Background: HopU1 ADP-ribosylates GRP7, suppressing plant immunity.
Results: The HopU1 structure has two novel loops required for GRP7 recognition, and HopU1 ribosylates GRP7 at an arginine in position 49 disrupting its function.
Conclusion: HopU1 targets a conserved arginine in GRP7, disabling its ability to bind immunity-related RNA.
Significance: The mechanistic details of how HopU1 recognizes its substrate reveal how HopU1 contributes to pathogenesis.
Keywords: Bacterial Pathogenesis, Host-Pathogen Interactions, Microbial Pathogenesis, Plant Defense, Plant Pathology, Protein Structure, RNA-binding Protein, Innate Immunity, Type III Effector
Abstract
The Pseudomonas syringae type III effector HopU1 is a mono-ADP-ribosyltransferase that is injected into plant cells by the type III protein secretion system. Inside the plant cell it suppresses immunity by modifying RNA-binding proteins including the glycine-rich RNA-binding protein GRP7. The crystal structure of HopU1 at 2.7-Å resolution reveals two unique protruding loops, L1 and L4, not found in other mono-ADP-ribosyltransferases. Site-directed mutagenesis demonstrates that these loops are essential for substrate recognition and enzymatic activity. HopU1 ADP-ribosylates the conserved arginine 49 of GRP7, and this reduces the ability of GRP7 to bind RNA in vitro. In vivo, expression of GRP7 with Arg-49 replaced with lysine does not complement the reduced immune responses of the Arabidopsis thaliana grp7-1 mutant demonstrating the importance of this residue for GRP7 function. These data provide mechanistic details how HopU1 recognizes this novel type of substrate and highlights the role of GRP7 in plant immunity.
Introduction
Pseudomonas syringae is a hemibiotrophic bacterial plant pathogen that injects a suite of type III effector proteins into host cells (1, 2). The majority of these type III effectors suppress the innate immune system of the plant (3, 4). Plants perceive pathogen- or microbe-associated molecular patterns (PAMPs4/MAMPs) through specific surface-localized transmembrane receptors and induce a response known as PAMP-triggered immunity (PTI). Plants can also perceive pathogen effectors intracellularly using nucleotide-binding site leucine-rich repeat immune receptors. These receptors either directly or indirectly recognize effectors and induce a response referred to as effector-triggered immunity (ETI) (5, 6). The downstream signaling networks of PTI and ETI have much in common although the ETI responses are more prolonged and robust than those of PTI and generally include the hypersensitive response (7), a form of programmed cell death. Common outputs of PTI and ETI include production of reactive oxygen species (ROS), transcription of pathogenesis-related genes, and deposition of lignin and callose in the cell wall.
P. syringae pv. tomato DC3000 contains ∼35 type III effectors (8). Although the majority of its type III effector inventory can suppress immune responses (9), only a few targets of P. syringae type III effectors have been identified. The diverse nature of type III effectors is illustrative of their multiple strategies to suppress host immunity. Thus far, their targets include immune receptor complexes (10–14), downstream MAPK cascades (15, 16), vesicle trafficking (17), and RNA metabolism (18). Other P. syringae effectors have been demonstrated to localize to the chloroplast and mitochondria to exert their effects (19, 20). One type III effector, HopU1, was identified as a mono-ADP-ribosyltransferase (mADP-RT) (18). This was the first report of an mADP-RT in a plant pathogen, and they have not yet been shown to exist in plants (21).
mADP-RTs are well described toxins of bacterial animal pathogens (22–24). They catalyze the hydrolysis of NAD+ and transfer an ADP-ribose moiety onto Arg, Cys, Asn, or diphthamide amino acid residues (25). Through this modification, the activity of proteins such as heterotrimeric GTP-binding proteins, actin, and elongation factor 2 are altered, resulting in the modulation of a wide array of processes including protein synthesis, actin polymerization, and electrolyte secretion (26). mADP-RTs are also present in mammals where the majority of them are extracellular membrane-associated mADP-RTs (ecto-ADP-RTs) that have regulatory functions, which can be reversed by ADP-ribose hydrolases (26). There are two broad groups of mADP-RTs: cholera toxin (CT) and diphtheria toxin (DT) groups. The DT group also includes poly(ADP-ribosyltransferases) known as poly(ADP-ribose)polymerases, which attach more than one ADP-ribose moiety onto an amino acid residue and are well distributed in eukaryotes including plants (26). HopU1 shares sequence similarity to mADP-RTs in animal pathogens and eukaryotes belonging to the CT group. Several structures of this large family of proteins have been determined (27–29).
A first hint at a potential mechanism for HopU1 virulence activity was that it targeted specific RNA recognition motif (RRM) RNA-binding proteins including the glycine-rich RNA-binding protein GRP7 (18). An Arabidopsis thaliana mutant lacking GRP7 was more susceptible to P. syringae and produced reduced amounts of callose in response to the flg22 PAMP indicating that GRP7 was a component of the plant's innate immune system (18). Subsequently GRP7 was implicated in other stress responses (30, 31) and flower development (32). Uncovering how HopU1 disables GRP7 would provide insights into how GRP7 functions in plant immunity.
Here, we present the structure of HopU1 at 2.7-Å resolution. It reveals two unique protruding loops, L1 and L4. These loops are not found in other mADP-RTs and are involved in recognition of GRP7. Through site-directed mutagenesis we identify residues critical for GRP7 binding and enzymatic activity. In addition, we found that HopU1 ADP-ribosylates Arg-49 of GRP7, which is a residue in the conserved ribonucleoprotein consensus sequence 1 motif within its RRM. A GRP7 derivative with Lys substituting for Arg at position 49 was unable to complement the susceptibility and immunity phenotypes exhibited by the Arabidopsis grp7-1 mutant, demonstrating the functional importance of the residue targeted by HopU1. Importantly, we show that overexpression of GRP7 enhances the plant immune system and resistance to P. syringae. These data uncover the molecular basis of the interaction between the HopU1 mADP-RT and its novel substrate GRP7; furthermore, they highlight the role of this RNA-binding protein in plant immunity.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Transgenic Arabidopsis Plants
Escherichia coli DH5α and BL21(DE3) were used for cloning and expression of recombinant proteins following standard protocols (33) and manufacturer's instructions. P. syringae pv. tomato strains were grown in King's broth at 30 °C (34). Antibiotics used include 100 μg/ml ampicillin, 20 μg/ml chloramphenicol, 10 μg/ml gentamicin, 50 μg/ml kanamycin, 100 μg/ml rifampicin, and 50 μg/ml spectinomycin. Arabidopsis plants used in the study include the wild-type Columbia-0 (Lehle Seeds, Round Rock, TX), and the grp7-1 mutant (a T-DNA insertion line SALK_039556.21.25.x). For complementation, a full-length GRP7 coding sequence derived from NM_127738.4 fused to the HA tag was placed under control of its native promoter resulting in construct pLN2325. A similar construct, pLN2678, was made resulting in a GRP7 derivative that substituted an arginine with a lysine residue in position 49. Both of these constructs were introduced into grp7-1 by Agrobacterium-mediated floral dip (35).
Protein Expression, Purification, and Crystallization
The hopU1 gene was amplified by PCR and then inserted into pGEX-6p-1 vector (GE Healthcare). All constructs were generated using PCR-based cloning strategies, and all mutants were generated by the QuikChange mutagenesis protocol (Stratagene). Protein expression, purification crystallization, and data collection have been described elsewhere (36). Briefly, GST-tagged HopU1 and its mutants were expressed at 15 °C in E. coli strain Rossetta cells, whereas GRP7 and its mutants were expressed in BL21(DE3) cells. Cells were harvested and resuspended in buffer containing 20 mm Tris-HCl (pH 8.0), 150 mm NaCl, supplemented with DNase and protease inhibitors. The soluble fraction of the cell lysate was loaded onto glutathione-Sepharose 4B column (GE Healthcare). The protein was eluted and further purified by ion-exchange chromatography (Source 15Q; GE Healthcare) followed by gel filtration (Superdex 200; GE Healthcare) and concentrated to 10 mg/ml for crystallization. The crystals of HopU1 were grown by the hanging-drop vapor-diffusion method at 4 °C by mixing 1 μl of protein solution and 1 μl of reservoir solution containing 0.1 m HEPES (pH 7.3), 5% PEG10000, 8% ethylene glycol. A selenomethionine derivative of HopU1 was expressed in M9 medium containing 60 mg liter−1 selenomethionine in BL21(DE3). The derivative crystals were obtained using similar conditions as those of native crystals.
Data Collection and Structure Determination
All crystals were flash-cooled in liquid nitrogen with cryoprotectant containing reservoir solution and 12% glycerol. Data collection was carried out at Beamline BL17A of Photon Factory (Japan) and Beamline BL17U at the Shanghai Synchrotron Radiation Facility. The program HKL2000 was used for data processing. Phases for HopU1 were initially determined by the single-wavelength anomalous dispersion, and automatic model building was performed by using the program package PHENIX (37). Data statistics for the SeMet single-wavelength anomalous dispersion dataset have been published previously (36). 16 selenium atoms were found when resolving the phase. After density modification, overall the figure of merit increased from 0.51 to 0.74. More than 50% of residues in one asymmetric unit were traced into the experimental electron density map. The remaining residues were manually built with COOT (38). The model was refined using native dataset to 2.7-Å resolution (supplemental Table S1) (36). All refinements were performed with the program package PHENIX (37). The final models were evaluated by PROCHECK, which showed a good stereochemistry according to the Ramachandran plot (39). The Rwork and Rfree for the highest resolution shell (2.80–2.71) are 21.3% and 27.1%, respectively. The structure similarity search was performed with DALI Server (40). All structure figures were generated by PyMOL.
HopU1-GRP7 Binding Studies
Isothermal titration calorimetry was used to obtain the binding affinity between of HopU1 and its substrate GRP7. Purified wild-type and HopU1 mutants (in cell) with concentration of 50 μm were titrated against purified GRP7 (in syringe) with concentration of 1 mm using a VP-ITC microcalorimeter (MicroCal) at 10 °C. All proteins were prepared in a buffer containing 10 mm HEPES (pH 8.0), 0.1 m NaCl. The data were fitted by software Origin 7.0.
ADP-ribosylation Assays
To determine the mADP-RT activity of wild type and mutants of HopU1, in vitro mADP-RT reactions were set up, and electrospray ionization-mass spectrometry was used to detect the molecular weight changes of HopU1 substrate, GRP7. The RRM RNA binding domain of GRP7 (residues 1–90) was used in these assays because it has been shown to be a substrate of HopU1 (18). Briefly, 20 ng of purified HopU1 recombinant proteins were incubated with 0.1 mm β-NAD and 2 μg purified recombinant GRP7-RRM in 5 mm HEPES (pH 7.2) buffer. Reactions were performed at 25 °C with total volume of 20 μl, and 20 μl of acetonitrile was added to stop the reaction after 1 h. Finally, the samples were subjected to electrospray ionization-mass spectrometry to detect the molecular mass changes.
HopU1-GRP7 Protein Complex Modeling
Based on the DALI search result and structure comparison, the structure of the eEF2-ExoA-NAD complex (Protein Data Bank (PDB) code 2ZIT) was used as a template for the modeling of HopU1-NAD complex. The binding site of NAD is highly conserved in both structures, hence the position of NAD in the HopU1-NAD complex was inferred. Because there was no available structure for GRP7, homology modeling of the GRP7 RRM domain was performed by SWISS-MODEL using the RRM domain of A18 hnRNP (PDB code 1X5S) as the template, which shares 45% sequence identity with the GRP7 RRM domain (41, 42). In the final step, the docking experiment for HopU1-NAD-GRP7-RRM complex was achieved on ZDOCK server with residues Gln-53, Gln-95, and Asp-96 of HopU1 and Arg-47 and Arg-49 of GRP7 used for constraint (43).
Identification of Amino Acids ADP-ribosylated by HopU1
To identify the amino acids within GRP7 that are ADP-ribosylated by HopU1, ADP-ribosylated GRP7 was separated by two-dimensional gel electrophoresis, and modified amino acids were identified by mass spectrometry essentially as described previously (18). Briefly, 3 μg of purified GRP7-GST was incubated with HopU1 or HopU1DD and separated on immobilized pH gradient strips with pH range of 5 to 8 in a Protean IEF Cell (Bio-Rad), followed by standard SDS-PAGE. All spots derived from GRP7-GST in the presence of HopU1 or HopU1DD were cut out and subjected to tandem mass spectrometry (MS/MS).
The stained spots were subjected to LC/MS as described (44). Briefly, gel pieces were digested by trypsin (V5111; Promega), and digested peptides were extracted in 5% formic acid (v/v)/50% acetonitrile (v/v) and separated using C18 reversed phase LC column (75 μm × 15 cm, BEH 130, 1.7 μm; Waters, Milford, MA). A Q-TOF Ultima tandem mass spectrometer coupled with a Nanoaquity HPLC system (Waters) with electrospray ionization was used to analyze the eluting peptides. The system was user controlled employing MassLynx software (version 4.1; Waters) in data-dependent acquisition mode with the following parameters: 0.9-s survey scan (380–1900 Da) followed by up to three 1.4-s MS/MS acquisitions (60–1900 Da). The instrument was operated at a mass resolution of 8000. The instrument was calibrated using the fragment ion masses of double-protonated Glu-fibrinopeptide. The peak lists of MS/MS data were generated using Distiller (Matrix Science, London, UK) using charge state recognition and deisotoping with the other default parameters for Q-TOF data. Database searches of the acquired MS/MS spectra were performed using Mascot (version 2.2.0; Matrix Science). A custom database containing the sequence of GRP7 was used for the data searches. Search parameters used were: no restrictions on protein molecular weight or pI, enzymatic specificity was set to trypsin with up to 3 missed cleavage sites, carbamidomethylation of C was selected as a fixed modification, and methionine oxidation along with ADP-ribosylation. Mass accuracy settings were 0.15 Da for peptide mass and 0.12 Da for fragment ion masses.
Inhibition of GRP7 RNA Binding by HopU1
Effects of ADP-ribosylation on the GRP7 RNA binding affinity were analyzed by first producing ADP-ribosylated GRP7 and then testing its RNA binding ability. Different concentrations of GRP7 were ADP-ribosylated by HopU1 (or HopU1DD as a negative control) in 25 mm Tris-HCl (pH 7.4), 10 mm DTT and 5 μm β-NAD, and a 32P-labeled RNA probe was added to the reaction mixture. The RNA probe used was 7-UTR_WT, which has been shown to bind GRP7 specifically (45). Bound and free RNA probes were separated on a native PAGE in 1× TBE and exposed to x-ray film. Gels were also exposed to PhosphorImager screens, and the radioactive signals were quantitated using a Storm 860 scanner and ImageQuant software (Molecular Dynamics). For analyses, ratios of bound to free probes were plotted against GRP7 concentrations in logarithmic scales, and the dissociation constant Kd was estimated from the x intercept of the resulting linear regression line (45).
Analyses of Immune Responses in Arabidopsis
Measurement of ROS has been described previously (46, 47). Briefly, disks of leaf tissue 0.4 cm in diameter were harvested and floated on water in 96-well plates overnight. The water was replaced with a solution containing 0.5 mm chemiluminescence probe L-012 (Wako, Japan), 10 mm MOPS/KOH (pH 7.4), and 1 μm flg22, 1 μm elf18 or 100 μg/ml shrimp chitin (Sigma). Luminescence was measured using a Synergy H4 Hybrid Multi-Mode Microplate Reader (Biotek). Detection of callose deposition was done as described by Guo et al. (9). Plants were infiltrated with 1 μm flg22, 1 μm elf18, or 100 μg/ml chitin and incubated for 16 h. Leaf samples were treated with alcoholic lactophenol solution to remove chlorophyll and were stained with 0.01% (w/v) aniline blue in a solution of 150 mm K2HPO4 (pH 9.5) for 30 min. The callose deposits were visualized with a fluorescence microscope (Zeiss Axionplan 2; Carl Zeiss, Oberkochen, Germany), and the number of callose deposits was determined using ImageJ (48).
In Planta Growth and Pathogenicity Assays
Wild-type Arabidopsis Columbia-0, grp7-1, or grp7-1 transgenic plants expressing GRP7 or a GRP7R49K derivative were grown in a growth chamber at 24 °C with 10-h days. Bacterial growth in plants was determined by spray-inoculating Arabidopsis leaves with DC3000, DC3000 type III defective hrcC mutant (49), or UNL227 (50) at a cell density of 2 × 108 cfu/ml with 0.02% Silwet L-77 (Lehle Seeds). The ability of bacteria to grow in planta was examined as described previously (51). Briefly, four leaf disks were harvested with a 0.4-cm2 cork borer 0, 2, and 4 days after inoculation. The samples were macerated in 250 μl of sterile water for 1 min and serially diluted. Twenty microliters of each dilution was plated on KB medium containing the appropriate antibiotics. The colonies recovered on plates after 48 h were enumerated.
RESULTS
HopU1 mADP-RT Structure
The structure of the HopU1 mADP-RT was determined by selenium single-wavelength anomalous diffraction with the final model refined to 2.7-Å resolution. The statistics of structure determination and refinement are summarized in supplemental Table S1. Two HopU1 molecules are found in the asymmetric unit (data not shown). However, mADP-RT assays on samples subjected to gel filtration chromatography indicated that HopU1 exists as a monomer in solution (36). Amino acid residues 1–28 corresponding to HopU1 type III secretion signal were not built in the model as they lacked electron density, possibly due to intrinsic disorder of the N terminus as indicated by secondary structure prediction (see the predict protein Web site) (52).
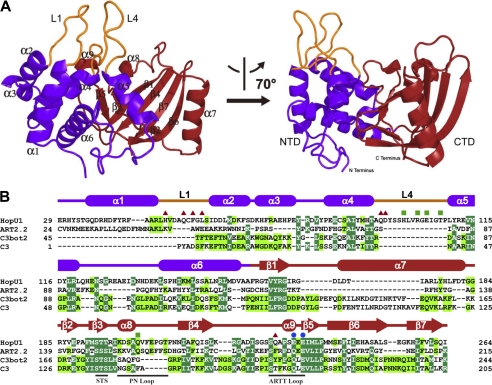
The structure of HopU1 adopts a typical Rossmann fold, commonly found in proteins that bind NAD+ (53). The overall dimensions of HopU1 are ∼40 × 45 × 50 Å3, and it resembles a bent right hand (Fig. 1A). The N-terminal domain is analogous to a thumb and palm and is made up of five α-helices (α1–α5), connected by four loops (L1–L4), which are packed against each other to form a compact core structure. The two protruding loops, L1 and L4, connect α1, α2 and α4, α5, respectively, and likely stabilize each other on one side. The C-terminal domain resembles the fingers and is composed of two perpendicular β-sheets, which form an atypical β-sandwich fold surrounded by three α-helices (α6 to α8). One of the β-sheets is formed by β-strands β3 and β5, and the other is formed by β-strands β1, β2, β4, β6, and β7. A central cleft located between the N- and C-terminal domains forms a potential NAD binding pocket (Fig. 1A).
FIGURE 1.
Structure of HopU1. A, overall structure of P. syringae pv. tomato DC3000 HopU1 in a ribbon representation. The N-terminal domain is colored in purple, C-terminal domain in red, and protruding loops L1 and L4 are shown in orange. Secondary structural elements are labeled, and two different views of the structure are shown. B, structure-based sequence alignment of HopU1 and three representative ADP-ribosyltransferases, rat ecto-ADP-ribosyltransferase ART2.2 (PDB code 1GXZ), ADP-ribosyltransferase C3bot2 from C. botulinum (PDB code 1R45), and C. limosum C3 exoenzyme (PDB code 3BW8). Identical residues are highlighted with dark green background, and highly conserved residues with light green background. Secondary structural elements are colored as in A and are shown above their corresponding sequences. Conserved motifs are indicated below their corresponding sequences. Residues that are critical for GRP7 recognition and enzymatic activity are indicated by red triangles, residues critical for enzymatic activity but not GRP7 recognition are indicated by blue dots, and those not critical for substrate recognition or activity are indicated by green squares.
Structure Comparison of HopU1 with Other ADP-ribosyltransferases
To begin to analyze the structure and function of HopU1, we performed a Dali search (40) of the HopU1 structure versus all known structures in the PDB. Among 451 entries with structural similarity (Z score higher than 2.0), the top three structures with Z score higher than 8.0 were selected for the following structural comparison. These are the rat ecto-ADP-RT ART2.2 (PDB code 1GXZ), the Clostridium botulinum mADP-RT C3bot2 (PDB code 1R45), and the Clostridium limosum C3 exoenzyme (PDB code 3BW8). HopU1 and these three proteins superimposed well with a root mean square deviation of 3.69 Å for 94 Cα atoms (for 1GXZ), 2.12 Å for 86 Cα atoms (for 1R45), and 2.4 Å for 48 Cα atoms (for 3BW8), respectively (supplemental Fig. S1). In general, the C-terminal core folds of HopU1 and the three proteins compared are similar, adopting a mixed α/β-fold with a characteristic four β-stranded core. However, there are obvious differences in the structure of the N terminus of HopU1 and the three mADP-RTs (supplemental Fig. S1E). Unique features of HopU1 are the two protruding loops L1 and L4 (Fig. 1B), which are substantially shorter in the corresponding regions of the other mADP-RTs (supplemental Fig. S1E). These two loops may be involved in substrate recognition, which will be discussed below.
Amino acid Residues of HopU1 Important for Substrate Recognition and mADP-RT Activity
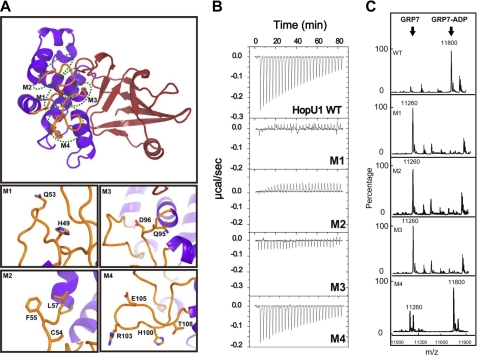
To understand the molecular mechanism of how HopU1 recognizes and ADP-ribosylates GRP7, we introduced alanine substitutions at specific residues in L1, L4, and near the NAD+ binding pocket (Fig. 2A and supplemental Fig. S2A). A qualitative measure of the binding of HopU1 (and HopU1 derivatives) with purified GRP7 was measured by isothermal titration calorimetry. In vitro ADP-ribosylation of GRP7 was detected by mass spectrometry. Wild-type HopU1 showed a binding affinity with a Kd of about 30 μm (Fig. 2B) and possessed strong mADP-RT activity (Fig. 2C). HopU1 mutants M1 (HopU1H49A/Q53A) and M2 (HopU1C54A/F55A/L57A), with substitutions in loop L1, abolished the ability of HopU1 to bind to GRP7 and its mADP-RT activity (Fig. 2B and C), indicating that the L1 loop is essential for substrate recognition. The M3 mutant (HopU1Q95A/D96A) with substitutions in the L4 loop was unable to bind GRP7 and did not possess detectable mADP-RT activity (Fig. 2, B and C). However, the M4 mutant (HopU1H100A/R103A/E105A/T108A), which also contains substitutions in the L4 loop, only slightly decreased the HopU1 binding affinity and possessed approximately 70% of its mADP-RT activity based on integration of substrate and product peak areas (Fig. 2, B and C). Amino acid residue Asp-96 is near the NAD+ binding pocket of HopU1 and interacts directly with Gln-229 through a hydrogen bond (2.8 Å) likely stabilizing the ARTT loop and L4 loop. Consistent with this, the HopU1 M8 mutant (HopU1Q229A) lost its ability to interact with GRP7 and no longer possessed mADP-RT activity (supplemental Fig. S2, B and C). However, the limited effect on substrate binding and mADP-RT activity of the M4 mutant (HopU1H100A/R103A/E105A/T108A) suggests that residues adjacent to Q95A/D96A in the L4 loop do not directly interact with GRP7.
FIGURE 2.
Effect of HopU1 mutations on activity and GRP7 recognition. A, structure of HopU1 indicating the location of HopU1 mutations M1–M4 (detailed in bottom panels). The residues changed to Ala are highlighted and shown as stick representations. The structure was rotated ∼90° about the horizontal axis compared with the view presented on the left side of Fig. 1A. B, isothermal titration calorimetry of wild-type HopU1 and HopU1 derivatives M1–M4 with GRP7, indicating the extent that the HopU1 derivatives can interact with GRP7. C, electrospray ionization-mass spectrometry results of mADP-RT activity for wild-type HopU1 and HopU1 derivatives. The peak of substrates and products of each enzymatic reaction are indicated. The molecular mass of GRP7 (1–90 with 4 additional residues (Gly, Pro, His, and Met) left over after affinity tag cleavage) is 11,260 Da and GRP7 with an ADP-ribose modification is 11,800 Da. The mutations of each HopU1 mutant are as follows: M1, HopU1H49A/Q53A; M2, HopU1C54A/F55A/L57A; M3, HopU1Q95A/D96A; and M4, HopU1H100A/R103A/E105A/T108A.
It has been shown for other mADP-RTs that the ARTT loop is essential for NAD+ binding and substrate recognition (54). Two conserved glutamate residues within the ARTT loop are required for NAD+ binding and mADP-RT activity. The mutant M9 (HopU1E233A/E235A) with substitutions in the corresponding amino acids Glu-233 and Glu-235 showed decreased GRP7 binding, suggesting that the ARTT loop may also play a role in GRP7 recognition (supplemental Fig. S2, B and C). The PN loop of HopU1 is likely not required for interaction with GRP7 because mutant M7 (HopU1Q202A), which substitutes a glutamine for an alanine in the PN loop, retained the ability to interact with GRP7 even though it no longer possessed mADP-RT activity (supplemental Fig. S2, B and C). Mutants M5 (HopU1R112/E113A) and M6 (HopU1R119A), containing amino acid substitutions in the α5-helix, showed little reduction in their ability to bind GRP7 (supplemental Fig. S2B), but were required for mADP-RT activity (supplemental Fig. S2C), suggesting that they are not essential for substrate recognition but critical for enzymatic activity. Because α5 and the PN loop are important for NAD+ binding, mutations in these regions may affect NAD+ interaction and thus decrease NAD+-dependent ADP-ribosylation. Taken together, loop L1, the N terminus of loop L4, and the ARTT loop are important for both GRP7 interaction and mADP-RT activity (supplemental Fig. S2D). In the HopU1 structure, these three regions are close together and form a major binding surface for interaction with GRP7.
Arginine 49 within GRP7 Is ADP-ribosylated by HopU1
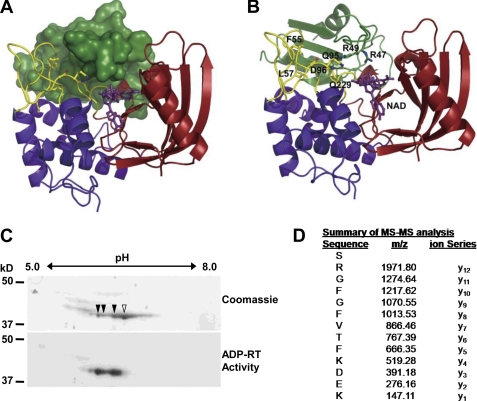
We showed previously that arginines 47 and 49 within the RRM RNA binding domain of GRP7 were required for HopU1 to ADP-ribosylate GRP7 (18). Thus far, we have been unable to determine the structure of GRP7 bound to HopU1. However, the structures of several RRM proteins are known (55), which allowed the modeling of the GRP7 RRM with the HopU1 structure. Arg-47 and Arg-49 are predicted to be near the NAD+ binding pocket of HopU1 (Fig. 3, A and B), and therefore both represent candidate ADP-ribosylation sites. To determine which arginine residue was ADP-ribosylated by HopU1, we employed a proteomic approach. In vitro ADP-ribosylation reactions containing purified GRP7 and HopU1 were resolved by isoelectric focusing and SDS-PAGE. Purified GRP7 was composed of at least four isoforms separated by pI value of ∼0.2, three of which were ADP-ribosylated (Fig. 3C). It is important to note that the different isoforms likely represent unmodified GRP7 and one isoform of ADP-ribosylated GRP7 exhibiting pI heterogeneity due to changes in molecular charge.
FIGURE 3.
Arginine 49 of GRP7 is ADP-ribosylated by HopU1. A, structure model of HopU1-NAD-GRP7-RRM complex. HopU1 is shown in ribbon representation, and the N-terminal domain is colored in blue, C-terminal domain in red, and protruding loops L1 and L4 are shown in yellow. GRP7-RRM (dark green) is shown in surface representation, and NAD (purple) is shown in stick representation. B, structure model with GRP7-RRM shown as ribbon representation. Critical HopU1 residues for interaction with GRP7 and potential ADP-ribosylation sites Arg-47 and Arg-49 are indicated in stick representation. C, two-dimensional PAGE gels of in vitro mADP-RT reactions containing purified HopU1-His, GRP7-GST, and 32P-labeled NAD stained with Coomassie Blue to visualize total protein or exposed to autoradiography film to identify 32P-labeled proteins. Protein spots labeled with 32P corresponding to ADP-ribosylated proteins are marked with filled arrowheads, and the unlabeled spot is marked with an open arrowhead. D, mass spectrometric analyses of tryptic peptides derived from the above mADP-RT reactions using nonradioactive NAD. All spots corresponding to the indicated spots in B were cut out and sent for MS/MS. One fragment shown contained a higher molecular mass than predicted. The molecular mass of the arginine (y12) corresponds to Arg-49 of GRP7 and was equal to ADP-ribosylated arginine, indicating that this residue was ADP-ribosylated by HopU1.
All GRP7-related spots were analyzed using MS/MS to sequence individual tryptic peptides. A double charged ion of m/z 1029.92 was observed in the spectra of the gel digest. The molecular mass of this peptide, 2057.82 Da, matches the ADP-ribosylation of the peptide (Ser-48–Lys-60), which results in a mass shift of 541.06 Da in the peptide mass. MS/MS of this double charged species produced a nearly complete y series of ions through y11 that show no shift in mass, indicating that residues 50–60 are not modified. When Arg-49 was present on the peptide the molecular mass increased by 697.16 Da, consistent with the addition of an ADP-ribosylated Arg residue (Fig. 3D). Ions supporting ADP-ribosylation of this peptide observed in the MS/MS spectrum are protonated adenine (m/z 136), AMP (m/z 348), and ADP (m/z 428) (Fig. 3D and supplemental Fig. S3). Additionally, trypsin specifically cleaves at the carboxyl side of Arg and Lys residues, and we were unable to identify trypsin cleavage sites adjacent to Arg-49, suggesting that ADP-ribosylation of Arg-49 likely prevented cleavage by trypsin. Arg-49 is located in the conserved ribonucleoprotein consensus sequence 1 (RNP-1) motif of the RRM domain, and in other RRM proteins it has been shown to interact directly with RNA by forming a salt bridge with the phosphate group of bound RNAs (55). Thus, HopU1 targets an amino acid that is likely critical for GRP7 to bind RNA.
ADP-ribosylation Reduces the Ability of GRP7 to Bind RNA
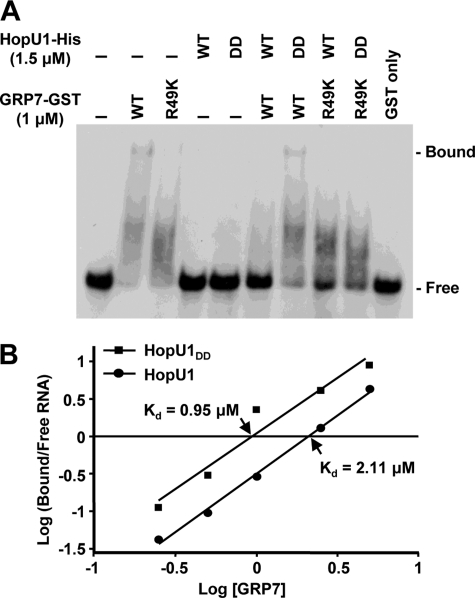
To test directly whether ADP-ribosylated GRP7 was less able to bind RNA, we performed gel-shift assays. Affinity-purified recombinant GRP7-GST either treated with HopU1-His or a catalytically inactive HopU1 derivative HopU1DD-His (HopU1E233D/E235D) (18) was incubated with the 32P-labeled RNA oligomer ATGRP7-UTR_WT, a known GRP7 target site within its own mRNA (45). After separation on a native polyacrylamide gel, a greater amount of RNA-protein complex was present when GRP7-GST was treated with HopU1DD-His compared with when it was treated with HopU1-His, indicating that ADP-ribosylated GRP7 was less efficient at binding its target RNA (Fig. 4A). Importantly, when a GRP7-HA derivative with Arg-49 replaced by lysine (GRP7R49K) was used less RNA-protein complex was detected, indicating that Arg-49 is important for RNA binding in vitro (Fig. 4A). There was no detectable higher molecular mass RNA-protein complex when GRP7R49K was used in the gel-shift assays. However, based on the lower molecular mass smear in the GRP7R49K lane (Fig. 4A), this protein retained some ability to bind RNA. To determine the equilibrium dissociation constant (Kd) for the binding of GRP7 to its target RNA we redid the assay with differing amounts of GRP7-GST (supplemental Fig. S4). The ratio of the GRP7-bound RNA to free RNA for each GRP7-GST concentration was plotted against the GRP7-GST concentration in logarithmic scales to obtain the Kd (Fig. 4B). The Kd value for ADP-ribosylated GRP7-GST preincubated with wild-type HopU1-His, was 2.11 μm, which was more than twice the Kd for the unmodified GRP7-GST, preincubated with HopU1DD-His (0.95 μm). The Kd value of the GRP7-GST treated with wild-type HopU1-His was similar to the previously reported Kd of GRP7R49Q mutant (this mutant is similar to GRP7R49K except that it has a Gln instead of Lys at position 49) (45), indicating that ADP-ribosylation of Arg-49 affected GRP7 binding kinetics similar to an Arg-49 substitution. Moreover, the Kd value of GRP7-GST treated with HopU1DD-His was similar to the previously reported Kd of the wild-type GRP7 (45), indicating that treatment of GRP7-GST with catalytically inactive HopU1 did not significantly affect its binding kinetics. Taken together, this clearly demonstrates that HopU1 action can interfere with the ability of GRP7 to bind its target RNA.
FIGURE 4.
ADP-ribosylation by HopU1 reduces ability of GRP7 to bind RNA. A, electrophoretic mobility shift assay of an RNA probe with GRP7-GST after treatment with HopU1-His or its catalytic inactive mutant HopU1DD-His. Standard ADP-ribosylation reactions were performed with GRP7-GST or GRP7-GSTR49K in the presence of HopU1 or HopU1DD, and a 32P-labeled probe (ATGRP7 UTR WT) was added to each reaction mix. These were run on native polyacrylamide gels and exposed to x-ray films. B, similar assays done with differing amounts of GRP7-GST. The protein-bound and free RNA probes were quantified using a PhosphorImager scanner. The ratio of protein-bound and free forms of RNA was plotted against the concentration of GRP7 using a logarithmic scale. The x intercept allowed estimation of Kd for both HopU1 and HopU1DD as indicated. The autoradiogram used to calculate the kDa is shown in supplemental Fig. S4.
Arabidopsis grp7-1 Mutant Is Complemented by Wild-type GRP7 but Not GRP7R49K
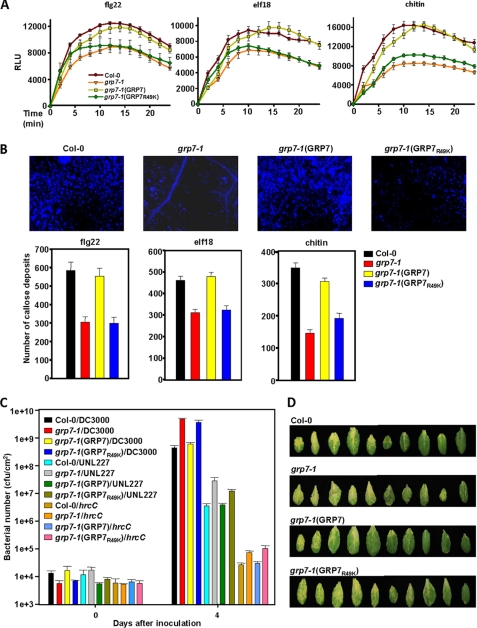
We previously reported that two independent Arabidopsis grp7 mutants were compromised in callose deposition in response to the 22 amino acid epitope (flg22) from the flagellin PAMP (18). To characterize GRP7 involvement in PTI further we tested the response of the grp7-1 mutant to other PAMPs. Callose deposition was also reduced in the grp7-1 mutant compared with wild-type plants when treated with elf18, the 18-amino acid epitope from the EF-Tu PAMP, and the chitin PAMP (Fig. 5B). Furthermore, production of ROS was reduced in the grp7-1 mutant compared with wild-type plants in response to all three treatments (Fig. 5A). Both phenotypes were complemented by expressing GRP7 fused to the HA epitope from its native promoter in grp7-1 (Fig. 5, A and B). Expression of GRP7-HA was confirmed with immunoblot analysis (supplemental Fig. S5). Notably, expression of a GRP7R49K did not complement the reduced PTI phenotype of the grp7-1 mutant (Fig. 5, A and B). Arg-49 has been shown to be important for the ability of GRP7 to bind RNA (45) (Fig. 4A), and ADP-ribosylation at this site by HopU1 reduces the ability of GRP7 to bind RNA (Fig. 4A).
FIGURE 5.
An Arabidopsis grp7 mutant is defective in PTI responses and more susceptible to P. syringae, and these phenotypes are complemented by wild-type GRP7 but not by a GRP7 derivative that is reduced in its ability to bind to RNA. A and B, ROS levels in relative light units (RLU) (A) and callose (B) in wild-type Arabidopsis Columbia-0 (Col-0), the Columbia-0 grp7-1 mutant, the grp7-1 mutant complemented with wild-type GRP7 (grp7-1(GRP7)), and the grp7-1 mutant complemented with a GRP7 derivative containing a lysine instead of an arginine at position 49 (grp7-1(GRP7R49K)) after treatment with flg22, elf18, or chitin. C, bacterial growth in the plants listed in A of wild-type P. syringae pv. tomato DC3000, a DC3000 polyeffector mutant lacking about one third of its type III effector inventory (UNL227), and a DC3000 mutant with a defective T3SS (hrcC). D, disease symptoms at day 4 on Arabidiopsis plants described in A after infection with wild-type DC3000.
To determine the susceptibility of grp7 mutant plants we infected them with wild-type P. syringae pv. tomato DC3000, a DC3000 hrcC mutant defective in type III secretion, and UNL227, a DC3000 polyeffector mutant lacking about one third of its type III effector inventory including HopU1 (50). DC3000 grows well in Arabidopsis because it injects about 35 type III effectors that suppress PTI. The DC3000 hrcC mutant cannot inject type III effectors, and therefore its growth is inhibited by PTI. UNL227 grows intermediately in Arabidopsis compared with DC3000 and the hrcC mutant because it injects fewer type III effectors. We reported previously that the grp7-1 mutant was more susceptible to DC3000 and the hrcC mutant compared with wild-type Arabidopsis (18). Here, we show that ectopic expression of GRP7-HA, but not GRP7-HAR49K, was able to restore the levels of bacterial growth in the grp7-1 mutant to those observed in the in wild-type plants (Fig. 5, C and D). Collectively, these data indicate that the RNA binding ability of GRP7 is required for its role in PTI.
DISCUSSION
The crystal structure of HopU1 reveals both conserved and unique features of mADP-RTs. HopU1 adopts a conserved four β-stranded core with characteristic ARTT and PN loops, which have been demonstrated to be essential for NAD+ binding and ADP-ribosylation (29). Thus, the mADP-RT family of proteins is conserved for both structure and catalytic mechanism not only between animal pathogens and eukaryotes, but also for plant pathogens. However, HopU1 possesses two protruding loops, L1 and L4, which are not found in other mADP-RTs. Functional analyses revealed that residues within these loops are required for substrate recognition. Tsuge et al. reported that specific residues of loop II of the Clostridium perfringens iota toxin mADP-RT were required for substrate interaction (56). These residues superimpose with residues Gln-95 and Asp-96 in the L4 loop of HopU1, suggesting that this region may play a role in substrate recognition for other mADP-RTs (data not shown). The ARTT and PN loops are important for substrate recognition in the majority of mADP-RTs studied, and this was also the case for HopU1. Thus, the HopU1 structure reveals the basis for recognition of the RNA-binding protein GRP7, a novel type of substrate.
Recently, another DC3000 type III effector, HopF2, was shown to be an mADP-RT. HopF2 targets a MAPK kinase MKK5 and RIN4, both signal transduction components of innate immunity (15, 57). The structure of a HopF2 homolog, AvrPphF/HopF1 from P. syringae pv. phaseolicola has been determined (58). HopU1 and AvrPphF do not superimpose well (supplemental Fig. S6), sharing limited homology in the C-terminal catalytic region and no similarity elsewhere in the proteins. The HopU1 structure belongs in the CT group of mADP-RTs whereas AvrPphF belongs to the DT group. Thus, our data show that P. syringae has representatives of both major groups of mADP-RTs that clearly target different substrates involved in plant immunity.
HopU1 ADP-ribosylated GRP7 at Arg-49, which is a residue within the conserved RNP1 motif of the RRM. Structure-function analysis of several RRM proteins including the heterogeneous nuclear ribonucleoprotein hnRNPA1, the spliceosomal protein U1A, and the Drosophila sex-determination factor SEX-LETHAL have shown that arginines in the analogous position contact the phosphate backbone of their respective RNA targets via salt bridges (59–61). Indeed, mutation of Arg-49 into glutamine previously has been shown to reduce the binding affinity of GRP7 for its RNA targets about 6-fold and to interfere with the in vivo function of GRP7 (45). We show here that ADP-ribosylation of GRP7 Arg-49 reduces the ability of GRP7 to bind an RNA target in vitro. This is the first example of a type III effector that modifies an RNA-binding protein interfering with its ability to interact with RNA and it suggests HopU1 subverts the immune system of the plant by interfering with processing of immune-related transcripts.
To investigate whether Arg-49 is crucial for plant immunity we performed a functional analysis in the A. thaliana grp7-1 T-DNA mutant that shows reduced PAMP induced callose deposition and ROS production and enhanced susceptibility to DC3000. Ectopic expression of wild-type GRP7 but not of the GRP7R49K variant complements these innate immunity and enhanced susceptibility phenotypes. Thus, it appears that HopU1 evolved to target an amino acid in a protein that plays a crucial role in plant immunity. Moreover, HopU1 targets a highly conserved amino acid in the RRM, one that could not easily be changed by evolutionary pressure. How could this heightened level of specificity have evolved? There are several examples of type III effectors mimicking the activity of host proteins (62). However, plants are not known to possess mADP-RTs, and GRP7 is not known to be regulated by mADP-RTs. Thus, it is unlikely this is a case of effector mimicry. Importantly, because RRM-containing RNA-binding proteins are well represented in prokaryotes and mADP-RTs can be used for regulation in bacteria (63, 64), it is possible that bacteria may have originally evolved these effectors to regulate bacterial processes and later adapted them to act inside plant cells.
GRP7 has been implicated in circadian rhythms, stress responses, and flower transition (18, 30–32, 65). Microarray analyses revealed that ectopic overexpression of GRP7 affects the steady-state abundance of a suite of cellular transcripts. Among those are circadian-regulated transcripts and transcripts influenced by abiotic and biotic stress (66). Because GRP7 interacts with translational machinery components it is likely that GRP7 plays a role in translation.5 Thus, it appears that GRP7 is involved in several plant processes and, therefore, likely has many different RNA cargoes. A subset of these RNA cargoes must play important roles in plant immunity and it is likely that the activity of HopU1 interferes with the translation of these RNAs thereby weakening the innate immune system of the plant.
Supplementary Material
Acknowledgments
We thank all the members of the D.S., Y.X. and J.R.A. laboratories and Mark Wilson for fruitful discussions and comments on the manuscript. This research was supported by the National Institutes of Health grant 1R01AI069146-01A2 and funds from the Center for Plant Science Innovation at the University of Nebraska (to J.R.A.); by the German Research Foundation grants STA 653/2 and STA 653/3 (to D.S.); and National Basic Research Program of China grant 2009CB918600 and the International Collaboration Program from Science and Technology Commission of Shanghai Municipality grant 10430709300 (to Y.X.).
This work was supported, in whole or in part, by National Institutes of Health Grant 1R01AI069146-01A2 (to J. R. A.). This work was also supported by Center for Plant Science Innovation at the University of Nebraska (to J. R. A.), German Research Foundation Grants STA 653/2 and STA 653/3 (to D. S.), and National Basic Research Program of China Grant 2009CB918600 and the International Collaboration Program from Science and Technology Commission of Shanghai Municipality Grant 10430709300 (to Y. X.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1 and Figs. S1–S6.
The atomic coordinates and structure factors (code 3U0J7) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
V. Nicaise, B.-r. Jeong, A. Joe, J. R. Alfano, and C. Zipfel, unpublished observations.
- PAMP
- pathogen-associated molecular pattern
- CT
- cholera toxin
- DT
- diphtheria toxin
- ecto-ADP-RT
- extracellular membrane-associated mADP-RT
- ETI
- effector-triggered immunity
- mADP-RT
- mono-ADP-ribosyltransferase
- MS/MS
- tandem mass spectrometry
- PDB
- Protein Data Bank
- PTI
- PAMP-triggered immunity
- RNP
- ribonucleoprotein
- ROS
- reactive oxygen species
- RRM
- RNA recognition motif.
REFERENCES
- 1. Büttner D., He S. Y. (2009) Plant Physiol. 150, 1656–1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Alfano J. R., Collmer A. (2004) Annu. Rev. Phytopathol. 42, 385–414 [DOI] [PubMed] [Google Scholar]
- 3. Hann D. R., Gimenez-Ibanez S., Rathjen J. P. (2010) Curr. Opin. Plant Biol. 13, 388–393 [DOI] [PubMed] [Google Scholar]
- 4. Block A., Alfano J. R. (2011) Curr. Opin. Microbiol. 14, 39–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Boller T., Felix G. (2009) Annu. Rev. Plant Biol. 60, 379–406 [DOI] [PubMed] [Google Scholar]
- 6. Jones J. D., Dangl J. L. (2006) Nature 444, 323–329 [DOI] [PubMed] [Google Scholar]
- 7. Tsuda K., Katagiri F. (2010) Curr. Opin. Plant Biol. 13, 459–465 [DOI] [PubMed] [Google Scholar]
- 8. Lindeberg M., Cartinhour S., Myers C. R., Schechter L. M., Schneider D. J., Collmer A. (2006) Mol. Plant Microbe Interact. 19, 1151–1158 [DOI] [PubMed] [Google Scholar]
- 9. Guo M., Tian F., Wamboldt Y., Alfano J. R. (2009) Mol. Plant Microbe Interact. 22, 1069–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rosebrock T. R., Zeng L., Brady J. J., Abramovitch R. B., Xiao F., Martin G. B. (2007) Nature 448, 370–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Xiang T., Zong N., Zou Y., Wu Y., Zhang J., Xing W., Li Y., Tang X., Zhu L., Chai J., Zhou J. M. (2008) Curr. Biol. 18, 74–80 [DOI] [PubMed] [Google Scholar]
- 12. Shan L., He P., Li J., Heese A., Peck S. C., Nürnberger T., Martin G. B., Sheen J. (2008) Cell Host Microbe 4, 17–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Göhre V., Spallek T., Häweker H., Mersmann S., Mentzel T., Boller T., de Torres M., Mansfield J. W., Robatzek S. (2008) Curr. Biol. 18, 1824–1832 [DOI] [PubMed] [Google Scholar]
- 14. Gimenez-Ibanez S., Hann D. R., Ntoukakis V., Petutschnig E., Lipka V., Rathjen J. P. (2009) Curr. Biol. 19, 423–429 [DOI] [PubMed] [Google Scholar]
- 15. Wang Y., Li J., Hou S., Wang X., Li Y., Ren D., Chen S., Tang X., Zhou J. M. (2010) Plant Cell 22, 2033–2044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang J., Shao F., Li Y., Cui H., Chen L., Li H., Zou Y., Long C., Lan L., Chai J., Chen S., Tang X., Zhou J. M. (2007) Cell Host Microbe 1, 175–185 [DOI] [PubMed] [Google Scholar]
- 17. Nomura K., Debroy S., Lee Y. H., Pumplin N., Jones J., He S. Y. (2006) Science 313, 220–223 [DOI] [PubMed] [Google Scholar]
- 18. Fu Z. Q., Guo M., Jeong B. R., Tian F., Elthon T. E., Cerny R. L., Staiger D., Alfano J. R. (2007) Nature 447, 284–288 [DOI] [PubMed] [Google Scholar]
- 19. Block A., Guo M., Li G., Elowsky C., Clemente T. E., Alfano J. R. (2010) Cell. Microbiol. 12, 318–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jelenska J., van Hal J. A., Greenberg J. T. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 13177–13182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fieldhouse R. J., Merrill A. R. (2008) Trends Biochem. Sci. 33, 546–556 [DOI] [PubMed] [Google Scholar]
- 22. Deng Q., Barbieri J. T. (2008) Annu. Rev. Microbiol. 62, 271–288 [DOI] [PubMed] [Google Scholar]
- 23. Aktories K., Wilde C., Vogelsgesang M. (2004) Rev. Physiol. Biochem. Pharmacol. 152, 1–22 [DOI] [PubMed] [Google Scholar]
- 24. Holbourn K. P., Shone C. C., Acharya K. R. (2006) FEBS J. 273, 4579–4593 [DOI] [PubMed] [Google Scholar]
- 25. Barbieri J. T., Burns D. L. (2003) in Bacterial Protein Toxins (Burns D. L., Barbieri J. T., Iglewski B. H., Rappuoli R. eds) pp. 215–228, American Society for Microbiology Press, Washinton, D. C [Google Scholar]
- 26. Hottiger M. O., Hassa P. O., Lüscher B., Schüler H., Koch-Nolte F. (2010) Trends Biochem. Sci. 35, 208–219 [DOI] [PubMed] [Google Scholar]
- 27. Mueller-Dieckmann C., Ritter H., Haag F., Koch-Nolte F., Schulz G. E. (2002) J. Mol. Biol. 322, 687–696 [DOI] [PubMed] [Google Scholar]
- 28. Okazaki I. J., Moss J. (1996) Rev. Physiol. Biochem. Pharmacol. 129, 51–104 [DOI] [PubMed] [Google Scholar]
- 29. Ménétrey J., Flatau G., Boquet P., Ménez A., Stura E. A. (2008) Protein Sci. 17, 878–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schmidt F., Marnef A., Cheung M. K., Wilson I., Hancock J., Staiger D., Ladomery M. (2010) Mol. Biol. Rep. 37, 839–845 [DOI] [PubMed] [Google Scholar]
- 31. Kim J. S., Jung H. J., Lee H. J., Kim K. A., Goh C. H., Woo Y., Oh S. H., Han Y. S., Kang H. (2008) Plant J. 55, 455–466 [DOI] [PubMed] [Google Scholar]
- 32. Streitner C., Danisman S., Wehrle F., Schöning J. C., Alfano J. R., Staiger D. (2008) Plant J. 56, 239–250 [DOI] [PubMed] [Google Scholar]
- 33. Sambrook J., Russell D. W. (2001) Molecular Cloning: A Laboratory Manual, 3rd Ed., Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 34. King E. O., Ward M. K., Raney D. E. (1954) J. Lab. Clin. Med. 44, 301–307 [PubMed] [Google Scholar]
- 35. Bechtold N., Ellis J., Pelletier G. (1993) C. R. Acad. Sci. Paris 316, 1194–1199 [Google Scholar]
- 36. Lin Y., Wang P., Yang H., Xu Y. (2010) Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 66, 932–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Terwilliger T. C., Adams P. D., Read R. J., McCoy A. J., Moriarty N. W., Grosse-Kunstleve R. W., Afonine P. V., Zwart P. H., Hung L. W. (2009) Acta Crystallogr. D Biol. Crystallogr. 65, 582–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 39. Laskowski R. A., Moss D. S., Thornton J. M. (1993) J. Mol. Biol. 231, 1049–1067 [DOI] [PubMed] [Google Scholar]
- 40. Holm L., Kääriäinen S., Rosenström P., Schenkel A. (2008) Bioinformatics 24, 2780–2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Arnold K., Bordoli L., Kopp J., Schwede T. (2006) Bioinformatics 22, 195–201 [DOI] [PubMed] [Google Scholar]
- 42. Arnold K., Kiefer F., Kopp J., Battey J. N., Podvinec M., Westbrook J. D., Berman H. M., Bordoli L., Schwede T. (2009) J. Struct. Funct. Genomics 10, 1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chen R., Li L., Weng Z. (2003) Proteins 52, 80–87 [DOI] [PubMed] [Google Scholar]
- 44. Kayser J. P., Vallet J. L., Cerny R. L. (2004) J. Biomol. Tech. 15, 285–295 [PMC free article] [PubMed] [Google Scholar]
- 45. Schöning J. C., Streitner C., Page D. R., Hennig S., Uchida K., Wolf E., Furuya M., Staiger D. (2007) Plant J. 52, 1119–1130 [DOI] [PubMed] [Google Scholar]
- 46. Asai S., Ohta K., Yoshioka H. (2008) Plant Cell 20, 1390–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mavrodi D. V., Joe A., Mavrodi O. V., Hassan K. A., Weller D. M., Paulsen I. T., Loper J. E., Alfano J. R., Thomashow L. S. (2011) J. Bacteriol. 193, 177–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Abramoff M. D., Magelhaes P. J., Ram S. J. (2004) Biophoton. Int. 11, 36–42 [Google Scholar]
- 49. Yuan J., He S. Y. (1996) J. Bacteriol. 178, 6399–6402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Crabill E., Joe A., Block A., van Rooyen J. M., Alfano J. R. (2010) Plant Physiol. 154, 233–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Espinosa A., Guo M., Tam V. C., Fu Z. Q., Alfano J. R. (2003) Mol. Microbiol. 49, 377–387 [DOI] [PubMed] [Google Scholar]
- 52. Rost B., Yachdav G., Liu J. (2004) Nucleic Acids Res. 32, W321–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Rao S. T., Rossmann M. G. (1973) J. Mol. Biol. 76, 241–256 [DOI] [PubMed] [Google Scholar]
- 54. Han S., Tainer J. A. (2002) Int. J. Med. Microbiol. 291, 523–529 [DOI] [PubMed] [Google Scholar]
- 55. Maris C., Dominguez C., Allain F. H. (2005) FEBS J. 272, 2118–2131 [DOI] [PubMed] [Google Scholar]
- 56. Tsuge H., Nagahama M., Oda M., Iwamoto S., Utsunomiya H., Marquez V. E., Katunuma N., Nishizawa M., Sakurai J. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 7399–7404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wilton M., Subramaniam R., Elmore J., Felsensteiner C., Coaker G., Desveaux D. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 2349–2354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Singer A. U., Desveaux D., Betts L., Chang J. H., Nimchuk Z., Grant S. R., Dangl J. L., Sondek J. (2004) Structure 12, 1669–1681 [DOI] [PubMed] [Google Scholar]
- 59. Ding J., Hayashi M. K., Zhang Y., Manche L., Krainer A. R., Xu R. M. (1999) Genes Dev. 13, 1102–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Nagai K., Oubridge C., Jessen T. H., Li J., Evans P. R. (1990) Nature 348, 515–520 [DOI] [PubMed] [Google Scholar]
- 61. Lee A. L., Volkman B. F., Robertson S. A., Rudner D. Z., Barbash D. A., Cline T. W., Kanaar R., Rio D. C., Wemmer D. E. (1997) Biochemistry 36, 14306–14317 [DOI] [PubMed] [Google Scholar]
- 62. Galán J. E. (2009) Cell Host Microbe 5, 571–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Cléry A., Blatter M., Allain F. H. (2008) Curr. Opin. Struct. Biol. 18, 290–298 [DOI] [PubMed] [Google Scholar]
- 64. Ludden P. W., Roberts G. P. (1989) Curr. Top. Cell Regul. 30, 23–56 [DOI] [PubMed] [Google Scholar]
- 65. Heintzen C., Nater M., Apel K., Staiger D. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 8515–8520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Streitner C., Hennig L., Korneli C., Staiger D. (2010) BMC Plant Biol. 10, 221–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.