Background: The enzyme soluble guanylyl cyclase (sGC) converts the NO signal into cGMP.
Results: Stoichiometric NO forms an unstable sGC-NO complex, which is stabilized by extra NO, GTP, or substitution of βHis-107.
Conclusion: Exchange of the proximal heme ligand contributes to sGC activation and desensitization.
Significance: Understanding the dynamics of sGC/NO interaction and its functional consequences is necessary to understand the process of NO/cGMP signaling.
Keywords: Cyclic GMP (cGMP), Guanylate Cyclase (Guanylyl Cyclase), Heme, Nitric Oxide, Signal Transduction, Soluble Guanylyl Cyclase
Abstract
Accumulating evidence indicates that the functional properties of soluble guanylyl cyclase (sGC) are affected not only by the binding of NO but also by the NO:sGC ratio and a number of cellular factors, including GTP. In this study, we monitored the time-resolved transformations of sGC and sGC-NO complexes generated with stoichiometric or excess NO in the presence and absence of GTP. We demonstrate that the initial five-coordinate sGC-NO complex is highly activated by stoichiometric NO but is unstable and transforms into a five-coordinate sGC-2 state. This sGC-2 rebinds NO to form a low activity sGC-NO complex. The stability of the initial complex is greatly enhanced by GTP binding, binding of an additional NO molecule, or substitution of βHis-107. We propose that the transient nature of the sGC-NO complex, the formation of a desensitized sGC-2 state, and its transformation into a low activity sGC-NO adduct require βHis-107. We conclude that conformational changes leading to sGC desensitization may be prevented by GTP binding to the catalytic site or by binding of an additional NO molecule to the proximal side of the heme. The implications of these observations for cellular NO/cGMP signaling and the process of rapid desensitization of sGC are discussed in the context of the proposed model of sGC/NO interactions and dynamic transformations.
Introduction
Nitric oxide is a signaling molecule that affects various physiological or pathological processes, depending on the amounts produced and targets involved (1). Although NO reacts with a number of different proteins, inducing changes in their properties, soluble guanylyl cyclase (sGC)3 is the main physiological target of NO (2). sGC is a member of a larger guanylyl cyclase family, which converts GTP into cGMP. Functional sGC is a heterodimer of α- and β-subunits, both of which are required for activity (3). sGC is also a heme protein with exceptional specificity and affinity for NO rather than CO or O2 (4–6). Transduction from ligand (NO) binding into production of a second messenger (cGMP) shows substantial amplification and rapid deactivation after ligand removal (7). Thus, sGC is often referred to as the NO receptor, with both the sensor and effector components built into one functional unit.
Similar to microbial two-component gaseous sensors (8–10), the sGC sensor component consists of the β-subunit heme-binding domain, also called the H-NOX (heme-nitric oxide- and oxygen-binding) domain. The C-terminal halves of both subunits are necessary (3) and sufficient (11) to form a functional cGMP-producing catalytic domain or the effector component. Upon binding of NO to sGC heme, the cGMP-forming capacity of the sGC catalytic domain is increased several hundred times above the basal activity (12).
sGC isolated from native and heterologous expression sources contains a five-coordinate high-spin ferrous heme (5C sGC) with His-105 as the proximal ligand and a Soret band at 431 nm (13, 14). Mechanistic studies of NO interaction with sGC heme revealed a two-step process (15, 16), which can be summarized by the following scheme: 5C His-105 heme + NO ↔ 6C His-105 heme-NO ↔ His-105 + 5C heme-NO. In the first step, 5C sGC heme binds NO with an essentially diffusion-limited rate (6, 14, 15) to form a six-coordinate complex (6C sGC-NO) characterized by a 420 nm Soret band. This 6C sGC-NO complex is transient, as it converts into a 5C sGC-NO form (399 nm Soret band) after cleaving the bond between heme iron and His-105 (15). Although the second step seems to be a simple first-order intramolecular conversion, experimental data demonstrate that this is a NO concentration-dependent process (14, 15), suggesting that an additional NO-binding step takes place.
Direct measurements of NO produced in different cells (17, 18) and calculations of NO available for sGC activation suggest that the physiological level of NO reaches subnanomolar concentrations (19). At the same time, it has been estimated that intracellular concentrations of sGC, at least in platelets and cerebellar astrocytes, are in the micromolar range (20). These assessments indicate that, under normal conditions, the interaction between NO and sGC occurs at substoichiometric ratios. Accumulating experimental evidence suggests that a number of environmental factors affect the dynamics of formation of the sGC-NO complex and its activity. Several studies reported that the sGC-NO complex formed with a stoichiometric amount of NO has a much lower cGMP-forming activity in comparison with sGC-NO formed with excess NO (21, 22). It was also reported that if sGC reacts with stoichiometric NO in the presence of GTP, the high cGMP-forming activity is restored (22).
Although the importance of the NO:sGC ratio in NO/cGMP signaling is well recognized, our knowledge of the kinetics of the sGC interaction with NO under physiological conditions is limited. In this study, we investigated the process of interaction between NO and sGC by recording time-resolved changes in the UV-visible spectra of sGC heme upon binding stoichiometric and excess amounts of NO in the presence and absence of GTP. We characterized the effects of GTP and different amounts of NO on the dynamics of sGC/NO interaction, the stability of the complexes obtained, and their transformations and cGMP-forming activities. The data presented in this work provide new mechanistic details of sGC function and reveal the intricate dynamics of sGC-NO complex transformation and catalytic activity. These data allow us to propose a comprehensive scheme of sGC function that reflects the multiple sGC-NO complexes formed in the cellular milieu. These data also provide direct insights into the mechanism of rapid desensitization of sGC by the NO ligand.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
The wild-type and mutant sGC enzymes were expressed in Sf9 cells co-infected with viruses expressing α1- and β1-subunits as described previously (14). To generate β1H107L mutant sGC (with His-107 replaced by Leu), the pBacPAK8 vector carrying the full-length open reading frame of the sGC β1-subunit was subjected to standard site-directed mutagenesis. The construct was then recombined with BaculoGold DNA (BD Biosciences) according to the manufacturer's protocol to produce the baculovirus expression construct for the β1H107L mutant. The wild-type and mutant sGC enzymes were expressed in Sf9 cells co-infected with viruses expressing α1- and β1-subunits as described previously (14). To purify the protein samples, the 100,000 × g supernatant obtained from 6–8 liters of Sf9 suspension culture was applied to a 30-ml DEAE-FF Sepharose column. sGC-containing fractions were eluted with 350 mm NaCl and immediately loaded onto a nickel-agarose column. After a 50 mm imidazole wash, sGC enzyme was eluted with 175 mm imidazole and captured on a 1-ml RESOURCE Q column (GE Healthcare). The RESOURCE Q column was then washed with 50 mm triethanolamine (TEA) buffer (pH 7.4) containing 0.5 mm EDTA and 0.5 mm EGTA, and the proteins were separated by a linear 50–500 mm NaCl gradient. The sGC-containing fractions were concentrated on a Centricon YW-100 concentrator (Millipore, Bedford, MA). The sample was supplemented with 1 mm MgCl2 and stored at −80 °C in 25% glycerol. The preparations were at least 95% pure as judged by SDS-PAGE with Coomassie Blue staining and immunoblotting as described previously (16). To prepare an anaerobic sample, the enzyme was placed in a glass tonometer and subjected to five cycles of vacuum (30 s)/argon (5 min) replacement on an anaerobic train.
Preparation of NO Solutions
To analyze the kinetics of sGC/NO interaction, 99.9% pure NO gas (Matheson) was further purified by passing it through a U-shaped tube filled with dry KOH pellets. 10 ml of anaerobic 50 mm TEA buffer (pH 7.4) was made by bubbling nitrogen gas for 10 min. It was then bubbled for an additional 10 min with purified NO to obtain an ∼2 mm solution of NO, which was kept in a gas-tight glass tonometer and stored in an anaerobic chamber (Model 110V, Coy Laboratory Products, Inc.).
The exact concentration of NO in the stock solution was determined using the HbO2 assay as described previously (23). Working NO solutions of different concentrations were prepared by adding the necessary amount of NO stock to a plastic gas-tight syringe containing anaerobic 50 mm TEA buffer (pH 7.4) and were used immediately for measurements.
Time-resolved Measurement of sGC Heme-NO
sGC heme-NO complex formation experiments were conducted at room temperature (24 °C) using a Bio-SEQUENTIAL DX-18MV stopped-flow instrument (Applied Photophysics, Leatherhead, United Kingdom) equipped with a rapid scan photodiode array detector. The entire working area was located in the anaerobic chamber filled with 10% H2 in N2 and fitted with a palladium-based O2 scrubber. A recently calibrated gas analyzer (Model 10, Coy Laboratory Products, Inc.) tracked both the hydrogen and oxygen levels to ensure that the latter was 0 ppm during the sample handling and kinetic measurements.
To measure the dynamics of the sGC-NO complex formation in the presence of GTP, 4 μm anaerobic sGC solution was first mixed with an equal volume of 2 mm Mg2+-GTP and incubated for 100 ms in the aging loop of the stopped-flow instrument (see schematics in supplemental Fig. S1). Subsequently, the sGC-GTP complex was further mixed with an equal volume of 2 μm NO solution at 24 °C. To measure NO binding in the absence of GTP, 4 μm anaerobic sGC solution was first mixed with buffer containing 50 mm TEA (pH 7.4) and 2 mm MgCl2 and then with an equal volume of NO solution at 2 μm (stoichiometric conditions) or 10 μm (excess amount). The changes in the UV-visible spectra between 350 and 700 nm were recorded at 400 scans/s for the first second. To monitor the transformations of the sGC-NO complex that took place 1 s after mixing with NO, UV-visible spectra were either recorded continuously for up to 4.5 min or probed at different intervals for at least 3 s at a rate of 400 scans/measurement.
Assay of sGC Activity by Rapid Chemical Quenching
Enzyme activity was assayed by the formation of [32P]cGMP from [α-32P]GTP at 37 °C. To measure the activity of the sGC-NO complex formed 1 s or 5 min after exposure to NO, 90 μl of 2 μm sGC sample (±1 mm GTP) was mixed with equal volume of 1, 2, or 10 μm NO in the stopped-flow instrument controlled by the double-push program using two knurled-screw plungers to preset the delivery volume of each push. The reaction mixture after the first push was incubated in the aging loop of the instrument for 1 s or 5 min (supplemental Fig. S1). The entire process was carried out in the anaerobic chamber, vesting the whole sample handling unit of the stopped-flow instrument. The samples were then expelled from the loop by the second push with 280 μl of 50 mm TEA buffer (pH 7.4) and collected into a tube containing 540 μl of reaction buffer (12.5 mm GTP/[α-32P]GTP in 25 mm TEA (pH 7.5), 1 mg/ml BSA, 1 mm cGMP, 3 mm MgCl2, 0.05 mg/ml creatine phosphokinase, and 5 mm creatine phosphate) prewarmed to 37 °C and placed in a 37 °C bath. For each reaction, 100-μl aliquots were withdrawn every 10 s and mixed with 400 μl of 100 mm zinc acetate, followed by 500 μl of 120 mm Na2CO3 to stop the reaction. The sample was then processed to determine the amount of cGMP as described previously (24). The specific activity of each complex was determined from the linear segment of the cGMP accumulation plots obtained from two independent measurements.
To compare the activity and stability of the cGMP-forming sGC-NO complexes of wild-type and αβH107L sGC, 500 μl of Sf9 lysates expressing these enzymes was mixed with 1.5 ml of reaction buffer (see above) and incubated at 37 °C. The reaction was initiated by the addition of 500 μl of GTP/[α-32P]GTP mixture to a final concentration of 1 mm. 100-μl aliquots were withdrawn every 10 s throughout the entire experiments. To monitor the response to NO activation, the NO donor 2-(N,N-diethylamino)diazenolate-2-oxide diethylammonium salt (DEA-NO; 10 μm) was added 100 s after initiation of the reaction. To compare the stability of the active sGC-NO complex, 80 s after the addition of DEA-NO, 120 μm HbO2 was added to scavenge free NO. Aliquots were withdrawn for an additional 120 s. The reaction in each aliquot was stopped as described above and processed to determine the amount of generated cGMP as described previously (24).
RESULTS
Dynamics of sGC/NO Interaction at a Stoichiometric Ratio
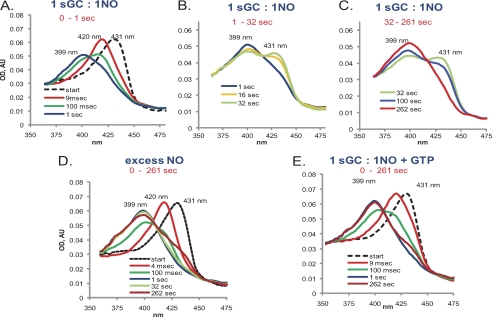
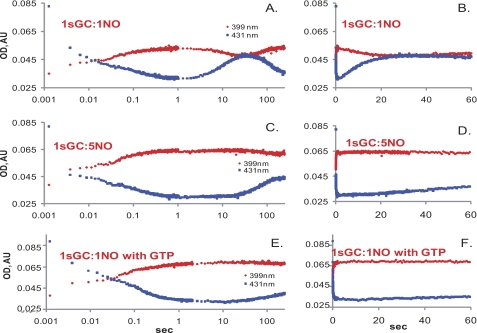
Mixing a stoichiometric amount of NO and purified human recombinant sGC yielded a multistage reaction recorded using a rapid scan stopped-flow instrument (Fig. 1). In the first stage, a 6C sGC-NO complex was formed (420 nm) (Fig. 1A) within the dead time of the stopped-flow instrument (<2 ms). In the second stage, the 420 nm 6C sGC-NO complex converted into a 399 nm species corresponding to the 5C sGC-NO complex (Fig. 1A). Under the tested experimental conditions (2 μm sGC versus 2 μm NO), the 6C sGC-NO → 5C sGC-NO transition was completed within 1 s (Fig. 1A) at a rate of ∼8.5 s−1. This 5C sGC-NO complex generated at a 1:1 NO:sGC ratio was unstable, and within seconds, a significant portion of it converted to a new 431 nm species at a rate of ∼0.12 s−1 (Fig. 1B). We will refer to this new state of sGC, which has the spectral features of ferrous sGC, as sGC-2, whereas the initial sGC will be called the sGC-1 state.
FIGURE 1.
Time-dependent optical spectral changes of α1β1-NO complex formation. 4 μm sGC prepared anaerobically as described under “Experimental Procedures” was mixed with an equal volume of anaerobic 50 mm TEA buffer (pH 7.4) without (A–D) or with (E) 2 mm Mg2+-GTP, incubated for 100 ms in the aging loop of the stopped-flow instrument, and mixed with equimolar (A–C and E) or 5-fold excess (D) NO. Spectra were recorded for different time intervals (1 s (A), 32 s (B), and 262 s (C–E)) at a rate of 400 spectra per time interval. Spectra for the initial sGC (dashed lines) were obtained in separate measurements in which NO solution was replaced with 50 mm TEA buffer (pH 7.4). The base line for all spectra was adjusted based on the 700 nm value. Representative spectra out of at least four independent experiments with different sGC preparations are shown. AU, absorbance unit.
The conversion to the sGC-2 state is not due to oxidation, as no increase in the 393 nm component of the spectra was observed, and the sGC-2 enzyme converted further to a 399 nm species typical for the 5C sGC-NO complex. However, in contrast to the very fast diffusion-limited formation of the first 5C sGC-NO complex (Fig. 1A), generation of this second 5C sGC-NO complex from the sGC-2 state was very slow (∼0.01 s−1) and was completed after several minutes (Fig. 1C). The time-resolved absorbance changes at 399 and 431 nm demonstrate the distinct multiphasic dynamics of sGC/NO interaction at a stoichiometric amount of NO (Fig. 2, A and B). The sGC-NO complex behaved similarly when stoichiometric NO was reacted with sGC in the presence of 1 mm DTT (supplemental Fig. S2).
FIGURE 2.
Kinetic changes at 431 and 399 nm during the sGC reaction with NO. The absorbance changes at 431 nm (blue) and 399 nm (red) for the anaerobic stopped-flow experiment during a reaction between 4 μm sGC and stoichiometric (A and B) or 5-fold excess (C and D) NO or stoichiometric NO plus 1 mm GTP (E and F) recorded over 0–5-min intervals. The data are composite measurements collected over the 0–1-s and 0–262-s time intervals. Changes in the absorbance are shown over a logarithmic (A, C, and E) or linear (B, D, and F) time scale. AU, absorbance unit.
Dynamics of sGC/NO Interaction with Excess NO
To determine the effect of excess NO on the behavior of the sGC-NO complex, we monitored the spectral changes of sGC mixed with 5-fold excess NO. Formation of the 5C sGC-NO complex in this case was also a two-stage process, with fast formation of a transient 6C sGC-NO complex (Fig. 1D) as observed previously (14–16). The subsequent 6C sGC-NO → 5C sGC-NO transition was slightly faster (∼10 s−1) than in the reaction with stoichiometric NO. In stark contrast to the unstable 5C sGC-NO complex formed with stoichiometric NO, the 5C sGC-NO complex formed with excess NO was much more stable. A measurable amount of sGC-2 was detected only after >1 min of observation (Figs. 1D and 2, C and D). The observed rate of conversion from the 5C sGC-NO complex to the sGC-2 state in the presence of excess NO was ∼0.01 s−1. The addition of DTT to the reaction yielded similar results (supplemental Fig. S2)
Effect of the GTP Substrate on the Dynamics of sGC/NO Interaction
A number of reports have indicated that the GTP substrate affects the stability (21, 25) of the 5C sGC-NO complex. Thus, we monitored the effect of GTP on the kinetics of the 5C sGC-NO complex generated with stoichiometric NO. Under this conditions, we observed that even if sGC was preincubated with GTP (see “Experimental Procedures”) prior to mixing with a stoichiometric amount of NO, the same early two-stage NO binding took place. However, in the presence of GTP, the 5C sGC-NO complex was much more stable, and only a small fraction of the complex converted slowly into sGC-2 at 0.005 s−1 (Figs. 1E and 2, E and F). Similar results were obtained when sGC was replaced with DTT (supplemental Fig. S2).
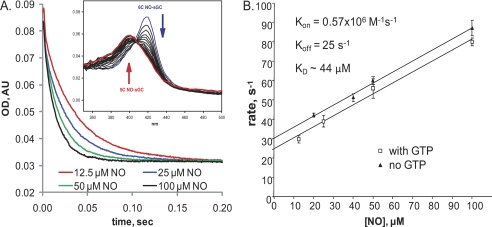
We further tested if binding of GTP to the catalytic site affects the rate of the 6C sGC-NO → 5C sGC-NO transition and its dependence on NO concentration (Fig. 3A). We found an almost identical linear [NO] dependence up to 100 μm in the absence or presence of GTP (Fig. 3). These data indicate that GTP does not affect the binding of NO to the secondary sGC-binding site. From the secondary plot in Fig. 3B, we calculated the observed secondary NO association (kon) and dissociation (koff) rate constants as 0.57 × 106 m−1 s−1 and 25 s−1, respectively, and the equilibrium constant (KD as the ratio koff/kon) as ∼44 μm.
FIGURE 3.
GTP effect on the conversion from 6C sGC-NO to 5C sGC-NO at various NO concentrations. 4 μm sGC was mixed anaerobically with an equal volume of 2 mm Mg2+-GTP in 50 mm TEA buffer (pH 7.4); incubated for 100 ms in the aging loop; and mixed with 12.5, 25, 50, or 100 μm NO. A, changes in the absorbance at 420 nm (6C sGC-NO) recorded at a rate of 400 points/0.2 s. Inset, selected spectra collected between 2 and 50 ms after sGC was supplied with GTP mixed with 100 μm NO. AU, absorbance unit. B, [NO] dependence of the observed rate (kobs) obtained by single-exponential fits to the traces from the experiments in A under pseudo-first-order conditions. Data are presented as means ± S.D. from at least three independent measurements for each concentration. The secondary plot between kobs and [NO] was used to determine koff (ordinate intercept of linear regression), kon (slope of linear regression), and KD (koff/kon).
Catalytic Activity of sGC-NO Complexes
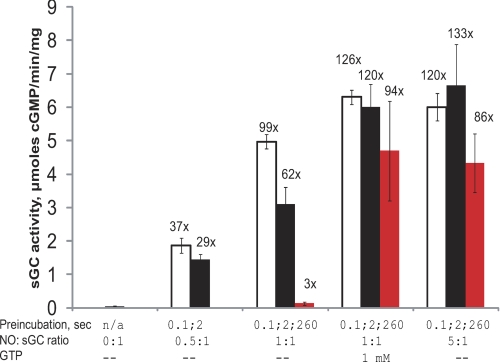
Because the dynamics of sGC/NO interaction suggest the existence of several kinetically distinct 5C sGC-NO complexes, depending on the presence of GTP or NO stoichiometry (Fig. 1), we investigated the cGMP-forming ability of these 5C sGC-NO complexes. We evaluated the cGMP-forming activity of sGC-NO complexes generated at different time points after mixing with 0.5, 1, or 5 eq of NO using the stopped-flow instrument (see schematics in supplemental Fig. S1). Previous reports demonstrated that sGC-NO complexes formed over several minutes with excess NO donor (21, 26) or with stoichiometric NO donor in the presence of GTP (21, 22) have the high cGMP-forming activity expected from fully activated sGC. Our measurements corroborated these observations and demonstrated that high cGMP-forming activity was retained for up to 260 s after the sGC-NO complex was formed with excess NO gas or with stoichiometric NO in the presence of GTP (Fig. 4). However, the activity of the sGC-NO complex formed with stoichiometric NO varied depending on when it was determined after exposure to NO. When the activity was probed ∼4 min after exposure to NO, the sGC-NO complex was only a few -fold more active than the resting sGC (Fig. 4), consistent with previous findings that stoichiometric NO does not lead to full activation of sGC (21, 22, 26). This low activity was not due to sGC protein aggregation during preincubation with NO. If protein aggregation were to develop during incubation, increased turbidity (upward drift at 600 nm) and a slanted base line, especially in the near-UV region, would develop over time. Time-resolved measurements of sGC incubated with different amounts of NO (±GTP) over a period of 260 s showed only an insignificant 354 nm drift, which was identical in all samples, and no change for 600 nm (supplemental Fig. S3), ruling out sGC inactivation by protein aggregation. However, if the activity was probed 100 ms after exposure to stoichiometric amounts of NO gas, the sGC-NO complex had a specific activity comparable with that of the sample exposed to excess NO. This high activity diminished if the sample was probed 2 s after complex formation (Fig. 4). Decreasing the amount of NO to a 0.5 molar ratio while maintaining preincubation times at 0.1 or 2 s also led to a lower activity, consistent with a smaller fraction of the 5C sGC-NO complex formed as evidenced by the optical spectral data (supplemental Fig. S4).
FIGURE 4.
Activity of the 5C sGC-NO complex obtained at different NO ratios with and without the GTP substrate. 4 μm sGC was mixed anaerobically with the indicated equivalents of NO and incubated in the aging loop for 100 ms (open bars), 2 s (black bars), or 260 s (red bars) in the stopped-flow instrument. The reaction mixture was then expelled from the loop and combined with [α-32P]GTP/GTP, and the amount of generated cGMP was determined as described under “Experimental Procedures.” Parallel sGC samples were premixed with 1 mm GTP prior to mixing with stoichiometric NO. The numbers above the bars represent the average -fold activation relative to the basal activity. n/a, not applicable.
Dynamics of Heme/NO Interaction in α1β1H107L Mutant sGC
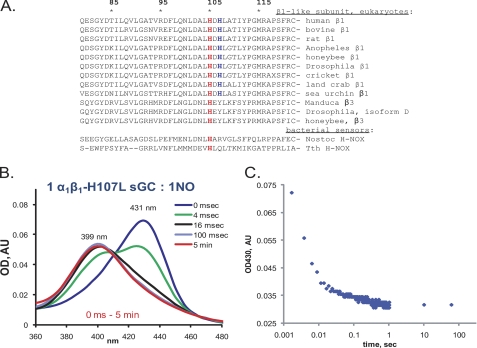
The dynamics of sGC/NO interactions at a 1:1 ratio suggest that the kinetic behavior of the resting sGC, or sGC-1 (Fig. 1A), is significantly different from that of the sGC-2 state produced by the transformation of the 5C sGC-NO complex (Fig. 1C). Although the spectral properties of sGC-1 and sGC-2 species are similar, typical for a heme complex with a proximal histidyl ligand (431 nm Soret peak and 399 nm nitrosyl-heme peak), their overall rates of 5C sGC-NO complex formation are significantly different (∼8.5 versus 0.01 s−1). Sequence alignment of the β1-subunits from different organisms revealed the presence of an additional His-107 residue next to the heme His-105 proximal ligand (Fig. 5A). This His-107 residue is conserved among the β1-like subunits of all mammals and in many eukaryotes, but not in the alternative β-subunits of insects (Fig. 5A). We substituted His-107 with leucine, a non-polar residue of similar size yet incapable of heme iron coordination. We tested the dynamics of NO interaction with the α1β1H107L enzyme. In contrast to the wild-type sGC, the reaction of the α1β1H107L mutant with stoichiometric NO formed a very stable 5C sGC-NO complex (Fig. 5, B and C) reminiscent of the 5C sGC-NO complex formed by the wild-type sGC in the presence of GTP or excess NO (Fig. 1, D and E). A rapid scan at 400 scans/s failed to detect the accumulation of the 420 nm intermediate, indicating a very fast 6C sGC-NO → 5C sGC-NO transition for this mutant.
FIGURE 5.
αβH107L sGC forms a stable 5C sGC-NO complex even with stoichiometric NO. A, sequence comparison of the area surrounding the heme-coordinating His-105 for various mammalian sGC β1-subunits, β1-like insect isoforms, and bacterial H-NOX sensors. Numbering corresponds to the human sGC β1-subunit sequence, with His-105 (red) and His-107 (blue) indicated. B, time-resolved spectral changes of 2 μm anaerobic αβH107L sGC mixed with a stoichiometric amount of NO. Data were recorded at a rate of 400 spectra/s for the first second and then for at least 3, 15, and 60 s after mixing with NO. Representative spectra are shown. AU, absorbance unit. C, time-resolved changes in the 431 nm trace collected from B over the indicated interval.
Decay of the cGMP-forming Activity of the sGC-NO Complex
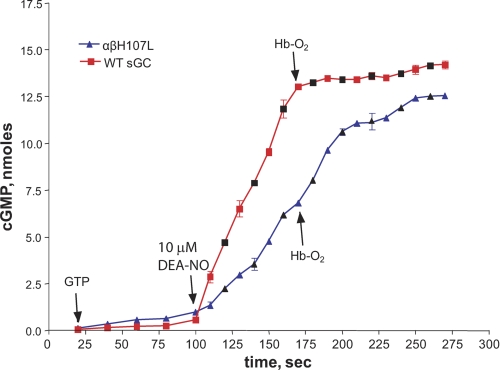
In addition to monitoring the stability of the 5C α1β1H107L-NO complex optically, we compared the rate of decay of the cGMP-forming activity of the wild-type and αβH107L mutant sGC. The cGMP-forming activity of both the wild-type and α1β1H107L sGC increased upon addition of the NO donor DEA-NO, although the mutant had a lower rate of cGMP synthesis (Fig. 6). However, a significant difference between these enzymes was observed after free NO was scavenged by excess HbO2. The wild-type sGC immediately lost its high cGMP-forming activity, consistent with the <0.5 s half-life estimated previously for the active sGC-NO complex (7). The cGMP-forming activity of the α1β1H107L-NO complex decayed rather slowly with a half-life of ∼22 s (Fig. 6), indicating a much more stable complex with NO, corroborating the extended life time of the 399 nm species (Fig. 5, B and C).
FIGURE 6.
Different stabilities of the cGMP-forming sGC-NO complex for wild-type and αβH107L sGC. Sf9 lysates expressing wild-type or α1β1H107L sGC were incubated at 37 °C in reaction buffer containing 1 mm GTP/[α-32P]GTP (see “Experimental Procedures”). Aliquots were taken at different time points to determine basal activity prior to activating the samples with 10 μm DEA-NO. To scavenge free NO, 120 μm HbO2 was added, and sampling was continued for an additional 2 min for activity measurements. The amount of synthesized cGMP was determined as described under “Experimental Procedures.” Combined data from two independent experiments (colored and black symbols) performed in triplicates are presented as the mean ± S.D.
DISCUSSION
Dynamics of NO Binding to sGC
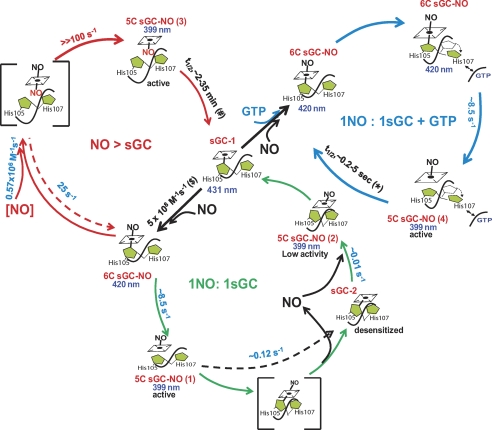
Direct measurements of physiological amounts of NO (17–19) and estimation of intracellular concentrations of sGC (20) indicate that, under normal conditions, cellular sGC encounters and is stimulated by substoichiometric NO. The goal of this study was to investigate the dynamics of NO interaction with sGC and the functional outcome of such interaction. For this purpose, we used a stoichiometric amount of NO gas, rather than NO donors, and all experiments were performed in a strictly anaerobic environment to ensure the stability of NO. The data collected using this approach allow us to propose a more conclusive model of interaction between NO and sGC and its functional outcomes, which we summarized as three pathways in a comprehensive scheme shown in Fig. 7. The green path illustrates what happens under conditions of stoichiometric NO, the blue path reflects the processes occurring in the presence of GTP, and the red path shows the behavior of sGC when NO is in excess.
FIGURE 7.
Dynamics of sGC/NO interaction. Shown is a schematic representation of the dynamic process of sGC/NO interaction. Upon binding of NO to sGC, a transient 6C sGC-NO complex is formed, which converts into the 5C sGC-NO(1) complex. When only stoichiometric NO is present (green path), the active 5C sGC-NO(1) complex transforms to sGC-2, which binds NO very slowly, presumably due to conformational changes involving His-107, to form an inactive 5C sGC-NO(2) complex. When NO is in excess (red path), the second NO binds to 5C sGC-NO(1) to concurrently disrupt the iron–His ligation and form an unstable bisNO-sGC complex, which releases the distal NO to form an active 5C sGC-NO(3) complex. The proposed binding site of the second NO to the proximal site of the heme prevents the formation of sGC-2. In the presence of GTP (blue path), 5C sGC-NO(1) does not convert to sGC-2, presumably due to the conformational effects induced by GTP, which prevent the repositioning of His-107. Notice that three 5C sGC-NO intermediates, 5C sGC-NO(1), 5C sGC-NO(2), and 5C sGC-NO(4), have NO at the distal site of the heme, but the distinction is at the protein conformation, which is not explicitly expressed by our simplified graphic representation. The reaction rates determined in these studies are shown in blue, the number in red is the theoretical estimation of the minimal reaction rate, and the numbers in black are from the literature: *, values from Refs. 21, 25, and 57; #, values from Refs. 21, 34, and 58; $, value from Ref. 15.
Our data demonstrate that the 5C sGC-NO complex formed with stoichiometric NO (Fig. 7, green path) in the absence of GTP is rather labile (Fig. 1, B and D). Despite the presence of NO in the observation cell and the diffusion-limited binding of NO to sGC, we still observed the decay of the 5C sGC-NO complex (Fig. 1, B and C). Such a transition indicates a reduced NO-binding activity of sGC-2. These transformations, which occur at a 1:1 ratio of sGC and NO, are reflected in the multiphasic dynamics of sGC-NO complex formation and decay (Fig. 2). These dynamic changes were not observed in previous studies using excess NO gas (14, 15). The reports that dealt with the reaction of sGC with NO at a 1:1 ratio also failed to observe these cycles of complex formation and decomposition, primarily due to a slow rate of data sampling and the use of NO donors as the source of NO (22, 27). Because the NO-releasing kinetics of NO donors are much slower than the fast dynamics of sGC/NO interaction (DEA-NO, t½ ∼ 900 s; PROLI-NO, t½ > 2 s at 25 °C), they are not well suited to monitor the fast processes of 5C sGC-NO formation and decomposition. Our use of a NO gas solution under strictly anaerobic condition guarantees quantitative NO delivery in a millisecond time range.
We showed that the unstable 5C sGC-NO(1) complex generated after NO binding (Fig. 7, green path) is stabilized by an excess of NO (Fig. 2, A and B versus C and D; Fig. 7, red path). We also confirmed previous observations (14, 15) that an excess of NO accelerates the 6C-sGC-NO → 5C sGC-NO transition in a [NO]-dependent manner (Fig. 3, A and B). These data clearly support the notion that a second NO-binding event takes place, yet its nature is uncertain, as both protein thiols and the heme moiety are possible binding targets. In vitro and in vivo studies clearly demonstrate that thiol nitrosylation affects sGC activity (28–31). However, the formation of nitrosocysteine requires oxygen to generate reactive nitrogen species as active nitrosating agents (32, 33) or to induce oxidative modification of cysteine capable of directly reacting with NO (28). Because our experiments were done in anaerobic conditions, cysteine oxidation or the formation of reactive nitrogen species was prohibited. Moreover, the rate of thiol nitrosylation is much slower (34) than the observed binding of a second NO molecule (Fig. 3) (14, 15). The fast kinetics indicate that the secondary NO binds either to the heme or a protein radical. However, no sGC radical intermediate(s) was observed by EPR,4 leaving heme as the most likely target. Direct evidence to support this mechanism was obtained recently.5
The observed linear [NO] dependence of the 6C sGC-NO → 5C sGC-NO transition (Fig. 3) provides additional information regarding the binding mechanism of a second NO molecule. If dissociation of His-105 is required for binding of the second NO, then the intrinsic rate of His-105 dissociation, estimated at 8.5 s−1 (Fig. 1), would be rate-limiting. However, we observed linear [NO] dependence at a rate >90 s−1 at 100 μm NO (Fig. 3B) without any sign of saturation. This indicates that the binding of the second NO follows a concerted, rather than a sequential, mechanism with NO binding and His-105 dissociation from the heme occurring simultaneously. The strain to the iron–His bond is enhanced by the attack of the second NO at the heme iron, facilitating the rupture of the iron–His bond, reflected by an enhanced rate. We thus propose that binding of an additional NO molecule to the proximal site of the heme in the 5C sGC-NO(1) complex is quickly followed by the loss of the distal NO (Fig. 7, red path). The rate for the bisNO-heme → 5C sGC-NO(3) conversion should be much greater than the rate of bisNO-heme formation (≫100 s−1) to account for the failure to observe a bisNO-heme intermediate. Furthermore, the bisNO-heme → 5C sGC-NO(3) conversion must be irreversible, as we never observed a reverse 5C sGC-NO(3) → bisNO-heme conversion even in the presence of 0.1 mm NO (Fig. 3A, inset). Our data imply that binding of NO to the proximal side in the 5C sGC-NO(3) complex significantly impedes the reformation of the iron–His bond (Fig. 7, red path), thus stabilizing the nitrosyl-heme. Binding of NO to the proximal side of heme is well documented for cytochrome c′ from the bacterium Alcaligenes xylosoxidans (AXCP) (35, 36).
Role of His-107 in Destabilization and Desensitization
Although the spectral feature of sGC-2 formed after the decay of the 5C sGC-NO(1) complex (Fig. 7, green path) points to a histidyl proximal ligand, kinetic data demonstrate that this is a distinct state different from the starting sGC-1. Our measurements demonstrate that, in the sGC-2 state, the NO-binding rate is 0.65 × 104 m−1 s−1, which is several orders slower than the ∼108 m−1 s−1 rate of NO binding to resting sGC-1. Our data show that substitution of His-107 with Leu results in stabilization of the 5C sGC-NO complex at stoichiometric NO both in spectral (Fig. 5, B and C) and functional (Fig. 6) analyses. The lack of a 6C sGC-NO (420 nm) intermediate for the αβH107L mutant (Fig. 5B) suggests a much faster transition from the 6C sGC-NO complex to the 5C sGC-NO complex and may reflect either a weakened iron–His-105 bond or an easier access of the proximal side for the second NO. Future resonance Raman and structural studies will test this assumption.
The closest model for the sGC heme-binding domain is derived from the structure of Nostoc sp. H-NOX protein (9), which has the highest sequence identity (33%) to sGC among all known heme sensors. This model positions His-107 in α-helix located just below the heme and suggests that it may be exposed to the solvent (9, 37). Recent studies demonstrated the importance of Asp-102 located in the same region for the mediation of signal transduction that leads to activation of sGC by NO (37). Our data suggest that His-107 plays an important role in mediating conformational changes that lead to destabilization of 5C sGC-NO(1) and formation of the sGC-2 state desensitized to NO (Fig. 7, green path). These changes may depend on the interaction of His-107 with other elements of the protein, e.g. the catalytic domain. The binding of GTP to the catalytic domain or of additional NO to heme may interfere with this destabilizing interaction(s), resulting in the observed stable NO activation (Fig. 4). The exact nature of these conformational changes remains to be determined, and future structural studies on a full-length sGC may provide a definitive answer.
Another possibility is that His-107 may act as an alternative but less efficient heme ligand than His-105. Although a heme proximal ligand switch was not previously described for sGC, such events are well documented for other heme proteins. For example, the CO-sensing transcription factor CooA from the bacterium Rhodospirillum rubrum is coordinated by Cys-75 in the ferric form, but, upon reduction to the ferrous form, His-77 becomes the ligand (38, 39). Similarly, ligand switching was described for the bacterial phosphodiesterase EcDOS (40) and the transcriptional regulator RcoM-2 from the bacterium Burkholderia xenovorans (41). In all cases, the properties of the affected proteins changed. Some previous studies support the notion that an alternative coordination of sGC heme may be conceivable. For example, we have previously reported that, upon heme reconstitution, the mutant αβCys-105 sGC displayed spectral properties characteristic of a histidine-ligated hemoprotein, although the heme-coordinating His-105 was mutated to cysteine (42). These data suggest that the heme proximal ligand in the αβH105C mutant is not Cys-105 but rather an alternative histidine, perhaps His-107. Other studies indicate that, depending on the method of in vitro heme reconstitution, two different states of sGC may be achieved (43).
Effect of GTP on the Stability of the sGC-NO Complex
As demonstrated in Figs. 1E and 2E, conversion of the 5C sGC-NO(1) complex into sGC-2 is prevented not only by excess NO but also by the substrate GTP (Fig. 7, blue path). At first glance, these results seem to contradict the previous observation that GTP decreases the half-life of the 5C sGC-NO complex from minutes (21, 34) to seconds (25, 44). However, the half-life of the 5C sGC-NO complex was determined by following the decay of the 5C sGC-NO complex after scavenging free NO (25, 44), whereas our observations were performed anaerobically, in the absence of any NO scavengers. Under such conditions, the stoichiometric NO used to generate the 5C sGC-NO(1) complex remains present in the system and may rebind sGC after the sGC-NO complex decays. Such an enclosed system allows investigation of the dynamic behavior of sGC-NO complexes under conditions mimicking the cellular milieu, where the absence of strong scavengers should allow the rebinding of NO to sGC enzyme available at in higher concentrations. Specifically, it allowed us to observe the instability of the 5C sGC-NO(1) complex due to its conversion to sGC-2 (Fig. 7, green path) and the stabilization of 5C sGC-NO by GTP (5C sGC-NO(4) in Fig. 7, blue path) or excess NO (5C sGC-NO(3) in Fig. 7, red path). Previously reported resonance Raman studies demonstrated a difference between sGC-NO complexes obtained in excess or stoichiometric NO (26) and subtle changes in the EPR spectrum due to varying g2 values (45). The addition of GTP induced a much more pronounced difference in the EPR spectra of the sGC-NO complex (45). Corroborating these findings, our EPR measurements also showed a changed line shape in the presence of GTP (supplemental Fig. S5). Thus, this spectroscopic evidence suggests that certain conformational changes take place and may account for the differences in the kinetic and functional properties of various 5C sGC-NO complexes observed in this study.
Our results also indicate that the GTP substrate does not cause obvious changes in the kinetics of the formation of 6C sGC-NO and its subsequent conversion to 5C sGC-NO (Figs. 2 and 3). These observations are in contrast to previously reported experiments done with the DEA-NO donor, where GTP appeared to prevent the formation of 6C sGC-NO (22). The most probable explanation is that the previously reported 420 nm species observed 14 s after the addition of the DEA-NO donor, believed to be 6C sGC-NO (22), was in fact a mixture of sGC-1, 5C sGC-NO(1), sGC-2, and perhaps even 5C sGC-NO(2), which added up to yield a 420 nm peak. As discussed above, the relatively slow kinetics of NO release from DEA-NO and slow data sampling are not optimal for monitoring the process of NO interaction with sGC and may account for the discrepancy of our data and the reported observations.
Activity of Various sGC-NO Complexes and Implication for NO/cGMP Signaling
The dynamics of sGC/NO interaction and subsequent complex transformations reported here provide additional insights into the function of cellular sGC and the mechanism of its desensitization. Previous observations that 1 eq of NO does not fully activate sGC (21, 22) gave rise to the hypothesis that, after its formation, the cellular sGC-NO complex is in a “tonically” activated state, which requires additional NO for full activation and efficient NO/cGMP signaling (21, 22, 27). However, under normal non-inflammatory conditions, the sGC concentration may be several orders of magnitude higher than the NO concentration (20), and only a fraction of sGC may convert into the sGC-NO complex. Thus, under the premises of the “tonic” activation hypothesis, under physiological conditions, NO/cGMP signaling should be very inefficient. This conclusion is inconsistent with experimental observations of an effective cGMP synthesis in response even to picomolar puffs of NO (20).
To resolve this discrepancy, we evaluated the cGMP-forming activities of several kinetically distinct 5C sGC-NO complexes observed in our studies (Figs. 1 and 7). In corroboration with previous reports (21, 22), the complexes obtained with stoichiometric NO in the presence of GTP (5C sGC-NO(4)) or with excess NO (5C sGC-NO(3)) were highly activated by NO and showed a high cGMP-forming activity (Fig. 4). However, our measurement of sGC-NO complexes obtained with stoichiometric NO in the absence of GTP yielded results different from those reported previously (21, 22, 26). Our data indicate that the cGMP-forming activity of the 5C sGC-NO(1) complex formed with stoichiometric NO is high, if tested shortly after exposure to NO (100 ms). However, this activity was time-sensitive and declined even 2 s after NO exposure. If the activity was measured 260 s after exposure to NO, the enzyme was only marginally activated (Fig. 4), despite the spectrally observed sGC-NO complex (Figs. 1C and 2A). These time-dependent changes in the catalytic properties of the sGC-NO complex with stoichiometric NO are consistent with the observed transformations into sGC-2 and 5C sGC-NO(2) (Figs. 1C and 2A). Thus, our data show that the sGC-NO complex formed with stoichiometric NO is active for at least several seconds (Fig. 4), indicating that NO/cGMP signaling can proceed if GTP molecules are available to prevent the formation of sGC-2 and/or low activity 5C sGC-NO(2). The low cGMP-forming activity of the sGC-NO complex present 260 s after stoichiometric NO binding is similar to the low activity of stoichiometric sGC-NO complexes reported previously (21, 22, 26). In those studies, stoichiometric sGC-NO complexes were obtained by sGC titration with a NO donor (21) or after removal of unreacted excess NO donor by gel filtration (22) or multiple membrane filtration procedures (26). Because these approaches led to a considerable delay between formation of the sGC-NO complex and evaluation of its activity, we believe that it was the activity of 5C sGC-NO(2) accumulated over time that was in fact measured in those studies.
Mechanistic Implications for Rapid Desensitization of sGC
The tendency of the sGC-NO complex formed with stoichiometric NO to decompose in the absence of GTP and to form the sGC-2 state with lower NO-binding properties may be important for sGC desensitization. It has been reported in different cells and tissues (46–49) that treatments with various NO donors or nitrovasodilators diminish or abolish the responsiveness of sGC to subsequent NO stimulations. Such desensitization was reported not only when high NO concentrations were applied but even when repeated pulses of low NO doses were used (50).
A number of processes directly affecting the function of sGC are attributed to sGC desensitization. These include oxidation of sGC heme and subsequent protein degradation (51, 52) and reduced sGC activity in response to S-nitrosylation (30, 31). Although it is clear that these mechanisms play an important role in desensitization of sGC signaling under chronic oxidative conditions, their contribution to rapid desensitization of sGC in response to NO (46) is unclear. Decreased sGC activity in response to phosphorylation by cGMP-dependent kinase (53) remains the only reported mechanism of rapid sGC desensitization. Our data suggest that, in addition, the transformation of the highly active 5C sGC-NO(1) complex into sGC-2 with slow NO binding, followed by formation of low activity 5C sGC-NO(2), may be an important factor contributing to rapid desensitization. The estimated intracellular concentration of GTP is in the range of 100–200 μm (54, 55), which is close to the Km(GTP) value for purified sGC (56). Thus, robust NO-induced cGMP synthesis in cells with micromolar sGC levels (20) may temporarily decrease the intracellular levels of GTP and promote accumulation of sGC-2 and/or inactive 5C sGC-NO(2).
In summary, our studies demonstrate that the behavior and properties of the 5C sGC-NO complex obtained after the initial interaction of sGC with NO are strongly affected by the availability of the GTP substrate or NO excess. We have demonstrated that the 5C sGC-NO complex formed with stoichiometric NO is enzymatically active, but in the absence of GTP, it may lose NO and transition into a form with lower affinity for NO, followed by formation of a low activity sGC-NO complex. These conformational transformations likely involve the participation of His-107, which mediates interaction with other sGC domains or acts as an alternative heme ligand. We propose that these protein conformational changes might be prevented by the binding of the GTP substrate or excess NO. These processes may control the amount of intracellular active sGC and contribute to the rapid sGC desensitization observed under physiological conditions.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants HL088128 and HL088128S (to E. M.) and HL095820 (to A.-L. T.) from NHLBI. This work was also supported by Grant-in-aid 09GRNT2060182 from the American Heart Association, South Central Affiliate (to E. M.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5.
V. Berka, A.-L. Tsai, and E. Martin, unpublished data.
E. Martin, V. Berka, I. Sharina, and A.-L. Tsai, manuscript submitted.
- sGC
- soluble guanylyl cyclase
- 5C
- five-coordinate
- 6C
- six-coordinate
- TEA
- triethanolamine
- DEA-NO
- 2-(N,N-diethylamino)diazenolate-2-oxide diethylammonium salt
- PROLI-NO
- Disodium 1-[2-(Carboxylato)pyrrolidin-1-yl]diazen-1-ium-1,2diolate.
REFERENCES
- 1. Davis K. L., Martin E., Turko I. V., Murad F. (2001) Annu. Rev. Pharmacol. Toxicol. 41, 203–236 [DOI] [PubMed] [Google Scholar]
- 2. Moncada S., Higgs E. A. (2006) Handb. Exp. Pharmacol. 176, 213–254 [DOI] [PubMed] [Google Scholar]
- 3. Kamisaki Y., Saheki S., Nakane M., Palmieri J. A., Kuno T., Chang B. Y., Waldman S. A., Murad F. (1986) J. Biol. Chem. 261, 7236–7241 [PubMed] [Google Scholar]
- 4. Boon E. M., Marletta M. A. (2005) Curr. Opin. Chem. Biol. 9, 441–446 [DOI] [PubMed] [Google Scholar]
- 5. Makino R., Park S. Y., Obayashi E., Iizuka T., Hori H., Shiro Y. (2011) J. Biol. Chem. 286, 15678–15687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Martin E., Berka V., Bogatenkova E., Murad F., Tsai A. L. (2006) J. Biol. Chem. 281, 27836–27845 [DOI] [PubMed] [Google Scholar]
- 7. Russwurm M., Mergia E., Mullershausen F., Koesling D. (2002) J. Biol. Chem. 277, 24883–24888 [DOI] [PubMed] [Google Scholar]
- 8. Karow D. S., Pan D., Tran R., Pellicena P., Presley A., Mathies R. A., Marletta M. A. (2004) Biochemistry 43, 10203–10211 [DOI] [PubMed] [Google Scholar]
- 9. Ma X., Sayed N., Beuve A., van den Akker F. (2007) EMBO J. 26, 578–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nioche P., Berka V., Vipond J., Minton N., Tsai A. L., Raman C. S. (2004) Science 306, 1550–1553 [DOI] [PubMed] [Google Scholar]
- 11. Derbyshire E. R., Fernhoff N. B., Deng S., Marletta M. A. (2009) Biochemistry 48, 7519–7524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Derbyshire E. R., Marletta M. A. (2009) Handb. Exp. Pharmacol. 191, 17–31 [DOI] [PubMed] [Google Scholar]
- 13. Stone J. R., Marletta M. A. (1994) Biochemistry 33, 5636–5640 [DOI] [PubMed] [Google Scholar]
- 14. Martin E., Berka V., Tsai A. L., Murad F. (2005) Methods Enzymol. 396, 478–492 [DOI] [PubMed] [Google Scholar]
- 15. Zhao Y., Brandish P. E., Ballou D. P., Marletta M. A. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 14753–14758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Makino R., Matsuda H., Obayashi E., Shiro Y., Iizuka T., Hori H. (1999) J. Biol. Chem. 274, 7714–7723 [DOI] [PubMed] [Google Scholar]
- 17. Hall C. N., Garthwaite J. (2009) Nitric Oxide 21, 92–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sato M., Nakajima T., Goto M., Umezawa Y. (2006) Anal. Chem. 78, 8175–8182 [DOI] [PubMed] [Google Scholar]
- 19. Chen K., Popel A. S. (2006) Free Radic. Biol. Med. 41, 668–680 [DOI] [PubMed] [Google Scholar]
- 20. Batchelor A. M., Bartus K., Reynell C., Constantinou S., Halvey E. J., Held K. F., Dostmann W. R., Vernon J., Garthwaite J. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 22060–22065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cary S. P., Winger J. A., Marletta M. A. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 13064–13069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Russwurm M., Koesling D. (2004) EMBO J. 23, 4443–4450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Feelisch M., Noack E. A. (1987) Eur. J. Pharmacol. 139, 19–30 [DOI] [PubMed] [Google Scholar]
- 24. Martin E., Lee Y. C., Murad F. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 12938–12942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kharitonov V. G., Russwurm M., Magde D., Sharma V. S., Koesling D. (1997) Biochem. Biophys. Res. Commun. 239, 284–286 [DOI] [PubMed] [Google Scholar]
- 26. Ibrahim M., Derbyshire E. R., Soldatova A. V., Marletta M. A., Spiro T. G. (2010) Biochemistry 49, 4864–4871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cary S. P., Winger J. A., Derbyshire E. R., Marletta M. A. (2006) Trends Biochem. Sci. 31, 231–239 [DOI] [PubMed] [Google Scholar]
- 28. Fernhoff N. B., Derbyshire E. R., Marletta M. A. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 21602–21607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mayer B., Kleschyov A. L., Stessel H., Russwurm M., Münzel T., Koesling D., Schmidt K. (2009) Mol. Pharmacol. 75, 886–891 [DOI] [PubMed] [Google Scholar]
- 30. Sayed N., Baskaran P., Ma X., van den Akker F., Beuve A. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 12312–12317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sayed N., Kim D. D., Fioramonti X., Iwahashi T., Durán W. N., Beuve A. (2008) Circ. Res. 103, 606–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Handy D. E., Loscalzo J. (2006) Arterioscler. Thromb. Vasc. Biol. 26, 1207–1214 [DOI] [PubMed] [Google Scholar]
- 33. Hess D. T., Matsumoto A., Kim S. O., Marshall H. E., Stamler J. S. (2005) Nat. Rev. Mol. Cell Biol. 6, 150–166 [DOI] [PubMed] [Google Scholar]
- 34. Kharitonov V. G., Sharma V. S., Magde D., Koesling D. (1997) Biochemistry 36, 6814–6818 [DOI] [PubMed] [Google Scholar]
- 35. Lawson D. M., Stevenson C. E., Andrew C. R., Eady R. R. (2000) EMBO J. 19, 5661–5671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Martí M. A., Capece L., Crespo A., Doctorovich F., Estrin D. A. (2005) J. Am. Chem. Soc. 127, 7721–7728 [DOI] [PubMed] [Google Scholar]
- 37. Baskaran P., Heckler E. J., van den Akker F., Beuve A. (2011) Biochemistry 50, 4291–4297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lanzilotta W. N., Schuller D. J., Thorsteinsson M. V., Kerby R. L., Roberts G. P., Poulos T. L. (2000) Nat. Struct. Biol. 7, 876–880 [DOI] [PubMed] [Google Scholar]
- 39. Shelver D., Thorsteinsson M. V., Kerby R. L., Chung S. Y., Roberts G. P., Reynolds M. F., Parks R. B., Burstyn J. N. (1999) Biochemistry 38, 2669–2678 [DOI] [PubMed] [Google Scholar]
- 40. Kurokawa H., Lee D. S., Watanabe M., Sagami I., Mikami B., Raman C. S., Shimizu T. (2004) J. Biol. Chem. 279, 20186–20193 [DOI] [PubMed] [Google Scholar]
- 41. Marvin K. A., Kerby R. L., Youn H., Roberts G. P., Burstyn J. N. (2008) Biochemistry 47, 9016–9028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Martin E., Sharina I., Kots A., Murad F. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 9208–9213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Vogel K. M., Hu S., Spiro T. G., Dierks E. A., Yu A. E., Burstyn J. N. (1999) J. Biol. Inorg. Chem. 4, 804–813 [DOI] [PubMed] [Google Scholar]
- 44. Bellamy T. C., Garthwaite J. (2001) J. Biol. Chem. 276, 4287–4292 [DOI] [PubMed] [Google Scholar]
- 45. Derbyshire E. R., Gunn A., Ibrahim M., Spiro T. G., Britt R. D., Marletta M. A. (2008) Biochemistry 47, 3892–3899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bellamy T. C., Wood J., Goodwin D. A., Garthwaite J. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 2928–2933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Davis J. P., Vo X. T., Sulakhe P. V. (1997) Biochem. Biophys. Res. Commun. 238, 351–356 [DOI] [PubMed] [Google Scholar]
- 48. Mülsch A., Busse R., Bassenge E. (1988) Eur. J. Pharmacol. 158, 191–198 [DOI] [PubMed] [Google Scholar]
- 49. Waldman S. A., Rapoport R. M., Ginsburg R., Murad F. (1986) Biochem. Pharmacol. 35, 3525–3531 [DOI] [PubMed] [Google Scholar]
- 50. Halvey E. J., Vernon J., Roy B., Garthwaite J. (2009) J. Biol. Chem. 284, 25630–25641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Meurer S., Pioch S., Pabst T., Opitz N., Schmidt P. M., Beckhaus T., Wagner K., Matt S., Gegenbauer K., Geschka S., Karas M., Stasch J. P., Schmidt H. H., Müller-Esterl W. (2009) Circ. Res. 105, 33–41 [DOI] [PubMed] [Google Scholar]
- 52. Stasch J. P., Schmidt P. M., Nedvetsky P. I., Nedvetskaya T. Y., H S A. K., Meurer S., Deile M., Taye A., Knorr A., Lapp H., Müller H., Turgay Y., Rothkegel C., Tersteegen A., Kemp-Harper B., Müller-Esterl W., Schmidt H. H. (2006) J. Clin. Invest. 116, 2552–2561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhou Z., Sayed N., Pyriochou A., Roussos C., Fulton D., Beuve A., Papapetropoulos A. (2008) Arterioscler. Thromb. Vasc. Biol. 28, 1803–1810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hatakeyama K., Harada T., Kagamiyama H. (1992) J. Biol. Chem. 267, 20734–20739 [PubMed] [Google Scholar]
- 55. Otero A. D. (1990) Biochem. Pharmacol. 39, 1399–1404 [DOI] [PubMed] [Google Scholar]
- 56. Schmidt P., Schramm M., Schröder H., Stasch J. P. (2003) Eur. J. Pharmacol. 468, 167–174 [DOI] [PubMed] [Google Scholar]
- 57. Bellamy T. C., Wood J., Garthwaite J. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 507–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Brandish P. E., Buechler W., Marletta M. A. (1998) Biochemistry 37, 16898–16907 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.