Abstract
CENP-W was originally identified as a putative oncogene, cancer-upregulated gene 2 (CUG2) that was commonly up-regulated in many cancer tissues. Recently, CENP-W has also been identified as a new centromeric component that interacts with CENP-T. As a complex with CENP-T, CENP-W plays crucial roles in assembly of the functional kinetochore complex. In this study, the subnuclear localization of CENP-W was extensively analyzed using various approaches. We found that ectopically expressed CENP-W primarily accumulated in the nucleolus and remained substantially associated with the nucleolus in stable cells. The following fractionation study also showed that CENP-W is associated with RNA as well as DNA. Moreover, a considerable amount of CENP-W was found in the nuclear mesh-like structure, nuclear matrix, possibly indicating that CENP-W participates in diverse subnuclear activities. Finally, biochemical affinity binding analysis revealed that CENP-W specifically interacts with the nucleolar phosphoprotein, nucleophosmin (B23). Depletion of cellular B23 by siRNA treatment induced a dramatic decrease of CENP-W stability and severe mislocalization during prophase. Our data proposed that B23 may function in the assembly of the kinetochore complex by interacting with CENP-W during interphase.
Keywords: Centromeres, Nuclear matrix, Nucleolus, Nucleus, Oncogene, RNA, CENP-T, CENP-W, Kinetochore, Nucleophosmin (B23)
Introduction
Mammalian nucleus is a highly organized well compartmentalized organelle, and each nuclear compartment is considered a distinct functional domain that performs various nuclear processes (1). Nuclear proteins participating in specific cellular activities are targeted toward specific sites, and the integration of functionally related proteins may support efficient coordination of nuclear activities (2, 3).
The nucleolus is one of the most easily observed amorphous subnuclear structures in mammalian cells during interphase (4). Even though the function of the nucleoli is primarily associated with ribosome biogenesis, several recent investigations have revealed that nucleoli regulate many essential cellular processes such as cell cycle control, sequestering of regulatory molecules, and assembly of ribonucleoprotein (5, 6). The composition of nucleoli can change dynamically under different cellular conditions (7, 8). Furthermore, several recent studies have also revealed that there are some proteins found in nucleoli with no identifiable function related to the ribosome biogenesis or other recently identified novel functions of the nucleolus (9). This observation led us to consider this compartment as an interesting multifunctional organelle, and not simply an RNA factory. Indeed, determining the roles of temporal components of the nucleoli is a topic of current interest (9).
Nucleophosmin/B23 is a multifunctional nuclear phosphoprotein that primarily exists in the nucleoli (10). B23 participates in diverse cellular processes, including ribosome biogenesis, ARF-p53 interaction, apoptotic cell death, and centrosome amplification (11, 12). B23 is frequently overexpressed, mutated, or deleted in human cancer and is believed to play a crucial role in the regulation of cell growth, proliferation, and transformation. Protein B23 was also observed in a nuclear matrix abundant in malignant cells and proliferating B lymphocytes (13). The nuclear matrix is defined as the insoluble structural framework of the nucleus that remains after sequential extraction of the nuclei with non-ionic detergents, nucleases, and high salt buffers (14). Although the nuclear matrix mainly consists of a nuclear lamina protein shell, numerous studies have reported the presence of other components that participate in essential nuclear events, including DNA- and RNA-binding proteins (15). Functionally, the matrix is believed to act as a dynamic support for many essential nuclear functions such as organization of chromatin, DNA replication, and RNA synthesis (16–18).
CENP-W is initially identified as CUG2,3 which is commonly up-regulated in various types of human cancers and highly oncogenic when expressed in mouse fibroblast cell line (19). Recently, CUG2 was re-named CENP-W (20) based on the new finding that it forms a complex with another novel centromere component, CENP-T, which was found to be a member of the CENP-A centromeric nucleosome-associated complex (21). Hori et al. (20) reported that the CENP-W-CENP-T complex functions upstream of other components and that depletion of this complex induced severe defects in the recruitment of other kinetochore components, such as CENP-H, -O, and -S. Although many previous reports showed evidences of a close functional relationship between the centromere and nucleolus, Foltz et al. (21, 22) recently discovered that B23 is present in the CENP-A nucleosome, suggesting that B23 may play a sequestering role in the formation of centromeric protein complex during interphase.
In the present study, we analyzed the biochemical characteristics of CENP-W to gain insight into the undiscovered aspects of this newly identified putative oncoprotein. To our surprise, CENP-W was present in many of the important nuclear subcompartments. Furthermore, this new centromeric component specifically interacts with a nucleolar protein, B23.
EXPERIMENTAL PROCEDURES
Cloning and Antibodies
A549 and HeLa cells purchased from American Type Culture Collection were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum under the standard cell culture condition. Cell fractionation studies of CENP-W were conducted using either transiently expressed 293T cells or stable cells generated in A549 and HeLa cells with pcDNA-3FLAG-CENP-W (23). Transient transfection of 293T cells was performed using either EffecteneTM (Qiagen) or polyethylenimine reagent (Sigma-Aldrich) according to the vendor's instructions. The full-length cDNA gene of B23 and fibrillarin was isolated by PCR-based amplification using a human stomach cancer library (Invitrogen) and then inserted into the pDsRed-N1 plasmid (Clontech) with XhoI and EcoRI. For co-immunoprecipitation, the ORF of B23 is also reconstructed to the pcDNA3–6Myc vector (23) using NcoI and XhoI restriction sites. A series of nuclear localization sequence (NLS) mutants of EGFP-CENP-W (23) were generated by PCR-based mutagenesis using an EZchange site-directed mutagenesis kit (Enzynomics) according to the manufacturer's instructions. The small interfering RNAs (siRNA) against B23 (B23–1; 5′-AACACCACCAGUGGUCUUAAG-3′ and B23–2; 5′-GAAAAUGAGCACCAGUUAU-3′) (24) was synthesized from Bioneer referring to the previous report and transfected to cells using Lipofectamine (Invitrogen).
Anti-FLAG, anti-GFP, anti-GST, and anti-Myc antibodies were purchased from Sigma-Aldrich, and anti-B23, CENP-H, CENP-B, cyclin B1, and Mad1 antibodies were purchased from Santa Cruz Biotechnology. Anti-CENP-T antiserum production was performed by Aprogen using recombinant hexahistidine-fused CENP-T proteins purified using Ni-NTA Superflow™ resin (Qiagen).
Identification of CENP-W-interacting Nucleolar Proteins
To identify CENP-W-interacting proteins in nucleolus, immunoprecipitation was performed with 293T cells transfected with FLAG-CENP-W. After nucleoli purification (25), the nucleoli were lysed in a protein extraction buffer (50 mm Tris, pH 7.5, 150 mm NaCl, 0.5% Nonidet P-40, and 0.5% Triton X-100), incubated with 1 μg of anti-FLAG antibody (Sigma-Aldrich), and then treated with protein A-agarose (Amersham Biosciences). Next, the samples were washed and the bound proteins were then eluted and subjected to SDS-PAGE. After the proteins were visualized using a silver staining kit (Peptron), the protein bands that were only found in the CENP-W-bound fraction were cut from the gel and sent for subsequent identification by mass spectroscopy (Genomine). For the GST pulldown assay, the lysate containing GST-CENP-W (23) was incubated with 30 μl of 50% slurry of glutathione-agarose beads (Peptron). After three washes, eluents were subjected to SDS-PAGE, and the further analysis methods were identical with those of immunoprecipitation. Total RNAs from H293 cells or Escherichia coli were extracted using a GenEluteTM total RNA miniprep kit (Sigma-Aldrich), and the mRNA fraction was further purified with an Oligotex mRNA midi kit (Qiagen) from H293 cells.
Cell Fractionation
Cell fractionation experiments were conducted as previously described, with slight modification (26). For isolation of the cell nuclei, cells were resuspended in CSK buffer (10 mm PIPES, pH 6.8, 100 mm NaCl, 300 mm sucrose, 3 mm MgCl2, 0.5 mm phenylmethylsulfonyl fluoride, 1 mm DTT) supplemented with 0.5% (v/v) Triton X-100. After 5 min of incubation on ice, the sample was sedimented by centrifugation at 7,500 rpm for 4 min. The pellet, which contained the chromatin DNA, was then digested by incubation with 1 unit/μl RNase-free DNase I (Sigma) or 100 μg/ml RNase A (Sigma) at 37 °C for 15 min. Next, the samples were centrifuged, and the supernatant and pellet were designated fractions S1 and P1, respectively. The pellet was then extracted with (NH4)2SO4 in CSK buffer at a final concentration of 0.25 m, and the supernatant (S2) was collected by centrifugation. After the pellet was further washed with the CSK buffer containing 0.25 m (NH4)2SO4, the remaining insoluble fractions were dissolved in 8 m urea and designated as the P3 fraction.
Nuclear Matrix Isolation
High salt isolation of the nuclear matrix was performed as previously described (27). Briefly, cells were extracted in CSK buffer supplemented with 0.5% Triton X-100 for 5 min at 4 °C. After being separated from the soluble proteins by centrifugation, the pellet was subsequently incubated with DNase I (1 unit/μl) to remove the chromatin. Ammonium sulfate solution was then added to the sample to a final concentration of 0.25 m, and the solution was spun down after 5 min of incubation. The pellet was further extracted by the addition of 2 m NaCl in CSK buffer, after which the soluble proteins were finally harvested by centrifugation. The remaining insoluble pellet was dissolved with urea buffer containing 8 m urea and considered to be the nuclear matrix fraction. Where indicated, cells were pretreated with hydrogen peroxide (2 mm), Trichostatin A (30 μm), or camptothecin (10 μm) for 24 h before fractionation. Anti-TBP (TFIID) (Santa Cruz Biotechnology), anti-Lamin A/C (Santa Cruz Biotechnology), and anti-α-Tubulin antibodies (Sigma) were used to verify the fractionation.
RNA Binding Assay
Sucrose fractionation was performed as previously described (28). The purified nuclei of HeLa-FLAG-CENP-W stable cells were resuspended in buffer A (25 mm Tris, pH 7.5, 100 mm KCl, 1 mm DTT, 2 mm EDTA) supplemented with 0.05% Nonidet P-40, 1 mm NaF, protease inhibitor mixture, and 0.1 unit/ml of RNase inhibitor (New England Biolabs). Next, the samples were sonicated several times for 15 s. The cleared supernatants by centrifugation were overlaid on top of 5 ml of 10–30% (w/w) sucrose gradients in buffer A. The samples were then centrifuged at 38,000 rpm for 3.5 h and 4 °C in a Beckman SW55Ti rotor. Where indicated, the nuclear extracts were pretreated with RNase A (250 μg/ml) for 20 min at 30 °C prior to sedimentation. Finally, the fractions were collected from the top with a capillary pipette and labeled as fraction 1. RNA was separately purified using TRI reagent (MRC) and resolved by electrophoresis with 1.2% agarose gel containing 0.6 m formaldehyde.
Fluorescence Microscopy
Localization of the EGFP-conjugated wild-type and mutant CENP-W proteins was observed with an Olympus IX70 fluorescence microscope (200 × magnification) after transient transfection to HeLa or SKOV-3 cells. The immunostaining was performed as described before (23) using either fluorescein isothiocyanate (FITC)-linked anti-mouse secondary antibodies (Vector Laboratories) or Cy3-linked anti-rabbit antibodies (Jackson ImmunoResearch). Imaging was done either using an Olympus IX70 fluorescence microscope or an LSM5-Pascal confocal imaging system (Zeiss).
For the nuclear matrix in situ immunofluorescence, CENP-W stable cells grown on the coverslips were fixed and permeabilized with ice-cold CSK buffer containing 0.5% Triton X-100 for 10 min at 4 °C. Then, cells were further incubated with DNase I (1 unit/μl) or RNase A (200 μg/ml) in digestion buffer (50 mm NaCl, 10 mm PIPES, pH 6.8, 3 mm MgCl2, 1 mm EGTA, and 0.5% Triton X-100) for 30 min. After extraction with 0.25 m (NH4)2SO4 in CSK buffer, the coverslips were fixed and immunostained with anti-FLAG antibody.
RESULTS
Nucleolar Localization of CENP-W
We previously reported that CENP-W contains a putative NLS sequence and is exclusively localized in the nucleus in mouse fibroblast NIH3T3 cells (19). To elucidate the subnuclear distribution of CENP-W in human cells, we transiently transfected EGFP-conjugated CENP-W to HeLa cells and examined its localization using fluorescence microscopy and comparison with EGFP alone as a control. In HeLa cells, the green fluorescence of EGFP-CENP-W was detected as several clumped dots inside the nucleus (Fig. 1A), whereas EGFP was dispersed throughout the entire cell in the control group. This distribution pattern did not agree with our earlier findings in CENP-W stable cells established in A549, MCF-7, or HeLa cells with FLAG-tagged CENP-W (23). Rather, CENP-W in stable cells was seen as numerous nuclear foci throughout the nucleus. Therefore, we considered the possibility that the unexpected localization of CENP-W was an artifact of high expression levels. However, the earliest images, which were observed at around 6 h after transfection when the expression levels were low, showed almost identical subnuclear distribution as previous observations.
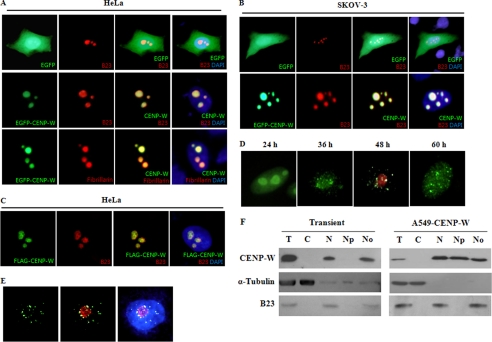
FIGURE 1.
Subnuclear distribution of CENP-W. A, localization of EGFP-tagged CENP-W in HeLa cells. After EGFP-CENP-W was transfected to HeLa cells, the green fluorescence images were captured using fluorescence microscopy at 200× magnification. As nucleolar marker proteins, B23 and fibrillarin were conjugated with RFP and co-transfected with CENP-W. DAPI signals were also applied to localize the nucleus of each cell. B, localization of EGFP-CENP-W in SKOV-3 cells. C, localization of FLAG-CENP-W in HeLa cells. After HeLa cells were transfected with the FLAG-tagged CENP-W, CENP-W was visualized using anti-FLAG and FITC-labeled anti-mouse secondary antibody and B23 using anti-B23 and Cy3-labeled anti-rabbit antibody. D, localization of FLAG-CENP-W in time course. After transfection of FLAG-CENP-W in HeLa cells, the distribution of CENP-W was analyzed at different time points using immunofluorescence microscopy. The endogenous B23 (red) was also stained at 48 h. E, localization of CENP-W in A549 stable cells. After extraction of soluble proteins with 0.5% Triton X-100, cells were immunostained with anti-FLAG antibody. F, nucleolar fractionation of CENP-W-expressing cells. The nucleolus fraction was obtained from either transiently expressed 293T cells or A549-FLAG-CENP-W stable cells. Each fraction was analyzed by Western blotting with anti-FLAG, anti-α-tubulin, and anti-B23 antibodies. T, total cell extract; C, cytoplasmic fraction; N, nuclear fraction; Np, nucleoplasmic fraction; No, nucleolar fraction.
This subnuclear pattern suggested that CENP-W is localized in the nucleolus. To determine if this were the case, we transfected EGFP-CENP-W along with RFP-B23 fusion protein as a nucleolar marker and then evaluated the samples to determine if these two proteins were co-localized in cells. In contrast with the EGFP controls, EGFP-CENP-W accumulated in the specific subnuclear regions, which were well merged with the B23 protein signals (Fig. 1A). To confirm the nucleolar localization of CENP-W, we visualized the nucleolus using another well known nucleolar marker protein, fibrillarin. The results of this experiment also indicated that these two proteins clearly co-localized in the nucleoli (Fig. 1A). To determine if this nucleolar localization of CENP-W was a HeLa cell-specific phenomenon, we transfected EGFP-CENP-W and RFP-B23 plasmids into the human ovarian adenocarcinoma cell line, SKOV-3. A clear co-localization of CENP-W and B23 was also observed in this cell line (Fig. 2B), which excluded the possibility that nucleolar accumulation of EGFP-CENP-W is HeLa cell-specific. Finally, we considered the possibility that an EGFP tag might induce the nucleolar localization of CENP-W. To test this, we transfected the FLAG-tagged version of CENP-W, which is the same construct used for establishment of stable cell lines, and the immunostained CENP-W with anti-FLAG antibody. As shown in Fig. 1C, FLAG-tagged CENP-W also co-localized with endogenous B23 in the nucleoli, confirming the nucleolar localization of CENP-W.
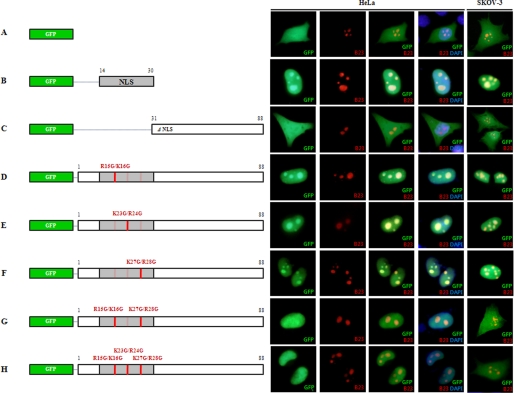
FIGURE 2.
Identification of crucial determinants in CENP-W for nuclear and nucleolar localization. After cells were transfected with EGFP-conjugated CENP-W mutants along with RFP-B23, the photo-images of green and red fluorescence were captured and merged for the analysis. A, EGFP control; B, EGFP conjugated with CENP-W putative NLS sequence (amino acids 14–30); C, CENP-W C terminus region (amino acids, 31–88); D, CENP-W NLS mutant (R15G/K16G); E, CENP-W (K23G/R24G); F, CENP-W (K27G/R28G); G, CENP-W (R15G/K16G/K27G/R28G); and H, CENP-W (R15G/K16G/K23G/R24G/K27G/R28G).
Because the subnuclear distribution of CENP-W in stable cells differed from that of the transiently expressed protein, we traced the localization of FLAG-CENP-W over time after transfection in HeLa cells using immunofluorescence microscopy. CENP-W was localized in the nucleolus in most of the transfected cells at 24 h after transfection. Approximately 48 h after transfection, the initial nucleolar accumulation tended to gradually spread out to become small nuclear dots, which were still nucleolus-associated (Fig. 1D). Finally, numerous foci patterns were observed in most cells at 3 days after transfection.
These observations promptly raised the question of whether the CENP-W proteins were still associated with the nucleolus in stable cells. To address this question, we first selected an A549 stable cell line in which the expression level of FLAG-CENP-W was as low as the endogenous level by RT-PCR. We then visualized FLAG-CENP-W in these stable cells by eliminating the soluble proteins inside the nucleus by pretreatment with 0.5% Triton X-100. When the soluble nucleoplasmic proteins were removed in the A549-CENP-W cells, CENP-W did not appear randomly scattered throughout the nucleus, but rather substantially associated with the nucleolus (Fig. 1E). This nucleolar association was observed in ∼70% of the tested cells. To confirm these findings, a biochemical subcellular fractionation experiment was conducted using both A549-CENP-W stable cells and transiently transfected 293T cells. The nucleolar fractionation results clearly demonstrated that CENP-W was present in the nucleoli of stable cells, being distributed in approximately the same level in both the nucleoplasm and nucleoli, whereas most transiently expressed CENP-W was localized in the nucleolus (Fig. 1F). These results demonstrated that even though CENP-W appears to be scattered throughout the nucleus, it is still associated with the nucleoli in stable cells. The integrity of the cell fractionation was verified by immunodetection of α-tubulin and B23. Taken together, these results indicated that CENP-W, the new component of the centromeric complex, is associated with the nucleolus in human cells.
Identification of the Nucleolar Determinants of CENP-W
Based on a web-based searching program, we previously reported that there is one predicted NLS sequence of 17 amino acids (14KRKAPRGFLKRVFKRKK30) with a 98% probability score (19). In comparison to NLS, no consensus sequence for nucleolar localization has been established to date, except for the finding that such sequences are frequently enriched with arginine and lysine residues (29). Because the predicted NLS sequence of CENP-W is highly enriched in arginine and lysine residues, we evaluated this sequence to determine if it also functions as the primary determinant of nucleolar targeting of CENP-W. To accomplish this, we reconstructed partial cDNAs of CENP-W fused to EGFP. We then tested the localization of EGFP fused with the predicted NLS sequence (amino acid residues 14–30) in both HeLa and SKOV-3 cells and compared these findings with the results obtained using an RFP-conjugated B23 nucleoli marker. The results revealed that this putative NLS alone was able to target the recombinant protein to the nucleoli in both cell lines (Fig. 2B), even though some of this protein was dispersed in the nucleus in low levels. Whereas, EGFP alone (Fig. 2A) or EGFP conjugated with the C terminus region without NLS (residues 31–88) (Fig. 2C) diffused throughout the entire cell.
To determine the key element inside this NLS, we next generated site-directed mutants of CENP-W based on the finding that there are three basic amino acid stretches in this putative NLS. Two basic amino acids from each stretch were substituted for glycine residues (R15G/K16G, K23G/R24G, or K27G/R28G) in the EGFP-fused construct, and the green fluorescence signals of each mutant protein were monitored after transfection in both HeLa and SKOV-3 cells. When we altered the basic amino acids in the three different stretches separately, each mutant protein was relatively well accumulated in the nucleolus (Fig. 2, D–F). However, when two sites (R15G/K16G and K27G/R28G) were changed together (Fig. 2G), or all three were mutated (Fig. 2H), the nucleolar accumulation in HeLa cells and even the nuclear localization in SKOV-3 was disturbed. B23 conjugated with RFP was co-expressed as a nucleolar control, and no disturbance was observed in the nucleolar targeting of B23 by the dislocalization of the mutant CENP-Ws. The failure of these mutants to drive EGFP into the nucleolus suggested that the predicted NLS sequence of CENP-W plays a major role in nuclear and nucleolar targeting and that its three basic stretches may function together in the direction of this protein.
CENP-W May Form a Complex with RNA
Based on the finding that CENP-W is localized in the nucleolus, we evaluated CENP-W to determine if it was associated with RNA. To accomplish this, transiently transfected 293T cells or A549- FLAG-CENP-W stable cells were subjected to a chromatin fractionation experiment. Initially, the soluble fraction of the cells was extracted by permeabilizing the cell with Triton X-100 and then washed, after which the remaining sample was further incubated with DNase I or RNase A. We found that a portion of the CENP-W proteins was released by DNase I and RNase A digestion (S1 fractions of Fig. 3, A and B) and further extracted by the addition of salt (S2 fractions). Because the release of CENP-W is not observed in the S1 fraction of the control samples without nuclease, the solubilization of CENP-W appears to be nuclease-specific. To confirm this finding, we gradually increased the amount of nuclease and then harvested the soluble CENP-W. As shown in Fig. 3 (C and D), the amount of CENP-W released was proportional to the amount of nuclease added. This observation may indicate that CENP-W forms a complex with DNA and RNA.
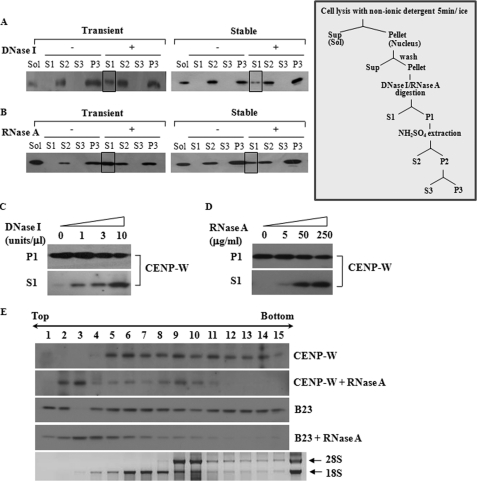
FIGURE 3.
CENP-W may form a complex with RNA as well as DNA. A, CENP-W is released by DNase I treatment. A549-FLAG-CENP-W stable cells or 293T cells transfected with FLAG-CENP-W were fractionated according to the protocols depicted below (For details, see under “Materials and Methods”). After the cells were digested DNase I (1 unit/μl), the released proteins were isolated (S1), and the remaining precipitation was further extracted using 0.25 m ammonium sulfate. The control experiment was also performed under the same conditions without nuclease. Each fraction was analyzed by Western blot analysis using anti-FLAG antibody. The boxed fraction of CENP-W corresponds to the protein released by the DNase I digestion. B, CENP-W is released by RNase A digestion. The solubilized portion of CENP-W by RNase A (100 μg/ml) digestion is highlighted as a box. C, DNase I-dependent release of CENP-W. After cells were incubated with increasing amounts of DNase I (0, 1, 3, and 10 units/μl), the supernatant was collected by centrifugation. D, RNase A-dependent release of CENP-W. After increasing amounts of RNase A (0, 5, 50, and 250 μg/ml) was added to the cells, the supernatant was separated from remaining sample. E, RNA binding assay in sucrose gradient centrifugation. The nuclear extracts isolated from HeLa-FLAG-CENP-W stable cells were incubated with or without RNase A (250 μg/ml). The resulting sample was then analyzed by 10–30% sucrose gradient sedimentation. CENP-W and B23 were identified using Western blotting with anti-FLAG and anti-B23 antibodies, respectively.
To demonstrate the previous finding more clearly, a sucrose gradient sedimentation assay was conducted using HeLa-CENP-W stable cells. To accomplish this, the nuclear extracts were overlaid on top of the sucrose gradients and then fractionated by high speed sedimentation. If necessary, RNase A digestion was conducted prior to loading. Because it is a well known RNA-binding protein, endogenous B23 was also detected to confirm the fidelity of the RNase A treatment. Without RNase treatment, both CENP-W and B23 were widely distributed, even in the high density fractions and the fractions that were co-fractionated with the RNA (Fig. 3E). Conversely, preincubation with RNase A shifted the peak position of both proteins to the low density fractions, although some CENP-W remained in the high density area. These findings showed that RNA digestion caused the CENP-W to be released into the soluble fraction, supporting the notion that some of the CENP-W is present in a nuclear complex with RNA.
CENP-W Is Nuclear Matrix-associated
During the chromatin fractionation experiment, we noticed that a considerable amount of CENP-W remained in the insoluble fraction after full digestion of the DNA or RNA (P3 fraction in Fig. 3, A and B). Indeed, even after the cells were subjected to double digestion with DNase I and RNase A, the insoluble portion was still observed in the last pellet, and the amount did not differ greatly from the amount observed during the single digestion (data not shown). Considering that the remaining insoluble portion of the nuclei that are resistant to the salt extraction corresponds to the nuclear matrix, these findings indicated that some CENP-W was associated with components of the nuclear matrix. To test this, we fractionated CENP-W-expressing stable A549 cells according to the widely accepted high salt nuclear matrix preparation method (27) and then compared the findings with the results from the transiently expressed CENP-W. As a first step, soluble proteins were obtained by detergent treatment (Lane 1 in Fig. 4A). Next, chromatin-associated proteins were released by DNase I digestion followed by ammonium sulfate addition (Lane 2), after which they were further extracted with 2 m NaCl (Lane 3). The resulting pellet was then collected and dissolved in 8 m urea (Lane 4), which contains the enriched nuclear matrix and matrix-associated proteins (27). The fractionation results showed that CENP-W was evenly distributed, with approximately one-third being in the soluble, chromatin-enriched and nuclear matrix fractions. In addition, a similar pattern was observed in transient cells with a slight decrease in the chromatin fraction. The fractionation was verified by immunodetection of endogenous marker proteins such as α-tubulin as a cytoplasmic protein marker, Lamin A/C as a nuclear matrix protein, and TATA-binding protein (TBP) as a matrix-associated transcription factor (30). Furthermore, the association of CENP-W with the nuclear matrix appeared to be stable and unresponsive to environmental changes, because no significant proportional changes were observed upon the treatment of various chemicals, such as trichostain A, hydrogen peroxide, and camptothecin (Fig. 4B).
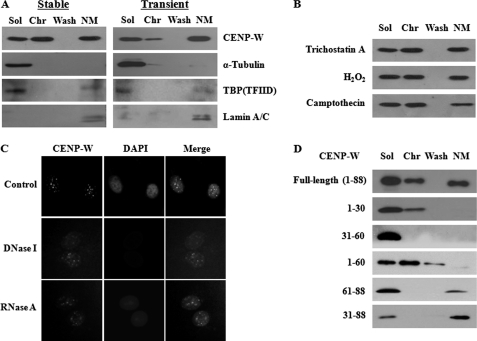
FIGURE 4.
A fraction of CENP-W is associated with nuclear matrix. A, nuclear matrix fractionation of CENP-W-expressing cells. Either transient 293T or stable A549 cells was fractionated by the traditionally conducted high salt nuclear matrix preparation method as described under “Materials and Methods.” FLAG-CENP-W or other subcellular marker proteins were visualized by Western blot analysis. Sol, soluble cytoplasmic fraction after 0.5% Triton X-100 treatment; Chr, chromatin-enriched fraction released after DNase I and 0.25 m ammonium sulfate extraction; Wash, supernatant after 2 m NaCl high salt washing; NM, nuclear matrix fraction finally obtained insoluble fraction solubilized by 8 m urea. B, association of CENP-W with nuclear matrix is stable. CENP-W A549 stable cells were pretreated with trichostatin A (30 μm), H2O2 (2 mm), or camptothecin (10 μm) for 24 h before cell fractionation. C, in situ cell extraction experiment. After extraction of soluble proteins, A549-CENP-W stable cells grown in coverslips were incubated with DNase I (1 unit/μl) or RNase A (200 μg/ml) and further extracted with 0.25 m ammonium sulfate. The resulting sample was fixed and then immunostained with anti-FLAG antibody, followed by FITC-conjugated anti-mouse antibody to observe the remaining CENP-W. D, domain mapping of CENP-W for matrix association. 293T cells expressing various CENP-W domains were fractionated by the high salt nuclear matrix preparation method and analyzed by Western blotting.
The biochemical fractionation results were further supported by in situ indirect immunodetection of CENP-W. Coverslips harboring A549 CENP-W stable cells were incubated with detergent and then subjected to DNase I or RNase A extraction. The resulting samples were used for immunostaining of CENP-W with anti-FLAG antibody. Although the signals were greatly attenuated after nuclease digestion, some CENP-W, which is resistant to nuclease extraction, was still observed by in situ immunodetection (Fig. 4C). These data confirm that some of the CENP-W associates with the nuclear matrix in human cells. To determine which domain of CENP-W was important for targeting to the nuclear matrix, the FLAG-tagged deletion mutants of CENP-W were expressed in 293T cells and then fractionated. Western analysis showed that the C terminus of CENP-W (amino acids 61–88) rather than N terminus region (amino acids 1–60) functions more critically for the incorporation into the matrix (Fig. 4D). These findings suggest that this basic N-terminal region, which is responsible for nuclear and nucleolar targeting of CENP-W, is not relevant to matrix association.
CENP-W Interacts with B23
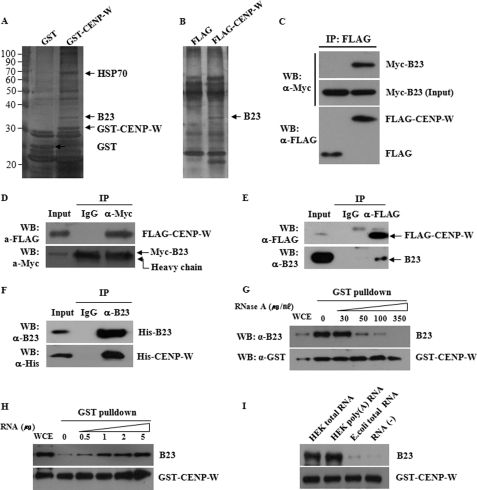
In an attempt to search for new binding partners of CENP-W and understand how CENP-W is associated with nucleoli, the nucleolar proteins present in the complex with CENP-W were analyzed in an affinity binding experiment. Specifically, we transfected 293T cells with GST- or FLAG-tagged CENP-W and then performed GST pulldown assay or co-immunoprecipitation with anti-FLAG antibody. Proteins that co-eluted with CENP-W were identified by SDS-PAGE and compared with proteins bound to controls. Several protein bands that appeared to be present only in the CENP-W fraction were excised and identified by mass spectroscopy. Based on the GST pulldown assay, we found that CENP-W interacts with heat shock protein, Hsp70, and B23 (Fig. 5A). The interaction of B23 with CENP-W became evident when the FLAG-CENP-W co-complex also contained B23 in a separately performed co-immunoprecipitation experiment (Fig. 5B). To confirm the in vivo interaction between CENP-W and B23, we conducted co-immunoprecipitation using ectopically expressed Myc-B23 and FLAG-CENP-W. Immunoprecipitation with anti-FLAG antibody co-fractionated Myc-B23 in a complex with the FLAG-CENP-W protein (Fig. 5C). Conversely, the presence of FLAG-CENP-W was detected in the Myc-B23 complex when immunoprecipitation was conducted using anti-Myc antibody (Fig. 5D). We also tested this interaction in the endogenous level using HeLa stable cells expressing FLAG-CENP-W. The endogenous B23 was found in the FLAG-CENP-W fraction in the stable cells (Fig. 5E). Finally, the specific interaction between these proteins was confirmed using recombinant proteins expressed by the in vitro cell-free expression system (Fig. 5F). Taken together, these results demonstrate that CENP-W specifically interacts with nucleolar protein B23.
FIGURE 5.
CENP-W interacts with B23. A, affinity purification of CENP-W-interacting proteins. Nucleoli fraction obtained from cells expressing GST-CENP-W were subjected to GST pulldown analysis. Isolated proteins were then analyzed by SDS-PAGE and identified by mass spectroscopy. B, identification of CENP-W-interacting protein by using the immunoprecipitation method. C, co-immunoprecipitation between CENP-W and B23. Cell lysates transfected with Myc-B23 and FLAG-CENP-W were subjected to immunoprecipitation with anti-FLAG antibody. D, reciprocal immunoprecipitation between FLAG-CENP-W and Myc-B23 was performed using anti-Myc antibody or normal rabbit IgG. E, GST pulldown assay for endogenous B23. HeLa stable cells expressing FLAG-CENP-W were used for immunoprecipitation and endogenous B23 in complex with CENP-W was detected using anti-B23 antibody. F, in vitro binding assay. After cloning into the pET15b bacterial expression vector (Novagen), the His-tagged recombinant B23 and CENP-W was expressed in vitro using the E. coli S30 T7 protein expression system (Promega). Then, co-immunoprecipitation was carried out with anti-B23 antibody. G, GST pulldown analysis after RNase treatment. RNase was added at the indicated concentration to the lysate of H293 cells transfected GST-CENP-W, and the mixture was incubated at 25 °C for 20 min before GST pulldown analysis. Endogenous B23 was monitored using anti-B23 antibody. H, GST pulldown assay with RNA addition. After endogenous RNA is digested with 200 μg/ml RNase, RNA fractions extracted from H293 cells were added before GST pulldown assay. I, binding assay with different kinds of RNA. Three different kinds of RNA preparation (1 μg) were added to the RNase-treated sample before GST pulldown assay.
To further investigate the interaction between B23 and GST-CENP-W, the lysate was pretreated with RNase before GST pulldown analysis. The amount of co-isolated endogenous B23 with GST-CENP-W is dramatically decreased in proportion to the increasing amount of RNase (Fig. 5E). Then, to test if the purified RNA can restore the interaction of B23 and CENP-W, we digested endogenous RNA of the lysate with 200 μg/ml RNase and then added RNA fraction purified from H293 cells. As shown in Fig. 5F, the B23-CENP-W complex was gradually restored by addition of RNA, indicating that RNA is essential to the formation of B23 and CENP-W complex. We then tested which type of RNA is most effective in this complex formation. When the same amount of RNA was added to the sample, both total RNA and mRNA preparation from H293 cells functioned effectively, but RNA from E. coli did not restore the interaction between B23 and CENP-W.
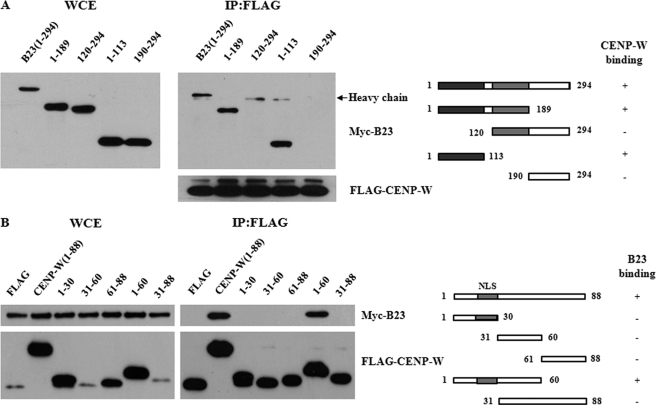
To determine the crucial domains for the interaction between B23 and CENP-W, we first generated the B23 deletion mutants. After we confirmed that all designed constructs were expressed successfully in 293T cells, the binding activity of each deletion mutant was determined by co-immunoprecipitation with FLAG-CENP-W. The results showed that the dimerization domain of B23 located in the N terminus (amino acids 1–113) of the protein was sufficient for binding with CENP-W (Fig. 6A). In contrast, the C-terminal domain, which is reportedly involved in histone and nucleic acid binding (10), did not bind with CENP-W. Conversely, most small CENP-W deletion mutants did not interact with B23 (Fig. 6B), whereas the N terminus domain (amino acids 1–60) showed high protein stability and high binding affinity for B23. These findings suggest that, because CENP-W is a relatively small protein, the N-terminal two-thirds of the protein may participate in interaction with B23 or that the three-dimensional structure sustained by this N-terminal domain is highly required for maintaining the stability of the entire protein and B23 binding.
FIGURE 6.
Determination of domains required for the interaction between CENP-W and B23. A, domain mapping of B23 for binding with CENP-W. Deletion mutants of Myc-B23 were generated and transfected into 293T cells with FLAG-CENP-W. Co-immunoprecipitation was conducted using anti-FLAG antibody, followed by Western blot analysis with anti-Myc antibody. B, domain mapping of CENP-W for binding with B23. Immunoprecipitation between FLAG-CENP-W mutant proteins and Myc-B23 was conducted using anti-FLAG antibody and the co-fractionated B23 was detected using Western blot analysis.
CENP-W Is Stabilized by B23
Given that CENP-W has RNA-binding activity and is associated with B23, we next evaluated CENP-W to determine if it was involved in ribosomal RNA biogenesis or processing. To accomplish this, we first tested the localization of CENP-W after treatment with actinomycin D, considering that the localization of many nucleolar proteins that are involved in RNA biogenesis are highly dependent on a transcriptionally active nucleolus (29). Unfortunately, the nucleolar localization of CENP-W was not disturbed by this inhibition of ribosomal RNA transcription, whereas B23 clearly became dispersed throughout the nucleus (data not shown). We also conducted RT-PCR analysis of the pre-RNA sequence (31) to monitor the RNA synthesis over time following transfection with CENP-W. Although treatment with a low concentration of actinomycin D induced a sharp decrease in pre-rRNA production, no significant changes were induced by CENP-W deregulation (data not shown). These findings may indicate that CENP-W is not closely related to the activities of ribosomal RNA production.
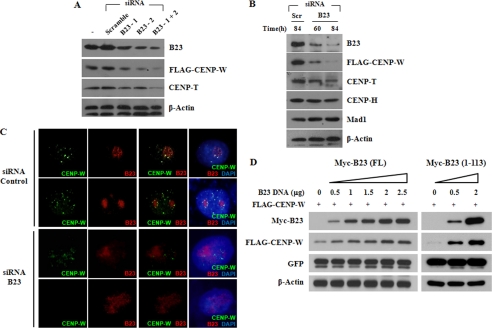
Based on earlier reports that demonstrated B23 has molecular chaperone-like activities (32), we tested whether the depletion of B23 altered CENP-W protein stability. The reduction of cellular B23 by siRNA treatment induced a dramatic decrease in the CENP-W protein level in HeLa-CENP-W stable cells (Fig. 7, A and B). To exclude the possibility that CENP-W level was reduced by cell cycle arrest upon B23 knockdown, CENP-W was monitored during various stages of the cell cycle. No significant changes were found in the mRNA and protein levels of CENP-W during the cell cycle (supplemental Fig. S1, A and B). When several other centromere components were monitored (Fig. 7B), the influence of B23 reduction was the most effective on CENP-W protein stability. CENP-T, known to interact with CENP-W, is also reduced when B23 is depleted. But no significant changes were found in the CENP-H and Mad1 protein level. Immunostaining also revealed that CENP-W signals were clearly diminished by the suppression of B23 in B23 siRNA-treated cells when compared with untreated controls (Fig. 7C). To further demonstrate the effects of B23 on CENP-W stability, 293T cells were transiently transfected with FLAG-CENP-W combined with increasing amounts of Myc-B23. GFP was also co-expressed as a transfection control. Although the same amount of DNA was transfected, CENP-W expression increased proportionally to the amount of co-transfected Myc-B23 when tested with the full-length B23 (Fig. 7D). The stabilization of CENP-W by B23 co-expression was clearly demonstrated when CENP-W was co-transfected with the plasmid encoding the N-terminal domain (amino acids 1–113) of B23, which was previously identified to be sufficient for interaction with CENP-W. Taken together, these findings indicate that the new centromeric protein, CENP-W, is stabilized via interaction with B23.
FIGURE 7.
CENP-W is stabilized by interaction with B23 (A) B23 knockdown induced destabilization of CENP-W. Two different kinds of B23 siRNA or combined siRNA were transfected into HeLa-FLAG-CENP-W stable cells, and the protein level of endogenous B23 and CENP-W was examined by Western analysis. B, stability of kinetochore components upon B23 depletion. Western analysis was performed to monitor the protein stability of several centromere proteins after B23 siRNA treatment. C, immunofluorescence staining of B23-depleted cells. Cellular distribution of B23 and CENP-W was visualized by immunostaining in B23-depleted HeLa-CENP-W cells. D, CENP-W is stabilized by B23. Increasing amounts of B23 construct encoding either full-length or N-terminal 1–113 amino acids were co-transfected with FLAG-CENP-W in 293T cells, and the protein expression level was examined by Western blot analysis. GFP plasmid was also co-transfected and used as a transfection control.
B23 Depletion Induces Mislocalization of CENP-W during Mitotic Prophase
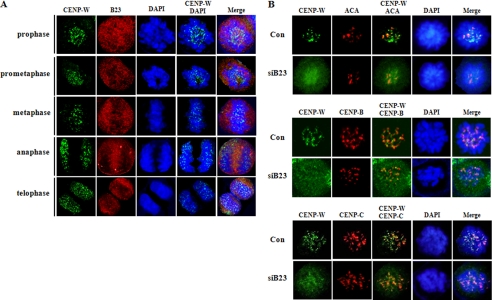
Because B23 was suggested to function in the formation of the pre-kinetochore complex, we examined if B23 depletion may affect the recruitment of CENP-W to centromere during prophase. Unlike B23, which was dispersed during mitosis, CENP-W was clearly identified in centromere region during mitosis in normal cell (Fig. 8A and supplemental Fig. S2). Double immunostaining showed that CENP-W is co-localized with the centromere proteins during prophase in normal cells (Fig. 8B). However, when B23 was depleted from cells, CENP-W was not co-localized with the centromere proteins, but rather was highly dispersed during prophase (Fig. 8B), indicating that B23 functions in the recruitment of CENP-W to the centromere complex during the assembly stage of kinetochore at prophase.
FIGURE 8.
B23 functions in the recruitment of CENP-W to mitotic centromere during prophase. A, distribution of CENP-W and B23 in mitotic cells. After synchronization with nocodazole (100 ng/ml) for 12 h, HeLa-CENP-W cells were harvested at 30-min time intervals. Cells were then co-immunostained for CENP-W and B23 using anti-FLAG and anti-B23 antibodies, respectively. B, B23 depletion inhibits the centromeric recruitment of CENP-W. After incubation with B23 siRNA or control siRNA for 72 h, HeLa-CENP-W cells were incubated with nocodazole for 12 h. At 30 min after release, cells were harvested and CENP-W was immunostained. Anti-CENP-B, anti-CENP-C, and anti-centromere antibody (ACA, Cortex Biochem) were also used to visualize the centromere.
DISCUSSION
CENP-W was originally identified as a putative oncogene that is specifically up-regulated in many human cancer tissues (19). Two recent papers have shed light on the cellular role of CENP-W (20, 23). Specifically, this putative oncoprotein was identified as a centromeric component that plays an important role in the formation of a proper kinetochore complex during cell mitosis. Due to the unavailability of specific antibodies for CENP-W, we established FLAG-CENP-W stable cells in many different mammalian cell lines. In these stable cells, CENP-W proteins were seen as nuclear foci during interphase, some of which are co-localized with anti-human centromere antibody (23). However, we also found minor signals in other regions of the nucleus, which indicated that some CENP-W proteins are not centromeric. Based on these observations, we evaluated CENP-W to determine if it targeted other nuclear subdomains in addition to the centromere. To address this issue, we started a localization study of CENP-W from the simple approach of tracing EGFP-tagged protein. Surprisingly, the EGFP-CENP-W was predominantly localized to the nucleolus in various human cells, and altering tags did not change this nucleolar localization (Fig. 1, A–C). This phenomenon did not seem to be an artifact of abrupt expression when we tested stable cells that express CENP-W at an almost endogenous level. Indeed, ∼50% of total proteins were fractionated with the nucleoli, indicating that a significant amount of CENP-W is associated with the nucleoli even when it is expressed in low levels.
There is evidence of a close functional relationship between the centromere and nucleolus. Specifically, human autoantigens constituting of the centromere are frequently found in the nucleolar periphery or inside nucleoli during interphase (33). Affinity purification of a centromeric protein, CENP-C, has demonstrated that it specifically interacts with nucleolar transcription factors UBF1 and UBF2 (34).
Recently, Foltz et al. successfully isolated several undiscovered centromeric components using TAP-tagged CENP-A. CENP-T, which was later found to form a complex with CENP-W, was first identified in that study as a proximal CENP-A nucleosome component. In the same study, they published the unexpected result that B23 was present in the CENP-A nucleosome, not in the corresponding H3-nucleasome. These findings suggested that B23 play a sequestering role in the formation of centromeric protein complex and chromatin components during interphase, which may be needed for the assembly of a functional kinetochore (22). In the present study, we found that CENP-W specifically interacts with B23, and CENP-W protein is highly stabilized by addition of B23. Moreover, B23 knockdown clearly promoted CENP-W mislocalization during prophase. Our findings suggest that the proposed role B23 on the assembly of kinetochore complex (22) may be accomplished through a direct interaction with CENP-W, thereby increasing its stability and sequestering it during interphase, which facilitates the recruitment of CENP-W to kinetochore during prophase. We failed to detect any binding activity between ectopically expressed CENP-T and B23 using co-immunoprecipitation, supporting the specific interaction between CENP-W and B23.
Although the existence of nuclear matrix structures is still in question, nuclear matrix has traditionally been considered to be an extensive RNA/protein fibrillar network that functions as a platform to facilitate the functional assembly of nuclear components for specialized nuclear activities (16, 17), which is envisioned as a nuclear mesh-like architecture with numerous round-shaped granules connected by filaments (35). Despite the rigid skeletal images, it has been proposed that the nuclear matrix is a dynamic sponge-like open structure that allows the free diffusion of soluble nuclear materials (36). B23 has been identified as a matrix component in diverse human cell lines and tissues quite often, and Hsp70 is also found in the matrix (15). The fact that these previously documented matrix constituents were all co-purified with CENP-W in the GST pulldown analysis (Fig. 5A) supports that CENP-W is associated with the nuclear matrix. Several centromeric components (CENP-B, -C, and -F) have previously been detected in biochemically extracted nuclear matrix (37, 38). Additionally, immunoelectron microscopic analysis with autoantibodies that recognize several centromeric proteins revealed that each pre-kinetochore complex forms a stable, but highly dynamic association with the filaments of the nuclear matrix, proposing that the strikingly mobile properties of the centromeric complex may be facilitated by anchoring to the matrix network (39).
Historically, alterations in the composition, number, size, and intranuclear localization of nucleoli were used as an early indication of cancer and to distinguish a few types of tumor cells from normal cells (40, 41). Alterations in the nuclear matrix composition are also known to be cell type- and tumor-specific and have been used for the development of protein biomarkers of cancer (42). Given that CENP-W was originally identified as a putative oncogene, its subnuclear localization in nucleoli and nuclear matrix might be related to the tumorigenic process of human cells. The results of the present study indicate that the new centromeric component, CENP-W, is not solely localized in the centromere, but rather widely distributed in various subnuclear compartments, suggesting that it might be involved in a variety of crucial nuclear activities. The spatial or temporal alteration of the subnuclear distribution of CENP-W in these or other nuclear areas in accordance with neoplasmic cell transformation will be topics of our future research.
Supplementary Material
This work was supported by grants funded by Korean Government, the Basic Science Research Program through National Research Foundation (Grant 20100005431), and the Korea Healthcare Technology R and D Project through the Ministry for Health, Welfare and Family Affairs (Grant A080588-21).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
- CUG2
- cancer-upregulated gene 2
- NLS
- nuclear localization sequence
- RFP
- red fluorescence protein.
REFERENCES
- 1. Leonhardt H., Cardoso M. C. (1995) Int. Rev. Cytol. 162B, 303–335 [DOI] [PubMed] [Google Scholar]
- 2. Zaidi S. K., Young D. W., Javed A., Pratap J., Montecino M., van Wijnen A., Lian J. B., Stein J. L., Stein G. S. (2007) Nat. Rev. Cancer 7, 454–463 [DOI] [PubMed] [Google Scholar]
- 3. Hager G. L., Elbi C., Becker M. (2002) Curr. Opin. Genet. Dev. 12, 137–141 [DOI] [PubMed] [Google Scholar]
- 4. Carmo-Fonseca M., Mendes-Soares L., Campos I. (2000) Nat. Cell Biol. 2, E107–E112 [DOI] [PubMed] [Google Scholar]
- 5. Olson M. O., Dundr M., Szebeni A. (2000) Trends Cell Biol. 10, 189–196 [DOI] [PubMed] [Google Scholar]
- 6. Boisvert F. M., van Koningsbruggen S., Navascués J., Lamond A. I. (2007) Nat. Rev. Mol. Cell Biol. 8, 574–585 [DOI] [PubMed] [Google Scholar]
- 7. Andersen J. S., Lam Y. W., Leung A. K., Ong S. E., Lyon C. E., Lamond A. I., Mann M. (2005) Nature 433, 77–83 [DOI] [PubMed] [Google Scholar]
- 8. Ahmad Y., Boisvert F. M., Gregor P., Cobley A., Lamond A. I. (2009) Nucleic Acids Res. 37, D181–D184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pederson T., Tsai R. Y. (2009) J. Cell Biol. 184, 771–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Grisendi S., Mecucci C., Falini B., Pandolfi P. P. (2006) Nat. Rev. Cancer 6, 493–505 [DOI] [PubMed] [Google Scholar]
- 11. Korgaonkar C., Hagen J., Tompkins V., Frazier A. A., Allamargot C., Quelle F. W., Quelle D. E. (2005) Mol. Cell. Biol. 25, 1258–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ma Z., Kanai M., Kawamura K., Kaibuchi K., Ye K., Fukasawa K. (2006) Mol. Cell. Biol. 26, 9016–9034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Feuerstein N., Mond J. J. (1987) J. Immunol. 139, 1818–1822 [PubMed] [Google Scholar]
- 14. Berezney R., Coffey D. S. (1974) Biochem. Biophys. Res. Commun. 60, 1410–1417 [DOI] [PubMed] [Google Scholar]
- 15. Albrethsen J., Knol J. C., Jimenez C. R. (2009) J. Proteomics 72, 71–81 [DOI] [PubMed] [Google Scholar]
- 16. Ciejek E. M., Tsai M. J., O'Malley B. W. (1983) Nature 306, 607–609 [DOI] [PubMed] [Google Scholar]
- 17. Anachkova B., Djeliova V., Russev G. (2005) J. Cell. Biochem. 96, 951–961 [DOI] [PubMed] [Google Scholar]
- 18. Biggiogera M., Cisterna B., Spedito A., Vecchio L., Malatesta M. (2008) Differentiation 76, 57–65 [DOI] [PubMed] [Google Scholar]
- 19. Lee S., Gang J., Jeon S. B., Choo S. H., Lee B., Kim Y. G., Lee Y. S., Jung J., Song S. Y., Koh S. S. (2007) Biochem. Biophys. Res. Commun. 360, 633–639 [DOI] [PubMed] [Google Scholar]
- 20. Hori T., Amano M., Suzuki A., Backer C. B., Welburn J. P., Dong Y., McEwen B. F., Shang W. H., Suzuki E., Okawa K., Cheeseman I. M., Fukagawa T. (2008) Cell 135, 1039–1052 [DOI] [PubMed] [Google Scholar]
- 21. Foltz D. R., Jansen L. E., Black B. E., Bailey A. O., Yates J. R., 3rd, Cleveland D. W. (2006) Nat. Cell Biol. 8, 458–469 [DOI] [PubMed] [Google Scholar]
- 22. Mellone B., Erhardt S., Karpen G. H. (2006) Nat. Cell Biol. 8, 427–429 [DOI] [PubMed] [Google Scholar]
- 23. Kim H., Lee M., Lee S., Park B., Koh W., Lee D. J., Lim D. S., Lee S. (2009) Mol. Cells 27, 697–701 [DOI] [PubMed] [Google Scholar]
- 24. Weng J. J., Yung B. Y. (2005) Biochem. Biophys. Res. Commun. 335, 826–831 [DOI] [PubMed] [Google Scholar]
- 25. Scherl A., Couté Y., Déon C., Callé A., Kindbeiter K., Sanchez J. C., Greco A., Hochstrasser D., Diaz J. J. (2002) Mol. Biol. Cell 13, 4100–4109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Okuwaki M., Tsujimoto M., Nagata K. (2002) Mol. Biol. Cell 13, 2016–2030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Reyes J. C., Muchardt C., Yaniv M. (1997) J. Cell Biol. 137, 263–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Strezoska Z., Pestov D. G., Lau L. F. (2000) Mol. Cell. Biol. 20, 5516–5528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Meder V. S., Boeglin M., de Murcia G., Schreiber V. (2005) J. Cell Sci. 118, 211–222 [DOI] [PubMed] [Google Scholar]
- 30. Kurg R., Sild K., Ilves A., Sepp M., Ustav M. (2005) J. Virol. 79, 10528–10539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Huang M., Ji Y., Itahana K., Zhang Y., Mitchell B. (2008) Leuk. Res. 32, 131–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Szebeni A., Olson M. O. (1999) Protein Sci. 8, 905–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ochs R. L., Press R. I. (1992) Exp. Cell Res. 200, 339–350 [DOI] [PubMed] [Google Scholar]
- 34. Pluta A. F., Earnshaw W. C. (1996) J. Biol. Chem. 271, 18767–18774 [DOI] [PubMed] [Google Scholar]
- 35. Nickerson J. A., Krockmalnic G., Wan K. M., Penman S. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 4446–4450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Peterson J. T., Li H., Dillon L., Bryant J. W. (2000) Cardiovasc. Res. 46, 307–315 [DOI] [PubMed] [Google Scholar]
- 37. Liao H., Winkfein R. J., Mack G., Rattner J. B., Yen T. J. (1995) J. Cell Biol. 130, 507–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. He D., Zeng C., Brinkley B. R. (1995) Int. Rev. Cytol. 162B, 1–74 [DOI] [PubMed] [Google Scholar]
- 39. He D., Brinkley B. R. (1996) J. Cell Sci. 109, 2693–2704 [DOI] [PubMed] [Google Scholar]
- 40. Weber J. D., Taylor L. J., Roussel M. F., Sherr C. J., Bar-Sagi D. (1999) Nat. Cell Biol. 1, 20–26 [DOI] [PubMed] [Google Scholar]
- 41. Oudes A. J., Campbell D. S., Sorensen C. M., Walashek L. S., True L. D., Liu A. Y. (2006) BMC Genomics 7, 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sjakste N., Sjakste T., Vikmanis U. (2004) Exp. Oncol. 26, 170–178 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.