Background: Selenoprotein K (SelK) is an endoplasmic reticulum (ER) protein of unknown function.
Results: SelK associates with proteins involved in elimination of misfolded proteins from the ER.
Conclusion: SelK is a functional component of ER homeostasis.
Significance: This study defines a new family of selenoproteins and links it to an ER-associated degradation.
Keywords: Endoplasmic Reticulum (ER), ERAD, Protein Evolution, Protein Misfolding, Redox Regulation, Selenium, Selenocysteine, Selenoprotein, Thioredoxin Reductase
Abstract
Selenoprotein K (SelK) is an 11-kDa endoplasmic reticulum (ER) protein of unknown function. Herein, we defined a new eukaryotic protein family that includes SelK, selenoprotein S (SelS), and distantly related proteins. Comparative genomics analyses indicate that this family is the most widespread eukaryotic selenoprotein family. A biochemical search for proteins that interact with SelK revealed ER-associated degradation (ERAD) components (p97 ATPase, Derlins, and SelS). In this complex, SelK showed higher affinity for Derlin-1, whereas SelS had higher affinity for Derlin-2, suggesting that these selenoproteins could determine the nature of the substrate translocated through the Derlin channel. SelK co-precipitated with soluble glycosylated ERAD substrates and was involved in their degradation. Its gene contained a functional ER stress response element, and its expression was up-regulated by conditions that induce the accumulation of misfolded proteins in the ER. Components of the oligosaccharyltransferase complex (ribophorins, OST48, and STT3A) and an ER chaperone, calnexin, were found to bind SelK. A glycosylated form of SelK was also detected, reflecting its association with the oligosaccharyltransferase complex. These data suggest that SelK is involved in the Derlin-dependent ERAD of glycosylated misfolded proteins and that the function defined by the prototypic SelK is the widespread function of selenium in eukaryotes.
Introduction
The endoplasmic reticulum (ER)3 is an important organelle for folding and maturation of secreted and membrane proteins. Most secretory proteins synthesized in the ER are modified by N-linked glycosylation. The oligosaccharyltransferase (OST) complex covalently attaches glucose3-mannose9-N-acetylglucosamine2 oligosaccharide to an asparagine residue in Asn-Xaa-Thr/Ser motifs of nascent polypeptides entering the ER (1). During the calnexin-calreticulin cycle (2, 3) proteins adopt their native structure and exit the ER in route to their final destination. If glycoproteins and nonglycosylated proteins fail to acquire their native conformation, they undergo ER-associated degradation (ERAD).
ERAD is a multi-step process involving dozens of proteins. The key events include (i) recognition of misfolded proteins (ERAD substrates) in the ER lumen; (ii) delivery of misfolded proteins from the ER lumen to the cytosol through a channel; (iii) extraction of proteins from the ER; and (iv) targeting them to the cytosolic proteasome for degradation (4–7).
The identity of the ERAD channel/translocon remains controversial because a number of multi-spanning ER-membrane proteins have been proposed to serve this function, including Sec61α; Derlin-1, -2, and -3; and E3 ubiquitin ligases (3, 4). Numerous data support the idea that Derlins could form a channel. First, these proteins are involved in or are required for degradation of many ERAD substrates (8–14). Second, Derlin-1 and -2 were found to interact with lumenal (8, 11), membrane (15, 16) and cytosolic (9, 14, 17) proteins required for retrotranslocation. Finally, Derlins are capable of forming homo- and heterooligomers (11, 15, 16, 18), which is consistent with their ability to form large complexes and serve as channels. Extraction of many misfolded, ubiquitinated proteins to the cytosol requires the presence of a p97 (also known as VCP) complex, a hexameric, cytosolic ATPase containing two co-factors (Ufd1 and Npl4) (19). This complex may be associated with the site of ubiquitination on the ER membrane by interaction with either ER membrane integral ubiquitin ligases, for instance, GP78 and HRD1 (16, 20), or with other integral proteins, such as Derlin-1 (16); erasin, also known as UBXD2 (21); and selenoprotein S (SelS) (22), also known as VIMP (14–16).
Many fundamental studies on the ERAD mechanism were performed using baker's yeast, Saccharomyces cerevisiae, as a model organism. Although ERAD is conserved across eukaryotes (4–7), mammals have many additional proteins that are involved in retrotranslocation of misfolded proteins. For example, these organisms contain a much larger set of ubiquitin ligases (4) and have three Derlins (7) instead of the single yeast Der1p. The mammalian ERAD complex also includes SelS, whereas fungi lack selenoproteins and the associated selenoprotein synthesis machinery. It is unclear why mammalian cells use a selenoprotein, which is absent in many species in the evolutionary conserved pathway.
Selenoproteins constitute a diverse group of proteins that contain the 21st naturally occurring amino acid, selenocysteine (Sec, one letter U). Sec is encoded by the codon, UGA, that usually serves as a termination signal. Decoding UGA during translation as a codon for Sec insertion requires a specific stem-loop structure in the 3′-UTR of eukaryotic selenoprotein mRNAs, designated the selenocysteine insertion sequence (SECIS) element, and a machinery to synthesize Sec on its own tRNA and insert it into proteins in response to the UGA codon (23–26). One of the reasons why nature maintained this machinery is the high reactivity of the Sec residue in the active sites of selenoenzymes. Seven of 25 human selenoproteins (22) are ER-resident selenoproteins; selenoprotein K (SelK) is one of them (27, 28). Among eukaryotic selenoproteins, SelK is one of the most widespread selenoproteins (29), but despite this evolutionary significance (29, 30) and ubiquitous expression in mammalian tissues (27, 31), the biochemical function of SelK is unknown. No light has been shed on the molecular basis of SelK function by the recent findings that (i) SelK is an important protein for promoting effective Ca2+ flux during immune cell activation (28); (ii) SelK is a target of calpains, proteases that modulate apoptosis, proliferation, and migration and are involved in regulation of inflammation and immune response (32); and (iii) SelK is regulated by ER stress, and its knockdown aggravates cell death and apoptosis induced by ER stress agents (3), as well as other reports on SelK (27, 34).
Here, we applied bioinformatics approaches to define a new protein family that involves SelK and SelS. We provide evidence that SelK, the prototypic protein of this family, associates with multi-protein complexes of the ER membrane. We also show its involvement in the Derlin-dependent ERAD of soluble glycosylated proteins and maintenance of ER homeostasis. This work defines the molecular role of the trace element selenium in these processes.
EXPERIMENTAL PROCEDURES
Bioinformatics Analyses
Previously identified SelK and SelS proteins were used as seed sequences in the computational searches, using BLASTP and TBLASTN for additional SelK/SelS homologs in both the nonredundant protein data base and sequenced eukaryotic genomes. Approximately 80 SelK and 50 SelS protein sequences from different organisms were collected. These proteins were analyzed manually, and a set of criteria (see “Results”) for identification of SelK and SelS proteins was developed.
The complete sets of predicted proteins of several sequenced eukaryotic organisms were retrieved from NCBI (ftp://ftp.ncbi.nih.gov/genomes/). The following model organisms were selected: Caenorhabditis elegans, Chlamydomonas reinhardtii, Cyanidioschyzon merolae, Drosophila melanogaster, Homo sapiens, Leishmania major, Plasmodium falciparum, S. cerevisiae, Takifugu rubripes, Tetraodon nigroviridis, and Thalassiosira pseudonana. Prediction of the transmembrane domain for each query protein was performed using TMHMM (Trans Membrane prediction using Hidden Markov Models) server tool (35). Only those containing a single N-terminal transmembrane region were selected for further analysis. Perl scripts were developed to extract proteins that satisfy the predefined SelK/SelS criteria. A subsequent analysis of secondary structure patterns and the occurrence of identified proteins in multiple organisms defined the SelK/SelS protein family.
Antibodies and Reagents
Mouse monoclonal antibodies to β-actin, KDEL, and rabbit polyclonal antibodies to FLAG tag were from Abcam. Anti-BiP (glucose-regulated protein 78) rabbit polyclonal antibodies were from Santa Cruz Biotechnology. Anti-Derlin-1, anti-Derlin-2, anti-Derlin-3, anti-HA, anti-ribophorin I (RPN I), anti-SelK Prestige, and anti-SelS rabbit affinity-isolated antibodies were from Sigma; anti-p97 mouse affinity-isolated antibodies were from BioLegend. Anti-HA-agarose, protein G-agarose, and tunicamycin were from Sigma. Highly cross-adsorbed Alexa Fluor® 488 goat anti-rabbit, Alexa Fluor® 647 goat anti-mouse antibodies, and DAPI FluoroPureTM grade were from Molecular Probes. Proteasome inhibitor MG132 and high purity water-soluble digitonin were from Calbiochem. RNase B, N-glycanase, and TEV protease were from New England Biolabs, Prozyme, and Invitrogen, respectively. Unless otherwise stated, all remaining reagents were from Sigma.
Constructs
cDNAs for mouse SelS, human SelK, mouse SelK, transcriptional variant 2 of human Derlin-3 (Derlin-3b), human Derlin-1, and human Derlin-2 were purchased as cDNA clones (IMAGE ID 3983625, 4345594, 2645414, 30338943, NM_024295, and NM_016041) from Thermo Scientific Open Biosystems.
Human Derlins with the FLAG epitope at the N terminus were cloned into the XhoI/XbaI restriction sites of pCI-neo vector (Promega). Full-length ORFs of human and mouse selenoproteins were cloned into the pCI-neo-based vector pCI-Toxo-SECIS developed in our laboratory. pCI-Toxo-SECIS contains a highly efficient Toxoplasma gondii SelT SECIS element (36) that was cloned downstream of the multiple cloning site into the NotI restriction site.
To overexpress mouse HA-tagged SelK and SelS, ORFs of these selenoproteins were inserted into the NheI/SmaI restriction sites of pCI-Toxo-SECIS. HA tag followed by EcoRI restriction site was inserted into the ORFs of mouse selenoproteins between the transmembrane and cytosolic regions through PCR and subcloning. Sec-to-Cys and Sec-to-Stop point mutations in the mouse selenoproteins were generated by site-directed mutagenesis using QuikChange kit (Stratagene).
To overexpress human SelK, its ORF was cloned into the XbaI/SalI restriction sites of pCI-Toxo-SECIS, yielding pCI-SelK-Toxo-SECIS. The HA-TEV tag was cloned into EcoRI/XbaI restriction sites of pCI-SelK-Toxo-SECIS, giving the construct, HT-SelK, coding for HA-TEV-tagged SelK. For efficient ER translocation, we generated a construct in which the signal sequence from mouse MHC class I heavy chain H-2Kb sequence peptide was fused to the N terminus of HA-TEV tag. The H-2Kb-HA-TEV tag was cloned into the EcoRI/XbaI restriction sites of pCI-SelK-Toxo-SECIS, yielding HHT-SelK. A Sec-to-Stop point mutation in the SelK sequence was generated by site-directed mutagenesis resulting in the construct HHT-SelKtr. Constructs expressing an HA-tagged form of ribophorin I (RPN332, a truncated form of ribophorin I containing 332 amino acids of its N-terminal end) and HA-tagged null Hong Kong mutant variant of α1-antitrypsin (NHK) were kind gifts from Dr. R. R. Kopito (Stanford University).
Cell Culture, Transfection, and RNA Interference
Human HEK 293 cells were grown in high glucose DMEM supplemented with 10% calf serum and antibiotic/antimycotic solution (all from Invitrogen) at 37 °C in 5% CO2. Human HeLa cells were grown in the same conditions, except that the medium was supplemented with 10% fetal bovine serum (Invitrogen). The cells were treated with tunicamycin or MG132 at concentrations and for the duration of time indicated in specific experiments.
Transient transfection of HeLa cells was carried out using FuGENE HD (Roche Applied Science) according to the manufacturer's recommendations. HEK 293 cells were transfected by the calcium phosphate co-precipitation technique. When co-transfections were performed, the constructs were co-transfected in a 1:1 ratio. SelK siRNA-mediated knockdown in HEK cells was performed using a set of three Stealth siRNA oligonucleotides (Invitrogen). Expression of human Derlins was decreased using siGENOME SMART pool siRNA (Thermo Scientific). Cells transfected with the Stealth scramble siRNAs with the high GC content (Invitrogen) were used as a control. RNAi was performed using DharmaFECT 1 transfection reagent according to the manufacturer's protocol. Transfection of constructs in siRNA-transfected cells was performed by the calcium phosphate method, 60–70 h after siRNA transfection. Knockdown of siRNA-targeted proteins was detected 96–106 h after transfection, and overexpression of recombinant proteins was analyzed 36–51 h after transfection. The cells were lysed in PBS containing complete protease inhibitor tablets (Roche Applied Science), 5 mm N-ethylmaleimide, and 1% Triton X-100.
Immunofluorescence Microscopy
HeLa cells were seeded 42 h after transfection onto Lab-Tek II four-chamber slides and allowed to attach for 9 h. After fixation with 4% paraformaldehyde (Polysciences) for 15 min at room temperature and two washes with ice-cold PBS, the cells were permeabilized with 0.1% Triton X-100 for 15 min at room temperature. The cells were washed three times with PBS and blocked in 10% goat serum (Invitrogen) for 1 h at room temperature. The cells were then incubated with primary antibodies diluted in 10% goat serum to 5 μg/ml (anti-KDEL) and/or 0.4 μg/ml (anti-SelK Prestige) antibodies for 9 h at 4 °C. After three washes with 1% goat serum for 10 min, the cells were incubated with secondary antibodies (dilution 1:300) in 10% goat serum for 1 h at room temperature in the dark. After three washes with 1% goat serum, the cells were then incubated with 300 ng/ml nuclear staining DAPI for 10 min in the dark, rinsed with PBS, and mounted on coverslips.
Immunofluorescence was analyzed on a confocal LSM700 microscope (Carl Zeiss) using a Plan-Apochromat 63 × 1.4NA oil objective (Carl Zeiss) at room temperature with ZEN2009 software. DAPI, Alexa Fluor® 488, and Alexa Fluor® 647 fluorescence was induced by exciting with 405-, 488-, and 639-nm lasers. Minimal adjustment of brightness and contrast was applied to all images in ZEN2009 software.
75Se Metabolic Labeling and Immunoprecipitation
75Se metabolic labeling of 2 × 107 HEK 293 cells was performed as previously described (38) for 38–40 h. The cells were lysed in 700 μl of ice-cold TNM buffer (1.8% digitonin, 25 mm Tris-HCl, pH 7.4, 150 mm NaCl, 5 mm MgCl2) containing complete protease inhibitor tablets (Roche Applied Science) and 2.5 mm N-ethylmaleimide for 40 min. Lysates were cleared of debris by centrifugation at 16,000 × g for 15 min at 4 °C. The supernatant was incubated with or without specific antibodies for 2 h at 4 °C. Then 30 μl of 50% protein G-agarose slurry was added for overnight incubation at 4 °C. The beads were washed five times with TNE buffer (0.2% digitonin, 25 mm Tris-HCl, pH 7.4, 150 mm NaCl, and 5 mm MgCl2). Bound proteins were eluted with denaturing 1× SDS-PAGE sample buffer (Invitrogen) by boiling for 5 min. The 75Se radioactivity pattern was detected using a PhosphorImager system (GE Healthcare) as described (37).
Cells co-transfected with the constructs coding for HA-tagged mouse SelK/SelS and FLAG-tagged human Derlins were lysed 40 h after transfection in TNM buffer and subjected to the same procedure with the only difference being that the anti-HA agarose beads were used instead of specific antibodies and protein G-agarose. Cells transfected with HA-tagged ERAD substrates were lysed 40 h after transfection in PBS containing complete protease inhibitor tablets, 5 mm N-ethylmaleimide, and 1% Triton X-100 and subjected to immunoprecipitation (IP) with anti-HA-agarose beads according to the manufacturer's protocol.
Anti-HA Affinity Purification
5 × 108 HEK 293 cells transfected with tagged human SelK constructs were collected in PBS buffer 61 h after transfection. Untransfected HEK 293 cells were used as a control. Affinity purification of HA-TEV-tagged SelK proteins was performed as described in Ref. 38. The SelK proteins and their binding proteins were eluted from HA-agarose beads with 100 units of TEV protease in 400 μl of TNE buffer for 14 h at 4 °C. The beads were washed with an additional 400 μl of TNE buffer. The eluted materials and washes were pooled, concentrated, and desalted in 50 mm BisTris, pH 7.4, 30 mm NaCl, 0.2% digitonin, using Amicon Ultra-4 10K (Millipore). The final volume of concentrated materials was 100 μl. An aliquot of 45 μl was immediately frozen in liquid nitrogen and used for liquid chromatography-mass spectrometry/mass spectrometry (LC-MS/MS) analyses. Three independent purifications of HA-TEV-tagged SelK were performed and subjected to mass spectrometry analyses.
Mass Spectrometry Analyses
TCA-precipitated proteins were trypsinized, purified with Empore C18 extraction media (3 m), and analyzed via LC-MS/MS with a hybrid linear ion trap-Orbitrap mass spectrometer (LTQ-Orbitrap XL; Thermo Fisher Scientific) with a 20-cm × 100-μm (ID) C18 column and a 83-min 6–25% acetonitrile gradient. Spectra were acquired using a data-dependent top 10 method. The resulting spectra were recorded for each run and then searched against a target decoy data base of human tryptic peptides (39) using the Sequest algorithm (version 28; Thermo Fisher Scientific) and a 1.0% false discovery rate. The list of identified proteins was compared with the list of proteins found in the control sample. The resulting list of proteins for two parallel mass spectral studies was loaded into CompPass (40) to assign DN and Z scores. Proteins with a DN score more than 1 and a Z score more than 3.5 were considered to be SelK-associated proteins.
Deglycosylation Assay
Aliquots of 12.5 μl of the sample obtained during affinity purification of the truncated form of HHT-SelK and 5 μg of RNase B in 50 mm Bis-Tris, pH 7.4, 30 mm NaCl, were treated with 0.1% SDS and 50 mm β-mercaptoethanol at 100 °C for 5 min. Deglycosylation was performed by addition of 1.75 units of N-glycanase and 0.75% Nonidet P-40 at 37 °C followed by incubation for 17 h. Deglycosylation was terminated by the addition of denaturing 3× SDS PAGE buffer.
Blue Native PAGE, SDS-PAGE, and Western Blotting
Nondenaturing Blue Native PAGE was performed using a NativePAGETM Novex Bis-Tris Gel System (Invitrogen) according to the manufacturer's recommendations. Briefly, 7.5 ml of NativePAGETM sample buffer (4×) were added immediately prior to electrophoresis to 21.5 μl of affinity-purified SelK-tagged proteins and their control. Thirty microliters of each sample in 50 mm BisTris-HCl, pH 7.4, 40 mm NaCl, 5% glycerol, 0.01% Ponceau S, and 0.14% digitonin were loaded per lane of NativePAGETM Novex 3–12% BisTris gel. The gel was run using light blue cathode buffer (50 mm Tricine, 15 mm BisTris-HCl, pH 7.0, 0.002% Coomassie® G-250) and anode buffer (50 mm Tricine, 15 mm BisTris-HCl, pH 7.0) at +4 °C. The gel was run at constant voltage (150 V) for 1 h, and then the voltage was increased to 200 V for the reminder of the run (1 h). Proteins in blue-colored gel were destained and fixed in 40% methanol, 10% acetic acid for 1 h of incubation on shaker. The proteins were detected using Pierce® silver stain kit (Thermo Scientific) according to the manufacturer's recommendations. SDS-PAGE was performed as previously described (37). The proteins were detected using Coomassie Brilliant Blue Staining or silver staining (Pierce) according to the manufacturer's recommendations. Western blotting was performed as previously described (37). Dilution of antibodies was used according to the manufacturer's recommendations.
RESULTS
SelK and SelS Define a New Eukaryotic Protein Family
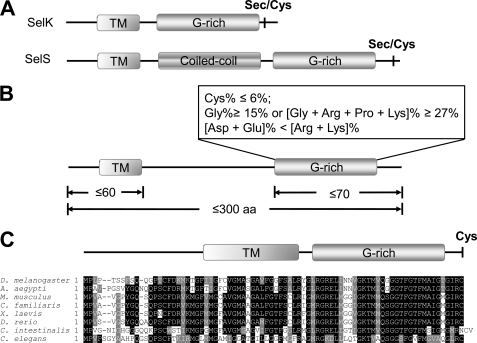
There is no sequence similarity between SelK and SelS. However, both selenoproteins lack a thioredoxin-like fold found in the majority of selenoenzymes and have a very similar domain organization. They are type III transmembrane proteins (30, 34) that contain a short N-terminal ER lumenal sequence (∼20 amino acids), a transmembrane domain (∼20 amino acids) and a longer C-terminal cytosolic sequence (∼53 amino acids in SelK and ∼141 amino acids in SelS). Moreover, the cytosolic tails of SelK and SelS possess a region termed glycine-rich or G-rich region that does not have a pronounced secondary structure and is extremely rich in glycine, proline, and positively charged amino acids (Fig. 1A). Both selenoproteins have a Sec residue located in the second or third positions from the C terminus. To assess the significance of the unusual organization of SelK and SelS, we searched both nucleotide and protein databases for additional SelK/SelS proteins and developed a set of features that define these proteins: (i) the length of proteins is ≤300 amino acids; (ii) the proteins contain a single N-terminal transmembrane domain; (iii) the predicted isoelectric point value of the proteins is ≥8.3; and (iv) the G-rich region of the proteins has the amino acid composition that fits the criteria reported in Fig. 1B.
FIGURE 1.
SelK and SelS form a novel SelS/SelK family of proteins. A, schematic representation of domain organization of SelK and SelS proteins. B, criteria for the identification of eukaryotic proteins belonging to the SelS/SelK family. C, multiple alignment of a novel member of the SelK/SelS family, Romo1. The following elements of domain structure are marked: transmembrane region (TM); a region rich in glycine, proline, and positively charged amino acids (G-rich); and a coiled-coil domain (Coiled-coil) of SelS. The locations of Sec or Cys residues are shown in bold type.
We further examined model eukaryotic organisms containing nearly 200,000 protein sequences for occurrence of proteins that satisfy these homology-independent criteria and identified 108 proteins whose homologs were present in more than one organism (Table 1). Interestingly, some of the detected proteins were also selenoproteins previously not known to be related to SelS and SelK, such as plasmodial selenoproteins Sel1 and Sel4 (41); these two proteins were conserved across Plasmodia and showed no sequence similarity to proteins in other organisms, but they satisfied the SelK/SelS search criteria. Moreover, Sec in these proteins was located in the C-terminal penultimate position, followed by the C-terminal glycine or arginine. Likewise, a selenoprotein previously found in Euplotes crassus (42) and named Ep22 was found to belong to the SelK/SelS family. It also had a C-terminal penultimate Sec. In addition, several uncharacterized selenoproteins in Aureococcus anophagefferens (43) satisfied the search criteria and could be renamed as SelK/SelS-like proteins. Additional proteins detected in the search formed a Cys-containing SelK/SelS-like subfamily, reactive oxygen species modulator 1 (Romo1) (Table 1). These proteins were present in a wide range of organisms from fungi to mammals. It has been reported that Romo1 induces reactive oxygen species production in mitochondria, which may play an important role in redox signaling in cancer cells (44, 45). A multiple alignment of these proteins is shown in Fig. 1C.
TABLE 1.
Occurrence of SelK/SelS-like proteins in eukaryotes
The first two columns show sequenced eukaryotic organisms and the number of predicted proteins that were retrieved from NCBI, respectively. The third column shows proteins that pass the criteria presented in Fig. 2B. Proteins that were found in multiple organisms were considered as members of the SelK/SelS family (shown in the fourth, fifth, and sixth columns).
| Organism | Total proteins | Initial candidates | SelK/SelS family |
Romo1 subfamily | |
|---|---|---|---|---|---|
| Known SelK and SelS proteins | SelK/SelS-like proteins | ||||
| D. melanogaster | 17,878 | 68 | SelK, SelK (Cys) | 5 | 1 |
| C. reinhardtii | 15,256 | 113 | SelK | ||
| P. falciparum | 5,363 | 49 | SelK, SelS | 2 | |
| C. merolae | 5,014 | 20 | |||
| L. major | 8,280 | 27 | SelK | 4 | |
| C. elegans | 25,811 | 172 | SelK (Cys), SelS (Cys) | 10 | 1 |
| S. cerevisiae | 10,814 | 79 | 6 | ||
| H. sapiens | 38,806 | 261 | SelK, SelS | 33 | 1 |
| T. nigroviridis | 27,918 | 204 | SelK, SelS | 16 | 1 |
| T. rubripes | 26,721 | 151 | SelK, SelS | 12 | 1 |
| T. pseudonana | 11,397 | 85 | 6 | ||
| Total | 193,258 | 1,229 | 14 | 94 | 5 |
SelK Interacts with Components of the ERAD Machinery
Initially, SelS was identified as a protein that interacts with Derlin-1, p97, and retrotranslocating substrates (14). Later, SelS was also found to bind Derlin-2 (11, 15). Because SelS and SelK belong to the same protein family, we tested whether SelK, like SelS, associates with Derlins. HEK 293 cells were metabolically labeled with [75Se]selenite, and their digitonin lysates were subjected to immunoprecipitation with anti-Derlin-1 and anti-Derlin-2 antibodies. Only two (of 25) human selenoproteins co-precipitated with Derlins (Fig. 2A). Western blotting with specific antibodies revealed the identities of radioactive bands as SelK and SelS (Fig. 2A). Immunoprecipitates that were obtained from cells treated with an inhibitor of N-linked glycosylation and ER stress inducer, tunicamycin, showed an increased level of SelK associated with Derlins (Fig. 2A, compare SelK bands in lanes 2 and 3 with lanes 4 and 5). Recently, an increased SelK expression in response to ER stress agents was also shown in the human hepatocellular carcinoma cell line, HepG2 (33). Interestingly, SelS preferentially co-precipitated with Derlin-2 (SelS bands in lanes 2 and 3 versus lines 4 and 5, Fig. 2A). This is consistent with the previously reported affinity of SelS (15). In contrast, we found that SelK had a higher affinity for Derlin-1 (Fig. 2A, SelK bands in lanes 2 and 4 versus lanes 3 and 5).
FIGURE 2.
Association of SelK with Derlins, SelS, and p97. A, HEK 293 cells were metabolically labeled with 75Se for 40 h. During the labeling procedure, the cells were either untreated or treated with 10 μg/ml tunicamycin (shown as Tm at the top of the panel) for 24 h. Digitonin lysates (Input) were subjected to IP with Derlin-1 and Derlin-2 antibodies. The selenoprotein pattern was visualized using a PhosphorImager system, and protein occurrence was analyzed by Western blotting (WB) with the indicated antibodies. Migration and molecular weights of SelS, SelK, and major endogenous selenoproteins, thioredoxin reductase 1 (TR1), and glutathione peroxidase 1 (GPx1) are indicated by arrows. B, lysates of HEK 293 cells labeled with 75Se for 38 h were subjected to IP with SelS (lane 1), SelK (lane 2), and β-actin (lane 3) antibodies. Radioactivity pattern on the membrane was detected by a PhosphorImager system (upper panel). The same membrane was subjected to Western blotting with p97 antibody (lower panel). C, schematic representation of constructs coding for the HA-tagged mouse SelK or SelS used for co-transfection with the constructs coding for FLAG-tagged human Derlins. Sec-to-Cys and Sec-to-Stop mutants of HA-tagged mouse SelK/SelS are shown by abbreviations Cys and tr, respectively. D, HEK 293 cells were co-transfected in a 1:1 ratio with the constructs coding for HA-tagged Cys-containing (Cys) or truncated (tr) forms of mouse SelK and the constructs coding for three FLAG-tagged human Derlins. Lanes 1 and 2, HA-tagged SelK and FLAG-tagged Derlin-1; lanes 3 and 4, HA-tagged SelK and FLAG-tagged Derlin-2; lanes 5 and 6, HA-tagged SelK and FLAG-tagged Derlin-3b; lane 7, HA-tagged Cys-containing form of SelK (control); and lane 8, untransfected cells (control). The cells were lysed 40 h after transfection and subjected to IP with anti-HA agarose. The IP samples were analyzed by Western blotting with the indicated antibodies. An asterisk marks the position of FLAG-tagged Derlin-3b; two asterisks mark the position of FLAG-tagged Derlin-1 and -2. E, HEK 293 cells were subjected to the same procedure as in D with the only difference being that the constructs coding for the HA-tagged mouse SelS were used instead of the HA-tagged SelK.
To test whether endogenous SelK interacts with other proteins, we subjected lysates of 75Se labeled HEK 293 cells to immunoprecipitation with anti-SelK antibodies (anti-SelS and anti-actin antibodies were used as a positive and negative control, respectively). Co-precipitation of SelK with SelS (Fig. 2B, upper panel, lane 1) and co-precipitation of SelS with SelK (Fig. 2B, upper panel, lane 2) were readily observed. Approximately half of SelS was tightly associated with SelK, and approximately half of SelK was associated with SelS. We also found that the SelK-SelS complex binds the cytosolic p97 (Fig. 2B, lower panel, lane 2).
We further examined whether SelK and SelS interact with the longer form of Derlin-3, Derlin-3b (11). HA-tagged mouse SelK or SelS were co-expressed with the FLAG-tagged human Derlins in HEK 293 cells. In this experiment, HA tag was inserted into the ORFs of mouse SelK or SelS between their transmembrane and cytosolic regions (Fig. 2C). The constructs expressing Cys-containing and truncated forms of selenoproteins were used, because expression of selenoproteins is significantly lower than the expression of their Sec mutants. Digitonin extracts of co-transfected cells (Fig. 2D, lanes 1–6), cells transfected only with the HA-tagged SelK (lane 7), or untransfected cells (lane 8) were subjected to immunoprecipitation with anti-HA-agarose. Western blotting revealed that SelK interacts with all three Derlins and has a higher affinity for Derlin-1 (Fig. 2D, compare lanes 1 and 2 with lanes 3–6). We also used the HA-tagged mouse SelS in a concurrent experiment (Fig. 2E). The higher affinity of SelS for Derlin-2 was observed once again, and a new interaction of SelS with Derlin-3b was found.
Isolation and Identification of Proteins Associated with SelK
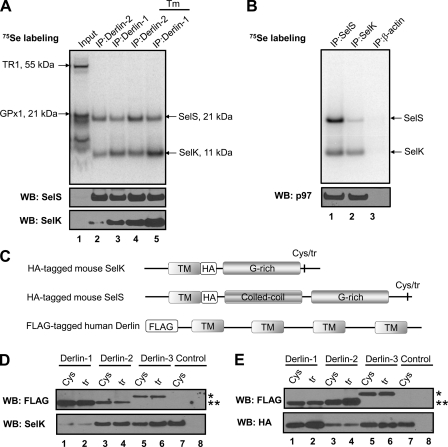
Because the expression levels of many selenoproteins are low in mammalian cells and many of them, including SelK, could not be seen in the selenoprotein pattern (Fig. 2A, lane 1), we performed a large scale affinity purification of human SelK. Two constructs coding for Sec-containing SelK (H-2Kb-HA-TEV-tagged SelK, HHT-SelK (Fig. 3A), and HA-TEV-tagged SelK, HT-SelK (Fig. 3B)) and one construct coding for the Sec-to-Stop mutant or truncated form of SelK (HHT-SelKtr) were used for the expression of different forms of SelK. Because the H-2Kb signal peptide is digested by host proteases (46), all expressed mature proteins contained only the N-terminal HA-TEV tag. To verify that expression of the HA-TEV-tagged SelK does not change the ER morphology or cause SelK dislocation, we first examined SelK localization. Immunofluorescence with SelK antibodies demonstrated the diagnostic reticular ER staining pattern and co-localization of SelK with the ER marker KDEL (Fig. 3). To characterize proteins associated with SelK, Sec-containing and truncated forms of the HA-TEV-tagged SelK and proteins associated with SelK proteins were isolated by immunoprecipitation with anti-HA agarose from digitonin lysates of HEK 293 cells. Proteins eluted with TEV protease were subjected to SDS-PAGE (Fig. 4A) and LC-MS/MS analysis (Table 2).
FIGURE 3.
Tagged human SelK localizes to the ER. A, HeLa cells were transfected with the construct coding for the ER-targeted H-2Kb-HA-TEV-tagged SelK (upper panel). Transfected cells were stained with antibodies to SelK (green) and the ER marker, KDEL (red). The nuclei staining with DAPI (blue) is shown in the merged image (lower panel). B, HeLa cells were transfected with the construct coding for the non-targeted HA-TEV-SelK (upper panel) and subjected to the same procedure as in A. The merged image represents co-localization of SelK with the ER-marker.
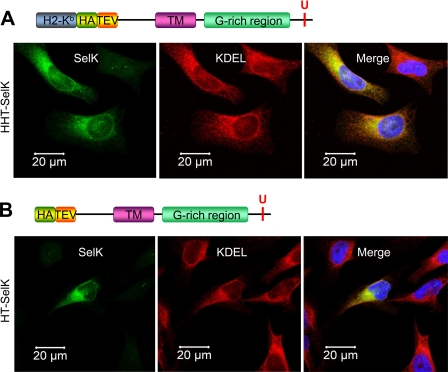
FIGURE 4.
Isolation and identification of SelK-associated proteins. A–C, HEK 293 cells (control) and HEK 293 cells transfected with the constructs coding for HHT-SelK, HT-SelK, or HHT-SelKtr were lysed in 1.8% digitonin, and the lysates were subjected to immunoprecipitation with anti-HA beads. Bound material was eluted from the beads with TEV protease and separated on SDS-PAGE (A) or Blue Native PAGE (B) or subjected to Western blotting (WB) analysis with the indicated antibodies (C). Proteins were visualized by silver staining (A and B). Migration of TEV protease and a band that was cut and subjected to LC-MS/MS (marked as TEV and p97 complex) is shown on the right for A and B, respectively. Migration of SelK and the glycosylated SelK (SelK*) is displayed by arrows in C. D, sequence of the truncated form of SelK (Sec-to-Stop mutation) that occurs following expression of the construct coding for HHT-SelKtr, protein isolation, and treatment with TEV protease. Two amino acid residues in the N-terminal region upstream of the N-terminal methionine residue (shown in bold type) are those that remain after the cleavage with TEV protease; the transmembrane domain is shown in bold type; and the Asp-Ser-Ser (NSS) sequon is displayed in bold and underlined type. E, Western blotting analysis with SelK antibodies of samples prepared in the deglycosylation assay. Lane 1, 5 μg of RNase B treated with N-glycanase (control for deglycosylation); lane 2, 5 μg of RNase B; lane 3, sample prepared during affinity purification of the truncated form of SelK (as for C) and treated with N-glycanase; lane 4, untreated sample from lane 3. Migration of deglycosylated SelK (SelK) and glycosylated SelK (SelK*) is shown on the right. SelK mobility in treated and untreated samples is different because of detergent addition in the glycosylation assay. F, Coomassie Brilliant Blue (CBB) staining of the same membrane. Migration and mass of N-glycanase, TEV protease, glycosylated RNase B (RNAseB*), and deglycosylated RNase B are pointed out on the right.
TABLE 2.
LC-MS/MS analyses of affinity-purified proteins
| Detected proteins | Known function/role | Constructs used for transfection in HEK 293 cells |
||||
|---|---|---|---|---|---|---|
| Total (unique) peptides |
HHT-SelKtr |
|||||
| HHT-SelK | HT-SelK | Total (unique) peptides | Protein coverage | Mass | ||
| % | kDa | |||||
| p97 | Ref. 19 | 62 (32) | 72 (34) | 88 (40) | 56.7 | 97 |
| SelS | ERAD | 2 (2) | 4 (3) | 7 (5) | 31.8 | 21 |
| SelK | Refs. 28, 32, and 33 and this study | 3 (2) | 5 (3) | 38 (10) | 33.8 | 11 |
| Derlin-1 | ERAD | 7 (5) | 16 (5) | 12 (4) | 17.5 | 22 |
| Derlin-2 | ERAD | 3 (2) | 7.9 | 21 | ||
| Ribophorin 1 | N-Linked glycosylation | 10 (6) | 2 (2) | 13 (9) | 16.3 | 69 |
| Ribophorin 2 | N-Linked glycosylation | 2 (2) | 3 (2) | 4.0 | 69 | |
| OST48 | N-Linked glycosylation | 3 (3) | 3.8 | 49 | ||
| STT3A | N-Linked glycosylation | 3 (2) | 4.2 | 81 | ||
| Calnexin | Lectin-like chaperone | 31 (17) | 39 (16) | 25.2 | 67 | |
Proteins already identified by immunoprecipitation as SelK interactors (Derlin-1, Derlin-2, SelS, and p97) were detected by mass spectrometry (Table 2), except Derlin-3, which is expressed at a low level in kidney cells (11). Again, Derlin-1 appeared to be more abundant than Derlin-2 in the SelK immunoprecipitates as evidenced by protein coverage and the number of peptides observed (Table 2). Previous studies utilizing mass spectrometry for the identification of ERAD proteins failed to detect SelK, probably because SelK (i) is expressed at low levels in mammalian cells (Fig. 2A) and (ii) is a small protein that has multiple lysines and arginines (Figs. 1A and 4D), sites for trypsin digestion. Additional proteins, not previously known as SelK-associated proteins, including integral components of the OST complex (RPN I, RPN II, OST48, and STT3A) and an ER chaperone, calnexin, were also detected.
To observe multi-protein complexes that include SelK, nondenaturing Blue Native PAGE (Fig. 4B) was performed. A sharp band (∼600 kDa) detected only in the samples of purified SelK was cut and subjected to LC-MS/MS. The major component of this complex was a hexameric form of p97 (121 total peptides including 59 unique peptides were detected), but Derlin-1 and SelK were also associated with p97 (one unique peptide for each protein). We did not observe intact hetero-oligomeric OST complexes by Blue Native PAGE, probably because their efficient detection by this method (47) requires conditions different from those applied for SelK purification.
However, Western blotting analysis confirmed that RPN I, a core protein of the multi-protein OST complex, was present in the purified SelK sample (Fig. 4C). We also obtained indirect evidence that SelK interacts with other components of the endogenous OST complex by analysis of the purified SelK sample. Two SelK bands were detected in this sample by Western blotting (Fig. 4C). The upper band has never been observed in vivo. We hypothesized that it could correspond to the glycosylated form of SelK as the Asp-Ser-Ser motif that matches the sequon for N-linked glycosylation was identified in the SelK sequence (Fig. 4D). Indeed, we found that the upper band originated by N-linked glycosylation (Fig. 4, E and F). Glycosylation of SelK in vivo seems unlikely because SelK has the C-terminal tail exposed to the cytosol (34), whereas OST glycosylates proteins co-translationally or post-translationally (48) in the ER lumen. The OST active site glycosylates asparagine residues positioned more than 12–14 residues above the luminal ER membrane surface (1). The glycosylation site of SelK is located ∼12 amino acids from the transmembrane domain (Fig. 4D) and should be accessible for glycosylation. Because SelK overexpression did not lead to the accumulation of the glycosylated form (Fig. 5, A and C), we suggest that the disruption of the ER membrane and the lipid bilayer rearrangement during HA-TEV-tagged SelK purification caused SelK glycosylation in vitro. Thus, SelK glycosylation indirectly revealed a close association of SelK with the OST complex rather than a biologically relevant form of the selenoprotein.
FIGURE 5.
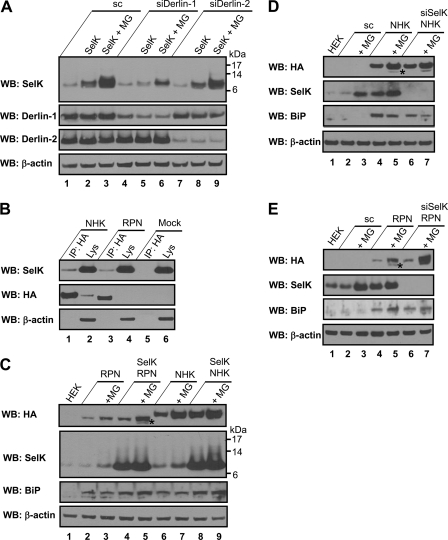
Role of SelK in the Derlin-dependent ERAD. A, HEK 293 cells were transfected with scramble siRNA (lanes 1–3), Derlin-1 siRNA (lanes 4–6), or Derlin-2 siRNA (lanes 7–9). These cells were untransfected or transfected with the construct coding for HHT-SelK (marked as SelK at the top of the panel) 70 h after siRNA transfection. Starting 24 h after HHT-SelK transfection, the cells were untreated or treated with 10 μm proteasome inhibitor MG132 (MG) for 12 h. Cell lysates were analyzed by Western blotting (WB) with the indicated antibodies. B, HEK 293 cells transfected with HA-tagged NHK (lanes 1 and 2), HA-tagged RPN332 (lanes 3 and 4), or mock transfected (lanes 5 and 6) were lysed with Triton X-100 and subjected to IP with anti-HA agarose. Lysates (Lys) and IP samples (IP: HA) were analyzed by Western blotting with the indicated antibodies. C, HEK 293 cells were untransfected (lane 1) or co-transfected in a 1:1 ratio with the construct coding for the HA-tagged RPN332 and empty vector (lanes 2 and 3); constructs coding for human SelK and HA-tagged RPN332 (lanes 4 and 5); constructs coding for the HA-tagged NHK and empty vector (lanes 6 and 7); and constructs coding for human SelK and the HA-tagged NHK (lanes 8 and 9). The cells were untreated or treated with MG132 (MG) for 7 h (lanes 3, 5, 7, and 9), starting 30 h after transfection. Protein expression levels were analyzed by Western blotting with the indicated antibodies. D, HEK 293 cells were untransfected (lane 1) or transfected with scramble siRNA (lanes 2–5) or with SelK siRNA (lanes 6 and 7). The cells were again either transfected (lanes 4–7) or not with HA-tagged NHK 61 h after siRNA transfection. The cells were treated (lanes 3, 5, and 7) or not with 10 μm proteasome inhibitor MG132 (MG) 91 h after siRNA transfection for 12 h, lysed, and analyzed by Western blotting with antibodies indicated on the left. E, HEK 293 cells were subjected to the same procedure as in D with the only difference being that the HA-tagged RPN332 was used instead of the HA-tagged NHK.
Characterization of SelK as an ERAD Component
It was shown that SelS is not an ERAD substrate, because a p97 mutant, p97AA, defective in substrate binding co-precipitated with SelS (14). To check whether SelK serves as a substrate for the Derlin-dependent ERAD, we decreased the levels of Derlin-1 or Derlin-2 by RNAi in HEK 293 cells either transfected with the HHT-SelK construct or not and examined protein expression by Western blotting (Fig. 5A). Approximately 60% reduction in Derlin-1 and 80% in Derlin-2 levels were achieved (Fig. 5A). The level of endogenous SelK was not affected by Derlin-1 (Fig. 5A, compare lanes 1 and 4) and Derlin-2 (compare lanes 1 and 7) deficiency. Moreover, a decrease in Derlin-1 expression resulted in the concomitant reduction in the HA-TEV-tagged SelK levels (Fig. 5A, compare lane 5 and 2). Because Derlin deficiency did not cause the accumulation of the endogenous and recombinant SelK, SelK could not be a substrate for the Derlin-dependent ERAD. Instead, concurrent reduction in Derlin-1 and HA-TEV-tagged SelK expression appears to reflect co-occurrence of Derlin-1 and SelK in the same complex. When the proteasome activity was blocked by inhibitor MG132, the HA-TEV-tagged SelK level was significantly increased in the scramble siRNA-transfected (Fig. 5A, compare lanes 2 and 3), Derlin-1 siRNA-transfected (compare lanes 5 and 6) and Derlin-2 siRNA-transfected (compare lanes 8 and 9) cells, demonstrating that accumulation of misfolded proteins in the ER leads to SelK induction.
To examine whether SelK has a role in the degradation of model ERAD substrates, we first showed that endogenous SelK co-precipitates with the well characterized, soluble glycosylated ERAD substrates, HA-tagged RPN332 and HA-tagged NHK (Fig. 5B). Next, a nontagged human SelK was overexpressed in HEK 293 cells together with the substrates, and protein expression was analyzed by Western blotting (Fig. 5C). Treatment of cells expressing the ERAD substrates with MG132 blocked degradation of both substrates (Fig. 5C, compare lanes 3 and 2 and lanes 7 and 6). SelK overexpression caused an accumulation of glycosylated forms of HA-tagged RPN332 (compare lanes 4 and 2) and HA-tagged NHK (compare lanes 8 and 6). A significant accumulation of glycosylated and deglycosylated forms of HA-tagged RPN332 (more than 2-fold) was found if SelK was overexpressed in MG132-treated cells (compare lanes 5 and 3), indicating that the ERAD of HA-tagged RPN332 includes SelK. Only a slight increase in the expression of the glycosylated HA-tagged NHK was observed in cells overexpressing SelK and treated with MG132 (compare lanes 9 and 7).
SelK knockdown in HEK 293 cells coupled with overexpression of ERAD substrates gave contradicting results (Fig. 5, D and E). Depletion of SelK in cells expressing ERAD substrates did not cause an accumulation of the HA-tagged NHK (Fig. 5D, compare lanes 6 and 4) and slightly increased the amount of glycosylated HA-tagged RPN332 (Fig. 5E, compare lanes 6 and 4). Inclusion of a proteasome inhibitor in the experiment led to an accumulation of the deglycosylated forms of substrates (Fig. 5, D and E, compare lanes 7 and 5), whose N-linked glycans are removed by a cytosolic N-glycanase attached to Derlin-1 (17). Knockdown of SelK in MG132-treated cells did not affect clearance of the HA-tagged NHK (Fig. 5D, compare lanes 7 and 5) and significantly increased expression of glycosylated and deglycosylated HA-tagged RPN332 (Fig. 5E, compare lanes 7 and 5), again indicating that SelK is involved in the ERAD of the truncated form of HA-tagged ribophorin I. Interestingly, overexpression of both ERAD substrates increased the level of endogenous SelK more than 2-fold (Fig. 5, D and E, compare lanes 4 and 2), further demonstrating that an accumulation of misfolded proteins in the ER induces SelK expression.
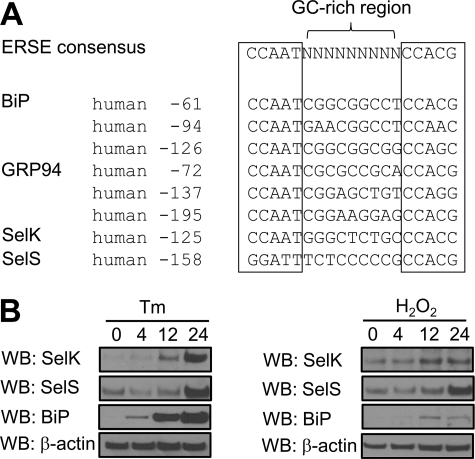
SelK Promoter Region Contains an ER Stress Response Element
Transcriptional regulation of genes coding for mammalian ER chaperones, ER proteins and ERAD components is mediated by the cis-acting elements present in their promoter regions (49, 50). The ER stress response element (ERSE), the canonical sequence of which is CCAAT(N9)CCAC(G/A) (50), was previously found in the promoter regions of ER chaperones (50) and SelS (51). We analyzed the SelK promoter region and found the same evolutionarily conserved element (Fig. 6A). Consistent with the presence of ERSE, we detected the increased expression of endogenous SelK in HEK 293 and HeLa cells under conditions that cause accumulation of misfolded proteins in the ER, such as inhibition of the proteasome (Fig. 5, D and E, compare lanes 3 and 2), overexpression of ERAD substrates (Fig. 5, D and E, compare lanes 4 and 2), and treatment with tunicamycin or hydrogen peroxide (Fig. 6B).
FIGURE 6.
Human SelK promoter carries a functional ER stress response element. A, alignment of human SelK ERSE with the consensus ERSE-I of human ER chaperones and SelS. B, HeLa cells were treated with 10 μg/ml tunicamycin (Tm) or 500 μm H2O2 for 4, 12, and 24 h. Protein expression levels were analyzed by Western blotting (WB) with the indicated antibodies.
DISCUSSION
In this study, we used bioinformatics approaches to define a new selenoprotein family, the SelK/SelS family, and then found that its prototypic member, human SelK, is involved in the regulation of ER homeostasis and may play a role in ER-associated degradation of many misfolded proteins, including components of the OST complex. SelK and SelS show highly variable sequences, thereby even animal orthologs of these proteins are difficult to detect by standard homology-based approaches. We noticed a similar domain organization of SelK and SelS (Fig. 1A), the presence of a disordered region in the C-terminal tail that is rich in glycine, proline, and basic amino acids, and the characteristic location of their Sec residues. With this information, we designed an algorithm (Fig. 1B) for the identification of similarly organized proteins. Interestingly, our searches yielded a limited number of proteins, but the protein list was highly enriched in selenoproteins. Besides SelK and SelS, it had selenoproteins Sel1 and Sel4 from Plasmodia, selenoproteins from E. crassus and A. anophagefferens, and several other selenoproteins. In all of these proteins, Sec was located within the last three residues in a predicted flexible tail. Overall, with the exception of thioredoxin reductases and selenoprotein O homologs, nearly all eukaryotic selenoproteins having Sec within the last three residues could be assigned to the SelK/SelS family. The resulting set of SelK/SelS-like selenoproteins becomes the most widespread eukaryotic selenoprotein family, which is present in essentially every organism that has selenoproteins. In addition to selenoproteins, we found Cys homologs of SelS and SelK in some eukaryotes, such as insects and plants, and identified a similarly organized protein, Romo1, that contained Cys as the C-terminal residue (Fig. 1C). Romo1 proteins occur in plants and animals (including mammals) and are involved in generation of reactive oxygen species in mitochondria (44, 45).
Based on our bioinformatics analyses, we suggest that a human selenoprotein of unknown function, SelK, is involved in the same general process as the characterized member of the SelK/SelS family, SelS (14–16). We found an association of SelK not only with Derlins and p97 (Fig. 2, A, B, and D), but also with SelS (Fig. 2, B and D). An interaction of SelK with the cytosolic part of SelS was largely abolished,4 suggesting that the SelK/SelS or SelK/Derlin/SelS interactions are mediated by their transmembrane domains. Interestingly, two p97 co-factors (Ufd1 and Npl4) and many proteins interacting with Derlins and required for ERAD (e.g. protein disulfide isomerase (8), ER degradation enhancing α-mannosidase-like protein (11), E3-ubiquitin ligases HRD1 (15, 16), SEL1L (15), and GP78 (16)) were not bound by SelK (Table 2), suggesting that we identified a transient ER-membrane associated complex that exists at early stages of ERAD and includes only selenoproteins, Derlins, and p97. Among all of the known selenoproteins, this is the first example when two selenoproteins share the same binding proteins.
We found that SelK had a higher affinity for Derlin-1 (Fig. 2, A and D, and Table 2), whereas SelS had a higher affinity for Derlin-2 (Fig. 2, A and E). Our observation that SelK is specific for Derlin-1 adds SelK to the list of candidates that mediate the association of p97 with the ER membrane proteins, including GP78, HRD1 (16, 20), erasin (21), and SelS (14–16). Furthermore, the different SelK and SelS specificity for Derlins could determine the nature of the substrate that is translocated through the specific channel. For example, Derlin-1 was involved in retrotranslocation of pro-alpha factor (13) and cholera toxin (8); Derlin-1 and -2 were implicated in the escape of polyomaviral-encoded proteins from the ER and in establishing an infection (10, 12); Derlin-2 and -3 were specifically required for the degradation of HA-tagged NHK (11). Thus, SelK associated with different Derlins could participate in binding different substrates.
We have shown that SelK was not degraded through the Derlin-dependent ERAD pathway, because Derlin-1 and Derlin-2 knockdown did not cause an accumulation of the endogenous SelK (Fig. 5A, lanes 4 and 7 versus lane 1). On the other hand, inhibition of the proteasome significantly increased the expression of endogenous SelK (Fig. 5, D and E, compare lanes 3 and 2) and the expression of recombinant HA-TEV-tagged SelK (Fig. 5A, compare lanes 3 and 2), suggesting that SelK (i) could be dislocated through ERAD or (ii) could serve as a chaperone interacting with misfolded proteins and assisting in their dislocation for proteasomal degradation. The possibility that SelK is an ERAD substrate competing with other ERAD substrates for degradation is unlikely because SelK knockdown in tunicamycin-treated cells was associated with significant accumulation of HA-tagged RPN332 (Fig. 5E, compare lanes 5 and 7), suggesting that SelK is involved in ERAD of this substrate as a component assisting in substrate degradation. The possibility that SelK serves as a chaperone for degradation of some ERAD substrates is more relevant because SelK has a functional ERSE element in the promoter region (Fig. 6A). Transcriptional induction of many ER chaperones (50, 51) and ERAD components including SEL1L, HRD1, HERP, Derlins, and SelS (11, 15, 49–54) involved in maintaining ER homeostasis is a well known process that depends on the ERSE type (53, 54). In addition, we have shown that SelK co-precipitates with glycosylated soluble ERAD substrates (Fig. 5B) and that the expression of endogenous SelK is induced by overexpression of ERAD substrates (Fig. 5, D and E). To summarize, the data suggest that SelK is overexpressed under conditions that disrupt ER homeostasis because it binds misfolded proteins and could target them to ERAD through the Derlin-selenoprotein-p97 complex.
Our data did not suggest a universal role of SelK in ERAD, because SelK had different effects on the degradation of two soluble glycosylated ERAD substrates. SelK overexpression and knockdown led to the accumulation of the HA-tagged RPN332 (Fig. 5, C and E), whereas SelK overexpression induced an accumulation of the HA-tagged NHK (Fig. 5C), and its knockdown did not affect the levels of this substrate (Fig. 5D). We suggest that the differential effect of SelK knockdown on the degradation of tested proteins was caused by a different composition of the Derlin-selenoprotein-p97 complex required for their translocation. Because SelS or Derlin-3, but not SelK, could be the proteins important for HA-tagged NHK degradation, there was no effect of SelK knockdown on the degradation of this protein. A common effect of SelK overexpression in the degradation of ERAD substrates is more consistent with the idea that SelK interacts with misfolded proteins and targets them for the Derlin-dependent ERAD. An accumulation of HA-tagged NHK and RPN332 upon SelK overexpression reflects, for example, an accumulation of the SelK-ERAD substrate complex that targets the substrate for degradation. Because SelK is unlikely to be involved in the critical steps of ERAD, such as ubiquitination and extraction, its overexpression could not facilitate degradation of ERAD substrates adjacent to the Derlin channel in the form of the SelK-ERAD substrate complex.
We identified components of the multi-protein OST complex interacting with SelK, in addition to the components of the ERAD complex (Table 2). Components of the OST complex (RPN I, RPN II, and OST48), the ER lumenal (BiP, calreticulin) and integral (calnexin) chaperones, were previously found in the purified sample of the HA-TEV-tagged US11 (9, 15), cytomegalovirus protein targeting class I MHC heavy chain for ERAD. US11 was also found interacting with MHC class I, Derlin-1, and SelS (9, 14–16). SelK and SelS bound to US11 could be responsible for the reported interaction of US11 with the components of the OST complex, because there is no data that this interaction could be mediated by Derlin-1 or class I MHC heavy chain. Co-precipitation of SelK with OST subunits could not be an artifact of purification, because the same procedure applied for purification of the HA-TEV-tagged SEL1L did not reveal SEL1L as the OST-binding protein (38).
Selenoproteins have never been shown to associate with stable and functional OST complexes, perhaps reflecting a transient nature of this interaction. In addition, low coverage of OST proteins in purified SelK samples (Table 2) suggests that the SelK-OST complex is not abundant. It is hard to predict how and why selenoproteins interact with the heterooligomers of the OST complex, until the specific role of selenoproteins in N-linked glycosylation or ERAD of OST proteins is clarified. One possibility is that RPNI and RPNII, two high confidence (Z score > 3.5) interactors (Table 2), are glycosylated transmembrane proteins (1) that could be degraded through the Derlin-dependent ERAD. A possible function of selenoproteins in this process could be the same as that of other SelK/SelS interacting substrates, for example, association of ribophorins with the Derlin channel.
In summary, this study identified SelK as a new member of the mammalian ERAD complex composed of Derlins, SelS, and p97. SelK was involved in the degradation of glycosylated soluble ERAD substrates through interaction with these proteins and the ERAD complex. SelK had the ERSE, and the expression of this protein was up-regulated by elevated levels of misfolded proteins, the ERAD substrates in particular, reflecting a chaperone-like function of SelK in the ER clearance. We have shown that two selenoproteins, SelK and SelS, are prototypic selenoproteins that define the most widespread eukaryotic selenoprotein family, involved in ERAD. SelK and SelS are two selenoproteins sharing similar structural characteristics, binding proteins, and roles in a fundamental biological process.
This work was supported, in whole or in part, by National Institutes of Health Grant GM061603 (to V. N. G.). This work was also supported by the Intramural Research Program of the Center for Cancer Research, NCI, National Institutes of Health (to D. L. H.).
V. A. Shchedrina and V. N. Gladyshev, unpublished data.
- ER
- endoplasmic reticulum
- SelS
- selenoprotein S
- SelK
- selenoprotein K
- ERAD
- ER-associated degradation
- OST
- oligosaccharyltransferase
- Romo1
- reactive oxygen species modulator 1
- RPN
- ribophorin
- NHK
- null Hong Kong variant of α1-antitrypsin
- TEV
- Tobacco Etch Virus
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- IP
- immunoprecipitation
- Sec
- selenocysteine
- SECIS
- selenocysteine insertion sequence
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine
- ERSE
- ER stress response element
- H2-Kb
- signal sequence from mouse MHC class I heavy chain H2-Kb sequence peptide
- HT-tagged
- HA-TEV-tagged
- HHT-tagged
- H-2Kb-HA-TEV-tagged
- LC-MS/MS
- liquid chromatography-mass spectrometry/mass spectrometry.
REFERENCES
- 1. Kelleher D. J., Gilmore R. (2006) Glycobiology 16, 47R–62R [DOI] [PubMed] [Google Scholar]
- 2. Ellgaard L., Helenius A. (2003) Nat. Rev. Mol. Cell Biol. 4, 181–191 [DOI] [PubMed] [Google Scholar]
- 3. Hebert D. N., Bernasconi R., Molinari M. (2010) Semin. Cell Dev. Biol. 21, 526–532 [DOI] [PubMed] [Google Scholar]
- 4. Bagola K., Mehnert M., Jarosch E., Sommer T. (2011) Biochim. Biophys. Acta 1808, 925–936 [DOI] [PubMed] [Google Scholar]
- 5. Hegde R. S., Ploegh H. L. (2010) Curr. Opin. Cell Biol. 22, 437–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hirsch C., Gauss R., Horn S. C., Neuber O., Sommer T. (2009) Nature 458, 453–460 [DOI] [PubMed] [Google Scholar]
- 7. Meusser B., Hirsch C., Jarosch E., Sommer T. (2005) Nat. Cell Biol. 7, 766–772 [DOI] [PubMed] [Google Scholar]
- 8. Bernardi K. M., Forster M. L., Lencer W. I., Tsai B. (2008) Mol. Biol. Cell 19, 877–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lilley B. N., Ploegh H. L. (2004) Nature 429, 834–840 [DOI] [PubMed] [Google Scholar]
- 10. Lilley B. N., Gilbert J. M., Ploegh H. L., Benjamin T. L. (2006) J. Virol. 80, 8739–8744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Oda Y., Okada T., Yoshida H., Kaufman R. J., Nagata K., Mori K. (2006) J. Cell Biol. 172, 383–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schelhaas M., Malmström J., Pelkmans L., Haugstetter J., Ellgaard L., Grünewald K., Helenius A. (2007) Cell 131, 516–529 [DOI] [PubMed] [Google Scholar]
- 13. Wahlman J., DeMartino G. N., Skach W. R., Bulleid N. J., Brodsky J. L., Johnson A. E. (2007) Cell 129, 943–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ye Y., Shibata Y., Yun C., Ron D., Rapoport T. A. (2004) Nature 429, 841–847 [DOI] [PubMed] [Google Scholar]
- 15. Lilley B. N., Ploegh H. L. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 14296–14301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ye Y., Shibata Y., Kikkert M., van Voorden S., Wiertz E., Rapoport T. A. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 14132–14138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Katiyar S., Joshi S., Lennarz W. J. (2005) Mol. Biol. Cell 16, 4584–4594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Crawshaw S. G., Cross B. C., Wilson C. M., High S. (2007) Mol. Membr. Biol. 24, 113–120 [DOI] [PubMed] [Google Scholar]
- 19. Ye Y. (2006) J. Struct. Biol. 156, 29–40 [DOI] [PubMed] [Google Scholar]
- 20. Ballar P., Shen Y., Yang H., Fang S. (2006) J. Biol. Chem. 281, 35359–35368 [DOI] [PubMed] [Google Scholar]
- 21. Liang J., Yin C., Doong H., Fang S., Peterhoff C., Nixon R. A., Monteiro M. J. (2006) J. Cell Sci. 119, 4011–4024 [DOI] [PubMed] [Google Scholar]
- 22. Kryukov G. V., Castellano S., Novoselov S. V., Lobanov A. V., Zehtab O., Guigó R., Gladyshev V. N. (2003) Science 300, 1439–1443 [DOI] [PubMed] [Google Scholar]
- 23. Gromer S., Eubel J. K., Lee B. L., Jacob J. (2005) Cell. Mol. Life Sci. 62, 2414–2437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hatfield D. L., Carlson B. A., Xu X. M., Mix H., Gladyshev V. N. (2006) Prog. Nucleic Acid Res. Mol. Biol. 81, 97–142 [DOI] [PubMed] [Google Scholar]
- 25. Papp L. V., Lu J., Holmgren A., Khanna K. K. (2007) Antioxid. Redox Signal. 9, 775–806 [DOI] [PubMed] [Google Scholar]
- 26. Reeves M. A., Hoffmann P. R. (2009) Cell Mol. Life Sci. 66, 2457–2478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lu C., Qiu F., Zhou H., Peng Y., Hao W., Xu J., Yuan J., Wang S., Qiang B., Xu C., Peng X. (2006) FEBS Lett. 580, 5189–5197 [DOI] [PubMed] [Google Scholar]
- 28. Verma S., Hoffmann F. W., Kumar M., Huang Z., Roe K., Nguyen-Wu E., Hashimoto A. S., Hoffmann P. R. (2011) J. Immunol. 186, 2127–2137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lobanov A. V., Fomenko D. E., Zhang Y., Sengupta A., Hatfield D. L., Gladyshev V. N. (2007) Genome Biol. 8, R198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shchedrina V. A., Zhang Y., Labunskyy V. M., Hatfield D. L., Gladyshev V. N. (2010) Antioxid. Redox Signal. 12, 839–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hoffmann P. R., Höge S. C., Li P. A., Hoffmann F. W., Hashimoto A. C., Berry M. J. (2007) Nucleic Acids Res. 35, 3963–3973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Huang Z., Hoffmann F. W., Norton R. L., Hashimoto A. C., Hoffmann P. R. (2011) J. Biol. Chem. 286, 34830–34838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Du S., Zhou J., Jia Y., Huang K. (2010) Arch. Biochem. Biophys. 502, 137–143 [DOI] [PubMed] [Google Scholar]
- 34. Chen C. L., Shim M. S., Chung J., Yoo H. S., Ha J. M., Kim J. Y., Choi J., Zang S. L., Hou X., Carlson B. A., Hatfield D. L., Lee B. J. (2006) Biochem. Biophys. Res. Commun. 348, 1296–1301 [DOI] [PubMed] [Google Scholar]
- 35. Sonnhammer E. L., von Heijne G., Krogh A. (1998) Proc. Int. Conf. Intell. Syst. Mol. Biol. 6, 175–182 [PubMed] [Google Scholar]
- 36. Novoselov S. V., Lobanov A. V., Hua D., Kasaikina M. V., Hatfield D. L., Gladyshev V. N. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 7857–7862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Shchedrina V. A., Novoselov S. V., Malinouski M. Y., Gladyshev V. N. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 13919–13924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mueller B., Klemm E. J., Spooner E., Claessen J. H., Ploegh H. L. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 12325–12330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Elias J. E., Gygi S. P. (2007) Nat. Methods 4, 207–214 [DOI] [PubMed] [Google Scholar]
- 40. Sowa M. E., Bennett E. J., Gygi S. P., Harper J. W. (2009) Cell 138, 389–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lobanov A. V., Delgado C., Rahlfs S., Novoselov S. V., Kryukov G. V., Gromer S., Hatfield D. L., Becker K., Gladyshev V. N. (2006) Nucleic Acids Res. 34, 496–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Turanov A. A., Lobanov A. V., Fomenko D. E., Morrison H. G., Sogin M. L., Klobutcher L. A., Hatfield D. L., Gladyshev V. N. (2009) Science 323, 259–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gobler C. J., Berry D. L., Dyhrman S. T., Wilhelm S. W., Salamov A., Lobanov A. V., Zhang Y., Collier J. L., Wurch L. L., Kustka A. B., Dill B. D., Shah M., VerBerkmoes N. C., Kuo A., Terry A., Pangilinan J., Lindquist E. A., Lucas S., Paulsen I. T., Hattenrath-Lehmann T. K., Talmage S. C., Walker E. A., Koch F., Burson A. M., Marcoval M. A., Tang Y. Z., Lecleir G. R., Coyne K. J., Berg G. M., Bertrand E. M., Saito M. A., Gladyshev V. N., Grigoriev I. V. (2011) Proc. Natl. Acad. Sci. U.S.A. 108, 4352–4357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chung Y. M., Kim J. S., Yoo Y. D. (2006) Biochem. Biophys. Res. Commun. 347, 649–655 [DOI] [PubMed] [Google Scholar]
- 45. Na A. R., Chung Y. M., Lee S. B., Park S. H., Lee M. S., Yoo Y. D. (2008) Biochem. Biophys. Res. Commun. 369, 672–678 [DOI] [PubMed] [Google Scholar]
- 46. Gewurz B. E., Ploegh H. L., Tortorella D. (2002) J. Biol. Chem. 277, 11306–11313 [DOI] [PubMed] [Google Scholar]
- 47. Shibatani T., David L. L., McCormack A. L., Frueh K., Skach W. R. (2005) Biochemistry 44, 5982–5992 [DOI] [PubMed] [Google Scholar]
- 48. Ruiz-Canada C., Kelleher D. J., Gilmore R. (2009) Cell 136, 272–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Adachi Y., Yamamoto K., Okada T., Yoshida H., Harada A., Mori K. (2008) Cell Struct. Funct. 33, 75–89 [DOI] [PubMed] [Google Scholar]
- 50. Roy B., Lee A. S. (1999) Nucleic Acids Res. 27, 1437–1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Gao Y., Feng H. C., Walder K., Bolton K., Sunderland T., Bishara N., Quick M., Kantham L., Collier G. R. (2004) FEBS Lett. 563, 185–190 [DOI] [PubMed] [Google Scholar]
- 52. Kaneko M., Yasui S., Niinuma Y., Arai K., Omura T., Okuma Y., Nomura Y. (2007) FEBS Lett. 581, 5355–5360 [DOI] [PubMed] [Google Scholar]
- 53. Yamamoto K., Suzuki N., Wada T., Okada T., Yoshida H., Kaufman R. J., Mori K. (2008) J. Biochem. 144, 477–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yamamoto K., Yoshida H., Kokame K., Kaufman R. J., Mori K. (2004) J. Biochem. 136, 343–350 [DOI] [PubMed] [Google Scholar]