Abstract
A versatile and stable liposomal platform is developed for rapid optimization of its peripheral composition. The platform is based on polydiacetylene lipids terminated with alkynyl groups. Conditions for copper-catalyzed azide–alkyne cycloaddition (a “click” reaction) are optimized for rapid attachment of azides with controlled composition onto the liposomes.
Liposomes modified with multiple targeting and imaging agents at their periphery have widely been used for diagnostic imaging.1 A thorough optimization of the composition of functional moieties on the periphery is necessary for each specific application. The conventional process involves preparation of liposomes individually from a mixture of lipids with varied ratios. This process is tedious and time-consuming, and greatly hampered by the inherent instability of the liposomes.2–4 Furthermore, aggregation and phase segregation of functional lipids often occurs, which limits the range of compositions on the liposome.2–4 To overcome these limitations, in this work we demonstrate the use of “clickable”, polymerized liposomes (CPL) as a general liposomal platform for rapid attachment of functional moieties to their peripheries via click chemistry5 (Scheme 1).
Scheme 1.
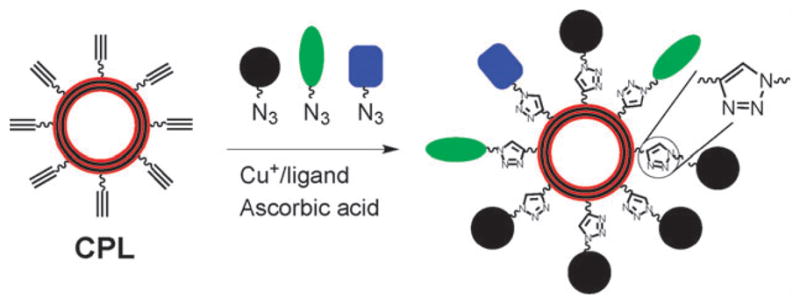
Illustration of using robust, alkynyl-terminated liposomes as a versatile platform for rapid attachment of multiple functional moieties with controlled composition via click reactions.
The CPL are based on polydiacetylene liposomes presenting alkynyl groups on their peripheries. We chose copper-catalyzed azide–alkyne cycloaddition5 (CuAAC, a “click” reaction) for bioconjugation on the CPL, since the reaction is compatible with biomolecules, and generally not affected by the substituents.5 Hence, multiple functional moieties including matrices such as poly(ethylene glycol)s with a controllable composition can be attached onto the CPL in a one-pot reaction (Scheme 1). CuAAC reactions have been performed on non-polymerized liposomes.6,7 However, these liposomes have a limited stability against fusion and breakage. In addition, due to the fluidity of lipids in non-polymerized liposomes, phase segregation may occur post modification. These problems are overcome by our CPL that are uniform in size and highly stable against fusion and breakage.4 Nevertheless, the physically robust CPL, like other unsaturated liposomes, are prone to decomposition by peroxy radicals8 that can be generated under conventional CuAAC reaction conditions.5 Herein, we show that this problem can be solved by the use of a modified copper ligand and optimized CuAAC conditions that greatly reduce the reaction time and the usage of copper and ascorbic acid for the reaction.
The CPL were prepared from the alkyne-terminated diacetylene lipid 1 diluted with the lipid 2 (Scheme 2, see the ESI for the detailed procedure†). Briefly, an aqueous solution of the lipids was extruded 20 times through a 100 nm filter. The resultant liposomes were polymerized under 254 nm UV at 0 °C to form the CPL with diameters of ~110 nm measured by dynamic light scattering (ESI†). To facilitate optimization of the click reaction, the non-fluorescent coumarin azide 3 was used, which forms highly fluorescent triazole-coumarin derivatives on the CPL upon the reaction (Scheme 3).9 The yield of the reaction could then be estimated by the fluorescence intensity with a calibration (ESI†).
Scheme 2.
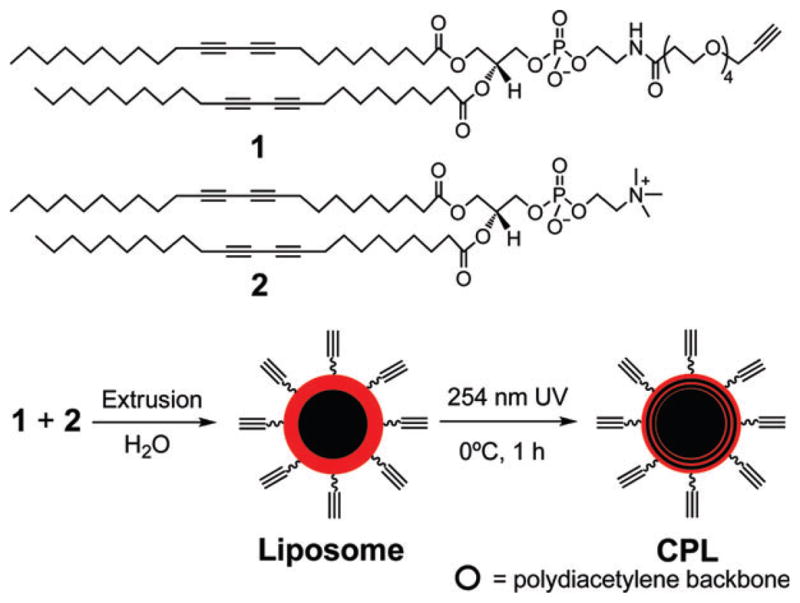
Synthesis of CPL. The ethynyl groups inside the CPL are not indicated.
Scheme 3.
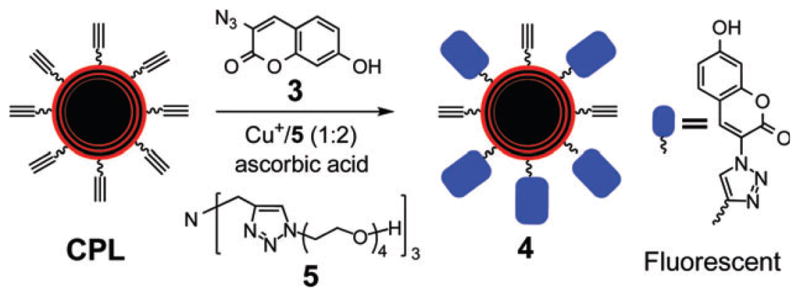
Optimization of CuAAC reaction on CPL.
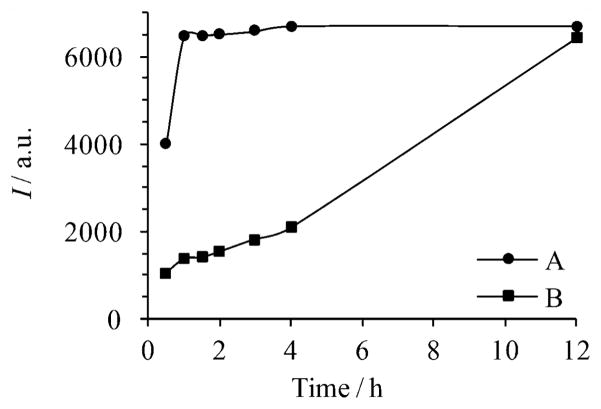
Fokin and co-workers have demonstrated that CuAAC reactions are greatly accelerated by appropriate Cu+ ligands, e.g. the commercially available tris(benzyltriazolylmethyl)-amine (TBTA).10 However, TBTA did not perform well in our system, likely due to its sparse solubility in water. The most commonly used water soluble ligand to promote the CuAAC reaction is sulfonated bathophenanthroline.11 However, its copper complex is highly sensitive to air oxidation. To address these issues, we tested a series of triazole derivatives tethering oligo(ethylene glycol) side chains to render them water soluble (unpublished results). Among them, the tridentate ligand 5 proved to be particularly efficient for CuAAC reactions on the liposomes. Moreover, its copper complex is quite stable in air, allowing the reactions to be performed under ambient conditions.12 Thus, as shown in Fig. 1, the presence of the ligand 5 greatly accelerated the CuAAC reaction, shortening the reaction time by 12 fold.
Fig. 1.
Time dependent fluorescence intensities (I) of 4 (λex=360 nm, λem = 465 nm) formed by CuAAC reaction of CPL (1 mg mL−1) with azide 3 (5 mM) in the presence of (A) 312.5 μM Cu2+, 6.25 mM ascorbic acid and 625 μM ligand 5, and (B) 2 mM Cu2+, 50 mM ascorbic acid.
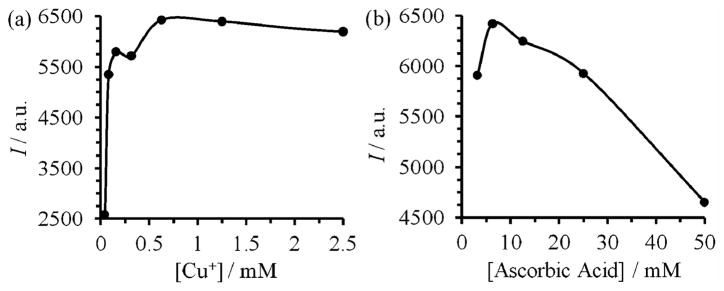
The effects of the concentrations of copper and ascorbic acid were then investigated, while keeping a constant CPL concentration (1 mg mL−1), a Cu+/ligand ratio of 1 : 213 and a reaction time of 1 h. As shown in Fig. 2a, the most efficient copper concentration was in the range of 300–600 μM. Below it, the reaction rate dropped steeply. This result was in agreement with the reported second order kinetics for copper concentrations in the sub-millimolar range.13 In the millimolar range, the yield of the reaction slowly decreased with copper concentration. At a fixed copper concentration (0.3 mM), the optimum concentration of ascorbic acid was around 6 mM (Fig. 2b). Although air oxidation of Cu+ was retarded by the ligand 5, an excess of ascorbic acid was still needed to reduce Cu2+ back to catalytically active Cu+. However, a high concentration of copper and ascorbic acid promotes the formation of peroxy radicals5b that decomposes not only the fluorescent product (as indicated by the decreased fluorescence in Fig. 2) but also the unsaturated liposomes via autooxidation.5,8 Indeed, under such conditions we observed leakage of dye encapsulated inside the CPL (ESI†).
Fig. 2.
Fluorescence intensities (I) of 4 (λex=360 nm, λem=465 nm) formed after 1 h reaction of CPL with the azide 3 in the presence of (a) 25 mM of ascorbic acid, CuSO4 with concentrations varied from 0.039 to 10 mM, and the ligand 5 with a 5/CuSO4 ratio of 2 : 1, and (b) 312.5 μM of CuSO4 and 625 μM of 5 and ascorbic acid with concentrations varied from 3.125 to 50 mM.
Overall, the ligand 5 and the optimized conditions greatly reduced the use of copper by 6.4 times (2.0 mM to 312.5 μM) and ascorbic acid by 8 times (50 mM to 6.25 mM). Our reaction conditions are far superior than the previously reported conditions6,7 and ensure a fast reaction (1 h vs. 6 h) utilizing smaller amounts of copper (0.3 mM vs. 2.3 mM) and ascorbic acid (6.25 mM vs. 50 mM). After the reaction, the copper species and ascorbic acid were easily removed by treatment with EDTA followed by filtering through a size exclusion column (PD SpinTrap™ G-25, GE Healthcare). Remarkably, under the optimized conditions we did not observe any decomposition of CPL at completion of the reaction (ESI†). The overall yield from the lipid 1 to the fluorescent liposome 4 (Schemes 2 and 3) was 23%. Note that half of the lipids are on the inner surface of the liposomal bilayer, and are thus not available for the reaction. Also, self-quenching of fluorescence by the dyes on the liposome may lead to an underestimation of the yield. Further, loss of material occurred during preparation of the CPL involving extrusion 20 times.
Efficient optimization of the peripheral composition on the CPL was demonstrated using the azides 6 and 714 (Scheme 4). The GRGD peptide in 6 targets the integrin receptors on a cell membrane, while the FITC dye in 7 serves as the imaging agent. After attaching 6 and 7 to the CPL via click reactions using the above optimized conditions, the resultant liposomes 8 were incubated with rat osteogenic cells (UMR-106) for 1 h. Fluorescence images showed that the liposomes bound to the cell membrane (Fig. 3). To optimize the fluorescence intensity on the targeted cells, the composition of the targeting and imaging agents on the liposome 8 was varied. This process was efficiently performed via the click reactions in a 96 well plate (Scheme 4) with varied concentrations of the GRGD azide 6 while keeping the concentrations of the azide 7 constant at 0.5 mM. After reaction for 1 h, the low molecular weight reagents were removed by centrifuge through size exclusion columns. The targeting efficiency of the liposomes was evaluated with an anti-adhesion assay. Thus, UMR-106 cells were incubated with the liposomes in a 96-well plate coated with vitronectin that binds the cells (ESI†). Competitive binding of the GRGD-presenting liposomes 8 to the cells reduced the cell attachment to the GRGD epitopes on the vitronectin. After staining with crystal violet, the amount of cells attached to the vitronectin was quantified by the absorbance (ESI†).15 Fig. 4a plots the absorbance vs. concentration of the GRGD azide 6 used to prepare the liposome 8. A decrease of the absorbance corresponds to an increase of the binding efficiency of the liposomes to the cells. Thus, the binding efficiency initially increased with the concentration of 6 below 0.15 mM. However, it saturated at a concentration above 0.2 mM, presumably because the density of GRGD on 8 had exceeded the density of GRGD-integrin binding sites on the cell. Fig. 4b plots the fluorescence signal intensities for imaging the immobilized UMR-106 cells using the liposomes 8 obtained with varied concentrations of the GRGD azide 6 (Scheme 3). Liposomes with a too low or too high ratio of targeting/imaging agents (6/7) exhibited a low imaging signal. A ratio of 6/7 higher than 1/10 was needed for efficient targeting of the cells. The optimum ratio of 6/7 ~2/5 for signal intensity coincided with the one for binding efficiency (see above). At a higher 6/7 ratio, the binding efficiency saturated while the fluorophore density decreased, thus decreasing the signal intensity.
Scheme 4.
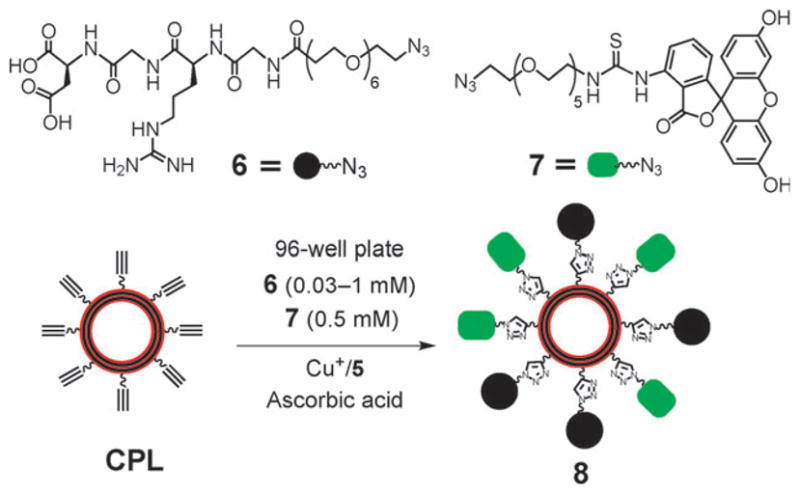
Attaching GRGD-N3 (6) as a targeting agent and FITCN3 (7) as an imaging agent to CPL to form the multifunctional liposomes 8 with varied ratios of GRGD and FITC in a 96 well plate.
Fig. 3.
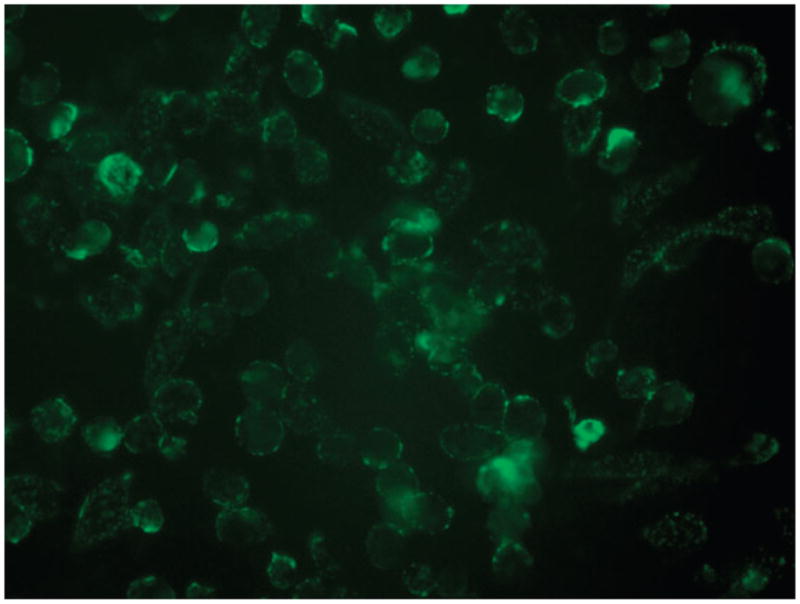
Fluorescence image of UMR-106 cells targeted by the liposome 8 carrying GRGD and FITC dye, obtained with a 60× objective.
Fig. 4.
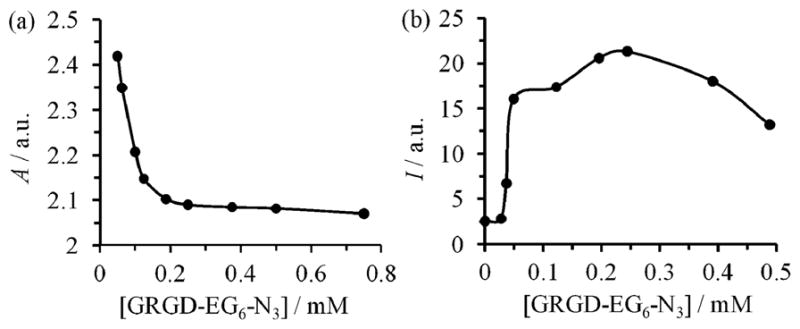
Measurement of efficiencies for the binding (a) and imaging (b) of UMR-106 cells with the liposome 8 prepared by click reaction of the CPL in 0.5 mM FITC-N3 (7) with various concentrations of the GRGD-EG6-N3 (6, Scheme 4). (a) Anti-adhesion assay plotting the absorbance (A) (λex 530 nm) of the crystal violet-stained cells attached to the vitronectin plate, which were not blocked by the liposome 8. (b) Fluorescence intensity (I) (λex 490 nm; λem 520 nm) of the immobilized cells after incubation with the liposomes 8 obtained at various concentrations of 6.
In summary, click reaction conditions and copper catalysts were optimized for CPL. The current platform allows rapid attachment of multiple azido-labeled functional molecules without decomposition of the liposomes. In addition, CPL is robust and can be stored for a long period of time. Therefore, CPL is a versatile and efficient platform for optimization of multifunctional liposomes with a wide range of compositions for targeted imaging in specific systems.
Supplementary Material
Acknowledgments
We thank the Welch Foundation (grant E-1498), National Science Foundation (grant DMR-0706627), the Institute of Biomedical Imaging Science and the National Institutes of Health (grant EY018303) for support of this work.
Footnotes
Electronic supplementary information (ESI) available: Experimental procedures for liposome modification, synthesis and characterization of all compounds used. See DOI: 10.1039/c0cc00784f
Notes and references
- 1.(a) Torchilin VP. Adv Drug Delivery Rev. 2006;58:1532–1555. doi: 10.1016/j.addr.2006.09.009. [DOI] [PubMed] [Google Scholar]; (b) Brandwijk R, Mulder WJM, Nicolay K, Mayo KH, Thijssen V, Griffioen AW. Bioconjugate Chem. 2007;18:785–790. doi: 10.1021/bc060316h. [DOI] [PubMed] [Google Scholar]; (c) Kluza E, van der Schaft DWJ, Hautvast PAI, Mulder WJM, Mayo KH, Griffioen AW, Strijkers GJ, Nicolay K. Nano Lett. 2010;10:52–58. doi: 10.1021/nl902659g. [DOI] [PubMed] [Google Scholar]; (d) Kamaly N, Kalber T, Thanou M, Bell JD, Miller AD. Bioconjugate Chem. 2009;20:648–655. doi: 10.1021/bc8002259. [DOI] [PubMed] [Google Scholar]; (e) Seo JW, Zhang H, Kukis DL, Meares CF, Ferrara KW. Bioconjugate Chem. 2008;19:2577–2584. doi: 10.1021/bc8002937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allen TM. Biochim Biophys Acta, Biomembr. 1981;640:385–397. doi: 10.1016/0005-2736(81)90464-8. [DOI] [PubMed] [Google Scholar]
- 3.Fendler JH. Science. 1984;223:888–894. doi: 10.1126/science.223.4639.888. [DOI] [PubMed] [Google Scholar]
- 4.Spevak W, Nagy JO, Charych DH, Schaefer ME, Gilbert JH, Bednarski MD. J Am Chem Soc. 1993;115:1146–1147. [Google Scholar]
- 5.(a) Rostovtsev VV, Green LG, Fokin VV, Sharpless KB. Angew Chem, Int Ed. 2002;41:2596. doi: 10.1002/1521-3773(20020715)41:14<2596::AID-ANIE2596>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]; (b) Hong V, Presolski SI, Ma C, Finn MG. Angew Chem, Int Ed. 2009;48:9879–9883. doi: 10.1002/anie.200905087. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Meldal M, Tornoe CW. Chem Rev. 2008;108:2952–3015. doi: 10.1021/cr0783479. [DOI] [PubMed] [Google Scholar]
- 6.Cavalli S, Tipton AR, Overhand M, Kros A. Chem Commun. 2006:3193–3195. doi: 10.1039/b606930d. [DOI] [PubMed] [Google Scholar]
- 7.Hassane FS, Frisch B, Schuber F. Bioconjugate Chem. 2006;17:849–854. doi: 10.1021/bc050308l. [DOI] [PubMed] [Google Scholar]
- 8.(a) Bittner O, Gal S, Pinchuk I, Danino D, Shinar H, Lichtenberg D. Chem Phys Lipids. 2002;114:81–98. doi: 10.1016/s0009-3084(01)00208-0. [DOI] [PubMed] [Google Scholar]; (b) Gal S, Lichtenberg D, Bor A, Pinchuk I. Chem Phys Lipids. 2007;150:186–203. doi: 10.1016/j.chemphyslip.2007.08.001. [DOI] [PubMed] [Google Scholar]; (c) Gal S, Pinchuk I, Lichtenberg D. Chem Phys Lipids. 2003;126:95–110. doi: 10.1016/s0009-3084(03)00096-3. [DOI] [PubMed] [Google Scholar]
- 9.Sivakumar K, Xie F, Cash BM, Long S, Barnhill HN, Wang Q. Org Lett. 2004;6:4603–4606. doi: 10.1021/ol047955x. [DOI] [PubMed] [Google Scholar]
- 10.Chan TR, Hilgraf R, Sharpless KB, Fokin VV. Org Lett. 2004;6:2853–2855. doi: 10.1021/ol0493094. [DOI] [PubMed] [Google Scholar]
- 11.Lewis WG, Magallon FG, Fokin VV, Finn MG. J Am Chem Soc. 2004;126:9152–9153. doi: 10.1021/ja048425z. [DOI] [PubMed] [Google Scholar]
- 12.Sen Gupta S, Kuzelka J, Singh P, Lewis WG, Manchester M, Finn MG. Bioconjugate Chem. 2005;16:1572–1579. doi: 10.1021/bc050147l. [DOI] [PubMed] [Google Scholar]
- 13.Rodionov VO, Presolski SI, Diaz DD, Fokin VV, Finn MG. J Am Chem Soc. 2007;129:12705–12712. doi: 10.1021/ja072679d. [DOI] [PubMed] [Google Scholar]
- 14.Santos CM, Kumar A, Zhang W, Cai CZ. Chem Commun. 2009:2854–2856. doi: 10.1039/b821148e. [DOI] [PubMed] [Google Scholar]
- 15.Montet X, Funovics M, Montet-Abou K, Weissleder R, Josephson L. J Med Chem. 2006;49:6087–6093. doi: 10.1021/jm060515m. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.