Abstract
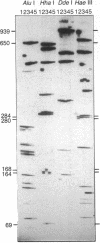
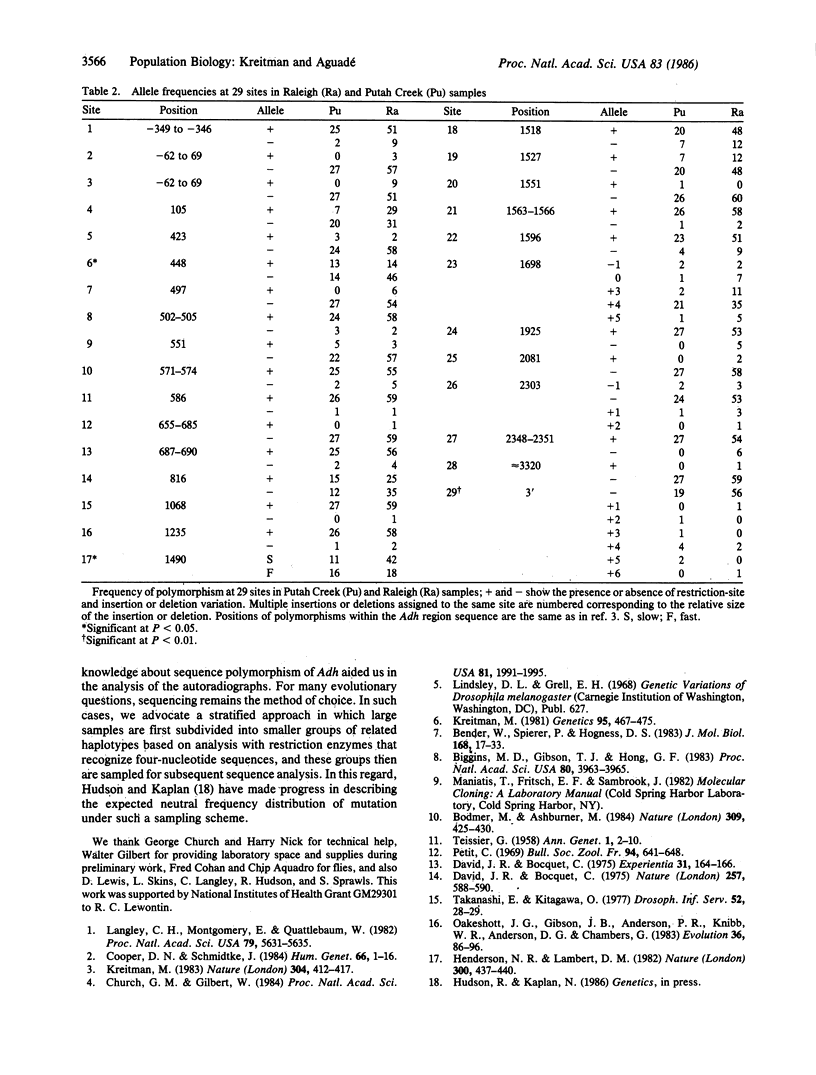
A filter hybridization method is described for identifying restriction-site and insertion/deletion variation by using restriction enzymes that recognize four-nucleotide sequences and denaturing polyacrylamide gels for separating fragments. Eighty-seven lines of Drosophila melanogaster representing two natural populations were surveyed over a 2.7-kilobase region encompassing the alcohol dehydrogenase locus. Fifty distinct haplotypes were identified from 17 restriction-site and 11 insertion/deletion polymorphisms and from one allozyme polymorphism. There was no evidence for genetic differentiation between an East-Coast and a West-Coast (North American) sample. This technique has widespread applications in screening for DNA polymorphism.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bender W., Spierer P., Hogness D. S. Chromosomal walking and jumping to isolate DNA from the Ace and rosy loci and the bithorax complex in Drosophila melanogaster. J Mol Biol. 1983 Jul 25;168(1):17–33. doi: 10.1016/s0022-2836(83)80320-9. [DOI] [PubMed] [Google Scholar]
- Biggin M. D., Gibson T. J., Hong G. F. Buffer gradient gels and 35S label as an aid to rapid DNA sequence determination. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3963–3965. doi: 10.1073/pnas.80.13.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodmer M., Ashburner M. Conservation and change in the DNA sequences coding for alcohol dehydrogenase in sibling species of Drosophila. 1984 May 31-Jun 6Nature. 309(5967):425–430. doi: 10.1038/309425a0. [DOI] [PubMed] [Google Scholar]
- Church G. M., Gilbert W. Genomic sequencing. Proc Natl Acad Sci U S A. 1984 Apr;81(7):1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper D. N., Schmidtke J. DNA restriction fragment length polymorphisms and heterozygosity in the human genome. Hum Genet. 1984;66(1):1–16. doi: 10.1007/BF00275182. [DOI] [PubMed] [Google Scholar]
- David J. R., Bocquet C. Evolution in a cosmopolitan species: genetic latitudinal clines in Drosophila melanogaster wild populations. Experientia. 1975 Feb 15;31(2):164–166. doi: 10.1007/BF01990682. [DOI] [PubMed] [Google Scholar]
- David J. R., Bocquet C. Similarities and differences in latitudinal adaptation of two Drosophila sibling species. Nature. 1975 Oct 16;257(5527):588–590. doi: 10.1038/257588a0. [DOI] [PubMed] [Google Scholar]
- Kreitman M. Assessment of variability within electromorphs of alcohol dehydrogenase in Drosophila melanogaster. Genetics. 1980 Jun;95(2):467–475. doi: 10.1093/genetics/95.2.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreitman M. Nucleotide polymorphism at the alcohol dehydrogenase locus of Drosophila melanogaster. Nature. 1983 Aug 4;304(5925):412–417. doi: 10.1038/304412a0. [DOI] [PubMed] [Google Scholar]
- Langley C. H., Montgomery E., Quattlebaum W. F. Restriction map variation in the Adh region of Drosophila. Proc Natl Acad Sci U S A. 1982 Sep;79(18):5631–5635. doi: 10.1073/pnas.79.18.5631. [DOI] [PMC free article] [PubMed] [Google Scholar]