Background: Alterations of the APP pathway or DNA damage induce neuronal cell death.
Results: Alterations of the APP pathway or DNA damage increase TAp73 expression and reduce ΔNp73 protein levels.
Conclusion: A tight control of the expression of p73 isoforms participates in neuronal cell death.
Significance: p73 isoforms may play a role in neurodegenerative diseases such as Alzheimer and in the neurotoxicity of anticancer drug therapies.
Keywords: Alzheimer Disease, Cell Death, DNA Damage, Neurons, p73
Abstract
Genetic ablations of p73 have shown its implication in the development of the nervous system. However, the relative contribution of ΔNp73 and TAp73 isoforms in neuronal functions is still unclear. In this study, we have analyzed the expression of these isoforms during neuronal death induced by alteration of the amyloid-β precursor protein function or cisplatin. We observed a concomitant up-regulation of a TAp73 isoform and a down-regulation of a ΔNp73 isoform. The shift in favor of the pro-apoptotic isoform correlated with an induction of the p53/p73 target genes such as Noxa. At a functional level, we showed that TAp73 induced neuronal death and that ΔNp73 has a neuroprotective role toward amyloid-β precursor protein alteration or cisplatin. We investigated the mechanisms of p73 expression and found that the TAp73 expression was regulated at the promoter level. In contrast, regulation of ΔNp73 protein levels was regulated by phosphorylation at residue 86 and multiple proteases. Thus, this study indicates that tight transcriptional and post-translational mechanisms regulate the p73 isoform ratios that play an important role in neuronal survival.
Introduction
Given that over 50% of human tumors exhibit mutations or inactivation of p53 (1), this gene has been extensively studied for its major role in tumor suppression. DNA damage, oncogene activation, and also various other cellular stresses (hypoxia, radical oxidized species production, etc.) increase p53 protein levels leading to cell growth arrest or apoptosis through up-regulation of its target genes (p21, Bax, GADD 45, MDM2, or DR5 (2)). Beside its tumor suppressor gene function, p53 plays a role in other physiological or pathological situations. In the nervous system, p53 has been shown to participate in terminal neuronal maturation as well as neuronal apoptosis (3–9). Importantly, p53 activation has been observed in various neurodegenerative diseases, such as ALS, Alzheimer, Parkinson, and Angelman syndrome (9–15). However, the signaling pathways leading to p53 activation or the target genes of p53 in neurons have never been documented.
More recently, p63 and p73, two homologs of p53, have been identified (16). Similarly to p53, these genes play important roles in the embryonic development of various organs (brain, skin, limb, immune system, etc.). In particular, p73−/− mice exhibit defects in the nervous system, notably in the hippocampus and the olfactory bulbs (17). Analysis of the contribution of p73 to neuronal differentiation and apoptosis is complicated by the existence of multiple isoforms. These isoforms vary in their C terminus due to alternative splicing (TAp73α, -β, -δ, and -γ) or in their N terminus due to alternate promoter usage that skips an N-terminal transactivation domain (ΔNp73α and -β) (18, 19). TAp73s are transcription factors with positive or negative effects on gene expression (20, 21). ΔNp73 isoforms, which lack the N-terminal transactivation domain (TA),6 were shown to exert a dominant inhibitory effect on the full-length isoforms. TAp73 isoforms induce neuronal differentiation in SYH5 cells (22) and participate in oligodendrocyte differentiation (23). ΔNp73 isoforms favor neuronal survival (24, 25). More recently, a complex regulation of p73 and p63 has also been identified in cortical neurons following ischemia, with TAp73 being down-regulated while TAp63 is induced (26).
Neuronal apoptosis has been proposed to be a part of the processes involved in neurodegenerative diseases, such as ALS, Alzheimer, Parkinson, or even neuropathy following anticancer treatments. Alzheimer disease is characterized by the pathological accumulation of amyloid-β plaques resulting from mutations in the amyloid-β precursor protein (APP) or the γ-secretases, PS1 and PS2. Interestingly, p73 expression is increased in neurons from Alzheimer patients, and the TAp73 isoforms are induced by treatment with the amyloid-β peptide (27, 28). It has also recently been shown that the deletion of the p73 gene favors neuronal aging and the appearance of Tau phosphorylation, a characteristic of Alzheimer disease (29). These studies suggest that p73 may be playing a role in the neuronal apoptosis induced by alteration of the APP processing.
To clarify the role of the p73 gene in the regulation of the balance between survival and apoptosis in neurons, we undertook a comparative study of the TAp73 and ΔNp73 isoforms in primary cultures of neurons submitted to activation of the β-amyloid polypeptide precursor and cisplatin, a DNA-damaging drug. Using these models, we previously showed a regulation of MDMX and MDM2, two proteins that control p53 and p73 activity (30). The data obtained here clearly indicate that both p73 isoforms exert opposite effects on neuronal survival. Furthermore, we found that the ratio between the two types of isoforms was altered by a variety of neurotoxic insults, including the activation of the APP pathway. Transcriptional and post-transcriptional mechanisms contribute to tight regulation of this ratio, critical for neuronal survival.
EXPERIMENTAL PROCEDURES
Cell Culture
Cortical neurons were dissected from E16 murine embryos as described previously (30). Briefly, after enzymatic and mechanical dissociation, cells were plated at a density of 500 cells/mm2 on 0.1 mg/ml polyornithine precoated culture dishes and were grown at 37 °C in a humidified atmosphere (5% CO2, 95% air). Plating culture medium contained Dulbecco's modified Eagle's medium (DMEM; Invitrogen) supplemented with 10% heat-inactivated horse serum (Invitrogen), 100 nm insulin (Invitrogen), and 50 mg/ml gentamycin (Invitrogen). 48 h after plating, cells were switched to defined medium containing DMEM supplemented with 10 nm insulin, 100 mg/ml human transferrin, 60 mm putrescine, 30 nm sodium selenite, and 50 mg/ml gentamycin. Neurons were considered as immature up to 2 days after plating and as mature 5 days after plating. N2A cells were obtained from the ATCC and grown in DMEM with 10% fetal bovine serum.
Colorimetric MTT Assay
Cells were cultured in 96-well culture dishes (Costar). A modified procedure of the original method (31) was used to measure mitochondrial activity (MTT assay) as described previously (32). Briefly, cultures were rinsed with DMEM and incubated for 1 h at 37 °C in freshly prepared culture medium containing 0.5 mg/ml of MTT (Sigma). Medium was then removed, and dark blue crystals formed during reaction were dissolved by adding 100 μl/well of 0.04 m HCl in isopropyl alcohol. Plates were stirred at room temperature to ensure that all crystals were dissolved and read on a Bio-Rad 680 micro-ELISA plate reader, using a test wavelength of 490 nm and a nonspecific wavelength of 650 nm for background absorbency. Results are given as percentage of survival, taking cultures grown in HK medium as 100%.
Expression Vectors, Reporter Genes, and siRNA
HA-TAp73β and HA-ΔNp73β expression vectors were described previously (33). pCMV cyclin A, pCMV cyclin D, pCMV CDK4, and pCMV CDK5 expression vector were a gift from Dr. K. Okamoto (Cancer Center, Tokyo). pCMV HA-JNK was kindly provided by Dr. Ron Prywes. pcDNA constructs encoding CDK1 or CDK2 proteins were a gift from Dr. S. van den Heuvel. pCMV-HA ΔNp73β T86A was generated using the Stratagene PCR mutagenesis kit. CMV wild type APP, APP KM670/671NL (APPSW), AICD, PS1, and PS1 A79V (PS1μt) were generated by RT-PCR and subcloned in pcDNA3 vector and site-directed mutagenesis. pBax-Luc has been described (34). pBax-min-Luc was kindly provided by Dr. C. DiComo (Aureon Biosciences) and contains a duplex oligonucleotide encoding the p53-responsive cis-acting element from bax, cloned upstream from the minimal c-fos promoter (−53 to +42) in pGL3-OFLUC. The DR5 promoter reporter plasmid was obtained from Dr. T. Sakai (Kyoto, Japan). The Noxa luciferase construct was a generous gift of Dr. Tanigushi (Tokyo, Japan).
Stealth siRNA annealed duplexes were obtained from Invitrogen and targeted the following 5′ to 3′ DNA sequences: siTAp73#1, GCCATCCGAAACACCAATGAGTACA; siTap73#2, GCAGGAAAGGACTCATGAAATCATA; siCt, sequence control GC 50% (Invitrogen). Cotransfection experiments with siRNA and GFP-expressing plasmid were performed as described previously (35).
Transfection and Luciferase Assays
Cells were transfected by a polyethyleneimine-based protocol as described previously (36). Cells were grown in DMEM, 10% FBS and transfected with various amounts of DNA. DNA and polyethyleneimine (0.33 μl of 0.1 m/μg DNA) were mixed and added to the cells. Plates were centrifuged at 1500 rpm for 5 min and placed in the incubator for 1 h, after which fresh medium was added. For luciferase assays, cells were seeded in 12-well, 3.8-cm2 plates and transfected with the expression vectors (200 ng of each) and the reporter constructs (250 ng of each). Luciferase activity was measured in each well 24 h later, and results were normalized with a CMV-driven reporter gene.
Transfection and Apoptosis Assay
Neurons were grown on coverslips coated with polyornithine in 24-well plates. Cells were cotransfected with the expression vectors (200 ng/well), siRNA (50 nm), and the GFP expression vector (50 ng/well) as described previously (35). Cells were treated or not for 18 h with the neurotoxic agents. The cells were then washed with PBS and fixed with 4% paraformaldehyde for 15 min. After two washes, cells were incubated for 10 min with the Hoechst nuclei staining agent (1 μg/ml). GFP-positive cells were then observed with a microscope to assess the morphology of the nuclei and the neurites.
Preparation of Whole Cell Extracts and Immunoblotting
Neurons were lysed as described previously (37) in 150 μl of lysis buffer (TEGN: 20 mm Tris-HCl, pH 8, 1 mm EDTA, 0.5% Nonidet P-40, 150 mm NaCl, 1 mm DTT, 10% glycerol, protease inhibitors (Sigma)), sonicated for 30 s, and centrifuged at 13,000 rpm for 12 min. Protein concentrations were determined using a colorimetric assay (Bio-Rad). Sample buffer (38) was added to 20–75 μg of proteins, and samples were heated to 95 °C for 3 min followed by electrophoresis through a 10% SDS-polyacrylamide gel. Proteins were transferred to nitrocellulose membranes (Schleicher & Schuell) and visualized by enhanced chemiluminescence detection (Amersham Biosciences). CO73A and TA73A were produced and purified using a differential purification with peptides located in the N-terminal part and in the core domain. Bax was detected with the anti-Bax (N20; Santa Cruz Biotechnology, 1:2000) and DR5 with the anti-DR5 (M20; Santa Cruz Biotechnology, 1:200) antibodies. Actin detection was done with an anti-actin antibody from Dr. Aunis (Strasbourg, France).
Immunopurification of p73 Proteins, in Vitro Phosphorylation, and Degradation Assays
Neurons or N2A cells were plated in 6-cm diameter plates and treated in duplicates or triplicates subsequently mixed. N2A cells were transfected with ΔNp73β expression vectors, and after 48 h cells were harvested. Cells were lysed in 100 μl of lysis buffer (TEGN: 20 mm Tris-HCl, pH 8, 1 mm EDTA, 0.5% Nonidet P-40, 150 mm NaCl, 1 mm DTT, 10% glycerol, 5 mm PMSF, 100 μm benzamidine, 300 μg/ml leupeptin, 10 μg/ml bacitracin, 100 μg/ml, α-macroglobulin, and phosphatase inhibitors (cocktails I and II, Sigma)), and the extracts were centrifuged at 13,000 rpm for 12 min to remove cell debris. p73 proteins (400–750 μg of whole cell extract) were immunoprecipitated at 4 °C for 3 h with 30 μl of 50% slurry protein A-Sepharose beads cross-linked to an anti-HA antibody (12CA5) or with a mixture of protein A-Sepharose beads and anti-pan p73 antibody. After incubation, the samples were washed four times with 1 ml of wash buffer (20 mm Tris-HCl, pH 8, 150 mm NaCl, 1 mm EDTA, 0.5% Nonidet P-40, 1 mm DTT, 10% glycerol). For immunoblotting, the excess of liquid was aspirated, and 35 μl of sample buffer was added to the beads before heating at 95 °C for 3 min. After centrifugation (3 min at 13,000 rpm), samples were separated by electrophoresis through 10% SDS-PAGE. For in vitro phosphorylation, dried beads were resuspended in 40 μl of kinase reaction mixture containing radioactive γ-ATP and the immunopurified cyclin A-CDK1 complex. After a 30-min incubation period at 37 °C, beads were washed, resuspended in sample buffer, heated, and loaded on a 10% SDS-PAGE. For in-tube degradation assay, immunoprecipitation of ΔNp73 proteins was done in the absence of protease inhibitors, and purified p73 proteins were incubated 2 h with extracts from neurons prepared in TEGN buffer in the absence of protease inhibitors. After electrophoresis, proteins were transferred to nitrocellulose membranes (Schleicher & Schuell). For HA detection, the 16B12 monoclonal HA antibody (Babco, 1 mg/ml) was used at 1:1000. Thr-86 phosphorylation was followed by an antigen-purified phospho-Thr-86 p73 rabbit polyclonal antibody used at a dilution of 1:1000 (33). p73 detection was performed with the mouse monoclonal ER15 (New England Biolabs), the rabbit polyclonal PAN (CO73A), or the rabbit polyclonal specific for TAp73 isoforms (TA73A) antibodies. CO73A and TA73A were produced and purified using a differential purification with peptides located in the N-terminal part and in the core domain. Bax was detected with the anti-Bax (N20; Santa Cruz Biotechnology, 1:2000) and DR5 with the anti-DR5 (M20; Santa Cruz Biotechnology, 1:200) antibodies. Actin detection was done with an anti-actin antibody from Dr. Aunis (Strasbourg, France). Proteins were then visualized with an enhanced chemiluminescence detection system (Amersham Biosciences).
Real Time PCR
Cells were grown in 6-well plates. After treatment, cells were harvested, and total RNA was extracted using a Qiagen kit and treated with DNase (RQ1 RNase-free DNase, Promega). Reverse transcriptions were performed with 1 μg of RNA using the IScript kit (Bio-Rad). The IQ SYBR Green supermix kit (Bio-Rad) was used for real time PCRs. For each gene analyzed, a standard curve (from 2 to 0.032 μl) was done with a mix of all RT reactions. Based on this curve, 0.5 μl of RT reaction was analyzed for each condition with all sets of oligonucleotides (250 nm). Real time PCR protocol included in all experiments a melting curve that assessed the specificity of the products. Data were taken in account only if the efficiency was between 85 and 105% and if the percentage of correlation (r) was better than 0.995. Real time PCRs and analysis were performed on an ICycler (Bio-Rad). The following primers were used: mouse ΔNp73 (forward, TGC CTG GCC CTG TCT TTG; reverse, CAG CTC CAG CAT GAG TGC); mouse TAp73 (forward, TGG CCT CTC TTG GCC TCC AAC TTG T; reverse, TTG ACC AGG CCT TGG AGA GCT C); mouse TBP (forward, CTT CGT GCA AGA AAT GCT GA; reverse, CCC ACC ATG TTC TGG ATC TT); mouse Noxa (forward, CAG ATG CCT GGG AAG TCG; reverse, TGA GCA CAC TCG TCC TTC AA); mouse Bax (forward, ATG CGT CCA CCA AGA AGC TGA; reverse, AGC AAT CAT CCT CTG CAG CTC C); mouse DR5 (forward, GTC CAG CTG GCC TAC AGC; reverse, GCT TGC AGT TCC CTT CTG AC).
CHIP Assay
Cells were fixed with 1% (v/v) formaldehyde for 10 min at room temperature and quenched with 0.125 m glycine for 5 min. ChIP experiments were carried out using the EZ-Magna ChIPTM chromatin immunoprecipitation kit (Millipore). Sheared cross-linked chromatin from ∼106 cells was incubated overnight at 4 °C with 3 μg of mouse anti-CHOP (Santa Cruz Biotechnology) or preimmune mouse IgG (Millipore). Input corresponds to nonimmunoprecipitated sheared cross-linked chromatin from ∼105 cells (1%). PCR analysis was performed with 1/25th of immunoprecipitated DNA as template and primers of the mouse Noxa promoter (forward, GAA GTT TCC CTC CCA CCT TC; reverse, GAC GTC ATG TGA CGA CAT CC).
RESULTS
Neurotoxic Stresses Induce TAp73β and Down-regulate ΔNp73β Protein Levels
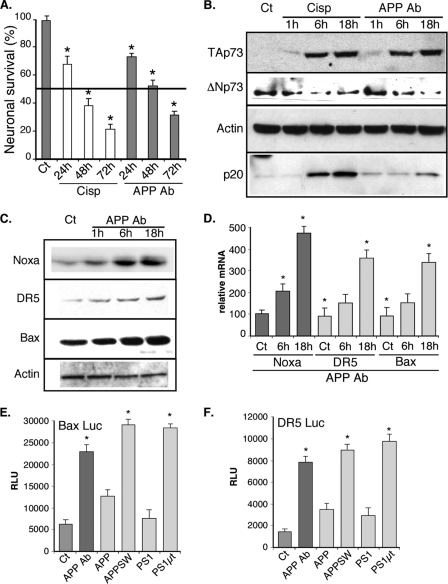
To analyze the potential contribution of the p73 gene to neuronal apoptosis, we first examined the expression of its isoforms following neurotoxic treatments in primary cultures of murine cortical neurons. These neurons were treated with either the DNA-damaging drug cisplatin, which causes neuropathy and has already been shown to induce p73 expression (39, 40), or an antibody that binds to the extracellular part of APP and has been shown to induce neuronal cell death (41–43). Both of these treatments induced neuronal cell death as revealed by an MTT test (Fig. 1A) or by detection of the active fragment of caspase 3 (p20, Fig. 1B). Under the same conditions, we monitored by Western blot the protein levels of p73 after 1, 6, and 18 h of treatment using new polyclonal antibodies that we had generated to detect the various isoforms of p73. The antibody TA73A was raised against a peptide located in the N-terminal part of the TAp73 isoforms. The antibody CO73A was raised against a peptide sequence of the core domain of the p73 proteins common to the TA and the ΔN isoforms (see supplemental data 1). In mouse cortical neurons, the TA73A antibody detected a signal around 65 kDa corresponding to TAp73β, whereas the CO73A antibody revealed a band migrating slightly below corresponding to ΔNp73β (Fig. 1B and supplemental data 1). The lack of detection of the TA isoforms by the CO73A antibody might be due to its weaker sensitivity (as observed with the commercial ER15 antibody) toward the TA isoforms (supplemental data #1). Other p73 isoforms (TAp73α, -δ, and -γ and ΔNp73α), which display noticeable size difference with TAp73β and ΔNp73β, were not detected in mouse cortical neurons, and immunoblotting with commercial antibodies also suggested that these cells mostly express the β isoforms of p73 (Fig. 1B and supplemental data). As reported in cancer cells, cisplatin increased TAp73β protein levels (44–46). Treatment with the APP-directed antibody also led to an increase of protein levels of TAp73β. In contrast, ΔNp73β protein levels progressively decreased upon both neurotoxic treatments (Fig. 1B).
FIGURE 1.
Cisplatin and APP alteration induce a shift in p73β isoform protein levels in cortical neurons leading to the activation of p53 target genes. A, primary cultures of mouse cortical neurons were treated for the indicated time with cisplatin (Cisp, 20 μm) or the APP-directed antibody (APP Ab, 5 μg/ml). Neuronal survival was evaluated using an MTT test. Results are presented in % compared with the control condition (Ct). Bars represent means ± S.D. of 8 wells from one experiment out of four independent experiments. The thick black line corresponds to 50% of viability (IC50). B and C, cortical neurons were treated for the indicated time with cisplatin (20 μm) or the APP Ab (5 μg/ml). Cells were lysed, and proteins (75 μg) were separated on a 10% SDS-PAGE and immunoblotted with the TA73A, CO73A, actin, p20 (active fragment of Caspase 3), Noxa, DR5, or Bax antibodies. D, cortical neurons were treated for the indicated time with the APP Ab (5 μg/ml). Cells were lysed, and total RNA were extracted and reverse-transcribed (1 μg). Quantitative PCR for Noxa, DR5, Bax, and TBP genes was performed. Bars correspond to means of normalized (TBP) and calibrated (no APP Ab = 100) values with standard deviation of triplicates from one experiment out of three independent experiments. E and F, cortical neurons were transfected with luciferase reporter genes containing either the Bax or the DR5 promoter. Where indicated, cells were treated with the APP Ab antibody (5 μg/ml) or cotransfected with expression vectors coding for either APP or PS1 variants (wild type or harboring mutations found in Alzheimer patients). Bars represent means ± S.D. (relative light units (RLU)) of 3 wells from one experiment out of three independent experiments. * indicates statistically significant differences tested by a one-way analysis of variance followed by a Neuman-Keuls test by pairs (p < 0.05).
This set of experiments indicates that in cortical neurons the respective levels of the TA and ΔN p73β isoforms are inversely regulated upon various neurotoxic stresses, including those associated with neurodegenerative diseases. In addition, the balance between the p73 isoforms induced (TA) and down-regulated (ΔN) isoforms shifts toward a probable activation of the p53/p73 target genes in neurons.
Activation of the APP Pathway Leads to p53/p73 Target Gene Expression
To test the possibility that a shift between the TA and the ΔN p73 isoforms might lead to the induction of p53/p73 target genes, we followed the expression of such genes involved in apoptosis, namely Noxa, Bax, and DR5 (47–49). Indeed, protein levels as well as mRNA levels of the three genes progressively increased after treatment with the APP-directed antibody (Fig. 1, C and D). Transcriptional activities of reporter genes containing the Noxa, Bax, or the DR5 promoter upstream of the luciferase gene were also stimulated by the APP-directed antibody (Figs. 1, E and F, and 2A).
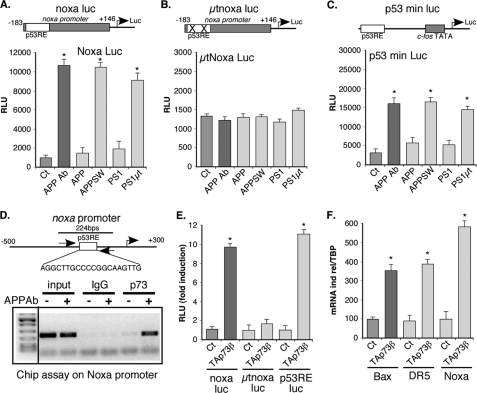
FIGURE 2.
Alteration of function of APP leads to induction of Noxa through TAp73. A–C, cortical neurons were transfected with luciferase reporter genes containing either the Noxa promoter (wild type, noxa Luc; mutated in the p53-binding sites, μtnoxa Luc) or a chimerical promoter containing p53-binding sites (p53RE) in front of the c-fos promoter (p53 min Luc). Where indicated, cells were treated with the APP Ab antibody (5 μg/ml) or cotransfected with expression vectors coding for either APP or PS1 variants. Bars represent means ± S.D. (relative light units (RLU)) of 3 wells from one experiment out of three independent experiments. D, chromatin immunoprecipitation of p73 on the Noxa promoter. Control (−) or APP Ab-treated (5 μg/ml, +) cortical neuron lysates were used for chromatin immunoprecipitation with control (normal IgG) or TA73A antibodies. Agarose gel showing PCR amplification (35 cycles) of a Noxa promoter fragment using inputs (1% of chromatin used for ChIP) or ChIPs as templates. E, cortical neurons were transfected with luciferase reporter genes containing either the Noxa promoter (wild type, noxa Luc; mutated in the p53-binding sites, μtnoxa Luc) or a chimerical promoter containing p53-binding sites in front of the c-fos promoter (p53 min Luc). Cells were cotransfected with an empty vector (Ct) or with an expression vector coding for TAp73β. Bars represent means ± S.D. (relative light units (RLU)) of 3 wells from one experiment out of three independent experiments. F, N2A neuroblastoma cells were transfected with an empty vector (Ct) or an expression vector encoding TAp73β. After 36 h of expression, cells were lysed, and total RNA were extracted and reverse-transcribed (1 μg). Quantitative PCR for Noxa, DR5, Bax, and TBP genes were performed. Bars correspond to means of normalized (TBP) and calibrated (Ct = 100) values with standard deviation of triplicates from one experiment out of three independent experiments. * indicates statistically significant differences tested by a one-way analysis of variance followed by a Neuman-Keuls test by pairs (p < 0.05).
APP-dependent neuronal apoptosis has been linked to mutations in the APP or its processing proteases, the γ-secretase presenilins (50). These mutations have been found in patients with early onset familial Alzheimer disease. To confirm that the alteration of the APP function leads to activation of these genes, we expressed in cortical neurons mutated version of the APP (K670N/M67L, APPSW) and presenilin 1 (A79V, PS1μt) proteins (51). The mutations correspond to the ones found in familial Alzheimer disease. As expected, expression of the mutated APP and presenilin proteins induced the promoter activity of Noxa, Bax, and DR5 (Figs. 1, E and F, and. 2A and supplemental data 4). Similar results were obtained with a minimal synthetic reporter gene (p53 min Luc, Fig. 2C), suggesting that the p53 response element mediated the induction seen by alteration of the APP function. Accordingly, no stimulation was observed when the p53-binding sites of the Noxa promoter were mutated (Fig. 2B).
To further confirm that alteration of the APP function in cortical neurons induced Noxa expression through p73 activation, we performed chromatin immunoprecipitation assays using the TA73A antibody. As shown on Fig. 2D, treatment of cortical neurons with the APP Ab led to an increased binding of TAp73 to the Noxa promoter. The ability of p73 to stimulate the Noxa, as well as Bax and DR5 promoters in cortical neurons was confirmed by using reporter genes or analyses of the mRNA levels (Fig. 2, E and F). Altogether, the data are consistent with the activation of the Noxa, Bax, DR5 promoters by the APP-directed antibody being mediated by p73. Thus, the switch of expression between the p73 isoforms observed upon activation of the pro-apoptotic APP pathway correlates with the stimulation of the p53/p73-dependent transcription of pro-apoptotic genes.
TAp73 and ΔNp73 Isoforms Regulate Neuronal Survival
To evaluate the functional repercussion of the shift between p73β isoforms, we analyzed the ability of these isoforms to regulate the balance between survival and death in primary culture of neurons.
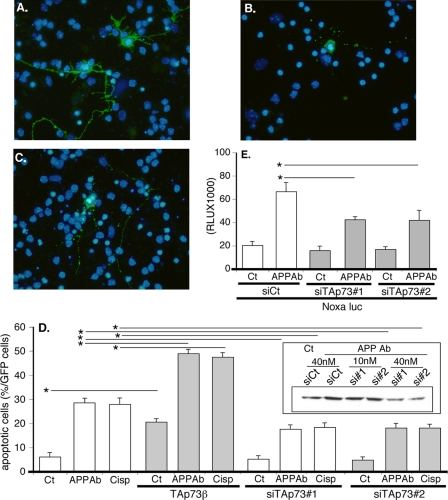
The functional impact of TAp73β in neurons was first examined by overexpression. TAp73β was coexpressed with GFP to specifically monitor the morphology (fragmentation or condensation of the nucleus and integrity of the neurites) of the transfected cells. Overexpression of TAp73β in cortical neurons induced nuclear changes indicative of neuronal cell death (Fig. 3, A–C). Concomitant pro-neurotoxic treatment of the neurons with the APP-directed antibody or cisplatin further increased cell death (Fig. 3D and supplemental data 4).
FIGURE 3.
TAp73β induces pro-apoptotic gene expression and neuronal apoptosis. A–D, cortical neurons were transfected with a GFP expression vector and either with the TAp73β expression vector or TAp73 siRNA duplexes (siTAp73#1 or siTAp73#2). After 24 h, cells were left untreated (Ct) or treated with APP Ab (5 μg/ml) or cisplatin (Cisp) (20 μm) for 24 h. Cells were then fixed, stained with Hoechst, and observed with a fluorescence microscope. A shows two GFP-positive control cells (untreated). B shows a dead GFP-positive cell treated with APP Ab. C shows a dead cortical neuron transfected with TAp73β. D, graph represents the percentage of dead cells among the GFP-positive cells counted. Bars correspond to means of three wells of a representative experiment with S.D. The inset illustrates the silencing of TAp73β by TAp73 siRNAs (Western blot with TA73A and actin antibodies). E, cortical neurons were cotransfected with the noxa Luc reporter construct and either control (siCt) or TAp73 siRNAs for 36 h. Then cells were treated with APP Ab for 8 h or not (Ct). The siRNA concentration used was 40 nm. Bars represent means ± S.D. (relative light units (RLU)) of 3 wells from one experiment out of three independent experiments. * indicates statistically significant difference tested by a one-way analysis of variance followed by a Neuman-Keuls test by pairs (p < 0.05).
To assess if a TAp73 isoform contributes to the apoptosis induced by the APP-directed antibody or cisplatin in cortical neurons, we used siRNA to specifically silence TAp73 expression (Fig. 3D). The inset in Fig. 3D shows the knockdown of TAp73 expression obtained by the transfection of two different TAp73 siRNA duplexes. Apoptosis induced by the APP-directed antibody or cisplatin was partially blocked by the silencing of TAp73 expression (Fig. 3D). In addition, silencing of TAp73 by siRNA significantly reduced the stimulation of the Noxa reporter construct caused by the APP-directed antibody (Fig. 3E). Thus, neuronal apoptosis induced by alteration of the APP function is partially achieved through stimulation of pro-apoptotic genes that are regulated by an increased level of TAp73.
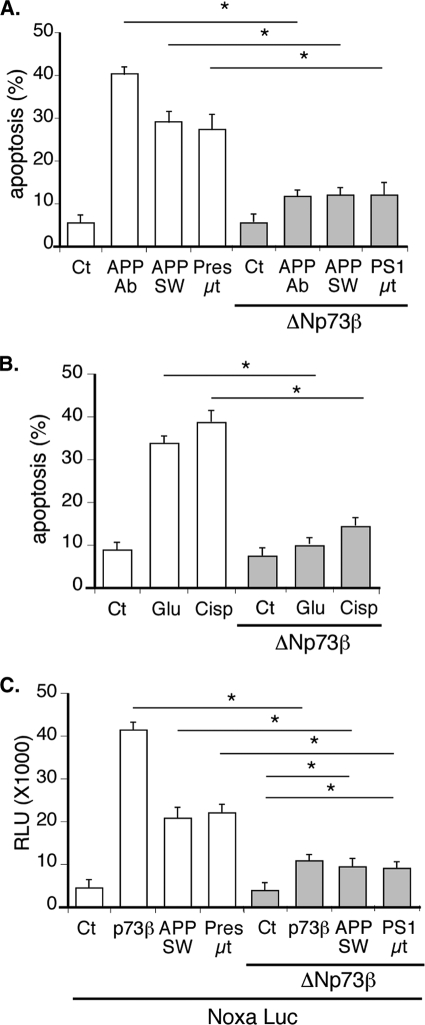
We used similar approaches to assess the role of ΔNp73 in neuronal survival. In contrast with TAp73β, overexpression of ΔNp73β significantly reduced the apoptosis of cortical neurons obtained by cisplatin, treatment with the APP Ab, or expression of mutated APP or presenilin 1 (Fig. 4A). Note that as observed previously (52), overexpression of wild type APP induced also a slight increase in neuronal cell death. In cortical neurons, apoptosis induced by cisplatin or excitotoxic stress induced by glutamate was also decreased upon overexpression of ΔNp73β (Fig. 4B). In addition, overexpression of ΔNp73β significantly reduced the activity of the Noxa reporter gene generated by the mutated forms of APP or presenilin 1 (Fig. 4C).
FIGURE 4.
Overexpression of ΔNp73 antagonizes neuronal apoptosis induced by various kinds of neurotoxic stresses. A and B, cortical neurons were cotransfected with the indicated combination of ΔNp73β, mutated APP (APP SW), or mutated presenilin 1 (Pres μt) and GFP expression vectors. After 24 h of expression, cells were left untreated (Ct) or treated with APP Ab (5 μg/ml), cisplatin (Cisp, 20 μm) or glutamate (Glu, 250 μm) for 24 h. Cells were then fixed, stained with Hoechst, and observed with a fluorescence microscope. Bars represent the percentage of dead cells among the GFP-positive cells counted. Bars are means of 3 wells of a representative experiment with S.D. C, cortical neurons were cotransfected with the noxaLuc reporter gene and the indicated combination of TAp73β, ΔNp73β, mutated APP, or mutated presenilin 1 expression vectors. Bars represent means ± S.D. (relative light units (RLU)) of 3 wells from one experiment out of three independent experiments. * indicates statistically significant differences tested by a one-way analysis of variance followed by a Neuman-Keuls test by pairs (p < 0.05).
Taken together these observations indicate that in cortical neurons the TA and ΔN isoforms of p73β exert opposite effects on neuronal apoptosis in response to alteration of APP function or other neurotoxic stresses. The induced expression of TAp73 isoforms could account for part of the neuronal cell death caused by activation of the APP pathway. On the contrary, ΔNp73β exerts protective effect against various neurotoxic stresses.
Neurotoxic Stresses Increase Both TAp73 and ΔNp73 mRNA Levels
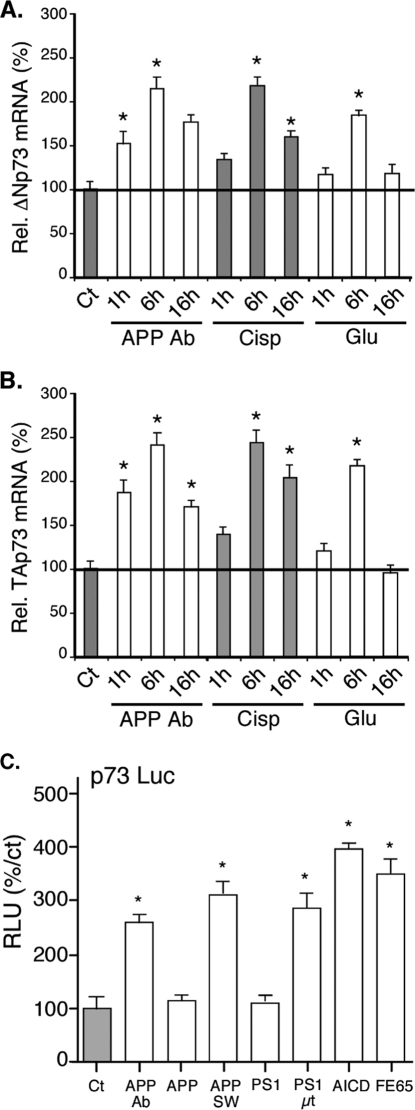
To analyze the molecular mechanisms leading to changes in the protein levels of p73β isoforms, we monitored by real time PCR the mRNA levels of TAp73 and ΔNp73 after activation of the APP pathway in cortical neurons (Fig. 5, A and B). As expected from the protein data, mRNA levels of TAp73 isoforms were augmented by the APP-directed antibody, suggesting a regulation at the promoter level. The mRNA levels of ΔNp73 isoforms were also increased, which contrasted with the data obtained by Western blot (Fig. 1B).
FIGURE 5.
Neurotoxic stresses induce both TAp73 and ΔNp73 isoforms mRNAs. A and B, cortical neurons were left untreated (Ct) or treated with the APP Ab (5 μg/ml), cisplatin (Cisp, 20 μm) or glutamate (Glu, 250 μm) for the indicated time before preparation of total mRNAs. After reverse transcription, real time PCRs were performed using TBP and either ΔNp73 (A) or TAp73 (B) isoform-specific primers. For standardization, TBP values were used. Graphs represent the percentage of induction relative to the control condition (Ct). Bars are means of three wells of a representative experiment with S.D. C, cortical neurons were transfected with the p73-Luc reporter construct along with different expression vectors (empty, Ct; encoding APP, wild type (APP), mutated as in Alzheimer patients (APPSW) or the AICD cytoplasmic fragment (AICD); encoding the APP cofactor FE65 (FE65)) or treated with the APP-directed antibody (5 μg/ml). Graphs represent means ± S.D. (relative light units (RLU)) of 3 wells from one experiment out of three independent experiments. * indicates statistically significant differences (analysis of variance, Neuman-Keuls multiple comparison test, p < 0.05).
To confirm the regulation of p73 expression by transcriptional mechanisms, we used a luciferase reporter gene controlled by the p73 promoter (53). Treatment with the APP-directed antibody, overexpression of a mutated form of APP, or a mutated PS1 led to a stimulation of the p73 promoter activity (Fig. 5C). Interestingly, overexpression of the cytoplasmic AICD fragment of APP or the APP cofactor FE65 also stimulated the activity of the p73 promoter.
These results suggest that the increase of TAp73β protein levels upon neurotoxic stresses is mediated through transcriptional activation of the p73 promoter. However, in the case of ΔNp73β, the regulation at the mRNA level could not explain the disappearance of ΔNp73β proteins after neurotoxic treatments.
APP Signaling and Neurotoxic Stresses Induce the Degradation of ΔNp73β through Multiple Proteases
To understand the mechanisms involved in the down-regulation of the ΔNp73β proteins that contrasted with the stimulation at the mRNA level, we tested the possibility that ΔNp73β proteins could be degraded in the course of neural apoptosis. To this end, neurotoxic treatments were performed in the presence of various proteases inhibitors that were found to be involved in neuronal apoptosis (54). As shown in Fig. 6A, ΔNp73β protein level in cortical neurons slightly increased in the presence of caspases (VAD) and proteasome (ALLN) inhibitors. More strikingly, these inhibitors reversed in part the loss of ΔNp73β caused by the APP-directed antibody as shown by the increased difference between control and the ALLN or VAD conditions in presence of APP Ab (Fig. 6A and supplemental data 3).
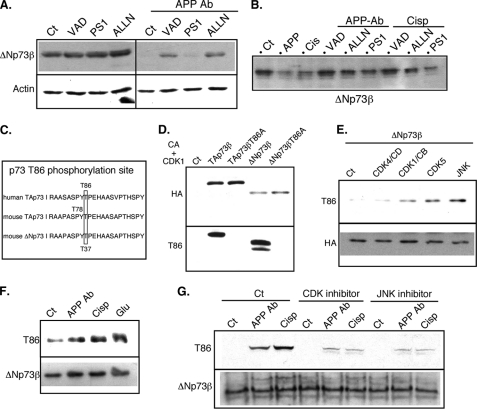
FIGURE 6.
Neurotoxic stresses lead to ΔNp73 degradation by proteases and phosphorylation at threonine 86. A, cortical neurons were left untreated or treated for 18 h with APP Ab (5 μg/ml) in the presence or absence of protease inhibitors (VAD, 50 μm; PS1, 25 μm; ALLN, 15 μm). Cells were lysed, and proteins (75 μg) were separated on a 10% SDS-PAGE and immunoblotted with the p73 CO73A and actin antibodies. B, overexpressed ΔNp73β was immunoprecipitated from N2A protein extracts and incubated with extracts of cortical treated with the APP-directed antibody (APP Ab, 5 μg/ml) or cisplatin (Cisp, 20 μm) in the presence or absence of protease inhibitors. Proteins were then separated on a 10% SDS-PAGE and immunoblotted with an HA-directed antibody. C, schematic representation and comparison between mouse and human sequences for TAp73 and ΔNp73. D, TAp73β wild type, TAp73β mutated in the Thr-86 site (TAp73βT86A), ΔNp73β wild type, and ΔNp73β mutated in the Thr-86 site (ΔNp73βT86A) were expressed in N2A cells. Proteins were extracted with a TEGN buffer containing no phosphatase inhibitor, and p73 proteins were immunoprecipitated as described. Immunoprecipitated p73 proteins were submitted to an in vitro kinase assay using a purified cyclin A-CDK1 complex (CA+CDK1) before SDS-PAGE and successive immunoblotting with the phospho-specific Thr-86 antibody (Thr-86, lower panel) and the HA antibody (upper panel) to detect the level of immunoprecipitated p73 proteins. E, N2A cells were cotransfected with HA-ΔNp73β, cyclin D (CD), and CDK4, cyclin B (CB), and CDK1, CDK5, or JNK expression vectors as indicated. HA-immunoprecipitated p73 proteins were separated by SDS-PAGE and successively immunoblotted with the anti-phospho-Thr-86 antibody (Thr-86, upper panel) and the HA antibody (lower panel). F and G, cortical neurons were treated with the APP-directed antibody (APP Ab, 5 μg/ml), cisplatin (Cisp, 20 μm), or glutamate (Glu, 250 μm) for 24 h. Co-treatments with CDK inhibitor (roscovitine, 5 μm) or JNK inhibitor (PB1557, 5 μm) was performed in G. p73 proteins were immunoprecipitated from the protein extract (2 mg) using the COP73A p73 antibody, separated by SDS-PAGE, and successively immunoblotted using the anti-phospho-Thr-86 antibody (Thr-86, upper panel) and the COP73A antibody (ΔNp73β, lower panel) to check the levels of immunoprecipitated p73 proteins. Images presented are from a representative experiment out of at least three independent experiments.
To dismiss the possibility that the increase in ΔNp73β protein levels was due to an indirect effect of these protease inhibitors, we tested their ability to antagonize the degradation of ΔNp73β protein in vitro. ΔNp73β was expressed in the N2A cell line and immunoprecipitated. After multiple washes, the immunoprecipitated ΔNp73β was incubated with various cellular extracts in the presence or absence of protease inhibitors. Extracts from cortical neurons treated by the APP-directed antibody or cisplatin induced the degradation of the immunopurified ΔNp73β proteins (Fig. 6B and supplemental data 3). Remarkably, addition of the caspase inhibitor or the proteasome inhibitor reversed this degradation. These results demonstrate that the diminution of ΔNp73β protein levels observed after neurotoxic treatments resulted from a degradation induced by multiple proteases.
Neurotoxic Stress-induced Degradation of ΔNp73β Depends on Phosphorylation of Thr-86 by JNK and CDK
Protein degradation is often controlled by phosphorylation events, as has been shown for p53. In previous studies (33), we have found that the TAp73 proteins can be phosphorylated at Thr-86 by CDKs. This phosphorylation site is also present in ΔNp73 isoforms. Given that CDK have been implicated in neural cell death (55), we hypothesized that neurotoxic stresses may induce CDK activity leading to phosphorylation of ΔNp73β at Thr-86, as this site is conserved between human and mouse, and between TA and ΔN isoforms (Fig. 6C).
We first analyzed whether Thr-86 of ΔNp73β could be phosphorylated in vitro by CDKs. For this, ΔNp73β was immunoprecipitated from transfected N2A cells and then incubated with a purified cyclin A-CDK1 complex. Western blot analysis using an antibody specifically directed against p73 proteins phosphorylated at Thr-86 indicated Thr-86 phosphorylation for wild type ΔNp73β but not for a T86A mutant of ΔNp73β (Fig. 6D). Of note, ΔNp73β in vitro phosphorylation at Thr-86 by cyclin A/CDK1 appeared stronger than that of TAp73β. Next, we verified that ΔNp73β was the target of CDKs in intact N2A cells. ΔNp73β was coexpressed with various CDK complexes, including CDK5 that has been extensively studied for its role in Alzheimer disease (56). As we have already shown for TAp73, CDK complexes containing cyclin A or B associated with CDK1 or CDK2 phosphorylated ΔNp73β (Fig. 6E). In contrast, complexes containing cyclin E or D associated with CDK4 did not. Finally, we found that CDK5 and JNK were able to induce substantial phosphorylation of Thr-86 (Fig. 6E). Thr-86 is also located in a potential JNK phosphorylation site, and the importance of JNK in neuronal apoptosis has been largely demonstrated (57). In particular, we have shown that JNK is a mediator of the cell death induced by the APP-directed antibody (43).
Next, we monitored the Thr-86 phosphorylation status of endogenous ΔNp73β in cortical neurons submitted to various neurotoxic stresses. p73 proteins were immunoprecipitated and analyzed by Western blot using the phosphospecific Thr-86 antibody. As shown in Fig. 6F, treatment with the APP-directed antibody or other neurotoxic treatment (cisplatin or glutamate) increased Thr-86 phosphorylation. Using specific kinase inhibitors, we then determined whether CDKs or JNKs were involved in Thr-86 phosphorylation. Both inhibitors led to a significant but partial reversion of the Thr-86 phosphorylation induced by neurotoxic stresses, suggesting that both types of kinases target this site in neurons (Fig. 6G). Taken together, these data suggest that increased Thr-86 phosphorylation of ΔNp73β by CDKs and JNKs upon neurotoxic stress enhances its degradation through caspases or the proteasome pathway.
Phosphorylation of Thr-86 Regulates the Stability ΔNp73β and Modulates Its Neuroprotective Effect
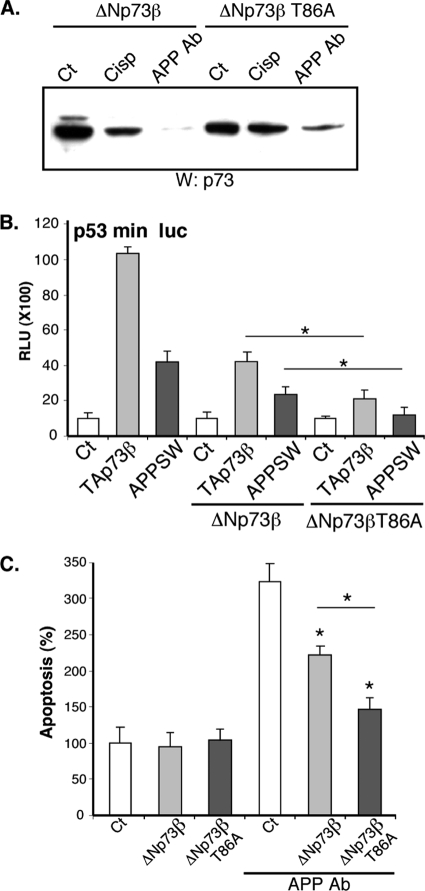
To test the importance of the phosphorylation of ΔNp73β at Thr-86, we compared the activity of wild type and mutant T86A ΔNp73β on the p53 min luciferase reporter gene. Overexpression of TAp73β or mutant APP induced the activity of this reporter gene in cortical neurons (Fig. 7B). This activation was inhibited by coexpression of ΔNp73β. Interestingly, the T86A mutant ΔNp73β displayed a more robust inhibition than wild type ΔNp73β (Fig. 7B). When we studied the relative protein levels of the ΔNp73β variants, we observed that the mutant T86A was expressed at a slightly higher level (Fig. 7A). From these results, we hypothesized that the stability of the T86A mutant could be different from the wild type. Therefore, we tested the sensitivity of wild type and T86A mutant ΔNp73β toward extracts from cortical neurons treated with the APP-directed antibody. As observed in Fig. 7A, extracts from neurons treated with neurotoxic stresses induced the degradation of wild type ΔNp73β. In contrast, the T86A mutant was significantly more resistant (Fig. 7A).
FIGURE 7.
Thr-86 phosphorylation regulates and destabilizes ΔNp73 proteins. A, overexpressed ΔNp73β and ΔNp73βT86A were immunoprecipitated from N2A cellular extracts and incubated with extracts of cortical neurons left untreated (Ct), treated with the APP-directed antibody (APP Ab, 5 μg/ml), or cisplatin (Cisp, 20 μm). Proteins were then separated on a 10% SDS-PAGE and immunoblotted with a p73 antibody. B, cortical neurons were cotransfected with the p53 min luciferase reporter construct, and the expression vectors coding for APP SW, TAp73β, and ΔNp73β (wild type or T86A mutant) as indicated. Bars represent means ± S.D. (relative light units (RLU)) of 3 wells from one experiment out of three independent experiments. C, cortical neurons were cotransfected with combination of ΔNp73β or ΔNp73βT86A and GFP expression vectors. After 24 h of expression, cells were treated with APP Ab (5 μg/ml) for 24 h. Cells were then fixed, stained with Hoechst, and observed with a fluorescence microscope. Graphs represent the percentage of apoptotic cells among the GFP-positive cells counted. Bars are means of three wells of a representative experiment with S.D. * indicates statistically significant differences (analysis of variance, Neuman-Keuls Multiple comparison test, p < 0.05).
We further analyzed the physiological relevance of the JNK and CDK-dependent phosphorylation of ΔNp73β at Thr-86 by testing its ability to rescue neurons from cell death. Wild type or T86A ΔNp73β proteins were coexpressed with GFP in cortical neurons treated with the APP-directed antibody. GFP-positive cells were observed, and cell death was assessed on the morphology of the nucleus and the neurites. As illustrated in Fig. 7C, overexpression of wild type ΔNp73β partially rescued neurons from apoptosis induced by the APP-directed antibody (Fig. 7C). However, the rescue induced by the T86A ΔNp73β variant was more pronounced, confirming the importance of the Thr-86 phosphorylation site for regulation of the function of ΔNp73β in neurons.
DISCUSSION
Neuronal cell death is a cellular fate inherent to the development of an integrated neural network, but it may however also be inadequately activated by multiple stresses associated with neurodegenerative disorders such as Alzheimer, Parkinson, ALS, etc. Several key molecular mechanisms that relay external stresses or genetic alterations and generate neuronal cell death have been identified. They involve several specific pathways such as JNK/c-Jun, CDKs/E2F1, or p53, which often induce caspase 3 cleavage and nucleus alterations. The elucidation of these signaling pathways is a necessary step to design highly specific and efficient therapies against neurodegenerative disorders. However, it is still extremely difficult to have a precise and complete scheme on how each individual stress among their vast diversity is linked to specific apoptotic mechanisms. In this study, we have investigated in detail the contribution of p73 to neuronal cell death induced by several kinds of neurotoxic stresses in cortical neurons. We have also identified mechanisms that regulate the expression and the activity of several p73 isoforms harboring opposite functions. Our results indicate a convergence of various neurotoxic stresses toward common molecular mechanisms that affect p73 expression and might therefore represent interesting therapeutic targets.
p73, Ying and Yang in Neuronal Apoptosis?
Altogether, our data showed interesting opposite regulations and effects during neuronal survival for TAp73 and ΔNp73, which highlight the essential role of p73 in the CNS. This role was already seen in the p73 knock-out mice that have neurological defects (17) but was mainly attributed to ΔNp73 isoforms. However, as all isoforms were inactivated through this approach, the interpretations about the respective role of TA and ΔN isoforms were limited de facto. Furthermore, a possible compensation by the other members of the p53 family, p53 and p63, might also complicate the understanding of the role of each individual member in neuronal apoptosis. Recently, such complexity was further illustrated through the realization of ΔNp73 transgenic mice that display more severe embryonic defects than the individual p53, p73, or p63 knock-out mice (58). Therefore, in vitro studies performed with primary cultures of neurons are useful to decipher the intricate roles of the p53 family members.
Our analysis of p73 expression in cortical neurons clearly showed that TAp73 and ΔNp73 isoforms were coregulated, with the ratio of both types of isoforms depending on the physiological conditions (Fig. 1). The expression level was regulated by diverse apoptotic conditions such as DNA damages (cisplatin) or alterations of the APP pathway (Fig. 1), leading to increased expression of p53/p73 target genes, such as Noxa (Figs. 1 and 2). This regulation suggested that both ΔNp73 and TAp73 were playing a role in neuronal survival. Indeed, overexpression of TAp73 induced apoptosis, although ΔNp73 was protecting neurons from stresses (Figs. 3 and 4).
The neuroprotective role of ΔNp73 might be linked to the inactivation of p53, as reported previously for the survival of derived root ganglia neurons following NGF treatment (24) or CNS neurons (25). This study shows that TAp73 isoforms are expressed and play a role in cortical neuron apoptosis in response to alteration of the APP pathway or DNA damages, indicating that ΔNp73 can also exert its neuroprotective activity by inhibiting TAp73 isoforms. Furthermore, recent studies have also shown that p63 is involved in other neurotoxic stress responses in cortical neurons, such as ischemia (26). Therefore, the situation in cortical neurons is rather complex, and the exact role of each p53 family member remains to be established because they potentially share similar cellular functions and target genes. In vivo functional studies using appropriate transgenic animals, as well as the precise identification of the specific pool of target genes regulated by each p53 protein in specific neurotoxic conditions, would bring crucial information for understanding the respective role of each member. The molecular mechanism of the dominant negative activity of ΔNp73 also remains to be determined. Obviously, ΔNp73 might repress some pro-apoptotic p53 family members through heterodimerization, as described for TAp73 and p63 (59), which still remains controversial for p73 and wild type p53 (59). Another possibility is that ΔNp73 exerts its dominant negative activity through the competition for p53-binding sites. However, the fact that the various p53 family members display different binding affinities might limit the dominant negative effect of ΔNp73 on p53 target genes (60). Alternatively, the high level of ΔNp73 proteins might have an effect on its own, targeting a specific subset of genes linked to neuronal survival.
Finally, it is important to note that ΔNp73 did not rescue all neurons from apoptosis, even if a significant proportion was protected. The death of these unprotected neurons might be attributed to the limit of the experimental approach that did not allow an accurate correlation between the expression level of ΔNp73 expression in each neuron and its fate. Obviously, it may also correspond to the involvement of mechanisms independent of p53 family members.
Conversely, we found that TAp73 was important for cisplatin- and APP-induced neuronal cell death (Fig. 3). Overexpression of TAp73 induced apoptosis, whereas siRNAs that knock down specifically TAp73 isoforms partially rescued neurons from apoptosis (Fig. 3). Furthermore, we identified pro-apoptotic genes that were up-regulated by TAp73 in cortical neurons, namely Noxa, Bax and DR5 (Fig. 1). This indicates that TAp73 can directly regulate the expression of genes linked to cell death in neurons (61, 62). These genes are also targeted by p53 and might therefore be coregulated by p53 and TAp73 in neuronal stress conditions that induce both transcription factors, such as APP stimulation, DNA damage, or neurotoxic stress (Figs. 1 and 3) (30, 49). Nevertheless, our data about TAp73 provide some molecular explanations to the observation that p73 proteins are induced in neurons of Alzheimer patients (27, 28).
Regulation of p73 Isoforms in Neurons
We found that the protein levels of TAp73 and ΔNp73 were oppositely affected by neurotoxic stresses (Figs. 1 and 3). The molecular mechanisms involved were different for both types of isoforms.
TAp73 expression was increased at both the protein and the mRNA levels (Figs. 1, 3, and 5), suggesting that its expression was partially regulated through its promoter. Indeed, we found that the APP pathway activated the p73 promoter (Fig. 5). The transcription factors involved remain to be identified, but one possible player is E2F1 as its expression is induced by the β-amyloid peptide (63).
Our analysis of ΔNp73 expression revealed a post-translational mechanism. Inhibitors of calpain, caspases, and proteasome rescued the loss of ΔNp73 expression after neurotoxic treatments. All three kinds of proteases played a role, but the proteasome and the caspases seemed to be predominant (Fig. 6). These proteases have been described for their important role in neuronal apoptosis (54). Calpain cleavage and proteasome-dependent degradation of p73 have already been described and might account for the regulations we observed here (64–66). It remains to be established whether these mechanisms are recruited in neurons to directly degrade ΔNp73 or if specific neuronal cofactors, such as ubiquitin ligases, step in.
The degradation of ΔNp73 was tightly controlled by phosphorylation at threonine 86 (Fig. 6). Neurotoxic stresses induced this phosphorylation through JNK and CDK (Fig. 6). These kinases (especially CDK5 and JNK) were frequently associated with neuronal apoptosis and Alzheimer disease (41, 56, 57). The exact mechanisms that link Thr-86 phosphorylation to ΔNp73 degradation remain to be established. These mechanisms may involve changes in ΔNp73 protein conformation, its localization, or its association with partner proteins that control its degradation.
It is likely that additional post-translational modifications, such as other phosphorylation, acetylation, sumoylation, or isomerization by Pin1, participate in the control of ΔNp73 and TAp73 activity and stability during neuronal cell death (67, 68). In particular, the tyrosine kinase c-Abl could be an interesting candidate as it has been shown to phosphorylate and stabilize TAp73 (44) and has also been proposed to play a role in tauopathies and Alzheimer disease (69). Additional studies will allow us to further detail the cascades of events that link p73 to neuronal cell death and in association with neurodegenerative diseases.
p53 Family and Neurodegenerative Diseases
Our results show a significant role of the p73 gene in the control of neuronal fate, with TAp73 being a neuronal cell death mediator, although ΔNp73 has an interesting potential for rescuing neurons from various stresses. p73 appears to be a sensor of various neurotoxic stresses, making it an ideal intracellular relay for genetic or sporadic neurodegenerative diseases. In particular, we have shown that alteration of the APP pathway using APP Ab or mutated APP leads to changes in p73 isoform expression (Fig. 1). Moreover, silencing of TAp73 reduced the neurotoxic effect of APP signaling pathway alteration (Fig. 3). Even if the exact APP processing cascade involved in these observations is not yet clear, these data support previous observations. Indeed, p73 expression is increased in the brain of Alzheimer patients (27, 28), but as p53 and p63 are also involved in neuronal fate (30, 70, 71), it might not be the only member of the p53 family to be involved in neurodegenerative diseases. p53 has already been associated with several neurodegenerative disorders such as ALS (72), Alzheimer (15), HIV-mediated dementia (73), or Parkinson (74). In particular, the AICD domain of APP that can act as a transcription factor (75) has been shown to regulate p53 (76). The transcriptional activity of the AICD might also explain the ability of an altered APP signaling pathway to regulate the p73 promoter (Fig. 5). Several key questions remain unanswered, such as whether p53 family members share common target genes or whether they interact in specific physiological situations. However, our study shows that deleterious neurodegenerative-associated mechanisms can be reduced through a modulation of the p53-like dependent function, which suggests that the p53 family represents in its whole a potential target for a therapeutic approach against neurodegenerative diseases.
Supplementary Material
Acknowledgments
We thank Drs. T. Tanigushi and T. Sakai for providing us the p73 and Noxa vectors.
This work was supported by CNRS, Université de Strasbourg, Association pour la Recherche Contre le Cancer Grant 3288, La Ligue Contre le Cancer (Comité du Haut-Rhin, Moselle, and Meurthe-et-Moselle), ANR, Institut National du Cancer, and CONECTUS Alsace, Fond National pour la Recherche (Luxembourg, FNR).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–4.
- TA
- transactivation domain
- APP
- amyloid-β precursor protein
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide
- CDK
- cyclin-dependent kinase
- AICD
- amyloid precursor protein intracellular cytoplasmic/C-terminal domain.
REFERENCES
- 1. Prives C., Hall P. A. (1999) J. Pathol. 187, 112–126 [DOI] [PubMed] [Google Scholar]
- 2. Menendez D., Inga A., Resnick M. A. (2009) Nat. Rev. Cancer 9, 724–737 [DOI] [PubMed] [Google Scholar]
- 3. Eizenberg O., Faber-Elman A., Gottlieb E., Oren M., Rotter V., Schwartz M. (1996) Mol. Cell. Biol. 16, 5178–5185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ferreira A., Kosik K. S. (1996) J. Cell Sci. 109, 1509–1516 [DOI] [PubMed] [Google Scholar]
- 5. Migliorini D., Lazzerini Denchi E., Danovi D., Jochemsen A., Capillo M., Gobbi A., Helin K., Pelicci P. G., Marine J. C. (2002) Mol. Cell. Biol. 22, 5527–5538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sakhi S., Bruce A., Sun N., Tocco G., Baudry M., Schreiber S. S. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 7525–7529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Morrison R. S., Wenzel H. J., Kinoshita Y., Robbins C. A., Donehower L. A., Schwartzkroin P. A. (1996) J. Neurosci. 16, 1337–1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jordán J., Galindo M. F., Prehn J. H., Weichselbaum R. R., Beckett M., Ghadge G. D., Roos R. P., Leiden J. M., Miller R. J. (1997) J. Neurosci. 17, 1397–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Trimmer P. A., Smith T. S., Jung A. B., Bennett J. P., Jr. (1996) Neurodegeneration 5, 233–239 [DOI] [PubMed] [Google Scholar]
- 10. Jiang Y. H., Armstrong D., Albrecht U., Atkins C. M., Noebels J. L., Eichele G., Sweatt J. D., Beaudet A. L. (1998) Neuron 21, 799–811 [DOI] [PubMed] [Google Scholar]
- 11. de la Monte S. M., Sohn Y. K., Wands J. R. (1997) J. Neurol. Sci. 152, 73–83 [DOI] [PubMed] [Google Scholar]
- 12. Ohyagi Y., Asahara H., Chui D. H., Tsuruta Y., Sakae N., Miyoshi K., Yamada T., Kikuchi H., Taniwaki T., Murai H., Ikezoe K., Furuya H., Kawarabayashi T., Shoji M., Checler F., Iwaki T., Makifuchi T., Takeda K., Kira J., Tabira T. (2005) FASEB J. 19, 255–257 [DOI] [PubMed] [Google Scholar]
- 13. Zhang Y., McLaughlin R., Goodyer C., LeBlanc A. (2002) J. Cell Biol. 156, 519–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Seidl R., Fang-Kircher S., Bidmon B., Cairns N., Lubec G. (1999) Neurosci. Lett. 260, 9–12 [DOI] [PubMed] [Google Scholar]
- 15. Kitamura Y., Shimohama S., Kamoshima W., Matsuoka Y., Nomura Y., Taniguchi T. (1997) Biochem. Biophys. Res. Commun. 232, 418–421 [DOI] [PubMed] [Google Scholar]
- 16. Arrowsmith C. H. (1999) Cell Death Differ. 6, 1169–1173 [DOI] [PubMed] [Google Scholar]
- 17. Yang A., Walker N., Bronson R., Kaghad M., Oosterwegel M., Bonnin J., Vagner C., Bonnet H., Dikkes P., Sharpe A., McKeon F., Caput D. (2000) Nature 404, 99–103 [DOI] [PubMed] [Google Scholar]
- 18. De Laurenzi V., Costanzo A., Barcaroli D., Terrinoni A., Falco M., Annicchiarico-Petruzzelli M., Levrero M., Melino G. (1998) J. Exp. Med. 188, 1763–1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Casciano I., Ponzoni M., Lo Cunsolo C., Tonini G. P., Romani M. (1999) Cell Death Differ. 6, 391–393 [DOI] [PubMed] [Google Scholar]
- 20. Yang A., Kaghad M., Wang Y., Gillett E., Fleming M. D., Dötsch V., Andrews N. C., Caput D., McKeon F. (1998) Mol. Cell 2, 305–316 [DOI] [PubMed] [Google Scholar]
- 21. Grob T. J., Novak U., Maisse C., Barcaroli D., Lüthi A. U., Pirnia F., Hügli B., Graber H. U., De Laurenzi V., Fey M. F., Melino G., Tobler A. (2001) Cell Death Differ. 8, 1213–1223 [DOI] [PubMed] [Google Scholar]
- 22. De Laurenzi V., Raschellá G., Barcaroli D., Annicchiarico-Petruzzelli M., Ranalli M., Catani M. V., Tanno B., Costanzo A., Levrero M., Melino G. (2000) J. Biol. Chem. 275, 15226–15231 [DOI] [PubMed] [Google Scholar]
- 23. Billon N., Terrinoni A., Jolicoeur C., McCarthy A., Richardson W. D., Melino G., Raff M. (2004) Development 131, 1211–1220 [DOI] [PubMed] [Google Scholar]
- 24. Pozniak C. D., Barnabé-Heider F., Rymar V. V., Lee A. F., Sadikot A. F., Miller F. D. (2002) J. Neurosci. 22, 9800–9809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lee A. F., Ho D. K., Zanassi P., Walsh G. S., Kaplan D. R., Miller F. D. (2004) J. Neurosci. 24, 9174–9184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bui T., Sequeira J., Wen T. C., Sola A., Higashi Y., Kondoh H., Genetta T. (2009) PLoS ONE 4, e4373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wilson C., Henry S., Smith M. A., Bowser R. (2004) Neuropathol. Appl. Neurobiol. 30, 19–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li Q., Athan E. S., Wei M., Yuan E., Rice S. L., Vonsattel J. P., Mayeux R. P., Tycko B. (2004) BMC Med. Genet. 5, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wetzel M. K., Naska S., Laliberté C. L., Rymar V. V., Fujitani M., Biernaskie J. A., Cole C. J., Lerch J. P., Spring S., Wang S. H., Frankland P. W., Henkelman R. M., Josselyn S. A., Sadikot A. F., Miller F. D., Kaplan D. R. (2008) Neuron 59, 708–721 [DOI] [PubMed] [Google Scholar]
- 30. Benosman S., Gross I., Clarke N., Jochemsen A. G., Okamoto K., Loeffler J. P., Gaiddon C. (2007) Cell Death Differ. 14, 2047–2057 [DOI] [PubMed] [Google Scholar]
- 31. Mosmann T. (1983) J. Immunol. Methods 65, 55–63 [DOI] [PubMed] [Google Scholar]
- 32. Meng X., Leyva M. L., Jenny M., Gross I., Benosman S., Fricker B., Harlepp S., Hebraud P., Boos A., Wlosik P., Bischoff P., Sirlin C., Pfeffer M., Loeffler J. P., Gaiddon C. (2009) Cancer Res. 69, 5458–5466 [DOI] [PubMed] [Google Scholar]
- 33. Gaiddon C., Lokshin M., Gross I., Levasseur D., Taya Y., Loeffler J. P., Prives C. (2003) J. Biol. Chem. 278, 27421–27431 [DOI] [PubMed] [Google Scholar]
- 34. Sohm F., Gaiddon C., Antoine M., Boutillier A. L., Loeffler J. P. (1999) Oncogene 18, 2762–2769 [DOI] [PubMed] [Google Scholar]
- 35. Gross I., Armant O., Benosman S., de Aguilar J. L., Freund J. N., Kedinger M., Licht J. D., Gaiddon C., Loeffler J. P. (2007) Cell Death Differ. 14, 1802–1812 [DOI] [PubMed] [Google Scholar]
- 36. René F., Monnier D., Gaiddon C., Félix J. M., Loeffler J. P. (1996) Neuroendocrinology 64, 2–13 [DOI] [PubMed] [Google Scholar]
- 37. Gaiddon C., de Tapia M., Loeffler J. P. (1999) Mol. Endocrinol. 13, 742–751 [DOI] [PubMed] [Google Scholar]
- 38. Silhavy T. J., Berman M. L., Enquist L. W. (1984) Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 39. Kedar A., Cohen M. E., Freeman A. I. (1978) Cancer Treat. Rep. 62, 819–821 [PubMed] [Google Scholar]
- 40. Steeghs N., de Jongh F. E., Sillevis Smitt P. A., van den Bent M. J. (2003) Anti-Cancer Drugs 14, 443–446 [DOI] [PubMed] [Google Scholar]
- 41. Mbebi C., Sée V., Mercken L., Pradier L., Müller U., Loeffler J. P. (2002) J. Biol. Chem. 277, 20979–20990 [DOI] [PubMed] [Google Scholar]
- 42. Bouron A., Mbebi C., Loeffler J. P., De Waard M. (2004) Eur. J. Neurosci. 20, 2071–2078 [DOI] [PubMed] [Google Scholar]
- 43. Mbebi C., González de Aguilar J. L., Sée V., Dupuis L., Frossard N., Mercken L., Pradier L., Larmet Y., Loeffler J. P. (2005) Neurobiol. Dis. 19, 129–141 [DOI] [PubMed] [Google Scholar]
- 44. Gong J. G., Costanzo A., Yang H. Q., Melino G., Kaelin W. G., Jr., Levrero M., Wang J. Y. (1999) Nature 399, 806–809 [DOI] [PubMed] [Google Scholar]
- 45. Irwin M. S., Kondo K., Marin M. C., Cheng L. S., Hahn W. C., Kaelin W. G., Jr. (2003) Cancer Cell 3, 403–410 [DOI] [PubMed] [Google Scholar]
- 46. Urist M., Tanaka T., Poyurovsky M. V., Prives C. (2004) Genes Dev. 18, 3041–3054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kiryu-Seo S., Hirayama T., Kato R., Kiyama H. (2005) J. Neurosci. 25, 1442–1447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Di Como C. J., Gaiddon C., Prives C. (1999) Mol. Cell. Biol. 19, 1438–1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wu G. S., Burns T. F., McDonald E. R., 3rd, Jiang W., Meng R., Krantz I. D., Kao G., Gan D. D., Zhou J. Y., Muschel R., Hamilton S. R., Spinner N. B., Markowitz S., Wu G., el-Deiry W. S. (1997) Nat. Genet. 17, 141–143 [DOI] [PubMed] [Google Scholar]
- 50. Pardo L. M., van Duijn C. M. (2005) Mutat. Res. 592, 89–101 [DOI] [PubMed] [Google Scholar]
- 51. Brouwers N., Sleegers K., Van Broeckhoven C. (2008) Ann. Med. 40, 562–583 [DOI] [PubMed] [Google Scholar]
- 52. Nishimura I., Uetsuki T., Dani S. U., Ohsawa Y., Saito I., Okamura H., Uchiyama Y., Yoshikawa K. (1998) J. Neurosci. 18, 2387–2398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ding Y., Inoue T., Kamiyama J., Tamura Y., Ohtani-Fujita N., Igata E., Sakai T. (1999) DNA Res. 6, 347–351 [DOI] [PubMed] [Google Scholar]
- 54. Yuan J., Lipinski M., Degterev A. (2003) Neuron 40, 401–413 [DOI] [PubMed] [Google Scholar]
- 55. Freeman R. S., Estus S., Johnson E. M., Jr. (1994) Neuron 12, 343–355 [DOI] [PubMed] [Google Scholar]
- 56. Monaco E. A., 3rd, Vallano M. L. (2005) Front. Biosci. 10, 143–159 [DOI] [PubMed] [Google Scholar]
- 57. Xia Z., Dickens M., Raingeaud J., Davis R. J., Greenberg M. E. (1995) Science 270, 1326–1331 [DOI] [PubMed] [Google Scholar]
- 58. Hüttinger-Kirchhof N., Cam H., Griesmann H., Hofmann L., Beitzinger M., Stiewe T. (2006) Cell Death Differ. 13, 174–177 [DOI] [PubMed] [Google Scholar]
- 59. Gaiddon C., Lokshin M., Ahn J., Zhang T., Prives C. (2001) Mol. Cell. Biol. 21, 1874–1887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lokshin M., Li Y., Gaiddon C., Prives C. (2007) Nucleic Acids Res. 35, 340–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Martin L. J., Chen K., Liu Z. (2005) J. Neurosci. 25, 6449–6459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Vogel M. W. (2002) Cerebellum 1, 277–287 [DOI] [PubMed] [Google Scholar]
- 63. Giovanni A., Wirtz-Brugger F., Keramaris E., Slack R., Park D. S. (1999) J. Biol. Chem. 274, 19011–19016 [DOI] [PubMed] [Google Scholar]
- 64. Munarriz E., Bano D., Sayan A. E., Rossi M., Melino G., Nicotera P. (2005) Biochem. Biophys. Res. Commun. 333, 954–960 [DOI] [PubMed] [Google Scholar]
- 65. Asher G., Tsvetkov P., Kahana C., Shaul Y. (2005) Genes Dev. 19, 316–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Rossi M., De Laurenzi V., Munarriz E., Green D. R., Liu Y. C., Vousden K. H., Cesareni G., Melino G. (2005) EMBO J. 24, 836–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Oberst A., Rossi M., Salomoni P., Pandolfi P. P., Oren M., Melino G., Bernassola F. (2005) Biochem. Biophys. Res. Commun. 331, 707–712 [DOI] [PubMed] [Google Scholar]
- 68. Lunghi P., Costanzo A., Mazzera L., Rizzoli V., Levrero M., Bonati A. (2009) Clin. Cancer Res. 15, 6495–6502 [DOI] [PubMed] [Google Scholar]
- 69. Lebouvier T., Scales T. M., Williamson R., Noble W., Duyckaerts C., Hanger D. P., Reynolds C. H., Anderton B. H., Derkinderen P. (2009) J. Alzheimers Dis. 18, 1–9 [DOI] [PubMed] [Google Scholar]
- 70. Jacobs W. B., Govoni G., Ho D., Atwal J. K., Barnabe-Heider F., Keyes W. M., Mills A. A., Miller F. D., Kaplan D. R. (2005) Neuron 48, 743–756 [DOI] [PubMed] [Google Scholar]
- 71. Jacobs W. B., Walsh G. S., Miller F. D. (2004) Neuroscientist 10, 443–455 [DOI] [PubMed] [Google Scholar]
- 72. González de Aguilar J. L., Gordon J. W., René F., de Tapia M., Lutz-Bucher B., Gaiddon C., Loeffler J. P. (2000) Neurobiol. Dis. 7, 406–415 [DOI] [PubMed] [Google Scholar]
- 73. Garden G. A., Guo W., Jayadev S., Tun C., Balcaitis S., Choi J., Montine T. J., Möller T., Morrison R. S. (2004) FASEB J. 18, 1141–1143 [DOI] [PubMed] [Google Scholar]
- 74. Mogi M., Kondo T., Mizuno Y., Nagatsu T. (2007) Neurosci. Lett. 414, 94–97 [DOI] [PubMed] [Google Scholar]
- 75. Cao X., Südhof T. C. (2001) Science 293, 115–120 [DOI] [PubMed] [Google Scholar]
- 76. Alves da Costa C., Sunyach C., Pardossi-Piquard R., Sévalle J., Vincent B., Boyer N., Kawarai T., Girardot N., St George-Hyslop P., Checler F. (2006) J. Neurosci. 26, 6377–6385 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.