Background: Acid-sensing ion channels (ASICs) are activated by extracellular protons and are inhibited by amiloride.
Results: Amiloride activates and sensitizes ASIC3 channels depending on the nonproton ligand sensing domain.
Conclusion: ASICs can sense nonproton ligands in addition to protons.
Significance: The results indicate caution in the use of amiloride for studying ASIC physiology and in the development of amiloride-derived ASIC inhibitors for treating pain syndromes.
Keywords: Acid-sensing Ion Channels (ASIC), Membrane Proteins, Neurobiology, Pain, Signaling, Amiloride, Channel Gating, Nonproton Ligand Sensor
Abstract
Acid-sensing ion channels (ASICs), which belong to the epithelial sodium channel/degenerin family, are activated by extracellular protons and are inhibited by amiloride (AMI), an important pharmacological tool for studying all known members of epithelial sodium channel/degenerin. In this study, we reported that AMI paradoxically opened homomeric ASIC3 and heteromeric ASIC3 plus ASIC1b channels at neutral pH and synergistically enhanced channel activation induced by mild acidosis (pH 7.2 to 6.8). The characteristic profile of AMI stimulation of ASIC3 channels was reminiscent of the channel activation by the newly identified nonproton ligand, 2-guanidine-4-methylquinazoline. Using site-directed mutagenesis, we showed that ASIC3 activation by AMI, but not its inhibitory effect, was dependent on the integrity of the nonproton ligand sensing domain in ASIC3 channels. Moreover, the structure-activity relationship study demonstrated the differential requirement of the 5-amino group in AMI for the stimulation or inhibition effect, strengthening the different interactions within ASIC3 channels that confer the paradoxical actions of AMI. Furthermore, using covalent modification analyses, we provided strong evidence supporting the nonproton ligand sensing domain is required for the stimulation of ASIC3 channels by AMI. Finally, we showed that AMI causes pain-related behaviors in an ASIC3-dependent manner. These data reinforce the idea that ASICs can sense nonproton ligands in addition to protons. The results also indicate caution in the use of AMI for studying ASIC physiology and in the development of AMI-derived ASIC inhibitors for treating pain syndromes.
Introduction
Acid-sensing ion channels (ASICs)3 are members of the sodium-selective cation channels belonging to the epithelial sodium channel/degenerin family. They act as receptors for extracellular protons (1–3). Accumulating evidence implicates ASICs in ischemia (4–7), neurotransmission (8), epilepsy (9), mechanosensation (10–13), chemosensation (14), and pain perception (15–21). The physiological roles of ASICs are studied using both genetically manipulated mice and pharmacological tools such as amiloride (AMI) (1, 3, 22). AMI is a common blocker for all members of the epithelial sodium channel/degenerin family (23). It inhibits acid-induced currents from most known ASIC channels (1–3), and the AMI blockade is often used to demonstrate the physiological function of ASICs. A premise for its use in the study of ASIC physiology is that AMI does not directly or indirectly affect ASIC functions other than inhibiting channel activity; however, some studies have suggested that AMI may paradoxically stimulate ASICs. In addition to its blocking activity, AMI was found to stimulate the “Deg” mutation (at Gly-430) but not the wild-type (WT) ASIC2a channel (24) through exposing extracellular membrane-proximal residues in a similar fashion as that by protons (25). Based on the finding that AMI inhibition was voltage-dependent, but stimulation was not, it was suggested that AMI stimulates ASIC2a channels by binding to an extracellular site, whereas it inhibits the channels by interacting at a site within the channel pore. However, the submolecular motifs involved in these regulations have not been studied. In addition, the sustained window current of ASIC3 channels was enhanced by AMI (6), further arguing for a stimulatory effect of AMI on specific ASIC subtypes.
Recently, synthetic compounds such as 2-guanidine-4-methylquinazoline (GMQ) and an endogenous ligand, agmatine, were shown to cause persistent activation of ASIC3 at the neutral pH (26, 27). Using GMQ as a probe and combining mutagenesis and covalent modification analysis, we identified a novel nonproton ligand sensing domain (26). This domain is lined by residues near Glu-423 and Glu-79 of the extracellular palm domain (28). Both GMQ and agmatine are small molecules containing basic groups that are believed to form tight contacts with the carboxyl groups of acidic residues through hydrogen bonds (H-bonds) and/or electrostatic interactions, leading to activation of the ASIC3 channel (26). Because AMI has an amidino group (Fig. 1A) analogous to the guanidinium group of GMQ, and both contain a heterocyclic ring (29), we reasoned that AMI may activate ASIC channels in a manner similar to GMQ. Here, we show that the stimulatory action of AMI is similar to that of GMQ. Furthermore, using site-directed mutagenesis, electrophysiological recording, and covalent modification, we demonstrate that the nonproton ligand sensing domain is required for stimulation of ASIC function by AMI.
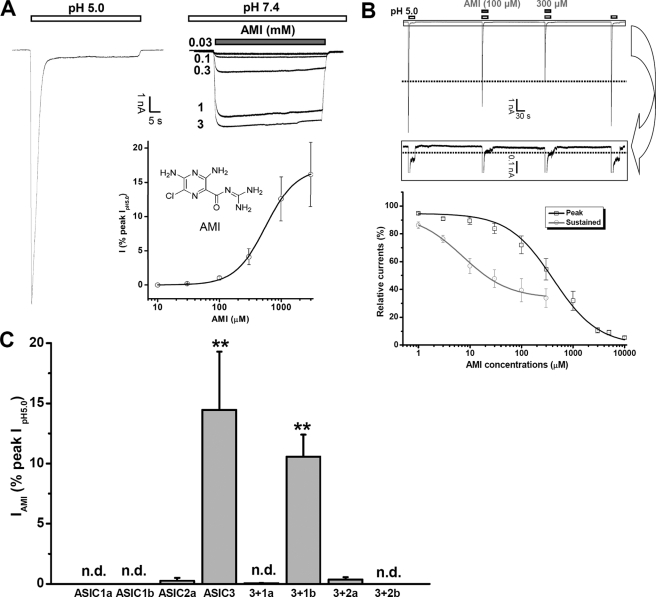
FIGURE 1.
Effects of AMI on ASIC3 channels. A, chemical structure of AMI and AMI-activated ASIC3 currents. AMI dose-dependently evoked inward currents in ASIC3-transfected (right panel) but not in nontransfected CHO cells (data not shown). Although the acid (pH 5.0)-induced current desensitized rapidly (left panel), little desensitization was observed for the AMI-evoked currents (above right panel). Similar results were obtained in six other experiments. Concentration-response relationship of AMI is shown below right panel. Each point is the mean ± S.E. of seven measurements normalized to the pH 5.0-induced peak currents, and the solid line was the fit to the Hill equation. The EC50 and n values of AMI are 0.56 ± 0.02 mm and 1.76 ± 0.07, respectively. B, effects of AMI on the activation of ASIC3 channels by acid (pH 5.0). AMI dose-dependently inhibited the pH 5.0-induced currents (i.e. both peak and sustained components) of ASIC3 channels as shown in the representative example (upper panel). Data points are means ± S.E. of four to eight measurements normalized to the currents induced by pH 5.0 in the absence of AMI, and the solid lines were the fit to the Hill equation giving an IC50 value of 7.2 ± 1.4 μm (n = 0.97 ± 0.22) for the sustained (gray) and 412.2 ± 2.0 μm (n = 0.86 ± 0.11) for the peak component (black), respectively. C, subunit selectivity of AMI action at the neutral pH. Shown is the summary of AMI (1 mm)-induced currents in CHO cells transfected with one or two different ASIC subunits. AMI activated ASIC3 homomeric channels and heteromeric ASIC3 + ASIC1b channels but not ASIC1a, ASIC1b, or ASIC2a homomeric channels, or heteromeric ASIC3 + 1a, ASIC3 + 2a, or ASIC3 + 2b channels at the neutral pH. Data are means ± S.E. n = 3–5. n.d., no detectable response to AMI. **, p < 0.001 versus base-line level.
EXPERIMENTAL PROCEDURES
Cell Culture and Transfection
All constructs were expressed in CHO cells as described previously (26). In brief, CHO cells were cultured at 37 °C in a humidified atmosphere of 5% CO2 and 95% air. The cells were maintained in F-12 medium (Invitrogen) supplemented with 1 mm l-glutamine, 10% fetal bovine serum, 50 units/ml penicillin, and 50 μg/ml streptomycin. Transient transfection of CHO cells was carried out using LipofectamineTM 2000 (Invitrogen). Electrophysiological measurements were performed 24–48 h after transfection.
Solutions and Drugs
The ionic composition of the incubation solution was as follows (in mm): 124 NaCl, 24 NaHCO3, 5 KCl, 1.2 KH2PO4, 2.4 CaCl2, 1.3 MgSO4, and 10 glucose, aerated with 95% O2/5% CO2 to a final pH of 7.4. The standard external solution contained (in mm) the following: 150 NaCl, 5 KCl, 1 MgCl2, 2 CaCl2, and 10 glucose, buffered to various pH values with either 10 mm HEPES for pH 6.0–7.4 or 10 mm MES for pH < 6.0. The pipette solution for whole-cell patch recording was as follows (in mm): 120 KCl, 30 NaCl, 1 MgCl2, 0.5 CaCl2, 5 EGTA, 2 Mg-ATP, and 10 HEPES, adjusted to pH 7.2 with Tris base. The osmolarities of all solutions were maintained at 300–325 mOsm (Advanced Instrument, Norwood, MA).
Solutions with different composition were applied using a rapid application technique termed the “Y-tube” method throughout the experiments (26). This system allows a complete exchange of external solution surrounding a cell within 20 ms.
Site-directed Mutagenesis
The cDNA of rat ASIC3 was subcloned into the pEGFPC3 vector (Promega Corp., Madison, WI). Each mutant was generated with the QuikChange® mutagenesis kit (Stratagene, La Jolla, CA) in accordance with the manufacturer's protocol using high performance liquid chromatography-purified or PAGE-purified oligonucleotide primers (Sigma Genosys). Individual mutations were verified by DNA sequence analysis, and the predicted amino acid sequences were determined by computer analysis.
Electrophysiology
The electrophysiological recordings were performed using the conventional whole-cell patch recording configuration under voltage clamp conditions. Patch pipettes were pulled from glass capillaries with an outer diameter of 1.5 mm on a two-stage puller (PP-830, Narishige Co., Ltd., Tokyo, Japan). The resistance between the recording electrode filled with pipette solution and the reference electrode was 3–5 megohms. Membrane currents were measured using a patch clamp amplifier (Axon 200B, Axon Instruments, Foster City, CA) and were sampled and analyzed using a Digidata 1440A interface and a computer running the Clampex and Clampfit software (version 10.0.1, Axon Instruments). In most experiments, series resistance was compensated at 70–90%. Unless otherwise noted, the membrane potential was held at −60 mV throughout the experiment under voltage clamp conditions. All experiments were carried out at room temperature (23 ± 2 °C).
Pain-related Behavioral Assays
Animals were acclimatized for 30 min before experiments. A total volume of 10 μl of either saline (0.9% NaCl) or AMI-containing solution (in 0.9% NaCl) was injected intraplantarly using a 30-gauge needle, and paw-licking behavior was quantified for 30 min (26).
Data Analysis
Results were expressed as means ± S.E. Except where noted otherwise, statistical comparisons were made with the Student's t test. *, p < 0.05 or **, p < 0.001 was considered significantly different. Concentration-response relationships for AMI-dependent activation of ASIC3 channels were obtained by measuring currents in response to AMI with graded concentrations. Each AMI concentration was tested on at least three CHO cells, and all results used to generate a concentration-response relationship were from the same group. The data were fit to the Hill Equation 1,
where I is the normalized current (to that induced by pH 5.0) at a given concentration of AMI; Imax is the maximal normalized current; EC50 is the concentration of AMI yielding 50% of the maximal current, and n is the Hill coefficient. The concentration-response relationships for AMI-dependent inhibition of ASIC3 channels were obtained by measuring currents in response to agonists in the presence of AMI with graded concentrations and the data were fit to the Hill Equation 2,
where I is the normalized current (to the current induced by agonists in the absence of AMI) in the presence of a given concentration of AMI; Imax is the maximal normalized current, IC50 is the concentration of AMI yielding 50% of the maximal current, and n is the Hill coefficient.
RESULTS
AMI Induces Sustained Activation of ASIC3 Channels at Neutral pH
Traditionally, AMI acts as a wide spectrum channel blocker for most ASICs (1–3). In CHO cells expressing the ASIC3 subunit, AMI inhibits channel activation induced by acid (Fig. 1B) or nonproton activators (supplemental Fig. 1) (26). However, AMI directly evoked inward currents in a dose-dependent manner. The AMI currents displayed little or no desensitization (Fig. 1A), reminiscent of the sustained activation by GMQ (26). The washout of higher concentrations of AMI often results in a rebound current (data not shown, see also Fig. 6, C and D), consistent with the notion that AMI both activates (Fig. 1A) and inhibits (Fig. 1B) ASIC3 channels. We noted that AMI was less effective in activating ASIC3 than GMQ (37.5 ± 3.1% of the GMQ response at 1 mm concentration; n = 4, p < 0.001, AMI versus GMQ). We ascribed the differences in their half-maximal activation concentration (EC50) (0.56 ± 0.02 mm for AMI versus 1.08 ± 0.09 mm for GMQ; n = 7–8, p < 0.001) as well as the extent of activation (16.2 ± 4.7% for AMI versus 84.4 ± 7.4% for GMQ of the pH 5.0-induced peak current; n = 7–8, p < 0.001) partially to the paradoxical effects of AMI (i.e. both stimulatory and inhibitory). Further studies on CHO cells expressing homomeric ASIC3 or various heteromeric ASIC subunit combinations revealed that AMI is a specific agonist for ASIC3 homomers and heteromers (ASIC3 + ASIC1b in particular) (Fig. 1C). This subunit specificity is consistent with the finding that ASIC3 channels are the only ASIC subtype activated by GMQ and agmatine (26, 27). AMI-induced activation was also observed in HEK293 and COS-7 cells transfected with ASIC3 (data not shown), indicating that the stimulatory effect of AMI is independent of the host cell type. Collectively, these results suggest that AMI specifically activates ASIC3 channels at neutral pH in a manner similar to GMQ and agmatine.
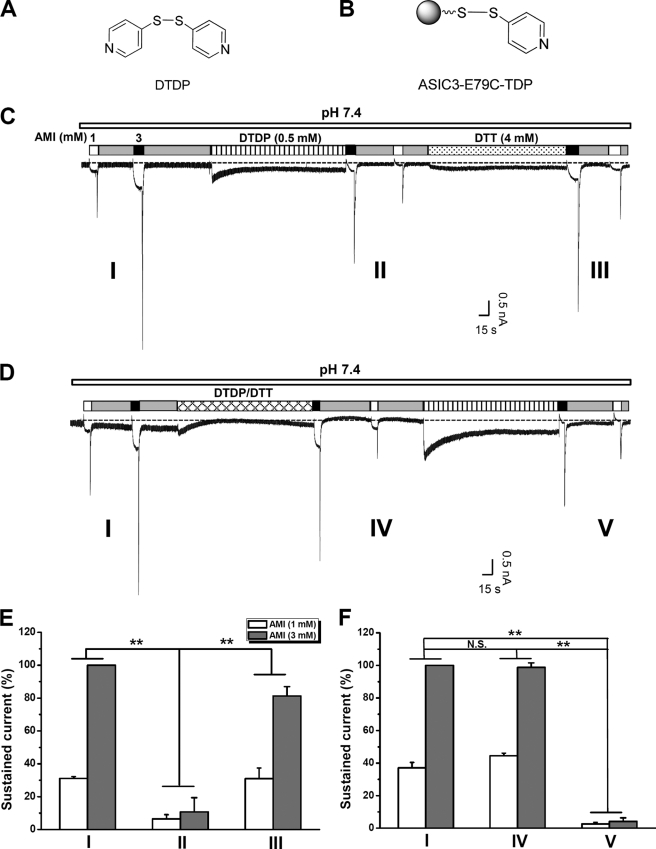
FIGURE 6.
Effects of DTDP on AMI stimulation of ASIC3E79C channels. A, structure of DTDP. B, illustration of TDP covalently linked to E79C via a mechanism of Ellman's reaction. C, typical recording showing the effect of DTDP (0.5 mm, pH 7.4) on the AMI stimulation of ASIC3E79C channels. AMI at 1 and 3 mm evoked inward currents from CHO cells expressing ASIC3E79C. Following DTDP treatment, a successive administration of AMI failed to induce ASIC3E79C channel activation because of the formation of E79C-S-S-TDP complexes preventing AMI binding. DTT (4 mm, pH 7.4, and 5 min) restores channel activation by AMI presumably by breaking the disulfide bond in the E79C-S-S-TDP complexes, rendering the channel responsive to subsequent AMI application. D, cotreatment of DTDP (0.5 mm, pH 7.4) and DTT (4 mm, pH 7.4) preserves channel activation by AMI. After washout of DTDP/DTT, DTDP treatment again abolished AMI activation of ASIC3E79C channel because of steric effects. E and F, statistical analysis of effects of DTDP on the sustained currents induced by AMI. Shown in E and F are pooled data from experiments in C and D, respectively. The dashed lines in C and D represent the base-line level. Data points are means ± S.E. of six measurements of sustained currents normalized to that induced by AMI (3 mm) before DTDP treatment. **, p < 0.001, demonstrating the significant difference in the sustained currents induced by both 1 and 3 mm AMI between the two different groups as indicated. N.S., not significant.
AMI Facilitates Sustained Activation of ASIC3 Channels Induced by Modest pH Decreases
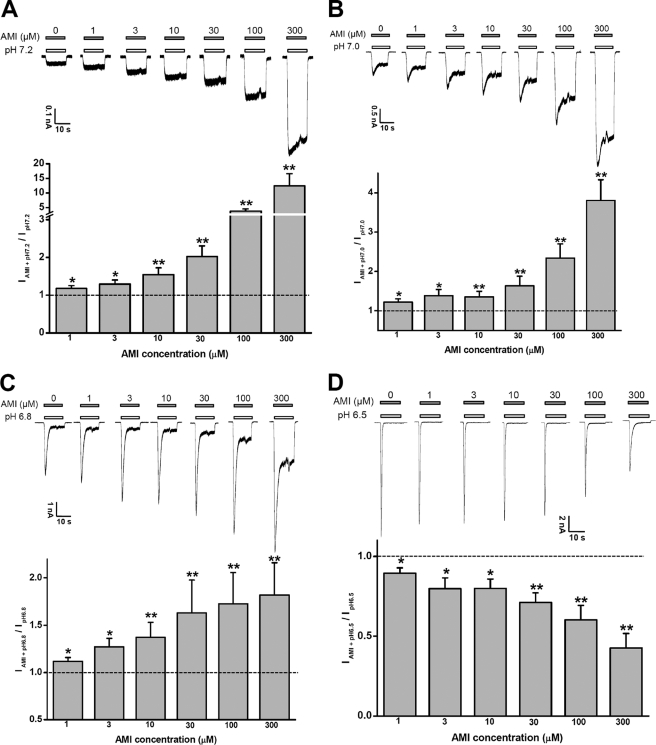
Similar to the modulatory profile of GMQ and agmatine (26, 27), submillimolar concentrations of AMI significantly sensitized ASIC3 channels to pH 7.2–6.8 (Fig. 2, A–C). However, at pH 6.5 (Fig. 2D) or below (Fig. 1B), it inhibited the peak response induced by acid. Although lower concentrations (∼μm) of AMI facilitate the activation of ASIC3 channels upon modest pH decreases (Fig. 2, A–C), high AMI concentrations above 1 mm antagonize the currents induced by pH 7.2 to 6.8 (data not shown). Thus, AMI action on ASIC channels depends on pH as well as its own concentration, consistent with previous observations (6).
FIGURE 2.
AMI sensitizes ASIC3 channels under mild acidosis. AMI at low micromolar concentrations (1–300 μm) sensitizes the response to mild acidosis (A, pH 7.2; B, pH 7.0; and C, pH 6.8) but not to a more extreme acidosis (D, pH 6.5). Upper panels show representative current traces at −60 mV, and lower panels are means ± S.E. of four to seven measurements normalized to the currents induced by acidosis alone (control; dashed line). *, p < 0.05; **, p < 0.001 versus controls.
Stimulatory Action of AMI Is Determined by Nonproton Ligand Sensing Domain
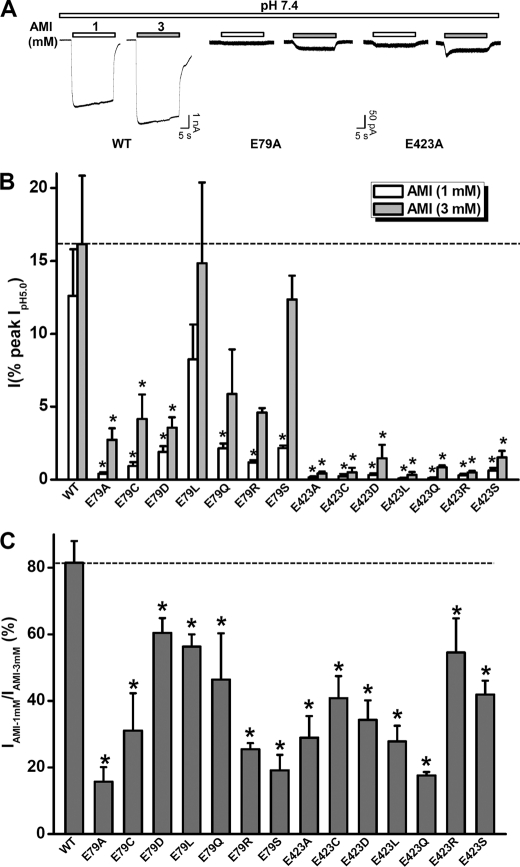
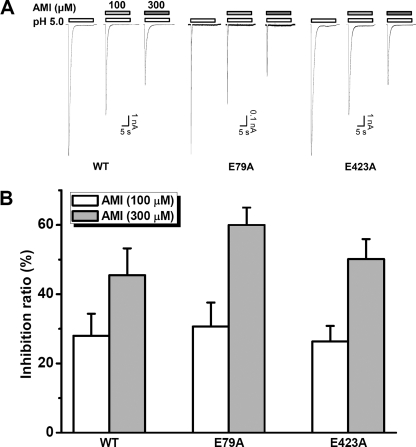
Previously, we have shown that the nonproton ligand sensing domain (26, 27) plays a critical role in mediating agmatine and GMQ effects on ASIC3. Considering the similarities in the chemical structures among AMI (Fig. 1A), GMQ, and agmatine (29) and their shared actions on ASIC3 channels, we hypothesized that AMI also activates ASIC3 channels through interacting with the nonproton ligand sensing domain. To test this hypothesis, we expressed the E79A and E423A mutants of ASIC3 in CHO cells. We have shown previously that Glu-79 and Glu-423 are the most critical residues in coordinating GMQ binding to the nonproton ligand sensing domain, and when mutated to Ala, the mutated ASIC3 channels became insensitive to GMQ (26). Similarly, AMI at millimolar concentrations failed to activate either ASIC3E79A or ASIC3E423A channels (Fig. 3A and supplemental Table 1). To distinguish the effects of mutations of the nonproton ligand sensing domain on AMI binding affinity from that on the efficacy of AMI as a channel activator, we recorded the responses at two AMI concentrations (1 and 3 mm), both of which induced a near maximum response in WT ASIC3 channels (Fig. 3A). We reasoned that an increase or decrease of the current ratio (IAMI-1 mM/IAMI-3 mM) might indicate a leftward or rightward shift of the AMI dose-response curve, hence a change in potency, but not necessarily a change in the coupling pathway or subsequent channel gating (efficacy). We used current ratio for these two concentrations because the full dose-response curve of AMI action on ASIC3 mutants was not feasible due to its low potency and the potential inhibition at higher concentrations. We found that mutations at Glu-79 or Glu-423 markedly impaired the stimulatory action of AMI with significant decreases in both current amplitudes (Fig. 3B and supplemental Table 1) and current ratios (Fig. 3C). Together, these results favor an essential role of the nonproton ligand sensing domain in mediating the stimulatory actions of AMI on ASIC3 channels.
FIGURE 3.
Activation of ASIC3 channels by AMI is determined by residues (Glu-79 and Glu-423) of the nonproton ligand sensing domain. A, representative current traces illustrating the effects of E79A and E423A mutations on 1 and 3 mm AMI-induced currents. B, pooled data from experiments as in A and similar results from other mutations of Glu-79 or Glu-423. Data points are means ± S.E. of four to seven measurements normalized to the pH 5.0-induced peak currents. Note: for most mutations, the maximal pH 5.0-induced currents were generated following pH 9.0 pretreatment to restore the steady-state desensitization of acid-dependent currents at pH 7.4. *, p < 0.05 versus wild-type (WT). The dashed line represents the current amplitude induced by AMI (3 mm) from WT ASIC3 channels. C, ratio of current amplitude induced by 1 mm AMI to that by 3 mm AMI. The data were generated from B and are means ± S.E. *, p < 0.05 versus WT (dashed line).
As reported previously (26, 30), the mutants at Glu-79 or Glu-423 exhibited significant steady-state desensitization to protons. To exclude the possibility that the reduced potency of AMI on these mutants resulted from channel desensitization, we pretreated cells with an alkaline pH condition (pH 9.0) to restore the ASIC3 mutants from desensitization, and then we tested the AMI response at neutral pH. Although pretreatment with the alkaline pH (pH 9.0) largely restored pH 5.0-induced channel activation, the AMI-induced current was still reduced in the Glu-79 or Glu-423 mutants (data not shown) as occurred without pretreatment by the alkaline pH, suggesting that the reduced response to AMI was not due to the pH-dependent desensitization associated with the mutations.
Inhibitory Action of AMI Is Not Determined by the Nonproton Ligand Sensing Domain
To test whether the reduced activation by AMI of the Glu-79 and Glu-423 mutants was a result of an increased inhibition associated with these mutations, we investigated the inhibition by AMI of the acid-induced currents in these mutants. As shown in Fig. 4, AMI inhibited the acid-induced currents in WT as well as E79A or E423A mutant channels to a similar extent. Thus, an increase in AMI-mediated inhibition does not account for the reduced activation by AMI in the Glu-79 and Glu-423 mutants.
FIGURE 4.
Constant efficacy of AMI inhibition on acid (pH 5.0)-induced currents from ASIC3 mutants at Glu-79 or Glu-423. A, representative current traces showing AMI inhibition of the pH 5.0-induced currents from WT, E79A, and E423A mutant channels. B, pooled data from experiments in A. The inhibition ratio of AMI was quantitatively calculated as follows: ((1 − IAMI + pH 5.0/IpH 5.0)·100) where IAMI + pH 5.0 and IpH 5.0 are the pH 5.0-induced currents measured in the presence or absence of AMI. Data points are means ± S.E. of four to seven measurements. There is no significant difference in the degree of inhibition by either 100 or 300 μm AMI among WT, E79A, and E423A groups. The WT data, taken from Fig. 1B, are regraphed again for comparison.
Differential Pharmacophore Requirement of AMI for Stimulation or Inhibition of ASIC3 Channels
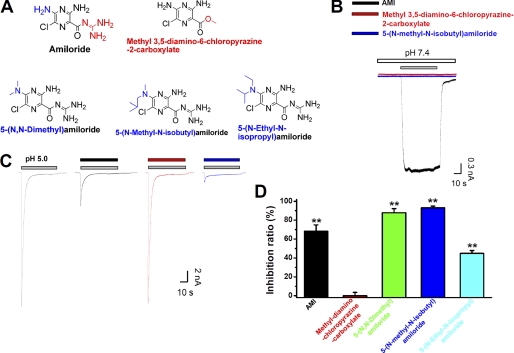
To explain structural bases of AMI underlying the stimulation or inhibition of ASIC3 channels, we electrophysiologically screened commercially available small molecules similar to AMI (Fig. 5A). When the guanidinium group of AMI was removed by replacing a methoxyl group, the resulting compound-induced channel activation (Fig. 5B, red) as well as the inhibition efficacy against the acid-induced channel activation (Fig. 5, C and D, red) was totally abolished. The important role of the guanidinium group of AMI in the activating ASIC3 channel was consistent with the previous structure-activity relationship studies on the GMQ-induced channel activation (26). Furthermore, the validation of the essential role of the guanidinium group in AMI-induced channel activation as well as channel blockade made the first step to study the AMI-ASIC3 interaction at the atomic level (28). By contrast, when the 5-amino group (at the ortho position of the 6-chloro group) was replaced with a N,N-dimethyl, N-methyl-N-isobutyl, or N-ethyl-N-isopropyl group (Fig. 5A), the resulting compounds as exemplified by 5-(N-methyl-N-isobutyl)amiloride still exerted a blocking effect (Fig. 5, C and D, blue) but lost the activation efficacy (Fig. 5B, blue), implying the interaction of the 5-amino group of AMI with residues of nonproton ligand sensing domain in the extracellular region but not with that of the pore domain in the transmembrane region. The differential requirement of the 5-amino group in AMI for its paradoxical effects raises a possibility to distinguish the stimulatory or inhibitory effects in the application and development of chemical modulators of ASIC3 channels in the future (see below).
FIGURE 5.
Structure-activity relationship of AMI derivatives. A, schematic demonstration for structure-activity relationship of AMI. B and C, representative current traces showing the channel activation under neutral pH (B) or inhibition of pH 5.0-activated currents (C) from ASIC3 expressing CHO cells induced by compounds as indicated. Every compound was used at the concentration of 1 mm. Different colored traces represent the currents induced in the presence of different compounds, in which black is for AMI; red is for methyl-3,5-diamino-6-chloropyrazine-2-carboxylate; and blue is for 5-(N-methyl-N-isobutyl)amiloride, respectively. The stimulatory and inhibitory results from 5-(N,N-dimethyl)- or 5-(N-methyl-N-isobutyl)amiloride (data not shown) were similar to that of 5-(N-ethyl-N-isopropyl) amiloride (blue). D, pooled data from experiments in C. The inhibition ratio was defined similarly as shown in Fig. 4B. Data points are means ± S.E. of four to seven measurements. **, p < 0.001 versus zero.
Covalent Modification at Glu-79 Prevents ASIC3 Channel Activation by AMI
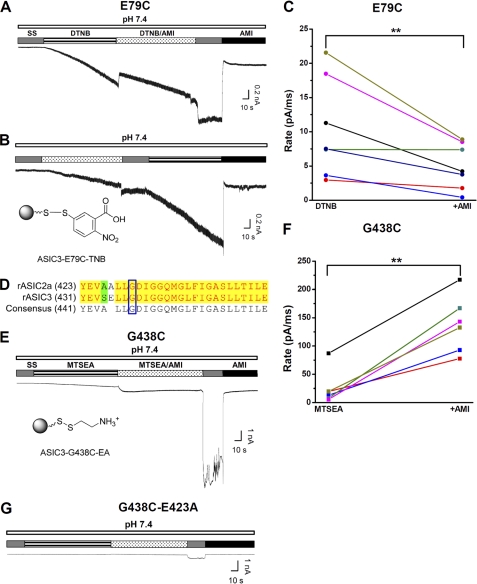
To ascertain that AMI activates ASIC3 via interactions with Glu-79 and Glu-423, we substituted these two residues with cysteine with the intention of blocking the interactions by covalent modification. To avoid the activation of ASIC3 channels through single covalent modification at E79C by 5,5′-dithiobis(2-nitrobenzoic acid) (DTNB) (26), we used 4,4′-dithiodipyridine (DTDP) (Fig. 6, A and B), a DTNB analog, as the redox donor. Consistent with the previous observation (26), bath application of DTDP (0.5 mm at pH 7.4) to CHO cells expressing ASIC3E79C resulted in a small inward current that gradually declined to a steady state (Fig. 6, C and D). Like DTNB, DTDP did not induce inward currents in CHO cells expressing the WT ASIC3, and it had no effect on either acid- or AMI-induced currents (data not shown), suggesting that specific covalent modification at the E79C site is responsible for its activation effect on ASIC3E79C. Taking advantage of the steady-state activation by the persistent TDP-E79C interaction, we reasoned that directly linking TDP to Cys-79 via a covalent bond should prevent further ASIC3 channel activation by AMI at the neutral pH, given that the activation of ASIC3 by nonproton ligands (GMQ and agmatine) relies on polar, steric, and electrostatic interactions with the nonproton ligand sensing domain in the channel (26). To test this hypothesis, we applied AMI at neutral pH before or after DTDP treatment (Fig. 6, C and D). As expected, we found that AMI-dependent ASIC3 channel activation (demonstrated by the sustained current (Fig. 6, C–F) as well as the rebound current after washout of AMI (Fig. 6, C and D, and supplemental Fig. 2, A and B), which might represent a brief period of unopposed stimulation by AMI and a complex interaction between AMI-induced channel activation and inhibition) was largely prevented by the DTDP pretreatment. Furthermore, this attenuation was reversed (Fig. 6, C and E, and supplemental Fig. 2A) or blocked (Fig. 6, D and F, and supplemental Fig. 2B) by the reducing reagent, 1,4-dithiothreitol (DTT). Thus, we concluded that the integrity of the nonproton ligand sensing domain is required for AMI-induced ASIC3 channel activation.
AMI Prevents Covalent Modification-induced Activation of ASIC3E79C
Having demonstrated that covalent modification at Glu-79 prevents ASIC3 channel activation by AMI, we further examined how AMI could affect activation of ASIC3E79C by the thiol-reactive covalent probe, DTNB (26). As shown previously (26), bath application of DTNB (0.5 mm, pH 7.4) slowly activated the ASIC3E79C channel, presumably reflecting the time-dependent modification of the E79C site by the thiol-reactive probe (Fig. 7A). We used this experimental model to monitor the efficiency of the DTNB-E79C reaction in the presence or absence of AMI. We reasoned that if AMI binds to the nonproton ligand sensing domain that includes Glu-79, it should perturb the interaction and hence covalent bond formation between 5-thio-2-nitrobenzoic acid and Cys-79 (Fig. 7B, lower panel) because of steric effects. As expected, AMI markedly slowed down the development of DTNB-induced ASIC3E79C channel current, irrespective of the sequence of AMI and DTNB applications (Fig. 7, A and B). To quantify the effect of AMI, we measured the rate (pA/ms) of DTNB-induced activation of ASIC3E79C, defined as the ratio of maximal current (pA) and the duration (ms) of DTNB application. Because of the intrinsic inhibitory effect of AMI, we defined the maximal current as that seen after the AMI washout. Fig. 7C shows that the rate of DTNB-induced activation of ASIC3E79C is significantly higher in the absence than in the presence of AMI, demonstrating an inhibitory effect of AMI on the DTNB-dependent modification of the E79C site, which strengthens the support of the view that AMI directly interacts with the nonproton ligand sensing domain of ASIC3.
FIGURE 7.
Effects of AMI on covalently activated ASIC3 channels. A, typical recording showing the effect of AMI (1 mm) on 0.5 mm DTNB (pH 7.4)-induced ASIC3E79C activation as a result of a covalent modification (26). Notably, a successive coadministration of AMI (1 mm) and DTNB (0.5 mm at pH 7.4) slowed down the development of DTNB currents (see below) because of steric competition between AMI and DTNB. B, illustration of 5-thio-2-nitrobenzoic acid (TNB) covalently linked to E79C via a mechanism of Ellman's reaction (lower panel) and a typical recording showing the effect of AMI (1 mm) on DTNB-induced ASIC3E79C activation (upper panel) with a different drug application sequence from A. C, pooled data from the combination of experiments in A and B. The rate (pA/ms) is defined as the maximal current (pA) divided by the duration (ms) of covalent modification treatment as indicated. For AMI-treated, the maximal current during AMI washout (as indicated by SS) is measured to minimize the contribution of AMI-induced inhibition. Different colored points and lines represent paired measurements for individual cells. D, sequence alignment between rat ASIC2a (rASIC2a) and rat ASIC3 subunits. The blue open box indicates the conserved Deg site (Gly-430 versus Gly-438 in ASIC2a and ASIC3, respectively). E, illustration of 2-aminoethyl (EA) group covalently linked to G438C via a mechanism of Ellman's reaction (lower panel) and a typical recording showing the effect of AMI on 0.2 mm MTSEA (pH 7.4)-induced ASIC3G438C activation (upper panel). F, pooled data from the experiments in E. G, typical recording showing the attenuated effect of AMI on MTSEA-induced ASIC3G438C-E423A activation. Similar results were obtained in five other experiments.
To exclude any nonspecific effect of AMI, we performed similar experiments on the modification of another site distinct from the nonproton ligand sensing domain. Testing the extracellular accessibility of the G430C mutant of ASIC2a by methanethiosulfonate compounds, Adams et al. (24) have shown a stimulatory effect of AMI on the Deg mutant (at Gly-430) ASIC2a channel. We assumed that AMI might have a similar action on ASIC3 channels because the Deg mutation site is conserved (Fig. 7D). To test this possibility, we created a Deg mutant (G438C) ASIC3 channel. As shown in Fig. 7E and supplemental Fig. 3B, bath application of 2-aminoethyl methanethiosulfonate (MTSEA, 0.2 mm, pH 7.4) in the absence of AMI slightly activated the ASIC3G438C channel, similar to the effect seen with the ASIC2aG430C channel (24, 25). By contrast, MTSEA (0.2 mm) was ineffective in CHO cells expressing WT ASIC3 channels (supplemental Fig. 3A), strengthening the residue specificity of MTSEA modification. Coapplication of AMI with MTSEA elicited a larger current from the ASIC3G438C channel, which was further enhanced dramatically upon removal of AMI (Fig. 7E). This large increase in current observed upon AMI washout was due to the removal of AMI-induced inhibition as it was completely reversed by the reapplication of AMI (Fig. 7E). These data demonstrate that AMI facilitates the persistent activation of ASIC3G438C through covalent modification by MTSEA. The facilitatory effect of AMI on covalent modification of ASIC3G438C is consistent with the view that AMI exposes Gly-438 to extracellular solution, allowing more efficient interaction with thiol-reactive probes (Fig. 7F) (24). However, the site(s) of AMI interaction should not overlap with the modified Cys residue. This result is opposite from the inhibitory action of AMI on modification of ASIC3E79C, demonstrating specificity of the nonproton ligand sensing domain in AMI interaction with the ASIC3 channel. Furthermore, when an additional mutation (E423A) of the nonproton ligand sensing domain was introduced into the ASIC3G438C background, the effect of AMI on MTSEA-induced activation was largely attenuated (Fig. 7G), strongly arguing for the importance of the nonproton ligand sensing domain in mediating stimulation of ASIC3 channels by AMI.
AMI Activates ASIC3 Channels in Vivo
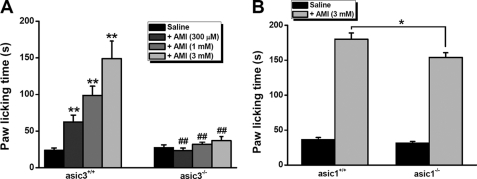
Finally, to gain insights into the in vivo consequence of paradoxical stimulation of ASIC3 channels by AMI, we performed pain-related behavioral tests (26, 27) following the injection of AMI with different concentrations into the right hind paw of asic3+/+ and asic3−/− mice, because previous studies have shown that ASIC3 channels mediate pain responses (15, 16, 31). We measured the total time that the animals spent licking the injected paw during a 30-min period. As shown previously (26), control asic3+/+ mice showed no noticeable increase in paw-licking time after AMI (100 μm) injection compared with saline-injected controls. In consideration of the requirement of higher concentration of AMI to activate ASIC3 channels (Fig. 1A), we re-evaluated the paradigms by injecting 300 μm and 1 and 3 mm AMI, respectively (Fig. 8A). As expected, asic3+/+ mice showed a significant increase in paw-licking time after AMI injection, in a dose-dependent manner (Fig. 8A). Furthermore, the reaction of asic3−/− mice to AMI was significantly reduced (Fig. 8A). Additionally, asic1 gene knock-out only slightly reduced the paw-licking time (Fig. 8B), indicating a specific interaction between AMI and ASIC3 channels in causing pain-like behaviors and strengthening the roles of heteromeric ASIC3 + ASIC1b channels (Fig. 1C) in responding to AMI in vivo. Thus, AMI is able to cause pain-related behaviors through activation of ASIC3 channels.
FIGURE 8.
AMI cause pain-related behaviors through activation of ASIC3 channels. A, pain-related behaviors as determined by the time spent for following saline or AMI (300 μm or 1 mm or 3 mm) injection (10 μl) in asic3+/+ and asic3−/− mice. Data are means ± S.E. n = 6–9. **, p < 0.001 versus saline; ##, p < 0.001, asic3+/+ versus asic3−/−. B, pain-related behaviors as determined by the time spent for following saline or AMI (3 mm) injection (10 μl) in asic1+/+ and asic1−/− mice. Data are means ± S.E. n = 10. *, p < 0.05, asic1+/+ versus asic1−/−.
DISCUSSION
In this study, we demonstrate that AMI activates and sensitizes ASIC3 channels, although AMI has been widely used as an inhibitor for many ion channels and transporters, such as Na+/H+ exchanger (32), low threshold calcium channel (33), glycine receptors (34), and the epithelial sodium channel/degenerin family (23), including ASIC channels (see also Figs. 1B and 2D) (2). Using mutagenesis and covalent modification analysis, we have revealed that the nonproton ligand sensing domain lined by residues around Glu-423 and Glu-79 (26) is required for the stimulatory action of AMI.
There is an increasing interest in the nonproton gating of the acid-sensitive ASIC channels (26, 27) because the rapid desensitization of proton-induced ASIC currents occurring over milliseconds to seconds (1–3) stands in stark contrast to the prolonged time course of the inflammatory pain and other pathological processes thought to involve ASICs (4–7, 9, 15, 16, 18, 20, 21). Following the discovery that synthetic compounds such as GMQ and endogenous agmatine activate ASIC3 channels at neutral pH through the nonproton ligand sensing domain (26, 27) in a nondesensitizing manner, we now add another example of a nonproton agent, AMI, activating ASIC3 channels in a similar manner and through a similar mechanism. This suggests a shared feature that may be applied to all nonproton activation of ASIC channels. The mechanisms underlying nondesensitizing gating of ASIC3 channels induced by nonproton activators, including AMI, are currently unknown but may involve interactions between two extracellular linker regions that control nondesensitizing activation of ASICs by protons (35). However, the chemical activation of ASICs independent of acidic pH seems to be a characteristic of ASIC3 but not ASIC1, or ASIC2 channels, because GMQ, agmatine, and AMI activate ASIC3 but not other channel subtypes at the neutral pH. Elucidating the molecular basis underlying these differences represents a substantial task in the future. A recent study (26) using molecular dynamics simulation and mutagenesis proposed that the nonconserved residues among different ASIC subunits shaping the nonproton ligand sensing domain play a part in defining subunit specificity of nonproton activators (i.e. GMQ) presumably including AMI through stabilizing the spatial conformation of the ligand-binding site in ASIC3 channels. However, given that AMI stimulates the Deg mutation (at Gly-430) of ASIC2a (24), it is likely that similar nonproton ligand sensing domains exist in other ASICs. Identification of more general and/or isoform-specific as well as more potent nonproton ligands of these ion channels should be possible with the better understanding of the nonproton ligands and their sensing domain(s).
Within the ASIC family, ASIC3 channels carry multiple unique features (22) distinct from other ASIC subtypes. For the acid-induced activation, ASIC3 is the only exception to the common features shared by most ASIC members that protons trigger a rapidly desensitizing transient current. Notably, ASIC3 mediates an extra sustained current that does not fully desensitize, whereas the extracellular pH remains acidic (36–39). Moreover, this sustained component of ASIC3 becomes markedly enhanced when the channel is activated in the presence of Phe-Met-Arg-Phe amide and related neuropeptides (40–42). In addition, ASIC3 channels open in a largely nondesensitizing manner (6) at more physiologically relevant pH values (pH 7.3–6.7). Furthermore, ASIC3 channels respond to nonproton ligands (i.e. AMI, GMQ, and agmatine) through the nonproton ligand sensing domain (26, 27). These properties together with its expression in sensory neurons as well as peripheral non-neuronal tissues (36–38) suggest that ASIC3 channels are well positioned to regulate multimodal sensory perception (22), and they are indeed proposed to be an integrator for multiple physiological stimuli, including nociceptive and inflammatory signals (27). The demonstration of the paradoxical action of AMI on ASIC3 channels raises the possibility that ASIC3-dependent sensory perception may be regulated by AMI derivatives or yet unknown AMI-like endogenous ligands.
Using a combination of mutagenesis, covalent modification, as well as functional analyses, we uncovered the requirement of the nonproton ligand sensing domain (26, 27) in mediating the stimulation effect of AMI on ASIC3 channels. Whereas our data identified key determinants essential for mediating AMI-ASIC3 interactions, there are at least three alternative explanations for the data observed here. First, AMI most likely binds specifically to a cavity around Glu-79 and Glu-423 to activate the ASIC3 channel. Second, a mutation-induced decrease in response to AMI could result from allosteric effects if Glu-79 and Glu-423 were in fact part of a linker region crucial for proton (30, 43, 44), and hence nonproton activation. Third, it is also possible that the cavity around Glu-79 and Glu-423 represents just one of the many AMI-binding sites (45).
We favor a direct and specific interaction between AMI and the cavity lined by Glu-79 and Glu-423 for the following reasons. First, a covalent linkage between TDP and Cys-79 largely reduced the channel activation by AMI (Fig. 6). Oppositely, AMI application reduced the rate of covalent modification-induced channel gating (Fig. 7, A–C). These results strongly suggest that the spatial and structural integrity of the cavity around Glu-79 and Glu-423 is crucial for ASIC3 channel activation by AMI. Second, mutations at the key residues of nonproton ligand sensing domain decreased the efficacy of AMI on ASIC3 channels (Fig. 3, A and B) and increased the apparent EC50 value as reflected by the reduced current ratio (IAMI-1 mM/IAMI-3 mM) (Fig. 3C) without affecting AMI inhibition of acid-induced currents (Fig. 4). Third, both Glu-79 and Glu-423 as the key residues of the nonproton ligand sensing domain lie in the middle of the β1 and β12 sheets, respectively (46). They are not located in the linker region between two β sheets as are Leu-85 (β1-β2 linker) (43) and Asn-415 (β11-β12 linker) (44), which are critical for allosteric gating by protons. Therefore, a similar allosteric effect is not sufficient to account for the mutation-induced decrease in the response to AMI. Finally, AMI shares a similar molecular configuration as GMQ. Linkage to the nonproton ligand sensing domain (E79C site) by GMQ dimer treatment directly induced ASIC3E79C channel opening, whereas a similar treatment on WT or ASIC3E423C channels did not induce channel gating (26). This result argues that ligand binding at the cavity around Glu-79 and Glu-423 is sufficient for ASIC3 channel gating. Therefore, even if other AMI-binding sites may exist (45) in ASIC3 channels, the nonproton ligand sensing domain most likely represents the site responsible for stimulation of channel opening.
The paradoxical stimulation of AMI on ASIC3 channels raises an interesting concern about the physiological significance of these findings. Traditionally, AMI is widely used as a nonselective ASIC blocking tool to examine the role of ASICs under pathophysiological conditions when the channel is activated. In this study, we demonstrate that AMI activates ASIC3 channels (Fig. 1A) at relatively high concentrations (EC50 = ∼560 μm) resulting in a sustained current, which would depolarize sensory neurons to a greater degree than the transient current induced by acid such as pH 5.0 (Fig. 1A). Moreover, AMI synergistically facilitates sustained activation of ASIC3 channels by mild acidosis (in a pH range between 7.2 and 6.8) at an initial effective concentration of ∼1 μm (Fig. 2, A–C). Based on the paradoxical property of AMI, we propose that whether activation or inhibition dominates should depend on the extent of physiological activation. If ASICs are activated largely as seen with stimulation at pH 6.5 or below (Figs. 1B and 2D), then the inhibitory effect would be stronger than activation under the commonly used concentrations (∼100 μm). However, when ASICs are closed or activated only by mild acidosis, AMI will exert a greater stimulatory effect than inhibition. Given that sensory ASIC3 channels have multiple models of activation and are demonstrated to have roles in multimodal sensory perception (22), we believe that the AMI-ASIC3 interaction would produce complex effects under different conditions. At least for modest pH changes that occur during myocardial ischemia (6), AMI-ASIC3 interaction would predict a stimulatory effect, an open question that relies on the development of specific chemical tools (Fig. 5).
In summary, we propose here that the stimulatory effect of the ASIC channel blocker AMI through the nonproton ligand sensing domain raises a concern for its use in pharmacological studies on ASIC3 function (16) and in developing AMI-derived ASIC inhibitors for the treatment of chronic pain (47). Given that AMI directly activates the ASIC3 channels, it is conceivable that AMI may also be used to generate activators of ASIC3 channels devoid of inhibitory actions in addition to inhibitors devoid of stimulatory actions (e.g. 5-(N-methyl-N-isobutyl)amiloride, see Fig. 5) in the future.
Supplementary Material
Acknowledgments
We thank all groups that provided us with ASIC cDNAs and thank Drs. John A. Wemmie, Margaret P. Price, and Michael J. Welsh for their kind gifts of asic3 and asic1 knock-out mice. We also thank Drs. Michael X. Zhu, James J. Celentano, and Yi-Zhi Wang for helpful comments on the manuscript.
This work was supported by National Natural Science Foundation of China Grant 30830035, National Basic Research Program of China Grant 2011CBA00408, and Shanghai Municipal Government Grant 09XD1404900.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3 and Table 1.
- ASIC
- acid-sensing ion channel
- AMI
- amiloride
- GMQ
- 2-guanidine-4-methylquinazoline
- DTDP
- 4,4′-dithiodipyridine
- TDP
- 4-thiodipyridine
- DTNB
- 5,5′-dithiobis(2-nitrobenzoic acid)
- MTSEA
- 2-aminoethyl methanethiosulfonate
- Deg
- degenerin.
REFERENCES
- 1. Wemmie J. A., Price M. P., Welsh M. J. (2006) Trends Neurosci. 29, 578–586 [DOI] [PubMed] [Google Scholar]
- 2. Waldmann R., Champigny G., Bassilana F., Heurteaux C., Lazdunski M. (1997) Nature 386, 173–177 [DOI] [PubMed] [Google Scholar]
- 3. Krishtal O. (2003) Trends Neurosci. 26, 477–483 [DOI] [PubMed] [Google Scholar]
- 4. Gao J., Duan B., Wang D. G., Deng X. H., Zhang G. Y., Xu L., Xu T. L. (2005) Neuron 48, 635–646 [DOI] [PubMed] [Google Scholar]
- 5. Sutherland S. P., Benson C. J., Adelman J. P., McCleskey E. W. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 711–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yagi J., Wenk H. N., Naves L. A., McCleskey E. W. (2006) Circ. Res. 99, 501–509 [DOI] [PubMed] [Google Scholar]
- 7. Xiong Z. G., Zhu X. M., Chu X. P., Minami M., Hey J., Wei W. L., MacDonald J. F., Wemmie J. A., Price M. P., Welsh M. J., Simon R. P. (2004) Cell 118, 687–698 [DOI] [PubMed] [Google Scholar]
- 8. Wemmie J. A., Chen J., Askwith C. C., Hruska-Hageman A. M., Price M. P., Nolan B. C., Yoder P. G., Lamani E., Hoshi T., Freeman J. H., Jr., Welsh M. J. (2002) Neuron 34, 463–477 [DOI] [PubMed] [Google Scholar]
- 9. Ziemann A. E., Schnizler M. K., Albert G. W., Severson M. A., Howard M. A., 3rd, Welsh M. J., Wemmie J. A. (2008) Nat. Neurosci. 11, 816–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lu Y., Ma X., Sabharwal R., Snitsarev V., Morgan D., Rahmouni K., Drummond H. A., Whiteis C. A., Costa V., Price M., Benson C., Welsh M. J., Chapleau M. W., Abboud F. M. (2009) Neuron 64, 885–897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Page A. J., Brierley S. M., Martin C. M., Price M. P., Symonds E., Butler R., Wemmie J. A., Blackshaw L. A. (2005) Gut 54, 1408–1415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Price M. P., McIlwrath S. L., Xie J., Cheng C., Qiao J., Tarr D. E., Sluka K. A., Brennan T. J., Lewin G. R., Welsh M. J. (2001) Neuron 32, 1071–1083 [DOI] [PubMed] [Google Scholar]
- 13. Price M. P., Lewin G. R., McIlwrath S. L., Cheng C., Xie J., Heppenstall P. A., Stucky C. L., Mannsfeldt A. G., Brennan T. J., Drummond H. A., Qiao J., Benson C. J., Tarr D. E., Hrstka R. F., Yang B., Williamson R. A., Welsh M. J. (2000) Nature 407, 1007–1011 [DOI] [PubMed] [Google Scholar]
- 14. Ziemann A. E., Allen J. E., Dahdaleh N. S., Drebot I. I., Coryell M. W., Wunsch A. M., Lynch C. M., Faraci F. M., Howard M. A., 3rd, Welsh M. J., Wemmie J. A. (2009) Cell 139, 1012–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chen C. C., Zimmer A., Sun W. H., Hall J., Brownstein M. J., Zimmer A. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 8992–8997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Deval E., Noël J., Lay N., Alloui A., Diochot S., Friend V., Jodar M., Lazdunski M., Lingueglia E. (2008) EMBO J. 27, 3047–3055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Deval E., Gasull X., Noël J., Salinas M., Baron A., Diochot S., Lingueglia E. (2010) Pharmacol. Ther. 128, 549–558 [DOI] [PubMed] [Google Scholar]
- 18. Duan B., Wu L. J., Yu Y. Q., Ding Y., Jing L., Xu L., Chen J., Xu T. L. (2007) J. Neurosci. 27, 11139–11148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Xu T. L., Duan B. (2009) Prog. Neurobiol. 87, 171–180 [DOI] [PubMed] [Google Scholar]
- 20. Mazzuca M., Heurteaux C., Alloui A., Diochot S., Baron A., Voilley N., Blondeau N., Escoubas P., Gélot A., Cupo A., Zimmer A., Zimmer A. M., Eschalier A., Lazdunski M. (2007) Nat. Neurosci. 10, 943–945 [DOI] [PubMed] [Google Scholar]
- 21. Sluka K. A., Price M. P., Breese N. M., Stucky C. L., Wemmie J. A., Welsh M. J. (2003) Pain 106, 229–239 [DOI] [PubMed] [Google Scholar]
- 22. Li W. G., Xu T. L. (2011) ACS Chem. Neurosci. 2, 26–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kellenberger S., Schild L. (2002) Physiol. Rev. 82, 735–767 [DOI] [PubMed] [Google Scholar]
- 24. Adams C. M., Snyder P. M., Welsh M. J. (1999) J. Biol. Chem. 274, 15500–15504 [DOI] [PubMed] [Google Scholar]
- 25. Adams C. M., Snyder P. M., Price M. P., Welsh M. J. (1998) J. Biol. Chem. 273, 30204–30207 [DOI] [PubMed] [Google Scholar]
- 26. Yu Y., Chen Z., Li W. G., Cao H., Feng E. G., Yu F., Liu H., Jiang H., Xu T. L. (2010) Neuron 68, 61–72 [DOI] [PubMed] [Google Scholar]
- 27. Li W. G., Yu Y., Zhang Z. D., Cao H., Xu T. L. (2010) Mol. Pain 6, 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yu Y., Li W. G., Chen Z., Cao H., Yang H., Jiang H., Xu T. L. (2011) J. Biol. Chem. 286, 24996–25006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bagriantsev S. N., Minor D. L., Jr. (2010) Neuron 68, 1–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cushman K. A., Marsh-Haffner J., Adelman J. P., McCleskey E. W. (2007) J. Gen. Physiol. 129, 345–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dubé G. R., Lehto S. G., Breese N. M., Baker S. J., Wang X., Matulenko M. A., Honoré P., Stewart A. O., Moreland R. B., Brioni J. D. (2005) Pain 117, 88–96 [DOI] [PubMed] [Google Scholar]
- 32. Frelin C., Barbry P., Vigne P., Chassande O., Cragoe E. J., Jr., Lazdunski M. (1988) Biochimie 70, 1285–1290 [DOI] [PubMed] [Google Scholar]
- 33. Tang C. M., Presser F., Morad M. (1988) Science 240, 213–215 [DOI] [PubMed] [Google Scholar]
- 34. Li Y. F., Li Y., Xu T. L. (2003) Neurosci. Lett. 345, 173–176 [DOI] [PubMed] [Google Scholar]
- 35. Springauf A., Bresenitz P., Gründer S. (2011) J. Biol. Chem. 286, 24374–24384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Babinski K., Lê K. T., Séguéla P. (1999) J. Neurochem. 72, 51–57 [DOI] [PubMed] [Google Scholar]
- 37. de Weille J. R., Bassilana F., Lazdunski M., Waldmann R. (1998) FEBS Lett. 433, 257–260 [DOI] [PubMed] [Google Scholar]
- 38. Waldmann R., Bassilana F., de Weille J., Champigny G., Heurteaux C., Lazdunski M. (1997) J. Biol. Chem. 272, 20975–20978 [DOI] [PubMed] [Google Scholar]
- 39. Salinas M., Lazdunski M., Lingueglia E. (2009) J. Biol. Chem. 284, 31851–31859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Askwith C. C., Cheng C., Ikuma M., Benson C., Price M. P., Welsh M. J. (2000) Neuron 26, 133–141 [DOI] [PubMed] [Google Scholar]
- 41. Catarsi S., Babinski K., Séguéla P. (2001) Neuropharmacology 41, 592–600 [DOI] [PubMed] [Google Scholar]
- 42. Chen X., Paukert M., Kadurin I., Pusch M., Gründer S. (2006) Neuropharmacology 50, 964–974 [DOI] [PubMed] [Google Scholar]
- 43. Li T., Yang Y., Canessa C. M. (2010) J. Biol. Chem. 285, 22706–22712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Li T., Yang Y., Canessa C. M. (2010) J. Biol. Chem. 285, 31285–31291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Qadri Y. J., Song Y., Fuller C. M., Benos D. J. (2010) J. Biol. Chem. 285, 9627–9635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Jasti J., Furukawa H., Gonzales E. B., Gouaux E. (2007) Nature 449, 316–323 [DOI] [PubMed] [Google Scholar]
- 47. Kuduk S. D., Di Marco C. N., Chang R. K., Dipardo R. M., Cook S. P., Cato M. J., Jovanovska A., Urban M. O., Leitl M., Spencer R. H., Kane S. A., Bilodeau M. T., Hartman G. D., Bock M. G. (2009) Bioorg. Med. Chem. Lett. 19, 2514–2518 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.