Abstract
Breast cancer metastasis suppressor 1 (BRMS1) has been reported to suppress metastasis without significantly affecting tumorigenicity in breast cancer and ovarian cancer. To investigate the role of BRMS1 in human melanoma progression and prognosis, we established tissue microarray and BRMS1 expression was evaluated by immunohistochemistry in 41 dysplastic nevi, 90 primary melanomas and 47 melanoma metastases. We found that BRMS1 expression was significantly decreased in metastatic melanoma compared with primary melanoma or dysplastic nevi (P=0.021 and 0.001, respectively, χ2 test). In addition, reduced BRMS1 staining was significantly correlated with American Joint Committee on Cancer stages (P=0.011, χ2 test), but not associated with tumor thickness, tumor ulceration and other clinicopathological parameters. Furthermore, BRMS1 expression was significantly correlated with disease-specific 5-year survival of melanoma patients (P=0.007, log-rank test). Multivariate Cox regression analysis revealed that BRMS1 staining was an independent prognostic factor for melanoma patients (relative risk=0.51; confidence interval =0.29–0.91; P=0.022). Moreover, we demonstrated that BRMS1 overexpression inhibited endothelial cell growth and tube formation ability by suppressing NF-κB activity and IL-6 expression in vitro. We also showed that knockdown of BRMS1 increased IL-6 expression and promoted endothelial cell growth and tube formation. In addition, our data revealed that the BRMS1-mediated IL-6 expression is dependent on NF-κB. Strikingly, our in vivo studies using nude mice confirmed that BRMS1 inhibited blood vessel formation and the recruitment of CD31-positive cells in matrigel plugs. Taken together, BRMS1 expression was decreased in metastatic melanomas, which resulted in deficient suppression of angiogenesis and contributed to melanoma progression. BRMS1 may serve an important prognostic marker and therapeutic target for melanoma patients.
Keywords: BRMS1, melanoma, tumor progression, prognosis, angiogenesis
Introduction
Cutaneous melanoma is the most lethal form of skin cancers and the incidence of melanoma has drastically increased during the past decades (Dauda and Shehu, 2005; Thompson et al., 2005). Although patients with low-risk primary melanoma can be completely cured by a successful operation, melanoma can spread to other organs very fast. Once metastatic melanoma develops, the conventional radio-, chemo- or immunotherapy are not effective. The 5-year survival rate for those patients with metastatic melanoma is only less than 5% (Grossman and Altieri, 2001; Ballo and Ang, 2003; Hersey, 2003; Soengas and Lowe, 2003; Tsao and Sober, 2005).
It is believed that metastasis is the major cause for the death of melanoma patients, and many factors are involved in the regulation of metastasis through diverse mechanisms (Kauffman et al., 2003). Among metastasis suppressors, breast cancer metastasis suppressor 1 (BRMS1) showed remarkable inhibition on metastasis in several models. The BRMS1 gene is located on chromosome 11q13.1-13.2 and comprised of 10 exons and 9 introns, spanning about 7 kb (Seraj et al., 2000b). The encoded BRMS1 protein is 246 amino acids long and carries two nuclear localization signals that target this protein to the nucleus (Samant et al., 2000; Seraj et al., 2000b). Another feature of BRMS1 protein is the large glutamic acid-rich region at the N-terminus, which is a putative acidic transcriptional transactivation domain and may stimulate transcription in various eukaryotic organisms (Struhl, 1995; Samant et al., 2000). Several metastasis-related genes were reported to be regulated by BRMS1, including epidermal growth factor receptor (Vaidya et al., 2008), osteopontin (Samant et al., 2007; Hedley et al., 2008), CXC chemokine receptor 4 (Yang et al., 2008), as well as microRNA-146 (Hurst et al., 2009). BRMS1 was also reported to interact with mSin3 chromatin remodeling complex and recruit histone deacetylases to suppress downstream gene expression (Meehan et al., 2004). Cicek et al. (2005, 2009) showed that BRMS1 physically interacts with RelA/p65 subunit of NF-κB and inhibits phosphorylation of IκBα, and thus negatively regulates NF-κB pathway. Through these mechanisms, BRMS1 has been shown to inhibit metastasis in xenograft models of breast cancer, melanoma and ovarian carcinoma (Seraj et al., 2000b; Shevde et al., 2002; Zhang et al., 2006). To further investigate the role of BRMS1 in melanoma progression and prognosis, we used tissue microarray technology and immunohistochemistry to evaluate BRMS1 expression in different stages of human melanocytic lesions. Our data demonstrated that BRMS1 expression was significantly decreased in melanoma metastases when compared with primary melanomas, and reduced BRMS1 staining was significantly correlated with American Joint Committee on Cancer (AJCC) stages and a worse survival of melanoma patients. In this study, we also showed that BRMS1 expression negatively regulated the growth and tube formation of endothelial cells in vitro, through suppressing NF-κB activity and IL-6 expression. Our in vivo data further demonstrated that BRMS1 exerted the inhibitory effect in blood vessel formation and CD31 positive cell recruitment in matrigel plugs.
Results
Decreased BRMS1 expression correlated with melanoma metastasis
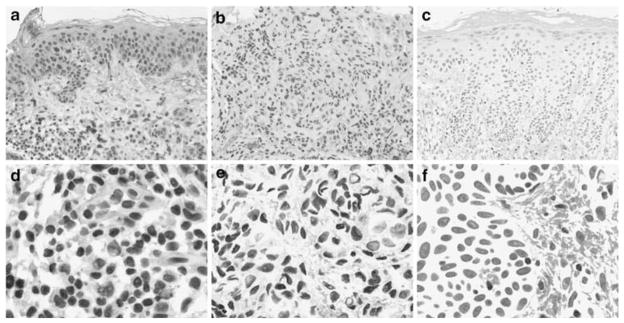
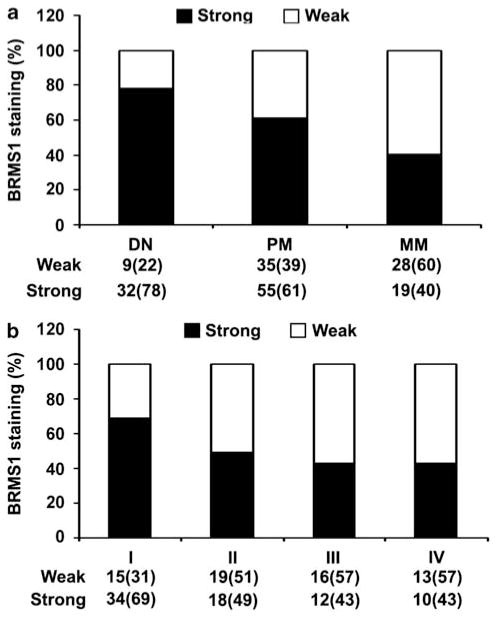
The clinicopathological features of 137 melanoma biopsies examined in this study were summarized in Table 1. Various levels of BRMS1 staining were observed in different melanocytic lesions, but the staining was generally located in the nucleus, which was attributed to the two nuclear localization signals that BRMS1 gene carries (Seraj et al., 2000b) (Figure 1). Strong BRMS1 staining decreased from 78% in dysplastic nevi to 61% in primary melanomas and further decreased to 40% in metastatic melanomas (Figure 2a). Significant differences for BRMS1 staining pattern were observed between metastatic melanoma and primary melanoma, or between metastatic melanoma and dysplastic nevi (P=0.021 and 0.001, respectively, χ2 test). However, there was no significant difference in BRMS1 staining between primary melanoma and dysplastic nevi.
Table 1.
Clinicopathologic characteristics of 137 melanomas
| Variables | No. of patients | % |
|---|---|---|
| Primary melanoma | ||
| Age | ||
| ≤58 | 44 | 49 |
| >58 | 46 | 51 |
| Gender | ||
| Male | 56 | 62 |
| Female | 34 | 38 |
| Tumor thickness | ||
| ≤2.0mm | 57 | 63 |
| >2.0mm | 33 | 37 |
| Ulceration | ||
| Absent | 72 | 80 |
| Present | 18 | 20 |
| Tumor subtype | ||
| Superficial spreading melanoma | 39 | 43 |
| Lentigo maligna melanoma | 16 | 18 |
| Nodular melanoma | 13 | 14 |
| Unspecified | 22 | 25 |
| Sitea | ||
| Sun-exposed | 18 | 20 |
| Sun-protected | 72 | 80 |
| Metastatic melanoma | ||
| Age | ||
| ≤58 | 23 | 49 |
| >58 | 24 | 51 |
| Gender | ||
| Male | 32 | 68 |
| Female | 15 | 32 |
| American joint committee on cancer stage | ||
| I | 49 | 36 |
| II | 37 | 27 |
| III | 28 | 20 |
| IV | 23 | 17 |
Sun-protected sites: trunk, arm, leg and foot; Sun-exposed sites: head and neck.
Figure 1.
Representative images of BRMS1 staining in human melanocytic lesions. (a, d) Dysplastic nevus with strong BRMS1 staining. (b, e) Metastatic melanoma with weak BRMS1 staining. (c, f) Negative control without the BRMS1 primary antibody. Magnification ×100 for a, b, c and ×400 for d, e, f.
Figure 2.
BRMS1 expression was correlated with melanoma progression. (a) Significant differences for BRMS1 staining pattern were observed between metastatic melanoma (MM) and primary melanoma (PM) (P=0.021, χ2 test), or between metastatic melanoma and dysplastic nevi (DN) (P=0.001, χ2 test). (b) BRMS1 expression was significantly decreased with the progression of AJCC stages in 137 melanomas. (P=0.011, χ2 test).
Decreased BRMS1 expression correlated with AJCC stage
In all 137 melanoma patients, we found that strong BRMS1 expression significantly decreased from 69% in AJCC stage I to 49% in stage II, and further deceased to 43% at stage III and IV (P=0.011, χ2 test; Figure 2b). However, we did not find any correlation between BRMS1 staining and age or gender in patients with primary melanoma or metastatic melanoma. In addition, we also assessed the correlations between BRMS1 expression and other clinicopathological parameters in primary melanoma patients, including tumor thickness, ulceration, histological subtype and anatomic site. We found no significant correlation between BRMS1 staining and these parameters.
Decreased BRMS1 expression correlated with a worse patient survival
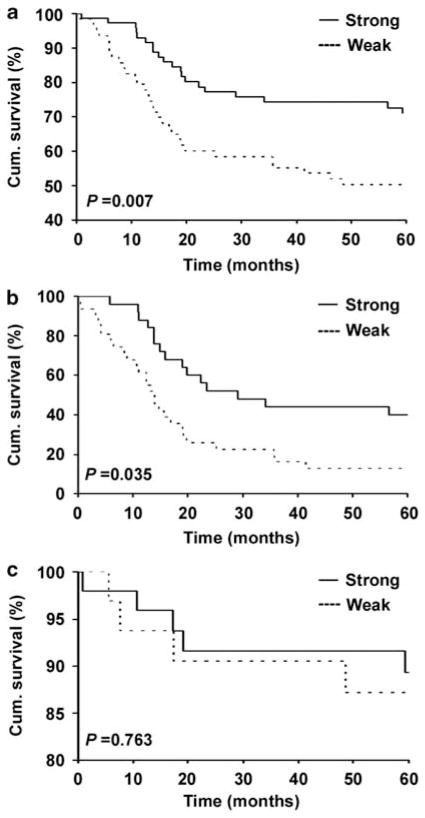
To study whether reduced BRMS1 expression is correlated with worse melanoma patient survival, we constructed Kaplan–Meier survival curves. Our data revealed that lower BRMS1 staining was significantly correlated with a poorer disease-specific 5-year survival of all melanoma patients. The survival rate dropped from 73%, in patients with strong BRMS1 expression, to 50% in those with weak BRMS1 expression (Figure 3a). Next, we examined whether BRMS1 expression was an independent marker for melanoma prognosis. We performed Multivariate Cox regression analysis to study the effect of BRMS1 expression in patient survival, together with AJCC stage as well as patient’s age and gender in all 137 melanomas. Our results indicated that BRMS1 expression was an independent prognostic factor for disease-specific 5-year survival (Table 2). As there is no reliable prognostic marker for advanced melanoma, we further investigated the correlation between BRMS1 staining and disease-specific 5-year survival in metastatic melanoma and late-stage primary melanoma with high metastatic potential (tumor thicker than 4mm with ulceration). We found that strong BRMS1 expression was significantly associated with better 5-year survival in this group of patients. The survival rate decreased from 40% in patients with strong BRMS1 expression to 12.9% in those with weak BRMS1 expression (Figure 3b). However, BRMS1 was not significantly correlated with 5-year patient survival of melanomas with low metastatic potential (≤4mmthick and>4mm without ulceration) (Figure 3c).
Figure 3.
Correlation between BRMS1 expression and 5-year disease-specific survival of melanoma patients. (a) Survival of all (primary plus metastatic) melanoma patients. (b) Survival of patients with metastatic melanoma and late stage primary melanoma. (c) Survival of primary melanomas with low metastatic potential. Cum., cumulative.
Table 2.
Multivariate Cox regression analysis of BRMS1 and AJCC stages on 5-year patient survival in 137 melanomas
| Variablea | Relative risk | 95%CI | P |
|---|---|---|---|
| BRMS1 | 0.51 | 0.29–0.91 | 0.02 |
| American joint committee on cancer stage | 4.34 | 2.38–8.33 | <0.01 |
| Age | 1.28 | 0.71–2.30 | 0.41 |
| Gender | 1.30 | 0.71–2.40 | 0.40 |
Abbreviation: CI, confidence interval.
Coding of variables: BRMS1 expression was coded as 1, weak; and 2, strong. AJCC stage was coded as 1, stage I–III; and 2, stage IV. Age was coded as 1, ≤58 years; and 2, >58 years. Gender was coded as 1, male; and 2, female.
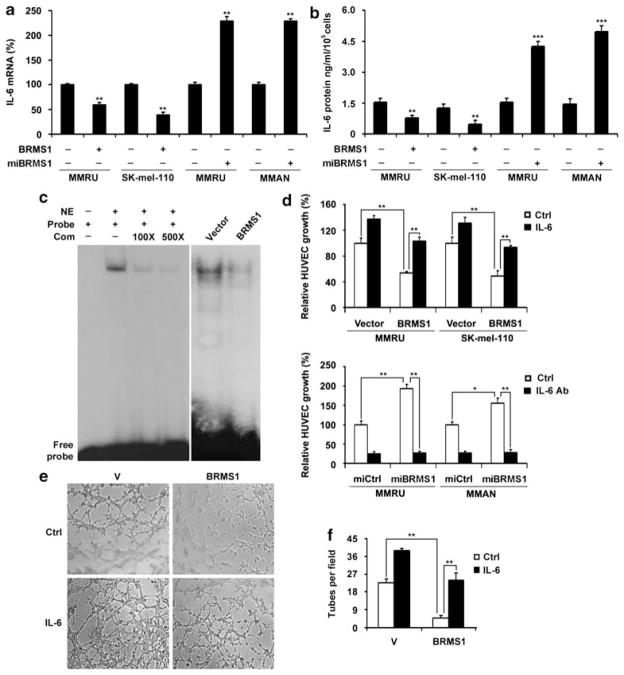
Expression of BRMS1 in melanoma cells inhibited growth and tube formation of human umbilical vein endothelial cells
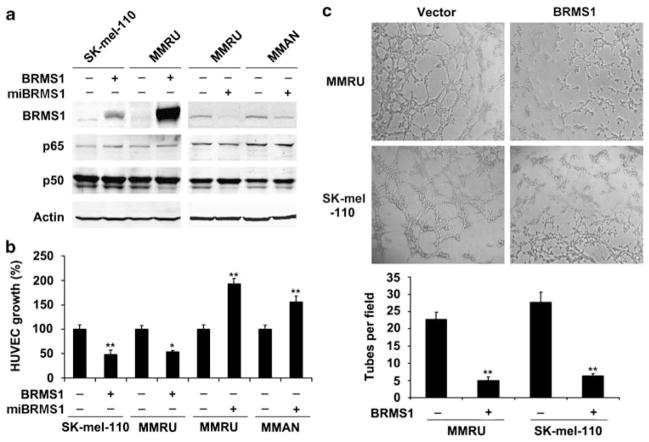
To test the effect of BRMS1 expression in melanoma cells on human umbilical vein endothelial cells (HUVECs) growth and tube formation, we overexpressed BRMS1 in MMRU and SK-mel-110 cells or knocked down BRMS1 in MMRU and MMAN cells. The expression of BRMS1 was then tested by both semi-quantitative PCR (Supplementary Figure S1) and western blot (Figure 4a). The expressions of p65 or p50 subunit of NF-κB were not affected by the manipulation of BRMS1 expression (Figure 4a). Then the conditioned medium was collected from melanoma cells and applied to either HUVEC growth assay or tube-formation assay. The growth of HUVECs in conditioned medium from BRMS1-overexpressing MMRU and SK-mel-110 cells was inhibited by 48 and 51%, respectively, when compared with the corresponding vector control (Figure 4b). BRMS1 knockdown in MMRU and MMAN promoted the HUVECs growth by 1.85- and 1.56-fold, respectively, compared with the control (Figure 4b). The average number of complete tubular structures formed by HUVECs was significantly decreased in conditioned medium from BRMS1-overexpressing MMRU and SK-mel-110 compared with vector controls (Figure 4c).
Figure 4.
Expression of BRMS1 in melanoma cells negatively regulated HUVECs growth and tube formation. (a) BRMS1 overexpression in MMRU and SK-mel-110 cells and knockdown in MMRU and MMAN cells were confirmed by western blot. (b) BRMS1 overexpression in melanoma cells inhibited HUVECs growth, whereas BRMS1 knockdown promoted HUVECs growth. Data were presented as means±s.d. from three independent experiments. (c) BRMS1 overexpression in melanoma cells inhibited HUVECs tube formation. Numbers of tubes formed per field were counted in five random fields for BRMS1-overexpressing and control groups (n=3/group). *P<0.05, **P<0.01; Student’s t-test.
BRMS1 inhibited IL-6 expression through suppressing NF-κB activity
We previously reported that NF-κB p50 subunit enhanced melanoma angiogenesis through upregulating IL-6 at both transcriptional and protein level (Karst et al., 2009). Therefore, we next investigated whether BRMS1 also exerts its inhibitory effect on melanoma angiogenesis through NF-κB/IL-6 pathway. Our data revealed that BRMS1 overexpression in MMRU and SK-mel-110 cells decreased IL-6 messanger (m)RNA expression by 45% and 41%, respectively, compared with vector control (Figure 5a). In contrast, BRMS1 knockdown in MMRU and MMAN cells resulted in elevated IL-6 mRNA level by 2.3-fold for both (Figure 5a). Furthermore, we found that ectopic expression of BRMS1 in MMRU and SK-mel-110 cells decreased the secreted IL-6 protein in the medium from 1.6 and 1.2 ng/ml/105 cells to 0.7 and 0.4 ng/ml/105 cells, respectively (Figure 5b). On the other hand, the IL-6 protein was increased by 2.8- and 3.5-fold in conditioned medium from BRMS1 knockdown MMRU and MMAN cells, respectively (Figure 5b). To explore whether BRMS1 regulates IL-6 expression through NF-κB pathway, we performed electrophoretic mobility shift assay and demonstrated that BRMS1 overexpression dramatically decreased the DNA binding activity of NF-κB p65 subunit to IL-6 promoter, whereas the unlabeled competitive sequence markedly inhibited this binding in a dosage-dependent manner (Figure 5c). To confirm the role of IL-6 in BRMS1-regulated melanoma angiogenesis, we performed IL-6 rescue and IL-6 blocking assays. The addition of 0.8 ng/ml recombinant IL-6 to BRMS1-overexpressing MMRU and SK-mel-110 cells rescued the cell growth of HUVECs to the similar level of the corresponding vector control cells, whereas the application of sufficient IL-6 antibody abrogated the elevated HUVECs growth by BRMS1-knockdown in MMRU and MMAN cells (Figure 5d). We also performed the HUVEC tube-formation assay after addition of IL-6 to the conditioned medium from BRMS1-overexpressing and vector-control MMRU cells. We found that the inhibited tubular structure formation can be rescued by addition of IL-6 (Figures 5e and f). A parallel tube-formation assay after addition of IL-6 was conducted in another cell line, SK-mel-110, and similar result was obtained (Supplementary Figure S2).
Figure 5.
BRMS1 expression in melanoma cells inhibited IL-6 expression and suppressed NF-κB activity. (a, b) BRMS1 suppresses IL-6 expression. MMRU and SK-mel-110 cells were transiently transfected with vector or Flag-BRMS1 plasmid for 48 h. In a parallel experiment, MMRU and MMAN cells were transfected with vector control or miBRMS1 plasmid for 72 h. IL-6 mRNA in the cells and secreted protein level in conditioned medium were determined with qRT–PCR (a) and ELISA (b). Data were presented as means±s.d. from three independent experiments. (c) BRMS1 overexpression suppressed the binding activity of NF-κB p65 subunit to IL-6 promoter. Competitive sequence inhibited the binding reaction in a dosage-dependent manner. (d) Addition of IL-6 recombinant protein (0.8 ng/ml) to the conditioned medium rescued BRMS1-suppressed endothelial cell growth, whereas application of IL-6 antibody (320 ng/ml) to neutralize the IL-6 in conditioned medium abrogated BRMS1 knockdown-enhanced endothelial cell growth (n=3/group). (e) Addition of IL-6 to conditioned medium rescued the tube formation inhibited by BRMS1 overexpression. (f) The numbers of tubes formed per field were counted for BRMS1-overexpressing and control groups in five random fields. *P<0.05, **P<0.01, ***P<0.001, Student’s t-test. Com, competitive sequence; NE, nuclear extract.
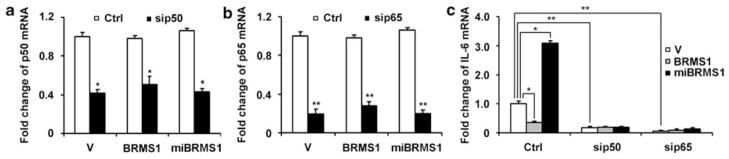
BRMS1-mediated IL-6 expression was dependent on p65 and p50 subunits of NF-κB
To investigate whether the regulation of IL-6 expression by BRMS1 is dependent on NF-κB, we overexpressed or knocked down BRMS1 together with or without silencing of NF-κB p65 or p50 subunit in MMRU cells. The mRNA expression of p65 and p50 was successfully decreased by 75 and 60% by specific small interfering RNA sequences (Figures 6a and b). However, overexpression or knockdown of BRMS1 did not change p50 or p65 expression. Next, we examined the impact of BRMS1, p65 or p50 on IL-6 mRNA. Our data revealed that knockdown of either p50 or p65 decreased IL-6 expression drastically (Figure 6c). The concomitant overexpression of BRMS1 together with silencing of p50 or p65 did not further reduce the IL-6 mRNA expression than knockdown of p50 or p65 alone (Figure 6c). Our data also demonstrated that p50 or p65 knockdown abrogated the elevated IL-6 mRNA expression by BRMS1 knockdown (Figure 6c), suggesting that the suppressive effect of BRMS1 on IL-6 expression is dependent on NF-κB. Similar results for the dependency of BRMS1-regulated IL-6 expression on NF-κB was also obtained in another cell line, SK-mel-110 (Supplementary Figure S3).
Figure 6.
BRMS1-mediated IL-6 expression in MMRU cells was dependent on p65 and p50 subunits of NF-κB. MMRU cells were transiently transfected with p50 or p65 siRNA for 24 h, then transfected with vector control, Flag-BRMS1 or miBRMS1 plasmid for 48 h. Total RNA was extracted and subjected to qRT–PCR. (a) p50 mRNA level. (b) p65 mRNA level. (c) IL-6 mRNA level. Data were presented as means±s.d. from three independent experiments. *P<0.05, **P<0.01; Student’s t-test.
Expression of BRMS1 in melanoma cells inhibited angiogenesis in vivo
We performed the in vivo matrigel plug assay to investigate whether BRMS1 expression could inhibit the new blood vessel formation in a mouse model. Visual examination revealed obviously less vascularization in matrigel plugs containing BRMS1-overexpressing MMRU cells than the control plugs (Figure 7a). CD31 staining demonstrated that the control plugs contained much denser neovessels with a sevenfold higher number of CD31-positive cells compared with BRMS1-overexpressing plugs (Figures 7b and c). We then investigated the level of tumor-derived IL-6 in those matrigel plugs. Quantitative RT–PCR data demonstrated that the IL-6 mRNA level was significantly decreased by 61% in matrigel plugs containing BRMS1-overexpressing MMRU cells compared with the control plugs (Figure 7d).
Figure 7.
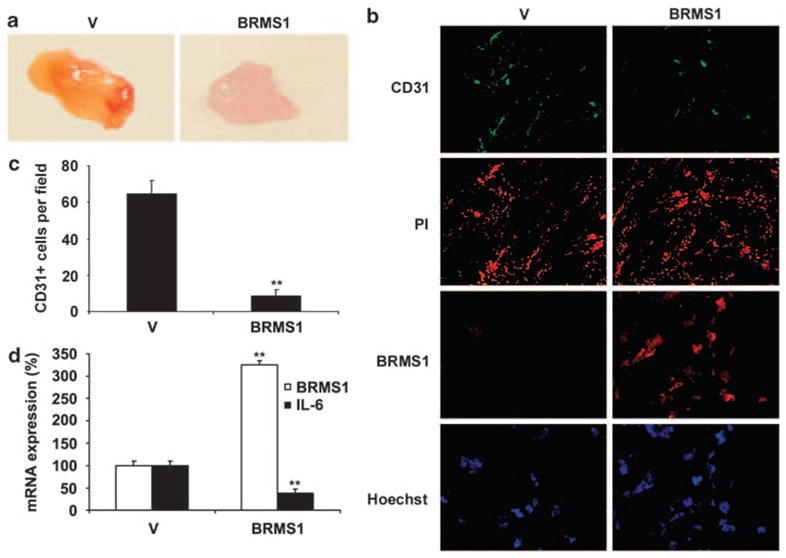
BRMS1 overexpression in melanoma cells inhibited blood vessels formation in vivo. (a) Photographs of matrigel plugs excised from mice after 10 days of growth in vivo. (b) The extent of host angiogenesis was examined by immunofluorescent staining for expression of CD31 and BRMS1 in matrigel plugs containing BRMS1-overexpressing or control MMRU cells. Propidium iodide and Hoechst nuclear staining indicated the overall cell density in each matrigel plug. (c) The number for CD31-positive cells was counted from five random fields for both vector control and BRMS1-overexpressing groups. (d) The expression of BRMS1 and IL-6 mRNA was examined in matrigel plugs (n=3/group). **P<0.01; Student’s t-test.
Discussion
BRMS1 was first identified by Seraj et al. (2000b) while studying the nonrandom amplifications and deletions in chromosome 11 using differential display. In this study, we investigated the role of BRMS1 in melanoma progression and the relevant mechanisms. Our data demonstrated that reduced BRMS1 expression was significantly correlated with melanoma metastasis, AJCC stage and poor patient survival. Further experiments revealed that BRMS1 inhibited endothelial cell growth and tube formation by decreasing IL-6 expression through suppressing NF-kB activity in vitro, as well as inhibited neovessels formation in mice in vivo. To our knowledge, this is the first study to examine the expression pattern of BRMS1 and its association with patient survival in a cohort of melanoma patients. It is also the first report on the role of BRMS1 in tumor angiogenesis.
We found a significant difference in BRMS1 expression between primary melanoma and metastatic melanoma, but not between dysplastic nevi and primary melanoma (Figure 2a), suggesting that loss of BRMS1 expression was mainly involved in melanoma metastasis instead of tumor growth. This is consistent with the findings that BRMS1 mRNA expression level was high in melanocytes, but barely detectable in metastatic melanoma cell lines (Shevde et al., 2002). Our (real-time) RT–PCR results also demonstrated reduced BRMS1 in melanoma cell lines compared with normal melanocytes (Supplementary Figure S4). It has been shown that re-introduction of BRMS1 into highly metastatic melanoma cell line C8161.9 significantly suppressed the metastatic potential in both experimental and spontaneous metastasis assays without affecting tumor growth after orthotopic injection (Shevde et al., 2002). The effect of BRMS1 to inhibit metastasis was also confirmed in other cancer models. BRMS1 was expressed fivefold higher in the metastasis-suppressed hybrid neo11/435 cells when compared with the highly metastatic breast-cancer cell line MDA-MB-435. Furthermore, BRMS1 transfected MDA-MB-435 cells showed significantly decreased incidence and number of metastases to the lung and regional lymph nodes when cells were injected orthotopically. However, the tumor growth rate of BRMS1-transfected MDA-MB-435 cells is similar to their parental controls, except a delay in growth for 1 week (Seraj et al., 2000b). BRMS1 was also shown to be expressed at lower level in a highly metastatic human bladder carcinoma cell line T24T compared with the less metastatic parental cell line T24 (Seraj et al., 2000a). The reduced expression of BRMS1 in metastatic melanoma can possibly be explained by the theory that deletion at chromosome 11q, which includes the BRMS1 gene, occurs at very high frequency in various human cancers (Welch and Goldberg, 1997; Meehan and Welch, 2003; Zainabadi et al., 2005). Another study recently reported that the methylation of BRMS1 promoter might account for the loss of BRMS1 in breast cancer cells. The same study also demonstrated a direct correlation between methylation status of BRMS1 promoter in the DNA isolated from those tissues with loss of BRMS1 by immunohistochemistry, concomitantly (Metge et al., 2008). These studies imply that the loss of the BRMS1 expression may be a common event in tumor metastasis.
We demonstrated that BRMS1 inhibited melanoma angiogenesis and this may at least partially explain the correlation between reduced BRMS1 expression and melanoma metastasis. A series of studies indicated angiogenesis was closely related to tumor metastasis in various tumors, including breast, lung, prostate, head and neck, as well as melanoma (Weidner et al., 1991; Weidner, 1995; Czubayko et al., 1996). Therefore, the deficient suppression on angiogenesis by loss of BRMS1 will be critical for melanoma to metastasize. In this study, we found that BRMS1 inhibited HUVECs growth and tube formation in vitro (Figures 4b and c, Supplementary Figure S2). We previously reported that p50 subunit of NF-κB increased IL-6 expression and melanoma angiogenesis (Karst et al., 2009). It has also been shown that BRMS1 suppressed NF-κB activity and thus regulated NF-κB downstream gene expression (Cicek et al., 2005). Therefore, we next investigated whether BRMS1 inhibits endothelial cell growth through NF-κB/IL-6 pathway. Further experiments showed that BRMS1 did not affect the expression level of p65 or p50 subunits of NF-κB (Figure 4a), but inhibited NF-κB DNA-binding activity (Figure 5c). BRMS1 overexpression decreased, whereas BRMS1 knockdown increased both IL-6 mRNA expression and protein secreted in the conditioned medium (Figures 5a and b). The role of IL-6 in BRMS1-regulated melanoma angiogenesis was further confirmed by IL-6 rescue and blocking assays (Figures 5d–f). These results indicated that the compensation of IL-6 loss caused by BRMS1 overexpression can neutralize the inhibitory effect of BRMS1, while blocking IL-6 activity by incubating with the IL-6 antibody can mimic the BRMS1 overexpression impact. Therefore, the reduction of IL-6 protein secreted by melanoma cells accounted for the inhibitory effect of BRMS1 on melanoma angiogenesis. To explore whether BRMS1 regulates IL-6 expression through NF-κB pathway, we concomitantly manipulated the expression of BRMS1 and p65 or p50 subunits of NF-κB, and then tested the expression of IL-6. We found that both overexpression of BRMS1 and knockdown of p65 or p50 alone decreased IL-6 mRNA expression, but combination of BRMS1 overexpression and p65 or p50 knockdown did not bring extra inhibitory effect. Silencing of BRMS1 caused higher IL-6 expression, which was abrogated by the knockdown of p65 or p50. These results indicated that BRMS1 inhibits IL-6 expression by suppressing NF-κB activity. Moreover, we showed that BRMS1 inhibited new blood vessels formation in mice in vivo (Figures 7a–c), and quantitative RT–PCR result further revealed that the BRMS1 overexpression was still present and IL-6 expression was decreased in matrigel plugs overexpressing BRMS1, indicating the consistency between our in vitro and in vivo data. Our results highlighted a novel role of BRMS1 in regulating tumor angiogenesis, which is critically important, as neovascularization is one of the essential steps required by tumor metastasis and progression (Folkman, 2002). Recently, Dai et al. (2009) showed that osteopontin induces angiogenesis by activating the PI3K/AKT pathway in human endothelial cells. Similar to IL-6, osteopontin is a secreted protein and also regulated by NF-κB (Diao et al., 2004; Standal et al., 2004). It was reported that BRMS1 inhibited the transcription of osteopontin by suppressing NF-κB activity (Samant et al., 2007). Whether osteopontin is indeed involved in BRMS1-mediated melanoma angiogenesis remains to be investigated.
We found that reduced BRMS1 staining was correlated with AJCC stages (Figure 2b), but not with other clinicopathological parameters. AJCC melanoma stage is evaluated by taking melanoma tumor thickness, ulceration, regional lymph node metastasis and distant metastasis into account and thus it is an integrated measure of the progression status and outcome of human melanoma patients (Balch et al., 2001). It is not surprising to see that BRMS1 expression is inversely correlated with AJCC stage because reduced BRMS1 expression showed close correlation with melanoma metastasis in this study. Furthermore, in primary melanoma patient group, BRMS1 expression showed significant difference between AJCC stages I and II (Figure 2b), but did not change significantly between thin melanoma (≤2.0mm) and thick melanoma (>2.0mm), or between melanoma with and without ulceration. These data suggested that BRMS1 staining was associated with the combination of tumor thickness and ulceration, which may be a better marker for melanoma metastatic potential than tumor thickness or ulceration alone.
Reduced BRMS1 expression was significantly correlated with melanoma metastasis in our study and with metastasis of breast and ovarian carcinomas from previous reports (Seraj et al., 2000b; Shevde et al., 2002; Zhang et al., 2006). Therefore, reduced BRMS1 expression is expected to correlate with poorer survival of melanoma patients because of the fact that metastasis is the major cause for the death of melanoma patients. Moreover, we found that strong BRMS1 expression is significantly correlated with better prognosis in those patients with thick (>4 mm) ulcerated primary melanoma and metastatic melanoma. As it was reported that both thick melanoma (≥4 mm) and presence of ulceration were associated with a high incidence of regional and distant metastasis (Mansfield et al., 1994; Ostmeier et al., 1999), our data suggested that BRMS1 could be one of the few prognostic markers for melanoma patients with existing metastases or with high metastatic potential. Possible explanations for the correlation between BRMS1 expression and better prognosis in this group of patients may be because of the inhibitory effect of BRMS1 on melanoma angiogenesis. Accumulated evidences nowadays indicated that the extent of angiogenesis is correlated with patient survival in many different cancers. The first study in this line revealed that vascular density functions as an independent prognostic marker in breast cancer (Weidner et al., 1991). This correlation was then extended to other types of malignancies including carcinoma of the prostate (Weidner et al., 1993; Brawer, 1996), lung (Yamazaki et al., 1994; Angeletti et al., 1996), stomach (Maeda et al., 1995), cervix (Wiggins et al., 1995), ovary (Hollingsworth et al., 1995) and head and neck squamous cell carcinoma (Gasparini et al., 1993), suggesting that angiogenesis is an indicator of both increased metastasis and decreased survival. Furthermore, our multivariate Cox regression analysis indicated that the correlation between reduced BRMS1 expression and worse patient survival was independent of age, gender and AJCC stage of the patients (Table 2), further underscoring the important role of BRMS1 in melanoma prognosis. Combined with the previous reports on the crucial metastasis suppressive functions of BRMS1, our findings implied that BRMS1 might serve as a promising prognostic marker for melanoma and restoration of BRMS1 may provide a novel strategy for the treatment of metastatic melanoma.
Materials and methods
Patients
Formalin-fixed and paraffin-embedded biopsies were obtained from the 1990–1998 archives of the Department of Pathology at Vancouver General Hospital. The use of human skin tissues in this study was approved by the Clinical Research Ethics Board of University of British Columbia and was performed in accordance with the Declaration of Helsinki guidelines. A total of 41 dysplastic nevi, 90 primary melanomas and 47 metastatic melanomas were evaluated for BRMS1 staining. Clinicopathological data were available for all melanoma cases.
Tissue microarray construction, immunohistochemistry and evaluation of immunostaining
Tissue microarray construction and immunohistochemistry staining were performed as described previously (Dai et al., 2005). We selected and marked the most representative tumor area on the hematoxylin and eosin-stained slide. The primary mouse anti-BRMS1 antibody (1:200 dilution) (Hicks et al., 2006) and the biotin-labeled secondary antibody (DAKO Diagnostics, Glostrup, Denmark) were used. Negative controls were included following the same procedure of test samples, except that BRMS1 antibody was omitted from the primary antibody incubation. The staining intensity and percentage of BRMS1-positive cells were evaluated in a blinded manner by three independent observers (including a dermatopathologist) simultaneously, and a consensus score was reached for each core. BRMS1 staining intensity was scored as 0–3 (0, negative; 1, weak; 2, moderate; 3, strong). The percentage of BRMS1-positive cells was scored into four categories: 1 (0–25%), 2 (26–50%), 3 (51–75%) and 4 (76–100%). In the cases with a discrepancy between duplicated cores, the higher score from the two tissue cores was taken as the final score. The level of BRMS1 staining was evaluated by immunoreactive score (Remmele and Stegner, 1987), which is calculated by multiplying the scores of staining intensity and the percentage of positive cells. On the basis of IRS, BRMS1 staining pattern was defined as: weak (IRS: 0–6) and strong (IRS: 8–12).
Statistical analyses
The SPSS version 11.5 software (SPSS, Chicago, IL, USA) was used for the statistical analysis and all tests of statistical significance were two-sided. We used the χ2 test to compare the BRMS1 staining level in different melanocytic lesions, as well as the correlation between BRMS1 staining and the clinicopathological parameters of the melanoma patients, including AJCC stage, age, gender, tumor thickness, ulceration, histological subtype and tumor location. The Kaplan–Meier survival curve and log-rank test were used to evaluate the correlations between BRMS1 expression and patient survival. Finally, a Cox regression model was used for multivariate analysis. P<0.05 was considered statistically significant.
Cell culture and transfection
Human melanoma MMRU and MMAN cell lines are kind gifts from Dr HR Byers (Boston University, Boston, MA, USA), SK-mel-110 cell line is kindly provided by Dr AP Albino (Memorial Sloan-Kettering Cancer Center, NY, USA). All melanoma cell lines were cultured in Dulbecco’s Modified Eagle’s Medium supplemented with 10% fetal bovine serum (Invitrogen, Burlington, Ontario, Canada). HUVECs were cultured in Kaighn’s Modified Ham’s F-12K medium (Media-tech, Manassas, VA, USA) supplemented with endothelial cell growth supplement (BD Biosciences, Mississauga, Ontario, Canada) and 10% fetal bovine serum. All cells were maintained in 5% CO2 atmosphere at 37 °C. Melanoma cells were grown to ~50% confluency and then transiently transfected with vector control, or Flag tagged BRMS1, or BLOCK-iT vector expressing microRNA-like sequences targeting BRMS1 (miBRMS1) with Effectene reagent (Qiagen, Mississauga, Ontario, Canada) according to the manufacturer’s instructions. small interfering RNA targeting NF-κB p65 and p50 subunits were synthesized (Qiagen) and were transfected overnight using SiLentFect (Bio-Rad, Mississauga, Ontario, Canada) according to the users manual. At 12 h after transfection, the medium containing transfection reagents was removed. The cells were rinsed with PBS and then incubated in fresh medium.
HUVEC growth and tube-formation assay
Melanoma cells were cultured in 60mm plate with fresh complete medium for 24 h, and 2ml of conditioned medium was collected. For HUVEC growth assay, the endothelial cells were seeded in a 24-well plate at 2×104 cells/well and cultured in fresh F-12K medium for 24 h, and then in 0.5 ml conditioned medium for another 24 h before sulforhodamine B staining was performed as previously described (Li et al., 1998). Basically the cells were fixed in 10% trichloroacetic acid before sulforhodamine B was applied for staining. The sulforhodamine B was then discarded and the plates were washed by 1% acetic acid and followed by dissolving and collecting sulforhodamine B dye in 10mM Tris (pH 10.5). For tube-formation assay, the 96-well plate was coated with Matrigel (BD Biosciences) and kept at 37 °C for 2 h. Then, 2×104 HUVECs were suspended in 100 μl conditioned medium and applied to the pre-coated 96-well plate. After incubation at 37 °C for another 24 h, photos were taken under microscope and the tubular structures formed in the matrigel were counted. For IL-6 rescue experiments, we applied 0.8 ng/ml recombinant IL-6 (eBioscience, San Diego, CA, USA) to the conditioned medium and conducted the HUVECs growth assay and tube-formation assay. For IL-6 blocking assay, we added 320 ng/ml IL-6 antibody (eBioscience) in the conditioned medium and performed HUVECs growth assay.
RT–PCR, western blot and enzyme-linked immunosorbent assay
Total RNA was extracted using TRIzol (Invitrogen). RT–PCR reactions were performed in triplicate with SYBR Green PCR Master Mix and using a 7900HT quantitative PCR system thermal cycler (Applied Biosystems, Foster City, CA, USA). For western blot, nuclear extracts were prepared as described previously (Garate et al., 2007) and then were separated on 12% SDS–PAGE gels and electroblotted onto polyvinylidene difluoride membranes (Bio-Rad). Membranes were blotted with the following primary antibodies: monoclonal mouse anti-Flag (Applied Biological Materials, Richmond, British Columbia, Canada), monoclonal mouse anti-BRMS1 (Phadke et al., 2008), polyclonal rabbit anti-p65 and p50 (Santa Cruz Biotechnology, Santa Cruz, CA, USA) and monoclonal mouse anti-Actin (Sigma-Aldrich, Oakville, Ontario, Canada). Signals were detected using an Odyssey infrared imaging system (LI-COR Biosciences, Lincoln, NE, USA). For enzyme-linked immunosorbent assay, the secreted IL-6 protein level in the conditioned medium was measured by a human IL-6 enzymelinked immunosorbent assay kit from eBioscience according to the manufacturer’s instructions.
Electrophoretic mobility shift assay
We performed electrophoretic mobility shift assay on nuclear extracts prepared as described previously (Garate et al., 2007) from either empty vector or BRMS1-overexpressing MMRU cells with the following double-strand oligonucleotide: 5′-CAAATGTGGGATTTTCCCATGAGTC-3′. The probe was labeled with a γ-32P phosphate at the 5′ end and another oligonucleotide with the same sequence but without labeling was used as a competitive sequence at 100- or 500-fold concentration. Binding reaction and detection procedure were carried out as described previously (Maffey et al., 2007).
In vivo angiogenesis assay and immunofluorescent staining
In vivo angiogenesis assay and immunofluorescent staining of mouse CD31 were performed as described previously (Karst et al., 2009). A total of 1×106 MMRU human melanoma cells were supported by 300 μl Matrigel and implanted subcutaneously into the flanks of 6-week-old male nude mice. After 10 days, the mice were killed and the implanted matrigel plugs were excised, photographed. Then the excised plugs were applied to the further embedding, sectioning and staining for mouse CD31. Finally, photos were taken and the number of CD31-positive cells was counted at five different fields for both vector control and BRMS1-overexpressing groups.
Supplementary Material
Acknowledgments
We thank Steve Hendy for technical assistance with EMSA. This work was supported by the Canadian Institutes of Health Research (MOP-84559 and MOP-93810) and Canadian Dermatology Foundation to GL. JL is a recipient of Postgraduate Scholarship from the Natural Sciences and Engineering Research Council of Canada and University of British Columbia Graduate Fellowship. Both JL and YC are recipients of CIHR Skin Research Training Centre Trainee Award.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc)
References
- Angeletti CA, Lucchi M, Fontanini G, Mussi A, Chella A, Ribechini A, et al. Prognostic significance of tumoral angiogenesis in completely resected late stage lung carcinoma (stage IIIA-N2). Impact of adjuvant therapies in a subset of patients at high risk of recurrence. Cancer. 1996;78:409–415. doi: 10.1002/(SICI)1097-0142(19960801)78:3<409::AID-CNCR5>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Balch CM, Buzaid AC, Soong SJ, Atkins MB, Cascinelli N, Coit DG, et al. Final version of the American Joint Committee on Cancer staging system for cutaneous melanoma. J Clin Oncol. 2001;19:3635–3648. doi: 10.1200/JCO.2001.19.16.3635. [DOI] [PubMed] [Google Scholar]
- Ballo MT, Ang KK. Radiation therapy for malignant melanoma. Surg Clin North Am. 2003;83:323–342. doi: 10.1016/S0039-6109(02)00096-8. [DOI] [PubMed] [Google Scholar]
- Brawer MK. Screening and early detection of prostate cancer will decrease morbidity and mortality from prostate cancer: the argument for. Eur Urol. 1996;29(Suppl 2):19–23. doi: 10.1159/000473832. [DOI] [PubMed] [Google Scholar]
- Cicek M, Fukuyama R, Cicek MS, Sizemore S, Welch DR, Sizemore N, et al. BRMS1 contributes to the negative regulation of uPA gene expression through recruitment of HDAC1 to the NFkappaB binding site of the uPA promoter. Clin Exp Metastasis. 2009;26:229–237. doi: 10.1007/s10585-009-9235-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cicek M, Fukuyama R, Welch DR, Sizemore N, Casey G. Breast cancer metastasis suppressor 1 inhibits gene expression by targeting nuclear factor-kappaB activity. Cancer Res. 2005;65:3586–3595. doi: 10.1158/0008-5472.CAN-04-3139. [DOI] [PubMed] [Google Scholar]
- Czubayko F, Schulte AM, Berchem GJ, Wellstein A. Melanoma angiogenesis and metastasis modulated by ribozyme targeting of the secreted growth factor pleiotrophin. Proc Natl Acad Sci USA. 1996;93:14753–14758. doi: 10.1073/pnas.93.25.14753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai DL, Martinka M, Li G. Prognostic significance of activated Akt expression in melanoma: a clinicopathologic study of 292 cases. J Clin Oncol. 2005;23:1473–1482. doi: 10.1200/JCO.2005.07.168. [DOI] [PubMed] [Google Scholar]
- Dai J, Peng L, Fan K, Wang H, Wei R, Ji G, et al. Osteopontin induces angiogenesis through activation of PI3K/AKT and ERK1/2 in endothelial cells. Oncogene. 2009;28:3412–3422. doi: 10.1038/onc.2009.189. [DOI] [PubMed] [Google Scholar]
- Dauda MM, Shehu SM. Malignant melanoma: a review. Niger Postgrad Med J. 2005;12:125–130. [PubMed] [Google Scholar]
- Diao H, Kon S, Iwabuchi K, Kimura C, Morimoto J, Ito D, et al. Osteopontin as a mediator of NKT cell function in T cell-mediated liver diseases. Immunity. 2004;21:539–550. doi: 10.1016/j.immuni.2004.08.012. [DOI] [PubMed] [Google Scholar]
- Folkman J. Role of angiogenesis in tumor growth and metastasis. Semin Oncol. 2002;29:15–18. doi: 10.1053/sonc.2002.37263. [DOI] [PubMed] [Google Scholar]
- Garate M, Campos EI, Bush JA, Xiao H, Li G. Phosphorylation of the tumor suppressor p33(ING1b) at Ser-126 influences its protein stability and proliferation of melanoma cells. Faseb J. 2007;21:3705–3716. doi: 10.1096/fj.07-8069com. [DOI] [PubMed] [Google Scholar]
- Gasparini G, Weidner N, Maluta S, Pozza F, Boracchi P, Mezzetti M, et al. Intratumoral microvessel density and p53 protein: correlation with metastasis in head-and-neck squamous-cell carcinoma. Int J Cancer. 1993;55:739–744. doi: 10.1002/ijc.2910550507. [DOI] [PubMed] [Google Scholar]
- Grossman D, Altieri DC. Drug resistance in melanoma: mechanisms, apoptosis, and new potential therapeutic targets. Cancer Metastasis Rev. 2001;20:3–11. doi: 10.1023/a:1013123532723. [DOI] [PubMed] [Google Scholar]
- Hedley BD, Welch DR, Allan AL, Al-Katib W, Dales DW, Postenka CO, et al. Downregulation of osteopontin contributes to metastasis suppression by breast cancer metastasis suppressor 1. Int J Cancer. 2008;123:526–534. doi: 10.1002/ijc.23542. [DOI] [PubMed] [Google Scholar]
- Hersey P. Adjuvant therapy for high-risk primary and resected metastatic melanoma. Intern Med J. 2003;33:33–43. doi: 10.1046/j.1445-5994.2002.00289.x. [DOI] [PubMed] [Google Scholar]
- Hicks DG, Yoder BJ, Short S, Tarr S, Prescott N, Crowe JP, et al. Loss of breast cancer metastasis suppressor 1 protein expression predicts reduced disease-free survival in subsets of breast cancer patients. Clin Cancer Res. 2006;12:6702–6708. doi: 10.1158/1078-0432.CCR-06-0635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingsworth HC, Kohn EC, Steinberg SM, Rothenberg ML, Merino MJ. Tumor angiogenesis in advanced stage ovarian carcinoma. Am J Pathol. 1995;147:33–41. [PMC free article] [PubMed] [Google Scholar]
- Hurst DR, Edmonds MD, Scott GK, Benz CC, Vaidya KS, Welch DR. Breast cancer metastasis suppressor 1 up-regulates miR-146, which suppresses breast cancer metastasis. Cancer Res. 2009;69:1279–1283. doi: 10.1158/0008-5472.CAN-08-3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karst AM, Gao K, Nelson CC, Li G. Nuclear factor kappa B subunit p50 promotes melanoma angiogenesis by upregulating interleukin-6 expression. Int J Cancer. 2009;124:494–501. doi: 10.1002/ijc.23973. [DOI] [PubMed] [Google Scholar]
- Kauffman EC, Robinson VL, Stadler WM, Sokoloff MH, Rinker-Schaeffer CW. Metastasis suppression: the evolving role of metastasis suppressor genes for regulating cancer cell growth at the secondary site. J Urol. 2003;169:1122–1133. doi: 10.1097/01.ju.0000051580.89109.4b. [DOI] [PubMed] [Google Scholar]
- Li G, Tang L, Zhou X, Tron V, Ho V. Chemotherapy-induced apoptosis in melanoma cells is p53 dependent. Melanoma Res. 1998;8:17–23. doi: 10.1097/00008390-199802000-00004. [DOI] [PubMed] [Google Scholar]
- Maeda K, Chung YS, Takatsuka S, Ogawa Y, Onoda N, Sawada T, et al. Tumour angiogenesis and tumour cell proliferation as prognostic indicators in gastric carcinoma. Br J Cancer. 1995;72:319–323. doi: 10.1038/bjc.1995.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maffey AH, Ishibashi T, He C, Wang X, White AR, Hendy SC, et al. Probasin promoter assembles into a strongly positioned nucleosome that permits androgen receptor binding. Mol Cell Endocrinol. 2007;268:10–19. doi: 10.1016/j.mce.2007.01.009. [DOI] [PubMed] [Google Scholar]
- Mansfield PF, Lee JE, Balch CM. Cutaneous melanoma: current practice and surgical controversies. Curr Probl Surg. 1994;31:253–374. doi: 10.1016/0011-3840(94)90025-6. [DOI] [PubMed] [Google Scholar]
- Meehan WJ, Samant RS, Hopper JE, Carrozza MJ, Shevde LA, Workman JL, et al. Breast cancer metastasis suppressor 1 (BRMS1) forms complexes with retinoblastoma-binding protein 1 (RBP1) and the mSin3 histone deacetylase complex and represses transcription. J Biol Chem. 2004;279:1562–1569. doi: 10.1074/jbc.M307969200. [DOI] [PubMed] [Google Scholar]
- Meehan WJ, Welch DR. Breast cancer metastasis suppressor 1: update. Clin Exp Metastasis. 2003;20:45–50. doi: 10.1023/a:1022542519586. [DOI] [PubMed] [Google Scholar]
- Metge BJ, Frost AR, King JA, Dyess DL, Welch DR, Samant RS, et al. Epigenetic silencing contributes to the loss of BRMS1 expression in breast cancer. Clin Exp Metastasis. 2008;25:753–763. doi: 10.1007/s10585-008-9187-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostmeier H, Fuchs B, Otto F, Mawick R, Lippold A, Krieg V, et al. Can immunohistochemical markers and mitotic rate improve prognostic precision in patients with primary melanoma? Cancer. 1999;85:2391–2399. doi: 10.1002/(sici)1097-0142(19990601)85:11<2391::aid-cncr14>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- Phadke PA, Vaidya KS, Nash KT, Hurst DR, Welch DR. BRMS1 suppresses breast cancer experimental metastasis to multiple organs by inhibiting several steps of the metastatic process. Am J Pathol. 2008;172:809–817. doi: 10.2353/ajpath.2008.070772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remmele W, Stegner HE. Recommendation for uniform definition of an immunoreactive score (IRS) for immunohistochemical estrogen receptor detection (ER-ICA) in breast cancer tissue. Pathologe. 1987;8:138–140. [PubMed] [Google Scholar]
- Samant RS, Clark DW, Fillmore RA, Cicek M, Metge BJ, Chandramouli KH, et al. Breast cancer metastasis suppressor 1 (BRMS1) inhibits osteopontin transcription by abrogating NF-kappaB activation. Mol Cancer. 2007;6:6. doi: 10.1186/1476-4598-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samant RS, Seraj MJ, Saunders MM, Sakamaki TS, Shevde LA, Harms JF, et al. Analysis of mechanisms underlying BRMS1 suppression of metastasis. Clin Exp Metastasis. 2000;18:683–693. doi: 10.1023/a:1013124725690. [DOI] [PubMed] [Google Scholar]
- Seraj MJ, Harding MA, Gildea JJ, Welch DR, Theodorescu D. The relationship of BRMS1 and RhoGDI2 gene expression to metastatic potential in lineage related human bladder cancer cell lines. Clin Exp Metastasis. 2000a;18:519–525. doi: 10.1023/a:1011819621859. [DOI] [PubMed] [Google Scholar]
- Seraj MJ, Samant RS, Verderame MF, Welch DR. Functional evidence for a novel human breast carcinoma metastasis suppressor, BRMS1, encoded at chromosome 11q13. Cancer Res. 2000b;60:2764–2769. [PubMed] [Google Scholar]
- Shevde LA, Samant RS, Goldberg SF, Sikaneta T, Alessandrini A, Donahue HJ, et al. Suppression of human melanoma metastasis by the metastasis suppressor gene, BRMS1. Exp Cell Res. 2002;273:229–239. doi: 10.1006/excr.2001.5452. [DOI] [PubMed] [Google Scholar]
- Soengas MS, Lowe SW. Apoptosis and melanoma chemoresistance. Oncogene. 2003;22:3138–3151. doi: 10.1038/sj.onc.1206454. [DOI] [PubMed] [Google Scholar]
- Standal T, Borset M, Sundan A. Role of osteopontin in adhesion, migration, cell survival and bone remodeling. Exp Oncol. 2004;26:179–184. [PubMed] [Google Scholar]
- Struhl K. Yeast transcriptional regulatory mechanisms. Annu Rev Genet. 1995;29:651–674. doi: 10.1146/annurev.ge.29.120195.003251. [DOI] [PubMed] [Google Scholar]
- Thompson JF, Scolyer RA, Kefford RF. Cutaneous melanoma. Lancet. 2005;365:687–701. doi: 10.1016/S0140-6736(05)17951-3. [DOI] [PubMed] [Google Scholar]
- Tsao H, Sober AJ. Melanoma treatment update. Dermatol Clin. 2005;23:323–333. doi: 10.1016/j.det.2004.09.005. [DOI] [PubMed] [Google Scholar]
- Vaidya KS, Harihar S, Phadke PA, Stafford LJ, Hurst DR, Hicks DG, et al. Breast cancer metastasis suppressor-1 differentially modulates growth factor signaling. J Biol Chem. 2008;283:28354–28360. doi: 10.1074/jbc.M710068200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidner N. Intratumor microvessel density as a prognostic factor in cancer. Am J Pathol. 1995;147:9–19. [PMC free article] [PubMed] [Google Scholar]
- Weidner N, Carroll PR, Flax J, Blumenfeld W, Folkman J. Tumor angiogenesis correlates with metastasis in invasive prostate carcinoma. Am J Pathol. 1993;143:401–409. [PMC free article] [PubMed] [Google Scholar]
- Weidner N, Semple JP, Welch WR, Folkman J. Tumor angiogenesis and metastasis–correlation in invasive breast carcinoma. N Engl J Med. 1991;324:1–8. doi: 10.1056/NEJM199101033240101. [DOI] [PubMed] [Google Scholar]
- Welch DR, Goldberg SF. Molecular mechanisms controlling human melanoma progression and metastasis. Pathobiology. 1997;65:311–330. doi: 10.1159/000164143. [DOI] [PubMed] [Google Scholar]
- Wiggins DL, Granai CO, Steinhoff MM, Calabresi P. Tumor angiogenesis as a prognostic factor in cervical carcinoma. Gynecol Oncol. 1995;56:353–356. doi: 10.1006/gyno.1995.1062. [DOI] [PubMed] [Google Scholar]
- Yamazaki K, Abe S, Takekawa H, Sukoh N, Watanabe N, Ogura S, et al. Tumor angiogenesis in human lung adenocarcinoma. Cancer. 1994;74:2245–2250. doi: 10.1002/1097-0142(19941015)74:8<2245::aid-cncr2820740807>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- Yang J, Zhang B, Lin Y, Yang Y, Liu X, Lu F. Breast cancer metastasis suppressor 1 inhibits SDF-1alpha-induced migration of non-small cell lung cancer by decreasing CXCR4 expression. Cancer Lett. 2008;269:46–56. doi: 10.1016/j.canlet.2008.04.016. [DOI] [PubMed] [Google Scholar]
- Zainabadi K, Benyamini P, Chakrabarti R, Veena MS, Chandrasekharappa SC, Gatti RA, et al. A 700-kb physical and transcription map of the cervical cancer tumor suppressor gene locus on chromosome 11q13. Genomics. 2005;85:704–714. doi: 10.1016/j.ygeno.2005.02.014. [DOI] [PubMed] [Google Scholar]
- Zhang S, Lin QD, Di W. Suppression of human ovarian carcinoma metastasis by the metastasis-suppressor gene, BRMS1. Int J Gynecol Cancer. 2006;16:522–531. doi: 10.1111/j.1525-1438.2006.00547.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.