Background: PKCα contains a unique PDZ ligand motif and is known to promote cellular migration.
Results: PKCα binds and phosphorylates the scaffold DLG1; both proteins are necessary for cellular migration in non-small cell lung cancer cells.
Conclusion: DLG1 coordinates PKCα signaling to promote cellular migration.
Significance: Control of PKCα signaling mediated by scaffolds is crucial to promoting its downstream functions.
Keywords: Lung Cancer, Migration, Protein Kinase C (PKC), Protein Phosphorylation, Scaffold Proteins, DLG1
Abstract
Protein scaffolds maintain precision in kinase signaling by coordinating kinases with components of specific signaling pathways. Such spatial segregation is particularly important in allowing specificity of signaling mediated by the 10-member family of protein kinase C (PKC) isozymes. Here we identified a novel interaction between PKCα and the Discs large homolog (DLG) family of scaffolds that is mediated by a class I C-terminal PDZ (PSD-95, disheveled, and ZO1) ligand unique to this PKC isozyme. Specifically, use of a proteomic array containing 96 purified PDZ domains identified the third PDZ domains of DLG1/SAP97 and DLG4/PSD95 as interaction partners for the PDZ binding motif of PKCα. Co-immunoprecipitation experiments verified that PKCα and DLG1 interact in cells by a mechanism dependent on an intact PDZ ligand. Functional assays revealed that the interaction of PKCα with DLG1 promotes wound healing; scratch assays using cells depleted of PKCα and/or DLG1 have impaired cellular migration that is no longer sensitive to PKC inhibition, and the ability of exogenous PKCα to rescue cellular migration is dependent on the presence of its PDZ ligand. Furthermore, we identified Thr-656 as a novel phosphorylation site in the SH3-Hook region of DLG1 that acts as a marker for PKCα activity at this scaffold. Increased phosphorylation of Thr-656 is correlated with increased invasiveness in non-small cell lung cancer lines from the NCI-60, consistent with this phosphorylation site serving as a marker of PKCα-mediated invasion. Taken together, these data establish the requirement of scaffolding to DLG1 for PKCα to promote cellular migration.
Introduction
Targeting of signaling proteins to specific intracellular locations via scaffolding proteins allows signals to be efficiently and selectively integrated, propagated, and regulated (1). Scaffolds serve a particularly important function in the organization of signaling by protein kinases because kinase substrate selectivity is determined by not only consensus motifs but also availability of substrates near the active site (2).
The PKC branch of the AGC kinase family tree has 10 family members that are regulated not only by phosphorylation and binding to lipid second messengers but also by interaction with binding partners. PKC isozymes are grouped into the following three classes based upon their cofactor dependence: the conventional PKC (cPKC)5 isozymes α, the alternatively spliced βI and βII, and γ, which depend on diacylglycerol and Ca2+ for their activity; the novel PKCs δ, ϵ, η, and θ, which depend on diacylglycerol; the atypical PKCs ζ and λ/ι, which rely mainly on protein-protein interactions for activation (3). Although the stimuli governing PKC activation have been extensively characterized, assigning specific biological roles to PKC isozymes has proved more difficult, partially because of the divergent roles of the different family members in various cellular processes (4). However, ever since the discovery that PKCs act as receptors for tumor-promoting phorbol esters (5), these enzymes have been hypothesized to positively regulate tumor progression and metastasis. Indeed, there is an increasing body of evidence implicating PKCα, in particular, in cancer cell survival and migration (6–10). In this regard, an antisense molecule targeting PKCα has been shown to have preclinical efficacy in various cancer models, including advanced non-small cell lung cancer (11, 12). Note, however, that the effects of PKCα signaling are complex because several types of cancer cells, including those generated in mouse models of colon cancer, show dramatically reduced levels of PKCα (13). Cell type-dependent differences in PKCα signaling are likely accounted for by differential interactions with key regulatory proteins, notably scaffold proteins.
PKC protein scaffolds have long been known to play essential roles in PKC function. Mochly-Rosen et al. (14) first described the role of protein scaffolds in directing the cellular function of PKC with the identification of proteins they named receptors for activated C kinase (RACKs). Since then numerous PKC-binding proteins have been identified and shown to regulate PKC in many ways, including 1) relieving PKC autoinhibition, 2) mediating PKC association with the actin cytoskeleton, 3) controlling the availability of upstream regulators of PKC, and 4) mediating PKC interaction with receptors, small GTPases, and other signaling proteins (10, 15, 16). These interactions play important roles in regulating PKC function, notably the transmission of signals from sites of cell-cell or cell-matrix contact to the cytoskeleton, with resulting effects on cell spreading and migration (2, 16). The key role of scaffolding in PKC signaling is epitomized by an elegant study by Zuker and co-workers showing that the PDZ (PSD-95, disheveled, and ZO1) domain-containing protein encoded by the inaD gene, which scaffolds PKC, is required for light-induced PKC signaling in the fly eye (17). The binding of Drosophila eye PKC to this scaffold is mediated by binding of a C-terminal PDZ ligand, which has the amino acid sequence ITII (17, 18). PDZ ligand interactions are powerful coordinators of cell signaling (19), yet their roles in signaling by mammalian PKC isozymes are relatively unexplored.
Of the eight diacylglycerol-regulated PKC isozymes, only PKCα contains a C-terminal PDZ ligand motif. The last four amino acids of this isozyme (QSAV) encode a class I PDZ ligand. PDZ ligands bind PDZ domains, which are relatively small globular domains (∼90 amino acid) that are abundant in the mammalian proteome; their canonical role is to bind short C-terminal peptide motifs (20). The only identified partner for the PDZ ligand of mammalian PKCα is the PDZ scaffold PICK1 (protein that interacts with C kinase 1) (21). The PKCα PDZ ligand has been shown to be necessary and sufficient for long term depression in cerebellar cultures (22). In vitro, peptides containing this motif bind numerous murine PDZ domains, including the third PDZ domain of all four members of the membrane-associated guanylate kinase (MAGUK) protein scaffold family. In rat brain extracts, PKCα has been shown to co-immunoprecipitate with the MAGUK SAP-102 (synapse-associated protein 102; also known as Discs large homolog 3) and PSD-95, with these interactions hypothesized to be mediated by the first and second PDZ domains of the MAGUKs (23). In addition, PKC has been shown to exist in a complex with protein kinase A, protein kinase A-anchoring protein 150, and PSD-95 or SAP-97 (synapse-associated protein 97; also known as DLG1 or Discs large homolog 1) (23, 24). Although the direct binding of the PDZ ligand of PKCα to these MAGUK proteins was not explored and no functional studies were performed to address the physiological importance of PKCα/MAGUK associations, these data suggest the possibility that PDZ interactions may coordinate the signaling of PKCα.
In this study we used a PDZ domain array to identify the third PDZ domains of DLG1/SAP-97 and DLG4/PSD95 as binding partners for the PDZ ligand of PKCα. Biochemical studies validated the interaction in cells, established that PKCα phosphorylates DLG1 on a novel site, Thr-656, and showed that both the PDZ ligand of PKCα and DLG1 are required for PKCα to promote cellular migration. Our data are consistent with a model in which DLG1 acts as a scaffold for PKCα to control PKC-dependent regulation of cellular migration.
EXPERIMENTAL PROCEDURES
Materials and Antibodies
Mitomycin C, Gö6983, Gö6976, phorbol myristate acetate (PMA), and calyculin A were purchased from Calbiochem. A control siRNA SmartPool and a siRNA SmartPool targeting human DLG1 were purchased from Dharmacon; the latter included the following sequences: #1, CCAAAAUGUAUAGAUCGUU; #2, CGAUGAGGUCGGAGUGAUU; #3, CCAGGAACAUAAAUUCAUU; #4, CCCACAAGUAUGUAUAUGA. A second siRNA (referred to as “DLG1 siRNA #2”) used to validate the effect of DLG1 depletion on wound healing was purchased from Sigma along with a universal negative control siRNA and had the targeting sequence CAGAGAAGAACUUAUCAGA. Two shRNAs against human PKCα and a non-targeting control lentivirus were obtained from Sigma. Sequences of the shRNAs were as follows (with target sequences underlined): PKCα #1, CCGGCAUGGAACUCAGGCAGAAAUUCUCGAGAAUUUCUGCC; PKCα #2, CCGGCGAGCTATTTCAGTCTATCATCTCGAGATGATAGACTGAAATAGCTCGTTTTT; non-targeting control, CCGGCAACAAGAUGAAGAGCACCAACUCGAG. PKCα shRNA #1 was used in all experiments except where otherwise noted. Primers used for PCR-based cloning were from IDT. All restriction enzymes were from New England Biolabs. Antibodies and dilutions used were: mouse anti-Myc (1:1000, 9E10; Covance), rat anti-HA (1:2000; Roche Diagnostics), rabbit anti-GST (1:1000; Sigma), mouse anti-SAP97/DLG1 (1:500 for Western blot, 1:200 for immunofluorescence (IF) staining; Stressgen), rabbit anti-PKCα (1:1000 for Western blot, 1:200 for IF staining; Santa Cruz), mouse anti-β-actin (1:1000; Sigma), and mouse anti-hsp70 (heat shock protein 70, 1:1000; BD Biosciences). Secondary antibodies used for immunofluorescence were goat anti-mouse-Alexa 568, goat anti-rabbit-Alexa 488, goat anti-mouse-Alexa 488, and goat anti-rabbit-Alexa 568, all from Invitrogen. The Thr(P)-656 antibody was raised by immunizing rabbits with an Ac-CKERARLK-T(PO3H2)-VKFN-NH2 peptide that was conjugated to keyhole limpet hemocyanin and was affinity-purified using the phosphopeptide antigen (NeoMPS). This antibody was used at 1:5000 for Western blot. All other materials were reagent grade.
Construction of Plasmids
A cDNA fragment encoding the last 25 amino acids of bovine PKCα was ligated into pGEX-6-P3 (Amersham Biosciences) to generate a GST fusion peptide (GST-PDZα). Sequences for bovine PKCα and PKCα lacking the last three amino acids (PKCαΔPDZ) were cloned into pcDNA3-HA by PCR, generating HA-PKCα and HA-PKCαΔPDZ. CFP-tagged DLG1/SAP97 (i3) was a generous gift from M. Dell'Acqua, and Myc-DLG1 was a generous gift from C. Garner. All mutagenesis was performed using a QuikChange kit (Stratagene) according to the manufacturer's instructions.
Purification of GST-tagged Proteins
The GST-PDZα construct was transformed into BL21 (DE3) cells, which were grown at 37 °C until their A600 reached 0.6 and then induced with 1 mm isopropyl 1-thio-β-d-galactopyranoside for 4 h at 25 °C. Cells were pelleted and homogenized in a buffer containing 50 mm Tris (pH 7.5), 1 mm EDTA, 1 mm DTT, 300 nm PMSF, 500 nm benzamidine, 500 ng/ml leupeptin, and 1 mg/ml lysozyme. The lysates were rocked for 30 min at 4 °C and briefly sonicated before treatment with DNase (100 μg/ml) and centrifugation at 14,000 × g for 30 min at 4 °C. The fusion peptide was purified from the filtered supernatant using the Profinia Protein Purification System (Bio-Rad) according to the manufacturer's specifications. The eluted pure protein was dialyzed against 20 mm HEPES (pH 7.5)/50 mm NaCl.
Peptide Overlay Array
An array of 96 PDZ domains was spotted onto membranes as described previously (25). Purified GST-PDZα (0.5 mg/ml) was overlaid onto the array and detected using a far Western blot approach, as previously described (26).
Dot Blot Validation of Thr(P)-656 Antibody
To analyze the specificity of the Thr(P)-656 antibody, phosphorylated (Ac-CKERARLK-T(PO3H2)-VKFN-NH2) and unphosphorylated (Ac-CKERARLK-TVKFN-NH2) peptides were synthesized by NeoMPS and spotted onto nitrocellulose membranes. Dot blots were incubated with various concentrations of the Thr(P)-656 antibody and analyzed by Western blot.
Cell Culture, PMA Stimulation Experiments, and Western Blotting
Unless otherwise noted, cells were maintained in DMEM (Cellgro) supplemented with 10% fetal bovine serum (FBS, Hyclone) and 1% penicillin/streptomycin (P/S), except for SNB-19, NCI-H322M, NCI-H23, A549, and HOP62 cells, which were cultured in RPMI 1640 (Cellgro) with 10% FBS and 1% P/S. Immortalized PKCα+/+ and PKCα−/− mouse embryonic fibroblasts (MEFs) were generous gifts from M. Leitges, primary astrocytes were isolated as described below, H1703 and SNB-19 cells were purchased from ATCC, and NCI-H322M, NCI-H23, A549, and HOP62 cells were gifts from the NCI. Cells were incubated at 37 °C, 5% CO2. For PMA stimulation experiments, cells were treated for the indicated times at 37 °C with PMA (200 nm), Gö6976 (500 nm), Gö6983 (250 nm), and/or calyculin A (100 nm). Unless otherwise noted, all stock solutions used were in DMSO, and a corresponding amount of DMSO was used as a control. The final concentration of DMSO in the culture media did not exceed 0.4% (v/v). For immunoblotting, primary astrocytes and H1703 cells were lysed in 1× Laemmli sample buffer, and the NCI-60 NSCLC lines and PKCα+/+ and PKCα−/− MEFs were lysed in buffer consisting of 50 mm Tris (pH 7.4), 150 mm NaCl, 1% Triton, 0.5% sodium deoxycholate, 0.1% SDS, 30 mm sodium pyrophosphate, 0.1 mm sodium vanadate, 200 mm benzamidine, 40 mg/ml leupeptin, and 1 mm PMSF. MEF and NSCLC lysate protein concentrations were quantified using a BCA protein assay kit (Thermo Scientific) and normalized. Levels of total and phosphorylated proteins were analyzed by SDS-PAGE and Western blotting.
Immunoprecipitation
HA-PKCα or HA-PKCαΔPDZ and Myc-DLG1 were transfected into HEK293T cells using Effectene (Qiagen) according to the manufacturer's instructions. Approximately 24 h later cells were lysed in immunoprecipitation buffer (50 mm Tris (pH 7.5), 10 mm sodium pyrophosphate, 50 mm NaF, 5 mm EDTA, 1% Triton, 1 mm DTT, 200 mm benzamidine, 40 mg/ml leupeptin, and 1 mm PMSF); the lysates were then cleared by centrifugation at 16,000 × g for 5 min at 22 °C and incubated with a anti-HA antibody (Covance; monoclonal; 1:450) overnight at 4 °C with rocking. In the morning, Ultra-Link Protein A/G beads (Thermo Scientific) were added to the immune complexes and incubated for 1 h at 4 °C with rocking. Beads were then washed with immunoprecipitation buffer followed by immunoprecipitation buffer containing 50 mm NaCl and analyzed by SDS-PAGE and Western blotting.
Primary Astrocyte Isolation
After isolation as described in Citro et al. (27), adherent astrocytes were washed twice with PBS, trypsinized, and plated at ∼3 × 105 cells per ml on lysine-coated six-well dishes. All astrocyte preparations that were subjected to wound-healing analysis had at least an 80% glial fibrillary acidic protein-positive cells by immunofluorescence, and experiments were performed on cells derived from three different preparations.
Wound-healing Assay
Primary murine astrocytes, SNB-19 glioblastoma cells, and H1703 NSCLC cells were plated on lysine-coated six-well dishes at consistent cell densities. Approximately 48 h after plating, confluent monolayers were treated with mitomycin C (10 μg/ml, stock solution dissolved in PBS) in serum-free DMEM or RPMI for 1 h to inhibit cell proliferation and then washed twice with PBS. Fresh DMEM or RPMI containing 10% FBS and either DMSO or Gö6976 (500 nm) was added to the cells. After 20 min of pretreatment, monolayers were scratched once with a 10-μl pipette tip, and pictures of the central region of the scratch were taken immediately and at various time points after scratching with a 5× objective lens. During the assay cells were maintained at 37 °C, 5% CO2. The wound area at the various time points was quantified using ImageJ (National Institutes of Health).
Immunofluorescence
Primary murine astrocytes were prepared and scratched as described above, except that they were plated on lysine-coated glass cover slips. Four hours after being scratched, cells were washed twice in cold PBS, fixed with 3% paraformaldehyde and 2% sucrose in PBS (pH 8.0) for 20 min, quenched in 50 mm NH4Cl in 10 mm PIPES (pH 6.8), 150 mm NaCl, 5 mm EGTA, 5 mm glucose, and 5 mm MgCl2 for 15 min, and blocked in blocking buffer (10% goat serum in PBS, 0.1% Triton) for 1 h before overnight incubation at 4 °C with primary antibodies diluted in blocking buffer. After washing, the indicated secondary antibodies were added at a dilution of 1:600 in PBS, 0.1% Tween for 1 h at 4 °C. Washed slides were mounted using Vectashield and photographed on a Zeiss Axiovert microscope (Carl Zeiss Microimaging, Inc.) using a MicroMax digital camera (Roper-Princeton Instruments) controlled by MetaFluor software (Universal Imaging, Corp.). Optical filters were obtained from Chroma Technologies. PKCα/DLG1 colocalization was assessed in a blinded fashion for at least 30 cells per condition per experiment (n = 5 experiments using 3 different astrocyte preparations).
Lentiviral shRNA Production and Infection
A shRNA targeting human PKCα was packaged into recombinant lentiviruses using the Invitrogen ViraPower™ Lentiviral Expression System according to the manufacturer's protocol. A non-targeting lentiviral RNAi (NT-RNAi) that recognizes no human genes was used as a negative control. For lentiviral shRNA infection, H1703 cells were seeded in 100-mm plates and grown to 70–80% confluency. The culture medium was removed from the cells, and 3 ml of complete culture media containing Polybrene (6 mg/ml) was added. After 5 min at 22 °C, 400 ml (multiplicity of infection ∼3) of viral supernatant was added. After a 24-h incubation at 37 °C, cells were washed and grown for 24 h in 10 ml of fresh culture medium. Populations of stably infected cells were selected in 5 mg/ml puromycin.
Re-expression of PKCα in Stable PKCα Knockdown Cells
HA-tagged bovine PKCα constructs, which are resistant to knockdown by our shRNA construct targeted against human PKCα, were transfected into NT shRNA- or PKCα shRNA-expressing H1703 cells as follows. Cells were plated into 24-well plates and, 3 h after plating, transfected with enhanced GFP alone or with HA-PKCα (wild type; 0.25 μg/well) or HA-PKCαΔPDZ (0.2 μg/well) using Jetprime transfection reagent (Polyplus) according to the manufacturer's instructions. Fresh culture media was added 4 h after transfection, and cells were incubated overnight before being replated onto lysine-coated 6-well plates at consistent cell densities. Wound-healing assays were performed ∼12 h after re-plating. At least 70% of cells were transfected (as marked by GFP fluorescence at the time of scratch), and cells were lysed in 1× Laemmli buffer immediately after completion of the scratch assay for validation of PKCα expression.
Transient siRNA Transfection
H1703 cells were transiently transfected with control or DLG1 siRNA (50 nm) and subjected to wound-healing analysis as described previously (28) with minor modifications. Briefly, cells were transfected using Lipofectamine 2000 (Invitrogen) in DMEM without FBS and penicillin/streptomycin 3 h after plating and incubated overnight before the addition of fresh media with 10% FBS and penicillin/streptomycin. At 48 h after transfection cells were replated onto lysine-coated 6-well dishes at consistent cell densities. Wound-healing assays were performed starting at ∼72 h after transfection, and cells were lysed in 1× Laemmli sample buffer immediately upon completion of the assay for verification of DLG1 knockdown by Western blot.
Identification and Alignment of PKCα Phosphorylation Site
A consensus PKCα/β/γ phosphorylation site in DLG1 was identified using Scansite, and a comparison of amino acid motifs in the human DLG family and in DLG1 isoforms from various species was performed using MegAlign.
Statistical Analyses
Differences among groups were analyzed using Student's t test or analysis of variance (ANOVA), with significance set at p < 0.05. For multiple comparisons, a post-hoc Tukey was applied after the ANOVA.
RESULTS
The PDZ Ligand of PKCα Mediates Binding to DLG1
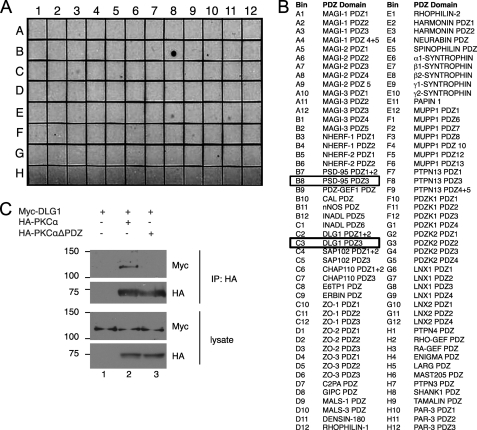
To identify potential binding partners for the PDZ ligand of PKCα, we generated a GST fusion of the last 25 amino acids of bovine PKCα and overlaid it onto a PDZ domain array containing 96 PDZ domains (mostly Type I PDZ domains) from various proteins (Fig. 1B). Far Western blotting for GST revealed that the C-terminal peptide of PKCα bound strongly to the third (but not first and second) PDZ domain of PSD-95 (also known as DLG4) (Fig. 1A, and B8). In addition, we identified weaker, but readily detectable binding to the third (but not first and second) PDZ domain of DLG1 (SAP97) (Fig. 1A, C3), β2-syntrophin (E8), PAPIN 1 (E11), and PTPN13 (F9). In contrast to PKCα, the last 25 amino acids of PKCζ, the only other PKC isozyme that contains a PDZ ligand motif, failed to bind to any of the PDZ domains on the array.6 Because the PKCζ PDZ ligand (EESV) is predicted to bind only Type III PDZ domains (20), this finding was not unexpected and points to a specific PDZ-based interaction of PKCα with DLG scaffolds. The binding of the PKCα PDZ ligand to the third PDZ domain of two DLG scaffolds prompted us to focus on this interaction. To validate the PKCα/DLG1 interaction, we overexpressed HA-tagged bovine PKCα and Myc-tagged rat DLG1 in HEK293T cells and asked if DLG1 was present in immunoprecipitates of PKCα. Fig. 1C shows that Myc-DLG1 was present in HA-PKCα immunoprecipitates (lane 2) but not control immunoprecipitates (lane 1). This interaction depended on the PDZ ligand of PKCα; DLG1 did not co-immunoprecipitate with a construct of PKCα lacking the last three amino acids (PKCαΔPDZ; lane 3). Similar results were obtained for the interaction between PKCα and PSD95 (data not shown). These results reveal that the PDZ ligand of PKCα mediates the binding of PKCα to the third PDZ domain of DLG1/SAP97 and DLG4/PSD95.
FIGURE 1.
PKCα interacts via its PDZ ligand with the PDZ domain scaffold DLG1/SAP97. A, PDZ domain array overlay is shown. The last 25 amino acids of PKCα were tagged with GST and overlaid on an array of 96 PDZ domains. B, shown is a list of PDZ domains in the array shown in A, with two positive interactions boxed. C, co-immunoprecipitation of PKCα and DLG1 is shown. Myc-tagged DLG1 was expressed alone (lane 1) or in combination with HA-tagged PKCα (lane 2) or HA-tagged PKCα lacking the last three amino acids (HA-PKCαΔPDZ; lane 3) in HEK293T cells, PKCα was immunoprecipitated (IP) using the HA tag, and immunoprecipitates were probed for Myc-DLG1 or HA-PKCα.
PKCα and DLG1 Colocalize at the Leading Edge of Migrating Cells
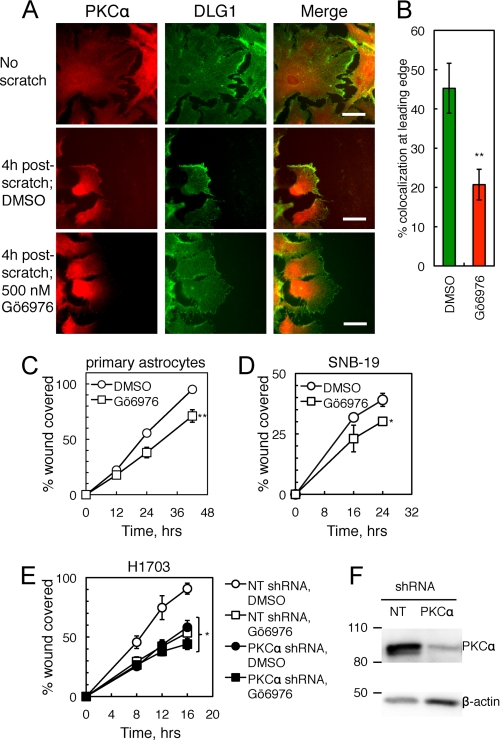
Given the scaffolding interaction between PKCα and DLG1, we asked whether these two proteins colocalize in cells. Specifically, we examined the distribution of the two proteins in primary murine astrocytes in a wound-healing assay (as described in Valster et al. (28)). Detection of endogenous PKCα or DLG1 by immunofluorescence revealed that the two proteins codistributed at the leading edge in ∼30% of migrating primary astrocytes 4 h after scratch (Fig. 2A, middle row) but not at the membranes of unscratched cells (Fig. 2A, top row). This scratch-induced codistribution depended on PKC activity because pretreatment with the cPKC inhibitor Gö6976 decreased codistribution (Fig. 2A, bottom row). Blinded scoring of DLG1 and PKCα colocalization in >30 cells over three different astrocyte preparations (n = 5 experiments) revealed that treatment with Gö6976 resulted in a ∼60% decrease in colocalization of the two proteins (Fig. 2B). These data indicate that PKCα and DLG1 codistribute at the leading edge of migrating cells and that this colocalization depends on cPKC activity.
FIGURE 2.
PKCα and DLG1 promote cell migration in wound-healing assays. A, PKCα and DLG1 colocalization at the leading edge of migrating astrocytes is shown. Primary murine astrocytes were pretreated with Gö6976 (500 nm; bottom row) or a corresponding amount of DMSO (middle row), scratched with a 10-μl pipette tip, fixed 4 h after scratch, and stained for endogenous PKCα and DLG1. Scale bars: 50 nm. B, shown is quantification of the percent of cells with colocalization of PKCα and DLG1 at the leading edge of migrating cells (n = 5 experiments with >30 cells per condition in each experiment). C–E, shown is the effect of cPKC inhibition on wound healing. Primary astrocytes (C), SNB-19 glioblastoma cells (D), and stable non-targeting shRNA-expressing (NT; open symbols) or PKCα shRNA-expressing (filled symbols) cell lines derived from H1703 NSCLC cells (E) were pretreated with mitomycin C (to inhibit proliferation) followed by DMSO (circles) or Gö6976 (squares), scratched with a pipette tip, and followed over 12–42,16–24, or 8–16 h, respectively. The area covered by migrating cells was quantified using ImageJ; data points represent the mean ± S.E. of at least three experiments. F, a Western blot shows PKCα knockdown efficiency for a representative experiment as in E. After completion of the scratch assay, cells were lysed, and lysates were probed for PKCα and the loading control β-actin. *, significantly different from control DMSO-treated cells, p < 0.05; **, p < 0.01.
PKCα Positively Regulates Wound Healing
Given the codistribution of PKCα and DLG1 at the leading edge of migrating cells, we next asked whether the activity of PKC enhances wound healing. Using the same wound-healing paradigm described above, we examined the effect of inhibition of cPKC activity by pretreatment with Gö6976 on the rate of migration of primary astrocytes (Fig. 2C), SNB-19 glioblastoma cells (Fig. 2D), or H1703 NSCLC cells (Fig. 2E). In all three cell types, Gö6976 (Fig. 2E, squares) reduced the rate of cellular migration by ∼25–40% compared with vehicle treatment (circles). To determine whether the Gö6976 sensitivity reflected exclusively inhibition of PKCα, H1703 cells were stably transfected with a NT shRNA (Fig. 2E, open symbols) or an shRNA targeting PKCα (filled symbols), and the sensitivity of cellular migration to Gö6976 was tested in wound-healing assays. Importantly, PKCα knockdown and Gö6976 treatment did not have an additive effect on the inhibition of cellular migration (Fig. 2E, filled squares). Furthermore, the rate of migration of control cells treated with the PKC inhibitor (open squares) was the same as that of cells depleted of PKCα (filled circles). Western blot analysis of lysates revealed that PKCα was knocked down by 70 ± 10% (Fig. 2F). To further validate the specific role of PKCα in wound healing, we depleted H1703 cells of PKCα using a second shRNA, resulting in 60 ± 10% inhibition of PKCα expression and a 70% decrease in wound healing (supplemental Fig. 1). The inability of Gö6976 to further inhibit migration in PKCα-depleted cells reveals that PKCα is the major cPKC promoting cell migration.
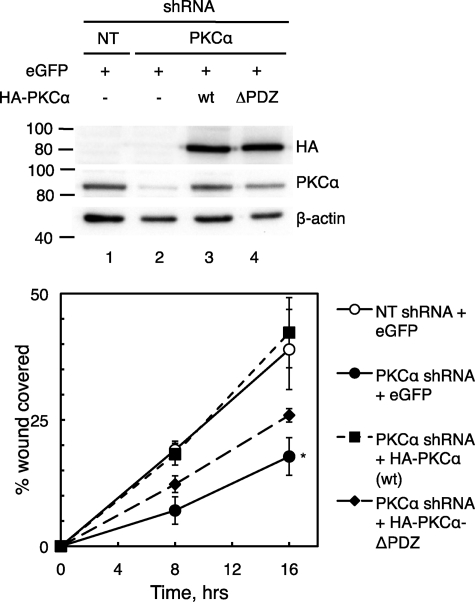
The PDZ Ligand of PKCα Is Necessary for Its Ability to Promote Wound Healing
We next attempted to rescue the effects of PKCα knockdown on wound healing by expressing an shRNA-resistant bovine form of PKCα in PKCα-depleted H1703 cells (Fig. 3). Re-expression of wild type PKCα at a level close to that of control cells (Fig. 3, compare lanes 1 and 3) resulted in a complete rescue of wound healing (Fig. 3, filled squares), whereas expression of a PKCα mutant lacking the last three amino acids (PKCαΔPDZ) only slightly increased the rate of wound healing (Fig. 3, filled diamonds). These data support the conclusion that the PKCα PDZ ligand, which mediates its interaction with DLG1, is required for a large portion of the positive effects of PKCα on cellular migration.
FIGURE 3.
Wild type PKCα but not PKCα lacking the last three amino acids rescues wound healing in PKCα-depleted H1703 cells. Upper panel, a Western blot shows re-expression of bovine wild type (wt) HA-tagged PKCα or PKCα lacking the last three amino acids (ΔPDZ) in H1703 cells stably expressing NT or PKCα shRNA in a wound healing assay as described below. After completion of the scratch assay, cells were lysed, and lysates were probed for PKCα, HA, and β-actin. Lower panel, shown is the effect of PKCα rescue on wound healing. The indicated H1703 cells were transiently transfected with enhanced GFP (eGFP; as a marker for transfection) with or without HA-PKCα or HA-PKCα-ΔPDZ. Cells were plated at equal densities and then pretreated with mitomycin C (to inhibit proliferation) and scratched with a pipette tip, and their migration was monitored over 8–16 h. The area covered by migrating cells was quantified using ImageJ, and data points represent the mean ± S.E. of four experiments. *, significantly different from the NT shRNA + eGFP condition, p < 0.05.
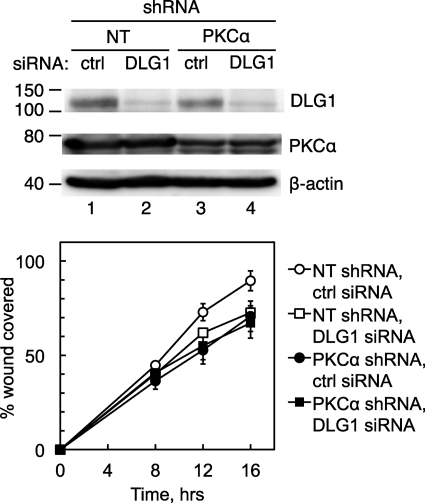
DLG1 Depletion Blocks the Ability of PKCα to Promote Cellular Migration
We next asked whether DLG1 is necessary for PKCα to promote cell migration by examining the effect of inhibiting conventional PKC activity on cell migration in control cells, cells lacking PKCα, or cells lacking PKCα and DLG1. Specifically, stably transfected NT or PKCα shRNA-expressing H1703 cells were transiently transfected with a control siRNA (ctrl siRNA; Fig. 4, circles) or a siRNA targeting human DLG1 (DLG1 siRNA; Fig. 4, squares), and cell migration was measured in wound-healing assays. DLG1 siRNA transfection resulted in an ∼50% reduction in DLG1 levels (n = 3; Fig. 4, upper panel); these cells displayed a reduction in migration that was similar to that observed in cells depleted of PKCα (Fig. 4, lower panel). To further validate the specific role of DLG1 in wound healing, we depleted H1703 cells of DLG1 using a second siRNA sequence, resulting in 64 ± 7% inhibition of DLG1 expression and a 20% decrease in wound healing, similar to that observed with the original siRNA (supplemental Fig. 2). Strikingly, no further reduction in migration was observed in cells depleted of both DLG1 and PKCα (Fig. 4, filled squares). These data suggest that the regulation of cellular migration by PKCα depends on DLG1.
FIGURE 4.
Interfering with DLG1 blocks the effects of PKCα knockdown on wound healing. Upper panel, a Western blot shows DLG1 and PKCα knockdown efficiencies for a representative wound-healing experiment as described below. After completion of the scratch assay, cells were lysed, and lysates were probed for PKCα, DLG1, and actin. Lower panel, shown is the effect of combined PKCα and DLG1 knockdown on wound healing. H1703 NSCLC cells stably expressing a NT or a PKCα shRNA were transiently transfected with a scrambled siRNA pool (ctrl) or a siRNA pool targeting DLG1, re-plated, and subjected to a wound healing assay as described for Fig. 3. Results reflect the means ± S.E. of three experiments.
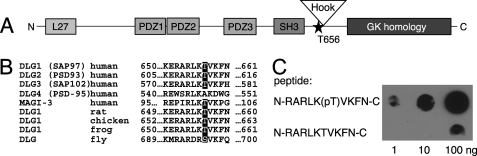
PKCα Phosphorylates DLG1 at Thr-656
Analysis of the sequence of full-length DLG for predicted phosphorylation sites (ScanSite) revealed the presence of a putative cPKC phosphorylation site at Thr-656. This residue lies between the SH3 and Hook domains of DLG1 (Fig. 5A). Alignment of DLG isoforms and MAGI-3 (Fig. 5B) revealed that this phosphorylation site is conserved in all of these proteins except PSD-95, which has an Ala at the potential phospho-acceptor site. Furthermore, this potential phosphorylation site is conserved among species, including Drosophila. To identify whether this site is phosphorylated in cells, we generated a phospho-specific antibody using a phosphorylated peptide corresponding to the region surrounding Thr-656. Fig. 5C shows that this antibody (Thr(P)-656) recognizes the phosphorylated peptide with almost 100-fold selectivity compared with the unphosphorylated peptide.
FIGURE 5.
DLG1 contains a conserved conventional PKC phosphorylation site. A, domain architecture of rat DLG1/SAP97 shows the location of a putative PKC phosphorylation site, identified using Scansite, at threonine 656. B, alignment shows conservation of the PKC phosphorylation site in three of four human DLG isoforms and DLG isoforms from lower organisms. C, shown is generation of an antibody specific for DLG1 phosphorylated at Thr-656. Various amounts of phospho- and de-phosphopeptides corresponding to residues 649–661 of human DLG1 were spotted onto nitrocellulose and overlaid with the phospho-specific (Thr(P)-656) antibody.
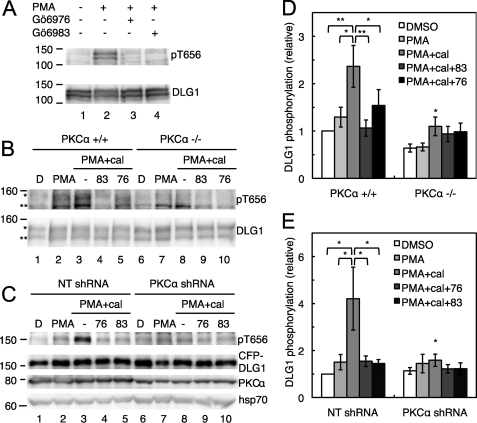
We next tested whether Thr-656 is phosphorylated in cells. Primary astrocytes, chosen because of their high level of expression of DLG1, were treated with 1) PMA, which acutely stimulates PKC activity, 2) Gö6983, which inhibits conventional and novel PKC isoforms, and 3) Gö6976, which inhibits only the conventional PKC isoforms. Western blot analysis of astrocyte lysates in Fig. 6A shows that an antibody for total DLG1 recognized a doublet that likely corresponds to splice variants of DLG1 (30). These bands were strongly labeled by the Thr(P)-656 antibody in lysates from cells treated with PMA (lane 2) but not vehicle alone (lane 1). Importantly, Thr(P)-656 labeling was abolished in cells pretreated with the PKC inhibitors Gö6976 (lane 3) or Gö6983 (lane 4) before PMA stimulation.
FIGURE 6.
PKCα phosphorylates DLG1 at Thr-656. A, cPKC-dependent phosphorylation of DLG1 is shown. Primary murine astrocytes were treated for 30 min with vehicle or PMA (lanes 2–4) and Gö6976 (lane 3) or Gö6983 (lane 4). Cells were lysed, and lysates were probed for levels of phospho- and total DLG1. B, DLG1 phosphorylation in the presence and absence of PKCα is shown. Immortalized wild type MEFs (PKCα+/+) or MEFs lacking the gene for PKCα (PKCα−/−) were pretreated with DMSO (denoted by D), Gö6983 (83), or Gö6976 (76) for 10 min before the addition of PMA for 20 min and the phosphatase inhibitor calyculin A (cal) for the last 2 min of PMA treatment. Cells were lysed, and lysates were probed for DLG1 phosphorylated at Thr-656 (pT656; relevant bands are marked with asterisks) and total DLG1 levels. C, shown are changes in DLG1 phosphorylation upon PKCα knockdown. H1703 NSCLC cells stably expressing a NT or a PKCα shRNA were transfected with CFP-tagged DLG1 and treated as described in B; lysates were probed for Thr(P)-656, DLG1, PKCα, and the loading control heat shock protein 70 (hsp70). D, shown is a bar graph representing the mean ± S.E. of 8 experiments as described in B. E, shown is a bar graph representing the mean ± S.E. of 7 experiments as described in C. Significantly different from control PMA+cal-treated cells; *, p < 0.05; **, p < 0.01.
To determine whether PKCα is the primary kinase that phosphorylates DLG1 at Thr-656, we examined the phosphorylation state of this residue in MEFs lacking the gene for PKCα (PKCα−/−) compared with that in their wild type counterparts (PKCα+/+). MEFs were treated with PMA to promote PKC-catalyzed phosphorylation and the PP1/PP2A inhibitor calyculin A to stabilize the phosphorylated state or pretreated with the PKC inhibitors Gö6983 and Gö6976 and then treated with PMA and calyculin A. The Western blot in Fig. 6B shows that DLG1 migrated as a doublet in the MEFs, similar to what we observed for DLG1 in astrocytes. The bands were only weakly labeled with the Thr(P)-656 antibody, but immunoreactivity increased upon PMA treatment, and cotreatment with calyculin A further enhanced labeling with this antibody. This PMA/calyculin A-stimulated phosphorylation of DLG1 was abolished in cells treated with Gö6983 and significantly reduced in cells treated with Gö6976, as shown by quantitation of the data from eight separate experiments, presented in Fig. 6D. Both basal and phorbol ester-stimulated phosphorylation of Thr-656 were significantly reduced in PKCα−/− MEFs; quantitation of 8 independent experiments (Fig. 6D) revealed that basal phosphorylation was reduced ∼50%, with no significant increase after phorbol ester treatment. Calyculin A treatment unmasked a very modest increase in phosphorylation of Thr-656, which was not significantly affected by PKC inhibitors, suggesting that inhibition of phosphatases allowed slight phosphorylation of this site by other kinases. These data reveal that PKCα is the predominant kinase catalyzing the phosphorylation of Thr-656 in MEFs.
We also examined whether DLG1 is phosphorylated by PKCα in H1703 NSCLC cells, which were used to examine the effects of PKCα and DLG1 on migration. Because of the low basal level of endogenous DLG1 phosphorylation in these cells, we overexpressed a CFP-tagged form of DLG1 in H1703 cells expressing a non-targeting shRNA or a PKCα shRNA and examined phosphorylation of Thr-656 under conditions of PKC activation and inhibition. The results (Fig. 6C) were very similar to those observed in the PKCα+/+ and PKCα−/− MEFs; depletion of PKCα (by 60 ± 10%) impaired DLG1 phosphorylation stimulated by treatment with PMA and calyculin A (compare lanes 3 and 8) as did pretreatment with Gö6976 (lane 4) or Gö6983 (lane 5). Quantitation of n = 7 experiments (Fig. 6E) showed that interference with PKCα expression or activity significantly decreased Thr-656 phosphorylation in H1703 cells, establishing PKCα as the primary kinase for this site in this NSCLC cell line.
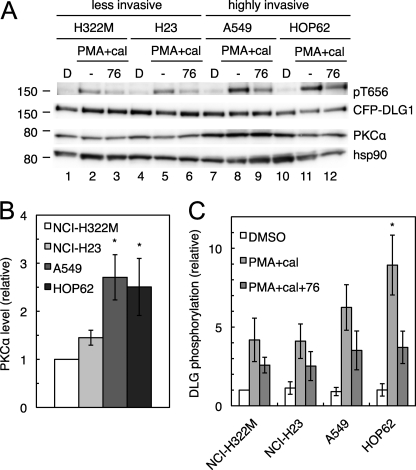
PKCα Activity at the DLG1 Scaffold Is Increased in Highly Invasive Cells
Given that PKCα mediates phosphorylation of DLG1 at Thr-656, we next examined whether DLG1-associated PKCα activity (as read out by Thr-656 phosphorylation) is correlated with invasiveness in human lung cancers. Taking advantage of a study profiling invasion by the 9 NSCLC lines in the NCI-60 (31), we chose two cell lines with a non-invasive phenotype (NCI-H322M and NCI-23; invasion scores of less than 1000) and two more highly invasive cell lines (A549 and HOP62; invasion scores of ∼4000) and examined PKCα expression and DLG1 phosphorylation. Similar to H1703 cells, these cell lines have very low basal levels of endogenous DLG1 phosphorylation, so we examined PKCα signaling at the DLG1 scaffold by overexpressing CFP-DLG1 and treating with PMA and calyculin A with or without pretreatment with the cPKC inhibitor Gö6976. Overall PKCα expression was significantly increased (2.5–3-fold) in A549 and HOP62 cells compared with H322M and H23 cells (Fig. 7, A and B), in accordance with data from reverse phase protein array profiling of the NCI-60 (32), which establish relative PKCα protein levels in these four cell lines as 1.0 (H322M), 1.3 (H23), 4.7 (A549), and 4.6 (HOP62). To read out PKCα activity at the scaffold, we examined the component of PMA-stimulated DLG1 phosphorylation that was reversed by Gö6976 treatment, corresponding to cPKC-mediated phosphorylation. This parameter was increased in the more highly invasive cell lines (Fig. 7A); quantitation of 8 separate experiments revealed increases of 1.7 ± 0.5- and 3.3 ± 0.7-fold in Gö6976-reversible Thr-656 phosphorylation in A549 and HOP62 cells, respectively, compared with H322M, although only the difference between the H322M and HOP62 cells was significant. As expected, the difference between H322M and H23 cells was negligible (Fig. 7C). These results show that DLG1-associated PKCα activity is correlated with invasion across several human lung cancer cell lines.
FIGURE 7.
PKCα signaling at the DLG1 scaffold is increased in highly invasive NSCLC lines relative to their less invasive counterparts. A, PKCα expression and phosphorylation of DLG1 at Thr-656 in NSCLC lines are shown. NCI-H322M, NCI-H23, A549, and HOP62 cells were transiently transfected with CFP-DLG1 and pretreated with DMSO (denoted by D) or the PKC inhibitor Gö6976 (76) for 10 min before the addition of PMA for 20 min and the phosphatase inhibitor calyculin A (cal) for the last 10 min of PMA treatment. Cells were lysed, and the lysates were probed for Thr(P)-656, DLG1, PKCα, and the loading control heat shock protein 90 (hsp90). B and C, bar graphs representing the mean ± S.E. of PKCα expression (B) and DLG1 phosphorylation at Thr-656 (C) in 8 experiments performed as described in A. Gö6976-reversible phosphorylation was calculated by subtracting the relative phosphorylation in the PMA+cal+76 condition from the relative phosphorylation in the PMA+cal condition. Significantly different from H322M cells; *, p < 0.05.
DISCUSSION
In this study we have identified the PDZ domain-containing scaffold DLG1 as a binding partner and substrate for PKCα that is required for PKCα to promote cell migration. Specifically, we show that the PDZ ligand of PKCα binds the third PDZ domain of DLG1, coordinating the phosphorylation at Thr-656 on DLG1. These two proteins colocalize at the leading edge of migrating cells, a dynamic region controlled by cytoskeletal interactions, where they promote scratch-induced migration in several different cell types. Key to our study was the finding that PKCα inhibition did not affect wound healing when DLG1 was depleted. Taken together, our results are consistent with a model in which the DLG1 scaffold coordinates signaling by PKCα to regulate cellular migration in two-dimensional culture and in which phosphorylation of DLG1 at Thr-656 marks PKCα activity at this scaffold.
Many studies have shown that PKCα plays roles in cell motility in vitro and in vivo (6); phorbol esters, which activate PKC, enhance cellular migration and have well characterized effects on the actin cytoskeleton (10), and PKCα-specific inhibitors and siRNA attenuate cell spreading, wound healing, metalloprotease activation, and metastasis in several models (33–40). Recently, microRNA profiling revealed that the presence of brain metastasis in NSCLC patients could be predicted in part by the expression of a microRNA whose forced expression results in up-regulation of PKCα levels, suggesting that PKCα plays an important role in metastasis in human lung cancer (41). Such effects on motility have been suggested to be mediated by several PKC binding proteins/substrates, including α6-tubulin (33), RhoGDP-dissociation inhibitor, syndecan-4 (35–37), ADAM-10 (a disintegrin and metalloprotease 10) (39), and fascin (34). However, the majority of these studies do not address the question of whether the PKCα-mediated effects on cellular migration are dependent on the presence of the substrate in question. Our results demonstrate a PKCα-DLG1 signaling pathway that positively affects migration and could play a pro-oncogenic role. Although DLG1 has historically been characterized as a tumor suppressor (42, 43), it varies in expression among malignancies (44) and is known to play different and sometimes opposing roles in various processes involved in tumor progression, including differentiation, cytokinesis, proliferation, cell migration, and control of the tumor microenvironment (43, 45). Moreover, a DLG1 mutant mouse in which the gene is disrupted starting after the third PDZ domain shows no increase in tumor development but does demonstrate a cleft palate phenotype that is consistent with failure of cell migration (46).
Previous studies have demonstrated a role for DLG1 in scratch-induced migration in primary astrocytes. Specifically, atypical PKC activity regulates the leading edge localization of DLG1, where it controls the localization of adenomatous polyposis coli and polarization of microtubules. Furthermore, DLG1 interacts with guanylate kinase anchoring protein along the microtubules to contribute to proper microtubule dynamics, which allow the cell to properly position the centrosome, establish cell polarity, and migrate into the scratch wound (47–49). These observations point to a critical role for DLG1 in astrocyte migration and highlight the importance of the guanylate kinase (GUK) domain in carrying out this function of DLG1.
The C-terminal moiety of DLG1 is composed of an SH3 domain and GUK domain separated by a Hook (hinge) region. In the fly, SH3-Hook mutants or mutations that otherwise disrupt SH3-GUK binding phenocopy DLG null mutants (50, 51); thus, this region is critical for the function of this scaffolding protein. The Hook domain of DLG1 cooperates with the SH3 domain to bind the GUK intramolecularly and regulate the accessibility of this domain to binding partners (52). Thus, phosphorylation at Thr-656, which is situated between the SH3 and Hook regions, could alter the interaction of DLG1 with partners whose binding site has been mapped to the SH3-GUK region, including the previously mentioned guanylate kinase-anchoring protein and ADAM-10, which bind to the DLG1 SH3 domain and also to PKCα (39, 53). Importantly, a GUK deletion mutant is unable to rescue the microtubule polarization defects upon DLG1 knockdown in migrating astrocytes (48), so this region is critical to DLG1 function in astrocyte migration. Despite the critical functions of this region and despite our finding that PKCα and DLG1 cooperate in regulating wound healing, mutation of the phospho-acceptor site had no effect on wound healing (data not shown). It is possible that DLG1 overexpression can compensate for mutation of this PKCα phosphorylation site. An alternative hypothesis is that DLG1 scaffolds PKCα to other substrates that are important for PKCα effects on wound healing, and therefore, phosphorylation of DLG1 at Thr-656 serves as a marker for PKCα activity at the scaffold but is not itself required to mediate the pro-migratory effects of PKCα. For example, the membrane-associated protein 4.1, which participates in membrane reorganization in multiple cell types, binds to DLG1 through the 4.1 conserved N-terminal FERM (four.one protein, ezrin, radixin, moesin) domain (54) and is phosphorylated by PKCα at a conserved (55) serine that is important for spectrin-actin association (56, 57). DLG1 may promote PKCα-mediated phosphorylation of 4.1 to promote membrane destabilization and allow cellular migration. Thus, although the specific functional consequences of Thr-656 phosphorylation remain to be elucidated, this phosphorylation event is correlated with increased invasiveness in four NSCLC cell lines, supporting a critical role for DLG1-associated PKCα activity in cancer cell motility.
In this study we connect PKCα, which is known to positively regulate cellular migration, with the scaffold DLG1, which acts to enable migration via interactions with numerous proteins involved in motility. Specifically, we identify a novel PDZ ligand interaction of PKCα that is necessary for it to facilitate cell migration.
Supplementary Material
Acknowledgments
We thank Dr. C. Garner for the Myc-DLG1 plasmid, Dr. M. Dell'Acqua for the CFP-DLG1 plasmid, the NCI for the NSCLC cell lines, and members of the laboratories of Dr. Newton and Dr. J. Brugge for helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grants GM43154 and P01 DK54441 (both to A. C. N.) and CA081436 (to A. P. F.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
M. T. Kunkel and R. A. Hall, unpublished data.
- cPKC
- conventional PKC
- NSCLC
- non-small cell lung cancer
- PSD-95
- postsynaptic density protein 95
- PDZ
- PSD-95, disheveled, and ZO1
- GUK
- guanylate kinase
- MAGUK
- membrane-associated GUK
- SAP-97
- synapse-associated protein 97
- DLG1
- discs large homolog 1
- MEF
- mouse embryonic fibroblast
- PMA
- phorbol 12-myristate-13-acetate
- SH3
- Src homology 3
- ADAM-10
- a disintegrin and metalloprotease 10
- NT
- non-targeting
- CFP
- cyan fluorescent protein
- eGFP
- enhanced GFP.
REFERENCES
- 1. Pawson C. T., Scott J. D. (2010) Nat. Struct. Mol. Biol. 17, 653–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rosse C., Linch M., Kermorgant S., Cameron A. J., Boeckeler K., Parker P. J. (2010) Nat. Rev. Mol. Cell Biol. 11, 103–112 [DOI] [PubMed] [Google Scholar]
- 3. Newton A. C. (2010) Am. J. Physiol. Endocrinol. Metab. 298, E395–E402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dempsey E. C., Newton A. C., Mochly-Rosen D., Fields A. P., Reyland M. E., Insel P. A., Messing R. O. (2000) Am. J. Physiol. Lung Cell Mol. Physiol. 279, L429–L438 [DOI] [PubMed] [Google Scholar]
- 5. Castagna M., Takai Y., Kaibuchi K., Sano K., Kikkawa U., Nishizuka Y. (1982) J. Biol. Chem. 257, 7847–7851 [PubMed] [Google Scholar]
- 6. Konopatskaya O., Poole A. W. (2010) Trends Pharmacol Sci. 31, 8–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fan Q. W., Cheng C., Knight Z. A., Haas-Kogan D., Stokoe D., James C. D., McCormick F., Shokat K. M., Weiss W. A. (2009) Sci. Signal. 2, ra4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Guo J., Ibaragi S., Zhu T., Luo L. Y., Hu G. F., Huppi P. S., Chen C. Y. (2008) Cancer Res. 68, 8473–8481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Holinstat M., Mehta D., Kozasa T., Minshall R. D., Malik A. B. (2003) J. Biol. Chem. 278, 28793–28798 [DOI] [PubMed] [Google Scholar]
- 10. Larsson C. (2006) Cell. Signal. 18, 276–284 [DOI] [PubMed] [Google Scholar]
- 11. Dean N., McKay R., Miraglia L., Howard R., Cooper S., Giddings J., Nicklin P., Meister L., Ziel R., Geiger T., Muller M., Fabbro D. (1996) Cancer Res. 56, 3499–3507 [PubMed] [Google Scholar]
- 12. Paz-Ares L., Douillard J. Y., Koralewski P., Manegold C., Smit E. F., Reyes J. M., Chang G. C., John W. J., Peterson P. M., Obasaju C. K., Lahn M., Gandara D. R. (2006) J. Clin. Oncol. 24, 1428–1434 [DOI] [PubMed] [Google Scholar]
- 13. Fields A. P., Murray N. R. (2008) Adv. Enzyme Regul. 48, 166–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mochly-Rosen D., Khaner H., Lopez J. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 3997–4000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mochly-Rosen D., Gordon A. S. (1998) FASEB J. 12, 35–42 [PubMed] [Google Scholar]
- 16. Poole A. W., Pula G., Hers I., Crosby D., Jones M. L. (2004) Trends Pharmacol. Sci. 25, 528–535 [DOI] [PubMed] [Google Scholar]
- 17. Tsunoda S., Sierralta J., Sun Y., Bodner R., Suzuki E., Becker A., Socolich M., Zuker C. S. (1997) Nature 388, 243–249 [DOI] [PubMed] [Google Scholar]
- 18. Adamski F. M., Zhu M. Y., Bahiraei F., Shieh B. H. (1998) J. Biol. Chem. 273, 17713–17719 [DOI] [PubMed] [Google Scholar]
- 19. van Ham M., Hendriks W. (2003) Mol. Biol. Rep. 30, 69–82 [DOI] [PubMed] [Google Scholar]
- 20. Lee H. J., Zheng J. J. (2010) Cell Commun. Signal. 8, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Staudinger J., Lu J., Olson E. N. (1997) J. Biol. Chem. 272, 32019–32024 [DOI] [PubMed] [Google Scholar]
- 22. Leitges M., Kovac J., Plomann M., Linden D. J. (2004) Neuron 44, 585–594 [DOI] [PubMed] [Google Scholar]
- 23. Lim I. A., Hall D. D., Hell J. W. (2002) J. Biol. Chem. 277, 21697–21711 [DOI] [PubMed] [Google Scholar]
- 24. Colledge M., Dean R. A., Scott G. K., Langeberg L. K., Huganir R. L., Scott J. D. (2000) Neuron 27, 107–119 [DOI] [PubMed] [Google Scholar]
- 25. Kunkel M. T., Garcia E. L., Kajimoto T., Hall R. A., Newton A. C. (2009) J. Biol. Chem. 284, 24653–24661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lau A. G., Hall R. A. (2001) Biochemistry 40, 8572–8580 [DOI] [PubMed] [Google Scholar]
- 27. Citro S., Malik S., Oestreich E. A., Radeff-Huang J., Kelley G. G., Smrcka A. V., Brown J. H. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 15543–15548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Valster A., Tran N. L., Nakada M., Berens M. E., Chan A. Y., Symons M. (2005) Methods 37, 208–215 [DOI] [PubMed] [Google Scholar]
- 29. Deleted in proof.
- 30. Roberts S., Calautti E., Vanderweil S., Nguyen H. O., Foley A., Baden H. P., Viel A. (2007) Exp. Cell Res. 313, 2521–2530 [DOI] [PubMed] [Google Scholar]
- 31. Hsu Y. C., Yuan S., Chen H. Y., Yu S. L., Liu C. H., Hsu P. Y., Wu G., Lin C. H., Chang G. C., Li K. C., Yang P. C. (2009) Clin. Cancer Res. 15, 7309–7315 [DOI] [PubMed] [Google Scholar]
- 32. Park E. S., Rabinovsky R., Carey M., Hennessy B. T., Agarwal R., Liu W., Ju Z., Deng W., Lu Y., Woo H. G., Kim S. B., Cheong J. H., Garraway L. A., Weinstein J. N., Mills G. B., Lee J. S., Davies M. A. (2010) Mol. Cancer Ther. 9, 257–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Abeyweera T. P., Chen X., Rotenberg S. A. (2009) J. Biol. Chem. 284, 17648–17656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Anilkumar N., Parsons M., Monk R., Ng T., Adams J. C. (2003) EMBO J. 22, 5390–5402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bass M. D., Roach K. A., Morgan M. R., Mostafavi-Pour Z., Schoen T., Muramatsu T., Mayer U., Ballestrem C., Spatz J. P., Humphries M. J. (2007) J. Cell Biol. 177, 527–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Elfenbein A., Rhodes J. M., Meller J., Schwartz M. A., Matsuda M., Simons M. (2009) J. Cell Biol. 186, 75–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Horowitz A., Tkachenko E., Simons M. (2002) J. Cell Biol. 157, 715–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kim J., Thorne S. H., Sun L., Huang B., Mochly-Rosen D. (2011) Oncogene 30, 323–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kohutek Z. A., diPierro C. G., Redpath G. T., Hussaini I. M. (2009) J. Neurosci. 29, 4605–4615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lin C. W., Shen S. C., Chien C. C., Yang L. Y., Shia L. T., Chen Y. C. (2010) J. Cell. Physiol. 225, 472–481 [DOI] [PubMed] [Google Scholar]
- 41. Arora S., Ranade A. R., Tran N. L., Nasser S., Sridhar S., Korn R. L., Ross J. T., Dhruv H., Foss K. M., Sibenaller Z., Ryken T., Gotway M. B., Kim S., Weiss G. J. (2011) Int. J. Cancer 129, 2621–2631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bilder D. (2004) Genes Dev. 18, 1909–1925 [DOI] [PubMed] [Google Scholar]
- 43. Humbert P. O., Grzeschik N. A., Brumby A. M., Galea R., Elsum I., Richardson H. E. (2008) Oncogene 27, 6888–6907 [DOI] [PubMed] [Google Scholar]
- 44. Cavatorta A. L., Fumero G., Chouhy D., Aguirre R., Nocito A. L., Giri A. A., Banks L., Gardiol D. (2004) Int. J. Cancer 111, 373–380 [DOI] [PubMed] [Google Scholar]
- 45. Unno K., Hanada T., Chishti A. H. (2008) Exp. Cell Res. 314, 3118–3129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Caruana G., Bernstein A. (2001) Mol. Cell. Biol. 21, 1475–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Etienne-Manneville S., Manneville J. B., Nicholls S., Ferenczi M. A., Hall A. (2005) J. Cell Biol. 170, 895–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Manneville J. B., Jehanno M., Etienne-Manneville S. (2010) J. Cell Biol. 191, 585–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Dujardin D. L., Vallee R. B. (2002) Curr. Opin. Cell Biol. 14, 44–49 [DOI] [PubMed] [Google Scholar]
- 50. Hough C. D., Woods D. F., Park S., Bryant P. J. (1997) Genes Dev. 11, 3242–3253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Newman R. A., Prehoda K. E. (2009) J. Biol. Chem. 284, 12924–12932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Funke L., Dakoji S., Bredt D. S. (2005) Annu. Rev. Biochem. 74, 219–245 [DOI] [PubMed] [Google Scholar]
- 53. Marcello E., Gardoni F., Mauceri D., Romorini S., Jeromin A., Epis R., Borroni B., Cattabeni F., Sala C., Padovani A., Di Luca M. (2007) J. Neurosci. 27, 1682–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lue R. A., Marfatia S. M., Branton D., Chishti A. H. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 9818–9822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Parra M., Gascard P., Walensky L. D., Gimm J. A., Blackshaw S., Chan N., Takakuwa Y., Berger T., Lee G., Chasis J. A., Snyder S. H., Mohandas N., Conboy J. G. (2000) J. Biol. Chem. 275, 3247–3255 [DOI] [PubMed] [Google Scholar]
- 56. Diakowski W., Grzybek M., Sikorski A. F. (2006) Folia Histochem Cytobiol. 44, 231–248 [PubMed] [Google Scholar]
- 57. Manno S., Takakuwa Y., Mohandas N. (2005) J. Biol. Chem. 280, 7581–7587 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.