Abstract
Exosomes are naturally occurring nanoparticles with unique structure, surface biochemistry and mechanical characteristics. These distinct nanometer sized bio-particles are secreted from the surface of oral epithelial cells into saliva, and are of interest as oral-cancer biomarkers. We use high- resolution AFM to show single vesicle quantitative differences between exosomes derived from normal and oral cancer patient’s saliva. Compared to normal exosomes (circular; 67.4 ± 2.9 nm), our findings indicate that cancer exosomes populations are significantly increased in saliva and display irregular morphologies, increased vesicle size (98.3 ± 4.6 nm) and higher inter-vesicular aggregation. At the single vesicle level, cancer exosomes exhibit significantly (P<0.05) increased CD63 surface densities. To our knowledge, it represents the first report detecting single exosome surface protein variations. Additionally, high- resolution AFM imaging of cancer saliva samples revealed discrete multi-vesicular bodies with intra-luminal exosomes enclosed. We discuss the use of quantitative, nanoscale ultra-structural and surface bio-molecular analysis of saliva exosomes, at the single vesicle and single protein level sensitivity, as a potentially new oral cancer diagnostic.
Keywords: saliva exosomes, AFM, nano-characterization, diagnostics, oral cancer
Introduction
Nanoparticles have potential applications in imaging, drug delivery and other types of therapy and diagnostics1. Exosomes are naturally occurring bio-nanoparticles, that have substantial interest for disease biomarker applications and their properties and applications are being investigated extensively2–4. Exosomes possess unique structural, surface biochemical and mechanical characteristics. Human saliva exosomes secreted by normal cells into saliva via exocytosis, are potential novel biomarkers showing tumor-antigen enrichment during oral cancer, the 6th most frequent cancer worldwide and represents 5% of newly diagnosed cancers in adult patients.5 In US, more than 50,000 new cases and 10,000 Head and Neck Squamous Cell Carcinomas (HNSCC) cancer-related deaths are projected annually.5 Although HNSCC is a potentially curable malignancy with early diagnosis; patients are often diagnosed at a locally advanced 3rd or 4th (less to more advanced stage and type of cancer) grade of disease. 5 Between 60–70% of those advanced-stage patients will develop loco-regional recurrences within 2 years. After standard therapy, including surgical resection and adjuvant radiation, the 5-year survival rate for advanced-stage disease is less than 30%6 and has remained unchanged over the past 20 years.6, 7 Survival rate is dependent on stage at diagnosis with a 5-year overall survival of 82% for localized disease, 48% for regional disease, and 26% for distant disease. These statistics clearly indicate an urgent need for discovery of new diagnostic modalities for oral cancer.
Non-invasive saliva biomarkers for diagnostic and prognostic assessments have been established in recent years.8 In particular, exosomes, small nano-vesicles (<100 nm in size) released from various cells including whole saliva, have been the subject of renewed interest, because these vesicles offer promising potential for diagnostics as well as therapeutics.9, 10 Exosomes, considered for a long time to be artifacts, are now recognized to be important in cell-cell communication and other significant biological processes, such as membrane trafficking, intercellular horizontal transfer of proteins and RNAs.11, 12 Malignancy and other diseases cause elevated exosome secretion and enrichment of tumor-antigen in exosomes is often associated with cancer cells.13, 14 Recent reports indicate that the presence of exosomes of likely tumor origin in plasma and other biological fluids from cancer patients.15–17 Recently, we have shown that human whole saliva contains secretory exosomes and identified their transcriptomic contents18 and biophysical characteristics.19 Exosomes possess highly specific surface composition and cargoes- proteins, RNA and DNA, it is likely that tumor-derived exosomes may differ from normal exosomes in certain physiological conditions, both in terms of structure and surface molecular characteristics. Exosomes released by normal and tumor cells have been suggested to differ in both functional and structural properties20 implicating their direct role in pathogenesis, which could consequently serve as an important cancer biomarker. Currently exosomes use for diagnostic and therapeutic purposes21, 22 mostly rely on bulk proteomic and genomic transcripts including previously reported over-expression of CD63, significantly associated with exosomes in biological fluids such as plasma of cancer patients.23 However, single molecular studies of exosomes derived from cancer patients remain elusive.
Although exosomes hold promise as biomarkers for cancer diagnosis21, 22, their nanoscale dimensions require sensitive, quantitative high-resolution detection tools for characterization on an individual basis24. Information on single exosomes via conventional semi-quantitative proteomic and transcriptional analytical methods prove ineffective due to sensitivity limitations. Current paradigms of exosomes analysis include Electron Microscopy (EM), Western blot, Flow Cytometry and Mass Spectrometry.25–27 Typically, EM imaging is used for identification of exosomes revealing apparent two-dimensional cup-shaped vesicles.27 Due to their sub-100nm size, exosomes are difficult to detect as discrete particles and require sizing beads which challenges the sensitivity limit of ~100nm for flow cytometers.28 AFM can characterize biological surfaces with sub-nanometer resolution and allows structural, biochemical and mechanical characterization of cells29, 30, biological molecules31 including vesicles32, 33. AFM based single-molecule force spectroscopy (SMFS) mode could be used as a rapid and reliable tool to provide information on the morphology as well as on the molecular recognition abilities of biological interfaces such as exosomes, down to the single-molecule level. Previously, we have shown the single vesicle structural and surface molecular details on normal human saliva exosomes using AFM and high resolution FESEM.19 The structural and bio-molecular changes in cancer saliva exosomes have not been reported. Here, we specifically report an AFM based assay for the identification of changes in exosome morphology and expression of bio-molecular surface receptor CD63 to detect single vesicle quantitative differences between normal and cancer human derived saliva exosomes. We present (1) Three dimensional ultra-structural characteristics and enumeration for isolated saliva exosomes using AFM imaging in air and (2) determine quantitative single exosomes surface receptor density of membrane marker CD63 under physiological buffer conditions, to elucidate structural and surface bio-molecular differences between the exosomes populations.
Experimental Section
Sample collection, exosome isolation and purification
Saliva samples were obtained from healthy volunteers or oral cancer patients from the Division of Otolaryngology, Head and Neck Surgery, at the Cancer Center, Medical University of South Carolina, Charleston, SC, in accordance with a protocol approved by the MUSC Institutional Review Board. For exosomes preparation, 1 ml of saliva was equally mixed with phosphate buffer saline and spun at 2600g for 15 min to remove cells. The supernatants were then sequentially centrifuged at 12 000g for 20 min and 120 000g for 3 h. The 120 000g pellet was re-suspended in PBS and used for AFM and Western blot analysis.
AFM imaging
Purified exosomes were diluted 1:200 in de-ionized water (stock 1mg/ml) and adsorbed to freshly cleaved mica sheets, rinsed with de-ionized water and air-dried. Dimension 5000 (Bruker Instruments) AFM under tapping mode and silicon probes (K~40 Nm−1; Bruker) was used. Constant force was maintained for imaging all samples. Topographic height, amplitude and phase images were recorded simultaneously at 512×512 pixels at a scan rate of 1Hz. Image processing was performed using SPIP™ software.
Single molecule Force spectroscopy: CD63 receptor mapping
Catalyst (Bruker) AFM was used with MLCT (Bruker) cantilevers with experimentally determined spring constants of 0.01 N/m and a tip radius < 20 nm. Exosome immobilization and AFM tips functionalization with antiCD63 antibodies were performed as previously.19 Force-separation curves were recorded at a ramp size of 1 μm and tip velocity of 1 μm/s under low forces (<500 pN).
Statistical Analysis
Data were expressed as mean ± s.e.m (std. error of mean), and the statistical significance of differences in mean values was assessed using a two-sample independent Student’s t-test at the 95% confidence level. Differences among means are reported using P values.
Results
Ten clinical samples, including five from oral cancer patients and five from normal healthy volunteers, obtained and purified from only 1ml of saliva using improved exosome isolation protocol, were used in this study.
Nanoscale structure of normal and oral cancer saliva exosomes
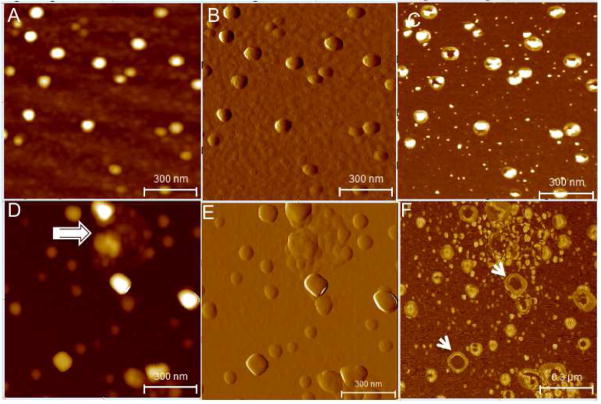
We measured the nanometer level quantitative three-dimensional structure and vesicular organization of salivary exosomes using AFM Tapping mode34- topographic (height), amplitude and phase images (allows mapping variations in material properties such as exosomes density and visco-elasticity). Figure 1 represents AFM images revealing morphologically distinct normal and cancer exosomes in terms of overall morphology, and ultra-structural characteristics. This data summarizes several thousand exosomes studied from all patient samples. Figure 1 A-correspond to the height, amplitude and phase images representative of exosomes morphological characteristics of normal saliva sample. The vesicles are homogeneous in morphology and size, 60–100nm in size (Figure 1A) and appear as discrete circular bulging vesicles (Figure 1B) without any apparent inter-vesicular fusion or aggregation. Single exosomes (Figure 1C) display a characteristic sub-vesicular structure with characteristic phase contrast suggesting the role of heterogeneous density and/or viscoelastic image contrast mechanisms. Interestingly, oral cancer exosomes represented in Fig 1D–F, show variable vesicles size distribution ranging from 20–400nm(Fig 1D) and are either circular/bulging vesicles or rather irregular in morphology (1E). The phase images (1F) show larger vesicles with a less dense core region compared to smaller vesicles. The large size vesicles are often seen surrounded by numerous small (20–50nm) vesicles or membrane debris (1F). Additionally, oral cancer patient saliva exosomes samples reveal aggregation of several individual vesicles to form bigger more extended agglomerates (Figure 1D). Exosomes size (vesicle diameter, nm) from saliva samples collected from healthy donors or patients with suspected oral cancer are shown in Figure 2. Only single vesicles with well-defined boundaries without any aggregation were used for analysis. The exosomes diameters ranged mostly from 40–100nm for normal exosomes with average diameters of 67.4±2.9 (n=486). For oral cancer exosomes, the average vesicle diameters were observed to be 98.3± 4.6 (n=482), significantly higher than normal saliva exosomes (P<0.05). It should be noted that vesicles smaller than 200 nm in diameter cannot be detected by confocal microscopy techniques35. The size of single exosomes is below the diffraction limit of light (typical lateral confocal imaging resolution lies between 200–300nm) and cannot be resolved via fluorescence imaging and has not been attempted in our study. Instead we have exploited the sub-nanometer resolution imaging capability of AFM to successfully investigate structural variations in cancer versus normal exosomes.
Figure 1.
Single vesicle structural characteristics of human salivary exosomes (A–C) AFM topographic (z 0–10nm range), amplitude and phase image of exosomes derived from saliva of normal healthy donors. The exosomes appear as homogeneous circular bulging vesicular structures with a distinct phase contrast between less dense vesicle periphery and more dense core region. Exosomes from oral cancer patient (D–F) show more irregular morphology with varying shapes and vesicle aggregation (arrow marked). The amplitude image (E) shows the clumping of several vesicles into an aggregate. In the phase image (F) the larger sized vesicles appear hollow (arrows) without the dense core region typically seen in normal exosomes. All images were obtained over mica substrates under ambient conditions.
Figure 2.
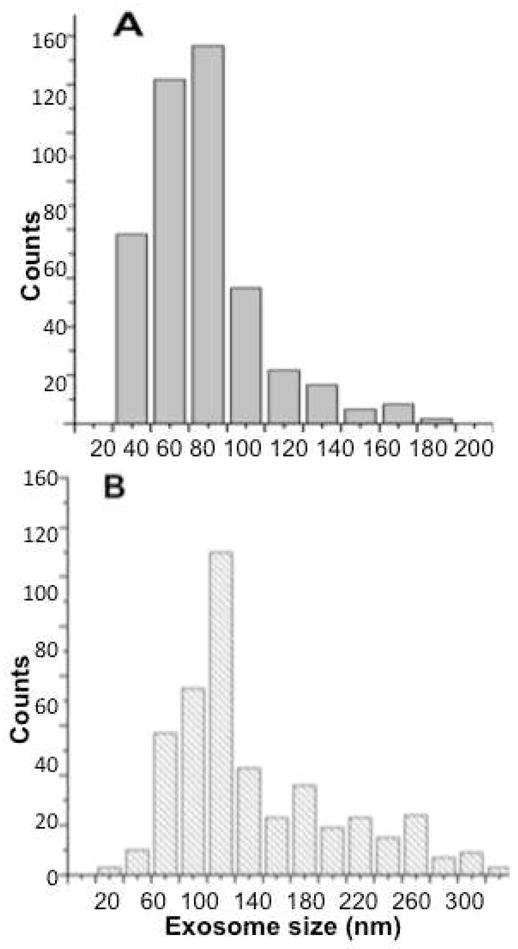
Size distribution comparison between single vesicle dimensions (diameter in nm) for (A) exosomes obtained from healthy donor’s (n=486) (B) oral cancer patient saliva samples (n=482). Individual vesicles with well-defined boundaries and showing no aggregation were analyzed from AFM topographic images. The average size of the exosomes is higher for oral cancer exosomes (P<0.05)
AFM based quantification of saliva exosomes
Other techniques, such as FACS (Fluorescence activated cell sorting) widely used to quantify exosomes population in biological samples completely rely on binding of the specific antibody to the surface or micron sized beads to capture the exosomes for quantifications. The capture of exosomes to the latex beads may lead to counting discrepancies due to latex beads surface- antibody binding and affinity limitations. Therefore the exosomes count is likely to be directly influenced by the affinity and protein density of the target probe, whereas attaching the exosomes to freshly cleaved mica surface for AFM imaging and subsequently counting the number of exosomes in specified scan area is independent of specific probe targeting. A two to four folds increase in exosome density in oral cancer patient samples compared to normal samples was observed (see Table 1).
Table 1.
Patient history and histopathology versus saliva exosome characteristics
| Case No. | Stage- Histological grade$ | Size (nm)** | Counts/64um2 | Vesicle Morphology | MVs^ | Treatment |
|---|---|---|---|---|---|---|
| 1. | 4a | 77.0± 3.7 | 345 | IR# | ++ | Chemotherapy |
| 2. | 4a | 138.6± 6.8 | 342 | IR | − | Chemotherapy |
| 3. | 4a | 102.7±6.7 | 414 | IR | + | Chemotherapy + Surgery |
| 4. | 1 | 92.5± 4.6 | 242 | IR | − | Surgery |
| 5. | 4a | NA% | Na | NA | NA | Chemotherapy |
| 6. | 2 | 80.9± 6.1 | 268 | IR | − | Surgery |
| 7. | Normal | 62.3± 3.4 | 128 | R& | − | None |
| 8. | Normal | 67.4± 2.6 | 137 | R | − | None |
| 9. | Normal | 66.4± 3.6 | 96 | R | − | None |
| 10. | Normal | 67.4± 2.9 | 193 | R | − | None |
| 11. | Normal | 71.6± 1.7 | 126 | R | − | None |
Grade 1–4 and a–c indicates less to more advanced stage and type of cancer;
Mean ± SEM;
Irregular
Regular circular;
Multi-vesicular bodies.
Not use for analysis due to sample serum contamination Normal and Cancer samples studied were sex (Males) and age (between 54–75yrs) matched.
Presence of Multi-vesicular bodies enclosing several exosomes
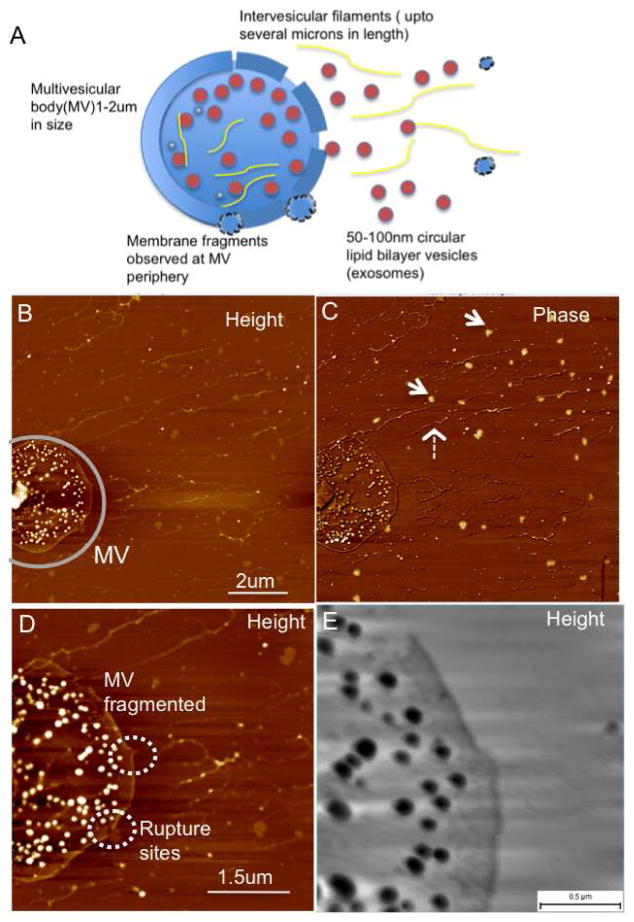
Exosomes biogenesis involves the tightly controlled process of inward budding from the limiting membrane of multi-vesicular bodies (MVBs) resulting in numerous distinct intra-luminal nano-sized vesicles contained within the lumen of MVBs 25, 36 (with an estimated diameter of 1–2um). Interestingly, in case of oral cancer saliva exosomes samples, multi-vesicular structures enclosing the exosomes within a limiting membrane were observed for the first time (Figure 3A). These multi-vesicular structures also reveal the presence of elongated nano-filaments, around 1–5um long and 20nm wide within the lumen of these MVs. While the lipid membrane constitution of the exosomes show striking contrast of the exosomes over mica substrates under AFM phase imaging (Figure 3B), the extra-vesicular filaments do not show such a phase variation and suggest that these are not lipid dense structures similar to lipid bilayers but more likely to be nucleic acids (RNA or DNA), proteins or filaments of the cellular cytoskeletal network. At higher resolution (Figure 3. C, D) the membrane shows ruptures at specific regions of the multi-vesicular body suggesting these to be sites for release of the exosomes from the MVBs as well as the extra-exosomal filamentous extensions. While two out of the five oral cancer exosomes samples (see Table 1. Case No. 1 & 3) show presence of these MVs, none were observed for any of the five normal salivary exosomes samples. In both normal and cancer saliva exosome samples, the vesicles are sometimes observed surrounded by a framework of elongated extra-vesicular nano-filaments, (similar to those observed in Figure 3.) The origins of these extensions are relatively unknown.
Figure 3.
Release of exosomes from multivesicular bodies (MVs) seen in oral cancer patient salivary exosomes (A) Schematics of a single MV membrane rupturing at multiple sites and release of several nanoscale vesicles, exosomes along with intervesicular filaments from the MV lumen (B) AFM topographic and (C) Phase image of a single multi-vesicular body filled with several exosome vesicles. Inter-vesicular filaments (broken arrow) without the characteristic exosome-like phase contrast (arrows) are observed. (D) At higher resolution the ruptures in the multivesicular body are seen clearly with membrane fragments appearing in the vicinity of the membrane breaks (short broken circles). Additionally the inter-vesicular filaments are also seen within the lumen of the MVB. A large rupture of the limiting membrane is seen in the top region at higher resolution in (E). Samples were imaged under ambient conditions.
Biochemical characterization of individual human saliva exosomes CD63 surface receptors via force spectroscopy
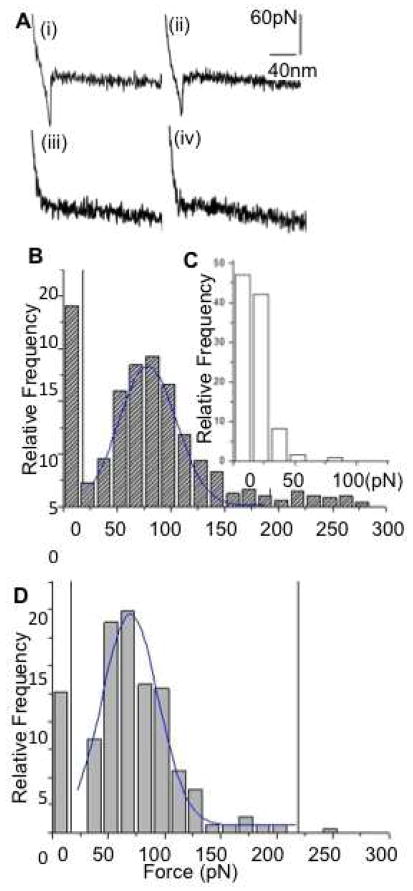
The exosomes possess a number of protein receptors on the outer membrane37, which are specific to their cellular origin and their targeted functions. We used Single molecule force spectroscopy (SMFS) to probe the membrane biochemical characteristics of saliva exosomes, using AFM tips functionalized with specific exosomes marker protein CD63 antibody. The anti-CD63 binding events were measured for both normal and cancer exosomes. Non-specific mouse IgGs coated AFM tips were used as controls. The highest rupture force was used as a measure for direct determination of the strength of the bond formed between one or multiple individual CD63 and antiCD63 pair. Specific antibody tip forces were in the 50 to 200pN range (Figure 4A(i) (ii). Much weaker adhesive forces (<50pN) only were observed for non-specific antibody tips (Figure 4A.iii.iv and Figure 4C), confirming the specificity of force measurements to antibody molecules. A series of ~1000 force-separation curves were quantified and plotted in a force histogram (Figure. 4B) showing the relative frequency of distribution of the rupture forces (0–300 pN). For normal exosomes, the mean interaction force from a normal distribution (horizontal markers) of the histogram was calculated from bins 30 to 200 pN as F = 70 ± 15 pN (mean ± SD), with 38% of the unbinding events falling between 50–100pNs, the characteristic force range for single antigen-antibody interactions19, 38, 39. Cancer exosomes measured F = 78 ± 10 pN (mean ± SD), with 58% unbinding events (50–100pNs). An increase of 47% in the number of observed specific rupture forces in case of cancer exosomes, occurring at the characteristic range of antigen-antibody interactions indicates presence of higher density of surface CD63 molecules in cancer exosomes. Besides single bio-molecular rupture events seen in the force retraction curves, multiple rupture peaks were observed indicating the possibility of multiple tetraspanin CD63 molecules constituting the vesicle surface.
Figure 4.
Biochemical characterization of human salivary exosomes via force spectroscopy showing quantitative differences between CD63 surface receptors density (a) Typical curves showing force (pN) as a function of separation (nm) for a single pull with (i) adhesive event between cancer exosomes and (ii) normal exosomes probed with antiCD63 functionalized tip. (iii)– (iv) show weak or no binding between nonspecific antibody functionalized tip with cancer and normal exosomes respectively. (b) Histogram of rupture events (relative frequency versus rupture force in pNs) shows specific CD63 antibody-induced forces were distributed in the range of 30–200 pN. Sampled forces (n~500 each) from six normal and six oral cancer patient saliva exosome had each data point representing a single force measurement at any position on the exosomes surface (bin size 15 pN) (c) Nonspecific interactions were observed mostly at <50 pN.
Western blotting of the exosomes isolated from the ten saliva samples showed the presence of CD63 molecules (Supplementary Figure 1). Relatively higher expression of CD63 molecules was observed for all five oral cancer samples compared to normal saliva exosomes consistent with the AFM single molecule force measurements. SMFS based single vesicle receptor mapping allows distinction between elevated exosomes density and over-expression of surface protein CD63. Surface mapping of vesicle receptors using AFM allow us to select, localize and measure receptors expressed on the outer surface of the exosomes. It enables inherent distinction of lumenal proteins from those present on the cytoplasmic side of exosomes40 which is a major advantage over 2D gel analysis where trans-membrane proteins are difficult to analyze.41 The high sensitivity detection of changes in the exosomes specific receptor densities (number of specific receptors differentially expressed on individual exosomes) of trans-membrane receptors such as CD63 represents to our knowledge, the first report on single exosome level surface receptor density variations.
Discussion
The characteristics of each sample used in this study (categorized on the basis of vesicle structure, morphology, density and structural organization); data collected from ten different clinical samples (positive for oral cancer n= 5; negative for oral cancer n=5) are individually summarized in Table 1. EM is the most commonly used technique for morphological characterization of exosomes. We have previously shown that electron dense staining, vacuum dehydration in EM imaging result in artifacts as cup shaped exosomes structure27 whereas Field Emission Scanning Electron Microscopy (FESEM) done at low voltages and low force AFM imaging of unstained, unfixed samples under ambient conditions without vacuum drying appear far less damaging to the exosomes and result in bulging round vesicles.19 Using tapping mode AFM in air, all normal saliva exosomes samples show mostly homogeneous circular morphology, and narrower size distribution localized between 40–80 nm however, oral cancer exosomes show morphological variations in shapes and sizes (ranging from 20–400nm) with some regions showing aggregated vesicles or clusters. We observed two folds increase in exosome density in oral cancer patient samples compared to normal samples. The observed increase in the density of the exosomes measured over mica surface may occur due to the differences in the binding affinity of the exosomes to the mica surface. If exosomes express more CD63 receptors on the surface it is possible that it results in some non-specific adhesion of the exosomes on the mica however, the variations in cancer exosomes morphology is clearly indicated. Several MVs filled with nanometer scale exosomes were observed in case of two cancer samples (Case 1 &3) while we did not observe any MVs in our AFM images obtained from normal saliva exosomes samples. Hence, we hypothesize that the observed morphological changes seen in saliva exosomes from oral cancer patients, either result due to inherent overall increase in the size of the secreted vesicles or aggregation/fusion of the secreted vesicles during or after excretion of the vesicles into the salivary fluid. The presence of increased exosomes counts, irregular morphology in patient saliva samples were observed irrespective of whether they solely received chemotherapy, surgery or both. The changes in vesicle size and morphology in all five cancer samples compared to normal salivary exosomes, clearly indicates the influence of subject patho-physiological condition reflected in exosomes structural and morphological aberrations at the single vesicle level.
The presence of extra-vesicular nano-filamentous structures observed in both normal and oral cancer patient samples, suggests a close association of these filaments with the exosomes both within the MVs (as seen in oral cancer samples) as well as outside the extra-vesicular milieu. The biological function of these filaments are not yet understood but based on the close resemblance to DNA/RNA structures42; we believe it would be interesting in future studies to test the possibility of mRNA or microRNA constituting all or part of these structures. The filaments have not been observed elsewhere and need further confirmatory work such as imaging in physiological buffer conditions.
CD63, also known as lysosome-associated antigen ME491, is a member of transmembrane4superfamily (TM4SF) which is a heterogenous membrane-bound glycoprotein associated with regulation of diverse cellular processes such as proliferation, adhesion, motility and differentiation43 and also a marker for late endosomal secretory vesicles44. CD63 was first discovered as an abundantly expressed surface antigen in early-stage melanoma cells45. A negative relationship between CD63 expression and increased malignancy or invasiveness has been reported in many tumors, including ovarian, lung, breast and colon cancers, as well as in melanoma 46. However, high CD63 expressions have been reported for other tumor subtypes such as pilocytic astrocytomas and glioblastomas47, similar to our observation of increase in the level of CD63 expression with advanced oral cancer stage using both AFM force spectroscopy and Western blots. CD63 is known to interact with many different proteins, including integrins and the Src family tyrosine kinases Lyn and Hck48, 49 and the biological implications of differential expression of CD63 molecules with respect to cancer aggressiveness need further investigation.
Conclusions
In summary, we have investigated the individual structural morphology and single receptor-ligand level- surface bio-molecular attributes, of saliva exosomes derived from normal and oral cancer patients, with nano-scale resolution. The cancer saliva samples show elevated exosomes-quantified via AFM scans on bare mica surface. Oral cancer saliva exosomes are generally larger in size and show more profound aggregation. Single vesicle receptor density comparisons show statistically verifiable increases in CD63 expression on exosomes derived from saliva of patients suffering from oral cancers. Our findings demonstrate that nanoscale exosomes characterization provides an excellent potential candidate for probing pre and early cancer progression via measurement of changes in the saliva exosomes at the single vesicle and single receptor level sensitivity in future diagnostic salivary tests. The results validate the AFM based approach to quantitatively detect exosomes structure; surface proteins constitution and content; and can be further extended to other cancer types in human biological fluids such as blood, urine or amniotic fluid.
Supplementary Material
Acknowledgments
Funding sources: This work was supported by California Nanosystems Institute (S.S.), MEXT, WPI Program: International Center for Materials Nanoarchitectonics MANA (NIMS) Japan (J.K.G) and NIH-NIDCR P20RR017696 and R00DE018165 grants (V.P.).
We acknowledge the use of the Scanning Probe Microscopes at the Nano and Pico Characterization Laboratory at the California NanoSystems Institute, UCLA.
Footnotes
Supporting Information: AFM Experimental section. Relative protein concentration and western blots. This information is available free of charge via the Internet at http://pubs.acs.org/.
References
- 1.Almeida JP, Chen AL, Foster A, Drezek R. Nanomedicine (Lond) 2011;6(5):815–35. doi: 10.2217/nnm.11.79. [DOI] [PubMed] [Google Scholar]
- 2.Al-Nedawi K, Meehan B, Rak J. Cell Cycle. 2009;8(13):2014–8. doi: 10.4161/cc.8.13.8988. [DOI] [PubMed] [Google Scholar]
- 3.Koga K, Matsumoto K, Akiyoshi T, Kubo M, Yamanaka N, Tasaki A, Nakashima H, Nakamura M, Kuroki S, Tanaka M, Katano M. Anticancer Res. 2005;25(6A):3703–7. [PubMed] [Google Scholar]
- 4.Skog J, Wurdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M, Curry WT, Jr, Carter BS, Krichevsky AM, Breakefield XO. Nat Cell Biol. 2008;10(12):1470–6. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Psyrri A, DiMaio D. Nat Clin Pract Oncol. 2008;5(1):24–31. doi: 10.1038/ncponc0984. [DOI] [PubMed] [Google Scholar]
- 6.Al-Sarraf M. Cancer Control. 2002;9(5):387–99. doi: 10.1177/107327480200900504. [DOI] [PubMed] [Google Scholar]
- 7.Neville BW, Day TA. CA Cancer J Clin. 2002;52(4):195–215. doi: 10.3322/canjclin.52.4.195. [DOI] [PubMed] [Google Scholar]
- 8.Wong DT. J Am Dent Assoc. 2006;137(3):313–21. doi: 10.14219/jada.archive.2006.0180. [DOI] [PubMed] [Google Scholar]
- 9.Delcayre A, Estelles A, Sperinde J, Roulon T, Paz P, Aguilar B, Villanueva J, Khine S, Le Pecq JB. Blood Cells Mol Dis. 2005;35(2):158–68. doi: 10.1016/j.bcmd.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 10.Garcia JM, Garcia V, Pena C, Dominguez G, Silva J, Diaz R, Espinosa P, Citores MJ, Collado M, Bonilla F. Rna. 2008;14(7):1424–32. doi: 10.1261/rna.755908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Nat Cell Biol. 2007;9(6):654–9. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 12.Keller S, Ridinger J, Rupp AK, Janssen JW, Altevogt P. J Transl Med. 2011;9:86. doi: 10.1186/1479-5876-9-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Andre F, Schartz NEC, Movassagh M, Flament C, Pautier P, Morice P, Pomel C, Lhomme C, Escudier B, Le Chevalier T, Tursz T, Amigorena S, Raposo G, Angevin E, Zitvogel L. The Lancet. 2002;360(9329):295–305. doi: 10.1016/S0140-6736(02)09552-1. [DOI] [PubMed] [Google Scholar]
- 14.Wolfers J, Lozier A, Raposo G, Regnault A, Thery C, Masurier C, Flament C, Pouzieux S, Faure F, Tursz T, Angevin E, Amigorena S, Zitvogel L. Nat Med. 2001;7(3):297–303. doi: 10.1038/85438. [DOI] [PubMed] [Google Scholar]
- 15.Valenti R, Huber V, Filipazzi P, Pilla L, Sovena G, Villa A, Corbelli A, Fais S, Parmiani G, Rivoltini L. Cancer Res. 2006;66(18):9290–8. doi: 10.1158/0008-5472.CAN-06-1819. [DOI] [PubMed] [Google Scholar]
- 16.Bard MP, Hegmans JP, Hemmes A, Luider TM, Willemsen R, Severijnen LA, van Meerbeeck JP, Burgers SA, Hoogsteden HC, Lambrecht BN. Am J Respir Cell Mol Biol. 2004;31(1):114–21. doi: 10.1165/rcmb.2003-0238OC. [DOI] [PubMed] [Google Scholar]
- 17.Andre F, Schartz NE, Movassagh M, Flament C, Pautier P, Morice P, Pomel C, Lhomme C, Escudier B, Le Chevalier T, Tursz T, Amigorena S, Raposo G, Angevin E, Zitvogel L. Lancet. 2002;360(9329):295–305. doi: 10.1016/S0140-6736(02)09552-1. [DOI] [PubMed] [Google Scholar]
- 18.Palanisamy V, Sharma S, Deshpande A, Zhou H, Gimzewski J, Wong DT. PLoS One. 2010;5(1):e8577. doi: 10.1371/journal.pone.0008577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sharma S, Rasool HI, Palanisamy V, Mathisen C, Schmidt M, Wong DT, Gimzewski JK. Acs Nano. 2010;4(4):1921–1926. doi: 10.1021/nn901824n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taylor DD, Lyons KS, Gercel-Taylor C. Gynecol Oncol. 2002;84(3):443–8. doi: 10.1006/gyno.2001.6551. [DOI] [PubMed] [Google Scholar]
- 21.Nilsson J, Skog J, Nordstrand A, Baranov V, Mincheva-Nilsson L, Breakefield XO, Widmark A. Br J Cancer. 2009;100(10):1603–7. doi: 10.1038/sj.bjc.6605058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Whiteside TL, Odoux C. Cancer Immunology Immunotherapy. 2004;53(3):240–248. doi: 10.1007/s00262-003-0468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Logozzi M, De Milito A, Lugini L, Borghi M, Calabro L, Spada M, Perdicchio M, Marino ML, Federici C, Iessi E, Brambilla D, Venturi G, Lozupone F, Santinami M, Huber V, Maio M, Rivoltini L, Fais S. PLoS One. 2009;4(4):e5219. doi: 10.1371/journal.pone.0005219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raimondo F, Morosi L, Chinello C, Magni F, Pitto M. Proteomics. 2011;11(4):709–20. doi: 10.1002/pmic.201000422. [DOI] [PubMed] [Google Scholar]
- 25.Thery C, Zitvogel L, Amigorena S. Nat Rev Immunol. 2002;2(8):569–79. doi: 10.1038/nri855. [DOI] [PubMed] [Google Scholar]
- 26.Hegmans JP, Gerber PJ, Lambrecht BN. Methods Mol Biol. 2008;484:97–109. doi: 10.1007/978-1-59745-398-1_7. [DOI] [PubMed] [Google Scholar]
- 27.Thery C, Amigorena S, Raposo G, Clayton A. Curr Protoc Cell Biol. 2006;Chapter 3(Unit 3):22. doi: 10.1002/0471143030.cb0322s30. [DOI] [PubMed] [Google Scholar]
- 28.Robert S, Poncelet P, Lacroix R, Arnaud L, Giraudo L, Hauchard A, Sampol J, Dignat-George F. Journal of Thrombosis and Haemostasis. 2009;7(1):190–197. doi: 10.1111/j.1538-7836.2008.03200.x. [DOI] [PubMed] [Google Scholar]
- 29.Suresh S. Acta Biomater. 2007;3(4):413–38. doi: 10.1016/j.actbio.2007.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cross SE, Jin YS, Rao J, Gimzewski JK. Nat Nano. 2007;2(12):780–783. doi: 10.1038/nnano.2007.388. [DOI] [PubMed] [Google Scholar]
- 31.Sharma S, Grintsevich EE, Phillips ML, Reisler E, Gimzewski JK. Nano Letters. 2011;11(2):825–7. doi: 10.1021/nl104159v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jena BP, Schneider SW, Geibel JP, Webster P, Oberleithner H, Sritharan KC. Proc Natl Acad Sci U S A. 1997;94(24):13317–22. doi: 10.1073/pnas.94.24.13317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baran J, Baj-Krzyworzeka M, Weglarczyk K, Szatanek R, Zembala M, Barbasz J, Czupryna A, Szczepanik A. Cancer Immunol Immunother. 2010;59(6):841–50. doi: 10.1007/s00262-009-0808-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Binnig G, Quate CF, Gerber C. Phys Rev Lett. 1986;56(9):930–933. doi: 10.1103/PhysRevLett.56.930. [DOI] [PubMed] [Google Scholar]
- 35.Thery C. F1000 Biol Rep. 2011;3:15. doi: 10.3410/B3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Muralidharan-Chari V, Clancy J, Plou C, Romao M, Chavrier P, Raposo G, D’Souza-Schorey C. Curr Biol. 2009;19(22):1875–85. doi: 10.1016/j.cub.2009.09.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jeffrey S, Schorey SB. Traffic. 2008;9(6):871–881. doi: 10.1111/j.1600-0854.2008.00734.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moy VT, Florin EL, Gaub HE. Science. 1994;266(5183):257–9. doi: 10.1126/science.7939660. [DOI] [PubMed] [Google Scholar]
- 39.van Oss CJ. J Immunoassay. 2000;21(2–3):109–42. doi: 10.1080/01971520009349531. [DOI] [PubMed] [Google Scholar]
- 40.Garin J, Diez R, Kieffer S, Dermine JF, Duclos S, Gagnon E, Sadoul R, Rondeau C, Desjardins M. J Cell Biol. 2001;152(1):165–80. doi: 10.1083/jcb.152.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Santoni V, Molloy M, Rabilloud T. Electrophoresis. 2000;21(6):1054–70. doi: 10.1002/(SICI)1522-2683(20000401)21:6<1054::AID-ELPS1054>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 42.Reed J, Mishra B, Pittenger B, Magonov S, Troke J, Teitell MA, Gimzewski JK. Nanotechnology. 2007;18(4):44032. doi: 10.1088/0957-4484/18/4/044032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hunziker W, Geuze HJ. Bioessays. 1996;18(5):379–89. doi: 10.1002/bies.950180508. [DOI] [PubMed] [Google Scholar]
- 44.Fukuda M. J Biol Chem. 1991;266(32):21327–30. [PubMed] [Google Scholar]
- 45.Hotta H, Ross AH, Huebner K, Isobe M, Wendeborn S, Chao MV, Ricciardi RP, Tsujimoto Y, Croce CM, Koprowski H. Cancer Res. 1988;48(11):2955–62. [PubMed] [Google Scholar]
- 46.Pols MS, Klumperman J. Exp Cell Res. 2009;315(9):1584–92. doi: 10.1016/j.yexcr.2008.09.020. [DOI] [PubMed] [Google Scholar]
- 47.Rorive S, Lopez XM, Maris C, Trepant AL, Sauvage S, Sadeghi N, Roland I, Decaestecker C, Salmon I. Mod Pathol. 2010;23(10):1418–28. doi: 10.1038/modpathol.2010.136. [DOI] [PubMed] [Google Scholar]
- 48.Jung KK, Liu XW, Chirco R, Fridman R, Kim HR. EMBO J. 2006;25(17):3934–42. doi: 10.1038/sj.emboj.7601281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Iida J, Skubitz AP, McCarthy JB, Skubitz KM. J Transl Med. 2005;3:42. doi: 10.1186/1479-5876-3-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.