induce splicing of Xbp-1, and co-treatment with Kif and MG-132 resulted in an increase in spliced Xbp-1 similar to that induced by treatment only with MG-132. In summary, Xbp-1 splicing was induced upon EerI but not by Kif treatment, suggesting that activation of the IRE1 pathway depends on the specific mechanism of ERAD inhibition.
FIGURE 7.
UPR activation in L444P GC fibroblasts treated with ERAD inhibitors. Cells were treated with EerI (2 and 6 μm), Kif (50 nm), and MG-132 (0.6 μm) for 24 h. A, Xbp-1 mRNA splicing was determined by RT-PCR followed by gel electrophoresis. B, spliced Xbp-1 band intensities were quantified with National Institutes of Health ImageJ analysis software. Relative mRNA expression levels of CHOP (p < 0.01) (C), ATF4 (p < 0.05) (D), and GC (p < 0.05) (E) were obtained by quantitative RT-PCR and calculated as described in the legend to Fig. 6. The data are reported as mean ± S.D. (error bars). F, Western blot analysis of cells treated with EerI (2 and 6 μm), Kif (50 nm), and MG-132 (0.6 μm) for 48 h using GC specific antibody. GAPDH expression was used as a loading control. G, Western blot band quantification. GC bands were quantified by National Institutes of Health ImageJ analysis software and corrected by GAPDH band intensities.
ATF4 expression was up-regulated 4.4-fold by treatment with EerI and 6.0-fold by co-treatment with EerI and MG-132 (p < 0.05), a clear indication of the PERK arm's activation in cells treated with EerI (Fig. 7D). A considerably lower increase in ATF4 expression was observed in cells treated with Kif (2.5-fold, p < 0.01). Moreover, co-treatment with Kif and MG-132 did not result in significant up-regulation of ATF4 expression compared with treatment only with MG-132, again suggesting that UPR activation depends on the mechanism of ERAD inhibition.
CHOP was found to be highly up-regulated upon EerI treatment (Fig. 7C). Specifically, 2 μm EerI resulted in a 4.2-fold increase in CHOP expression, 6 μm EerI resulted in an 18.5-fold increase, and co-administration of EerI (2 μm) and MG-132 resulted in a 24.0-fold increase (p < 0.01), indicating that EerI-mediated ERAD inhibition causes activation of the ATF6 pathway. Because CHOP plays a role in the induction of apoptotic pathways (32), these results also suggest that EerI treatment might activate UPR-induced apoptosis, which is analyzed below. Treatment with Kif alone or in combination with MG-132 led to 2.2- and 6.5-fold CHOP up-regulation, respectively, which are significantly lower than those observed upon EerI treatment and do not exceed those observed upon treatment with MG-132 (Fig. 7C). In summary, we demonstrated that EerI but not Kif, when used under conditions that promote rescue of L444P GC activity, is associated with dramatic activation of the UPR.
Up-regulation of the GC-encoding gene (GBA) as well as of other genes encoding for lysosomal proteins associated with the development of LSD was previously reported in cells treated with UPR-inducing proteostasis regulators (10). This was attributed to the general up-regulation of lipid metabolism that occurs during UPR in order to expand the size of the ER and dilute the load of misfolded proteins (30). We asked whether ERAD inhibition increases GC accumulation in the ER by up-regulating its transcription in addition to preventing its degradation. Quantitative RT-PCR was conducted to measure GC expression in L444P GC fibroblasts treated with EerI (2 and 6 μm), Kif (50 nm), and MG-132 (0.6 μm) (Fig. 7, E–G). EerI treatment was observed to enhance GC expression in a concentration-dependent fashion (2 μm EerI, 2.3-fold; 6 μm EerI, 3.3-fold; p < 0.01). Co-treatment with EerI (2 μm) and MG-132 (0.6 μm) resulted in a 4.4-fold increase in GC expression (p < 0.05), which is higher than what was observed using EerI alone (2.3-fold) but comparable with treatment with MG-132 only (4.0-fold). A lower increase in GC expression was measured in cells treated with Kif (1.9-fold), as expected, considering the modest UPR induction caused by Kif treatment. Similar to what was observed for EerI, the increase in GC expression observed upon co-treatment with MG-132 (2.8-fold) was lower than that detected in cells treated only with MG-132 (4.0-fold).
GC expression was also investigated by Western blot analyses (Fig. 7, F and G). It is important to notice that changes in protein accumulation detected by Western blot are attributable to both GC transcriptional modulation caused by ERAD inhibition-induced UPR and GC post-translational processing caused by ERAD inhibition-mediated protein rescue. L444P GC content was barely detectable in untreated cells, as expected, due to extensive ERAD (29), whereas treatment with either ERAD inhibitor enhanced GC accumulation level. GC accumulation increased in EerI-treated cells in a concentration-dependent fashion (cf. bands corresponding to 2 μm EerI and 6 μm EerI treatments). The addition of MG-132 further enhanced GC accumulation observed in EerI- and Kif-treated cells.
In summary, ERAD inhibition resulted in both an increase in GC expression and cellular accumulation. GC up-regulation was found to be proportional to the extent of UPR induction measured upon treatment with each specific ERAD inhibitor. However, co-treatment with an ERAD inhibitor and a proteostasis modulator, MG-132, which was demonstrated to have a synergistic effect on the rescue of mutated GC activity (Fig. 2, A and C), did not cause a corresponding synergistic increase in GC transcription, suggesting that rescue of mutated GC cannot be solely attributed to the effect of ERAD inhibitors on GC expression.
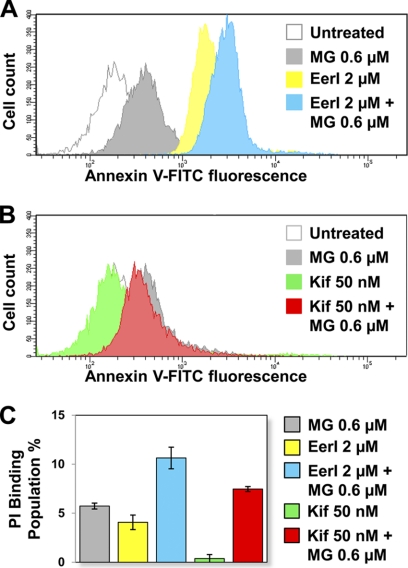
If ER stress persists, prolonged UPR activation typically leads to induction of apoptosis (30). We asked whether cell treatment with Eer and Kif influenced UPR-induced apoptosis. The CytoGLOTM annexin V-FITC apoptosis detection kit was used to detect membrane rearrangement (annexin V binding, a measurement of early apoptosis) and fragmentation (propidium iodide binding, a measurement of late apoptosis) in L444P GC fibroblasts treated with EerI (2 μm), Kif (50 nm), and MG-132 (0.6 μm) (Fig. 8, A–C). High annexin V binding and consequently dramatic increase in cell fluorescence were observed upon cell treatment with EerI compared with untreated cells (Fig. 8A). The addition of MG-132 to EerI-treated cells resulted in an even higher increase in annexin V binding. However, we did not detect any increase in annexin V binding in cells treated with Kif compared with untreated cells. Moreover, the addition of Kif and MG-132 resulted in annexin V binding indistinguishable from that observed in cells treated only with MG-132 (Fig. 8B). Measurements of propidium iodide binding, which is taken as an estimate of the dead cell population, showed similar results. A 4.1% increase in dead cells was observed upon EerI treatment compared with untreated cells, whereas a negligible increase (0.4%) was observed upon Kif treatment (Fig. 8C). These results demonstrate that although EerI treatment causes a dramatic increase in mutated GC activity at the cost of significant cell toxicity and apoptosis induction, treatment with Kif facilitates folding without induction of apoptosis.
FIGURE 8.
Apoptosis induction in L444P GC patient-derived fibroblasts treated with ERAD inhibitors. Flow cytometry histograms of annexin V-FITC fluorescence intensities (x axis, log scale) plotted against cell counts (y axis, linear scale) obtained from the analysis of untreated cells and cells treated with MG-132 (0.6 μm), EerI (2 μm), and EerI (2 μm) and MG-132 (0.6 μm) (A) or untreated cells and cells treated with MG-132 (0.6 μm), Kif (50 nm), and Kif (50 nm) and MG-132 (0.6 μm) (B) for 16 h. Three independent experiments were conducted, and results of one representative experiment are reported. C, propidium iodide (PI) binding population change (%) of cells treated with EerI (2 and 6 μm), Kif (50 nm), and MG-132 (0.6 μm) for 16 h compared with untreated cells (p < 0.01). The number of total counted cells was 10,000. The data are reported as mean ± S.D. (error bars).
DISCUSSION
The main therapeutic option for GD is currently enzyme replacement therapy (33). Although its safety and effectiveness has been demonstrated for several other LSD, including Fabry and Pompe disease, enzyme replacement therapy fails to provide economically sustainable treatment and efficiently address several aspects of the disease. Specifically, enzyme replacement therapy is limited to the treatment of non-neuronopathic symptoms due to inability of the intravenously injected recombinant enzyme to cross the blood-brain barrier (34, 35).
A number of highly prevalent alleles associated with LSD development contain non-inactivating, destabilizing mutations. Such protein variants retain function if forced to fold into their native structure. Hence, efforts have been recently devoted to the development of strategies to rescue folding, trafficking, and activity of unstable substrates, with particular attention to those associated with the manifestation of neuronopathic symptoms (8, 10, 11). We showed herein that inhibition of ERAD enhances folding, trafficking, and lysosomal activity of mutated enzyme variants that cause two clinically distinct LSD, Gaucher and Tay-Sachs disease. Results from this study suggest that ERAD limits the folding of secretory proteins containing misfolding, destabilizing mutations and provide proof of principle of ERAD inhibition as a viable strategy to rescue loss-of-function phenotypes in fibroblasts derived from patients with LSD.
ERAD inhibition was shown to promote folding of the two most common GC variants: L444P GC (Figs. 2 and 3), which is typically completely targeted to ERAD (7), and N370S GC (Fig. 4), a presumably less destabilized variant that is moderately resistant to ERAD and retains partial residual activity (4). ERAD inhibition was also observed to rescue folding of Tay-Sachs disease G269S HexA (Fig. 5), which, similar to N370S GC, retains partial activity (15). Interestingly, we consistently observed higher activity rescue in L444P GC cells, which normally display complete loss of activity, compared with N370S GC and G269S HexA cells. This suggests that the rescue in protein folding caused by treatment with ERAD inhibitors inversely correlates with the stability and residual activity of the mutated substrate.
Experimental evidence reported previously (6, 11) and in this study suggests that the single-site mutations considered in this study cause destabilization of the protein native, lowest free energy conformation. In order to quantify the stability of mutated variants, we conducted in silico analysis of folding free energy changes (ΔΔG) between WT and mutant proteins using PoPMuSiC software (36, 37). ΔΔG values of 0.92 and 4.17 kcal/mol were obtained for N370S and L444P GC, respectively, indicating that both mutations have a destabilizing effect on native folding, the L444P substitution causing significantly higher loss of stability. A similar analysis conducted for the HexA protein revealed the ΔΔG caused by the G269S substitution to be 0.49 kcal/mol, which is comparable with the ΔΔG of N370S GC and considerably lower than that of L444P GC. These values support the experimental results reported herein, suggesting a correlation between protein stability and degradation.
ERAD inhibition and proteostasis modulation resulted in synergistic rescue of lysosomal activity in patient-derived cells (Figs. 2, 4A, and 5), indicating that a larger pool of unstable proteins that escapes ERAD and can engage the ER folding pathway is rescued by combining ERAD inhibition with up-regulation of the cellular folding capacity. Interestingly, the EerI activity window was consistently shifted toward lower medium concentrations when EerI was combined with a proteostasis regulator, implying that ER retention needs to be carefully modulated to meet the capacity of the cellular folding machinery. It remains to be determined whether EerI treatment results in higher activity rescue than Kif treatment due to higher efficiency of the molecular mechanism involved (p97 versus ER mannosidase inhibition).
Detailed investigations of the molecular mechanism of ERAD inhibition and consequent changes in the cellular folding capacity were conducted in L444P GC fibroblasts treated to block two different steps of the ERAD pathway and prevent early recognition of misfolding intermediates (Kif) or retrotranslocation of irremediably misfolded substrates (EerI) (Fig. 1). ER stress and UPR normally observed upon accumulation of misfolded proteins were investigated (Figs. 6 and 7) and seemed to be highly dependent on the specific mechanism of ERAD inhibition. We speculate that by inhibiting retrotranslocation of irremediably misfolded proteins, EerI inevitably leads to significant accumulation of misfolded proteins and consequent induction of UPR and apoptosis. Kif, however, by preventing targeting of folding intermediates to the ERAD pathway, is expected to enhance retention of substrates that can still be assisted by the ER chaperone pathway and reach native folding. As a result, Kif-mediated ERAD inhibition, despite promoting significant ER retention and folding of mutated GC, particularly when used in combination with a proteostasis modulator (Figs. 2 and 3), does not cause ER stress, as demonstrated by investigating changes in ER chaperone expression (Fig. 6); nor does it cause activation of UPR (Fig. 7) and apoptosis (Fig. 8).
Up-regulation of the GC-encoding gene, which was previously suggested to contribute to the rescue of GC activity mediated by UPR inducing proteostasis regulators (10, 11), was also observed in this study in association with UPR activation (Fig. 7). Kif treatment, for instance, which did not cause significant UPR, was not associated with considerable increase in GC expression. Interestingly, co-treatment with Kif and MG-132, despite causing a dramatic increase in GC activity, did not result in UPR induction and GC up-regulation. These results, taken together, suggest that the rescue of GC folding observed in cells treated with ERAD inhibitors, alone or in combination with proteostasis regulators, cannot be solely attributed to UPR activation.
Finally, ERAD inhibitors led to dramatically different levels of apoptosis induction (Fig. 8). Specifically, Kif treatment did not cause cytotoxicity and did not increase MG-132 associated induction of apoptosis. Hence, we suggest that detailed investigations be conducted to identify the steps of the ERAD pathway that can be modulated for the rescue of degradation-prone substrates without dramatically compromising protein homeostasis and disrupting the functioning of the folding quality control system.
Supplementary Material

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Fig. S1.
- LSD
- lysosomal storage disorders
- BiP
- binding immunoglobulin protein
- CHOP
- C/EBP homologous protein
- CNX
- calnexin
- CRT
- calreticulin
- EerI
- eeyarestatin I
- Endo H
- endoglycosidase H
- ERAD
- ER-associated degradation
- GC
- glucocerebrosidase
- GD
- Gaucher disease
- HexA
- β-hexosaminidase A
- Kif
- kifunensine
- PERK
- double-stranded RNA-activated ER kinase
- UPR
- unfolded protein response
- ER
- endoplasmic reticulum.
REFERENCES
- 1. Futerman A. H., van Meer G. (2004) Nat. Rev. Mol. Cell Biol. 5, 554–565 [DOI] [PubMed] [Google Scholar]
- 2. Schueler U. H., Kolter T., Kaneski C. R., Zirzow G. C., Sandhoff K., Brady R. O. (2004) J. Inherit. Metab. Dis. 27, 649–658 [DOI] [PubMed] [Google Scholar]
- 3. Hruska K. S., LaMarca M. E., Scott C. R., Sidransky E. (2008) Hum. Mutat. 29, 567–583 [DOI] [PubMed] [Google Scholar]
- 4. Grace M. E., Newman K. M., Scheinker V., Berg-Fussman A., Grabowski G. A. (1994) J. Biol. Chem. 269, 2283–2291 [PubMed] [Google Scholar]
- 5. Schmitz M., Alfalah M., Aerts J. M., Naim H. Y., Zimmer K. P. (2005) Int. J. Biochem. Cell Biol. 37, 2310–2320 [DOI] [PubMed] [Google Scholar]
- 6. Ron I., Horowitz M. (2005) Hum. Mol. Genet. 14, 2387–2398 [DOI] [PubMed] [Google Scholar]
- 7. Grabowski G. A. (1997) Genet. Test. 1, 5–12 [DOI] [PubMed] [Google Scholar]
- 8. Mu T. W., Ong D. S., Wang Y. J., Balch W. E., Yates J. R., 3rd, Segatori L., Kelly J. W. (2008) Cell 134, 769–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lu J., Chiang J., Iyer R. R., Thompson E., Kaneski C. R., Xu D. S., Yang C., Chen M., Hodes R. J., Lonser R. R., Brady R. O., Zhuang Z. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 21665–21670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang F., Agnello G., Sotolongo N., Segatori L. (2011) ACS Chem. Biol. 6, 158–168 [DOI] [PubMed] [Google Scholar]
- 11. Wang F., Chou A., Segatori L. (2011) Chem. Biol. 18, 766–776 [DOI] [PubMed] [Google Scholar]
- 12. Muresan Z., Arvan P. (1998) Mol. Endocrinol. 12, 458–467 [DOI] [PubMed] [Google Scholar]
- 13. Jeyakumar M., Butters T. D., Dwek R. A., Platt F. M. (2002) Neuropathol. Appl. Neurobiol. 28, 343–357 [DOI] [PubMed] [Google Scholar]
- 14. Maegawa G. H., Tropak M., Buttner J., Stockley T., Kok F., Clarke J. T., Mahuran D. J. (2007) J. Biol. Chem. 282, 9150–9161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tropak M. B., Reid S. P., Guiral M., Withers S. G., Mahuran D. (2004) J. Biol. Chem. 279, 13478–13487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. McCracken A. A., Brodsky J. L. (1996) J. Cell Biol. 132, 291–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Vembar S. S., Brodsky J. L. (2008) Nat. Rev. Mol. Cell Biol. 9, 944–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nishikawa S. I., Fewell S. W., Kato Y., Brodsky J. L., Endo T. (2001) J. Cell Biol. 153, 1061–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hosokawa N., Tremblay L. O., You Z., Herscovics A., Wada I., Nagata K. (2003) J. Biol. Chem. 278, 26287–26294 [DOI] [PubMed] [Google Scholar]
- 20. Lederkremer G. Z., Glickman M. H. (2005) Trends Biochem. Sci. 30, 297–303 [DOI] [PubMed] [Google Scholar]
- 21. Fagioli C., Sitia R. (2001) J. Biol. Chem. 276, 12885–12892 [DOI] [PubMed] [Google Scholar]
- 22. Nakatsukasa K., Brodsky J. L. (2008) Traffic 9, 861–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fiebiger E., Hirsch C., Vyas J. M., Gordon E., Ploegh H. L., Tortorella D. (2004) Mol. Biol. Cell 15, 1635–1646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang Q., Shinkre B. A., Lee J. G., Weniger M. A., Liu Y., Chen W., Wiestner A., Trenkle W. C., Ye Y. (2010) PLoS ONE 5, e15479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Avezov E., Frenkel Z., Ehrlich M., Herscovics A., Lederkremer G. Z. (2008) Mol. Biol. Cell 19, 216–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ishii S., Chang H. H., Kawasaki K., Yasuda K., Wu H. L., Garman S. C., Fan J. Q. (2007) Biochem. J. 406, 285–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Maley F., Trimble R. B., Tarentino A. L., Plummer T. H., Jr. (1989) Anal. Biochem. 180, 195–204 [DOI] [PubMed] [Google Scholar]
- 28. Michelakakis H., Dimitriou E., Van Weely S., Boot R. G., Mavridou I., Verhoek M., Aerts J. M. (1995) J. Inherit. Metab. Dis. 18, 609–615 [DOI] [PubMed] [Google Scholar]
- 29. Sawkar A. R., Schmitz M., Zimmer K. P., Reczek D., Edmunds T., Balch W. E., Kelly J. W. (2006) ACS Chem. Biol. 1, 235–251 [DOI] [PubMed] [Google Scholar]
- 30. Schröder M., Kaufman R. J. (2005) Annu. Rev. Biochem. 74, 739–789 [DOI] [PubMed] [Google Scholar]
- 31. Ron D., Walter P. (2007) Nat. Rev. Mol. Cell Biol. 8, 519–529 [DOI] [PubMed] [Google Scholar]
- 32. Oyadomari S., Mori M. (2004) Cell Death Differ. 11, 381–389 [DOI] [PubMed] [Google Scholar]
- 33. Lim-Melia E. R., Kronn D. F. (2009) Pediatr. Ann. 38, 448–455 [DOI] [PubMed] [Google Scholar]
- 34. Rohrbach M., Clarke J. T. (2007) Drugs 67, 2697–2716 [DOI] [PubMed] [Google Scholar]
- 35. Sidransky E., LaMarca M. E., Ginns E. I. (2007) Mol. Genet. Metab. 90, 122–125 [DOI] [PubMed] [Google Scholar]
- 36. Gilis D., Rooman M. (2000) Protein Eng. 13, 849–856 [DOI] [PubMed] [Google Scholar]
- 37. Kwasigroch J. M., Gilis D., Dehouck Y., Rooman M. (2002) Bioinformatics 18, 1701–1702 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.